Abstract
Quantitative Structure Activity Relationship studies were carried out on arylpiperazine derivatives to investigate their anti-proliferate activity against prostate PC-3 cancer cell lines. The built model with statistical parameters; R2 = 0.8483, R2adj = 0.8078, Q2cv = 0.7122 and external validation (R2test) 0.6682 revealed that the anti-proliferate activities were strongly dependent on the descriptors: MATS7c, MATS3e, maxwHBa and WPSA-3. The Variance Inflation Factor of the descriptors were all greater than one but less than two and all descriptors were poorly correlated (r < 0.4). A graph of the experimental activities and predicted activities showed a high correlation and a William's plot showed the presence of only one outlier compound. These results are similar to those reported for stable and robust models with high predicting power. Molecular docking studies of compounds 5 (1-phenyl-4-(4-(2-(p-tolyloxy)ethyl)benzyl)piperazine) and 17 (4-(4-((4-phenylpiperazin-1-yl)methyl)phenethoxy)benzonitrile) with the androgen receptor gave binding affinities of −7.5 and −7.1 kcal/mol respectively. Compound 5 formed a more stable complex having hydrogen, electrostatic and hydrophobic bond interactions while compound 17 had hydrogen and hydrophobic bond interactions only. This study provides a roadmap to the design of more potent anti-prostate cancer compounds.
Keywords: Cancer research, Theoretical chemistry, Quantitative structure activity relationship, Molecular docking, Prostate cancer, Computational chemistry, Piperazine
Cancer research, Theoretical chemistry, Quantitative structure activity relationship; Molecular docking; Prostate cancer; Computational chemistry; Piperazine
1. Introduction
The most common non-cutaneous cancer diagnosed in men is Prostate cancer (PCa) (Center et al., 2012; Miller et al., 2016). Surveys have shown that 1 man in every 9 is diagnosed with PCa during his life time. It is also estimated that 1 man in every 39 between the ages 40 to 59 is diagnosed with PCa annually. Projections show that there would be 174,650 new cases of PCa in the US in 2019 and about 31,620 deaths from the cancer and in Brazil, 70.42 patients per 100,000 men are projected this year (ACS, 2016; Fernando and Ubirajara, 2018).
During the early stage of PCa, no symptoms are usually observed, however, in few cases, repeated urges to urinate, difficulty in initiating and maintaining urination, bloody urine, difficulty in attaining or sustaining erection etc., have been reported (Nordqvist, 2017). Though no therapy is ideal for all patients, common treatments for PCa are active surveillance, radical prostatectomy, External-Beam Radiation Therapy (EBRT), androgen deprivation etc. Reports showed treatment effects to be profound and prolonged, having adverse effects such as urinal and intestinal disorders, erectile dysfunction, urinary incontinence and in some cases death (Bell et al., 2014; Qaseem et al., 2013; Wolf et al., 2010). PCa is the leading cause of cancer related deaths in men worldwide and is estimated to be responsible for about 1–2% of all deaths in men (Gerhardt et al., 2015). Studies have shown that men of African descent are roughly twice more likely to develop PCa compared with Caucasian men. They are also approximately 2.4 times more likely to die from PCa than their Caucasian counterparts. This preferential attack of PCa on men of African ancestry is however yet to be understood (PCF, 2010).
Piperazine and its derivatives are of immense biological importance, they have been reported to be antidiarrheal, antipyretic, analgesic, antimicrobial, antitumor, anti-inflammatory, diuretic, antipsychotic, antimalarial and antidepressant properties (Arnatt et al., 2014; Baran et al., 2014; Szkaradek et al., 2013). Due to their wide spectrum of activity, piperazine and its derivatives are present in a lot of commercial drugs; piperazine is the third most frequently used nitrogen based pharmacophore among the U.S. Food and Drug Administration's approved drugs. It is the pharmacophore in over fifty nine (59) marketed drugs including Merck HIV protease inhibitor Crixivan, Proplomazine, thiethylperazine, Prazosin, Sildenafil, Cinnarizine etc (James et al., 2014; Vardanyan, 2017; Vitaku et al., 2014).
2. Computational methods
2.1. QSAR method
2.1.1. Dataset
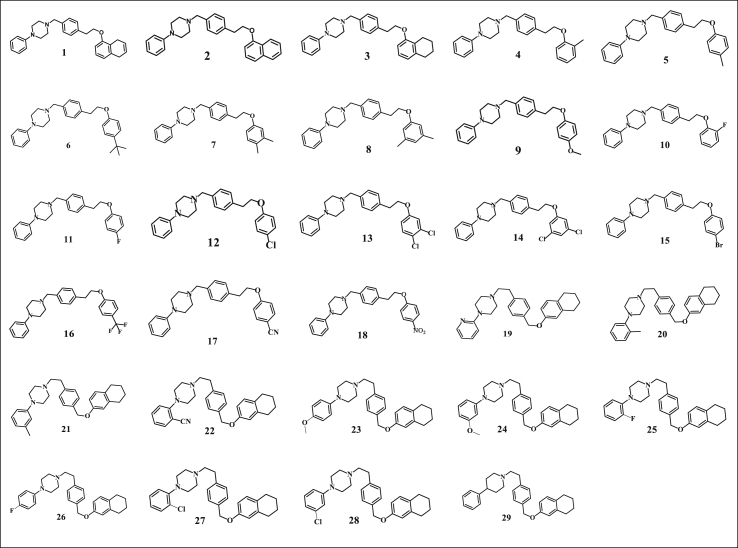
Twenty nine (29) arylpiperazine derivatives were used in this study. Their structures and activities were obtained from literature (Chen et al., 2018a, 2018b). Their activities were reported in IC50 (μM). The skew in the activities were reduced via normalization using the logarithmic scale formula . Figure 1 presents the 2D structures of the molecules used in this study while Table 1 presents the IUPAC nomenclature and anti-proliferate activity of the molecules.
