Summary
Distinguishing individuals from brain connectivity, and studying the genetic influences on that identification across different ages, improves our basic understanding of functional brain network organization. We applied support vector machine classifiers to two datasets of twins (adult, pediatric) and two datasets of repeat-scan individuals (adult, pediatric). Classifiers were trained on resting state functional connectivity magnetic resonance imaging (rs-fcMRI) data and used to predict individuals and co-twin pairs from independent data. The classifiers successfully identified individuals from a previous scan with 100% accuracy, even when scans were separated by months. In twin samples, classifier accuracy decreased as genetic similarity decreased. Our results demonstrate that classification is stable within individuals, similar within families, and contains similar representations of functional connections over a few decades of life. Moreover, the degree to which these patterns of connections predict siblings' data varied by genetic relatedness, suggesting that genetic influences on rs-fcMRI connectivity are established early in life.
Subject Areas: Biological Sciences, Neuroscience, Computational Bioinformatics
Graphical Abstract
Highlights
-
•
High-order functional brain networks play key role in functional fingerprints
-
•
Functional fingerprints are stable, even identifying pairs scanned months apart
-
•
Features of functional fingerprints are highly stable between children and adults
-
•
Classifier accuracy decreases as genetic similarity decreases
Biological Sciences; Neuroscience; Computational Bioinformatics
Introduction
One anticipated use of functional magnetic resonance imaging (fMRI) data is to identify reliable and clinically viable biomarkers for disease states. However, a more precise understanding of healthy functional brain network organization is necessary to achieve this goal. Resting state functional connectivity (rs-fcMRI) holds promise as a noninvasive method of reaching this goal, owing to its low behavioral demand, consistency within individuals, and stability over time (Poldrack et al., 2015). Furthermore, rs-fcMRI reveals inherent functional brain networks absent of a specific task state. These averaged networks are reliably observed across adult samples (Cohen et al., 2008, Power et al., 2011), and early functional network organization is readily identified early in life. By 6 months of age, early organizational patterns of common adult resting state networks such as the default mode network (DMN), somatosensory motor (SSM) network, and visual (VIS) network are identified (Mitra et al., 2017) and appear to become more adult-like in their organization and interaction during development (Gao et al., 2013, Lin et al., 2008).
Given the goal of identifying distinct biomarkers of disease, combined with the stability of rs-fcMRI brain networks, it is not surprising that a growing body of neuroimaging research has focused on defining functional “connectomes” or “fingerprints” of individuals based on their fMRI data (Byrge and Kennedy, 2019, Finn et al., 2015, Horien et al., 2019, Miranda-Dominguez et al., 2018, Miranda-Dominguez et al., 2014, Xu et al., 2016). A greater understanding of how functional connections become organized into reliable, functional fingerprints of an individual could give researchers and clinicians alike a quantifiable measure of individual change over time. In addition, a more precise understanding of the genetic influence on normative network organization throughout development may help pave the way for a more precise measure of the impact of environmental and disease variables on brain network organization. Extant research consistently reveals the presence of a functional fingerprint in rs-fcMRI data across multiple methodologies (Balsters et al., 2018, Finn et al., 2015, Haak et al., 2018, Kaufmann et al., 2017, Miranda-Dominguez et al., 2014). However, far less is known about the specific pattern of discrete functional connections that distinguish individuals from one another, specifically, whether these patterns differ at different ages or stages of development.
Studies using rs-fcMRI to identify individuals via functional fingerprint implicate the DMN and its connectivity to putative control networks, such as the cingulo-opercular (CO), dorsal attention, fronto-parietal (FP), and ventral attention networks, as the most highly distinctive between individuals (Finn et al., 2015, Horien et al., 2019, Miranda-Dominguez et al., 2018). Given previous evidence that the FP network coordinates other networks for task performance (Cole et al., 2013, Dixon et al., 2018, Marek and Dosenbach, 2018, Marek et al., 2015, Power et al., 2013, Zanto and Gazzaley, 2013), specific patterns of connectivity between the FP and other networks could be highly individualized during rest (Gordon et al., 2018), and the degree of network integration may be reflected in functional fingerprints. The network-integrating role of the FP network and the DMN's role in internal, self-focused mentation and mind-wandering (Buckner et al., 2008, Mason et al., 2007, Raichle, 2015) are presumably highly individualized and may explain their importance in the functional fingerprint, but this connection has not yet been explored directly. Despite the growing understanding of the functional networks involved in the makeup of a functional fingerprint, a consensus of specific functional connections that yield the most accurate identification of individuals has not yet been established.
