Abstract
Given the clinical effect of laminopathies, understanding lamin mechanical properties will benefit the treatment of heart failure. Here we report a mechano-dynamic study of LMNA mutations in neonatal rat ventricular myocytes (NRVM) using single cell spectroscopy with Atomic Force Microscopy (AFM) and measured changes in beating force, frequency and contractile amplitude of selected mutant-expressing cells within cell clusters. Furthermore, since beat-to-beat variations can provide clues on the origin of arrhythmias, we analyzed the beating rate variability using a time-domain method which provides a Poincaré plot. Data were further correlated to cell phenotypes. Immunofluorescence and calcium imaging analysis showed that mutant lamin changed NRVMs beating force and frequency. Additionally, we noted an altered microtubule network organization with shorter filament length, and defective hemichannel membrane localization (Connexin 43). These data highlight the interconnection between nucleoskeleton, cytoskeleton and sarcolemmal structures, and the transcellular consequences of mutant lamin protein in the pathogenesis of the cardiac laminopathies.
Keywords: Biological sciences, Cardiology, Biomechanics, Cytoskeleton, Mechanical property, Membrane, Lamin A/C, Cardiomyopathy, Atomic force microscopy, Cardiomyocytes, Beating, Cx43
Biological sciences; Cardiology; Biomechanics; Cytoskeleton; Mechanical property; Membrane; Lamin A/C; Cardiomyopathy; Atomic force microscopy; Cardiomyocytes; Beating; Cx43
1. Introduction
In order to maintain normal heart functionality, cardiomyocytes detect and react to mechanical stresses. With each heartbeat, these cells transfer (via mechanotransduction) contractile stress mainly via cytoskeleton fiber components. These fibers (actin, microtubule, intermediate filaments) act as structural “bridges” between the cell exterior and the nucleus via the nuclear lamina. If one of these connections is missing, anomalous or unbalanced, the global mechano-dynamic behavior of the cardiomyocyte is affected. One example in which the mechanical link between the cell environment and the nucleus is perturbed is demonstrated in mutations of the LMNA gene encoding for lamin A and C, main components of the nuclear lamina. These mutations cause diseases called laminopathies [1, 2, 3]. LMNA mutations are responsible for progressive myocardial dysfunction leading to heart failure frequently requiring cardiac transplantation, and lethal arrhythmias [4, 5, 6]. Laminopathies show a compromised mechanical cell performance [7, 8, 9], primarily manifesting in tissues subjected to high levels of mechanical stress, such as myocardial tissue. Although considerable progress has been made in understanding lamin structure and function, it is still not fully understood how different LMNA mutations affect both electrical function and contractility.
Nuclear lamins are intermediate filaments at the interface between the nuclear membrane and chromatin. They are critical for structural support of the nucleus, but evidence implies that nuclear lamins are also involved in other functions including nuclear envelope assembly, DNA synthesis, transcription, and apoptosis [10]. Furthermore, lamins, through specific proteins called “linkers of the nucleoskeleton and cytoskeleton (LINCs)” directly transmit forces from the extracellular matrix into the nucleus [11]. Genetic defects in lamin A/C (LMNA) cause numerous laminopathies, disorders affecting muscle, adipose tissue, nerve, bone, and skin, and are responsible for premature-aging diseases (Progeria) [12]. LMNA mutations modify lamin filament organization and nuclear mechanical properties [13]. In previous work [14], we investigated three human LMNA mutations Glu161Lys (E161K, rs28933093), Asp192Gly (D192G, rs57045855), and Asn195Lys (N195K, rs28933091). We selected these three LMNA mutations because of their clinical and cellular characteristics: E161K is a recurrent mutation [15, 16, 17], D192G and N195K were associated with disruption of nuclear envelope morphology and altered internal organization of cardiomyocytes [18, 19]. We showed [14, 20, 21] that expression of LMNAE161K, LMNAN195K, and LMNAD192G in neonatal rat ventricular myocytes (NRVM) increased nuclear stiffness compared to wildtype LMNA. More precisely, LMNAD192G expressing cells had the highest Young's modulus followed by LMNAN195K and LMNAE161K. These three LMNA mutations reduced the work of adhesion required to detach the spherical tip of an atomic force microscopy (AFM) cantilever from the cell membrane after indentation. In this case, LMNAD192G mutation was the most disruptive, with a reduction of 45% of adhesion compared to the control cells. We confirmed that the deleterious effects of LMNA mutations extended beyond altered nuclear mechanics, to include defective cell membrane adhesion work. At the same time, cells expressing these mutations were more viscous compared to control cells, and stored less elastic energy in their cytoskeleton components when subjected to a rapid mechanical stress [20].
Because arrhythmias and poor myocardial contractility are clinical features of laminopathies, we hypothesized that beating frequency and force could be significantly altered at the single-cell level in mutant LMNA cardiomyocytes, and that the assessment of cardiomyocyte mechano-dynamic properties in LMNA mutant models will provide insights into the process leading to cardiomyopathy [22].
Here, we report a mechano-dynamic study of LMNA mutants in neonatal rat ventricular myocytes (NRVM) using single cell spectroscopy with AFM to measure changes in force, frequency and contractile amplitude of their beating. We investigated NRVMs carrying the three aforementioned LMNA mutation (LMNAE161K, LMNAN195K, and LMNAD192G) and compared them to non-transfected (NT) and wild-type LMNA (WT) controls. We also analyzed the beating rate variability using a time-domain method and generated Poincaré plots. Data were further correlated to cell phenotypes using immunofluorescence and calcium imaging analysis, showing that mutant lamin changes NRVMs beating force and frequency, and lead to altered cytoskeletal structure and hemichannel localization.
2. Results
2.1. Beating behavior in LMNA mutant cardiomyocytes
To control our experimental conditions, such as temperature, pH and duration, the same protocol/methodology and the same model was used for all cell and AFM tests. Initially we checked how cell clustering affected cardiomyocyte behavior. Isolated beating cells and small clusters of beating cells were tested to assess variation in beating behavior. We found that isolated cells had significant mechano-dynamic variability during the acquisition time. Instead, control and mutant cell clusters showed a regular and stable beating pattern, providing more reliable data. Beating forces were obtained from AFM cantilever deflection data multiplied by the cantilever spring constant as detailed in the Method section. For instance, single NT cells beat with a force of 1.56 nanoNewtons (nN) (s.d. = 0.71, n = 26) while clusters beat with a force of 2.06 nN (s.d. = 0.93, n = 45, p = 0.01); similarly, single WT cells beat with a force of 1.47 nN (s.d. = 0.54, n = 26) while cluster WT beat with a force of 1.84 nN (s.d. = 0.76, n = 45, p = 0.02). Therefore, all further data reported in this study are related to selected cells within (in the center of) clusters obtained from NRVMs seeded at density of 20,000 cells/plate. To assess how the applied load influenced the beating property of our cells, we varied the applied AFM cantilever load. The signal remained constant for an applied force up to 4 nN; however; a 1 nN force was maintained on cells throughout all experiments.