Figure 1.
2D structure of dataset (Chen et al., 2018a, Chen et al., 2018b).
Table 1.
Structure and inhibitory concentration of arylpiperazine derivatives.
| S/No. | IUPAC Nomenclature | IC50 (μM) | pIC50 |
|---|---|---|---|
| 1 | 1-(4-(2-phenoxyethyl)benzyl)-4-phenylpiperazine | 8.20 | 5.09 |
| 2 | 1-(4-(2-(naphthalene-1-yloxy)ethyl)benzyl)-4-phenylpiperazine | 4.72 | 5.33 |
| 3 | 1-phenyl-4-(4-(2-((5,6,7,8-tetrahydronaphthalen-1-yl)oxy)ethyl)benzyl)piperazine | 17.70 | 4.75 |
| 4 | 1-phenyl-4-(4-(2-(o-tolyloxy)ethyl)benzyl)piperazine | 15.10 | 4.82 |
| 5b. | 1-phenyl-4-(4-(2-(p-tolyloxy)ethyl)benzyl)piperazine | 1.47 | 5.83 |
| 6 | 1-(4-(2-(4-(tert-butyl)phenoxy)ethyl)benzyl)-4-phenylpiperazine | 4.86 | 5.31 |
| 7b. | 1-(4-(2-(3,4-dimethylphenoxy)ethyl)benzyl)-4-phenylpiperazine | 6.72 | 5.17 |
| 8.b | 1-(4-(2-(3,5-dimethylphenoxy)ethyl)benzyl)-4-phenylpiperazine | 2.99 | 5.52 |
| 9 | 1-(4-(2-(4-methoxyphenoxy)ethyl)benzyl)-4-phenylpiperazine | 8.95 | 5.05 |
| 10 | 1-(4-(2-(2-fluorophenoxy)ethyl)benzyl)-4-phenylpiperazine | 48.80 | 4.31 |
| 11 | 1-(4-(2-(4-fluorophenoxy)ethyl)benzyl)-4-phenylpiperazine | 5.70 | 5.24 |
| 12b | 1-(4-(2-(4-chlorophenoxy)ethyl)benzyl)-4-phenylpiperazine | 3.59 | 5.44 |
| 13 | 1-(4-(2-(3,4-dichlorophenoxy)ethyl)benzyl)-4-phenylpiperazine | 23.00 | 4.64 |
| 14 | 1-(4-(2-(3,5-dichlorophenoxy)ethyl)benzyl)-4-phenylpiperazine | 10.10 | 5.00 |
| 15b | 1-(4-(2-(4-bromophenoxy)ethyl)benzyl)-4-phenylpiperazine | 6.69 | 5.17 |
| 16 | 1-phenyl-4-(4-(2-(4-(trifluoromethyl)phenoxy)ethyl)benzyl)piperazine | 4.96 | 5.30 |
| 17 | 4-(4-((4-phenylpiperazin-1-yl)methyl)phenethoxy)benzonitrile | 1.05 | 5.98 |
| 18 | 1-(4-(2-(4-nitrophenoxy)ethyl)benzyl)-4-phenylpiperazine | 9.23 | 5.03 |
| 19 | 1-(pyridin-2-yl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 29.20 | 4.53 |
| 20b | 1-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)-4-(o-tolyl)piperazine | 25.50 | 4.59 |
| 21b | 1-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)-4-(m-tolyl)piperazine | 56.80 | 4.25 |
| 22 | 3-(4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazin-1-yl)benzonitrile | 32.00 | 4.5 |
| 23 | 1-(4-methoxyphenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 76.50 | 4.12 |
| 24 | 1-(3-methoxyphenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 2.84 | 5.55 |
| 25 | 1-(2-fluorophenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 27.30 | 4.56 |
| 26b | 1-(4-fluorophenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 57.30 | 4.24 |
| 27b | 1-(2-chlorophenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 31.30 | 4.50 |
| 28 | 1-(3-chlorophenyl)-4-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperazine | 46.70 | 4.33 |
| 29 | 4-phenyl-1-(4-(((5,6,7,8-tetrahydronaphthalen-2-yl)oxy)methyl)phenethyl)piperidine | 10.50 | 4.98 |
Key:b = Molecules in the test set.
2.1.2. QSAR model
ChemDraw Ultra version 12.0 software was used to draw two-dimensional structures of the chemical compounds and converted to three-dimensional structures in Spartan 14.0 version 1.1.2 software. The equilibrium ground state geometry of the molecules were obtained by optimizing the structures at the DFT/B3LYP/6-31G* basis set in the abovementioned Spartan software (Arthur et al., 2018; Wavefunction, 2013; Becke, 1993). Pharmaceutical Data Exploration Laboratory (PaDEL) version 2.21 software was used to calculate the molecular descriptors of the optimized molecules. One thousand eight hundred and seventy five molecular descriptors were calculated per molecule. These descriptors are mathematical equivalents of spatial, topological, thermodynamic etc., properties of the molecule (Ogadimma and Adamu, 2016).
The calculated descriptors were pre-treated at a correlation coefficient cut-off of 0.8 using DTC Lab's Data Pre-treatment GUI 1.2 software to remove constant, non-informative and highly correlated descriptors (Abdullahi et al., 2019). Furthermore, the pre-treated descriptors were divided in to two independent sets (training and test set) using Kennard and Stone algorithm in the DTC Lab's Dataset Division GUI 1.29 software (Kennard and Stone, 1969; Arthur et al., 2018).
The training set was used in building the model using Biovia Material Studio version 8 software. In building the model, Genetic Function Algorithm - Multilinear Regression (GFA-MLR) analysis was employed (Ibrahim et al., 2018). The built model had the form shown in Eq. (1).
| (1) |
where Y is the inhibitory concentration; a1, a2, a3, …, an are regression coefficients; x1, x2, x3, …, xn are distinct molecular descriptors and c is the regression constant.
