Abstract
Background
Fabry disease (FD) is a recessive X-linked lysosomal storage disorder caused by α-galactosidase A (GLA) deficiency. Although the mechanism is unclear, GLA deficiency causes an accumulation of globotriaosylceramide (Gb3), leading to vasculopathy.
Methods
To explore the relationship between the accumulation of Gb3 and vasculopathy, induced pluripotent stem cells generated from four Fabry patients (FD-iPSCs) were differentiated into vascular endothelial cells (VECs). Genome editing using CRISPR-Cas9 system was carried out to correct the GLA mutation or to delete Thrombospondin-1 (TSP-1). Global transcriptomes were compared between wild-type (WT)- and FD-VECs by RNA-sequencing analysis.
Findings
Here, we report that overexpression of TSP-1 contributes to the dysfunction of VECs in FD. VECs originating from FD-iPSCs (FD-VECs) showed aberrant angiogenic functionality even upon treatment with recombinant α-galactosidase. Intriguingly, FD-VECs produced more p-SMAD2 and TSP-1 than WT-VECs. We also found elevated TSP-1 in the peritubular capillaries of renal tissues biopsied from FD patients. Inhibition of SMAD2 signaling or knock out of TSP-1 (TSP-1−/−) rescues normal vascular functionality in FD-VECs, like in gene-corrected FD-VECs. In addition, the enhanced oxygen consumption rate is reduced in TSP-1−/− FD-VECs.
Interpretation
The overexpression of TSP-1 secondary to Gb3 accumulation is primarily responsible for the observed FD-VEC dysfunction. Our findings implicate dysfunctional VEC angiogenesis in the peritubular capillaries in some of the complications of Fabry disease.
Funding
This study was supported by grant 2018M3A9H1078330 from the National Research Foundation of the Republic of Korea.
Keywords: Fabry disease, Human induced pluripotent stem cells (hiPSCs), Globotriaosylceramide (Gb3), Vascular dysfunction, Thrombospondin-1
Research in context.
Evidence before this study
In the treatment of Fabry disease, agalsidase beta (Fabrazyme) or agalsidase alpha (Replagal) enzyme replacement therapy (ERT) it typically used to clear Gb3 deposits from various cell types. Although the administration of therapeutic enzymes can ameliorate the pathophysiologic phenotypes of Fabry disease, these recombinant enzymes are unstable in the blood and their repeated administration can cause allergic reactions. Although early treatment with recombinant α-galactosidase leads to significant symptomatic improvements in Fabry disease patients, the effect of therapeutic enzymes is restricted to later-onset patients.
Added value of this study
We found that treatment with recombinant enzymes is ineffective in altering the functionality of FD-VECs in vitro. Instead, inhibition of SMAD2 signaling and/or downregulation of TSP-1 rescues the angiogenic dysfunction of FD-VECs.
Implications of all the available evidence
Our findings suggest FD patients can be treated with new drugs that can regulate TGF-beta signaling and/or TSP-1 expression.
Alt-text: Unlabelled box
1. Introduction
Fabry disease (OMIM #301500) is an X-linked recessive hereditary disorder caused by genetic defects in α-galactosidase A (GLA). Deficiency of GLA leads to the progressive accumulation of globotriaosylceramide (Gb3) in most cell types of Fabry disease patients, including vascular cells, cardiac cells, kidney epithelial cells, and neuronal cells. To date, more than 750 mutations in the GLA gene have been identified (http://www.hgmd.cf.ac.uk). Approximately 70% of these mutations are missense or nonsense mutations in the GLA coding region [1]. Many abolish GLA activity, but some of the missense mutations produce subclinical effects if there is sufficient residual (5%–10%) enzymatic activity to prevent severe Gb3 accumulation [2]. Although the incidence of Fabry disease is 1 in 117,000 males [3], making it a rare disease, the incidence is rising owing to increased newborn screening [4,5].
Children with FD typically present with angiokeratomas [6], but these progress to life-threatening complications like left ventricular hypertrophy, renal failure, and stroke in adult patients as they age [7,8]. These varied complications are all thought to be caused by capillary obstruction in the various tissues [9]. Currently, enzyme replacement therapy (ERT) is being used to clear Gb3 deposits in Fabry patients [10,11]. Although recombinant enzyme administration ameliorates the pathophysiologic phenotypes of Fabry disease, its therapeutic effects are limited long-term because it is unstable in the blood and leads to allergic reactions (http://www.rxlist.com/fabrazyme-drug.htm#CS). Furthermore, ERT is less effective in Fabry patients with advanced disease [1,12,13].
Studies on GLA knockout (KO) mice led to several important insights into the role Gb3 accumulation plays in endothelial dysfunction. GLA−/− mice show aberrant Gb3 accumulation in the caveolae of aortic endothelial cells [14]. In GLA−/− mice, the protein levels of thrombospondin-1, TGF-ß1, and VEGF were increased in the kidneys compared to WT-mice [15]. Also, this Gb3 induces dysfunction of the Kca 3.1 channel in GLA−/- endothelial cells, thereby producing a Fabry-associated vasculopathy [16]. Although the GLA−/- mice used as a Fabry disease model seem to have a normal, complication-free lifespan, GLA−/− mice expressing human Gb3 synthase (G3S) (GLA−/−/G3Stg mice) show the typical Fabry disease phenotype accompanied by body weight loss, neurological symptoms, and early lethality [17]. This result is consistent with the hypothesis that Gb3 accumulation is the primary factor leading to Fabry disease, but it also shows that the GLA−/−/G3Stg mouse model does not perfectly recapitulate the complications of human Fabry disease. This means the mechanisms by which GLA deficiency and Gb3 accumulation lead to the phenotypic complications of Fabry disease remain poorly understood. In addition to the existing mouse models, iPSCs generated from the somatic cells of Fabry patients (FD-iPSCs) can be useful in the study of Fabry disease in vitro [18]. FD-iPSC-derived cardiomyocytes show Gb3 accumulation and cardiac hypertrophy, which are similar to the pathophysiological defects observed in cardiac tissue biopsied from Fabry patients [19,20]. In conclusion, disease modeling through FD-iPSCs can overcome mouse model limitations.
Thrombospondin-1 (TSP-1) regulates vessel stabilization and cessation of vessel growth in a fibrillary network around vascular structures [21]. Overexpressed TSP-1 suppresses vascular growth and expands vessel diameter [22]. However, it remains elusive whether TSP-1 is associated with dysfunctional angiogenesis in FD.
Here, we propose a putative model whereby it is insufficient angiogenesis owing to Gb3 accumulation that gives rise to the progressive complications of FD patients as they age. Interestingly, vascular endothelial cells (VECs) differentiated from FD-iPSCs (FD-VECs) show various dysfunctional angiogenesis phenotypes: Gb3 accumulation in lysosomes, fewer tube-like structures, low expression of angiogenic factors, activated SMAD2 signaling, enhanced expression of the anti-angiogenic factor thrombospondin-1 (TSP-1), and increased oxygen consumption. Also, we observed reduced expression of angiogenic factors and enhanced expression of TSP-1 in renal tissues biopsied from FD patients compared with those of normal donors. While treatment with recombinant α-galactosidase (agalsidase-ß) fails to rescue the angiogenic dysfunctions of FD-VECs, we observed significant improvement in their endothelial dysfunction in vitro upon inhibiting SMAD2 signaling or knocking out (KO) TSP-1. Thus, the overexpression of TSP-1 and/or enhanced activity of SMAD2 signaling that are secondary to GLA mutation contribute to FD vasculopathy.
2. Materials and Methods
2.1. Generation of iPSCs from Fabry disease patients
Fabry disease (FD) fibroblasts were individually biopsied from four patients under the approval of the AMC Institutional Review Board (2011-0451). Studies using human materials including iPSCs were performed under the approval of the Institutional Review Board of KAIST (KH2016-52). Fibroblasts were cultured in DMEM (Welgene, Seoul, Korea) containing 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 1% penicillin-streptomycin (Invitrogen) at 37°C, 5% CO2 in air. Retroviruses expressing OCT4, SOX2, cMYC, and KLF4 were transfected into FD fibroblasts to generate iPSCs as previously described [23]. Wild type (WT)-iPSCs derived from foreskin fibroblasts (CRL-2097, ATCC, Manassas, VA) were used as a control. iPSCs were cultured in human embryonic stem cell culture medium containing DMEM/F12 (Invitrogen, Carlsbad, CA), 20% KnockOut™ Serum Replacement (Invitrogen), supplemented with 10 ng/mL bFGF2 (R&D systems, Minneapolis, MN), 1% non-essential amino acid (NEAA) (Invitrogen), 1% penicillin and streptomycin (Invitrogen) on mitomycin C-treated MEF-seeded culture plates at 37°C, 5% CO2 in air. After detachment with dispase (Invitrogen), human iPSCs were passaged to fresh plates every 6 d. The GLA mutations were identified from genomic DNAs extracted from FD-iPSCs and fibroblasts by direct sequencing. The primers used for detecting the mutations are listed in Table S1.
2.2. Bisulfite sequencing
Analysis of DNA methylation at the CpG sites on the promoters of the OCT4, REX1, and NANOG genes was performed as previously described. Briefly, genomic DNA was treated with sodium bisulfite using the EZ DNA Methylation-Gold Kit according to the manufacturer's protocol (Zymo Research, Irvine, CA). Then, bisulfite-treated DNA (25–50 ng) was amplified by PCR. The amplified PCR products were purified using the AccuPrep® plasmid Mini Extraction Kit (Bioneer, Daejeon, Korea) and subcloned into the pGEM-T EASY vector (Promega, Madison, WI). For each cell type, 5 clones were sequenced using the M13 primer and further analyzed using a web-based program (Blast-2) or a stand-alone software package (BiQ Analyzer, http://biq-analyzer.bioinf.mpi-inf.mpg.de/).
2.3. Differentiation of FD-iPSCs into vascular endothelial cells
Differentiation of human iPSCs into vascular endothelial cells (VECs) was accomplished as previously described [24]. Briefly, human iPSCs were transferred to Matrigel-coated (BD biosciences, Franklin Lakes, NJ) dishes and cultured in mTESR1 medium (STEMCELL Technologies, Vancouver, Canada) at 37 °C, 5% CO2 in air for 3 d. Then, the culture medium was changed to RPMI medium supplemented with 1% B27 (Invitrogen), 50 ng/ml Activin A (Peprotech, Rocky Hill, NJ), and 20 ng/ml BMP4 (Peprotech) at 37 °C, 5% CO2 in air. After 2 d of culture, the cells were incubated in RPMI medium containing 50 ng/ml VEGF-A (Peprotech) and 50 ng/ml bFGF at 37 °C, 5% CO2 in air for 3 d. Then, vascular progenitors (CD34+ cells), which can differentiate into endothelial cells and vascular smooth muscle cells, were isolated from the differentiated cells using CD34+ magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). To differentiate the CD34+ vascular progenitors into VECs, they were cultured in EGM-2 medium (Lonza, Basel, Switzerland) supplemented with 100 ng/ml VEGF-A and 100 ng/ml bFGF at 37 °C, 5% CO2 in air for 5–7 d. Endothelial cells were then seeded on culture dishes at a density of 2 × 104 cells/cm2 and maintained in EGM-2 media for 5 d. The number of VECs was calculated once every 2 d using a hemocytometer (Marienfeld, Lauda-Königshofen, Germany).
