Highlights
-
•
A novel pipeline for Parkinson`s disease structure segmentation is presented.
-
•
State-of-the-art fast multiatlas patch-based label fusion with systematic error correction is used to accurately and efficiently produce very competitive results in around 5 min.
-
•
The proposed pipeline works at high resolution (0.5 mm) but it can work also with standard resolution (1 mm) T2 images allowing the analysis of large legacy databases.
-
•
The proposed pipeline will be made publically available online through our volBrain platform.
Abstract
Parkinson is a very prevalent neurodegenerative disease impacting the life of millions of people worldwide. Although its cause remains unknown, its functional and structural analysis is fundamental to advance in the search of a cure or symptomatic treatment. The automatic segmentation of deep brain structures related to Parkinson`s disease could be beneficial for the follow up and treatment planning. Unfortunately, there is not broadly available segmentation software to automatically measure Parkinson related structures. In this paper, we present a novel pipeline to segment three deep brain structures related to Parkinson's disease (substantia nigra, subthalamic nucleus and red nucleus). The proposed method is based on the multi-atlas label fusion technology that works on standard and high-resolution T2-weighted images. The proposed method also includes as post-processing a new neural network-based error correction step to minimize systematic segmentation errors. The proposed method has been compared to other state-of-the-art methods showing competitive results in terms of accuracy and execution time.
1. Introduction
Parkinson's disease (PD) is the second most common neurodegenerative pathology after Alzheimer's disease. It is a slow progressive disease that affects about 0.3% of the total population and 1% in people over 60 years old, presenting about 100,000 new cases every year (Martinez-Fernandez et al., 2016).
The cause that triggers the disease is still unknown, but it is characterized mainly by the loss of dopaminergic neurons in the Substantia Nigra Pars Compacta (SNpc), causing mostly motor symptoms, although neuron losses have also been found in other areas of the brain. There are several hypotheses about the cause of the death of these neurons, but none of them have been proven, which explains why a cure has not yet been found (Kalia and Lang, 2015).
The disease is the cause of various symptoms, mostly motor such as bradykinesia, muscle stiffness and tremors, commonly known and caused by the loss of dopaminergic innervation. However, it also causes some non-motor symptoms such as cognitive and sleep problems, pain, fatigue and depression that, despite not being so well known, reduce in the same way or even more the quality of life of patients suffering from this disease (Williams-Gray and Worth, 2016).
Currently, there is no treatment that can cure or even slowdown the disease and current treatments are focused in the control of the symptoms (Williams-Gray and Worth, 2016). A typical example is the use of levodopa to alleviate motor symptoms. However, it has been seen that the adverse effects of the medications used can affect the quality of life of patients, so long-term treatments must be controlled. Another very common treatment for Parkinson's symptoms, especially for tremor, is neurostimulation or deep brain stimulation of the subthalamic nucleus due to its metabolic and electrophysiological activity. It is an alternative option when the patient does not respond to levodopa or simply to reduce the dose of the medication. It is a fairly widespread technique, but with a small risk in neurostimulatory implantation surgery (Guridi et al., 2009). Finally, a newer alternative therapy is the development of high intensity focal ultrasound (HIFU) guided by MRI, a non-invasive method that produces lesions in the subthalamic nucleus that ends with tremor (Martinez-Fernandez et al., 2016).
In this paper, we propose a novel method to segment three deep brain structures related with Parkinson's disease (substantia nigra (SN), subthalamic nucleus (STN) and red nucleus (RN)). As already commented, the substantia nigra plays a major role in the disease due to the degeneration of dopaminergic cells. On the other hand, subthalamic nucleus which is located next to SN is also affected as this structure is highly connected to SN and is part of the basal ganglia circuit controlling motor actions (Parent and Hazrati, 1995). In fact, in patients with Parkinson's, metabolic and electrophysiological hyperactivity is found in STN (Guridi et al., 2009). It is believed to be caused by the imbalance in neurotransmission that would cause the loss of neurons in the SN, although various theories have been considered. That is why an injury in this area would reduce this activity to normal levels, which makes the STN a therapeutic target. Finally, the red nucleus is a structure located also close to the SN and is also related to the progression of the disease since it can have a compensatory role as long as the disease progresses (Philippens et al., 2018).
During the last two decades, structures involved in Parkinson's disease have been studied for their special interest for early and objective diagnosis. That is why various automatic segmentation techniques have been developed in order to extract the volumes of these structures for diagnosis, as well as their location for a better treatment. Early methods for the automatic segmentation of structures involved in Parkinson`s disease were based on anatomical atlases, which consist of registering the case to segment to a template (atlas). In (Chakravarty et al., 2006) T1w images were used so the proposed method was limited to segment easily identifiable structures in this modality. However, soon it was found that using T2w or T2*w images are a better choice since structures involved in PD are more easily identifiable in those modalities (O'Gorman et al., 2011). Going a step forward, (Xiao et al., 2012) proposed to merge two modalities to further improve the results.