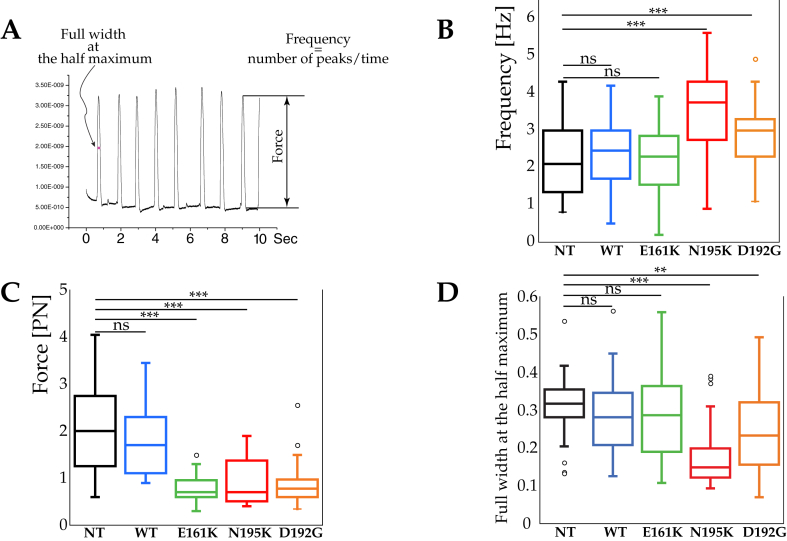
Figure 1A shows raw AFM data. Figure 1B shows the beating frequency of NT, WT and cells expressing the LMNA mutations.
Figure 1.
Mechanobiological cardiomyocyte properties: frequency, contraction force and peak duration (A) An example of the AFM trace during NT cardiomyocyte beating, (B) Box and whisker plots showing beating frequency of NT, WT and LMNAE161K, LMNAN195K, and LMNAD192G mutations. ***Represents p < 0.0001, (t-test) (C) Beating force. ***Represents p < 0.0001, (t-test) (D) Peak duration of LMNA mutant cardiomyocytes described as full width at the half-maximum (FWHM, namely the width of the beating pulse measured between those points on the y-axis which are half the maximum amplitude). ***Represents p < 0.0001, **p < 0.001, (t-test).
While there were no statistical differences between NT (2.23 beats/s, s.d. = 0.93, n = 45), WT (2.40 beats/s, s.d. = 0.89, n = 45) and LMNAE161K (2.21 beats/s, s.d. = 0.88, n = 54), both LMNAN195K (3.55 beats/s, s.d. = 1.14, n = 39) and LMNAD192G (2.87 beats/s, s.d. = 0.91, n = 36) mutations exhibited higher beating frequency as compared to NT.
Figure 1C shows the beating force for NT, WT and the three LMNA mutations. There were no statistical differences between NT (2.06 nN, s.d. = 0.93, n = 45) and WT (1.84 nN, s.d. = 0.76, n = 45) while LMNAE161K (0.76 n, s.d. = 0.27, n = 54), LMNAN195K (0.94 nN, s.d. = 0.49, n = 39) and LMNAD192G (0.88 nN, s.d. = 0.42, n = 36) had a lower beating force.
Figure 1D indicates the beat duration in terms of the full-width at half-maximum (FWHM). Values for NT, WT and LMNAE161K did not show any statistical differences (p = 0.20) while LMNAN195K and LMNAD192G had small FWHM measurements than NT (p < 0.0001 and p = 0.00094, respectively).
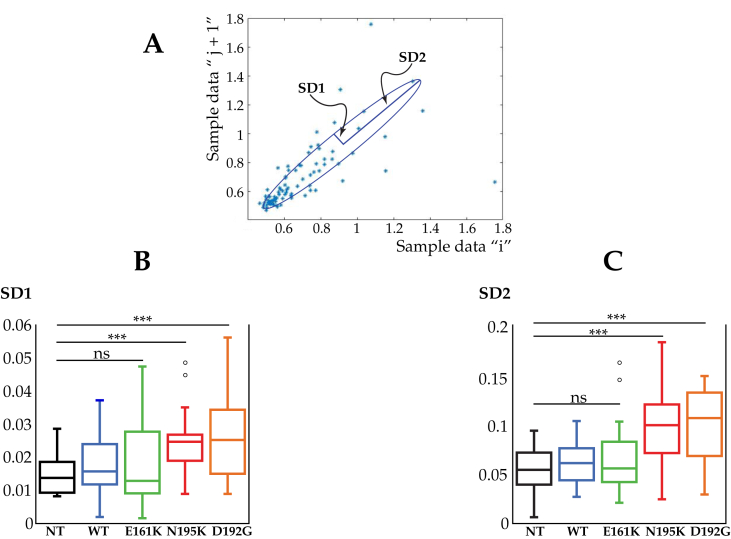
Investigations into beating rate variability (BRV) [23] utilized the Poincare' geometric nonlinear analysis. Figure 2A shows a typical Poincaré plot. A quantitative analysis of the Poincaré plot can be created by approximating it as an ellipse, where (i) SD1 (Standard Deviation 1), is the standard deviation of the short-term beat-to-beat interval variability (minor axis of the ellipse), (ii) SD2 (Standard Deviation 2) is the standard deviation of the long term interval variability (major axis of the ellipse) [24]. Figure 2B, C show the SD1 and SD2 value for the LMNA mutations. In this case, only LMNAN195K and LMNAD192G displayed a statistical difference for both SD1 and SD2 compared with the NT cells, suggesting an irregular, arrhythmic beat of the mutant cardiomyocytes.
Figure 2.
Beating rate variability by Poincaré plot of LMNA mutant cardiomyocytes. (A) A typical Poincaré Plot of RR intervals for NT cardiomyocyte. The peak interval (RR n) is displayed on the x-axis and the extent of the following interval (RR n+1) on the y-axis. All points described by successive beats of equal duration (RR n = RR n+1) are situated on the identity line. The points below this line represent all shortenings of the interval between 2 consecutive beats (RR n > RR n+1) while the points above the identity line correspond to all prolongations (RR n < RR n+1). (B) Analysis of the beating rate variability of LMNA mutant cardiomyocytes. SD1 (the standard deviation of the instantaneous - short term - beat-to-beat variability and (C) SD2 (the standard deviation of the long-term interval variability) for NT, WT and LMNAE161K, LMNAN195K, and LMNAD192G mutations. ***Represents p < 0.0001, (t-test).
2.2. Altered connexin 43 localization in mutant LMNA cardiomyocytes
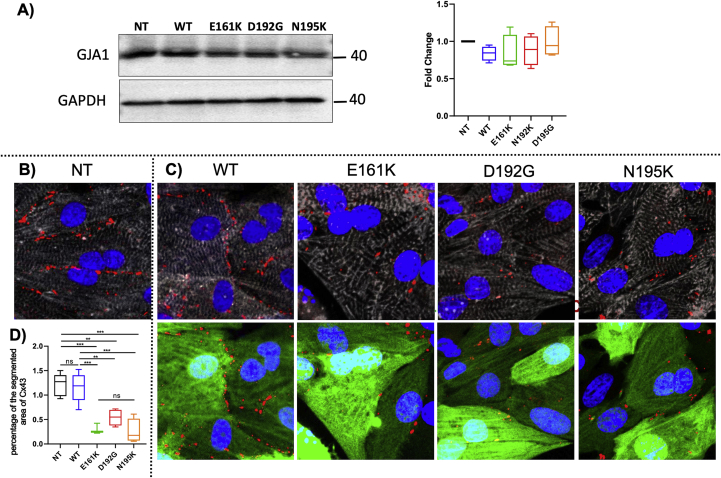
Since intercellular communication is a vital organizational feature of the heart [20, 25, 26] and disrupted ion channel function could affect contractile rates and forces, we examined the levels and localizations of gap-junctions through immunostaining for Connexin 43 (Cx43). Numerous studies have indicated that Cx43 organization is crucial for normal ventricular function and cellular impulse propagation in the healthy human heart [25, 27]. On the other hand, variation of the Cx43 organization might be one of the leading mechanisms for arrhythmias [28]. Figure 3A shows western blot results for Cx43; the results from four western blots indicate that there is no significant Cx43 difference between the groups. Figure 3B shows representative images of NRVMs stained for the cardiac marker α-actinin, Cx43, GFP and nuclei; data are represented as fold change over NT. Figure 3B reveals that mutant NRVMs presented significantly smaller with punctate staining for Cx43 (161: 0.24 ± 0.07; 192: 0.44 ± 0.11; 195: 0.21 ± 0.17 fold over NT) compared to controls (NT 1 ± 0.17-fold; WT 0.95 ± 0.22-fold over NT). The altered localization of Cx43 on the sarcolemma, in spite of preserved Cx43 protein expression, could be due to defective cytoskeletal structures anchoring to membrane proteins.