2.1.3. Model validation
The stability, robustness and predicting power of a built model is determined by subjecting the built model to certain statistical tests. The Co-efficient of determination (R2), Leave-One-Out (LOO) cross validation co-efficient of determination (Qcv2), Friedman's Lack of Fit (LOF), mean effect, Variance Inflation Factor (VIF), and Pearson's correlation tests were carried out on the built model to measure its fitness, robustness and predicting ability. Applicability domain as well as external validation of the model was also evaluated.
The R2 is a measure of the proportion of the variance in the anti-proliferate activity that can be explained/predicted by the molecular descriptors in the model. A good model usually has an R2 ≥ 0.6 (Veerasamy et al., 2011). Qcv2 is a model validation method that asses how the model will generalize to another independent dataset. Qcv2 values range from 0 – 1. A good model is reported to have Qcv2 ≥ 0.6 (Veerasamy et al., 2011). The LOF test measures the fitness of the model; if the prediction of the built model is significant. R2, Qcv2 and LOF values are automatically generated by Biovia Material Studio software while building the model. The mean effect of a molecular descriptor is the influence that molecular descriptor has on the anti-proliferate activity of the compounds. It measures the effect a unit change in the value of the descriptor would have on the anti-proliferate activity (Minovski et al., 2013). The mean effect was calculated using Eq. (2)
| (2) |
= coefficient of the jth descriptor in the QSAR model, = value of the jth descriptor for each training set molecule, n = number of molecules in the training set and m = number of descriptors in the model. VIF and Pearson's correlation tests were used to measure inter-correlation (or multicollinearity) among the descriptors. A good QSAR model is supposed to be made up of poorly correlated molecular descriptors. Thus, for a good model, the correlation coefficient (R) should be R < 0.5. VIF is calculated as (1 – R2)−1. VIF value of 1 means there is no inter-correlation, values of 2–5 means there is poor correlation and is generally acceptable while a value above 10 implies that there is significant inter-correlation between the molecular descriptors and thus, the model is unstable and should be discarded (Edache et al., 2017). The response surface in which the QSAR model makes reliable predictions is called its applicability domain. The applicability domain simply highlights the surface area where forecasts made by the model can be reliably useful (Netzeva et al., 2005). Thus, molecules within the applicability domain can be reliably used as ligands for QSAR based drug design. We employed leverage technique in evaluating the applicability domain using Eq. (3) (Abdullahi et al., 2019)
| (3) |
where is the leverage of the ith compound, is the 1 m descriptor row matrix of the ith compound, is an n k matrix made up of n rows of descriptor values and k molecules in the training set. The warning (or critical) leverage (h*) defines the boundary of the applicability domain and is calculated as: h* = 3 (n + 1)/k. where n is the number of descriptors in the model and k is the number of compounds in the training set.
The robustness of the built model was ascertained by subjecting the model to the test set. This external validation is aimed at measuring the predicting ability of the built model. External validation subjects the built model to compounds that were not initially part of the dataset from which the model was built. The coefficient of determination (Rtest2) is a measure of the predicting capacity of the model. Rtest2 ranges from 0 – 1, a good model should have Rtest2 ≥ 0.6 (Tropsha, 2010). Rtest2 is calculated as shown in Eq. (4) (Abdullahi et al., 2019)
| (4) |
is the experimental anti-proliferate activity of each molecule in the test set, is the corresponding predicted anti-proliferate activity and is the average experimental activity of the molecules in the training set.
2.2. Molecular docking studies
Molecular docking studies were used to explore the interaction between the molecules and the androgen receptor (AR). Compounds 5 and 17 were selected for the docking studies because they were the most potent compounds with pIC50 5.83 and 5.98 (IC50: 1.47 and 1.05 μM) respectively.
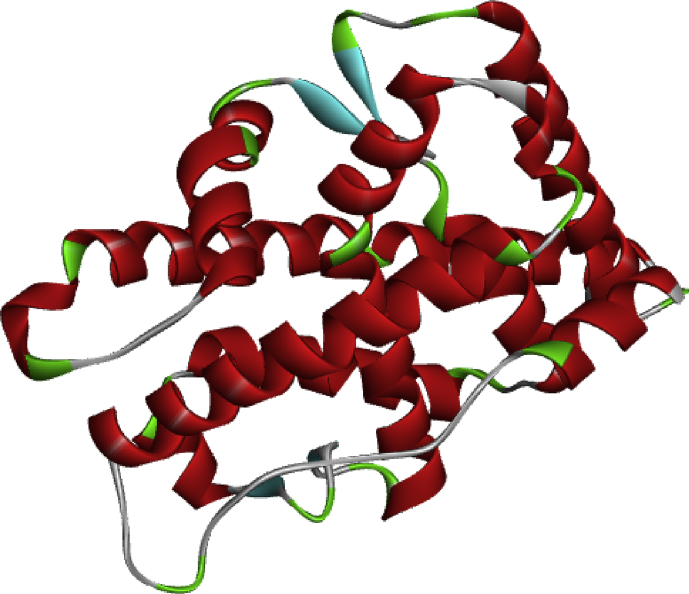
PCa is linked with alterations in AR functions (Tan et al., 2015). A three dimensional structure of the androgen receptor with PDB code 5T8E and resolution 2.71Ǻ deposited by Wilson et al. was obtained from the protein data bank. The heteroatoms, water molecules and cofactors were removed from the downloaded receptor when preparing it for docking studies using Biovia Discovery Studio Visualizer version 16.1.0 software (Veerasamy et al., 2011). The crystal structure of the prepared androgen receptor is presented in Figure 2. Drawn and optimized 3D structures of compounds 5 and 17 were prepared for molecular docking by converting them to the.pdb file format (Arthur et al., 2018).
