Abstract
Milk contains numerous proteins including bioactive molecules that may be important in human nutrition. Thanks to improvements in proteomic methods, hundreds of proteins identified in milk are available through open data from different publications. We gathered these public data to produce an atlas reporting the cow milk proteins. We aggregated data from 20 publications reporting milk proteome and produced an atlas of 4654 unique proteins detected in milk from healthy cows. In this atlas, proteins are categorized according to four milk fractions: skimmed milk, whey, milk fat globule membranes (MFGM) and exosomes; and five lactation stages: colostrum period, early lactation, peak of lactation, mid-lactation and drying-off. These 9 protein lists were compared and annotated by Gene Ontology (GO) terms to identify the pathways they contribute to, the molecular signatures of different milk fractions and lactation stages. This data article compiles the 4654 cow milk proteins. This atlas may be used by researchers on human nutrition interested in milk protein allergy and/or digestibility in humans, and for milk processing industry. The atlas may be useful to i) find molecular signatures of physiological adaptations of dairy cows, ii) facilitate the isolation of proteins of interest, thanks to the knowledge on their presence in milk fractions and their period of secretion during lactation.
Keywords: Milk, Proteome, Cow, Lactation stage, Milk fraction
Specifications Table
| Subject | Animal Science and Zoology |
| Specific subject area | We aggregated data from 20 publications reporting milk proteome and produced an atlas of 4654 unique proteins detected in milk from healthy cows. |
| Type of data | Table |
| How data were acquired | Systematic review of the literature |
| Data format | Raw data and analysed data |
| Parameters for data collection | From the 87 publications on cow milk proteome, we selected 20 publications based on the availability of sufficient information, such as accession of supplementary data, precision of days in milk (DIM) and health status of cows, in order to retrieve and annotate proteins. |
| Description of data collection | We collected data from the 20 publications on bovine milk proteome. Protein identifiers (ID) were retrieved from tables in Portable Document Format (PDF) or from supplementary data files. Data were extracted with Tabula. A computational workflow was used to aggregate data and produce an atlas of 4654 unique proteins. |
| Data source location | INRAE Paris France |
| Data accessibility | Repository name: Portail Data INRAE Data identification number: -Dataset 1 “Distribution of 4654 cows milk proteins among different milk fractions”, DOI:10.15454/SUJJSQ -Dataset 2 “Distribution of 4654 cows milk proteins among lactation stages”, DOI:10.15454/MKM1P4 -Dataset 3 “Gene ontology of proteins detected in different milk fractions and lactation stages of dairy cows”, DOI:10.15454/1RF3R2 Direct URLs to data: -Dataset 1: https://doi.org/10.15454/SUJJSQ -Dataset 2: https://doi.org/10.15454/MKM1P4 -Dataset 3: https://doi.org/10.15454/1RF3R2 |
| Related research article | Mylène Delosière, José Pires, Laurence Bernard, Isabelle Cassar-Malek & Muriel Bonnet Milk proteome from in silico data aggregation allows the identification of putative biomarkers of negative energy balance in dairy cows Scientific Reports 10.1038/s41598-019-46142-7 |
Value of the Data
|
1. Data
The three datasets supporting this article are available in the “Portail Data INRAE” online repository (https://data.inra.fr/). Each dataset contains one table file:
-
i)
The dataset “Distribution of 4654 cows milk proteins among different milk fractions” (doi.org/10.15454/SUJJSQ) contains the file: “milk_fraction_4654_proteins” (.tab format, possible to download with Excel software), which reports the comparison of the four milk fractions resulting in specific and common Gene Name (GN) lists. The sheet entitled “Milk fractions” reports the list of proteins (identified by GN) which are either specific of a milk fraction, or present in one, two, three or four milk fractions.
-
ii)
The dataset “Distribution of 4654 cows milk proteins among lactation stages” (doi.org/10.15454/MKM1P4) contains the file “lactation_stage_4654_proteins” (.tab format, possible to download with Excel software), which reports the comparison of the five lactation stages resulting in specific and common GN lists. The sheet entitled “Lactation stages” reported the lists of proteins (identified by GN), which are specific of a lactation stage and present in one, two, three, four or five lactation stages.