Figure 3.
Cx43 protein expression in LMNA mutant cardiomyocytes. (A) Western blot showing Cx43 expression (left) and the results from western blots (n = 4) indicating no significant difference between the groups (right), p = 0.5038. (B) Cx43 localization in control (NT) cardiomyocytes. Fluorescence images of Connexin 43 (red dots), sarcomeric α-actinin (gray), LMNA GFP (green) and Hoechst (blue) staining of NRVMs (C) Cx43 localization in (from left to right): wild-type LMNA, LMNAE161K, LMNAD192G, LMNAN195K. Upper panels: Sarcomeric alpha-actinin and connexin 43. Lower panels LMNA GFP expression and connexin 43. (D) Quantification of Cx43 gap junction area: significant differences on Cx43 gap-junction were observed between LMNAE161K, LMNAD192G and LMNAN195K and the non-transfected NT cells and WT. Data are presented as mean ± S.D. (n = 5). 161 vs non-treatment: ***p value: <0.0005; 192 vs non treatment: **p value: <0.0045; 195 vs non treatment: ***p value: <0.0001; 161 vs WT: ***p value: <0.0009; 192 vs WT: **p value: <0.0086; 195 vs WT: ***p value: <0.0002. Non-significant differences were observed between the mutant cells and between WT and NT. A full, non-adjusted image is provided in supplementary material (Figure S1).
2.3. Altered calcium transients in mutant LMNA cardiomyocytes
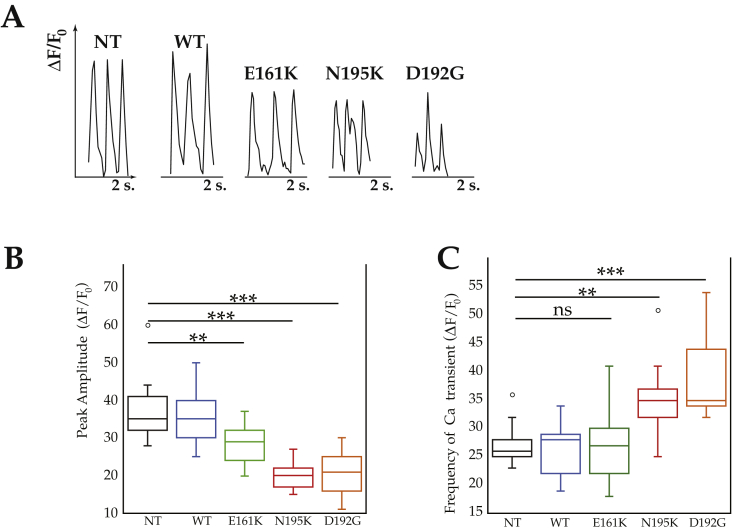
Calcium imaging was used to recapitulate the mechano-dynamic behavior assessed with AFM. Calcium transients were measured by the change in Fura Red fluorescence signal (Δ(F)/F0) according to Neher et al. [29] It is well known that calcium flux and contractile activity are related [30]. Wild-type cells exhibited rhythmic frequency and timing comparable to those of NT cardiomyocytes (Figure 4A). However, LMNAN195K and LMNAD192G cardiomyocytes exhibited a noisier spectrum for calcium spikes, suggestive of arrhythmic behavior (Figure 4A), and had significantly smaller intracellular Ca2+ transient amplitudes compared to those of the controls (p = 0.0001, Figure 4B). This finding was less pronounced in the case of LMNAE161K (p < 0.005). Likewise, the frequency of calcium transients was significantly higher in LMNAN195K and LMNAD192G (p = 0.00091 for LMNAN195K and p < 0.0001 for LMNAD192G respectively) compared to controls, while in the case of LMNAE161K it was not statistically significant (Figure 4C). This impaired ability to handle Ca2+ agrees with the finding of a weaker force production and therefore a lower contractility.
Figure 4.
Spontaneous calcium transients of NRVMs. (A) representative spontaneous calcium transient phenotype. From left to right: Control (NT), WT, LMNAE161K, LMNAN195K and LMNAD192G. (B) quantification of peak amplitude. (C) Quantification of Ca++ transient frequency. ***Represents p < 0.0001, **p < 0.001, (t-test).