In (Haegelen et al., 2013) three segmentation methods to segment basal ganglia and structures involved in PD were compared. Two of these methods were based on non-linear registration of manually labeled templates. The first is the Automatic Nonlinear Image Matching and Anatomical Labeling (ANIMAL) (Collins et al., 1995), the second is based on the Advanced Normalization Tools (ANTS) (Avants et al., 2008) registration package and the third is a patch-based label fusion technique (Coupé et al., 2011), which despite using only an affine registration demonstrated very good performance. In 2014, (Xiao et al., 2014) proposed a multiatlas method where multiple manually labeled templates were non-linearly registered to the case under study using ANTS and the multiple labels were combined using a majority voting rule. A year later, they proposed other method based on double-contrast patches which used multivariable cross-correlation and label fusion to segment the SN, the STN and RN (Xiao et al., 2015).
Recently, a Multimodal Image Segmentation Tool (MIST) method was proposed (Visser et al., 2016), which segments the three structures involved in PD. This method used MR images acquired at a 7T MR machine to calculate their QSM (Quantitative Susceptibility Mapping). This method detects the edges of the structures according to their intensity and shape profiles so it only requires a single reference mesh and a set of unlabeled training volumes, with which it learns in an unsupervised way. Unfortunately, 7T machines are not widely available which limits its applicability in common research and clinical settings.
In this paper, we propose a fast multiatlas patch-based label fusion segmentation method. Our method uses OPAL (Giraud et al., 2016) to produces fast and accurate segmentations. The proposed method is based on high resolution (0.5 × 0.5 × 0.5 mm3) T2w images but it also works on standard resolution T2w images (1 mm3) after a superresolution step (Manjón et al., 2010a; Coupe et al., 2013). The proposed method also includes a systematic error corrector method earlier introduced in (Romero et al., 2017) based on the use of a boosted ensemble of neural networks which reduces segmentation errors at post-processing.
2. Material and methods
2.1. Image data
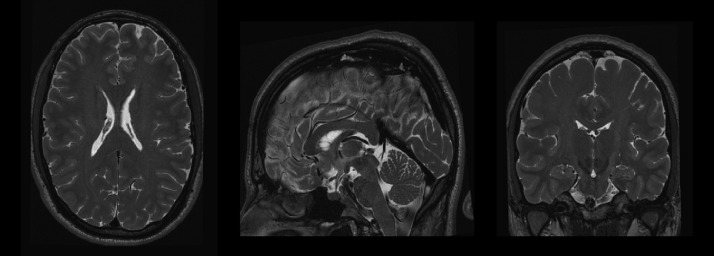
In this work, we have used a high-resolution MR dataset to develop the proposed method and to evaluate the results. This dataset contains 5 MR subjects with 0.3 × 0.3 × 0.3 mm3 high resolution T2-weighted images obtained by 2x interpolation of 0.6 × 0.6 × 0.6 mm3 acquisitions. The HR images are publicly available at the CoBrALab website (http://cobralab.ca/atlases). These MR images were taken from 5 healthy volunteers (2 males, 3 females, aged 29–57). High-resolution T2-weighted images were acquired using the 3D fast spin echo acquisition, FSE-CUBE (TE/TR=95.3 ms/2500 ms, ETL=100 ms, 2NEX, and isotropic resolution of 0.6 mm3). Reconstruction filters, ZIPX2 and ZIP512, were also used resulting in a final isotropic 0.3 mm3 dimension voxels. For more details see the original paper (Winterburn et al., 2013). This dataset was extended with 10 more cases using the same acquisition protocol and resolution kindly provided by the Dr. Chakravarty (See Fig. 1 for an example of one of these images).
Fig. 1.
Example of a HR T2 case from the Winterburn dataset.
2.2. Proposed method
Our proposed approach is a multiatlas segmentation method that uses a library of manually labeled atlases to perform the segmentation process.
2.2.1. Library construction
The images used to build the system library were preprocessed to locate them in a common intensity and coordinate space. To this end, we applied the following steps:
-
1
Registration: Affine registration to the Montreal Neurological Institute (MNI152) space using the Advanced Normalization Tools (ANTs) (Avants et al., 2008). This registration was estimated using a 0.5 × 0.5 × 0.5 mm3 resolution T2w MNI152 template when processing high resolution images and using the standard 1 mm3 resolution MNI152 template when dealing with standard resolution images. As we will describe later, the segmentation is always performed at 0.5 × 0.5 × 0.5 mm3 resolution.
-
2
Inhomogeneity correction: Intensity inhomogeneity correction is performed using the N4 bias field correction method (Tustison et al., 2010).
