Abstract.
In conventional retinal region detection methods for optical coherence tomography (OCT) images, many parameters need to be set manually, which is often detrimental to their generalizability. We present a scheme to detect retinal regions based on fully convolutional networks (FCN) for automatic diagnosis of abnormal maculae in OCT images. The FCN model is trained on 900 labeled age-related macular degeneration (AMD), diabetic macular edema (DME) and normal (NOR) OCT images. Its segmentation accuracy is validated and its effectiveness in recognizing abnormal maculae in OCT images is tested and compared with traditional methods, by using the spatial pyramid matching based on sparse coding (ScSPM) classifier and Inception V3 classifier on two datasets: Duke dataset and our clinic dataset. In our clinic dataset, we randomly selected half of the B-scans of each class (300 AMD, 300 DME, and 300 NOR) for training classifier and the rest (300 AMD, 300 DME, and 300 NOR) for testing with 10 repetitions. Average accuracy, sensitivity, and specificity of 98.69%, 98.03%, and 99.01% are obtained by using ScSPM classifier, and those of 99.69%, 99.53%, and 99.77% are obtained by using Inception V3 classifier. These two classification algorithms achieve 100% classification accuracy when directly applied to Duke dataset, where all the 45 OCT volumes are used as test set. Finally, FCN model with or without flattening and cropping and its influence on classification performance are discussed.
Keywords: image segmentation, image classification, fully convolutional networks, retina optical coherence tomography
1. Introduction
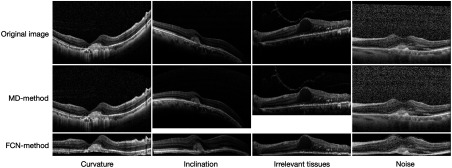
Over the past two decades, a majority of works related to optical coherence tomography (OCT)1,2 retinal image analysis have focused on two fields: segmentation3–21 and classification.22–34 Most of these works adopt a preprocessing process in order to make images have more attributes, which fit the needs of a follow-up procedure. This process can solve several key issues and prove to be very effective in practice. The first issue is that OCT images always include a large number of tissues outside the retinal area, which do not help the analysis of diseases, e.g., tissues under the retinal pigment epithelium (RPE) layer. Meanwhile, these irrelevant tissues and some background let the retina region-of-interest (RoI) area between internal-limiting membrane and RPE layers occupy only a small proportion of the entire B-scan. Thus, a preprocessing process is needed to wipe out these irrelevant areas so that follow-up procedures can focus on the key area. The second issue to be solved is speckle noise, which is caused by coherent processing of backscattered signals from multiple distributed targets that widely exist in most OCT images. Excessive noises are obviously disadvantageous for the analysis procedure, so we have a great demand to reduce their negative influences. A third issue of concern is that, due to idiosyncratic physiological structures of different patients and the inconsistency of imaging angles, OCT images of different instances always have different degrees of curvature and inclination angles. Characteristics formed by these differences do not contribute to the judgment of the disease, yet in some cases, they make it more difficult for the analysis process to achieve the desired purpose. Therefore, it is beneficial to flatten the retinal area according to the curvature of a retina and the angle of inclination, which can ensure that every instance has a relatively uniform form to avoid their harmful effects. The first row in Fig. 1 illustrates some OCT B-scans with the three previously mentioned key issues.
Fig. 1.
Performance of two preprocessing methods on three key issues.
Previous works that have concentrated on OCT image preprocessing for detecting retinal regions can be divided into two groups. One group aligns a retinal region by fitting a second-order polynomial to the RPE layer and then using it to flatten the retinal region.24,25,28,35 The problem with this type of method lies in its relying heavily on the segmentation of a clear RPE layer. When RPE layer has been severely distorted, where the disease is critical in some cases or some region of a retina is not imaged correctly, these algorithms will not be efficient since the right position of the RPE layer cannot be found. Another group aligns a retinal area by detecting the whole retinal region using morphological detection (MD) and fitting a second-order polynomial and/or a straight line to it.23,27,30 Using MD method to find the main part of a retinal region is a more robust work than segmenting a slender RPE layer, which could wipe out irrelevant areas that follow-up procedures do not need. However, this method also has some problems, which are mainly related to the precise detection of the retinal morphology. When speckle noise is heavy, or diseases cause some regions to have attributes that are similar to the background, or a detached tissue exists, it would detect a wrong morphology or include a number of unrelated parts, thus it could not detect the retinal region or wipe out demanding areas correctly, as is shown in the second row in Fig. 1, where the large size of images is displayed since the retinal regions are not detected correctly. The MD method could improve the detection quality of retinal regions by adjusting algorithm parameters, such as filter window size or binarization threshold, but it is still inaccurate and needs a lot of manual working.
Noise, irrelevant tissues, or diverse structure of retina B-scans persistently plague OCT image analysis and classification. It is difficult to handle complex situations with a unified process or stable parameters. But the advent of segmentation methods based on neural networks makes it possible to understand and process pictures at a semantic level. This attractive feature inspires us to propose a segmentation method for retinal OCT B-scans. Specifically, we use fully convolutional networks (FCN),36 a well-functioning deep learning segmentation framework to detect the RoI of an OCT image. FCN are based on convolutional neural networks (CNN), which have been widely used in computer vision (especially in image classification and image segmentation) since the ImageNet competition in 2012. Recently, several works have been done to simultaneously segment several retina layers from OCT images37–39 using deep learning or not, where they are devoted to segmenting different layers accurately. Here, we focus on segmenting the whole retinal area (i.e., RoI) to deal with the aforementioned preprocessing problems, and we discuss its effectiveness in improving automatic diagnosis performance of abnormal maculae.