2.4. Microtubule organization
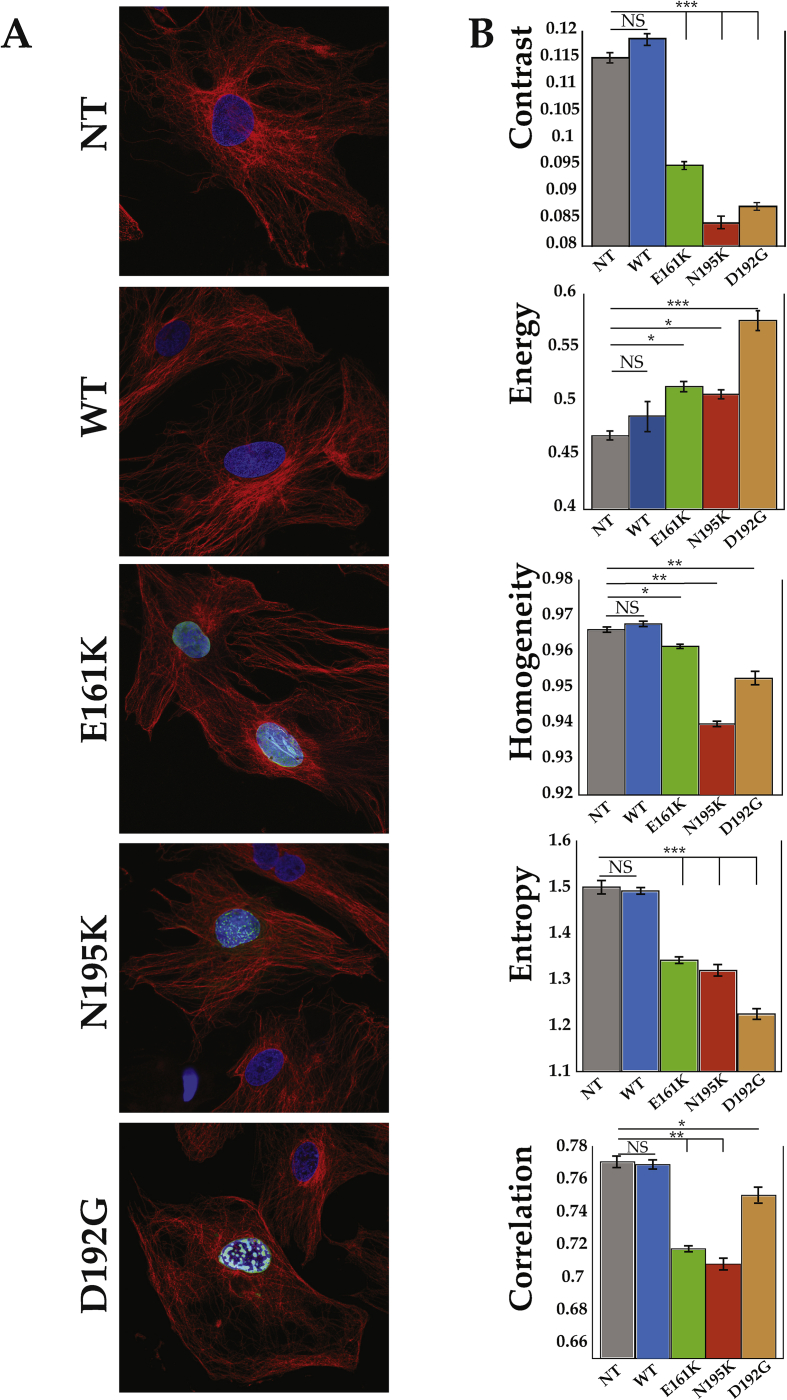
Figure 5 shows disarray of the tubulin network in mutant LMNA NRVMs compared to controls. To quantify the tubulin network organization in the different mutations (Figure 5A), we used the optical Haralick correlation metric. This optical method computes texture features at multiple offset distances and angles. As shown in Table 1 and Figure 5B, the three mutant LMNA cell models showed contrast, uniformity (homogeneity or angular second moment), local homogeneity (Inverse Difference Moment (IDM)), entropy and correlation significantly different from the control LMNA models, indicating less organized microtubule networks with shorter microtubule filaments length.
Figure 5.
Microtubule network organization. (A) Example of microtubule organization used to calculate the Haralick features. (B) Haralick texture features: Contrast, Angular second moment (energy), Inverse difference moment (homogeneity), Entropy and Correlation are shown only for an angle of O° (complete set of data in Table 1); Data are presented as mean ± S.D. (n = 4 independent experiments). *p < 0.05; **p < 0.005; ***p < 0.0001.
Table 1.
Haralick correlation metric to determine microtubule organization.
| Haralick Texture Features (O°) | NT | WT | % | E161K | % | N195K | % | D192G | % |
|---|---|---|---|---|---|---|---|---|---|
| Contrast | 0.1151 | 0.1187 | 3.13 | 0.0951 | 17.38 | 0.0843 | 26.74 | 0.0874 | 24.06 |
| Angular second moment (energy) | 0.4678 | 0.4864 | 3.98 | 0.514 | 9.88 | 0.5070 | 8.37 | 0.5755 | 23.00 |
| Inverse difference moment (homogeneity) | 0.9526 | 0.9399 | 0.29 | 0.9615 | 0.93 | 0.9678 | 1.62 | 0.9663 | 1.46 |
| Entropy | 1.5004 | 1.4921 | 0.55 | 1.3424 | 10.53 | 1.3199 | 12.03 | 1.2248 | 18.37 |
| Correlation |
0.7707 |
0.7689 |
0.23 |
0.7171 |
6.95 |
0.7077 |
9.08 |
0.7501 |
2.66 |
|
Haralick Texture Features (45°) |
NT |
WT |
% |
E161K |
% |
N195K |
% |
D192G |
% |
| Contrast | 0.1403 | 0.1369 | 2.42 | 0.1041 | 25.80 | 0.0897 | 36.08 | 0.1032 | 26.39 |
| Angular second moment (energy) | 0.4495 | 0.4561 | 1.47 | 0.484 | 7.69 | 0.5031 | 11.93 | 0.5626 | 25.16 |
| Inverse difference moment (homogeneity) | 0.9300 | 0.9344 | 0.47 | 0.949 | 2.04 | 0.9552 | 2.71 | 0.9484 | 1.97 |
| Entropy | 1.5717 | 1.5466 | 1.60 | 0.714 | 53.92 | 1.3369 | 14.94 | 1.2730 | 19.01 |
| Correlation |
0.7212 |
0.7258 |
0.64 |
0.709 |
1.69 |
0.7052 |
2.22 |
0.7112 |
1.39 |
|
Haralick Texture Features (90°) |
NT |
WT |
% |
E161K |
% |
N195K |
% |
D192G |
% |
| Contrast | 0.1929 | 0.1853 | 3.94 | 0.129 | 32.86 | 0.1273 | 34.01 | 0.1217 | 36.94 |
| Angular second moment (energy) | 0.3657 | 0.37 | 1.18 | 0.385 | 5.28 | 0.3819 | 4.42 | 0.4945 | 35.21 |
| Inverse difference moment (homogeneity) | 0.9038 | 0.9098 | 0.66 | 0.933 | 3.23 | 0.9364 | 3.60 | 0.9392 | 3.92 |
| Entropy | 1.8226 | 1.7961 | 1.45 | 0.877 | 51.88 | 1.6512 | 9.41 | 1.3871 | 23.90 |
| Correlation |
0.6388 |
0.6455 |
1.05 |
0.566 |
11.2 |
0.572 |
10.46 |
0.5042 |
21.04 |
|
Haralick Texture Features (135°) |
NT |
WT |
% |
E161K |
% |
N195K |
% |
D192G |
% |
| Contrast | 0.2109 | 0.2094 | 0.71 | 0.185 | 2.51 | 0.1745 | 17.23 | 0.1386 | 34.25 |
| Angular second moment (energy) | 0.3558 | 0.3646 | 2.47 | 0.372 | 4.55 | 0.3904 | 9.72 | 0.4839 | 36.02 |
| Inverse difference moment (homogeneity) | 0.8948 | 0.9031 | 0.93 | 0.9490 | 6.06 | 0.9127 | 2.00 | 0.9308 | 4.02 |
| Entropy | 1.8573 | 1.8373 | 1.08 | 1.388 | 25.23 | 1.7738 | 4.49 | 1.4246 | 23.30 |
| Correlation | 0.6037 | 0.6121 | 1.39 | 0.496 | 17.68 | 0.4791 | 20.62 | 0.4604 | 27.9 |
3. Discussion
In this study, we found that mutant lamin (LMNAD192G and LMNAN195K) produced abnormal contractile and beating behavior of NRVMs, as indicated by decreased beating force, increased and irregular beating frequency, decreased and irregular calcium transients and abnormal sarcolemma localization of Cx43 hemichannels. These data all point to a strong correlation between the nuclear envelope, global cardiomyocyte mechanotransduction and electrical function in LMNA mutant cardiomyocytes, highlighting that lamin A/C function and dysfunction extend beyond the nuclear membrane. Not surprisingly, LMNAE161K caused more subtle changes, which aligns with the phenotype of LMNAE161K patients found to be less severe than in LMNAD192G and LMNAN195K patients [14].