-
3
Intensity standardization: The images were intensity normalized so brain tissues have similar intensity levels across all the subjects of the library. For this purpose, we applied a histogram matching method (Nyúl and Udupa, 1999).
-
4
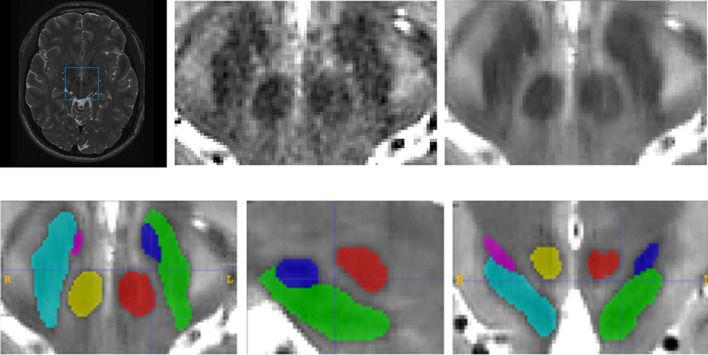
Cropping: To reduce the memory requirements and the computational cost, the image was cropped around the area of interest. For this purpose, a bounding box surrounding the different structures of interest was calculated (using a margin of 5 voxels in each direction) in the MNI152 space from the manual segmentations to ensure that all the manual segmentations were included in this bounding box (see Fig. 2).
-
5
Denoising: A denoising step using the PRI-NLPCA Filter (Manjón et al., 2015) is also done. We perform the denoising over the cropped region for efficiency reasons because applying it in the original volume would be too time consuming and totally unnecessary to analyze a small portion of the volume.
-
6
Manual labeling: The 6 structures of interest (we assigned different labels for left and right structures) were manually segmented in the 15 library cases using ITK-SNAP software (Yushkevich et al., 2006). Taking benefit from the symmetric properties of the brain, the library size was duplicated by including the axially mirrored cases which lead to a total library size of 30 cases. All the images were manually segmented by a trained person (AB) and supervised by an expert in brain anatomy (EL) (see Fig. 2).
-
7
Non-linear registration: A non-linear deformation was estimated between the cropped region of every subject and a reference cropped MNI152 template. The non-linear deformation was estimated using the Advanced Normalization Tools (ANTs) (Avants et al., 2008) using cross correlation metric and pyramidal framework at 8 × 4 × 2 × 1x scales and 200, 200, 200 and 0 iterations at each scale. For the library construction, the transformations were estimated and stored but no transformation was applied to the library cases since this operation is only done during the segmentation process as we will describe later.
Fig. 2.
Upper row shows an example of the cropping operation and the following denoising. Bottom row shows an example of the result of the manual labeling process.
2.2.2. New cases preprocessing
The proposed method can be applied to both HR data and standard resolution data. Depending on this, the preprocessing pipeline is slightly different. When dealing with HR data the preprocessing steps were the same than for the library construction (steps 1 to 5). However, when standard resolution the preprocessing pipeline is the following:
-
1
Denoising: We used the Spatially Adaptive Non-local Means Filter (Manjón et al., 2010b).
-
2
Registration: Affine registration to the Montreal Neurological Institute (MNI152) space using the Advanced Normalization Tools (ANTs) (Avants et al., 2008). This registration was estimated using as reference the standard 1 mm3 resolution T2w MNI152 template.
-
3
Inhomogeneity correction: Intensity inhomogeneity correction using the N4 bias field correction (Tustison et al., 2010).
-
4
Intensity standardization: The images were intensity normalized using a histogram matching method (Nyúl and Udupa, 1999).
-
5
Cropping: A standard resolution bounding box surrounding the different structures was applied to crop the region of interest at 1 mm3 resolution.
-
6
Superresolution: The cropped data must be upsampled to produce HR 0.5 × 0.5 × 0.5 mm3 resolution data. This is performed using a patch-based super-resolution technique called LASR (Coupé et al., 2013).
-
7
Non-linear registration: To achieve a better match between the different subjects' anatomy, a non-linear deformation was estimated between the cropped regions of every subject and the reference cropped MNI152 template. The non-linear deformation is estimated with the Advanced Normalization Tools (ANTs) (Avants et al., 2008) using cross correlation metric and pyramidal framework at 8 × 4 × 2 × 1x scales and 200, 200, 200 and 0 iterations at each scale. This transformation will be used to create a subject-specific library.