Figure 2.
Crystal structure of the androgen receptor (5t8e).
The interaction between the prepared receptor and ligands was carried out using autodock vina integrated into the PyRx – Python Prescription 0.8 software (Trott and Olson, 2010). Afterwards, the ligand-receptor interaction was observed using Discovery Studio software (Ibrahim et al., 2018).
3. Results and discussion
3.1. QSAR
The Genetic Function Algorithm was used in building four models for predicting the anti-proliferate activities of the chemical compounds. Each model contained four molecular descriptors and the fourth model was chosen as the best model as its statistical parameters best fits the criteria described in Section 2.1.3. The fourth model was also selected because it was the only model that gave an R2test > 0.6.
Model 1
| pIC50 = − 0.090295289 * ATSC3s − 6.625744745 * MATS5c − 0.254838435 * BCUTp-11 + 0.258170707 * RDF120m + 5.466086523 |
Model 2
| pIC50 = − 5.570729167 * AATS7e − 3.109356589 * MATS3s − 7.739104160 * VE1_Dzs +0.646311321 * RDF120m + 44.100000350 |
Model 3
| pIC50 = 1.859359226 * GATS3e − 0.838587442 * GATS7s − 6.532303653 * VE1_Dzs +0.577948965 * RDF120m + 2.200052020 |
Model 4
| pIC50= 5.031260843 * MATS7c − 7.803254787 * MATS3e − 4.545755956 * maxwHBa +0.078229414 * WPSA−3+ 12.277442821 |
The description and class of each descriptor in model 4 are presented in Table 2. The positive coefficients of descriptors MATS7c and WPSA-3 means an increase in the value of these descriptors would increase the anti-proliferate activity of the compounds against PC3 PCa cell lines while the negative coefficients of MATS3e and maxwHBa means increasing the value of the descriptors would decrease the activity of the compounds. Statistical parameters for each model are shown in Table 3 and these parameters are all in agreement with values reported in literature (Veerasamy et al., 2011; Tropsha, 2010). The values of these parameters show the stability and robustness of each model.
Table 2.
Description and class of descriptors.
| Descriptor | Description | Class |
|---|---|---|
| MATS7c | Moran autocorrelation - lag 7/weighted by charges | 2D |
| MATS3e | Moran autocorrelation - lag 3/weighted by Sanderson electronegativities | 2D |
| maxwHBa | Maximum E-States for weak Hydrogen Bond acceptors | 2D |
| WPSA-3 | Charge weighted partial positive surface area * total molecular surface area/1000 | 3D |
Table 3.
Validation parameters for built models.
| Validation parameter | Model 1 | Model 2 | Model 3 | Model 4 |
|---|---|---|---|---|
| Friedman LOF | 0.1700 | 0.1779 | 0.1859 | 0.1883 |
| R-squared | 0.8630 | 0.8566 | 0.8502 | 0.8483 |
| Adjusted R-squared | 0.8264 | 0.8184 | 0.8103 | 0.8078 |
| Cross validated R-squared | 0.7675 | 0.6783 | 0.6436 | 0.7122 |
| Significant Regression | Yes | Yes | Yes | Yes |
| Significance-of-regression F-value | 23.619191 | 22.401335 | 21.285013 | 20.968048 |
| Critical SOR F-value (95%) | 3.103976 | 3.103976 | 3.103976 | 3.103976 |
| Replicate points | 0 | 0 | 0 | 0 |
| Computed experimental error | 0 | 0 | 0 | 0 |
| Lack-of-fit points | 15 | 15 | 15 | 15 |
| Min expt. error for non-significant LOF (95%) | 0.149558 | 0.153001 | 0.156375 | 0.157374 |
The calculated molecular descriptors in model 4 were used to predicted anti-proliferate activity for each compound in the training and test set. The calculated descriptors, predicted anti-proliferate activities, experimental anti-proliferate activities and the residuals for the compounds in the training and test set are displayed in Tables 4 and 5. The difference between the experimental anti-proliferate activity and the corresponding predicted activity is equal to the residual, the low values obtained showed that the model had good predicting power.
Table 4.
Molecular descriptors, activities and residual for compounds in the training set.
| Molecule | MATS7c | MATS3e | maxwHBa | WPSA-3 | YPred. | YExp. | Residual |
|---|---|---|---|---|---|---|---|
| 28 | −0.00996 | −0.07502 | 2.3708 | 28.756 | 4.29 | 4.33 | 0.04 |
| 17 | −0.00979 | −0.11039 | 2.2367 | 35.084 | 5.67 | 5.98 | 0.31 |
| 9 | −0.09700 | −0.11748 | 2.2562 | 34.629 | 5.16 | 5.05 | −0.11 |
| 19 | −0.05012 | −0.05887 | 2.2596 | 31.262 | 4.66 | 4.53 | −0.13 |
| 25 | −0.01999 | 0.02512 | 2.2392 | 31.039 | 4.23 | 4.56 | 0.33 |
| 23 | −0.15786 | −0.07128 | 2.2631 | 32.690 | 4.31 | 4.12 | −0.19 |
| 13 | −0.04992 | −0.03405 | 2.2847 | 37.637 | 4.85 | 4.64 | −0.21 |
| 24 | 0.03529 | −0.07128 | 2.2620 | 33.181 | 5.32 | 5.55 | 0.23 |
| 4 | 0.01127 | −0.07922 | 2.2705 | 27.621 | 4.79 | 4.82 | 0.03 |
| 29 | 0.08565 | −0.08940 | 2.2858 | 27.378 | 5.16 | 4.98 | −0.18 |
| 14 | −0.04565 | −0.14332 | 2.3191 | 32.459 | 5.16 | 5.00 | −0.16 |
| 18 | −0.02349 | −0.07812 | 2.1992 | 29.622 | 5.09 | 5.03 | −0.06 |
| 10 | 0.02869 | 0.07070 | 2.1964 | 31.400 | 4.34 | 4.31 | −0.03 |
| 1 | −0.00437 | −0.11089 | 2.2653 | 29.462 | 5.13 | 5.09 | −0.04 |
| 22 | 0.00795 | −0.04089 | 2.3213 | 30.751 | 4.49 | 4.50 | 0.01 |
| 16 | 0.00484 | −0.03508 | 2.1624 | 35.533 | 5.53 | 5.30 | −0.23 |
| 6 | −0.11949 | −0.11486 | 2.2748 | 34.826 | 4.96 | 5.31 | 0.35 |
| 2 | 0.02322 | −0.07484 | 2.2746 | 34.950 | 5.37 | 5.33 | −0.04 |
| 11 | −0.03900 | −0.10688 | 2.2190 | 29.625 | 5.15 | 5.24 | 0.09 |
| 3 | 0.02246 | −0.04938 | 2.2949 | 31.074 | 4.77 | 4.75 | −0.02 |
Key: YPred = predicted anti-proliferate activity, YExp. = Experimental activity.