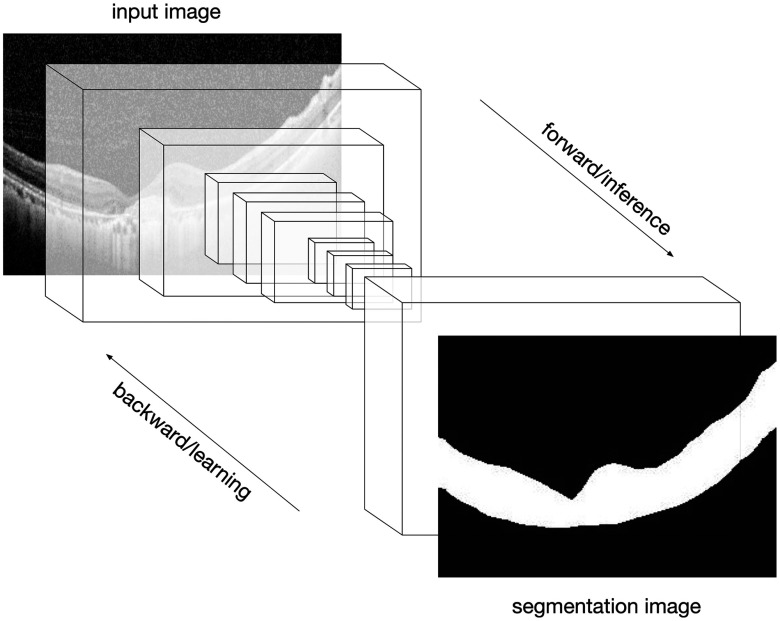
MD method mainly consists of three steps: denoising, such as median filter and block-matching; 3-D filtering (BM3D);40 binarizing by Otsu’s algorithm;41 and morphological opening-and-closing operation. In this method, a denoising step is necessary and a denoising method like BM3D not only has a high computation complexity but also blurs the RoI to some extent. By contrast, our proposed method (FCN method) utilizes a pretrained FCN model to segment a retinal region directly, as is shown in Fig. 2, where the brightness of pixels outside the retinal region is set to be zero. It needs neither adjusting parameters manually nor denoising preprocessing, and it is robust enough to adapt to various situations, as is illustrated in the third row in Fig. 1, where the output images are in small sizes compared to the second one, implying FCN method could deal with all the complex images well.
Fig. 2.
FCN efficiently made predictions on OCT image segmentation.
The rest of this paper is organized as follows. Sec. 2 describes our preprocessing method based on FCN in detail. Segmentation accuracy, performance evaluations in automatic diagnosis, and discussions are presented in Sec. 3, and Sec. 4 outlines conclusions.
2. Approach
2.1. Datasets
Two datasets are applied in our experiments. One of the datasets is a publicly available OCT dataset provided by the joint efforts from Duke University, Harvard University, and University of Michigan (Duke dataset). The other dataset is obtained from Beijing Hospital by Tsinghua University (THOCT dataset), using CIRRUS TM (Heidelberg Engineering Inc., Heidelberg, Germany) SD-OCT device.
Duke dataset:24 This dataset consists of retinal SD-OCT (Heidelberg Engineering Inc.) B-scans from 45 subjects [15 age-related macular degeneration (AMD), 15 diabetic macular edema (DME), and 15 normal (NOR)]. The number of OCT B-scans in each subject varies from 36 to 97. The image resolutions mainly consist of sizes , , and .
THOCT dataset: This dataset consists of 3000 retinal SD-OCT B-scans (1000 AMD, 1000 DME, and 1000 NOR), which is labeled by professional doctors. Each B-scan in this dataset comes from a single patient. There are no cases where multiple B-scans belong to the same patient and thus avoid learning the characteristics of a patient rather than a lesion. The image resolutions mainly consist of sizes and .
2.2. Detection Method of Retinal Region
Our preprocessing method could be divided into two steps. (1) Segmentation: segment the RoI from an OCT image using a pretrained FCN model and set the brightness of pixels outside the RoI to be zero. (2) Flattening and cropping: generate a straight line or curve that fits the RoI, use it to flatten the image, and then crop the image to retain only valuable information. The second step is optional.
2.2.1. Segmentation of retinal region
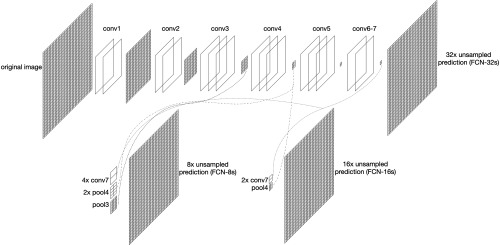
In 2015, Long et al.36 put forward FCN framework and extended CNN to semantic segmentation field. CNN puts several fully connected layer after convolutional layers, which intends to map the feature into a fixed-length feature vector and gives a possibility of classification. Unlike CNN, FCN replaces the fully connected layer by convolutional layers and uses deconvolution layers to upsample the feature map to the same size as the input image while preserving spatial relationships between pixels. It gives a prediction of classification for each pixel to achieve the purpose of segmenting the image. The concise structure of FCN is shown in Fig. 3.