3.1. Single cell spectroscopy unravels mechanical and electrical defects in LMNA mutant cardiomyocytes
We posited that valuation of the beating properties of mutant cardiomyocytes at the single-cell scale would help clarify the pathophysiology of heart failure and arrhythmias in laminopathies, and provide novel insights for the future development of new therapies. Moreover, our study at the single-cellular level offered unique advantages, such as the analysis of the inotropic/chronotropic behavior of cardiomyocytes with LMNA mutations without influences such as neuro-hormonal activities, or pre- and after-loads. Our findings are in line with single cell spectroscopy studies of other inherited cardiomyopathies, which likewise found altered beating frequency and force of mutant induced pluripotent stem cell (iPSC)-derived cardiomyocytes [31, 32, 33, 34].
As shown in Figure 1C, all three mutations showed a decreased force of beating suggesting a negative inotropic effect of the LMNA mutations. Furthermore, LMNAD192G and LMNAN195K mutations had a positive chronotropic effect, since the frequency of beating increased (Figure 1B). Finally, both the Poincaré plot, which analyzes beat-to-beat variability, and the Ca2+ transients data revealed an irregular beating compared to controls, indicating an arrhythmic behavior at the cellular level.
It should be noted that for our single-cell spectroscopy analysis, we used the spherical AFM probe, which is ideal for soft biological samples since the force is applied to a broader cell area rather than with a sharp tip, resulting in a lower pressure and less cell damage. Due to the fact that cells or tissues are very inhomogeneous and consist of different components (nucleus, cytoskeletal components, etc.) a spherical probe will return more reliable data than a sharp tip.
3.2. LMNA mutations alter hemichannel localization in cardiomyocytes
For correct and efficient cardiac function, inter-cellular communications are crucial. Electrical and mechanical activities need to be synchronized so that the work of individual cardiomyocytes collectively organizes into normal cardiac contraction. Electrical current propagation throughout the heart is mediated through precisely regulated ion movements coordinated by various gap junction proteins [35]. Connexins assemble in hemichannels to form gap junctions which facilitate communication and signaling between adjacent cells. Cx43 is an important protein primarily expressed in the heart and in functioning myocytes, which ensures coordinated contractile action [36]. Our study found altered Cx43 localization with unchanged overall cellular levels of Cx43, which aligns with prior published data on the LMNAN195K mutation [37]. Little is known concerning the molecular mechanisms of remodeling of Cx43 in pathological context. Trafficking of Cx43 is regulated in part by the microtubule network [38, 39] and in fact, we observed a partial variation of the microtubule organization compared to our control cells as indicated by the disarray of the microtubule network (Figure 5, Table 1), with less organized microtubule networks and shorter microtubule filaments length. This result is in agreement with data from Macquart et al. [40] who showed that microtubule instability leads to an aberrant localization of Cx43 in cells expressing a lamin A mutation. Another study [41] suggested that appropriate nuclear lamina organization and a correct microtubule network is necessary for a collaborative effort to maintain a correct nuclear morphology and function. Indeed, we have already reported that the three mutations studied show changes in nuclear morphology [14]. Moreover, several studies showed that microtubules adversely affect the beating rate of neonatal cardiomyocytes, in fact when microtubules in neonatal heart cells are depolymerized the beating rate increases [42, 43, 44, 45]. Using the Haralick optical technique of texture analysis (Table 1), we found that the microtubule network was compromised by the LMNA mutations, and at the same time, the beating rate was increased, providing a link between the defective nuclear envelope and the abnormal hemichannels localization.
Our findings suggest that in cardiomyocytes, the chain of transfer of electrical and mechanical signaling from the cell membrane to the nucleus must be intact, and if one of the components, such as LMNA, is defective, the whole mechanism is compromised.
3.3. Limitations of the study
Even though it is well known that rodents show differences with the human biology [46], they represent an established and reproducible in vitro model [21], since cardiac tissue from patients is difficult to obtain and does not survive in long-term culture. Our results show that AFM can be used to assess the mechano-dynamic behavior of the chosen cells: we realize that orientation of actin-myosin filaments within a cardiomyocyte is anisotropic and therefore different parts of the cardiomyocytes may show varied amounts of movement and contractile forces with each beat. Furthermore, lateral modes of the contraction may not be accounted by the simple z-movement of the AFM cantilever. However, since in our methods of cell handling, study protocol and methodology, remained consistent throughout the entire series of experiments, we assume that our calculated beating force can be compared for all specimens.
4. Conclusions
Our study shows that LMNA gene mutations cause impairment of force production, frequency and rhythm of spontaneous beating and calcium handling in a single-cardiomyocyte cell model. Although the in vitro model presents limitations, it is intriguing that the changes found in cardiomyocytes recapitulate at the cellular level the clinical phenotype of LMNA cardiomyopathy patients, who typically present with decreased cardiac contractile function and arrhythmias, eventually leading to heart failure and sudden cardiac death. Single-cell spectroscopy by atomic force microscopy represents a unique tool to complement cardiac cell biology investigations and elucidates the complex mechanisms leading to cardiac dysfunction in laminopathies.
5. Materials and methods
5.1. Isolation and culture of ventricular cardiomyocytes from neonatal rat (NRVMS)
The University of Colorado Denver institutional ethical committee gave approval for these experiments. All animal studies were performed according to the guidelines Animal Care and Use Committee. All animal experiments have been done using all possible methods to alleviate or minimize potential pain, suffering or distress, and enhance animal welfare. Animals have been provided with housing in an enriched environment, with at least some freedom of movement, food, water and daily care and cleaning. The well-being and state of health of experimental animals have been observed by competent persons, dedicated to the management of the Animal House, able to prevent pain or avoidable suffering, distress or lasting harm. Experiments have been performed solely by competent authorized persons. Primary cultures of neonatal rat ventricular cardiomyocytes (NRVMs) were isolated and cultured from 1-3 days old Sprague Dawley rat pups (Charles River), following decapitation by enzymatic digestion as previously described with minor modifications [28, 47, 48, 49]. Briefly, ventricles were separated from the atria using scissors and then dissociated in CBFHH (calcium and bicarbonate-free Hanks with Hepes) buffer containing 0.5 mg/ml of Collagenase type 2 (Worthington, Biochemical Corporation), and 1 mg/ml of Pancreatine (Sigma). Cardiomyocytes were enriched (>90% purity) over non-myocytes by a pre-plating step on 100-mm dishes in Minimum Essential Media (MEM), supplemented with 5% bovine calf serum and 2 mg/ml vitamin B12 (Sigma). Myocytes that were either in solution or lightly attached were then separated from the adherent stromal cells by gentle mechanical disaggregation and subsequently plated at a density of 2 × 105 cells/ml in primary petri dishes (Falcon) or in multi-chambered slides coated with 0.1% gelatin (Sigma) and cultured as previously described [28, 49]. After 18 h, the culture medium was changed and cells were subjected to infection with the relevant adenoviral-LMNA constructs.
5.2. Isolation adenoviral constructs and infection
Methods for adenoviral infection have been previously reported [20, 21]. In brief, shuttle constructs were generated in Dual CCM plasmid DNA containing GFP gene and human LMNA cDNA. Constructs were bicistronic with the two inserts (LMNA and GFP) driven by two different CMV promoters to identify cells expressing LMNA protein using GFP as a marker of cellular infection. NRVMs were infected by adenoviruses at a multiplicity of infection (MOI) of 50 in serum free medium; 6 h post-infection, complete medium was replaced and the cells were incubated at 37 °C and 5% CO2. Previous control experiments have shown that GFP transfection and expression did not affect endpoints of interest in NRVM in our experimental conditions [21]. Five experimental conditions were present: NT-non-transfected; WT-wild-type LMNA construct and the three LMNA mutant conditions (LMNAE161K, LMNAN195K, and LMNAD192G).