2.2.3. Online subject-specific library construction
In classical multi-atlas label fusion methods a common approach consists of non-linearly register all the manually labeled atlases to the new case to be segmented. However, this is a very time-consuming process, since it requires to estimate and apply N non-linear registrations (Wang et al., 2013). In our proposed method, we generate a subject specific library by concatenating the direct non-linear transformation of every library case (previously estimated) to the MNI152 cropped template with the inverse non-linear transformations of the case to be segmented to the same template. In this way, we move the entire library (N = 30) to the new case space estimating only a single non-linear transformation online as done in a previous work (Romero et al., 2017). This registration process introduces some blurring due to the interpolation used to apply the transformations. This has a negative impact in the segmentation step. For this reason, to enhance the images, we sharpened the images by adding the Laplacian of each image.
2.2.4. Labeling
Our segmentation method is based on the non-local patch-based label fusion technique (Coupe et al., 2011) where patches of the subject to be segmented are compared with patches of the training library to look for similar patterns within a defined search volume to assign the proper label v as can be seen in equation 1.
| (1) |
where Vi corresponds to the search area, N is the number of subjects in the template library, ys,j is a possible label from the voxel xs,j at the position j in the library subject s and w(xi, xs,j) is the patch similarity defined as:
| (2) |
| (3) |
where P(xi) is the patch centered at xi, P(xs,j) the patch centered at xj in the templates and ||.||2 is the normalized L2 norm (normalized by the number of elements) calculated from the distance between each pair of voxels from both patches P(xi) and P(xs,j). h is a normalization parameter that is estimated from the minimum of all patch distances within the search area.
However, exhaustive patch comparison process is very time consuming (even in reduced neighborhoods). To reduce the computational burden of this process, we have used a multiscale adaptation of the OPAL method (Giraud et al., 2016) previously proposed in (Romero et al., 2017) which takes benefit from the concept of Approximate Nearest Neighbor Fields (ANNF). We refer the interested reader to the original paper for more details.
2.2.5. Systematic error correction
Any segmentation method is subject to both random and systematic errors. The first error type can be typically minimized by using bootstrapped estimations. Luckily, non-local label fusion technique estimates the voxel label averaging the votes of many patches which naturally reduces the random classification error. Unfortunately, systematic errors cannot be reduced using this strategy since they are not random. However, this systematic bias can be learned and later use this knowledge to correct the segmentation output (Wang et al., 2011).
In (Romero et al., 2017), we proposed an error corrector method based on a patch-based ensemble of neural networks (PEC for Patch-based Ensemble Corrector) to increase the segmentation accuracy by reducing the systematic errors. Specifically, the neural network ensemble is trained with image patches of sizes 3 × 3 × 3 voxels (fully sampled) and 7 × 7 × 7 voxels (subsampled by skipping two voxels at each dimension) from T2w images, the automatic segmentations, a distance map value, and their x, y and z coordinates in MNI152 space. The distance map we used is calculated for the whole structure as the distance in voxels to the structure contour. This results in a feature vector of 112 features that are mapped to a patch of manual segmentations of size 3 × 3 × 3 voxels. We used a multilayer perceptron with two hidden layers of size 83 and 55 neurons resulting in a network with a topology of 112 × 83 × 55 × 27 neurons. An ensemble of 10 neural networks was trained using a boosting strategy. Each new network was trained with a different subset of data which was selected by giving a higher probability of appearance to the samples that were misclassified in the previous ensemble.
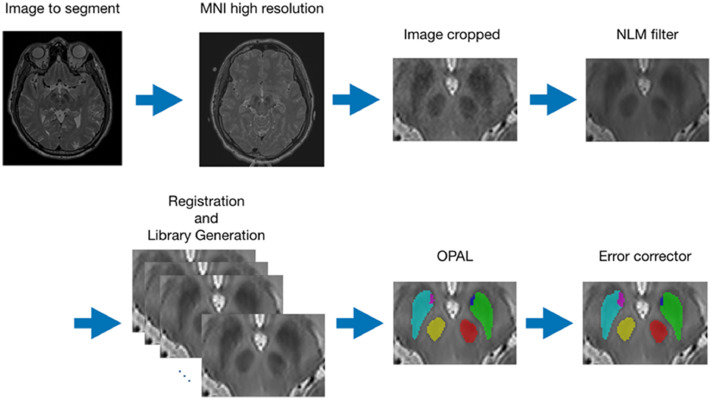
We have named the proposed pipeline as pBrain. In Fig. 3, an outline of the proposed pipeline is presented.
Fig. 3.
Summary of the HR pBrain segmentation pipeline. First, the HR image is registered to HR MNI152 space, inhomogeneity corrected and intensity normalized. Later, the image is cropped and denoised and the case-specific library is constructed. Finally, the OPAL method is used to produce an initial segmentation which is later refined using PEC method.