Fig. 3.
Concise structure of FCN.
In Fig. 3, due to max pooling step, the original image size is reduced to its 1/2 after conv1 and pool1, 1/4 after conv2 and pool2, and 1/32 after conv5 and pool5. After several times of convolution and pooling operation, the resolution of the output image is getting lower and lower. Since image details are lost gradually in this procedure, this makes the unsampled high-dimensional feature map obtained by the last convolution layer still inaccurate for segmentation. To solve this inaccuracy issue, FCN provides options to provide details with other convolutional layers before the last convolutional layer. Upsamplings with factor 32, 16, and 8 unsampled predictions are demonstrated in Fig. 3.36 For example, unsampled prediction (FCN-16s) is a fusion of prediction from conv4 with upsampling of prediction from conv7. Since unsampled prediction model is not accurate enough and unsampled prediction model has incorrect segmentation on some small-scale areas, we finally use the unsampled prediction model’s segmentation results in our OCT image segmentation application.
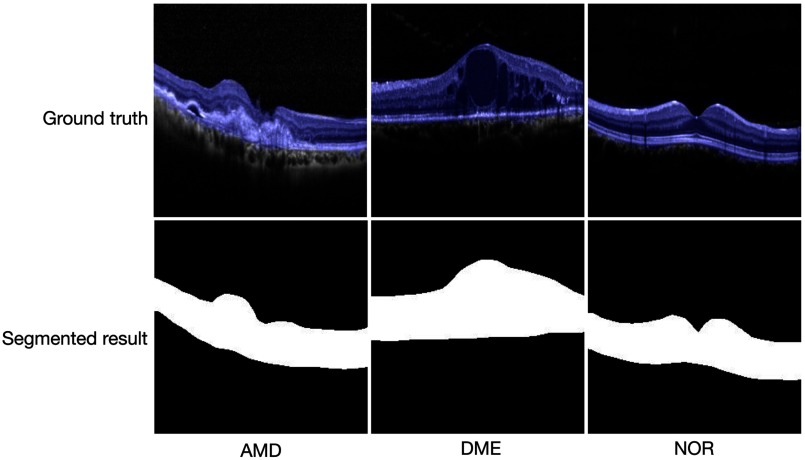
In our application, we first label OCT B-scans in training set, i.e., label the RoI as one category and mark the rest as the other category, then resize labeled OCT images to size (the same input size as that in vgg-net42) and migrate the weights of the vgg-net model to FCN model. Using transfer learning can greatly save training time and directly use diverse underlying features that are difficult to be trained well by a small or specified dataset. Since OCT images are all grayscale, we reshape all images to three channels to fit the input of the network. Image normalization is also required. Finally, we obtain a pretrained FCN model by using the OCT training images, where the output images are also of size . By resizing the output images to the original size of training images, we can directly get the segmentation result of an input OCT image. For any test OCT B-scan, we could get its RoI segmental result with this FCN model and set the brightness of pixels outside the RoI to be zero so that the background noise is removed. Some examples of the labeled AMD, DME, and NOR OCT B-scans (blue pixels as RoI ground truth) and their RoI segmented results by using our pretrained FCN model are presented in Fig. 4.
Fig. 4.
Ground truth (blue pixels) of RoI (first row) and segmented results by using our FCN model (second row).
2.2.2. Flattening and cropping
After finding the RoI of an OCT B-scan, the next step is to generate a straight line or quadratic curve that best fits the RoI. By aligning the B-scan with the fitting line, the B-scan could be transformed to get a flattened image. In this stage, how to select fitted points from RoI and what fitting line should be adopted for the selected points are two key issues.
There are two ways to choose the points: the midpoints between the top and bottom edges or the points at the bottom edge of the RoI. In general, the cross-section of the human eye is round, so the retina itself has a certain degree of curvature. In particular, a macular area is always hollower than other parts, thus, a normal retina appears to be high on both sides in the OCT image and low in the middle. In this case, an OCT image can be very well flattened by selecting the midpoints. But a lesion may change the shape of a retina. In some OCT B-scans with DME, we have found that edema causes the macular area to swell instead while the bottom is still flat. If we still use midpoints for fitting the retina, it may lead to an issue that the retina in flattened image may be bent upward. Our method, when choosing the sets of data points (the midpoints versus the bottom data points), performs a second-order polynomial fitting to the middle data points for judging: if the fitted parabola opens upward, then the midpoints are chosen; if the parabola opens downward, then the bottom data points are chosen. After given the selected points, we choose linear fitting or second-order polynomial fitting as done in Ref. 27.
It should be pointed out that since the bottom edge of an RoI detected by morphological opening-and-closing operation is not smooth enough usually, selecting the midpoints of the RoI is prior to selecting its bottom points for judging.27 Since bottom edge of an RoI detected by our FCN-based method is often smooth enough, the selecting sequence of the midpoints and the bottom points of an RoI maybe not be so important.
Another point needing attention is that when using a second-order polynomial fitting curve to flatten an OCT B-scan, the vertices of the curve may be outside the image. In this case, we just need to change the vertex to the extreme point on the left or right side of the retina, and then flatten the image.