Crucial to establishing a functional fingerprint is a better examination of the impact of individual variance in relation to expected functional network organization and the influence of heritable factors on rs-fcMRI. Gratton et al. (2018) recently revealed that averaged group differences in functional network organization are driven more by individual variability and common participant features, with only a modest influence of cognitive or task demands. This suggests that group-level results that do not explicitly account for individual differences in functional network organization may misattribute person-specific effects to group-level task performance. Similarly, evidence of distinct organizational variations from the expected group-level resting state functional network topography was found in a subset of the adult population (Gordon et al., 2017). Given that individual differences in functional network organization can be buried in group-averaged analyses, an individual variance framework is vital for accurately characterizing functional network organization across multiple samples.
Another factor to consider in characterizing a functional fingerprint is the genetic impact on rs-fcMRI network organization. Fu et al. (2015) observed a significant genetic influence on resting state organization across sensory, but not cognition-related, networks in a pediatric sample of twins (aged 12–19 years). The authors posited that the age of their sample and the protracted development of cognitive functions that require coordination across many networks might be why cognition-related networks showed a weaker genetic influence. Indeed, in genetic studies of adult twins, within-network connectivity of the DMN is found to be the most heritable (Glahn et al., 2010, Xu et al., 2017). Recent work has found evidence that an individual's functional fingerprint, calculated via a model-based approach to identifying a functional fingerprint called “connectotyping,” also shows a pattern of genetic influence in adults (Miranda-Dominguez et al., 2018). In this study, identification accuracy was highest in identical (monozygotic) twins, followed by fraternal (dizygotic) twins, with non-twin siblings showing the lowest accuracy. However, analyses that include larger and more age-diverse twin samples are necessary to identify specific patterns and developmental trajectories of highly heritable and individually unique functional connections within resting state data. The inclusion of genetically informative, age-varying neuroimaging datasets is important to gain a complete understanding of the genetic influence on individually unique rs-fcMRI connections.
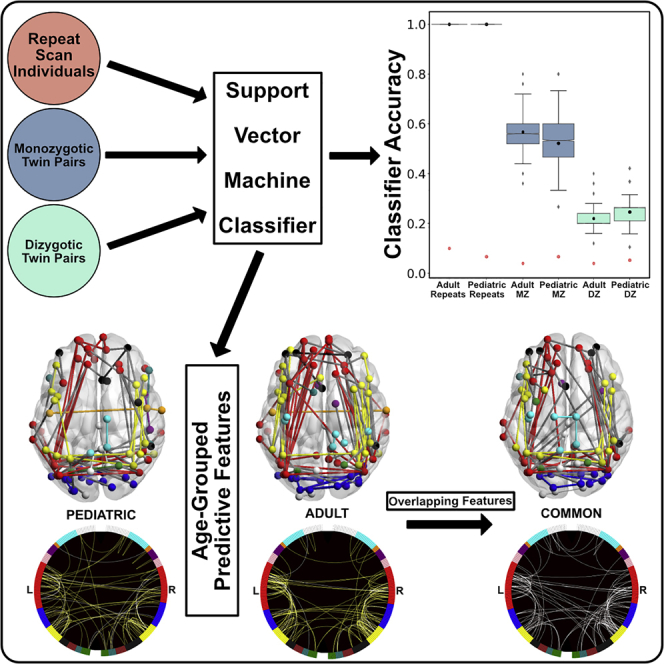
Based on mounting evidence from functional fingerprint research, we hypothesized that adapting resting state time courses to use ANOVA feature selection would benefit our analyses. Current resting state approaches often use all available features from a chosen region of interest (ROI) set, thus providing too many features that can overpower the signal with noise, or apply feature reduction methods that may be more appropriate for group membership classification (Dosenbach et al., 2010, Greene et al., 2016). This shift in resting state feature selection could provide a clean and straightforward method of isolating a core set of unique functional connections for each individual, which then contributes to the group feature set. In