-
iii)
The dataset “Gene ontology of proteins detected in different milk fractions and lactation stages of dairy cows” (doi.org/10.15454/1RF3R2) contains 10 files “GO_specific_fraction or lactation stage_4654_proteins” (.tab format, possible download with Excel software), which reports the GO enrichment analysis performed on the GN lists specific to a milk fraction or a lactation stage using the ProteINSIDE web service. As an example, the file entitled “GO_3_Whey” reported the GO enrichment of the lists of GN exclusively found in whey.
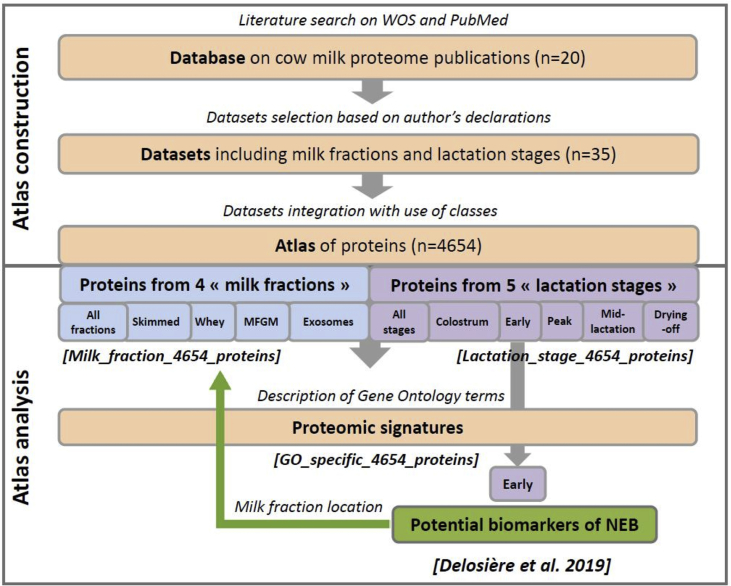
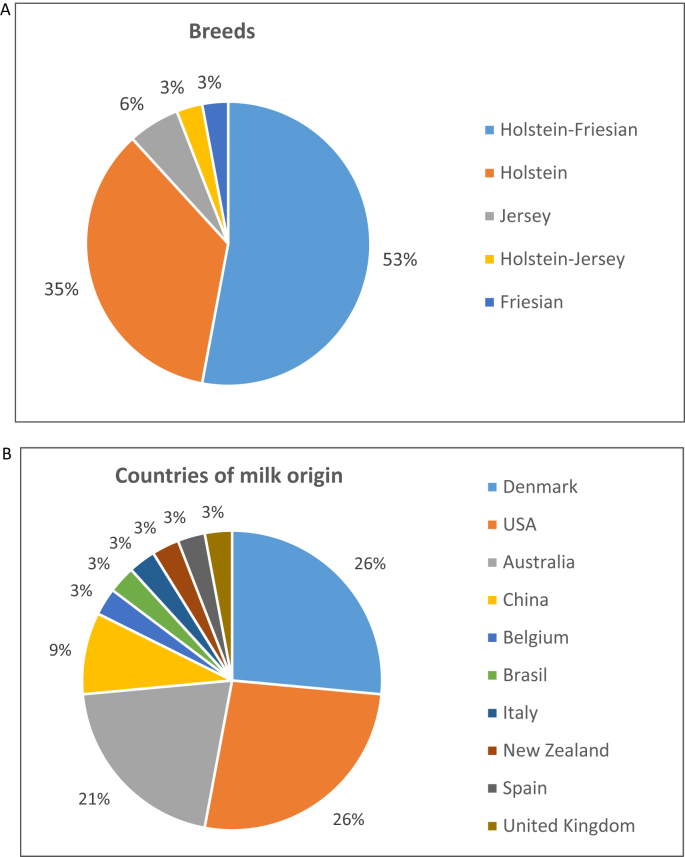
Fig. 1 is a flowchart reporting the workflow for the construction and analysis of the milk proteome atlas. Table 1 reports the list of the 20 publications on cow milk proteome used to build the atlas. Table 2 reports numbers of GN without duplicate, numbers of datasets (under parenthesis) and the associated references (with publication identifiers from Table 1 in superscript) depending on lactation stage and milk fraction. Fig. 2 describes the distribution of the cow breeds (A) and experimentation countries (B) of the 35 selected datasets.