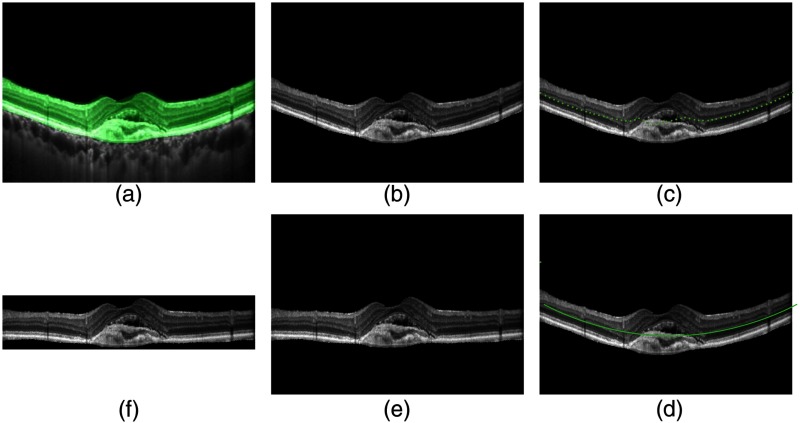
In order to keep the retinal region in a flattened OCT B-scan, we get rid of the background region by a cropping step. Specifically, we find the highest and the lowest point of the RoI area, and then cut-off all the areas above the highest point and below the lowest point of the flattened image to get a cropped image. Figure 5 illustrates steps of segmenting, flattening, and cropping an OCT image, where the midpoints of the RoI are selected and its second-order polynomial fitting curve is used.
Fig. 5.
Steps of segmenting, flattening, and cropping: (a) original image, (b) segmented result, (c) midpoints, (d) polynomial fitting, (e) flattened image, and (f) cropped image.
3. Experiments and Discussions
RoI segmentation performances and computation efficiency between our FCN-based model and previous MD method27 are evaluated and compared in Sec. 3.1. In Sec. 3.2, to show that our pretreatment method does improve the classification performance, the output of the two preprocessing methods (FCN method and MD method) into spatial pyramid matching based on sparse coding (ScSPM) OCT classifier27 is done to evaluate classification performance. The output of the FCN method and the RPE layer segmentation method (RPE method)28 into Inception V3 OCT classifier is also tested. Some problems concerned are discussed in Sec. 3.3. In our experiments, we divide THOCT dataset into two parts, the first part contains 1200 B-scans (400 AMD, 400 DME, and 400 NOR), which are used to train and test RoI segmentation performance; the second part contains 1800 B-scans (600 AMD, 600 DME, and 600 NOR), which are utilized to evaluate classification performance. Source code and preprocessed THOCT dataset are available at https://github.com/SJD095/OCT-Segmentation.
3.1. RoI Segmentation Performance
The 1200 OCT B-scans from THOCT dataset used in our experiments are all labeled with RoI manually; 900 B-scans (300 AMD, 300 DME, and 300 NOR) are used to train FCN model using transfer learning, and the other 300 B-scans (100 AMD, 100 DME, and 100 NOR) are used to test segmentation performance of the FCN model. The two metrics, pixel accuracy (PA) and mean intersection over union (meanIU),36 are used to compare the RoI segmentation performance between FCN method and MD method. PA computes the rate of pixels correctly classified to all pixels. MeanIU is a common criterion applied to evaluate the accuracy performance of segmentation in a specific dataset with ground truth. It calculates the rate of pixels correctly classified to all pixels in one category and then calculates mean rate with all categories. The higher the meanIU, the closer the predicted value to the actual value. Let be the number of pixels of class predicted belong to class , and there are total different classes, then we can calculate PA and meanIU as below:
| (1) |
| (2) |
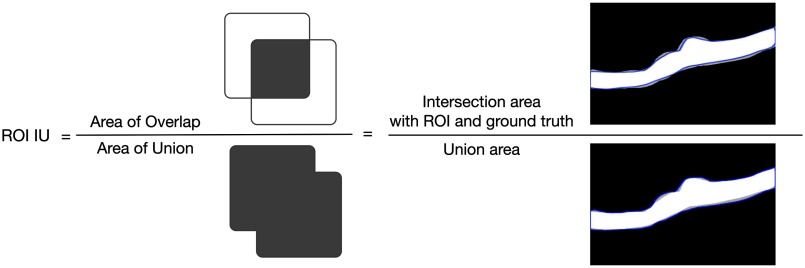
The calculation method of RoI IU for one OCT image is illustrated in Fig. 6. In our experiments, there are only foreground RoI and background for every OCT B-scan, and so, and .
Fig. 6.
Calculation method of RoI IU.
We compute PA and meanIU for every test image with the corresponding method, and then calculate their averages for all 300 test images separately. Similarly, we compute average segmentation time of the two preprocessing methods, not including flattening and cropping time. The experimental platform we use is a PC with an Ubuntu 18.04 64 bit operating system, an AMD Ryzen 1900x central processing unit (CPU), a Nvidia GeForce GTX 1080Ti graphics processing unit (GPU), and 32 GB of RAM. The FCN source code was downloaded from Ref. 43 and adopted in our own experiments. Our FCN model is implemented by Caffe framework, the version of Caffe framework used in the experiment is 1.0.0, and the version of Python is 3.6.5. The experimental results are shown in Table 1.
Table 1.