2.4. Tube-like structure assay in Matrigel
VECs were washed with PBS and treated with Accutase (eBioscience, Waltham, MA) for 4 min to dissociate them. Dissociated VECs (5 × 104 cells) were placed on Matrigel matrix (BD Biosciences) in EGM-2 medium supplemented with VEGF-A and incubated at 37 °C, 5% CO2 in air for 24 h. Then, images of vascular tube-like structures were observed on an inverted microscope (Olympus, Tokyo, Japan). Total tube length was measured using ImageJ and its Angiogenesis Analyzer plugin [25].
2.5. Measurement of α-galactosidase activity
GLA activity was measured in iPSCs and iPSC-VECs using a modified fluorogenic method as previously reported [26]. In brief, approximately 4 × 106 cells were gently sonicated in 200 µl of GLA assay buffer (100 mM sodium citrate, 200 mM sodium phosphate dibasic, pH 4.6) for 10 sec and centrifuged at 13,000 × g at 4 °C for 30 min. Then, the supernatant (cell lysate) was harvested. The GLA assay mixture was prepared with 30 µl of 200 mM N-acetyl-D-galactosamine (Sigma-Aldrich, St. Louis, MO), 6 µl of 50 mM 4-MU-α-D-galactopyranoside (Sigma-Aldrich), and 14 µl of GLA assay buffer. To measure GLA activity, 10 µl of supernatant and 60 µl of GLA assay mixture were placed on a Falcon® 96-well TC-treated imaging microplate (black with a clear, flat bottom and a lid, CORNING, Corning, NY), and incubated at 37 °C for 1 h. Then, 150 μl of GLA stop buffer (200 mM Glycine, pH 10.32) was added to each well to stop the fluorogenic reaction. Fluorescence was measured at Ex/Em = 355/460 nm using a Victor plate reader (Perkin Elmer, Waltham, MA).
2.6. Western blotting
Cells were lysed in PRO-PREPTM protein extraction solution (Intron Biotechnology), and then the extracted proteins were quantified using the Bradford protein assay (Bio-Rad, Hercules, CA). Total proteins (20 µg) were separated on a 12% SDS-PAGE gel (Elpis Biotech, Daejeon, Korea) and then transferred to a nitrocellulose membrane (Bio-Rad). The blotted membranes were blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST: 10 mM Tris-HCl, pH 7.5, 150 nM NaCl, and 0.1% Tween-20). The membranes were then incubated at 4 °C overnight with the following primary antibodies: SMAD2 (Cell Signaling Technology, Danvers, MA, 1:1000), p-SMAD2 (Cell Signaling Technologies, 1:1000), GLA (Abgent, San Diego, CA, 1:1000), GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, 1:3000), TSP-1 (Santa Cruz Biotechnology, 1:2000), VEGF (Cell Signaling Technologies, 1:1000), bFGF (Cell Signaling Technologies, 1:1000), KDR (Cell Signaling Technologies, 1:1000), eNOS (Cell Signaling Technologies, 1:1000). After washing with TBST, the samples were treated with an HRP-conjugated secondary antibody (Thermo Fisher Scientific, Waltham, MA) in blocking solution at room temperature (RT) for 1 h. After washing with TBST, specific bands on the membrane were visualized using an ECL system (Thermo Fisher Scientific) according to the manufacturer's protocol. The density of each protein band was quantified by imageJ.
2.7. Immunostaining
The cells were washed with PBS, fixed in 4% formaldehyde at RT for 15 min, and permeabilized with 0.1% Triton X-100 in PBS at RT for 20 min. After blocking with 4% normal donkey serum (Abcam, Cambridge, UK) at RT for 1 h, the samples were incubated at 4 °C overnight with primary antibodies against OCT4 (R&D Systems, 1:300), NANOG (Abcam, 1:300), SSEA-4 (R&D Systems, 1:300), Tra-1-81 (Millipore, Billerica, MA, 1:300), Tra-1-60 (Millipore, 1:300), TSP-1 (Santa Cruz Biotechnology, 1:300), KDR (Cell Signaling Technology, 1:400), VECAD (Abcam, 1:400), or Gb3 (GeneTex, Irvine, CA, 1:100) diluted with blocking solution. After rinsing several times with PBST (0.1% Tween-20 in PBS), the samples were incubated with Alexa-488- or Alexa-594-conjugated secondary antibodies (Invitrogen) at RT for 1 h. Then, the cells were counterstained with 4′-6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) and visualized on a fluorescence microscope (Olympus, Tokyo, Japan) or a Zeiss LSM 510 confocal microscope (Carl Zeiss, Oberkochen, Germany).
2.8. Quantitative RT-PCR
Total RNAs were extracted from cells by using the easy-BlueTM kit (Intron biotechnology), followed by treatment with DNase I (Invitrogen) according to the manufacturer's protocol. Total RNAs (1 μg) were reverse-transcribed into cDNA according to the manufacturer's protocol. The cDNAs were then analyzed via real-time quantitative RT-PCR (qRT-PCR) on a Bio-RAD iQ5 System (Bio-Rad) using Primer Q-Master Mix (GenetBio, Chungnam, Korea). PCR was carried out at 94 °C for 10 min to denature the DNA molecules and then for 40 cycles of 10 s at 94 °C, 25 s at 58 °C, and 30 s at 72 °C. A melt-curve analysis was performed to ensure specific amplification. All reaction sets were conducted in triplicate, and a non-template control (NTC; nuclease free water) was run for every assay. Gene expression was normalized to the endogenous reference gene GAPDH as an internal control and quantified using the comparative CT method (∆∆CT). The primers, which were designed with Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/), are listed in Table S1.
2.9. FACS analysis
The cells were dissociated by treatment with Accutase at 37 °C for 5 min. After centrifugation at 300 × g for 5 min, the cell pellets were resuspended in FACS buffer (PBS containing 2% FBS). The resuspended cells were then filtered through a cell strainer with a 40 μm pore size (SPL lifesciences, Pocheon, Korea). The cells were incubated with specific FACS antibodies against CD31, CD34, CD105, KDR, or isotype controls (Biolegend, San Diego, CA) at 4 °C for 30 min. After washing 5 times with FACS buffer, the samples were analyzed using a FACSCalibur flow cytometer (BD Biosciences). The proportion of cells expressing each respective EC marker was analyzed using the FlowJo software package (Tree star, Ashland, OR).
2.10. Immunohistochemistry of renal tissues biopsied from FD patients
Biopsied samples from 11 FD patients and 11 donors were used for this study, which was approved by the AMC Institutional Review Board (2016-0768). Renal biopsy tissues were obtained at Asan Medical Center (AMC) from FD patients from 2003 to 2018 and from normal donors in 2017. Formalin-fixed paraffin-embedded (FFPE) renal samples were cut into 4 μm-thick slices and immunostained using the Ventana Benchmark XT Automated Staining System (Ventana Medical Systems, Tucson, AZ) according to the manufacturer's protocol. The antibody information and dilution conditions are as follows: TSP1 (Abcam, 1:100); NOS3 (Santa Cruz Biotechnology, 1:100); ANG2 (BIOSS Antibodies, Woburn, MA, 1:50), KDR (Cell Signaling Technology, 1:400), and VECAD (Abcam, 1:400). The intensity of the staining for each respective marker was evaluated in the endothelial cells of peritubular capillaries. The intensity of the staining was arbitrarily scored on a scale of 0–3. The number of cells expressing each marker was counted and scored using the following formula: Score = (0 × No. of cells with intensity 0) + (1 × No. of cells with intensity 1) + (2 × No. of cells with intensity 2) + (3 × No. of cells with intensity 3) in two high power fields (× 400).
2.11. RNA-Sequencing analysis
Total RNAs were extracted from various samples using the easy-BlueTM kit. RNA-Sequencing and data analysis were carried out by EBIOGEN Inc. (Seoul, Korea). Briefly, RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Amstelveen, Netherlands). Library construction was performed using the QuantSeq 3’ mRNA-Seq Library Prep Kit (Lexogen, Inc., Greenland, NH) according to the manufacturer's protocol. High-throughput sequencing was performed as single-end 75 sequencing using a NextSeq 500 (Illumina, Inc., San Diego, CA). Analysis of differentially expressed genes and gene ontologies was carried out using the Excel-based Differentially Expressed Gene Analysis (ExDEGA) software package provided by EBIOGEN Inc. Heatmap generation and clustering analysis was performed using R (version 3.6.0) and MeV (version 4.9.0) for selected genes.
2.12. Measurement of oxygen consumption rate
VECs were seeded on gelatin-coated XFe96 microplates at a density of 40,000 cells/well and incubated in EGM2 medium overnight. The cells were retained in non-buffered assay medium (Seahorse Bioscience, North Billerica, MA) supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose in a non-CO2 incubator for 1 h before the assay. The Mito Stress Test kit (Seahorse Bioscience) was used to monitor the mitochondrial respiration rate under basal conditions. After sequential injections of an ATP synthase inhibitor (oligomycin, 3 µM), a mitochondrial uncoupling agent (carbonyl cyanide-4-(trifluoromethoxy)phenyl-hydrazone (FCCP), 1 µM), and respiratory chain inhibitors (antimycin A and rotenone, 1.5 µM), oxygen consumption rates (OCR) were measured using the Seahorse XFe96 analyzer (Seahorse Bioscience). Data are presented as means ± SEM from 8 wells/plate.
2.13. Correction of GLA mutation in FD-iPSCs by CRISPR/Cas9
To correct GLA gene mutations with CRISPR/Cas9, recombinant Cas9 protein was purchased from ToolGen, Inc (Seoul, Korea). The targeting sgRNA for the mutated GLA gene was synthesized via in vitro transcription using T7 RNA polymerase (New England Biolabs, Ipswich, MA) as described previously [27]. Briefly, the sgRNA template was synthesized by the annealing and extension of two oligonucleotides (Table S5). Then, sgRNAs were generated by in vitro transcription from the sgRNA template. In vitro transcription was performed using T7 RNA polymerase supplemented with NTPs and RNase inhibitor overnight at 37 °C. In vitro transcribed sgRNAs were then treated with DNase I for 30 min at 37 °C and purified using the MinElute Cleanup kit (Quagen, Hilden, Germany). The CRISPR/Cas9 RNP complex was prepared by incubating 20 µg of Recombinant Cas9 and 15 µg of sgRNA at RT for 5 min. FD-iPSCs (approximately 200,000 cells) were transfected with the CRISPR/Cas9 RNP complex and 100 pmol ssODN [28] using the Amaxa P3 Primary Cell 4D-Nucleofector Kit (Lonza, Allendale, NJ). Three days after transfection, FD-iPSCs were harvested and seeded on culture dishes coated with Matrigel (BD Biosciences). After 7–10 d of culture, FD-hiPSC colonies were detached using Dispase (STEMCELL Technologies) and transferred individually to 48-well culture dishes. Half of each colony was scraped for genotyping using targeted deep sequencing. The primers used for the targeted deep sequencing are listed in Table S4.
2.14. Genetic ablation of TSP1 in FD-iPSCs by CRISPR/Cas9
To introduce genetic mutations into the TSP1 gene of FD-iPSCs, 15 μg of Cas9 and 10 μg of the sgRNAs for each of two target sites (TSP-T1, TSP-T2) were incubated at RT for 5 min to form CRISPR/Cas9 RNP complexes. Then, each CRISPR/Cas9 RNP complex was introduced to 200,000 FD-iPSCs using the Amaxa P3 Primary Cell 4D-Nucleofector Kit (Lonza). Targeted deep sequencing was performed to check gene targeting efficiency and to establish TSP1-KO clones as described above.