3. Experiments and results
In this section, the parameters of the proposed method and its results are presented. The method parameters have been adjusted using a leave-two out cross validation strategy (removing the case being evaluated and its mirrored version). To evaluate the segmentation accuracy, we have used the DICE coefficient (Zijdenbos et al., 1994) measured in the MNI152 space. For the multiscale patch similarity we used patch sizes of 5 × 5 × 5 and 11 × 11 × 11 voxels, for each scale respectively and a mixing coefficient of 0.5. In OPAL, we used 64 independent Patch Matches with 4 iterations each.
3.1. High resolution results
In Table 1 we show the results of the proposed method for each structure and the overall performance. As can be noted, left and right results are consistent for each structure (being the STN the less stable probably due to its small size).
Table 1.
DICE results for each structure and overall result without using Patch-based Ensemble Corrector.
| RN left | RN right | SN left | SN right | STN left | STN right | Avg. |
|---|---|---|---|---|---|---|
| 0.9349 | 0.9340 | 0.8822 | 0.8840 | 0.8299 | 0.8349 | 0.8833 |
To evaluate the impact of the proposed error corrector, we performed a 3-fold validation. The 30 library cases were split in 3 subsets of 10 cases. Two sets (20 cases) were used to train the error corrector and the remaining set was used to test the error corrector. This operation was repeated 3 times. Therefore, the 30 cases were used to estimate the results of the whole dataset. For each training set the automatic segmentations were produced using only a library of 18 cases from this training set. Results are summarized in Table 2.
Table 2.
DICE results for each structure and overall result using Patch-based Ensemble Corrector.
| RN left | RN right | SN left | SN right | STN left | STN right | Avg. |
|---|---|---|---|---|---|---|
| 0.9489 | 0.9454 | 0.8996 | 0.9014 | 0.8563 | 0.8552 | 0.9012 |
As can be noted, the error corrector systematically improved the results of each structure and the overall performance.
3.2. Standard resolution results
The proposed method works optimally with HR MR images but these sequences are not always available in research or in clinical environments. Therefore, it would be desirable to be able to analyze legacy data at standard resolution. For this reason, we have evaluated the method over standard resolution (1 mm3) images upsampled to 0.5 × 0.5 × 0.5 mm3 using a super-resolution technique called LASR (Coupé et al., 2013). To do this, we reduced the resolution of the HR images by a factor 2 and later upsampled them using the SR method.
Table 3 shows the results of this experiment using the proposed method without PEC. As can be noticed, although the accuracy degrades from 0.88 to 0.86, pBrain can still produce competitive results when using standard resolution images.
Table 3.
DICE results for each structure and overall result on standard resolution data without using Patch-based Ensemble Corrector.
| RN left | RN right | SN left | SN right | STN left | STN right | Avg. |
|---|---|---|---|---|---|---|
| 0.9203 | 0.9214 | 0.8530 | 0.8616 | 0.8003 | 0.8047 | 0.8602 |
To evaluate the impact of the proposed error corrector on upsampled data, we performed a 3-fold validation but this time using super-resolved data instead of HR data. Results are summarized in Table 4. In this case, the improvement of the results was even bigger than in the HR data case with an overall improvement of 3% compared to the 2% obtained with the HR data. It is interesting to note that although SR images are not comparable to HR data, the use of PEC helped to increase the accuracy to almost reach the HR results (0.89 vs 0.90). This important result shows that the proposed framework can efficiently process common 1 mm3 MR data.
Table 4.
DICE results for each structure and overall result on standard resolution data using Patch-based Ensemble Corrector.
| RN left | RN right | SN left | SN right | STN left | STN right | Avg. |
|---|---|---|---|---|---|---|
| 0.9437 | 0.9400 | 0.8916 | 0.8957 | 0.8347 | 0.8452 | 0.8918 |
3.3. Method's meta-analysis
A direct comparison of the proposed method with related methods is not possible due to the use of different datasets. Therefore, we performed a method meta-analysis to locate then proposed method in its approximated context. We analyzed the proposed method pBrain with other related methods (both LR and HR results are provided). Specifically, we analyzed the methods proposed in (Xiao et al., 2012), three variants proposed by (Haegelen et al., 2013), another method from (Xiao et al., 2014), two variants of the proposed method in (Xiao et al., 2015) and finally the MIST method (Visser et al., 2016). We used published values provided by the authors in the corresponding papers. This analysis is only indicative since each method was evaluated in different conditions with different datasets and labeling protocols. However, we think it is important to provide this information to roughly locate the proposed method in its corresponding context. Table 5 shows the DICE index of the different methods included in the analysis.
Table 5.