Performance comparisons between FCN method and MD method.
| PA | MeanIU | Segmentation time | |
|---|---|---|---|
| MD method27 | |||
| FCN method |
It can be seen from Table 1 that, compared to traditional MD method, PA rate and meanIU of our method increase about 11% and 22%, respectively, and variance of our method is much smaller than those of traditional methods. This shows that the performance of our FCN method is more stable than that of MD method. As illustrated in Fig. 1, some complex OCT B-scans could not be detected correctly by the MD method but could be detected well by FCN method. In the aspect of computation efficiency, the average segmentation times of FCN method and MD method are about 0.12 and 6.14 s on each B-scan separately. MD method costs much mainly because the BM3D denoising used in it has a high computation complexity. Note: in our experiment, the segmentation of MD method was implemented only using a single core CPU; its computational performance may be improved by using multicore CPU.
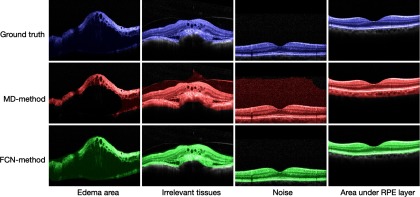
Figure 7 shows four OCT B-scan examples and their RoI segmentation results, where the first row demonstrates the original images and the blue pixels are labeled as retina region ground truth; the second row depicts the segmentation results by using MD method, marked by red pixels; the third row shows the segmentation results by using FCN method, labeled by shallow green pixels. It can be seen that the FCN method performs better than the MD method in segmentation accuracy.
Fig. 7.
Comparisons of segmentation accuracy between the two methods.
3.2. Classification Performance
This section is to validate the effectiveness of the proposed preprocessing method for OCT image classification applications. Here, FCN method, MD method, and RPE method are compared on Duke dataset and our clinic dataset by the following three experiments. Experiments 1 and 2 are designed to compare FCN method and MD method, where the preprocessed OCT images are obtained by using these two methods separately and then classified by ScSPM OCT classifier.27 The experiment 3 is designed to compare FCN method and RPE method on the clinic dataset, where their preprocessed OCT images are classified by Inception V3 model. The FCN model trained on THOCT dataset in Sec. 3.1 is used to all the experiments on both datasets.
3.2.1. Experiment 1 on Duke dataset with ScSPM classifier
Duke dataset is an open OCT image dataset; many researchers adopt it as a baseline to validate their methods. Here, we test our preprocessing method on this dataset. To compare our method fairly with those in Refs. 24 and 27, we also use leave-three-out cross-validation. In each experiment, 42 volumes are chosen as a training set and the other 3 volumes (one volume from each class) as a testing set so that each of the 45 SD-OCT volumes can be classified once. Since a volume contains many OCT B-scans of a specific subject, the majority voting of all predicted labels for each subject is treated as a class (AMD, DME, or NOR). The cross-validation results in Table 2 show that 100% of 45 OCT volumes are correctly classified with our proposed preprocessing method, which implies that FCN method is more precise than MD method in retinal RoI segmentation.
Table 2.
Fraction of volumes correctly classified with different methods on Duke dataset.
As pointed in Ref. 27, among the 15 normal OCT volumes, 14 volumes are correctly classified and 1 volume (i.e., normal volume 6) was misclassified into the DME class. The reason is that, for each B-scan in the normal volume 6, its large portion of insignificant area below the RPE layer visually resembling the pathological structures presented in the DME cases was retained by using MD method. Since our FCN-based segmentation method could extract the retina region correctly for all the normal volumes, the classification correctness rate is better.
3.2.2. Experiment 2 on THOCT dataset with ScSPM classifier
Each OCT image in THOCT dataset comes from an individual patient; thus, it includes more specific descriptors of AMD and DME diseases, and classification experiments on this dataset could enhance the persuasiveness of our method’s performance. In this experiment, the 1800 images in THOCT dataset are utilized.
For each category (AMD, DME, and NOR), we utilize accuracy, sensitivity, and specificity as indicators to evaluate the classification performance.25
To evaluate the classification performance of our pretrained FCN model used on retinal OCT B-scans, we conduct experiments on the 1800 OCT images in THOCT dataset with different proportion. First, we choose one-fourth B-scans of each class (150 AMD, 150 DME, and 150 NOR) for training and the rest (450 AMD, 450 DME, and 450 NOR) for testing. Then, we choose half of the B-scans of each class (300 AMD, 300 DME, and 300 NOR) for training and the rest (300 AMD, 300 DME and 300 NOR) for testing. To obtain reliable results, the experimental process repeats 10 times with different randomly selected OCT images for training and the rest for testing. Tables 3 and 4 detail the classification results in each category. From the results, we can see that the proposed OCT image preprocessing method significantly improves classification performance on all three indicators. The main reason is that, for an amount of OCT images difficult to deal with, FCN method could segment it well but MD method could not.
Table 3.
Classification performance comparisons of Skip classifier with different preprocessing methods (150 AMD, 150 DME, and 150 NOR images as the training set).
| Preprocessing | Classes | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| MD method27 | AMD | |||
| DME | ||||
| NOR | ||||
| FCN method | AMD | |||
| DME | ||||
| NOR |
Table 4.
Classification performance comparisons of ScSPM classifier with different preprocessing methods (300 AMD, 300 DME, and 300 NOR images as the training set).
| Preprocessing | Classes | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| MD method27 | AMD | |||
| DME | ||||
| NOR | ||||
| FCN method | AMD | |||
| DME | ||||
| NOR |
It can be seen from Tables 3 and 4, by using ScSPM classifier with sparse coding, the classification results with FCN method outperform those with MD method distinctly for all AMD, DME, and NOR categories, which reflects that FCN method is a better preprocessing method than MD method.