Table 5.
Calculated descriptors, activities and residual for test set compounds.
| Molecule | MATS7c | MATS3e | maxwHBa | WPSA-3 | YPred. | YExp. | Residual |
|---|---|---|---|---|---|---|---|
| 27 | −0.02542 | 0.00757 | 2.2927 | 30.538 | 4.06 | 4.50 | 0.44 |
| 26 | −0.11348 | −0.06624 | 2.2425 | 30.239 | 4.40 | 4.24 | −0.16 |
| 21 | −0.00299 | −0.06489 | 2.2981 | 31.621 | 4.80 | 4.25 | −0.55 |
| 20 | −0.04550 | −0.04596 | 2.2729 | 30.038 | 4.42 | 4.59 | 0.17 |
| 12 | −0.03117 | −0.12652 | 2.2740 | 35.946 | 5.58 | 5.44 | −0.14 |
| 8 | 0.00860 | −0.12149 | 2.2735 | 36.649 | 5.80 | 5.52 | −0.28 |
| 5 | −0.01263 | −0.11638 | 2.2686 | 32.988 | 5.39 | 5.83 | 0.44 |
| 7 | −0.00567 | −0.11612 | 2.2727 | 31.957 | 5.32 | 5.17 | −0.15 |
| 15 | −0.02748 | −0.13150 | 2.2764 | 33.115 | 5.41 | 5.17 | −0.24 |
Key: YPred = predicted anti-proliferate activity, YExp. = Experimental activity.
As earlier highlighted in section 2.1.3, the co-efficient of determination of the test set (R2test) is used to ascertain the stability, robustness and predicting power of the built model. R2test was calculated and obtained to be equal to 0.6682. This implies that the built model is able to explain 66.82% of the variation in the anti-proliferate activity of the molecules in the test set.
Table 6 shows the mean effect, correlation matrix and VIF of the descriptors in the model. The mean effect revealed that the descriptor maxwHBa had the highest effect on the activity of the molecule while descriptor MATS3e had the lowest. Descriptor pairs had low correlation coefficients (all <0.4) implying that no significant inter-correlation exists between the descriptors. VIF values obtained were less than 2 which show that there was little multicollinearity between the descriptors. The VIF values reveal that the descriptors are indeed poorly correlated and are therefore suitable for model building.
Table 6.
Pearson's correlation matrix, mean effect and VIF of the descriptors.
| MATS7c | MATS3e | maxwHBa | WPSA-3 | Mean Effect | VIF | |
|---|---|---|---|---|---|---|
| MATS7c | 1 | 0.01393 | 1.29861 | |||
| MATS3e | 0.28266 | 1 | −0.07242 | 1.22541 | ||
| maxwHBa | −0.00806 | −0.31990 | 1 | 1.39824 | 1.16618 | |
| WPSA-3 | −0.38535 | 0.00698 | −0.20256 | 1 | −0.33975 | 1.24057 |
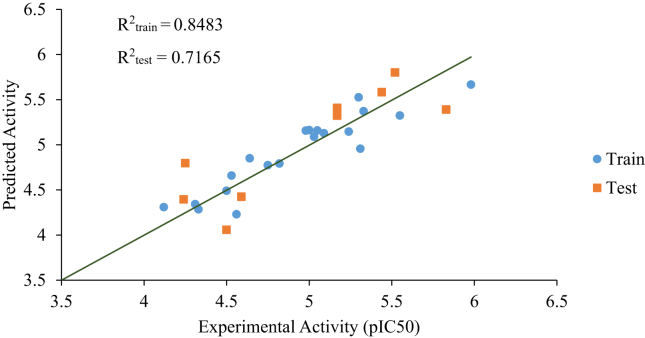
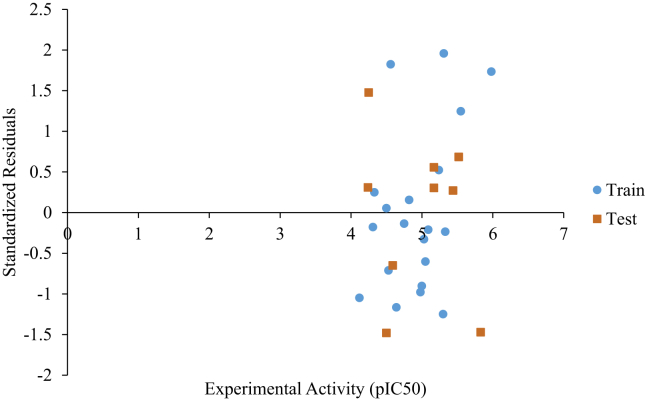
The predicted and experimental activity of the dataset are plotted on Figure 3. The R2 of the training and test set reflects the strong agreement between predicted and experimental activity, and affirms the reliability and predicting power of the built model. A graph of standardized residuals against experimental activity of the molecules is shown in Figure 4. The graph shows the absence of systematic errors because the standardized residuals are randomly distributed on either side of the zero mark.