2.15. Targeted deep sequencing for analyzing sgRNA off-target effects in FD-iPSCs
To examine the off-target mutation frequency of CRIPSR/Cas9 in FD-iPSCs, genomic DNA from CRIPSR/Cas9-transfected FD-iPSCs was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The on-target and off-target regions were amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs). PCR amplicons were amplified using HT Dual index-containing primers to generate deep sequencing libraries. The DNA libraries were then sequenced using an Illumina MiniSeq. The RGEN tool was used for analyzing the indel and HDR frequencies (Cas-analyzer) and for finding potential off-target sites (Cas-OFFinder) [29,30]. The primers used for amplification of the targeted genes are listed in Table S4.
2.16. Statistical analysis
All statistical analyses in this study were performed using Prism 5 (Graphpad software, Inc., CA, USA). All data are shown as means ± SEM. Statistical significance was analyzed using the Student's t-test. P values are represented as follows: * p < 0.05; ** p < 0.01**; *** p < 0.001.
3. Results
3.1. Genetic mutations and clinical records for four FD patients
Table 1 summarizes the clinical records for four FD patients. Each of these FD patients had different GLA mutations. A 37-year-old Fabry patient (FD-1) had a genomic deletion (c.803_806del) causing a frame shift in exon 6 and amino acid position 268. Another 50-year-old Fabry patient (FD-2) had a point mutation (c.658C>T) producing a stop codon in exon 5. A 28-year-old Fabry patient (FD-3) had a single point mutation (c.334C>T) that caused the insertion of an alternative amino acid in exon 2. Another 28-year-old Fabry patient (FD-4) had a single point mutation (c.1045T>C) that altered an amino acid in exon 7. The clinical records of each of these four patients with their distinct mutations showed extremely low GLA activity. Although treatment with agalsidase-β (Fabrazyme, Genzyme, A Sanofi company, Cambridge, MA) reduced the high levels of Gb3 in each of these patients to normal levels, they still exhibited classic Fabry disease symptoms, such as angiokeratoma, cornea verticillata, proteinuria, and acroparesthesia despite long-term (3–12 years) ERT. These clinical data suggest Gb3 reduction by recombinant α-galactosidase treatment does not repress the progressive etiology of Fabry disease.
Table 1.
Genetic mutations and clinical records of four Fabry patients.
| Patient Number | FD-1 | FD-2 | FD-3 | FD-4 | ||||
|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Male | Male | ||||
| Age | 37 yr | 28 yr | 50 yr | 28 yr | ||||
| Diagnosis (Giuliano and Pages) | 18 yr | 24 yr | 32 yr | 20 yr | ||||
| Phenotype | Classical | Classical | Classical | Classical | ||||
| GLA activity,1 % of Median Wild type | 0.0% | 0.6% | 0.0% | 0.6% | ||||
| GLA mutation c. | c.803_806del | c.658C>T | c.334C>T | c.1045T>C | ||||
| GLA mutation p. | p.L268fs*1 | p.Arg220* | p.Arg112Cys | p.Trp349Arg | ||||
| ERT duration (yrs) | 0 | 12 | 0 | 3 | 0 | 11 | 0 | 7 |
| Serum GL3 (3.9–9.9 ug/ml) | 18 | 4.8 | 16 | 8.3 | 10 | 5.5 | 12 | 5.6 |
| Angiokeratoma | (+) | (+) | (+) | (+) | (+) | (+) | (+) | (+) |
| Cornea verticillata | (+) | (+) | (+) | (+) | (+) | (+) | (+) | (+) |
| Sensorineural hearing loss | (+) | (+) | (-) | (+) | (+) | (+) | (-) | (-) |
| EKG abnormality | WPW | WPW | SB | SB | SB | SB | WPW | WPW |
| Cardiac hypertrophy | (-) | (-) | (-) | (-) | (+) | (+) | (-) | (-) |
| LV mass (g/m2) | n.a. | 89 | 89 | 98 | 126 | 153 | 94 | 105 |
| Anhidrosis/ Hypohydrosis | (+) | (+) | (+) | (+) | (+) | (+) | (+) | (+) |
| Acroparesthesia | (+) | (+) | (+) | (+) | (-) | (-) | (+) | (+) |
| Proteinuria (mg/day/m2) | 11 | 106 | 0.032 | 73 | 109 | 98 | 72 | 35 |
| Estimated glomerular filtration rate (ml/min/1.73m2) | 141 | 139 | 125 | 122 | 58 | 46 | 123 | 122 |
3.2. Generation of FD-iPSCs
To study human Fabry disease in vitro, we generated FD-iPSC lines from fibroblasts of each of four different patients. Like WT-iPSCs, these FD-iPSCs expressed pluripotency-associated markers (Supplementary Fig. 1a) and showed demethylation of the OCT4, NANOG, and REX1 promoters (Supplementary Fig. 1b). FD-iPSCs have normal karyotypes (Supplementary Fig. 1c). Unlike WT-iPSCs, the FD-iPSCs were Gb3-positive within lysosomes (Supplementary Fig. 1e, arrows in the left panel) and showed very little GLA activity (Supplementary Fig. 1f). Treatment of the FD-iPSCs with a recombinant α-galactosidase A protein (agalsidase-ß) reduced some of their Gb3 accumulation (Supplementary Fig. 1e, right panel). These results demonstrate that FD-iPSCs can retain pluripotency even in the presence of GLA deficiency-induced Gb3 accumulation.
3.3. Differentiation of FD-iPSCs into vascular endothelial cells
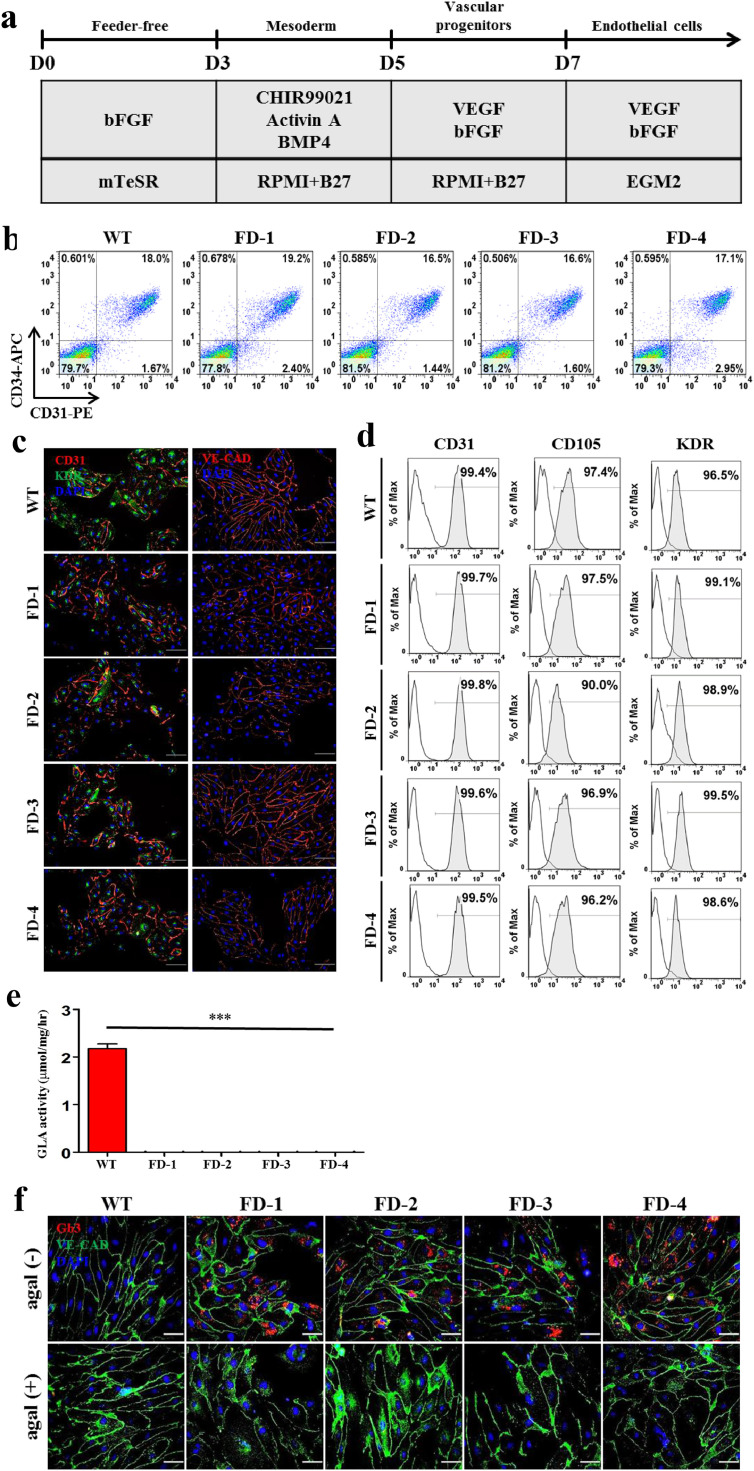
The progressive accumulation of Gb3 in vascular endothelial lysosomes causes the major FD complications such as renal failure, ischemic stroke, and hypertrophic cardiomyopathy [31]. To better understand how Gb3 accumulation influences angiogenesis in vitro, we induced the differentiation of FD-iPSCs into vascular endothelial cells (VECs). The protocol for this differentiation of iPSCs into VECs is described in Fig. 1a. Before achieving terminal differentiation into VECs, we isolated CD34+ progenitors at 7 d of differentiation from iPSCs by MACS sorting. We did not detect any difference in the proportion of endothelial cell progenitors (CD34+CD31+) among WT- and FD-derivatives (Fig. 1b). Then, we further differentiated isolated CD34+ progenitors into vascular endothelial cells for 7–9 d. Like WT, endothelial cells differentiated from FD-derivatives (FD-VECs) at 15 d of culture showed expression of the endothelial cell markers CD31, KDR, and VE-cadherin (Fig. 1c), as well as the endothelial cell surface markers CD31, CD105, and KDR (Fig. 1d). FD-VECs showed no GLA activity (Fig. 1e), thereby leading to the accumulation of Gb3 (Fig. 1f, upper row). But, this Gb3 accumulation in FD-VECs was reduced by agalsidase-β treatment (Fig. 1f, lower row). These results indicate that FD-iPSCs develop into VECs despite the presence of Gb3 accumulation in vitro.
Fig. 1.
Differentiation of FD-iPSCs into vascular endothelial cells (VECs) (a) A schematic protocol for the differentiation of FD-iPSCs into VECs. Detailed procedures are described in the Materials & methods. D, day. (b) Differentiation efficiency of FD-iPSCs into vascular endothelial progenitors (VEPs). Ratios of VEPs (CD34+CD31+) differentiated from FD-iPSCs ranged from ~16%–19%, which are similar to that of WT-iPSCs (18.0%). (c) Expression of EC markers in FD-VECs. Like WT-VECs, FD-VECs express the EC markers CD31, KDR, and VE-CAD. Scale bar, 100 μm. (d) Expression of EC surface makers in FD-VECs. More than 90% of FD-VECs are positive for CD31, CD105, and KDR. (e) Low GLA activity in FD-VECs compared with WT-VECs. The data are expressed as means ± SEM (n = 3). *** p < 0.001. (f) Gb3 accumulation in FD-VECs. FD-VECs accumulate Gb3, but this is reduced upon treatment with agalsidase-ß (agal). Scale bar, 50 μm.