DICE results for each structure and overall result for the different compared methods.
| Method | Mod. | RN left | RN right | SN left | SN right | STN left | STN right | Avg |
|---|---|---|---|---|---|---|---|---|
| Xiao2012 | T1+T2* | 0.830 | 0.790 | 0.560 | 0.600 | 0.710 | 0.670 | 0.690 |
| Haegelen2013 ANIMAL | T1 | 0.796 | 0.793 | 0.689 | 0.657 | 0.641 | 0.640 | 0.703 |
| Haegelen2013 ANTS | T1 | 0.781 | 0.782 | 0.671 | 0.640 | 0.626 | 0.640 | 0.690 |
| Haegelen2013 Patch-based | T2 | 0.790 | 0.783 | 0.696 | 0.653 | 0.631 | 0.575 | 0.688 |
| Xiao2014 | T1+T2 | 0.900 | 0.890 | 0.740 | 0.770 | 0.800 | 0.810 | 0.820 |
| Xiao2015 Single | T2* | 0.912 | 0.914 | 0.845 | 0.830 | 0.906 | 0.910 | 0.886 |
| Xiao2015 Double | T1+T2* | 0.922 | 0.929 | 0.877 | 0.900 | 0.932 | 0.947 | 0.918 |
| Visser2016 | QSM | 0.857 | 0.870 | 0.737 | 0.727 | 0.580 | 0.597 | 0.728 |
| pBrain (LR) | T2 | 0.944 | 0.940 | 0.892 | 0.896 | 0.835 | 0.845 | 0.892 |
| pBrain (HR) | T2 | 0.949 | 0.945 | 0.900 | 0.901 | 0.856 | 0.855 | 0.901 |
As can be noted, Xiao2012, Haegelen2013 and Visser2016 methods had a low performance (below 73%) compared with the rest of the methods. Xiao2014 results and specially Xiao2015 showed a much higher accuracy being the double contrast version (T1 + T2*) the best performing method overall. The proposed method had the second (HR) and third position (LR) and the first position when comparing with similar single contrast methods (0.901 vs 0.886). For specific structures, the proposed method was the best performing for RN and SN among the methods but not for STN which is the smaller and more complex structure to segment. Again, we want to highlight the fact that this ordering is just for orientation purposes since our only aim is to remark that the proposed method is well located among best performing methods in bibliography.
Regarding to the execution time, the whole HR pBrain pipeline takes less than 5 min (this time was estimated in a computer running windows 10 with an AMD Ryzen Threadripper CPU at 3,4 GHz and 64 GB RAM). The method proposed in Xiao et al. (2015) has been reported to take 28 min (not including the preprocessing and the library creation). The rest of the compared methods didn't supply their processing times.
3.4. Clinical data validation
When evaluating a new pipeline one aspect that sometimes is not analyzed is how the proposed method generalizes to other data sources different from the training/test dataset. To evaluate this aspect, we automatically analyzed 7 clinical cases from the Hospital La Fe of Valencia (Spain). Written informed consent was obtained for the use of the data and the hospital institutional ethical board approved its retrospective use. This dataset was composed of 7 Parkinson patients (4 males, 3 females) with an age range of 55–75 years. They were acquired in Phillips Achieva 3T MR scanner using a HR 3D T2 Spin Eco sequence (matrix size was 320 × 320 × 575 voxels at a resolution of 0.78 × 0.78 × 0.4 mm3).
The images were automatically segmented using the proposed pBrain pipeline and the results were compared with manually annotated labels from our expert. The results can be seen in Table 6. As can be noticed, the results are similar to those obtained in our training library despite the fact that these images belong to patients with Parkinson while the library was made exclusively with healthy subjects. This suggests that the proposed pipeline generalizes well to other data sources (including at least some current clinical acquisition protocols).
Table 6.
DICE results for each structure and overall result on the clinical dataset composed by PD patients.
| RN left | RN right | SN left | SN right | STN left | STN right | Avg. |
|---|---|---|---|---|---|---|
| 0.930 | 0.925 | 0.908 | 0.915 | 0.872 | 0.878 | 0.905 |
4. Discussion
In this paper we have presented a novel pipeline for Parkinson's disease related structure segmentation. The proposed pipeline is able to work with both HR and standard resolution MR T2 images. It is based on a state of the art multiatlas label fusion segmentation framework that uses a library of manually annotated templates to segment the substantia nigra, the red nucleus and the subthalamic nucleus.
We have compared our proposed pipeline with related methods previously proposed in bibliography. Although a direct comparison is not possible due to the use of different labeling protocols and datasets (and field strengths) we showed that the proposed pipeline produces results on pair and many times improving the results of related methods.
We believe that one of the reasons for the improved performance of the proposed framework is the fact that it works at high resolution (0.5 × 0.5 × 0.5 mm3). We think this is fundamental when dealing with such small structures were a single voxel error represents a 1 mm3 volume error (in MNI space) while the same one voxel error at HR only represents a 0.125 mm3 error. Although it is true that HR images are not widely available in clinic, we have shown that standard resolution can be effectively upsampled to HR providing very good results.