Fig. 1.
Flowchart reporting the workflow for the construction and analysis of the milk proteome atlas.
Table 1.
The list of the 20 publications on cow milk proteome used to build the atlas.
| ID | Authors | Title | Year | Journal | Volume | Pages | URL |
|---|---|---|---|---|---|---|---|
| 1 | Yang, M. et al. | Comparative proteomic analysis of milk-derived exosomes in human and bovine colostrum and mature milk samples by iTRAQ-coupled LC-MS/MS | 2017 | Food Research International | 92 | 17–25 | https://doi.org/10.1016/j.foodres.2016.11.041 |
| 2 | Samuel, M. et al. | Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth | 2017 | Scientific Reports | 7 | https://doi.org/10.1038/s41598-017-06288-8 | |
| 3 | Reinhardt, T. A. & Lippolis, J. D. | Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. | 2008 | Journal of Dairy Science | 91 | 2307–2318 | https://doi.org/10.3168/jds.2007-0952 |
| 4 | Murgiano, L. et al. | Comparison of Milk Fat Globule Membrane (MFGM) Proteins of Chianina and Holstein Cattle Breed Milk Samples Through Proteomics Methods | 2009 | Nutrients | 302–315 | https://doi.org/10.3390/nu1020302 | |
| 5 | Nissen, A. et al. | In-depth analysis of low abundant proteins in bovine colostrum using different fractionation techniques. | 2012 | Proteomics | 12 | 2866–2878 | https://doi.org/10.1002/pmic.201200231 |
| 6 | Nissen, A. et al. | Colostrum and milk protein rankings and ratios of importance to neonatal calf health using a proteomics approach. | 2017 | Journal of Dairy Science | 100 | 2711–2728 | https://doi.org/10.3168/jds.2016–11722 |
| 7 | Golinelli, L. P. et al. | Proteomic Analysis of Whey from Bovine Colostrum and Mature Milk. | 2011 | Brazilian Archives of Biology and Technology | 54 | 761–768 | |
| 8 | Yang, M. et al. | Comparative proteomic exploration of whey proteins in human and bovine colostrum and mature milk using iTRAQ-coupled LC-MS/MS. | 2017 | International Journal of Food Sciences and Nutrition | 68 | 671–681 | https://doi.org/10.1080/09637486.2017.1279129 |
| 9 | Lu, J. et al. | Changes in Milk Proteome and Metabolome Associated with Dry Period Length, Energy Balance, and Lactation Stage in Postparturient Dairy Cows. | 2013 | Journal of Proteome Research | 12 | 3288–3296 | https://doi.org/10.1021/pr4001306 |
| 10 | Zhang, L. et al. | Bovine Milk Proteome in the First 9 Days: Protein Interactions in Maturation of the Immune and Digestive System of the Newborn. | 2015 | Plos One | 10 | https://doi.org/10.1371/journal.pone.0116710 | |
| 11 | Nissen, A. et al. | Expanding the bovine milk proteome through extensive fractionation | 2013 | Journal of Dairy Science | 96 | 7854–7866 | https://doi.org/10.3168/jds.2013-7106 |
| 12 | Tacoma, R. et al. | Ratio of dietary rumen degradable protein to rumen undegradable protein affects nitrogen partitioning but does not affect the bovine milk proteome produced by mid-lactation Holstein dairy cows. | 2017 | Journal of Dairy Science | 100 | 7246–7261 | https://doi.org/10.3168/jds.2017-12647 |
| 13 | Tacoma, R. et al. | Characterization of the bovine milk proteome in early-lactation Holstein and Jersey breeds of dairy cows. | 2016 | Journal of Proteomics | 130 | 200–210 | https://doi.org/10.1016/j.jprot.2015.09.024 |