3.4. Defective angiogenesis of FD-iPSC-derived vascular endothelial cells (FD-VECs)
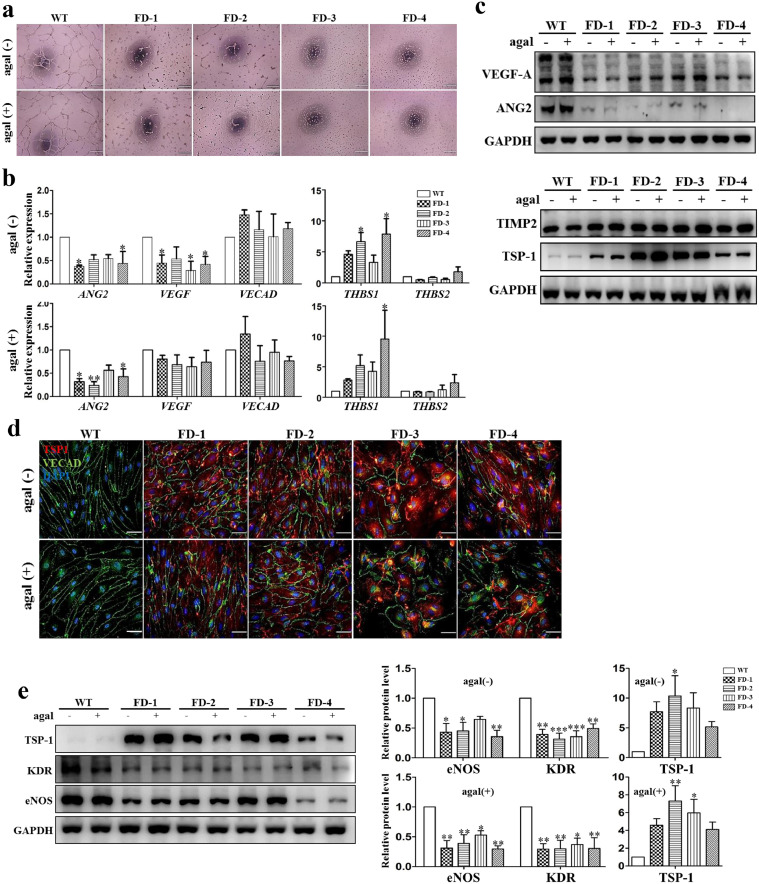
To test the functionality of FD-VECs in vitro, we carried out a tube-like structure assay. Early stage FD-VECs (3 d of EC development from isolated CD34+ progenitors) formed vascular tube-like structures in Matrigel (Supplementary Fig. 2a). Unlike WT-VECs, late stage FD-VECs (9 d of EC development) did not form tube-like structures even after agalsidase-β treatment (Fig. 2a). They also showed aberrant morphology with round shapes and reduced proliferative capacity (Supplementary Fig. 2b and 2c). This suggests the angiogenic ability of FD-VECs is gradually reduced during in vitro culture. When we compared FD-VECs and WT-VECs, we saw reduced expression of angiogenic genes like ANG2 and VEGF, and increased expression of the anti-angiogenic factor THBS1, regardless of agalsidase-β treatment. This was the case despite some variation in gene expression among the respective FD-VECs (Fig. 2b). We found, however, that the expression of the EC marker gene VECAD in both WT- and FD-VECs was similar. We then confirmed in WT- and FD-VECs that their protein expression profiles were analogous to their respective transcriptional activity profiles (Fig. 2c). Thus, it is likely that Gb3 accumulation is indirectly associated with the reduced tube-like structure observed in FD-VECs. In an immunofluorescent analysis, we found enhanced expression of TSP-1 in FD-VECs even after agalsidase-β treatment (Fig. 2d). TSP-1 binds to free- and or cell-associated VEGF [32] and scavenges ECM-associated FGF2 [33]. Thus, TSP-1 regulates the bioavailability of VEGF and FGF2 in each cell's microenvironment [34]. VEGF regulates the expression of KDR and FGF2 to modulate the expression of eNOS [35,36]. We were able to confirm that the enhanced TSP-1 in FD-VECs is associated with reduced expression of KDR and eNOS, despite little variation in protein expression levels among the FD-VECs originating from each patient (Fig. 2e). To determine whether Gb3 accumulation is associated with enhanced TSP-1 expression in FD, we treated WT-VECs with Lyso-Gb3 during EC culture. We found this treatment increased their expression of TSP-1 and reduced their expression of KDR and eNOS in a day-dependent manner (Supplementary Fig. 4c). These results suggest Gb3 accumulation indirectly contributes to the up-regulation of TSP-1 and the down-regulation of KDR and eNOS in FD. Thus, it is likely that a paucity of tube-like structure in FD-VECs is responsible for the down-regulation of angiogenic factors and the up-regulation of anti-angiogenic factors. In addition, agalsidase-β treatment does not seem to be effective for improving the angiogenic activity of FD-VECs.
Fig. 2.
Dysfunctional angiogenesis of FD-VECs. (a) Abnormal tube-like structure of FD-VECs. Unlike WT-VECs, FD-VECs do not form tubular structures regardless of enzyme treatment. Scale bar, 200 μm. (b) Transcriptional expression of angiogenic and anti-angiogenic factors in WT- and FD-VECs. The data are expressed as means ± SEM (n = 3). * p < 0.05; ** p < 0.01. (c) Expression of angiogenic and anti-angiogenic factors in FD-VECs. The angiogenic factors VEGF and ANG2 are down-regulated, while TSP-1 is up-regulated in FD-VECs compared to WT-VECs. (d) Immunostaining of TSP-1 in FD-VECs. TSP-1 is overexpressed in FD-VECs regardless of enzyme treatment when compared with WT-VECs. Scale bar, 50 μm. (e) Reduced expression of KDR and eNOS in FD-VECs. The relative protein ratios were quantified using the ImageJ software. The data are expressed as means ± SEM (n = 3). * p < 0.05; ** p < 0.01; *** p < 0.001.
3.5. Aberrant expression of angiogenesis-associated factors in renal biopsies of FD patients
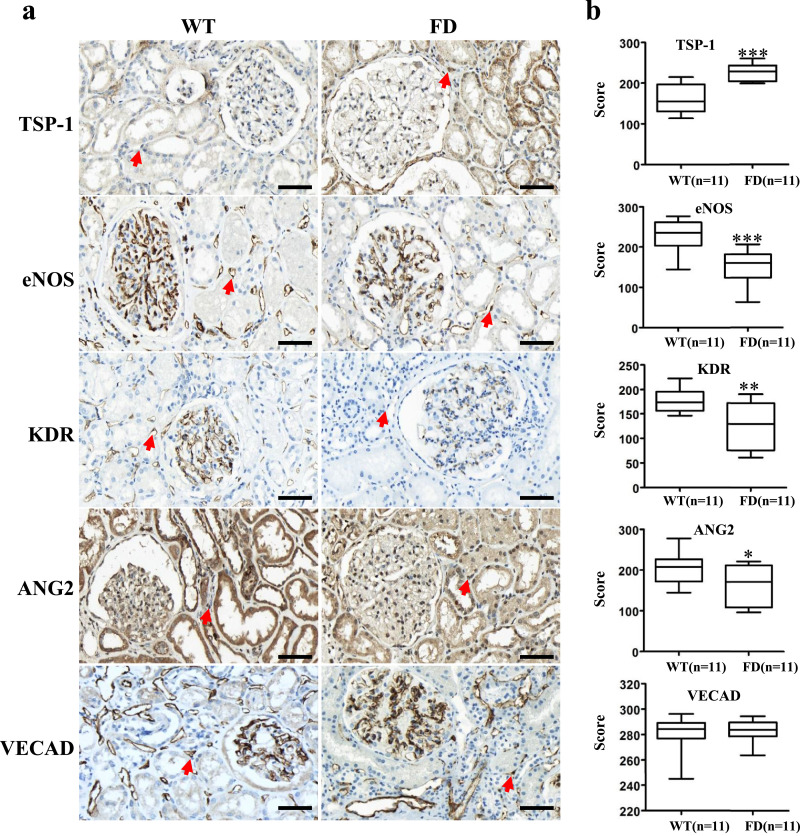
Using immunohistochemistry, we visualized the expression patterns of angiogenic and anti-angiogenic factors in the peritubular capillaries of renal tissues biopsied from 11 FD patients and 12 normal kidney donors (Fig. 3a). Red arrows indicate representative peritubular capillaries in each sample. We found significantly higher expression of the anti-angiogenic factor TSP-1 and reduced expression of the angiogenic factors eNOS, KDR, and ANG2 in the FD patient samples as compared to the normal kidney donor samples (Fig. 3b). Thus, the renal peritubular capillaries of FD patients show aberrant expression of angiogenesis-associated factors. Nonetheless, we found similar levels of expression of the EC marker VECAD in the peritubular capillaries of FD and donor samples (Fig. 3b). These results imply that although peritubular capillaries may form normally, they become dysfunctional in the kidneys of FD patients. Apart from the accumulation of Gb3, glomerular structure appeared normal in both FD and donor samples (Supplementary Fig. 3). These results from FD patient biopsies indirectly support the dysfunction of FD-iPSC-derived FD-VECs shown in Fig. 3.
Fig. 3.
