Abstract
This data article contains Raman experimental data, obtained with Centaur U Raman spectrometer (Russia), which can be used for rapid and early structure changes and biomarkers identification in individuals with cardiovascular decease (CVD) pathology in vitro. The data include analyzed Surface-Enhanced Raman Scattering (SERS) spectra of human platelets taken from healthy individuals and individuals with cardiovascular pathology. Data can provide information about characteristic maxima of different cell components and its changes in platelets.
Keywords: Raman spectroscopy, Platelet, SERS, Gold nanoparticles, Aminoacids
Specifications Table
| Subject | Chemistry, Physics |
| Specific subject area | Spectroscopy |
| Type of data | Table Figure |
| How data were acquired | Centaur U (LTD «NanoScanTechnology», Russia) Raman spectrometer |
| Data format | Analyzed Filtered |
| Parameters for data collection | 9 samples from patients with cardiovascular pathology and 11 samples from healthy patients were analyzed. Platelets were centrifuged three times at 4 °C to obtain platelet-rich plasma |
| Description of data collection | SERS spectra were obtained, using 532 Cobolt Samba 50 mW laser and homemade SERS substrates. Explanation of spectra has been performed using 400–1800 cm−1 «fingerprint» region and KnowItAll (Biorad) software. |
| Data source location | REC «Fundamental and Applied Photonics. Nanophotonics», Immanuel Kant Baltic Federal University, Kaliningrad, Russia |
| Data accessibility | With the article |
Value of the data
|
1. Data
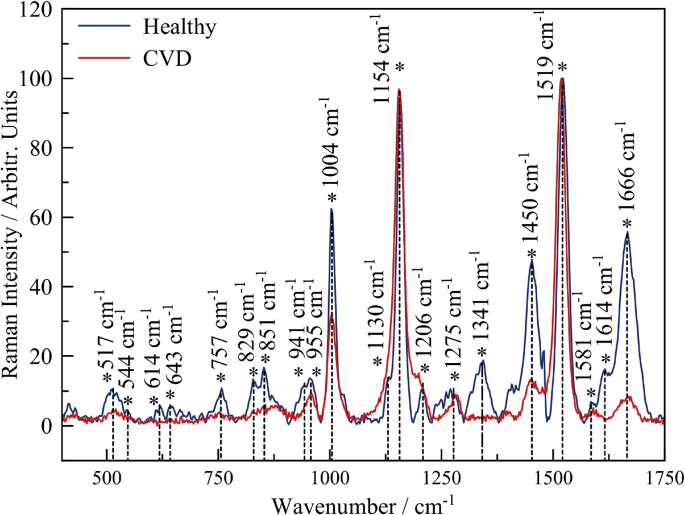
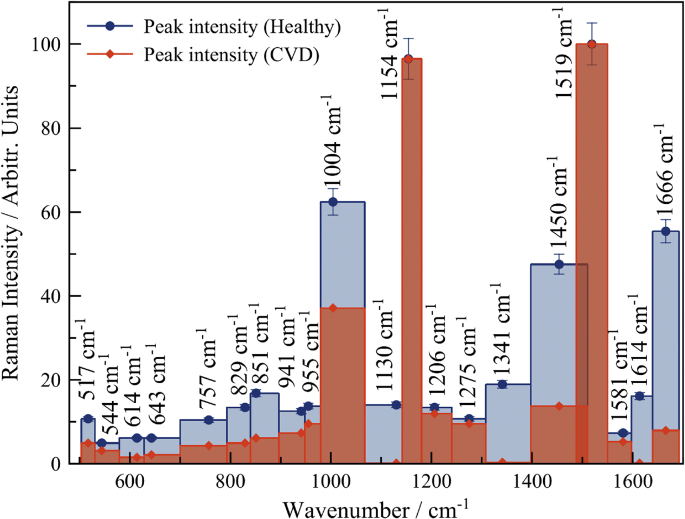
In this article, we present data on the human platelets SERS spectroscopy for healthy individuals and individuals with CVD. The presented data include SERS spectra for averaged and filtered data of both types (Fig. 2). SERS spectra intensities are shown on Fig. 3. The main vibrational bands are presented in Table 1. The main characteristic bands also are marked on Fig. 3. The presented data include 400–1750 cm−1 «fingerprint» spectral range.