addition, tracking these specific sets of connections across developmentally diverse samples and testing the degree of similarity within monozygotic (MZ) and dizygotic (DZ) twin pairs may advance our understanding of the influence of genetic similarity on the functional fingerprint and its stability into adulthood. The current work uses three in-house pediatric datasets and two publicly available adult datasets (see Table 1 for detailed subgroup demographics) to identify individuals and twin pairs via support vector machine (SVM) classifiers (De Martino et al., 2008, LaConte et al., 2005) applied to rs-fcMRI data. Starting from a common pre-defined ROI set (Power et al., 2011), we isolated subsets of functional connections via ANOVA feature selection that best discriminated between individuals within each group. Next, we identified individuals and twin pairs by training and testing fMRI pattern classifiers using those connections identified during feature selection. Groups of repeat scan individuals had one scan from that individual in the training set and a different scan of the same individual in the testing set. For twin groups, one twin's scan was in the training set and their co-twin's scan was in the testing set (see Transparent Methods in the Supplemental Information and Figure S1 for detailed information on feature selection and classification). We then mapped those functional connections consistently chosen as features to identify a functional fingerprint for each group. Finally, we tested the generalizability of commonly selected connections across our two age groups, by first defining an age-specific feature set for each age group and then applying pediatric features on adult groups during classification, and vice versa. The aim of this work was to extend the current body of emerging research by providing a better understanding of the impact of development and genetic similarity on the resting state functional fingerprint. Our research suggests the following: (1) individually unique patterns of functional connectivity are similar among family members and vary with genetic similarity and (2) the general pattern of resting state functional connections that are highly predictive of an individual translates across different ages and groups and appears stable within individuals during development and into adulthood.
Table 1.
Group Characteristics
| UT Repeat | MSC Repeat | UT MZ | HCP MZ | UT DZ | HCP DZ | UT SS | |
|---|---|---|---|---|---|---|---|
| Scan Count | 30 | 20 | 30 | 50 | 38 | 50 | 34 |
| "Pairs" (# Female) | 15 (9) | 10 (5) | 15 (24) | 25 (26) | 19 (16) | 25 (26) | 17 (12) |
| Age Range | 9.8–18.3 | 24–34 | 8.9–13.2 | 26–34 | 9.1–13.9 | 22–35 | 8.8–18.6 |
| Age (M ± SD) | 12.9 ± 2.6 | 29.1 ± 3.3 | 10.8 ± 1.3 | 29.8 ± 2.2 | 11.3 ± 1.3 | 29.2 ± 3.5 | 12.2 ± 2.7 |
| IQ/PMAT (M ± SD) | 102.3 ± 14.1 | 128.8 ± 6.6 | 106.3 ± 17.2 | 17.2 ± 4.3 | 105.2 ± 11.7 | 18.3 ± 4.0 | 108.3 ± 13.0 |
| Mean FD (M ± SD) | 0.10 ± 0.02 | 0.13 ± 0.02 | 0.10 ± 0.03 | 0.12 ± 0.02 | 0.10 ± 0.02 | 0.12 ± 0.02 | 0.10 ± 0.02 |
We included two scans for each participant in the repeat scan groups. All other groups consist of one scan per individual. HCP participants' intelligence score was calculated from the Raven's Progressive Matrices (PMAT); all other groups are intelligence quotient (IQ) scores. Framewise displacement (FD) means and standard deviations (in millimeters) are post-motion scrubbing. Age is reported in years.