Immunohistochemistry of renal tissues biopsied from FD patients (n = 11) and donors (n = 11). (a) Immunohistochemistry of renal tissues for TSP-1, eNOS, KDR, and ANG2. Red arrows indicate endothelial cells of peritubular capillaries. Scale bar, 50 μm. (b) Quantitative scoring of IHC in endothelial cells of peritubular capillaries. The data are expressed as means ± SEM. * p < 0.05; ** p < 0.01; *** p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Roles of SMAD2 signaling in the angiogenesis of FD-VECs
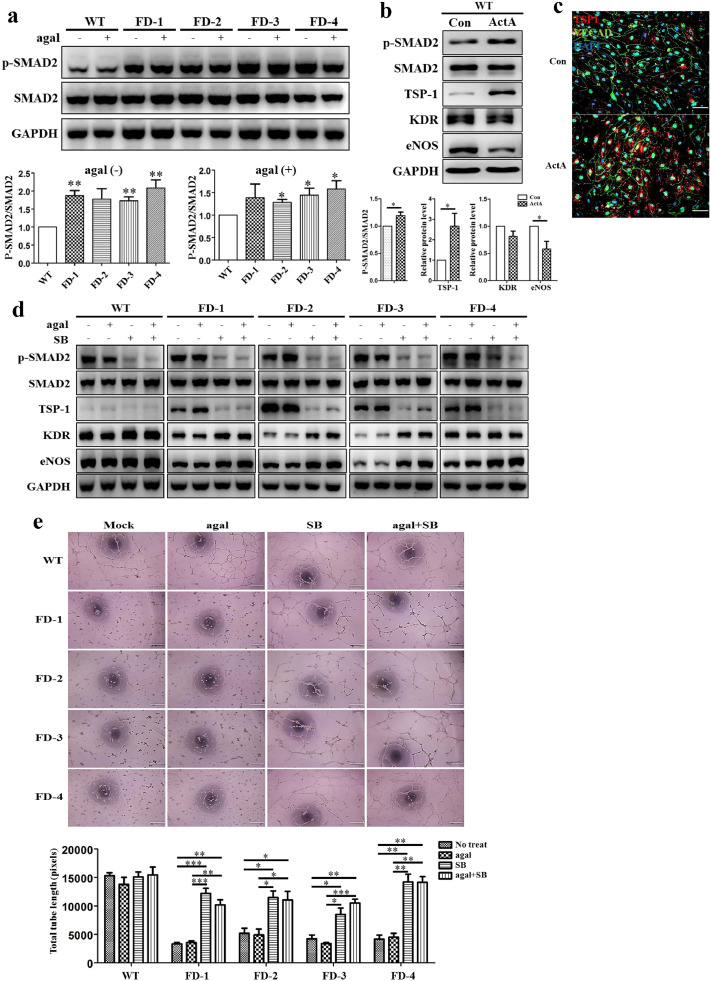
TSP-1 is a major regulator of TGF-β signaling that plays an important role in angiogenesis [37]. In the analysis of RNA sequencing data from FD-VECs, we identified 4 up-regulated GO terms related to SMAD signaling (Supplementary Fig. 8a). Hence, we wanted to explore whether SMAD signaling is associated with the dysfunctional angiogenesis observed for FD-VECs. As shown in Fig. 4a, FD-VECs produce higher levels of p-SMAD2 than WT-VECs regardless of agalsidase-β treatment, although the statistical significance of the difference in sample was different. To clarify whether SMAD2 activation enhances TSP-1 expression, we treated WT- CD34+ progenitors with Activin A (ActA) for 5 d. Indeed, we found ActA-treated WT-VECs show significantly increased expression of TSP-1 and reduced expression of eNOS (Fig. 4b), just like the FD-VECs shown in Fig. 2E. We confirmed with immunostaining that WT-VECs treated with ActA increase their expression of TSP-1 (Fig. 4c). Thus, overexpression of TSP-1 is associated with activation of SMAD2 signaling in FD-VECs. When we inhibited SMAD2 signaling in FD-VECs by treatment with SB431542 for 2 d, we found reduced expression of TSP-1 and increased expression of KDR and eNOS (Fig. 4d). This suppression of SMAD2 signaling also produced a morphological recovery in FD-VECs (Supplementary Fig. 5). The chemical-treated FD-VECs showed some tube-like structure (Fig. 4e). These results demonstrate that the dysfunctional activity of FD-VECs accounts for the hyperactivation of SMAD2 signaling and that inhibition of SMAD2 signaling rescues angiogenesis in FD-VECs. Thus, SMAD2 signaling functions as a key regulator leading to the dysfunction of angiogenesis in FD-VECs. Based on our findings, it seems likely that the modulation of SMAD2 signaling will be more effective in improving the angiogenic activity of FD-VECs than agalsidase-β treatment.
Fig. 4.
Role of SMAD2 signaling in FD-VECs. (a) Activation of SMAD2 signaling in FD-VECs. Western blot analysis showing increased p-SMAD2 in FD-VECs. The data are expressed as means ± SEM (n = 3). * p < 0.05; ** p < 0.01. (b) Enhanced SMAD2 signaling in Activin A-treated WT-VECs. Activation of SMAD2 signaling increases the expression of TSP-1. (c) Expression of TSP-1 in Activin A-treated WT-VECs. Scale bar, 200 μm. (d) Down-regulation of SMAD2 signaling in FD-VECs by TGF-β inhibition. Treatment of FD-VECs with SB (SB431542) reduces p-SMAD2 and TSP-1, but increases KDR and eNOS. (e) Rescue of angiogenic dysfunction in FD-VECs by inhibition of SMAD2 signaling. Tube-like structures formed by FD-VECs recovered upon treatment with SB. Scale bar, 100 μm. The data are expressed as means ± SEM (n = 3). * p < 0.05; ** p < 0.01; *** p < 0.001.
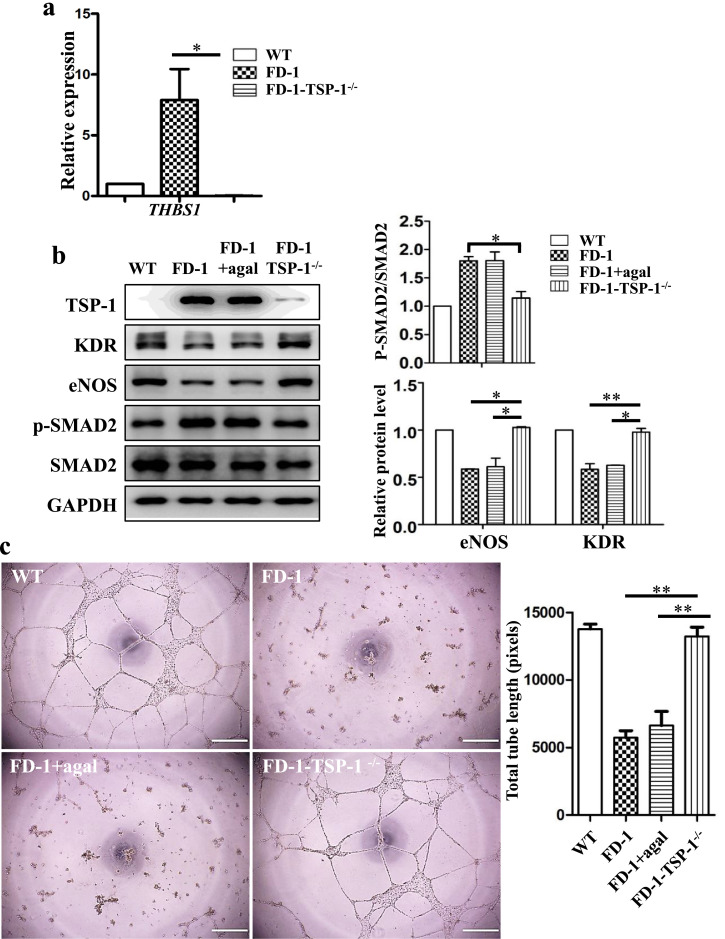
3.7. Knockout of TSP-1 in FD-1-iPSCs using CRISPR/Cas9
Next, we asked whether TSP-1 is also critical for angiogenesis in FD-VECs. We generated TSP-1-Knockout (KO) cells in FD-iPSCs using the CRISPR/Cas9 system. To disrupt TSP-1 expression in FD-1-iPSCs, we introduced two sgRNAs designed by Cas-Designer (http://Rgenome.net/cas-designer) into FD-1-iPSCs. The TSP-1 KO strategy and sgRNA target sequences are shown in Supplementary Fig. 6a. Of the two TSP-1 target sgRNAs (Supplementary Fig. 6b), the T2 sgRNA gave a higher indel efficiency in FD-1-iPSCs (Supplementary Fig. 6c). Thereafter, we transfected FD-1-iPSCs with this TSP-1-specific sgRNA (T2) and Cas9. Of 30 transfected clones, we found 3 TSP-1 KO clones using targeted deep sequencing. Of these three, only one clone successfully maintained pluripotency during subsequent cultures. FD-1-TSP-1−/− iPSCs showed biallelic mutations in the THBS-1 gene lacking 23 bp and 9 bp stretches, respectively (Supplementary Fig. 6d). Interestingly, unlike what we saw with FD-VECs and agalsidase-ß-treated FD-VECs, we were able to produce normal looking VECs from FD-1-TSP-1−/− iPSCs (Supplementary Fig. 6e). We did not observe any transcription of THBS1 in FD-1-TSP-1−/− VECs (Fig. 5a), nor did TSP-1−/− VECs show increased expression of the angiogenic factors KDR and eNOS or reduced p-SMAD2 levels (Fig. 5b). Also, unlike FD-1 and agalsidase-ß-treated FD-1 VECs, we found FD-1-TSP-1−/- VECs recovered the ability to form tubules (Fig. 5c). These results indicate enhanced TSP-1 plays a key role in the dysfunctional angiogenesis of FD-VECs.
Fig. 5.
Knock-out (KO) of TSP-1 in FD-1-iPSCs using the CRISPR/CAS9 system. (a) No transcriptional expression of THBS1 in FD-1-TSP-1−/− VECs. The data are expressed as means ± SEM (n = 3). * p < 0.05. (b) Decreased p-SMAD2 in FD-1-TSP-1−/− VECs. FD-1-TSP-1−/− VECs show enhanced expression of KDR and eNOS but reduced SMAD2 signaling. The data are expressed as means ± SEM (n = 2). * p < 0.05; ** p < 0.01. (c) Tube-like structure formation in FD-1-TSP-1−/− VECs. Knockout of TSP-1 restores tube-like structure formation in FD-1-VECs. Scale bar, 200 μm.
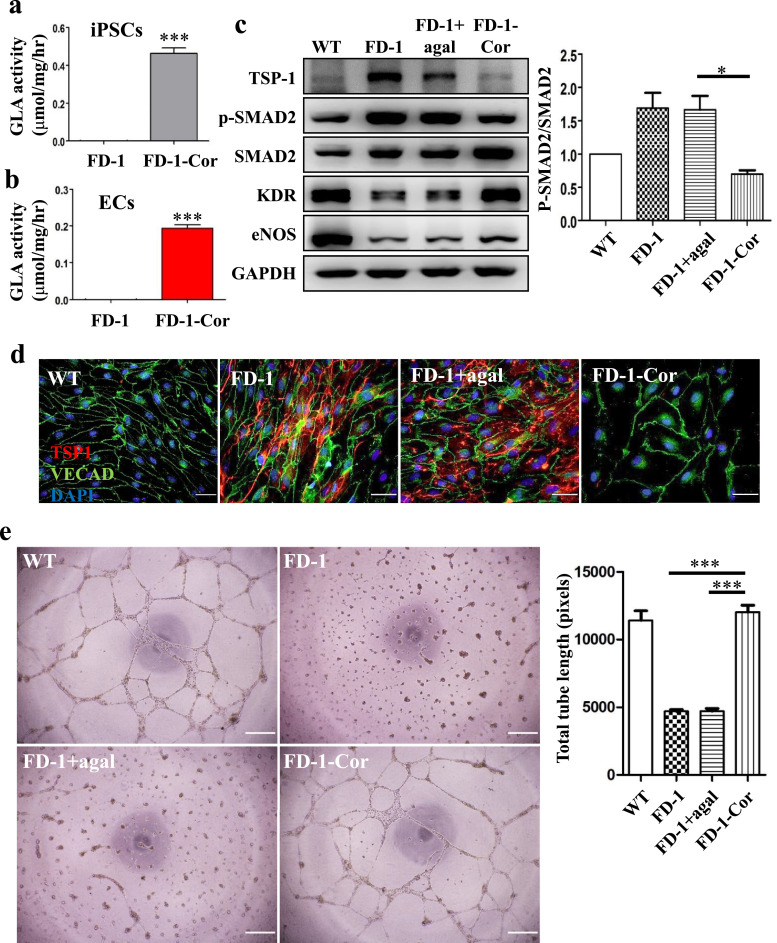
3.8. Correction of the mutated GLA gene using the CRISPR/Cas9 system in FD 1-iPSCs
To determine whether it is reduced GLA activity that interferes with the angiogenic activity of FD-VECs, we corrected a hemizygous GLA mutation in FD-iPSCs using the CRISPR/Cas9 genome editing tool. We performed the gene correction in FD-iPSCs via ribonucleoprotein (RNP) and single-stranded oligodeoxynucleotide (ssODN) delivery (Supplementary Fig. 7a). To discriminate the reference GLA sequence, we introduced four silent mutations in the ssODN (Supplementary Fig. 7b). We designed two sgRNAs near the mutated region to correct a deletion mutation in the GLA gene (Supplementary Fig. 7b). Of those sgRNAs, the T2 sgRNA showed a higher HDR frequency than the T1 sgRNA (Supplementary Fig. 7c). To assess off-target effects for the T2 sgRNA, we examined potential off-target sites that differed from the on-target site by up to three nucleotides using Cas-OFFinder [29]. Indel mutations for a total of 11 potential off-target sites for sgRNA#2 were examined in the genome of Cas9 protein-treated FD-1-iPSCs by targeted deep sequencing, but we found no evidence of off-target indels at those potential off-target sites (Supplementary Fig. 7f). These results indicate the T2 sgRNA specifically targets the mutated region of the GLA gene. We then seeded CRIPSR/Cas9-transfected FD-1-iPSCs as single cells to obtain gene-corrected iPSC clones. Of the 48 clones we analyzed by targeted deep sequencing, we identified one as gene-corrected by Sanger sequencing (Supplementary Fig. 7e). The resulting gene-corrected FD-1-Cor-iPSCs (Fig. 6a) and their differentiated FD-1-Cor-VECs (Fig. 6b) both showed significantly improved GLA activity when compared with their respective non-corrected controls. We also found enhanced expression of the angiogenic factors KDR and eNOS and reduced levels of TSP-1 and p-SMAD2 in FD-1-Cor-VECs (Fig. 6c). Using immunostaining, we confirmed FD-1-Cor-VECs produce low WT-like levels of TSP-1 (Fig. 6d). Furthermore, unlike FD-1 and agalsidase-ß-treated FD-1 VECs, FD-1-Cor-VECs were capable of forming tubules (Fig. 6e). These results suggest GLA mutation in FD-VECs leads to vascular dysfunction by enhancing the expression of anti-angiogenic factors.