Figure 3.
Predicted activity against experimental activity.
Figure 4.
Standardized residuals against experimental activity.
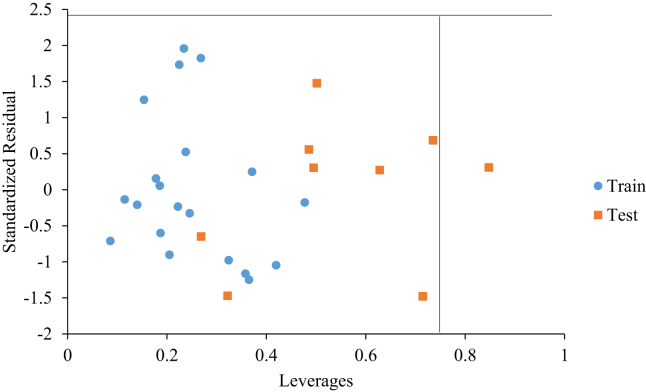
The leverage of each molecule in this study was calculated and plotted against the standardized residuals so as to identify outliers and influential compounds. This plot (the William's plot) is shown in Figure 5. The critical leverage (h*) was evaluated to be equal 0.75. Only one molecule had leverage above the warning leverage, further highlighting the reliability and predicting power of the built model.
Figure 5.
Applicability domain (William's plot) of the model.
3.2. Docking
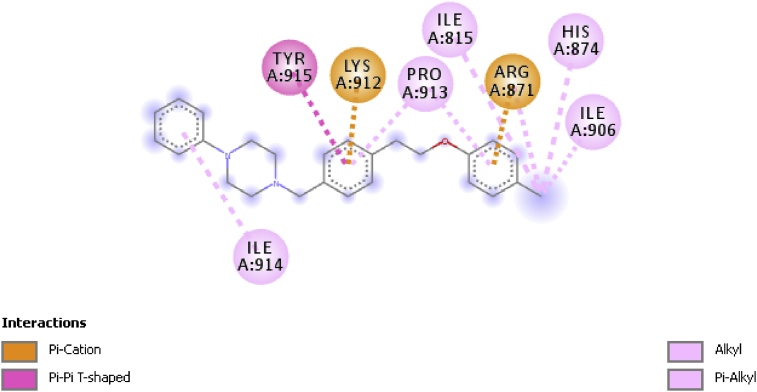
Findings from docking studies comprising of the binding affinity, amino acids, bond type, nature of interaction and bond distance of compounds 5 and 17 are presented in Table 7. Binding affinities of compounds 5 and 17 were found to be −7.5 and −7.1 kcal/mol respectively indicating that compound 5 formed a more stable complex with the ligand than did compound 17. The ligand-receptor interaction as observed from Biovia Discovery Studio revealed that compound 5 had hydrogen, electrostatic and hydrophobic interactions with the receptor. A 2D image of this interaction (Figure 6) revealed that the delocalized pi electrons in the benzene moieties formed a pi-cation bond with ARG871 (4.0679 Ǻ), LYS912 (4.4729 Ǻ) and a Pi-Pi T-shaped bond with TYR915 (4.84303 Ǻ).
Table 7.
Findings from docking studies of compounds 5 and 17 against the androgen receptor.
| Ligand | Binding Affinity (kcal/mol) | Amino Acids | Bond Type | Interaction | Distance (Ǻ) |
|---|---|---|---|---|---|
| 5 |
−7.5 |
ARG871 | Hydrogen Bond; Electrostatic | Pi-Cation; Pi-Donor Hydrogen Bond | 4.0679 |
| LYS912 | Electrostatic | Pi-Cation | 4.4729 | ||
| TYR915 | Hydrophobic | Pi-Pi T-shaped | 4.84303 | ||
| ILE815 | Hydrophobic | Alkyl | 5.28911 | ||
| ARG871 | Hydrophobic | Alkyl | 4.16664 | ||
| ILE906 | Hydrophobic | Alkyl | 4.65071 | ||
| HIS874 | Hydrophobic | Pi-Alkyl | 5.09449 | ||
| LYS912 | Hydrophobic | Pi-Alkyl | 5.08607 | ||
| PRO913 | Hydrophobic | Pi-Alkyl | 5.44251 | ||
| ILE914 | Hydrophobic | Pi-Alkyl | 4.87692 | ||
| ARG871 | Hydrophobic | Pi-Alkyl | 3.82029 | ||
| PRO913 | Hydrophobic | Pi-Alkyl | 3.82213 | ||
| 17 |
−7.1 |
SER853 | Hydrogen Bond | Carbon Hydrogen Bond | 3.36949 |
| TYR857 | Hydrophobic | Pi-Pi Stacked | 3.8868 | ||
| PHE856 | Hydrophobic | Pi-Pi T-shaped | 4.84843 | ||
| TYR857 | Hydrophobic | Pi-Pi T-shaped | 4.81343 | ||
| TYR857 | Hydrophobic | Pi-Alkyl | 5.04044 | ||
| LYS861 | Hydrophobic | Pi-Alkyl | 5.13623 | ||
| ARG854 | Hydrophobic | Pi-Alkyl | 5.4418 |
Figure 6.
2D Interaction between compound 5 and AR (5t8e).