UT, Developmental participants collected at The University of Texas at Austin; MSC, Midnight Scan Club adult participants; MZ, monozygotic twin pairs; DZ, dizygotic twin pairs; HCP, Human Connectome Project adult participants; SS, Developmental same-sex pairs (includes DZ twins and non-twin sibling pairs of the same sex).
Results
SVM Classifiers Predict Individuals and Co-twin Pairs
We applied ANOVA feature selection on segmented resting state time courses to identify features that best discriminated between individuals (see Transparent Methods for detailed description). We then applied the resulting feature mask on all full 5-min resting state time courses used for SVM classification (Figure 1). For all groups, the SVM classifier accurately predicted matched scans (individual repeat scans, co-twins, or same-sex siblings) at levels significantly above chance (Figure 2A). This set of accurate classifications was observed when the classifier was run with a single leave-one-group-out cross-validation (2-fold) scheme, as well as with our permutation average accuracy (Figure 2B), but only the permutation accuracies are reported (Table 2). The classifier accuracies for all groups are as follows: (1) Developmental Participants Collected at The University of Texas at Austin (UT) Repeats: accuracy = 1.0, chance = 0.06, p < .001; (2) Midnight Scan Club Adult Participants (MSC) Repeats: accuracy = 1.0, chance = 0.1, p < .001; (3) UT MZ Twins: accuracy = 0.52, chance = 0.06, p < .001; (4), Human Connectome Project Adult Participants (HCP) MZ Twins: accuracy = 0.56, chance = 0.04, p < .001; (5) UT DZ Twins: accuracy = 0.24, chance = 0.05, p < .001; (6) HCP DZ Twins: accuracy = 0.22, chance = 0.04, p < .001; (7) UT Same-Sex Sibling Pairs: accuracy = 0.35, chance = 0.06, p < .001.
Figure 1.
ANOVA Feature Selection and SVM Classification
(A) Individuals within the training set (twin A for twin groups or scan 1 for repeat-scan individuals) provided 5-min resting state time courses, extracted from each of the 255 ROIs.
(B) Time courses were then split into 20-s segments, and a correlation matrix was created for each segment. All correlation matrices were then converted into vectors of unique, off-diagonal correlation values.
(C) These vectors were then used for ANOVA feature selection to create the feature mask used by the classifier.
(D) The resulting feature mask is then applied to each full 5-min resting state time course, and the SVM classifier is trained on the same training set used for feature selection and tested on the held-out testing set (either the individual's second scan or co-twin data).
Figure 2.
SVM Classifiers Predict Individuals and Co-twin Pairs
(A) Mean SVM classifier accuracies, per group. Error bars are 95% confidence intervals. Both repeat visit subgroup accuracy scores were 1.0. Monozygotic (MZ) twins in both subgroups outperformed dizygotic (DZ) twins. Hatched box plots indicate when the opposite age group's feature mask was used for classification. All group accuracies are reduced when using the opposite mask, except for the Adult MZ twins. All groups' prediction scores were significantly better than chance, except the Adult DZ group when tested using the pediatric feature mask.
(B) Left: Distributions of accuracy scores in the twin groups, across age sets, show a similar distribution and mean accuracy score. Pediatric group distributions are outlined in black and the darker shade indicates overlap in age-grouped distributions. The spread of distributions reinforces the decision to shuffle twin labels to ensure that accidental group assignment bias does not influence group accuracy. Right: The pediatric same-sex sibling pair group outperformed pediatric DZ twins, suggesting an influence of sex on functional network organization.
(C) The area under the curve (AUC) values represent classifier sensitivity for each related scan pair. Repeat visit individuals are not plotted, as AUC values were 1.0 for all. A lower group average and larger distribution of AUC values in both DZ groups suggests that differences in DZ twin pairs' functional fingerprint resulted in more false-positives when classifying DZ twins than MZ twins. This is amplified in the adult DZ twins, compared with their pediatric counterparts.
Table 2.