Fig. 6.
Correction of the GLA mutation in FD-1-iPSCs using the CRISPR/CAS9 system. (a) GLA activity in FD-1-Cor-iPSCs. The data are expressed as means ± SEM (n = 3). *** p < 0.001. (b) GLA activity in FD-1-VECs. The data are expressed as means ± SEM (n = 3). *** p < 0.001. (c) Angiogenesis recovery in FD-1-Cor-VECs. Like WT-VECs, FD-1-Cor-VECs form tube-like structures. Scale bar, 200 μm. (d) Rescue of SMAD2 signaling in FD-1-Cor-VECs. Compared to their levels in FD-1-VECs, correction of the mutated GLA gene increases the expression of KDR and eNOS and reduces the levels of TSP-1 and p-SMAD2. The data are expressed as means ± SEM (n = 2). ** p < 0.01. (e) Immunostaining of TSP-1 in FD-1-Cor-VECs. Expression of TSP-1 is reduced in FD-1-Cor-VECs to a level similar to that of WT-VECs. Scale bar, 50 μm.
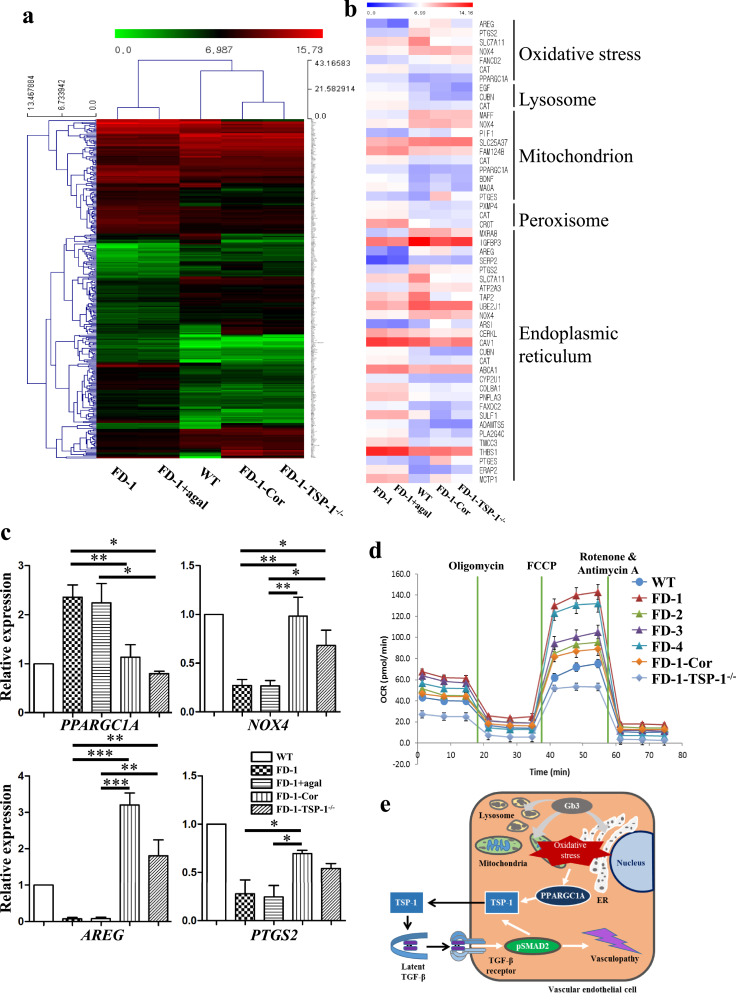
3.9. FD-VECs show altered transcriptional profiles in oxidative stress-related genes
In an RNA-sequencing (RNA-seq) analysis, we found global changes in the transcriptomes of FD-1-VECs and FD-1-VECs+agal that were mostly analogous to the changes in FD-1-Cor-VECs and FD-1-TSP-1−/− VECs when compared to WT-VECs (Fig. 7a). Next, because FD is a lysosomal storage disorder, we examined gene ontologies related to lysosomes and cell metabolism. We observed in FD-1-Cor-VECs and FD-1-TSP-1−/− VECs a recovery of the changes we observed in the differentially expressed genes (DEGs) related to lysosomes in FD-VECs to levels similar to WT-VECs (Fig. 7b). These results imply GLA mutations or TSP1 overexpression induce aberrant transcriptional activities in metabolic genes, including those related to oxidative stress, lysosomes, mitochondria, peroxisomes, and the endoplasmic reticulum. Furthermore, when we corrected the mutated GLA gene or knocked out TSP1 in FD-VECs, it rescued the expression of genes associated with angiogenesis, aging, cell migration, cell proliferation, the extracellular matrix, and the regulation of endothelial cell apoptosis to levels similar to those of WT-VECs (Supplementary Fig. 8b). Thus, it is conceivable that the enhanced expression of TSP-1 caused by GLA mutations induces the metabolic and cellular dysfunction of FD-VECs. We also confirmed the recovery of the expression of several oxidative stress-related genes (i.e., Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1A), NADPH oxidase 4 (NOX4), Amphiregulin, (AREG), and Prostaglandin-endoperoxide synthase 2 (PTGS2)) in FD-1-Cor-VECs and FD-1-TSP-1−/- VECs by RT-PCR (Fig. 7c). Furthermore, we found using a mitochondrial respiration assay that the increased oxidation consumption rate (OCR) of FD-VECs is reduced in FD-1-Cor-VECs and FD-1-TSP-1−/− VECs (Fig. 7d). Collectively, we have demonstrated that the correction of the GLA mutation in FD-VECs or knock-out of TSP1 rescues their metabolic dysfunction and reduces oxidative stress.
Fig. 7.
RNA-sequencing analysis. (a) Heat map and hierarchical clustering of the RNA-seq data from FD-1-VECs, FD-1-VECs+agal, WT-VECs, FD-1-Cor-VECs, and FD-1-TSP-1−/- VECs. (b) Heat map of the significant transcriptional changes in metabolism-associated genes. (c) Transcriptional expression of oxidative stress-associated genes as measured by qPCR. The data are expressed as means ± SEM (n = 2). * p < 0.05; ** p < 0.01**; *** p < 0.001. (d) Oxygen consumption rates in FD-VECs as measured with a Seahorse XF24 analyzer. (e) A schematic model for vasculopathy in FD-VECs.
4. Discussion
Here, we report for the first time that the overexpression of TSP-1 and/or the activation of SMAD2 signaling caused by Gb3 accumulation lead to endothelial cell dysfunction in FD. Gb3 accumulation contributes to several clinical complications for FD patients (i.e., angiokeratoma, cornea verticillata, hypertrophic cardiomyopathy, ischemic stroke, and renal failure) that all progress with age (www.bravecommunity.com). It is assumed that these clinical phenotypes are caused by Gb3 accumulation in the endothelium [38,39]. Thus, we suspected the progressive complications of FD patients may be caused by dysfunctional angiogenesis. To test this hypothesis, we induced the differentiation of FD-iPSCs into VECs and examined the functionality of the resulting FD-VECs. FD-VECs show no angiogenic capacity even after the transient removal of Gb3 by enzymatic treatment. Intriguingly, we found enhanced TSP-1 and p-SMAD2 levels in FD-VECs compared to WT-VECs. We also found that the knockout of TSP-1 or the suppression of p-SMAD2 activity rescues the angiogenic activity of FD-VECs. Based on our results, we propose a new model for the impaired angiogenesis of FD in which Gb3 accumulation caused by deficient GLA activity leads to enhanced expression of TSP-1. Then, this increased TSP-1 expression activates latent TGF-ß in FD-VECs, thereby disrupting angiogenesis.
Angiogenesis begins with the remodeling of tube-like structures formed from endothelial cell precursors. This allows vessels to grow as they form branches and sprout new vessels [40]. Complicated intercellular signaling in the initial tube-like structure recruits support cells such as pericytes, smooth muscle cells, and fibroblasts that help remodel the extracellular matrix [41,42]. The evolvement and remodeling of the vascular network is accomplished by interactions of angiogenesis-associated factors with their receptors and down-stream signaling networks [43]. We found FD-VECs are capable of forming tube-like structures at an early stage of differentiation (Supplementary Fig. 2a), but not at later stages of differentiation (Fig. 2a) because late stage-FD-VECs show aberrant expression of angiogenesis-associated factors (Fig. 2c and e). Thus, although FD-VECs can form vascular tubes at early stages, they seem to lose this ability as they differentiate. We also observed abnormal expression of angiogenesis-associated factors (except for VECAD) in the peritubular capillaries of renal biopsies from FD patients (Fig. 3). This weak angiogenic competence in late-stage FD-VECs suggests that the complications FD patients face as they age are likely caused by a progressive reduction in VEC functionality.