3.2.3. Experiment 3 on THOCT dataset with Inception V3 classifier
To compare with popular RPE method, we further do this test, as is done in experiment 2, the 1800 images in THOCT dataset are utilized. These images are preprocessed as did in Ref. 28, where the RPE is estimated as the same as in Ref. 24, and by our FCN method separately, then the processed images are resized to and the pixel value of the images is divided by 255 for normalization. These images are input into Inception V3 classifier with transfer learning and fine-tuning by using the Keras implementation. The test method is the same as the above experiment 2. Table 5 lists the classification results on each category, where the test method is the same as that used in Table 4.
Table 5.
Classification performance comparisons of Inception V3 classifier with different preprocessing methods (300 AMD, 300 DME, and 300 NOR images as the training set).
| Preprocessing | Classes | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| RPE method28 + Inception V3 | AMD | |||
| DME | ||||
| NOR | ||||
| FCN method + Inception V3 | AMD | |||
| DME | ||||
| NOR |
From Table 5, we can see that, by using Inception V3 for classification, FCN method outperforms RPE method for AMD and DME categories but is inferior to RPE method for NOR category. This is consistent with our expectation. In fact, by checking the preprocessed OCT images by RPE method, we find that about 21 AMD, 8 DME, and 2 normal OCT images in 1800 trained images are not detected successfully. This is not surprising because RPE layer detection is more suitable to NOR OCT images but may be failed for OCT images obtained from eye patients. By contrast, FCN-based preprocessing method could adapt to various situations.
Overall, classification results of different methods on clinic dataset are listed in Table 6.
Table 6.
Overall classification results of different methods on clinic dataset.
| Partition | Methods | Overall-Acc (%) | Overall-Se (%) | Overall-Sp (%) |
|---|---|---|---|---|
| 1/2 dataset | MD method + ScSPM | |||
| FCN method + ScSPM | ||||
| RPE method + Inception V3 | ||||
| FCN method + Inception V3 |
Table 6 shows that, for the same preprocessed OCT images with FCN method, Inception V3 outperforms ScSPM in classification results, and it is more stable; for the same Inception V3 classifier, our proposed FCN-method is almost the same as RPE method in average classification performance, yet it is more stable. Since RPE method needs costly BM3D denoising and many manually selected parameters, whereas FCN model does not, FCN method is more practical on the whole.
3.3. Discussion
Our FCN preprocessing method consists of segmenting retina region (RoI), flattening, and cropping steps. It could remove background by setting all brightness of pixels outside the RoI to be zero. Therefore, it needs no prior denoising, and so, many OCT image denoising methods40,44,45 are not needed in our preprocessing.
For our proposed automatic detection method of the retinal region, i.e., FCN method, it is just a generalized algorithm that works with both datasets. In fact, in our paper, the FCN model to segment retina region is just trained on 900 labeled OCT B-scans from THOCT dataset, then it is used to preprocess images from both datasets, i.e., Duke dataset and THOCT dataset (its second part, different from the training part). However, in our experiments, for each classifier ScSPM or Inception V3, it trained on the Duke dataset and the THOCT dataset separately; is it possible to have a generalized algorithm that works with both datasets? In this regard, we apply the classifiers FCN method + ScSPM and FCN method + Inception V3 trained on THOCT dataset in Table 6 to Duke dataset directly. Forty five SD-OCT volumes are used as a test set, and the voting majority is used for each volume. The experiments show that all 100% of 45 OCT volumes are correctly classified for both classification algorithms.
Another one question concerned is whether, without flattening and cropping, the proposed method can also achieve good classification performance. To answer this question, using our FCN method without flattening and cropping, we obtain the preprocessed OCT images on THOCT dataset, then the preprocessed OCT images are as the outputs of ScSPM classifier and Inception v3 classifier separately, where the test method is the same as that used in Table 4. Experimental results on classification performance are shown in Table 7.
Table 7.
Overall classification results on clinic dataset using FCN method without flattening and cropping.
| Partition | Methods | Overall-Acc (%) | Overall-Se (%) | Overall-Sp (%) |
|---|---|---|---|---|
| 1/2 dataset | FCN method + ScSPM | |||
| FCN method + Inception V3 |
Comparison of the same methods in Tables 6 and 7 shows that, without flattening and cropping, the proposed method also achieves good classification performance, but the classification performance of ScSPM classifier is deduced obviously, yet the classification performance of Inception V3 classifier makes no difference. So, to achieve the best abnormal retina detection performance, FCN model with flattening and cropping should be used for ScSPM classifier while FCN model without flattening and cropping could be utilized for Inception V3 classifier.
4. Conclusions
In this paper, we propose a retinal region detection method based on FCN and trained an FCN model by using our labeled OCT images. It has high segmentation accuracy, and its effectiveness in abnormal maculae recognition application is validated by extensive experiments on two OCT datasets. Compared with current widely used retina region detection methods, such as RPE layer segmentation and MD, our proposed FCN method works with no manually selected parameters, no prior denoising, and no layer segmentation. It also possesses many merits in performance, including high segmentation accuracy and high computational efficiency, and is robust to noises. These features improve results in the automatic diagnosis of abnormal maculae in OCT images.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Award No. 61671272), the Opening Project of Guangdong Province Key Laboratory of Big Data Analysis and Processing (Grant No. 201803), and the National Key Research Program of China (Grant No. 2016YFB1000600). The authors would like to thank the anonymous reviewers for their careful reading of our paper and their many insightful comments and suggestions that greatly improve the paper.