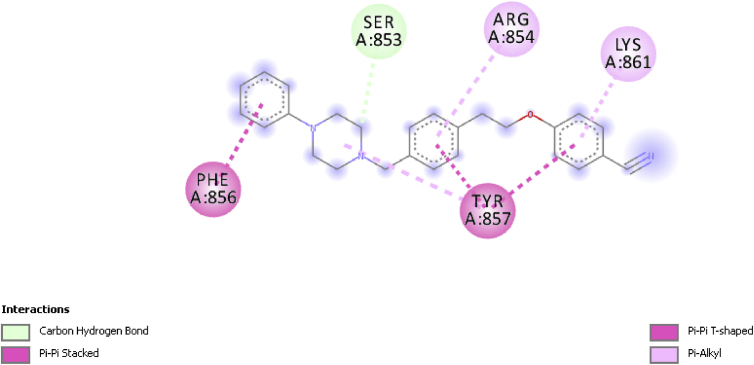
Compound 17 on the other hand was observed to form only hydrogen and hydrophobic interactions with the receptor. The piperazine ring in compound 17 formed a carbon-hydrogen bond interaction with the receptor, while it was also observed that benzene moieties in the compound formed a hydrophobic Pi-Pi T-shaped interaction with TYR857 (4.81343 Ǻ) and PHE 856 (4.84843 Ǻ) as seen in Figure 7.
Figure 7.
2D Interaction between compound 17 and AR (5t8e).
Results obtained revealed that both compounds mainly bind to AR (5t8e) via hydrogen and hydrophobic bond interactions. Chen et al. (2018a) reported some aryl piperazine derivatives to bind to the Ligand Binding Pocket (LBP) of AR through hydrogen bond interaction. Arjun et al. (2019) reported some Benzohydrazine derivatives to bind via hydrophobic interactions. Anti-prostate compounds have been reported to bind with ARG (arginine), ASN (Asparagine), PHE (Phenylalanine), LYS (Lysine), VAL (Valine), ILE (Isoleucine), LEU (Leucine) protein residues of the androgen receptor (McNerney and Onate, 2015; Chen et al., 2018a; Arjun et al., 2019). The binding affinity of compounds 5 and 17 were within the range reported Arjun et al. (2019).
4. Conclusion
A QSAR model for arylpiperazine was successfully developed which predicted the anti-proliferate activity against PC3 PCa cell lines by employing Genetic Function Algorithm method. Model 4 was the best model built; its R2, R2adj and Q2cv are 0.8483, 0.8078 and 0.7122 respectively. When tried on the test set, Model 4 gave an external validation (R2test) of 0.6682. Model 4 revealed that the activities of the compounds were strongly dependent on four descriptors; MATS7c, MATS3e, maxwHBa and WPSA-3. These descriptors were poorly correlated and had VIF values below 2. The robustness of the model was tested using the William's plot and only one outlier compound was obtained. Molecular docking studies of compounds 5 and 17 showed they had a binding affinity of −7.5 and −7.1 kcal/mol respectively. Compound 5 bound to the androgen receptor through electrostatic, hydrogen and hydrophobic bonds while compound 17 form hydrogen and hydrophobic bond interactions with the receptor. These findings provide a roadmap for the development of novel piperazine compounds with potent activity against PCa PC3 cell lines.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Author contribution statement
Fabian Adakole Ikwu: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Gideon A. Shallangwa: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Paul A. Mamza: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Adamu Uzairu: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Abdullahi M., Uzairu A., Shallangwa G.A., Mamza P., Arthur D.E., Ibrahim M.T. An Insilico modelling study on some C14-urea-Tetrandrine derivatives as potent anti-cancer against prostate (PC3) cell line. J. King Saud Univ. Sci. 2019;32(1):770–779. [Google Scholar]
- ACS (American Cancer Society) About Prostate cancer. Cancer. Org. 2016 https://www.cancer.org/content/dam/CRC/PDF/Public/8793.00.pdf 2016. [Google Scholar]
- Arjun H.A., Ramakrishnan E., Manikandan N., Lakshmithendral K., Muthiah R., Atanu B., Lokanath N.K., Senthamaraikannan K. Design, synthesis, and biological evaluation of (E)-N’-((1-Chloro-3,4-Dihydronaphthalen-2-yl)Methylene)Benzohydrazide derivatives as anti-prostate cancer agents. Front. Chem. 2019;7(474) doi: 10.3389/fchem.2019.00474. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arnatt C.K., Adams J.L., Zhang Z., Haney K.M., Li G., Zhang Y. Design, syntheses, and characterization of piperazine based chemokine receptor CCR5 antagonists as antiprostate cancer agents. Bioorg. Med. Chem. Lett. 2014;24(10):2319–2323. doi: 10.1016/j.bmcl.2014.03.073. [DOI] [PubMed] [Google Scholar]
- Arthur D.E., Uzairu A., Mamza P., Abechi S.E., Shallangwa G.A. Insilico modelling of quantitative structure-activity relationship of Pgi50 anticancer compounds on k-562 cell line. Cogent Chem. 2018;4(1) [Google Scholar]
- Baran M., Kepczynska E., Zylewski M., Siwek A., Bednarski M., Cegla M.T. Studies on novel pyridine and 2-pyridone derivatives of N-arylpiperazine as α-adrenoceptor ligands. Med. Chem. 2014;10(2):144–153. doi: 10.2174/0929867320999131122114922. [DOI] [PubMed] [Google Scholar]
- Becke A.D. Becke’s three parameter hybrid method using the LYP correlation functional. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- Bell N., Connor G.S., Shane A., Joffres M., Singh H., Dickinson J. Recommendations on screening for prostate cancer with the prostate-specific antigen test. Can. Med. Assoc. J. 2014;186(16):1225–1234. doi: 10.1503/cmaj.140703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center M.M., Jemal A., Lortet-Tieulent J., Ward E., Ferlay J., Brawley O., Bray F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012;61(6):1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Chen H., Wang C., Sun T., Zhou Z., Niu J., Tian X., Yuan M. Synthesis, biological evaluation and SAR of naftopidil-based arylpiperazine derivatives. Bioorg. Med. Chem. Lett. 2018;28:1534–1539. doi: 10.1016/j.bmcl.2018.03.070. [DOI] [PubMed] [Google Scholar]