5.3. Western blotting
Proteins were extracted from transduced NRVMs as published [50]. Briefly, transduced cells were lysed in a RIPA Buffer containing protease inhibitors and phosphatase inhibitor. Protein concentrations were determined by colorimetry RC-DC protein assay (Bio-Rad Laboratories). Equal amount of protein extracts was denatured and resolved on SDS-PAGE gels, and were then transferred to nitrocellulose membranes. Target proteins were detected using primary antibodies by overnight incubation at 4 °C. The respective horseradish linked secondary antibodies were incubated at room temperature for 1 h at concentration recommended by the manufacturer. The signal was amplified by an ECL chemiluminescence assay (GE Healthcare) and was detected using gel imager from Biorad. The primary antibodies used were as follows: rabbit polyclonal CX43 (Sigma, 1:1000) and mouse monoclonal GAPDH (ThermoFisher, 1:10000) as loading control.
5.4. Immunofluorescence
NRVMs were fixed in PBS containing 4% PFA for 15 min at room temperature. Cells were permeabilized at room temperature, with 1%Triton X-100 for 90 min, blocked with 2% BSA in PBS for 45 min and incubated overnight with alpha actinin 1:500 (Sigma) and Connexin 43 1:200 (Sigma) prepared in 2% BSA. Goat anti-mouse antibody conjugated to Cy5 (abcam) and Goat anti-rabbit antibody conjugated to Alexa Fluor 594 were used as secondary antibody 1:300 (Invitrogen). Each sample was stained with Hoechst 1:2000 (Thermo Fisher Scientific) to counter-staining the nuclei. Representative immunofluorescence images were acquired using Zeiss LSM780. Expression of human wild-type and mutant LMNA proteins were verified by western blot to confirm exogenous human LMNA in NRVMs together with the endogenous rat level expression.
5.5. Calcium imaging
To assess the electrical activity of cardiomyocytes with different mutations and controls, 2 days after infection, we recorded intracellular spontaneous calcium signaling. Cell-permeant Fura Red AM (Thermo Fisher Scientific) was added to each sample according to manufacturer instructions and incubated for 15 min. Samples were washed 3 times with warm media before imaging. Calcium transients were measured by the change in fluorescence divided by the resting fluorescence (ΔF/F0)29. Calcium transients during spontaneous cardiomyocytes beating were recorded using a Zeiss LSM780 confocal microscope and were measured for 21s (150 frames). Experiments were performed in triplicate from 3 independent experiments and averaged. Data was corrected for background epifluorescence.
5.6. Atomic force microscopy beating frequency and force
AFM is one of the methods for measuring cardiomyocyte mechano-dynamics properties [51]. In AFM, a small cantilever is brought in contact with the cell being measured. While the cell beats, the vertical movement of the cell is tracked producing the beating signal [52]. For instance, hPSC derived cardiomyocyte beating characteristics have been measured with AFM using this kind of protocol by Liu and colleagues [32].
In this work, a JPK NanoWizard 4a BioScience AFM was used to acquire the beating data. In AFM testing, it is very important to bear in mind that cells can be in different states and thus show distinct properties that can be difficult to compare. Therefore (i) AFM experiments were always performed on the same day after the adenoviral infection, (ii) multiple measurements from different cells have been collected to control for variability and ‘average’ data determined, and (iii) cells were monitored and their morphological details observed (an optical light microscope was used for cell selection throughout the tests). The AFM was equipped with PetriDishHeater™ tool to operate in liquid at controlled temperature (37 °C). A cantilever modified with a spherical gold probe (diameter of ~5.5 μm, CP-PNPL-Au-C, NanoAndMore GmbH) was used to precisely apply a compression force to the nucleus.
AFM probes were cleaned, prior to the indentation experiments, by embedding them in Tween (2% for 30 min) and then rinsed in milliQ water and ethanol, in order to remove contaminant molecules adsorbed on the probe surface.
All studies were performed on living, intact cells in cell culture medium. Only well-spread cells were investigated, those with a round shape and a dark edge were rejected. Furthermore, cardiomyocytes were selected based on the morphological aspect compared to the possible presence of fibroblasts and only if they were beating. When cluster cells were selected, based on cluster size, one or more cells were tested (usually 2–3 cells in every cluster).
The AFM tip was moved toward the cell with speed of 5 μm/s. The speed range was chosen to avoid cell movement (at low compression speed) or hydrodynamic force contribution (significant at high speed). All experiments were performed at the same velocity since these AFM tests are rate-dependent. Data were taken from a single point on the cell's surface on top of the nucleus.
The distal regions, away from the nucleus, were avoided since measurements performed around the nucleus are less affected by artifacts due to the substrate stiffness. For acquiring the beating data, the AFM tip was lowered until it touched the cell and the cantilever registered a deflection corresponding to 1 nN, then the z-piezo was locked for 30 s and beating frequency and probe travels were recorded. The beating forces were found by multiplying the probe displacements due to contractile activity and the cantilever's spring constant, assessed using the thermal noise method in liquid. The beating frequency of the cardiomyocytes was computed by quantifying the number of peaks in a measurement of a sufficiently long time (≥15 s). Peak duration was quantified in terms of full width at the half-maximum (FWHM, namely the width of the beating pulse measured between those points on the y-axis which are half the maximum amplitude). Data were processed using a self-developed MATLAB script. We used this approach either at single-cell resolution or on a cluster of cells which were beating synchronously. We assessed the beating force and frequency using at least 20 clusters for each condition (untreated/uninfected control: NT, wild type lamin: WT and three LMNA mutations: 161, 192 and 195). The test duration was never longer than 45–50 min to ensure cell viability.
5.7. Beating rate variability
Beating rate variability (BRV) was evaluated using the Poincaré plot, automatically generated during MATLAB processing. The Poincaré plot is a technique suggested by Tulppo et al. [53] for studying heart rate signals, as it helps to assess components of the beat variability related with short- and long-term connections of the signal [54, 55, 56]. It is a scatter graph of the peaks interval plotted against the preceding peaks interval (RR). The first peaks interval [i] represents the x-coordinate, the second interval [i+1] represents the y-coordinate. The quantitative analysis of the plot is realized by fitting an ellipse to the plot with its center coinciding with the centroid of the ellipse. The point where both ellipse axes intersect corresponds to the total mean of the intervals. The length of minor and major axis of the ellipse are 2SD1, 2SD2, where SD1, SD2 are the dispersion perpendicular to the line of identity (minor axis) and along the line of identity (major axis) respectively as proposed by [53]. SD1 is the standard deviation of the instantaneous (short term) beat-to-beat variability while SD2 is the standard deviation of the long-term interval variability.