Biographies
Zhongyang Sun received his BE degree in software engineering from Sun Yat-sen University in 2017 and was a research apprentice at the Department of Computer Science and Technology, Tsinghua University, in 2018, supported by the National Natural Science Foundation of China under Grant No. 61671272. He is currently a master’s student at the College of Engineering, Northeastern University, Boston, USA. His research interests are deep learning, medical image processing, and big data analytics.
Yankui Sun is an associate professor in the Department of Computer Science and Technology at Tsinghua University, Beijing, China. He received his PhD in 1999 from Beihang University, China. He visited Duke University as a scholar from September 2013 to September 2014. He has authored and coauthored more than 100 papers and 5 books. His current research interests include dictionary learning, deep learning, OCT image processing, scientific visualization, and wavelet analysis.
Disclosures
The authors declare that there are no conflicts of interest related to this article. The research data was acquired and processed from patients by authors unaffiliated with any commercial entity.
References
- 1.Huang D., et al. , “Optical coherence tomography,” Science 254, 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuman J. S., et al. , Optical Coherence Tomography of Ocular Diseases, Slack, Thorofare, New Jersey: (2004). [Google Scholar]
- 3.Abràmoff B. J., et al. , “A combined machine-learning and graph-based framework for the segmentation of retinal surfaces in SD-OCT volumes,” Biomed. Opt. Express 4(12), 2712–2728 (2013). 10.1364/BOE.4.002712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carass A., et al. , “Multiple-object geometric deformable model for segmentation of macular OCT,” Biomed. Opt. Express 5, 1062–1074 (2014). 10.1364/BOE.5.001062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu S. J., et al. , “Validated automatic segmentation of AMD pathology including drusen and geographic atrophy in SD-OCT images,” Invest. Ophthalmol. Visual Sci. 53, 53–61 (2012). 10.1167/iovs.11-7640 [DOI] [PubMed] [Google Scholar]
- 6.Chiu S. J., et al. , “Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation,” Opt. Express 18, 19413–19428 (2010). 10.1364/OE.18.019413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBuc D. C., et al. , “Reliability and reproducibility of macular segmentation using a custom-built optical coherence tomography retinal image analysis software,” J. Biomed. Opt. 14(6), 064023 (2009). 10.1117/1.3268773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández D. C., Salinas H. M., Puliafito C. A., “Automated detection of retinal layer structures on optical coherence tomography images,” Opt. Express 13(25), 10200–10216 (2005). 10.1364/OPEX.13.010200 [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa H., et al. , “Macular segmentation with optical coherence tomography,” Invest. Ophthalmol. Visual Sci. 46(6), 2012–2017 (2005). 10.1167/iovs.04-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang A., et al. , “Retinal layer segmentation of macular OCT images using boundary classification,” Biomed. Opt. Express 4(7), 1133–1152 (2013). 10.1364/BOE.4.001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer M. A., et al. , “Retinal nerve fiber layer segmentation on FD-OCT scans of normal subjects and glaucoma patients,” Biomed. Opt. Express 1(5), 1358–1383 (2010). 10.1364/BOE.1.001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra A., et al. , “Intra-retinal layer segmentation in optical coherence tomography images,” Opt. Express 17, 23719–23728 (2009). 10.1364/OE.17.023719 [DOI] [PubMed] [Google Scholar]
- 13.Mujat M., et al. , “Retinal nerve fiber layer thickness map determined from optical coherence tomography images,” Opt. Express 13, 9480–9491 (2005). 10.1364/OPEX.13.009480 [DOI] [PubMed] [Google Scholar]
- 14.Paunescu L. A., et al. , “Reproducibility of nerve fiber thickness, macular thickness, and optic nerve head measurements using StratusOCT,” Invest. Ophthalmol. Visual Sci. 45, 1716–1724 (2004). 10.1167/iovs.03-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shahidi M., Wang Z., Zelkha R., “Quantitative thickness measurement of retinal layers imaged by optical coherence tomography,” Am. J. Ophthalmol. 139, 1056–1061 (2005). 10.1016/j.ajo.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 16.Sun Y., et al. , “3D automatic segmentation method for retinal optical coherence tomography volume data using boundary surface enhancement,” J. Innov. Opt. Health Sci. 9, 1650008 (2016). 10.1142/S1793545816500085 [DOI] [Google Scholar]
- 17.Vermeer K., et al. , “Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images,” Biomed. Opt. Express 2, 1743–1756 (2011). 10.1364/BOE.2.001743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q., et al. , “Automated segmentation of outer retinal layers in macular OCT images of patients with retinitis pigmentosa,” Biomed. Opt. Express 2, 2493–2503 (2011). 10.1364/BOE.2.002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L. Y., et al. , “Segmentation based sparse reconstruction of optical coherence tomography images,” IEEE Trans. Med. Imaging 36(2), 407–421 (2017). 10.1109/TMI.2016.2611503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu P., Sun Y. K., “Automated extraction of the inner contour of the anterior chamber using optical coherence tomography images,” J. Innovative Opt. Health Sci. 5(4), 1250030 (2012). 10.1142/S1793545812500307 [DOI] [Google Scholar]