Another important factor that explains our competitive results is the use of a systematic error correction method early proposed in (Romero et al., 2017) which uses an ensemble of patch-based neural networks (PEC). The use of PEC consistently improved the results for all the structures in both high and standard resolution cases having an execution time overload of just few seconds.
The proposed pipeline is also very efficient with mean running times around 5 min (around 6 times faster than a similar reported method).
Our results on a small clinical dataset of patients with Parkinson`s disease reveal that the proposed method apparently provides similar results to those obtained in the training dataset, despite the fact that this dataset is solely formed with healthy cases with different MR machines/parameters. This suggests that the proposed method can be readily applied in different research and clinical conditions, although a much larger validation will be required. Structural brain imaging has been shown to be very useful for the accurate diagnosis of Parkinson's disease (Heim et al., 2017). In addition, the neurodegenerative process underlying the disease is thought to start at least years before motor symptoms (Ziegler et al., 2013), and thus our imaging method can also be helpful for early detection of the disease. Although most imaging studies have focused on the SN, the volume of the STN has been found to decrease with progression of Parkinson's disease, whereas that of the RN increases (Colpan and Slavin, 2010), probably as a result of the compensatory role named above (Philippens et al., 2018). In conclusion, our automatic segmentation method to measure the volume of the SN, STN and RN can be useful for the early diagnosis of the disease, as well as for the research of the dynamics of the brain structural alterations originated by Parkinson's disease.
Evidently, the proposed pipeline is not restricted to its use in Parkinson`s disease as it can be used to analyze the three segmented structures in many other neurological conditions as long as to study the normal brain aging.
We plan to deploy the proposed pipeline within our volBrain online platform (volbrain.upv.es) to make it freely accessible to the scientific community. As far as we know, this will be the first publically available pipeline for Parkinson-related structure segmentation and we hope that it will help the researchers to better analyze their data in an easy to use yet accurate and efficient manner.
Author contributions
J.M., R.V. and P.C. conceived the pipeline and designed the experiments. A.B. and E.L. generated the manually labeled library. F.A. provided the clinical dataset. J.R. and A.B. conducted the experiments and develop the pipeline. All authors discussed the results and contributed to the final manuscript.
Declaration of Competing Interest
None.
Acknowledgements
The authors want to thank Dr. Mallar Chakravarty for making accessible the HR MRI data used in the proposed pipeline. This research was supported by the Spanish DPI2017-87743-R grant from the Ministerio de Economia, Industria y Competitividad of Spain. This work also benefited from the support of the project DeepVolBrain of the French National Research Agency (ANR-18-CE45-0013). This study was achieved within the context of the Laboratory of Excellence TRAIL ANR-10-LABX-57 for the BigDataBrain project. Moreover, we thank the Investments for the future Program IdEx Bordeaux (ANR-10-IDEX- 03–02, HL-MRI Project), Cluster of excellence CPU and the CNRS.
References
- Avants B., Epstein C., Grossman M., Gee J. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty M., Bertrand G., Hodge C.P., Sadikot A.F., Collins D.L. The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage. 2006;30:359–376. doi: 10.1016/j.neuroimage.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Collins D., Holmes C., Peters T., Evans A. Automatic 3D model based neuroanatomical segmentation. Hum. Brain Mapp. 1995;3(3):190–208. [Google Scholar]
- Colpan M.E., Slavin K.V. Subthalamic and red nucleus volumes in patients with parkinson’s disease: do they change with disease progression? Parkinsonism Relat. Disord. 2010;16(6):398–403. doi: 10.1016/j.parkreldis.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Coupé P., Manjón J.V., Fonov V., Pruessner J., Robles M., Collins D.L. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage. 2011;54(2):940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Coupé P., Manjón J.V., Chamberland M., Descoteaux M., Hiba B. Collaborative patch-based super-resolution for diffusion-weighted images. Neuroimage. 2013;83:245–261. doi: 10.1016/j.neuroimage.2013.06.030. [DOI] [PubMed] [Google Scholar]
- Giraud R., Ta V.T., Papadakis N., Manjón J.V., Collins D.L., Coupé P., Initiative A.D.N. An optimized patchmatch for multi-scale and multi-feature label fusion. Neuroimage. 2016;124:770–782. doi: 10.1016/j.neuroimage.2015.07.076. [DOI] [PubMed] [Google Scholar]