| 14 | Boehmer, J. L. et al. | Proteomic Analysis of Differentially Expressed Proteins in Bovine Milk During Experimentally Induced Escherichia coli Mastitis. | 2008 | Journal of Dairy Science | 91 | 4206–4218 | https://doi.org/10.3168/jds.2008-1297 |
| 15 | Danielsen, M. et al. | Quantitative milk proteomics - Host responses to lipopolysaccharide-mediated inflammation of bovine mammary gland. | 2010 | Proteomics | 10 | 2240–2249 | https://doi.org/10.1002/pmic.200900771 |
| 16 | Alonso-Fauste, I. et al. | Proteomic characterization by 2-DE in bovine serum and whey from healthy and mastitis affected farm animals. | 2012 | Journal of Proteomics | 75 | 3015–3030 | https://doi.org/10.1016/j.jprot.2011.11.035 |
| 17 | Li, S. S. et al. | Effects of the processing methods of corn grain and soybean meal on milk protein expression profiles in dairy cows | 2015 | Animal | 9 | 267–274 | https://doi.org/10.1017/s1751731114002225 |
| 18 | Vincent, D. et al. | Milk Bottom-Up Proteomics: Method Optimization. | 2016 | Frontiers in Genetics | 6 | https://doi.org/10.3389/fphys.2015.00360 | |
| 19 | Boggs, I. et al. | Proteomics data in support of the quantification of the changes of bovine milk proteins during mammary gland involution. | 2016 | Data in brief | 8 | 52–55 | https://doi.org/10.1016/j.dib.2016.05.013 |
| 20 | Mudaliar, M. et al. | Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics | 2016 | Molecular Biosystems | 12 | 2748–2761 | https://doi.org/10.1039/c6mb00290k |
ID: Publication identifiers.
Table 2.
Numbers of GN without duplicate, numbers of datasets (under parenthesis) and the associated references (publication identifiers from Table 1 in superscript) depending on lactation stage and milk fraction.
| Colostrum | Early lactation | Peak lactation | Mid-lactation | Drying-off | Totala | |
|---|---|---|---|---|---|---|
| Exosomes | 3991 (4)1,2 | nd | nd | nd | nd | 3991 (4) |
| MFGM | 26 (1)3 | 246 (3)3,4,9 | nd | nd | nd | 246 (4) |
| Skimmed | 341 (4)5,6,7 | 318 (2)6,11 | 6 (2)12 | 122 (6)14−18 | nd | 496 (14) |
| Whey | 391 (3)5,8,10 | 223 (2)10,11 | 775 (4)12,13 | 548 (2)19,20 | 148 (2)19 | 1234 (13) |
| Totala | 4225 (12) | 594 (8) | 779 (6) | 577 (8) | 148 (2) | 4654b (35) |
nd: no data.
ID1 and ID2 grouping 3 datasets (24h, 48h, 72h after parturition); ID3, ID1, ID5, ID6, ID7 grouping 2 datasets (48h and 72h after parturition); ID8, ID9, ID10, ID11, ID12 grouping 2 datasets (different diets); ID13 grouping 2 datasets (different breeds); ID12, ID2, ID4, ID15, ID18 grouping 2 datasets (different breeds); ID19, ID20 grouping 2 datasets (different DIM during mid-lactation); ID19 grouping 2 datasets (3d and 8d after drying-off).
Total of unique GN after elimination of duplicates.
Total of unique GN after elimination of duplicates.
Fig. 2.
Distribution of the cow breeds (A) and cow's milk country of origin (B) of the 35 datasets.
2. Experimental design, materials, and methods
By a computational approach, we gathered and mined public data in the context of lactation, a complex and dynamic physiological process. The data collection was driven by the free access to large lists of proteins. These lists were organized according to the physiological states of cows and the milk fractions in which proteins were detected.