- 21.Sui X. D., et al. , “Choroid segmentation from optical coherence tomography with graph edge weights learned from deep convolutional neural networks,” Neurocomputing 237, 332–341 (2017). 10.1016/j.neucom.2017.01.023 [DOI] [Google Scholar]
- 22.Sun Y., Lei M., “Method for optical coherence tomography image classification using local features and earth mover’s distance,” J. Biomed. Opt. 14(5), 054037 (2009). 10.1117/1.3251059 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y.-Y., et al. , “Automated macular pathology diagnosis in retinal OCT images using multi-scale spatial pyramid and local binary patterns in texture and shape encoding,” Med. Image Anal. 15, 748–759 (2011). 10.1016/j.media.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan P. P., et al. , “Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images,” Biomed. Opt. Express 5, 3568–3577 (2014). 10.1364/BOE.5.003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang L., et al. , “Automatic classification of retinal three-dimensional optical coherence tomography images using principal component analysis network with composite kernels,” J. Biomed. Opt. 22(11), 116011 (2017). 10.1117/1.JBO.22.11.116011 [DOI] [PubMed] [Google Scholar]
- 26.Rasti R., Mehridehnavi A., “Macular OCT Classification using a multi-scale convolutional neural network ensemble,” IEEE Trans. Med. Imaging 37(4), 1024–1034 (2018). 10.1109/TMI.2017.2780115 [DOI] [PubMed] [Google Scholar]
- 27.Sun Y. K., Li S., Sun Z. Y., “Fully automated macular pathology detection in retina optical coherence tomography images using sparse coding and dictionary learning,” J. Biomed. Opt. 22(1), 016012 (2017). 10.1117/1.JBO.22.1.016012 [DOI] [PubMed] [Google Scholar]
- 28.Karri S., Chakraborty D., Chatterjee J., “Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration,” Biomed. Opt. Express 8, 579–592 (2017). 10.1364/BOE.8.000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kermany D. S., et al. , “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell 172, 1122–1131 (2018). 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 30.Ji Q. G., et al. , “Efficient deep learning based automated pathology identification in retinal optical coherence tomography images,” Algorithms 11, 88 (2018). 10.3390/a11060088 [DOI] [Google Scholar]
- 31.Wang Y., et al. , “Machine learning based detection of age-related macular degeneration (AMD) and diabetic macular edema (DME) from optical coherence tomography (OCT) images,” Biomed. Opt. Express 7, 4928–4940 (2016). 10.1364/BOE.7.004928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venhuizen F. G., et al. , “Automated age-related macular degeneration classification in OCT using unsupervised feature learning,” Proc. SPIE 9414, 94141I (2015). 10.1117/12.2081521 [DOI] [Google Scholar]
- 33.Lemaitre G., et al. , “Classification of SD-OCT volumes with LBP: application to DME detection,” in Medical Image Computing and Computer Assisted Intervention (MICCAI), Ophthalmic Medical Image Analysis Workshop (OMIA) (2015). [Google Scholar]
- 34.Rasti R., et al. , “Automatic diagnosis of abnormal macula in retinal optical coherence tomography images using wavelet-based convolutional neural network features and random forests classifier,” J. Biomed. Opt. 23(3), 035005 (2018). 10.1117/1.JBO.23.3.035005 [DOI] [PubMed] [Google Scholar]
- 35.Kafieh R., et al. , “Curvature correction of retinal OCTs using graph-based geometry detection,” Phys. Med. Biol. 58(9), 2925–2938 (2013). 10.1088/0031-9155/58/9/2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long J., et al. , “Fully convolutional network for semantic segmentation,” in IEEE Conf. Computer Vision and Pattern Recognit., pp. 3431–3440 (2015). [Google Scholar]
- 37.Fang L. Y., et al. , “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–4940 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A. G., et al. , “ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomed. Opt. Express 8(8), 3627 (2017). 10.1364/BOE.8.003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novosel J., et al. , “Joint segmentation of retinal layers and focal lesions in 3-D OCT data of topologically disrupted retinas,” IEEE Trans. Med. Imaging 36(6), 1276–1286 (2017). 10.1109/TMI.2017.2666045 [DOI] [PubMed] [Google Scholar]
- 40.Dabov K., et al. , “Image denoising by sparse 3-D transform-domain collaborative filtering,” IEEE Trans. Image Process. 16(8), 2080–2095 (2007). 10.1109/TIP.2007.901238 [DOI] [PubMed] [Google Scholar]
- 41.Otsu N., “Threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 42.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” CoRR abs/1409.1556 (2014).
- 43.Long J., et al. , “FCN,” https://github.com/shelhamer/fcn.berkeleyvision.org. (accessed 15 March 2018).
- 44.Fang L., et al. , “3-D adaptive sparsity based image compression with applications to optical coherence tomography,” IEEE Trans. Med. Imaging 34(6), 1306–1320 (2015). 10.1109/TMI.2014.2387336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang L., et al. , “Segmentation based sparse reconstruction of optical coherence tomography images,” IEEE Trans. Med. Imaging 36(2), 407–421 (2017). 10.1109/TMI.2016.2611503 [DOI] [PMC free article] [PubMed] [Google Scholar]