Fig. 2.
Raman spectra of platelets in 400–1750 cm−1 spectral region for healthy individuals (blue line) and individuals with cardiovascular pathology (red line).
Fig. 3.
Intensity of Raman spectra for platelets for healthy individuals (blue line) and individuals with cardiovascular pathology (red line).
Table 1.
Characteristic bands of platelets.
| CCD | Healthy | Vibrational modes | Component | Reference | P-value |
|---|---|---|---|---|---|
| 544 w | 544 w | S–S str vibrations | Cys | [1] | P < 0.05 |
| 643 w | 643 mw | C–C twist | Tyr | [1] | P < 0.05 |
| 757 w | 757 mw | Phosphate diester str | Phosphatidilethanolamine | [1] | P < 0.05 |
| 829 mw | 829 m | Ring br aromatic mode | Trp | [4] | P < 0.05 |
| 851 mw | 851 m | Ring br aromatic mode in Tyr/phosphate groups | Proteins, phospholipids | [1] | P < 0.05 |
| 941 mw | 941 m | C–C backbone vibration | Lipids, proteins | [1] | P < 0.05 |
| 955 mw | 955 m | C–C backbone vibration | Lipids, proteins | [1] | P < 0.05 |
| 1004 s | 1004 vs | Aromatic δ ring mode | Phe | [1] | P < 0.05 |
| 1130 m | 1130 m | ν(Cβ–methyl) | proteins | [2] | P < 0.05 |
| 1154 vs | 1154 vs | Bond stretching (v) C–C | proteins | [1] | P < 0.05 |
| 1275 m | 1275 m | = C–H in plane deformation vibrations | Unsaturated fatty acids | [1] | P < 0.05 |
| – | 1341 m | Aromatic ring mode | Trp | [3] | P < 0.05 |
| 1450 m | 1450 s | CH2 bend | Lipids | [1] | P < 0.05 |
| 1519 vs | 1519 vs | NH3-sym bend | proteins | [4] | P < 0.05 |
| 1581 m | 1581 m | Aromatic ring mode | Trp | [1] | P < 0.05 |
| – | 1614 m | Aromatic ring mode | Tyr | [1] | P < 0.05 |
| 1666 mw | 1666 s | Amide I, C C str | Proteins, cholesterol | [1] | P < 0.05 |
2. Experimental design, materials and methods
2.1. Subjects
11 healthy and 9 volunteers with CVD pathology were involved in study. All volunteers had signed informed consent and were approved by Immanuel Kant Baltic Federal University Independence Local Ethic Committee (Protocol № 8, May 16, 2019). All cardiological patients had a history of myocardial infarction, arterial hypertension, and were on antiplatelet therapy. Smoking patients were excluded from this study.
2.2. Sample preparation
Fresh venous blood samples were taken from healthy individuals and individuals with cardiovascular pathology. Blood samples were placed into centrifugal tubes containing EDTA to avoid blood coagulation. Then the fresh blood was centrifuged at 60 g for 15 min to separate platelet-rich plasma, and then the plasma at 60 g for 15 min was deposited onto the blood pellet. Platelets were finally collected by further centrifugation of the supernatant at 1500 g for 15 min. All the centrifugations were carried out at 4 °C. After platelet preparation the samples were immediately taken to be examined by SERS spectroscopy.
2.3. SERS substrates fabrication
Anodizing of titanium plates with a thickness of 0.1 mm was carried out on the laboratory hand-made equipment with a current source and a galvanic bath, in which titanium electrodes were immersed. An aqueous solution of KOH (5%) was used as the electrolyte. Anodizing was carried out at a current density of j = 30 mA/cm2 for 5 minutes. The titanium surface became a blue-colored after anodizing. Gold nanoparticles were deposited on these surfaces.
Gold nanoparticles were fabricated by the femtosecond laser ablation of gold plate in distilled water on the AVESTA unit, described in detail in Ref. [5]. The obtained nanoparticles have a plasmon resonance at λ = 530 nm.