A computational workflow (Fig. 1) was used to aggregate data from 20 publications reporting cow milk proteome and to produce an atlas of 4654 unique proteins. The atlas was categorized in lists according to four milk fractions: skimmed milk, whey, MFGM and exosomes; and five lactation stages: colostral period, early lactation, peak of lactation, mid-lactation and drying-off. These protein lists were compared depending on milk fractions and lactation stages and annotated by GO terms to identify pathways and molecular signatures of each milk fraction and lactation stage. Consequently, the present data article reports an atlas of 4654 unique proteins distributed according to i) four milk fractions; ii) five lactation stages, and iii) GO terms enrichment.
The data presented here were generated as part of an accompanying publication on the identification of putative biomarkers of negative energy balance in dairy cows by using milk proteome from in silico data aggregation [1].
3. Methods
-
a)
Publications database
First, we collected publications on bovine milk proteome whatever the effect studied by the authors. The thesaurus to collect the targeted publications combined a description of three terms or their related words: “bovine” with (bovine OR cows OR cattle), “milk” with (*milk OR dairy) and “proteome” with (proteom* OR protein* OR secretom* OR exosom* OR secreted OR biomarker* OR vesicle* NOT (content OR concentration OR production)). This thesaurus was submitted to the title section of the search engines of the PubMed.gov (NCBI) and Web of Science Core Collection (Clarivate Analytics) until February 2018. The 87 resulting publications were curated based on the availability of sufficient information, such as accession of supplementary data, precision of days in milk (DIM) and health status of cows, in order to retrieve and annotate proteins. Twenty publications (Table 1) were selected in order to extract proteins lists. A flowchart (Fig. 1) reports the workflow for the construction and analysis of the computational milk proteome atlas.
-
b)
Data extraction
Protein identifiers (ID) were retrieved from tables in Portable Document Format (PDF) or from supplementary data files of the 20 publications. Data were extracted with Tabula.
-
c)
ID conversion into Gene name
Protein ID were standardized and converted into the corresponding GN, as unique identifier to be free from species, by use of three tools: Retrieve/ID Mapping tool of the Uniprot database (The UniProt [2]), the Protein Identifier Cross-Reference service [3] and/or the ProteCONVERT tool of the ProteINSIDE web interface [4].
-
d)
Class definitions
We sorted data according to four milk fractions and five lactation stages of dairy cows, based on paper's statements on protein extraction and DIM. The milk fractions were: i) skimmed milk, aggregating proteins isolated by centrifugation under 100 000g combined with or without casein depletion by acidification; ii) whey, aggregating proteins isolated by centrifugation over 100 000g; iii) MFGM, aggregating proteins isolated from cream milk, and iv) exosomes, aggregating proteins isolated from skimmed milk by protocol based on sucrose gradient [5]. The lactation stages were: i) colostrum period, aggregating proteins from colostrum collected during the first 5 days post-partum; ii) early lactation, aggregating proteins from milk collection between 6 and 21 DIM; iii) peak lactation, aggregating proteins from milk collected between 22 and 80 DIM; iv) mid-lactation, aggregating proteins from milk collected after 81 DIM, and v) drying-off, aggregating proteins from milk collected at 3 and 8 days after stopping milking. This categorization provided 35 datasets from the 20 publications (Table 2): among them 14 datasets focused on skimmed milk, 13 on whey, 4 on MFGM and 4 on exosomes.
The number of datasets available in the literature decreased when the complexity for milk fractionation increased: only four datasets were available for the exosome fraction while 14 were available for skimmed milk fraction. According to lactation stages whatever the milk fraction, 12 datasets dedicated to colostrum's proteins, 8 to early lactation, 6 to peak lactation, 8 to mid-lactation and 2 to the drying-off period. From the 20 publications, the 35 datasets referred to experimentations carried out with different cow breeds and in various countries (Fig. 2).