TGF-β signaling plays important roles in vascular remodeling via its activation of endothelial cell proliferation and migration [44,45]. Additionally, TGF-β1 regulates the expression of KDR and cell surface VEGF binding capacity, thereby controlling the functionality of VECs [46]. Here, we found strong activation of SMAD2 signaling in FD-VECs regardless of whether they had been treated with agalsidase-β (Fig. 4a). We were also able to rescue the ability of FD-VECs to form tubules by inhibiting SMAD2 signaling (Fig. 4e). Thus, enhanced SMAD2 signaling causes FD-VEC dysfunction. In addition, agalsidase-β treatment cannot normalize SMAD2 signaling defects in FD-VECs (Fig. 4d and e). It is therefore likely that although agalsidase-β treatment transiently reduces Gb3 accumulation, it does not affect TGF-β signaling in FD patients. In a gene ontology analysis, we found 4 GO terms related to SMAD signaling that were particularly highly ranked in FD-VECs (Supplementary Fig. 8a). Still, the mechanisms by which Gb3 accumulation activates SMAD2 signaling remain unclear. Together, our findings imply activated TGF-β signaling is closely linked to FD-associated vasculopathy.
TSP-1 binds diverse ligands and receptors in the extracellular matrix [47]. TSP-1 bound to VEGF or FGF2 acts as an endogenous inhibitor of angiogenesis by inhibiting endothelial cell migration, growth, and permeability [48]. Enhanced TSP-1 levels reduce neovascularization in patients with peripheral artery disease [49]. Unexpectedly, in this study, we found FD-VECs show strongly enhanced TSP-1 expression compared to WT-VECs (Fig. 2c). We hypothesized that this enhanced TSP-1 expression contributes to their abnormal morphology (Supplementary Fig. 2b), reduced proliferation (Supplementary Fig. 2c), and defects in forming tube-like structures (Fig. 2a). We found that FD-VECs differentiated from TSP-1 KO-iPSCs show normal morphology (Supplementary Fig. 6e) and form normal tube-like structures (Fig. 5). We also found that the peritubular capillaries of renal biopsies from FD patients show enhanced TSP-1 expression when compared to control tissues (Fig. 3). Thus, our results suggest that the dysfunctional angiogenesis of FD patients is likely caused by enhanced TSP-1 expression in FD-VECs.
TGF-β is activated in bovine aortic endothelial cells when TSP-1 binds latent TGF-β [50] and causes a conformational rearrangement of the TGF-β LAP [51]. TGF-β1 stimulates the expression of angiogenic (VEGF-A) and antiangiogenic factors (TSP-1 and sFlt-1) in rat proximal tubular cells [52]. Until now, however, it remained unclear whether TGF-β signaling regulates the expression of TSP-1 in FD-VECs. Here, we report that the activation of TGF-β signaling enhances the expression of TSP-1 in FD-VECs. Activation of SMAD2 signaling enhances TSP-1 expression in WT-VECs (Fig. 4b and c), and, conversely, inhibition of SMAD2 signaling reduces TSP-1 in FD-VECs (Fig. 4d). In addition, TSP-1-KO FD-VECs show reduced p-SMAD2 (Fig. 5b), and correction of their mutated GLA gene down-regulates both the levels of p-SMAD2 and TSP-1 (Fig. 6c). Interestingly, the reduction of p-SMAD and/or TSP-1 rescues the functionality of FD-VECs (Figs. 4e, 5c, and 6e). Thus, elevated p-SMAD2 and TSP-1 contribute to FD-VEC dysfunction. Based on these results, it is possible that reciprocal interactions between SMAD2 signaling and TSP-1 expression are essential for maintaining VEC integrity.
Gb3 accumulation in FD patients may induce diverse metabolic changes, including the activation of oxidative stress, the production of inflammatory cytokines, and reduced production of nitric oxide [53]. We found when comparing FD-VECs to WT-VECs that the expression of eNOS (endothelial nitric oxide synthase) is significantly reduced (Fig. 2e) and the oxidative stress-associated genes NOX4, AREG, and PTGS2 are transcriptionally down-regulated (Fig. 7c). NOX4, an NADPH oxidase, plays a key role in intracellular ROS generation. AREG, an EGFR ligand, regulates numerous cellular functions. AREG KO mice show chronic liver damage because of dysregulation of the systems that deal with oxidative stress [54]. PTGS2 converts arachidonic acid to prostaglandin H2. PTGS2 inhibition reduces H2O2-induced increases in ROS generation in the endothelium [55]. Peroxisome proliferator-activated receptor gamma coactivator 1-α (PPARGC1A) acts as a crucial regulator of mitochondrial biogenesis and metabolism [56]. We observed in FD-VECs increased expression of PPARGC1A, which, in other cell types, is associated with GO terms like oxidative stress, aging, cell migration, and mitochondria (Fig. 7b and Supplementary Fig. 8b). FD-VECs produce more PPARGC1A transcripts than WT-VECs (Fig. 7c). Furthermore, we found that the oxygen consumption ratio (OCR) in FD-VECs is higher than that of WT-VECs (Fig. 7d). Thus, we have gathered conclusive evidence that Gb3 accumulation induces oxidative stress in FD-VECs. PPARGC1A buffers oxidative stress produced during myogenesis by enhancing the expression of antioxidant enzymes [57] and strongly suppresses the migration of VECs in vitro and angiogenesis by VECs in vivo [58]. Increased PPARC1A activates Notch1 signaling, which increases TSP-1 expression [58,59]. The transcriptional activity of the NEURL1B gene is obviously reduced in FD-VECs compared to WT-VECs and increased in FD-1-Cor-VECs and FD-1-TSP-1−/− VECs (Supplementary Fig. 8c). NEURL1B blocks Notch signaling by promoting the lysosomal degradation of Jagged1 [60]. Also, oxidative stress reportedly activates TSP-1 expression in VECs [61]. We found FD-VECs produce elevated levels of TSP-1 (Fig. 2), as do the renal tissues of FD patients (Fig. 3) when compared to controls. We also found that GLA mutation correction or TSP1 KO in FD-VECs rescues their expression of oxidative stress-associated genes (Fig. 7b and c) and their angiogenic tube-like structure forming capacity (Figs. 5c and 6e).
As depicted in Fig. 7e, we suggest a schematic model for the aberrant angiogenesis of FD-VECs in which Gb3 accumulation induces oxidative stress, which then enhances TSP-1 expression. This elevated TSP-1 activates TGF-β signaling, which then finally disrupts angiogenesis. This model provides new insight into how dysfunctional VEC angiogenesis in peritubular capillaries may lead to the progression of the complications associated with Fabry disease as patients age. Furthermore, our findings suggest chemicals or molecules that regulate SMAD2 signaling and/or TSP-1 should be screened for their potential as new therapeutic drugs in FD.
Acknowledgments
We thank Ms. Sora Oh for maintaining human iPSCs.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102633.
Contributor Information
Beom-Hee Lee, Email: bhlee@amc.seoul.kr.
Yong-Mahn Han, Email: ymhan@kaist.ac.kr.
Appendix. Supplementary materials
References
- 1.Desnick R.J., Ioannou Y.A., Eng C.M. α-Galactosidase A deficiency: Fabry disease. In: Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson KM, editors. The online metabolic and molecular bases of inherited disease. The McGraw-Hill Companies, Inc.; New York, NY: 2014. [Google Scholar]
- 2.Clarke J.R. Narrative review: Fabry disease. Ann Intern Med. 2007;146(6):425–433. doi: 10.7326/0003-4819-146-6-200703200-00007. [DOI] [PubMed] [Google Scholar]
- 3.Meikle P.J., Hopwood J.J., Clague A.E., Carey W.F. Prevalence of lysosomal storage disorders. JAMA. 1999;281(3):249–254. doi: 10.1001/jama.281.3.249. [DOI] [PubMed] [Google Scholar]
- 4.Spada M., Pagliardini S., Yasuda M., Tukel T., Thiagarajan G., Sakuraba H. High incidence of later-onset Fabry disease revealed by newborn screening. Am J Hum Genet. 2006;79(1):31–40. doi: 10.1086/504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H.Y., Chong K.W., Hsu J.H., Yu H.C., Shih C.C., Huang C.H. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ Cardiovasc Genet. 2009;2(5):450–456. doi: 10.1161/CIRCGENETICS.109.862920. [DOI] [PubMed] [Google Scholar]
- 6.Orteu C.H., Jansen T., Lidove O., Jaussaud R., Hughes D.A., Pintos-Morell G. Fabry disease and the skin: data from FOS, the Fabry outcome survey. Br J Dermatol. 2007;157(2):331–337. doi: 10.1111/j.1365-2133.2007.08002.x. [DOI] [PubMed] [Google Scholar]
- 7.Schiffmann R., Warnock D.G., Banikazemi M., Bultas J., Linthorst G.E., Packman S. Fabry disease: progression of nephropathy, and prevalence of cardiac and cerebrovascular events before enzyme replacement therapy. Nephrol Dial Transplant. 2009;24(7):2102–2111. doi: 10.1093/ndt/gfp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta A., Ricci R., Widmer U., Dehout F., de Lorenzo A.G., Kampmann C. Fabry disease defined: baseline clinical manifestations of 366 patients in the Fabry outcome survey. Eur J Clin Investig. 2004;34(3):236–242. doi: 10.1111/j.1365-2362.2004.01309.x. [DOI] [PubMed] [Google Scholar]
- 9.Cleeland C.S., Hilz M.J. Pathophysiology and assessment of neuropathic pain in Fabry disease. Acta Paediatr. 2002;91:33. doi: 10.1111/j.1651-2227.2002.tb03110.x. [DOI] [PubMed] [Google Scholar]
- 10.Eng C.M., Guffon N., Wilcox W.R., Germain D.P., Lee P., Waldek S. Safety and efficacy of recombinant human alpha-galactosidase A–replacement therapy in Fabry's disease. N Engl J Med. 2001;345(1):9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 11.Schiffmann R., Kopp J.B., Austin H.A., 3rd Sabnis S., Moore D.F., Weibel T. Enzyme replacement therapy in Fabry disease: a randomized controlled trial. JAMA. 2001;285(21):2743–2749. doi: 10.1001/jama.285.21.2743. [DOI] [PubMed] [Google Scholar]
- 12.Rombach S.M., Smid B.E., Bouwman M.G., Linthorst G.E., Dijkgraaf M.G.W., Hollak C.E.M. Long term enzyme replacement therapy for Fabry disease: effectiveness on kidney, heart and brain. Orphanet J Rare Dis. 2013;8:9. doi: 10.1186/1750-1172-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang E., Kim Y.M., Kim D.H., Yoo H.W., Lee B.H. Life-threatening bleeding from gastric mucosal angiokeratomas during anticoagulation A case report of Fabry disease. Medicine. 2017;96(6):3. doi: 10.1097/MD.0000000000006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu L., Shayman J.A. Caveolin-associated accumulation of globotriaosylceramide in the vascular endothelium of alpha-galactosidase A null mice. J Biol Chem. 2007;282(29):20960–20967. doi: 10.1074/jbc.M702436200. [DOI] [PubMed] [Google Scholar]
- 15.Lee M.H., Choi E.N., Jeon Y.J., Jung S.-C. Possible role of transforming growth factor-β1 and vascular endothelial growth factor in Fabry disease nephropathy. Int J Mol Med. 2012;30(6):1275–1280. doi: 10.3892/ijmm.2012.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S., Kim J.A., Joo K.Y., Choi S., Choi E.N., Shin J.A. Globotriaosylceramide leads to K(Ca)3.1 channel dysfunction: a new insight into endothelial dysfunction in Fabry disease. Cardiovasc Res. 2011;89(2):290–299. doi: 10.1093/cvr/cvq333. [DOI] [PubMed] [Google Scholar]