- Chen H., Yu Y.Z., Tian X., Wang C., Qian Y., Deng Z., Zhang J., Lv D., Zhang H., Shen J., Yuan M., Zhao S. Synthesis and biological evaluation of arylpiperazine derivatives as potential anti-prostate cancer agents. Bioorg. Med. Chem. 2018;27(1):133–143. doi: 10.1016/j.bmc.2018.11.029. [DOI] [PubMed] [Google Scholar]
- Edache E.I., Arthur D.E., Abdulfatai U. Quantitative structure-activity relationship analysis of the anti-tyrosine activity of some tetraketone and benzyl-benzoate derivatives based on genetic algorithm-multiple linear regression. J. Chem. Mater. Res. 2017;6(1):2–12. [Google Scholar]
- Fernando A., Ubirajara O. Current guidelines for prostate cancer screening: a systematic review and minimal core proposal. Rev. Assoc. Med. Bras. 2018;64(3):290–296. doi: 10.1590/1806-9282.64.03.290. [DOI] [PubMed] [Google Scholar]
- Gerhardt A., Chris P., Ros A.E., Fritz S., Scott A.T., Ian T., Charles G.D., Johann S.B. Prostate cancer. Seminar. 2015;387(10013):70–82. [Google Scholar]
- Ibrahim M.T., Uzairu A., Shallangwa G.A., Abdulqadir I. In-silico studies of some oxadiazoles derivatives as anti-diabetic compounds. J. King Saud Univ. Sci. 2018;32(1):423–432. [Google Scholar]
- James T., MacLellan P., Burslem G.M., Simpson I., Grant J.A., Warriner S., Sridharan V., Nelson A. A modular lead-oriented synthesis of diverse piperazine, 1,4-diazepane and 1,5-diazocane scaffolds. Org. Biomol. Chem. 2014;12(16):2584–2591. doi: 10.1039/c3ob42512f. [DOI] [PubMed] [Google Scholar]
- Kennard R.W., Stone L.A. Computer aided design of experiments. Technometrics. 1969;11(1):137–148. [Google Scholar]
- McNerney E.M., Onate S.A. New insights in the role of androgen-to-estrogen ratios, specific growth factors and bone cell microenvironment to potentiate prostate cancer bone metastasis. Nucl. Recept. Res. 2015;2 Article ID 101186. [Google Scholar]
- Miller A.B., Feld R., Fontana R., Gohagan J.K., Jatoi I., Lawrence W., Jr., Miller A., Prorok P.C., Rajput A., Sherman M., Welch G., Wright P., Yurgalevitch S., Albertsen P. Changes in and impact of the death review process in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Rev. Recent Clin. Trials. 2016;10(3):206–211. doi: 10.2174/1574887110666150730120752. [DOI] [PubMed] [Google Scholar]
- Minovski N., Župerl Š., Drgan V., Novič M. Assessment of applicability domain for multivariate counter-propagation artificial neural network predictive models by minimum Euclidean distance space analysis: a case study. Anal. Chim. Acta. 2013;759:28–42. doi: 10.1016/j.aca.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Netzeva T.I., Worth A., Aldenberg T., Benigni R. Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships. The report and recommendations of ECVAM Workshop 52. Altern. Lab. Anim. 2005;33(2):155–173. doi: 10.1177/026119290503300209. [DOI] [PubMed] [Google Scholar]
- Nordqvist C. Prostate cancer in detail. Med. News Today. 2017 https://www.medicalnewstoday.com/articles/150086.php [Google Scholar]
- Ogadimma A.I., Adamu U. Quantitative structure activity relationship analysis of selected chalcone derivatives as Mycobacterium tuberculosis inhibitors. Open Access Libr. J. 2016;3(3):1–13. [Google Scholar]
- PCF (Prostate Cancer Foundation) Prostate Cancer Foundation; Santa Monica: 2010. An Introduction to Prostate Cancer.https://www.pcf.org/guide/an-introduction-to-prostate-cancer/ [Google Scholar]
- Qaseem A., Barry M.J., Denberg T.D., Owens D.K., Shekelle P. Screening for prostate cancer: a guidance statement from the clinical guidelines committee of the American college of physicians. Ann. Intern. Med. 2013;158(10):761–769. doi: 10.7326/0003-4819-158-10-201305210-00633. [DOI] [PubMed] [Google Scholar]
- Szkaradek N., Rapacz A., Pytka K., Filipek B., Siwek A., Cegła M., Marona H. Synthesis and preliminary evaluation of pharmacological properties of some piperazine derivatives of xanthone. Bioorg. Med. Chem. 2013;21(2):514–522. doi: 10.1016/j.bmc.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Tan M.H., Li J., Xu E., Melcher K., Yong E. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharm. Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropsha A. Best practices for QSAR model development, validation and exploitation. Mol. Inf. 2010;29(6-7):476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31 doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardanyan R.S. Elsevier; US: 2017. Piperidine-based Drug Discovery. [Google Scholar]
- Veerasamy R., Rajak H., Jain A., Sivadasan S., Varghese C.P., Agrawal R.K. Validation of QSAR models-strategies and importance. Int. J. Drug Des. Discov. 2011;2(3):511–519. [Google Scholar]
- Vitaku E., Smith D.T., Njardarson J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014;57(24):10257–10274. doi: 10.1021/jm501100b. [DOI] [PubMed] [Google Scholar]
- Wavefunction, Inc . 2013. Spartan’14, version 1.1.2. Irvine, California, USA. [Google Scholar]
- Wolf A.M., Wender R.C., Etzioni R.B., Thompson I.M., D'Amico A.V., Volk R.J., Brooks D.D., Dash C., Guessous I., Andrews K., DeSantis C., Smith R.A. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA. Cancer J. Clin. 2010;60(2):70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.