- Guridi J., Rodríguez-Oroz M., Clavero P., Manrique M. Revisión crítica de la estimulación subtalámica en la enfermedad de parkinson. Neurocirugia. 2009;20(6):521–532. [PubMed] [Google Scholar]
- Haegelen C., Coupé P., Fonov V., Guizard N., Jannin P., Morandi X., Collins D. Automated segmentation of basal ganglia and deep brain structures in MRI of parkinson’s disease. Int. J. Comput. Assist. Radiol. Surg. 2013;8(1):99–110. doi: 10.1007/s11548-012-0675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim B., Krismer F., De Marzi R., Seppi K. Magnetic resonance imaging for the diagnosis of parkinson’s disease. J. Neural. Transm. 2017;124(8):915–964. doi: 10.1007/s00702-017-1717-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia L.V., Lang A.E. Parkinson’s disease. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- Manjón J.V., Coupé P., Buades A., Fonov V., Collins D.L., Robles M. Non-Local MRI upsampling. Med. Image Anal. 2010;14(6):784–792. doi: 10.1016/j.media.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Manjón J.V., Coupé P., Martí-Bonmatí L., Collins D.L., Robles M. Adaptive non-local means denoising of MR images with spatially varying noise levels. J. Magn. Reson. Imaging. 2010;31:192–203. doi: 10.1002/jmri.22003. [DOI] [PubMed] [Google Scholar]
- Martinez-Fernandez R., Gasca Salas C., Sanchez Ferro A., Obeso J. Actualización en la enfermedad de parkinson. Rev. Med. Clin. Condes. 2016;27(3):363–379. [Google Scholar]
- Nyúl L.G., Udupa J.K. On standardizing the MR image intensity scale. Magn. Reson. Med. 1999;42(6):1072–1081. doi: 10.1002/(sici)1522-2594(199912)42:6<1072::aid-mrm11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- O’Gorman R., Shmueli K., Ashkan K., Samuel M., Lythgoe D., Shahidi- ani A., Jarosz J. Optimal MRI methods for direct stereotactic targeting of the subthalamic nucleus and globus pallidus. Eur. Radiol. 2011;21(1):130–136. doi: 10.1007/s00330-010-1885-5. [DOI] [PubMed] [Google Scholar]
- Philippens I.H., Wubben J.A., Franke S.K., Hofman S., Langermans J.A. Involvement of the red nucleus in the compensation of parkinsonism may explain why primates can develop stable parkinson’s disease. Nature Sci. Rep. 2018;9:880. doi: 10.1038/s41598-018-37381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J.E., Coupé P., Manjón J.V. HIPS: a new hippocampus subfield segmentation method. Neuroimage. 2017;163:286–295. doi: 10.1016/j.neuroimage.2017.09.049. [DOI] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser E., Keuken M., Forstmann B., Jenkinson M. Automated segmentation of the substantia nigra, subthalamic nucleus and red nucleus in 7 T data at young and old age. Neuroimage. 2016;139:324–336. doi: 10.1016/j.neuroimage.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Suh J.W., Das S.R., Pluta J.B., Craige C., Yushkevich P.A. Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35(3):611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Gray C.H., Worth P.F. Parkinson’s disease. Medicine (Baltimore) 2016;44(9):542–546. [Google Scholar]
- Winterburn J., Pruessner J., Chavez S., Schira M., Lobaugh N., Voineskos A., Chakravarty M. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage. 2013;74:254–265. doi: 10.1016/j.neuroimage.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Bailey L., Chakravarty M., Beriault S., Sadikot A., Pike G., Collins D. Atlas-based segmentation of the subthalamic nucleus, red nucleus, and substantia nigra for deep brain stimulation by incorporating multiple MRI contrasts. In: IPCAI 2012. Lect. Note Comp. Sci. 2012;7330:135–145. [Google Scholar]
- Xiao Y., Jannin P., D’Albis T., Guizard N., Haegelen C., Lalys F., Collins D. Investigation of morphometric variability of subthalamic nucleus, red nucleus, and substantia nigra in advanced parkinson’s disease patients using automatic segmentation and PCA-Based analysis. Hum. Brain Mapp. 2014;35(9):4330–4344. doi: 10.1002/hbm.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Fonov V.S., Beriault S., Gerard I., Sadikot A.F., Pike G.B., Collins D.L. Patch-based label fusion segmentation of brainstem structures with dual-contrast MRI for parkinson’s disease. Int. J. Comput. Assist. Radiol. Surg. 2015;10(7):1029–1041. doi: 10.1007/s11548-014-1119-4. [DOI] [PubMed] [Google Scholar]
- Yushkevich P., Piven J., Hazlett H.C., Smith R., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Ziegler D.A., Wonderlick J.S., Ashourian P., Hansen L.A., Young J.C., Murphy A.J., Koppuzha C.K., Growdon J.H., Corkin S. Substantia nigra volume loss before basal forebrain degeneration in early parkinson disease. JAMA Neurol. 2013;70(2):241–247. doi: 10.1001/jamaneurol.2013.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijdenbos A.P., Dawant B.M., Margolin R.A., Palme A.C. Morphometric analysis of white matter lesions in MR images: method and validation. IEE Trans. Med. Imaging. 1994;13:716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]