5.8. Microtubule organization
Microtubule organization is calculated using Haralick features, which are pixel intensity-based algorithms for quantifying image texture [57, 58]. First, a gray-level co-occurrence matrix is calculated for given orientation and pixel pair offset distances. The co-occurrence matrix p calculates the frequency at which pixels within a specified intensity range are matched by spatially separated pixels of the same intensity. The result is a i × i matrix, where i is the number of gray-levels (or intensity bins) that are to be considered. From the co-occurrence matrix, several texture features can be measured. Haralick correlation is one of these features that measures the likelihood of finding two pixels of similar intensity separated by a given distance. Calculation of the co-occurrence matrix and corresponding Haralick correlation values was repeated at many orientation angles and spatial distances, resulting in an “m×n” matrix of Haralick correlation values, where m is the number of angles (values are symmetric about 180°) and n is the number of pixel offset. Other Haralick features that were considered in the comparison analysis include Contrast, Uniformity (also known as homogeneity or angular second moment), local Homogeneity (or Inverse Difference Moment (IDM)), Entropy and Correlation. We also measured a variance value, which is the sum of the Haralick contrast values at an offset distance of 1 pixel. We expected the variance value to be close to zero for homogenous images and close to one for patterned images.
5.9. Statistics
For all experiments, data were collected from at least three independent experiments. ImageJ was used for cell counting in 10 fields per each condition. For calcium transients, signals from myocytes were collected and analyzed from 10 fields per sample and averaged. Statistical significance between experimental groups was determined using ANOVA. ANOVA Power analysis was completed using GraphPad Prism7software. A p value of <0.05 was considered statistically significant.
Declarations
Author contribution statement
O. Sbaizero: conceived and designed the experiments; analyzed and interpreted the data; wrote the paper.
D. Borin: performed the experiments.
B. Pena and S. Chen: performed the experiments; analyzed and interpreted the data.
C. Long: conceived and designed the experiments; wrote the paper.
M. Taylor and L Mestroni: Conceived and designed the experiments; contributed reagents, materials, analysis tools or data; wrote the paper.
Funding statement
This work was supported by the Fondation Leducq Transatlantic Network of Excellence (14-CVD 03) and MIUR - PRIN2017.0004569.29-03 2018. The John Patrick Albright Foundation is gratefully acknowledged.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix ASupplementary data
The following is the supplementary data related to this article:
References
- 1.Broers J.L., Ramaekers F.C., Bonne G., Yaou R.B., Hutchison C.J. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 2006;86(3):967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 2.Isermann P., Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013;23(24):R1113–1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruenbaum Y., Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 4.Arbustini E., Pilotto A., Repetto A. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J. Am. Coll. Cardiol. 2002;39(6):981–990. doi: 10.1016/s0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 5.van Berlo J.H., de Voogt W.G., van der Kooi A.J. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. (Berl) 2005;83(1):79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S., Baldinger S.H., Gandjbakhch E. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J. Am. Coll. Cardiol. 2016;68(21):2299–2307. doi: 10.1016/j.jacc.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 7.Broers J.L., Peeters E.A., Kuijpers H.J. Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 2004;13(21):2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 8.Houben F., Ramaekers F.C., Snoeckx L.H., Broers J.L. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim. Biophys. Acta. 2007;1773(5):675–686. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Tamiello C., Halder M., Kamps M.A., Baaijens F.P., Broers J.L., Bouten C.V. Cellular strain avoidance is mediated by a functional actin cap - observations in an. J. Cell Sci. 2017;130(4):779–790. doi: 10.1242/jcs.184838. [DOI] [PubMed] [Google Scholar]
- 10.Machowska M., Piekarowicz K., Rzepecki R. Regulation of lamin properties and functions: does phosphorylation do it all? Open Biol. 2015;5(11) doi: 10.1098/rsob.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardi M.L., Lammerding J. Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 2011;39(6):1729–1734. doi: 10.1042/BST20110686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Captur G., Arbustini E., Bonne G. Lamin and the heart. Heart. 2018;104(6):468–479. doi: 10.1136/heartjnl-2017-312338. [DOI] [PubMed] [Google Scholar]
- 13.Pecorari I., Borin D., Sbaizero O. A perspective on the experimental techniques for studying lamins. Cells. 2017;6(4) doi: 10.3390/cells6040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurini E., Martinelli V., Lanzicher T. Biomechanical defects and rescue of cardiomyocytes expressing pathologic nuclear lamins. Cardiovasc. Res. 2018;114(6):846–857. doi: 10.1093/cvr/cvy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mewborn S.K., Puckelwartz M.J., Abuisneineh F. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sébillon P., Bouchier C., Bidot L.D. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J. Med. Genet. 2003;40(8):560–567. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrot A., Hussein S., Ruppert V. Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy. Basic Res. Cardiol. 2009;104(1):90–99. doi: 10.1007/s00395-008-0748-6. [DOI] [PubMed] [Google Scholar]
- 18.Sylvius N., Bilinska Z.T., Veinot J.P. In vivo and in vitro examination of the functional significances of novel lamin gene mutations in heart failure patients. J. Med. Genet. 2005;42(8):639–647. doi: 10.1136/jmg.2004.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwerger M., Roschitzki-Voser H., Zbinden R. Altering lamina assembly reveals lamina-dependent and -independent functions for A-type lamins. J. Cell Sci. 2015;128(19):3607–3620. doi: 10.1242/jcs.171843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanzicher T., Martinelli V., Long C.S. AFM single-cell force spectroscopy links altered nuclear and cytoskeletal mechanics to defective cell adhesion in cardiac myocytes with a nuclear lamin mutation. Nucleus. 2015;6(5):394–407. doi: 10.1080/19491034.2015.1084453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzicher T., Martinelli V., Puzzi L. The cardiomyopathy lamin A/C D192G mutation disrupts whole-cell biomechanics in cardiomyocytes as measured by atomic force microscopy loading-unloading curve analysis. Sci. Rep. 2015;5:13388. doi: 10.1038/srep13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suki B. Assessing the functional mechanical properties of bioengineered organs with emphasis on the lung. J. Cell. Physiol. 2014;229(9):1134–1140. doi: 10.1002/jcp.24600. [DOI] [PubMed] [Google Scholar]
- 23.Brennan M., Palaniswami M., Kamen P. Do existing measures of Poincaré plot geometry reflect nonlinear features of heart rate variability? IEEE Trans. Biomed. Eng. 2001;48(11):1342–1347. doi: 10.1109/10.959330. [DOI] [PubMed] [Google Scholar]
- 24.Lerma C., Infante O., Pérez-Grovas H., José M.V. Poincaré plot indexes of heart rate variability capture dynamic adaptations after haemodialysis in chronic renal failure patients. Clin. Physiol. Funct. Imaging. 2003;23(2):72–80. doi: 10.1046/j.1475-097x.2003.00466.x. [DOI] [PubMed] [Google Scholar]