-
e)
Dataset aggregation
The full lists of GN coming from the 35 datasets were aggregated in an atlas of 8841 GN. Redundancies were discarded in each class providing 7135 GN useful for the milk fractions comparison and 6323 GN for the lactation stages comparison. After aggregation and discarding redundancies, an atlas of 4654 unique milk proteins was produced.
-
f)
Comparison of classes
Venn diagram (Draw Venn Diagram tool from VIB/Ugent) were used to identify GN specifically identified in one and up to four milk fractions or lactation stages. The comparison of GN lists according to milk fractions identified 95 GN common to all four milk fractions whereas 93, 488, 15 and 3139 GN were unique to skimmed milk, whey, MFGM and exosomes fractions. Fourty-four GN were identified both in skimmed milk and whey, 2 in MFGM and skimmed milk, 95 in exosomes and skimmed milk, 7 in whey and MFGM, 407 in exosomes and whey, 65 in MFGM and exosomes. Fourteen GN were identified in MFGM, skimmed milk and whey; 142 in exosomes, skimmed milk and whey; 11 in exosomes, MFGM and skimmed milk; 37 between exosomes, MFGM and whey. The lists of GN in milk fractions are reported in the “milk_fraction_4654_proteins.tab” file.
The lists of GN according to lactation stages highlighted 105 GN present in all the lactation stages whereas 3288, 59, 185 and 155 GN were unique to colostrum period, early lactation, peak lactation and mid-lactation. One hundred ninety-seven GN were identified in both colostrum and early lactation milk; and 252 in both colostrum and peak lactation milk. Seventy-eight GN were identified in colostrum and in milk from early lactation, peak lactation and mid-lactation; 14 from colostrum and milk from early lactation, peak lactation and drying-off; 65 from colostrum and milk from early lactation and peak lactation; 9 from colostrum and milk from early lactation and drying-off. The lists of GN by lactation stage are reported in the “lactation_stage_4654_proteins.tab” file. The Venn diagrams compiling protein lists and the biological mining of protein categorization (are reported elsewhere [1]).
-
g)
Code availability
The code used for protein designation was the GN. Last conversion from ID to GN was February 2018 using tools described in the Methods section. The PDF extractor tool was Tabula (www.tabula.technology, Last update February 11, 2017). The used version of ProteINSIDE Workflow was 1.2 (last update November 17, 2016).
4. Data validation and quality control
Our search was based on a thesaurus conceived to target the milk bovine proteome and was submitted to two search engines of scientific publications (PubMed and Web of Science). From the resulting 87 publications, we applied exclusion criteria such as absence of protein ID access, species name, health status, or DIM of lactating cows; and selected only 20 publications for the atlas construction. These 20 publications referred to milk proteome from healthy cows characterized for breed, DIM and milk fraction. The main objective of this computational data aggregation is to obtain an overview of milk proteins independently of breed, age, country (Fig. 2), and whatever the methodologies of protein isolation and identification. Among those methodologies, iTRAQ labelling [[6], [7], [8], [9], [10], [11]], LCxLC-MS/MS detection [5,12] and 15 repetitions of nanoLC-MS/MS runs [13] allowed the detection of thousands of proteins. In order to verify the reliability of the atlas to mine pathways and biomarkers of the lactation processes, we combined Venn diagram to compare lists and annotations according to GO using ProteINSIDE web service [14]. ProteINSIDE was previously bench tested and the reliability and accuracy of GO annotations for ruminants species were published [14]. Lists of mined proteins were enriched for GO terms related to lactation process, and were composed of the major expected milk proteins as reported in Ref. [1], thus validating the atlas. The protein diversity may arise from the thousands of proteins identified from exosomes that are expected to derived from various cell types and found in the milk [5], and thus pave the molecular basis of the lactation process. However, recent publications prove the benefic wealth effect of minor milk components such as MFGM [15] that have bioactive properties [9], a potential to be markers of technological and sensorial milk qualities [16] and a role in the dynamics of digestion of human and bovine milk proteins for the improvement of infant formula [17]. During the building and analysis of our atlas, we found that proteins from the skimmed and the whey fractions were mostly different.