Mean Classifier Accuracies, per Group
| Group Chance | Mean Accuracy | Lower 95% CI | Upper 95% CI | Mean Accuracy (Opposite Age Mask) | Lower 95% CI | Upper 95% CI | |
|---|---|---|---|---|---|---|---|
| UT Repeat | 0.063 | 1.00a | – | – | 0.92a | 0.919 | 0.922 |
| MSC Repeat | 0.100 | 1.00a | – | – | 1.00a | – | – |
| UT MZ Twins | 0.066 | 0.52a | 0.518 | 0.525 | 0.32a | 0.322 | 0.327 |
| HCP MZ Twins | 0.041 | 0.57a | 0.565 | 0.571 | 0.35a | 0.353 | 0.359 |
| UT DZ Twins | 0.053 | 0.24a | 0.242 | 0.247 | 0.24a | 0.243 | 0.248 |
| HCP DZ Twins | 0.039 | 0.22a | 0.218 | 0.222 | 0.11 | 0.108 | 0.112 |
| UT SS Siblings | 0.057 | 0.35a | 0.349 | 0.354 | 0.23b | 0.232 | 0.236 |
Group chance values are empirical chance and are calculated by running the classifier after randomly shuffling scan pair labels (scan pairs were no longer correctly labeled with their matching scan). The left side of the table is running the classifiers normally, and the right side is running the classifier with the opposite age group's feature mask. (P values indicate classifier performance versus chance.).
p < 0.001.
p < 0.05.
Functional Fingerprints Are Similar across Age Groups
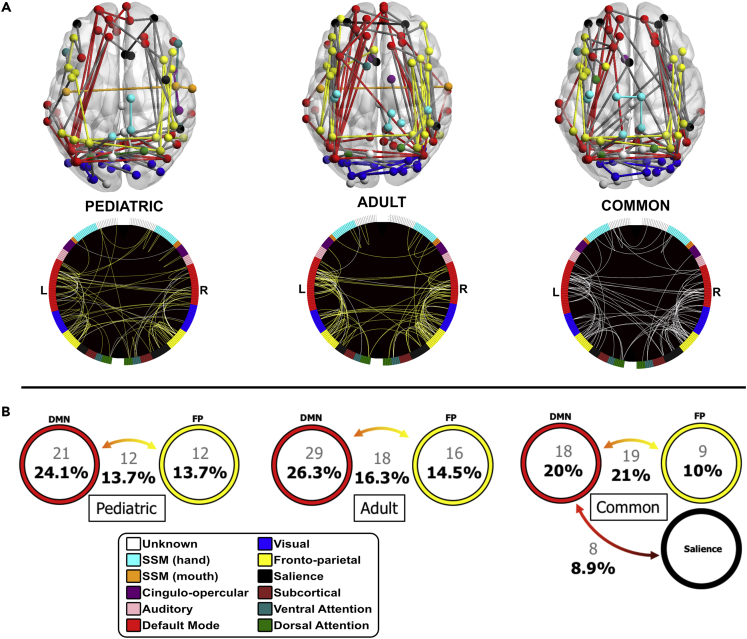
The summed fold-rank matrices for the pediatric (Figure S2) and adult (Figure S3) groups were examined to explore which functional connections were repeatedly chosen as features across subgroups. The data indicated that connections carrying the most consistently useful identifiers of an individual belonged to the FP network and the DMN (Figure 3A). Of the total 32,385 possible unique functional connections, our fold-rank matrix thresholding revealed 87 connections in the pediatric group and 110 in the adult group that were the most consistently representative of an individual in the SVM classifier. Both the pediatric and adult groups revealed a functional fingerprint mask dominated by longer, ipsilateral connections in the FP and DMN and short-range contralateral connections in the posterior DMN and VIS networks. Of the total 197 post-threshold features within both age groups, 13 features (6.59%) were identical across the pediatric and adult groups.
Figure 3.