- 25.Kostin S., Dammer S., Hein S., Klovekorn W.P., Bauer E.P., Schaper J. Connexin 43 expression and distribution in compensated and decompensated cardiac hypertrophy in patients with aortic stenosis. Cardiovasc. Res. 2004;62(2):426–436. doi: 10.1016/j.cardiores.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Severs N.J., Bruce A.F., Dupont E., Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res. 2008;80(1):9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Severs N.J., Coppen S.R., Dupont E., Yeh H.I., Ko Y.S., Matsushita T. Gap junction alterations in human cardiac disease. Cardiovasc. Res. 2004;62(2):368–377. doi: 10.1016/j.cardiores.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Martinelli V., Cellot G., Fabbro A., Bosi S., Mestroni L., Ballerini L. Improving cardiac myocytes performance by carbon nanotubes platforms. Front. Physiol. 2013;4:239. doi: 10.3389/fphys.2013.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neher E. The use of fura-2 for estimating Ca buffers and Ca fluxes. Neuropharmacology. 1995;34(11):1423–1442. doi: 10.1016/0028-3908(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 30.Huebsch N., Loskill P., Mandegar M.A. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. C Methods. 2015;21(5):467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun N., Yazawa M., Liu J. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012;4(130):130ra147. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J., Sun N., Bruce M.A., Wu J.C., Butte M.J. Atomic force mechanobiology of pluripotent stem cell-derived cardiomyocytes. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang W.T., Yu D., Lai Y.C., Lin K.Y., Liau I. Characterization of the mechanodynamic response of cardiomyocytes with atomic force microscopy. Anal. Chem. 2013;85(3):1395–1400. doi: 10.1021/ac3022532. [DOI] [PubMed] [Google Scholar]
- 34.Kiviaho A.L., Ahola A., Larsson K. Distinct electrophysiological and mechanical beating phenotypes of long QT syndrome type 1-specific cardiomyocytes carrying different mutations. Int. J. Cardiol. Heart Vasc. 2015;8:19–31. doi: 10.1016/j.ijcha.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suadicani S.O., Flores C.E., Urban-Maldonado M., Beelitz M., Scemes E. Gap junction channels coordinate the propagation of intercellular Ca2+ signals generated by P2Y receptor activation. Glia. 2004;48(3):217–229. doi: 10.1002/glia.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ai Z., Fischer A., Spray D.C., Brown A.M., Fishman G.I. Wnt-1 regulation of connexin43 in cardiac myocytes. J. Clin. Investig. 2000;105(2):161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mounkes L.C., Kozlov S.V., Rottman J.N., Stewart C.L. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum. Mol. Genet. 2005;14(15):2167–2180. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- 38.Shaw R.M., Fay A.J., Puthenveedu M.A., von Zastrow M., Jan Y.N., Jan L.Y. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128(3):547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth J.W., Hong T.T., Gao D. Limited forward trafficking of connexin 43 reduces cell-cell coupling in stressed human and mouse myocardium. J. Clin. Investig. 2010;120(1):266–279. doi: 10.1172/JCI39740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macquart C., Jüttner R., Le Dour C. Microtubule cytoskeleton regulates connexin 43 localization and cardiac conduction in cardiomyopathy caused by mutation in A-type lamins gene. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy227. [DOI] [PubMed] [Google Scholar]
- 41.Tariq Z., Zhang H., Chia-Liu A. Lamin A and microtubules collaborate to maintain nuclear morphology. Nucleus. 2017;8(4):433–446. doi: 10.1080/19491034.2017.1320460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster D.R., Patrick D.L. Beating rate of isolated neonatal cardiomyocytes is regulated by the stable microtubule subset. Am. J. Physiol. Heart Circ. Physiol. 2000;278(5):H1653–1661. doi: 10.1152/ajpheart.2000.278.5.H1653. [DOI] [PubMed] [Google Scholar]
- 43.Webster D.R. Microtubules in cardiac toxicity and disease. Cardiovasc. Toxicol. 2002;2(2):75–89. doi: 10.1385/ct:2:2:075. [DOI] [PubMed] [Google Scholar]
- 44.Lampidis T.J., Trevorrow K.W., Rubin R.W. Effects of colchicine on cardiac cell function indicate possible role for membrane surface tubulin. Exp. Cell Res. 1986;164(2):463–470. doi: 10.1016/0014-4827(86)90044-3. [DOI] [PubMed] [Google Scholar]
- 45.Lampidis T.J., Kolonias D., Savaraj N., Rubin R.W. Cardiostimulatory and antiarrhythmic activity of tubulin-binding agents. Proc. Natl. Acad. Sci. U. S. A. 1992;89(4):1256–1260. doi: 10.1073/pnas.89.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bracken M.B. Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 2009;102(3):120–122. doi: 10.1258/jrsm.2008.08k033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long C.S., Kariya K., Karns L., Simpson P.C. Sympathetic modulation of the cardiac myocyte phenotype: studies with a cell-culture model of myocardial hypertrophy. Basic Res. Cardiol. 1992;87(Suppl 2):19–31. doi: 10.1007/978-3-642-72477-0_3. [DOI] [PubMed] [Google Scholar]
- 48.Deng X.F., Rokosh D.G., Simpson P.C. Autonomous and growth factor-induced hypertrophy in cultured neonatal mouse cardiac myocytes. Comparison with rat. Circ. Res. 2000;87(9):781–788. doi: 10.1161/01.res.87.9.781. [DOI] [PubMed] [Google Scholar]
- 49.Martinelli V., Cellot G., Toma F.M. Carbon nanotubes promote growth and spontaneous electrical activity in cultured cardiac myocytes. Nano Lett. 2012;12(4):1831–1838. doi: 10.1021/nl204064s. [DOI] [PubMed] [Google Scholar]
- 50.Chen S.N., Gurha P., Lombardi R., Ruggiero A., Willerson J.T., Marian A.J. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ. Res. 2014;114(3):454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borin D., Pecorari I., Pena B., Sbaizero O. Novel insights into cardiomyocytes provided by atomic force microscopy. Semin. Cell Dev. Biol. 2018;73:4–12. doi: 10.1016/j.semcdb.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Domke J., Parak W.J., George M., Gaub H.E., Radmacher M. Mapping the mechanical pulse of single cardiomyocytes with the atomic force microscope. Eur. Biophys. J. 1999;28(3):179–186. doi: 10.1007/s002490050198. [DOI] [PubMed] [Google Scholar]
- 53.Tulppo M.P., Mäkikallio T.H., Takala T.E., Seppänen T., Huikuri H.V. Quantitative beat-to-beat analysis of heart rate dynamics during exercise. Am. J. Physiol. 1996;271(1 Pt 2):H244–252. doi: 10.1152/ajpheart.1996.271.1.H244. [DOI] [PubMed] [Google Scholar]
- 54.Tulppo M.P., Mäkikallio T.H., Seppänen T., Airaksinen J.K., Huikuri H.V. Heart rate dynamics during accentuated sympathovagal interaction. Am. J. Physiol. 1998;274(3 Pt 2):H810–816. doi: 10.1152/ajpheart.1998.274.3.H810. [DOI] [PubMed] [Google Scholar]
- 55.Karmakar C.K., Khandoker A.H., Gubbi J., Palaniswami M. Complex correlation measure: a novel descriptor for Poincaré plot. Biomed. Eng. Online. 2009;8:17. doi: 10.1186/1475-925X-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guzik P., Piskorski J., Krauze T., Wykretowicz A., Wysocki H. Heart rate asymmetry by Poincaré plots of RR intervals. Biomed. Tech. (Berl) 2006;51(4):272–275. doi: 10.1515/BMT.2006.054. [DOI] [PubMed] [Google Scholar]
- 57.Shapiro L.G., Haralick R.M. Decomposition of two-dimensional shapes by graph-theoretic clustering. IEEE Trans. Pattern Anal. Mach. Intell. 1979;1(1):10–20. doi: 10.1109/tpami.1979.4766871. [DOI] [PubMed] [Google Scholar]
- 58.Haralick R.M., Sternberg S.R., Zhuang X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987;9(4):532–550. doi: 10.1109/tpami.1987.4767941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.