The content and proportions of protein fractions have notable effects on the nutritional value and technological properties of milk [18]. This atlas allows the identification of fractions containing specific proteins that may be of interest for research and industry. This knowledge is useful for scientists working on the isolation of protein fractions in milk and dairy process. Particular interest concerned proteins from colostrum that reflect in part the physiological state of dairy cows in the 5 days of the post-partum period. Datasets on whey, largely studied in literature, covered all the lactation period, therefore allowing comparisons among lactation stages. Colostrum period provides thousands of proteins compared to hundreds in later lactation stages, which represents a limitation of our atlas. This observation encourage proteomic efforts onto the milk from late lactation stages.
Some limitations are reported. The first limitation of the computational approach was the use of only part of the 87 relevant publications on cows. Thus, 67 publications were unusable either because they concerned cows with mastitis or because of lacking information on sampling collection period, methods of milk fractionation, animal characteristics (phenotype, feeding, husbandry conditions …), or protein IDs. The second limitation is the conversion of protein ID into GN that led to the loss of some data, such as the protein isoforms. The number of proteins identified according to fractions (Table 2) is strongly imbalanced due to the diversity of the proteomic methods used. Indeed, thousands of proteins were identified for exosomes by 15 successive LC-MS/MS analyses, compared to only hundreds of proteins for the others fractions, determined either by gel-based or gel-free nanoLC-MS/MS proteomics. The atlas aggregates proteins that were identified in milk, whatever their abundance. Lastly, due to the nature of the protein identification algorithms, false-positives may be present in datasets because not all datasets are equally filtered.
5. Re-use potential
The atlas allow enhancing our knowledge of the diversity of cow milk proteins. The major benefit of making this atlas available is to provide information of interest to the scientific community. The applied output is for nutrition researchers interested in milk protein allergy and/or digestibility in humans, and for industrials working on milk processing.
The atlas may be used to identify potential biochemical properties of proteins/peptides in bovine milk, or to isolate proteins of interest. For example, according to our atlas, carbonic anhydrase 6 (CA6), an essential factor for development of gastrointestinal tract of the human new born, is present exclusively in the whey fraction of cow milk. This atlas provides information about the molecular signatures of metabolic adaptations that occur throughout lactation. As part of our research on dairy ruminants, the protein lists and the molecular signatures of the lactation stages provide information about secreted proteins and physiological adaptations of cows, which are a prerequisite to the identification of molecular biomarkers and the understanding of dairy cows adaptations to husbandry conditions.
Finally, this computational approach using publicly available data is an elegant alternative to animal experiments to obtain an atlas of milk protein, without conducting new experiments on ruminants, which is consistent with the principles of Replacement, Reduction and Refinement of “Three Rs” for the use of animals in research.
Funding
No applicable funding.
Authors’ contributions
M.D. and M.B. designed the work.
M.D. wrote the main text, curated, extracted and reformatted the original datasets, created the atlas and the three smoothed datasets.
J.P., L.B. and I.C.M. reviewed the text.
M.B. wrote part of the text and reviewed the text.
All authors approved the manuscript.
Acknowledgments
We would like to thank Dr. Isabelle Veissier for proof reading manuscript. This work was performed in the framework of the program “Investissement d'Avenir” (16-IDEX-0001 CAP 20-25) funded by the Agence Nationale de la Recherche of the French government.