Deposition of ablative gold nanoparticles on titanium rough surfaces was carried out by the following algorithm. First, the titanium substrate was immersed in the gold nanoparticles solution. Then the nanoparticles were deposited on the surface by evaporation of an aqueous colloidal gold solution at a temperature of 60 °C for 40 minutes.
Fig. 1 shows rough titanium surface with ablative gold nanoparticles SEM image obtained with Zeiss Cross Beam 540 electron microscope (FIB-SEM).
Fig. 1.
SEM image of rough titanium surface with ablative gold nanoparticles.
2.4. SERS experiment
SERS spectra were obtained by Centaur U («NanoScanTechnology» LTD, Russia) Raman spectrometer, using the 532 DPSS Cobolt Samba laser with 45 mW power on sample. The optical scheme included Olympus BX 41 microscope with 100X (NA 0.9) objective. Spectrometer had a focal length of 284 mm with 1200 g/mm diffraction grating and was equipped with an Andor IDus 401 CCD camera with 1024 × 256 pixels. Spectrometer had spectral resolution of 2,5 cm−1. The laser spot of 1 × 25 μm size was positioned at the platelets. Rayleigh scattering was eliminated by the notch filters.
Due to plasmon resonance generation availability, rough titanium surfaces with gold nanoparticles (530 nm and 570 nm for gold nanoparticles and rough Ti surface respectively) were used to enhance Raman signal up to 103 times.
5 μl droplet of platelet-rich plasma was put on substrate, dried for 1 minute at room temperature, and then placed to the microscope holder. Five three times averaged spectra in different places of the droplet have been collected from each sample. Signal acquisition time was 70 s. Each time before experiment, instrument was calibrated with silicon at a static spectrum centered at 520.1 cm−1 for 1 s. After registration, spectra were saved as.txt and specific format (.ngs) on PC, connected to the Raman unit. KnowItAll Vibrational Spectroscopy Edition (BioRad, USA) was used for linear baseline correction and normalization. Savitsky-Golay filtering algorithm was used for all registered spectra and further analysis of peaks position and their intensity. Averaged spectra from healthy and CVD patients are displayed on Fig. 2. To determine the normal distribution for both groups (healthy and CVD samples), data were analyzed by the Student's t-test, and p < 0.05 was considered statistically significant with a 95% confidence interval (95%; Statistics) for the comparisons of mean Raman peaks.
Acknowledgements
Research was supported by the Russian Science Foundation under grant No 19-15-00132.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2020.105145.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.García-Rubio D., de la Mora M., Badillo-Ramírez I., Cerecedo D., Saniger J., Benítez-Benítez J., Villagrán-Muniz M. Analysis of platelets in hypertensive and normotensive individuals using Raman and Fourier transform infrared-attenuated total reflectance spectroscopies. J. Raman Spectrosc. 2019;50:509–521. doi: 10.1002/jrs.5540. [DOI] [Google Scholar]
- 2.Wood B., Caspers P., Puppels G., Pandiancherri S., McNaughton D. Resonance Raman spectroscopy of red blood cells using near-infrared laser excitation. Anal. Bioanal. Chem. 2007;387(5):1691–1703. doi: 10.1007/s00216-006-0881-8. [DOI] [PubMed] [Google Scholar]
- 3.Lykina A., Artemyev D., Bratchenko I., Khristoforova Y., Myakinin O., Kuzmina T., Davydkin I., Zakharov V. CEUR Workshop Proceedings. 2017. Raman spectra analysis of human blood protein fractions using the projection on latent structures method; pp. 64–68. [DOI] [Google Scholar]
- 4.Zhu G., Zhu X., Fan Q., Wan X. Raman spectra of amino acids and their aqueous solutions. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011;78(3):1187–1195. doi: 10.1016/j.saa.2010.12.079. [DOI] [PubMed] [Google Scholar]
- 5.Bryukhanov V.V., Minaev B.M., Tsibul’nikova A.V., Slezhkin V.A. The effect of gold nanoparticles on exchange processes in collision complexes of triplet and singlet oxygen molecules with excited eosin molecules. Opt. Spectrosc. 2015;119(1):29–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.