- 17.Taguchi A., Maruyama H., Nameta M., Yamamoto T., Matsuda J., Kulkarni A.B. A symptomatic Fabry disease mouse model generated by inducing globotriaosylceramide synthesis. Biochem J. 2013;456:373–383. doi: 10.1042/BJ20130825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawagoe S., Higuchi T., Otaka M., Shimada Y., Kobayashi H., Ida H. Morphological features of iPS cells generated from Fabry disease skin fibroblasts using Sendai virus vector (SeVdp) Mol Genet Metab. 2013;109(4):386–389. doi: 10.1016/j.ymgme.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Itier J.M., Ret G., Viale S., Sweet L., Bangari D., Caron A. Effective clearance of GL-3 in a human iPSC-derived cardiomyocyte model of Fabry disease. J Inherit Metab Dis. 2014;37(6):1013–1022. doi: 10.1007/s10545-014-9724-5. [DOI] [PubMed] [Google Scholar]
- 20.Chou S.J., Yu W.C., Chang Y.L., Chen W.Y., Chang W.C., Chien Y. Energy utilization of induced pluripotent stem cell-derived cardiomyocyte in Fabry disease. Int J Cardiol. 2017;232:255–263. doi: 10.1016/j.ijcard.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 21.RayChaudhury A., Frazier W.A., D'Amore P.A. Comparison of normal and tumorigenic endothelial cells: differences in thrombospondin production and responses to transforming growth factor-beta. J Cell Sci. 1994;107(1):39–46. doi: 10.1242/jcs.107.1.39. [DOI] [PubMed] [Google Scholar]
- 22.Iruela-Arispe M.L., Luque A., Lee N. Thrombospondin modules and angiogenesis. Int J Biochem Cell Biol. 2004;36(6):1070–1078. doi: 10.1016/j.biocel.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 24.Park S.W., Koh Y.J., Jeon J., Cho Y.H., Jang M.J., Kang Y. Efficient differentiation of human pluripotent stem cells into functional CD34(+) progenitor cells by combined modulation of the MEK/ERK and BMP4 signaling pathways. Blood. 2010;116(25):5762–5772. doi: 10.1182/blood-2010-04-280719. [DOI] [PubMed] [Google Scholar]
- 25.Carpentier G. ImageJ News; 2012. ImageJ contribution: angiogenesis analyzer. [Google Scholar]
- 26.Benjamin E.R., Flanagan J.J., Schilling A., Chang H.H., Agarwal L., Katz E. The pharmacological chaperone 1-deoxygalactonojirimycin increases alpha-galactosidase A levels in Fabry patient cell lines. J Inherit Metab Dis. 2009;32(3):424–440. doi: 10.1007/s10545-009-1077-0. [DOI] [PubMed] [Google Scholar]
- 27.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweppe C.H., Bielaszewska M., Pohlentz G., Friedrich A.W., Buntemeyer H., Schmidt M.A. Glycosphingolipids in vascular endothelial cells: relationship of heterogeneity in Gb3Cer/CD77 receptor expression with differential Shiga toxin 1 cytotoxicity. Glycoconj J. 2008;25(4):291–304. doi: 10.1007/s10719-007-9091-7. [DOI] [PubMed] [Google Scholar]
- 29.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33(2):286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J.S., Meng X.L., Schiffmann R., Brady R.O., Kaneski C.R. Establishment and characterization of Fabry disease endothelial cells with an extended lifespan. Mol Genet Metab. 2007;92(1-2):137–144. doi: 10.1016/j.ymgme.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta K., Gupta P., Wild R., Ramakrishnan S., Hebbel R.P. Binding and displacement of vascular endothelial growth factor (VEGF) by thrombospondin: effect on human microvascular endothelial cell proliferation and angiogenesis. Angiogenesis. 1999;3(2):147–158. doi: 10.1023/a:1009018702832. [DOI] [PubMed] [Google Scholar]
- 33.Margosio B., Marchetti D., Vergani V., Giavazzi R., Rusnati M., Presta M. Thrombospondin 1 as a scavenger for matrix-associated fibroblast growth factor 2. Blood. 2003;102(13):4399–4406. doi: 10.1182/blood-2003-03-0893. [DOI] [PubMed] [Google Scholar]
- 34.Rusnati M., Urbinati C., Bonifacio S., Presta M., Taraboletti G. Thrombospondin-1 as a paradigm for the development of antiangiogenic agents endowed with multiple mechanisms of action. Pharmaceuticals. 2010;3(4):1241–1278. doi: 10.3390/ph3041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Morita I., Onodera M., Murota S.I. Induction of KDR expression in bovine arterial endothelial cells by thrombin: involvement of nitric oxide. J Cell Physiol. 2002;190(2):238–250. doi: 10.1002/jcp.10059. [DOI] [PubMed] [Google Scholar]
- 36.Mata-Greenwood E., Liao W.X., Zheng J., Chen D.B. Differential activation of multiple signalling pathways dictates eNOS upregulation by FGF2 but not VEGF in placental artery endothelial cells. Placenta. 2008;29(8):708–717. doi: 10.1016/j.placenta.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel C., Wiede J., Krutzsch H.C., Ribeiro S.M.F., Roberts D.D., Murphy-Ullrich J.E. Thrombospondin-1 is a major activator of TGF-beta in fibrotic renal disease in the rat in vivo. Kidney Int. 2004;65(2):459–468. doi: 10.1111/j.1523-1755.2004.00395.x. [DOI] [PubMed] [Google Scholar]
- 38.Elleder M., Bradová V., Smíd F., BudĚšínský M., Harzer K., Kustermann-Kuhn B. Cardiocyte storage and hypertrophy as a sole manifestation of Fabry's disease. Virchows Arch A. 1990;417(5):449–455. doi: 10.1007/BF01606034. [DOI] [PubMed] [Google Scholar]
- 39.Eng C.M., Banikazemi M., Gordon R.E., Goldman M., Phelps R., Kim L. A phase 1/2 clinical trial of enzyme replacement in Fabry disease: pharmacokinetic, substrate clearance, and safety studies. Am J Hum Genet. 2001;68(3):711–722. doi: 10.1086/318809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harry L.E., Paleolog E.M. From the cradle to the clinic: VEGF in developmental, physiological, and pathological angiogenesis. Birth Defects Res Part C Embryo Today Rev. 2003;69(4):363–374. doi: 10.1002/bdrc.10024. [DOI] [PubMed] [Google Scholar]
- 41.Avraamides C.J., Garmy-Susini B., Varner J.A. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8(8):604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gale NW, Yancopoulos GD. Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes Dev. 1999;13(9):1055–1066. doi: 10.1101/gad.13.9.1055. [DOI] [PubMed] [Google Scholar]
- 43.Cross M.J., Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci. 2001;22(4):201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- 44.Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 45.Oh S.P., Seki T., Goss K.A., Imamura T., Yi Y., Donahoe P.K. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci U S A. 2000;97(6):2626–2631. doi: 10.1073/pnas.97.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandriota S.J., Menoud P.A., Pepper M.S. Transforming growth factor beta 1 down-regulates vascular endothelial growth factor receptor 2/flk-1 expression in vascular endothelial cells. J Biol Chem. 1996;271(19):11500–11505. doi: 10.1074/jbc.271.19.11500. [DOI] [PubMed] [Google Scholar]
- 47.Rusnati M., Urbinati C., Bonifacio S., Presta M., Taraboletti G. Thrombospondin-1 as a paradigm for the development of antiangiogenic agents endowed with multiple mechanisms of action. Pharmaceuticals. 2010;3(4):1241. doi: 10.3390/ph3041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soto-Pantoja D.R., Kaur S., Roberts D.D. CD47 signaling pathways controlling cellular differentiation and responses to stress. Crit Rev Biochem Mol Biol. 2015;50(3):212–230. doi: 10.3109/10409238.2015.1014024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smadja D.M., d'Audigier C., Bieche I., Evrard S., Mauge L., Dias J.V. Thrombospondin-1 is a plasmatic marker of peripheral arterial disease that modulates endothelial progenitor cell angiogenic properties. Arterioscler Thromb Vasc Biol. 2011;31(3):551–559. doi: 10.1161/ATVBAHA.110.220624. [DOI] [PubMed] [Google Scholar]
- 50.Schultzcherry S., Murphyullrich J.E. Thrombospondin causes activation of latent transforming growth-factor-beta secreted by endothelial-cells by a novel mechanism. J Cell Biol. 1993;122(4):923–932. doi: 10.1083/jcb.122.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy-Ullrich J.E., Poczatek M. Activation of latent TGF-beta by thrombospondin-1: mechanisms and physiology. Cytokine Growth Factor Rev. 2000;11(1-2):59–69. doi: 10.1016/s1359-6101(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa T., Li J.H., Garcia G., Mu W., Piek E., Böttinger E.P. TGF-β induces proangiogenic and antiangiogenic factorsvia parallel but distinct Smad pathways1. Kidney Int. 2004;66(2):605–613. doi: 10.1111/j.1523-1755.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 53.Ravarotto V., Carraro G., Pagnin E., Bertoldi G., Simioni F., Maiolino G. Oxidative stress and the altered reaction to it in Fabry disease: a possible target for cardiovascular-renal remodeling? PLoS One. 2018;13(9):14. doi: 10.1371/journal.pone.0204618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berasain C., Garcia-Trevijano E.R., Castillo J., Erroba E., Santamaria M., Lee D.C. Novel role for amphiregulin in protection from liver injury. J Biol Chem. 2005;280(19):19012–19020. doi: 10.1074/jbc.M413344200. [DOI] [PubMed] [Google Scholar]
- 55.Munoz M., Sanchez A., Martinez M.P., Benedito S., Lopez-Oliva M.E., Garcia-Sacristan A. COX-2 is involved in vascular oxidative stress and endothelial dysfunction of renal interlobar arteries from obese Zucker rats. Free Radic Biol Med. 2015;84:77–90. doi: 10.1016/j.freeradbiomed.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 56.Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., Ciriolo M.R. Peroxisome proliferator-activated receptor gamma co-activator 1 alpha (PGC-1 alpha) and sirtuin 1 (SIRT1) reside in mitochondria possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285(28):21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baldelli S., Aquilano K., Ciriolo M.R. PGC-1 alpha buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 2014;5:13. doi: 10.1038/cddis.2014.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawada N., Jiang A., Takizawa F., Safdar A., Manika A., Tesmenitsky Y. Endothelial PGC-1 alpha mediates vascular dysfunction in diabetes. Cell Metab. 2014;19(2):246–258. doi: 10.1016/j.cmet.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rostama B., Turner J.E., Seavey G.T., Norton C.R., Gridley T., Vary C.P.H. DLL4/Notch1 and BMP9 interdependent signaling induces human endothelial cell quiescence via P27(KIP1) and thrombospondin-1. Arterioscler Thromb Vasc Biol. 2015;35(12):2626–2637. doi: 10.1161/ATVBAHA.115.306541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koutelou E., Sato S., Tomomori-Sato C., Florens L., Swanson S., Washburn M. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the notch ligand jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- 61.Hayashi H., Sakai K., Baba H., Sakai T. Thrombospondin-1 is a novel negative regulator of liver regeneration after partial hepatectomy through transforming growth factor-beta1 activation in mice. Hepatology. 2012;55(5):1562–1573. doi: 10.1002/hep.24800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.