Chosen Features of the Functional Fingerprint
(A) Pediatric and adult functional fingerprint mask connections that were selected across at least 5 of 6 possible folds are displayed on glass brains (viewed from above) to visualize anatomical location, and on circle graphs to visualize functional network involvement. Of a possible 32,385 unique functional connections, 87 pediatric and 110 connections survived at this threshold. Within the circle graphs, yellow lines represent presence of that connection across 5 testing folds and white lines represent presence in all 6 testing folds. A common set mask was created that represented connections that were chosen at least once within all six subgroups in the pediatric and adult groups; 90 functional connections survived this threshold in the common mask (white lines in common graph). See also Figures S2–S4.
(B) Ranking connections in each mask based on their network representation revealed that the largest percentage of connections were within the default mode (DMN) network, followed by connections between the DMN and fronto-parietal (FP) network, and those within the FP network. The common set mask revealed connections representing between FP and DMN, within DMN, within FP, and between the DMN and salience (SAL) network connections.
We then examined the functional network affiliation of the ROIs chosen as features in the pediatric and adult group. Post-threshold features were labeled by their functional network affiliation and ordered by frequency. This revealed a similar pattern of network representation between the two age groups (Figure 3B): The largest percentage of network representation was (1) within the DMN network (DMN to DMN) (Adults = 29 [26.3%]; Pediatric = 21 [24.1%]), followed by (2) between the DMN and FP network (Adults = 18 [16.3%]; Pediatric = 12 [13.7%]), and (3) within the FP network (FP to FP; Adults = 16 [14.5%]; Pediatric = 12 [13.7%]). After the top ∼50% connections, chosen features did not show any discernable pattern between the two groups and involved networks that often consisted of only a few connections.
To test the impact of the number of ROIs within each network on the observed patterns of network representation for each group, we simulated 10,000 feature sets for each group. Each simulated feature set consisted of the same number of features as our group fold-rank matrices (Adult = 110; Pediatric = 87), with all connections chosen at random (without replacement) from the list of all possible connection types. We then averaged how often the main connection types found in our group matrices were included in the 10,000 simulations and calculated the proportion of the feature set each accounted for. The average network representation in our simulated feature sets were (1) within DMN network (Adults = 5.4 [4.95%]; Pediatric = 4.3 [4.92%]), (2) between the DMN and FP network (Adults = 4.8 [4.40%]; Pediatric = 3.8 [4.38%]), and (3) within FP network (Adults = 1.0 [0.90%]; Pediatric = 0.8 [0.91%]).
Next, we examined the common set rank matrix (all connections chosen for at least one fold across all pediatric and adult subgroups) using the same steps outlined above. The data revealed a nearly identical representation of network assignment across the functional connections (Figure 3B). Of a total 90 connections in the common set rank matrix, 19 (21%) represented between FP and DMN network connections, 18 (20%) represented within DMN connections, 9 (10%) represented within FP connections, and 8 (9%) represented connections between the DMN and salience (SAL) networks. The same 10,000 simulated feature sets were created with 90 connections to compare with this common set rank matrix. The simulated sets showed an average of 3.9 (4.36%) between FP and DMN, 4.4 (4.92%) within DMN, 0.8 (0.09%) within FP, and 2.8 (3.16%) between DMN and SAL connections.
Common Feature Sets Have Classification Utility for Opposite Age Group
In an effort to quantify the similarity of pediatric-and adult-specific connections chosen as features, we used the summed fold-rank matrices as feature masks on the original timeseries and re-ran the SVM classifier permutation analysis. All connections with a value of five or six, after thresholding, were used as features. The pediatric groups were run with the adult feature mask (110 features), and the adult groups were run with the pediatric feature mask (87 features). There were 13 identical features that overlapped the pediatric and adult feature sets. Using the opposite age group's top-ranking features mask resulted in significant, although reduced, classifier accuracy for all groups except the MSC Repeats and resulted in non-significant accuracy for the HCP DZ twin subgroup (Figure 2A).