Conflict of Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Delosière M., Pires J., Bernard L., Cassar-Malek I., Bonnet M. Milk proteome from in silico data aggregation allows the identification of putative biomarkers of negative energy balance in dairy cows. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium T.U. 2017. UniProt: the Universal Protein Knowledgebase.http://www.uniprot.org/ [Google Scholar]
- 3.Wein S.P., Cote R.G., Dumousseau M., Reisinger F., Hermjakob H., Vizcaino J.A. Improvements in the protein identifier cross-reference service. Nucleic Acids Res. 2012;40:W276–W280. doi: 10.1093/nar/gks338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaspric N., Reichstadt M., Picard B., Tournayre J., Bonnet M. Protein function easily investigated by genomics data mining using the ProteINSIDE online tool. Genomics Comput. Biol. 2015;1:1–14. [Google Scholar]
- 5.Reinhardt T.A., Lippolis J.D., Nonnecke B.J., Sacco R.E. Bovine milk exosome proteome. J. Proteom. 2012;75:1486–1492. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Reinhardt T.A., Lippolis J.D. Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. J. Dairy Sci. 2008;91:2307–2318. doi: 10.3168/jds.2007-0952. [DOI] [PubMed] [Google Scholar]
- 7.Yang M., Cao X.Y., Wu R.N., Liu B.A., Ye W.H., Yue X.Q. Comparative proteomic exploration of whey proteins in human and bovine colostrum and mature milk using iTRAQ-coupled LC-MS/MS. Int. J. Food Sci. Nutr. 2017;68:671–681. doi: 10.1080/09637486.2017.1279129. [DOI] [PubMed] [Google Scholar]
- 8.Danielsen M., Codrea M.C., Ingvartsen K.L., Friggens N.C., Bendixen E., Rontved C.M. Quantitative milk proteomics - host responses to lipopolysaccharide-mediated inflammation of bovine mammary gland. Proteomics. 2010;10:2240–2249. doi: 10.1002/pmic.200900771. [DOI] [PubMed] [Google Scholar]
- 9.Ji X.X., Li X.S., Ma Y., Li D. Differences in proteomic profiles of milk fat globule membrane in yak and cow milk. Food Chem. 2017;221:1822–1827. doi: 10.1016/j.foodchem.2016.10.097. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y.X., Bu D.P., Zhao X.W., Sun P., Wang J.Q., Zhou L.Y. Proteomic analysis of cow, yak, buffalo, goat and camel milk whey proteins: quantitative differential expression patterns. J. Proteome Res. 2013;12:1660–1667. doi: 10.1021/pr301001m. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y., Zheng N., Zhao X.W., Zhang Y.D., Han R.W., Ma L. Proteomic characterization and comparison of mammalian milk fat globule proteomes by iTRAQ analysis. J. Proteom. 2015;116:34–43. doi: 10.1016/j.jprot.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Reinhardt T.A., Sacco R.E., Nonnecke B.J., Lippolis J.D. Bovine milk proteome: quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013;82:141–154. doi: 10.1016/j.jprot.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Tacoma R., Fields J., Ebenstein D.B., Lam Y.-W., Greenwood S.L. Characterization of the bovine milk proteome in early-lactation Holstein and Jersey breeds of dairy cows. J. Proteom. 2016;130:200–210. doi: 10.1016/j.jprot.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaspric N., Picard B., Reichstadt M., Tournayre J., Bonnet M. ProteINSIDE to easily investigate proteomics data from ruminants: application to mine proteome of adipose and muscle tissues in bovine foetuses. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard L., Bonnet M., Delavaud C., Delosière M., Ferlay A., Fougère H. Milk fat globule in ruminant: major and minor compounds, nutritional regulation and differences among species. Eur. J. Lipid Sci. Technol. 2018;120:1700039. [Google Scholar]
- 16.Nguyen H.T.H., Ong L., Hoque A., Kentish S.E., Williamson N., Ang C.S. A proteomic characterization shows differences in the milk fat globule membrane of buffalo and bovine milk. Food Biosci. 2017;19:7–16. [Google Scholar]
- 17.Zhang L.N., van Dijk A.D.J., Hettinga K. An interactomics overview of the human and bovine milk proteome over lactation. Proteome Sci. 2017;15 doi: 10.1186/s12953-016-0110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pegolo S., Mach N., Ramayo-Caldas Y., Schiavon S., Bittante G., Cecchinato A. Integration of GWAS, pathway and network analyses reveals novel mechanistic insights into the synthesis of milk proteins in dairy cows. Sci. Rep. 2018;8:566. doi: 10.1038/s41598-017-18916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]