When the adult subgroups were trained using the pediatric-specific feature mask (Table 2), classifier accuracies were as follows: MSC Repeats (accuracy = 1.0, chance = 0.1, p < .001); HCP MZ Twins (accuracy = 0.35, chance = 0.04, p < .001); HCP DZ Twins (accuracy = 0.11, chance = 0.04, p = .065). When the pediatric subgroups were trained using the adult-specific feature mask, classifier accuracies were as follows: UT Repeats (accuracy = 0.92, chance = 0.06, p < .001); UT MZ Twins (accuracy = 0.32, chance = 0.06, p < .001); UT DZ Twins (accuracy = 0.24, chance = 0.05, p < .001); UT Same-sex Sibling Pairs: (accuracy = 0.23, chance = 0.06, p = .015).
Discussion
In the current work, we address the stability and similarity of the functional fingerprint within pediatric and adult samples. Furthermore, we directly tested the ability of a functional fingerprint identified in our developmental sample to successfully identify adults, and vice versa. These results provide initial evidence that the functional fingerprint found in pediatric samples is remarkably similar to the functional fingerprint found in adulthood and contributes to a better understanding of the stability of the brain's functional fingerprints over age.
First, we identified the specific functional connections that were most consistently representative of an individual across multiple subgroup comparisons and tested their utility to predict individuals in the opposite-age datasets with substantial success. We then used SVM classifiers to identify an individual based on a previous scan or the scan of a genetic relative, across multiple pediatric and adult neuroimaging datasets. We also provided evidence that segmentation of resting state time courses is a viable means to provide multiple samples for ANOVA feature selection. We believe that this type of feature selection can identify highly individualized functional connections within a “functional fingerprint” of an individual. This process results in a more accurate characterization of functional connectivity within an individual's resting state signal and can identify a more discriminating set of functional connections than averaging the full time course during feature selection.
Our results also replicate and extend the generalizability of previous work that assessed fMRI-based functional fingerprints in adults, by highlighting the role of genetic similarity on functional fingerprinting. MZ twins in both the pediatric and adult subgroups exhibited similar and higher classifier accuracy of rs-fcMRI data than DZ twins. Including only same-sex siblings in our pediatric subgroup significantly improved classifier accuracy of ∼50% genetically similar individuals, but the classifier still did not reach the level of MZ classification accuracy. This suggests an influence of biological sex on functional fingerprint similarity.
SVM Classifier Accuracy Reveals a Genetic Impact on Functional Fingerprint
We observed higher classification accuracy for MZ co-twins than DZ co-twins in both the pediatric (MZ = 0.52; DZ = 0.24) and adult (MZ = 0.57; DZ = 0.22) subgroups. These results provide evidence of a genetic influence on the organization of highly individualized resting state functional connections. SVM classifier accuracy scores supported our prediction that classifier performance would be highest in subgroups with greater genetic similarity. It is notable that classifier accuracy was greater for repeat scans than for MZ twin subgroups, given that both groups include scans that share 100% genetic similarity. One possible explanation for reduced classifier accuracy (compared with repeat scan individuals) in both MZ twin subgroups is an increase of individual differences in network organization emerging with age (Freund et al., 2013). However, neither environmental influence nor age could be directly and accurately tested in the current dataset. The observed decrease in classifier accuracy from MZ to DZ twins also aligns with previous work that used the model-based connectotyping method (Miranda-Dominguez et al., 2018). Although the connectotyping method differs from our approach, our results provide corroborating evidence from a different methodology that a functional fingerprint in resting state network organization is, in fact, affected by genetic similarity. Furthermore, the current work emphasizes that this genetic impact is found in direct measures of resting state functional connectivity in both pediatric and adult participants. This similarity across age groups provides initial evidence that the pattern of organization within a functional fingerprint may be established early in life and is similar to that found in adulthood.
We expected the pediatric groups to exhibit overall lower classifier prediction accuracy due to ongoing organization of resting state functional networks throughout adolescence (Cai et al., 2018, Fair et al., 2007, Grayson and Fair, 2017, Power et al., 2010, Satterthwaite et al., 2013). Somewhat surprisingly, adult and pediatric twin groups showed similar prediction accuracy on the basis of zygosity. During development, individuals may share a level of common developmental change as a result of age-related stages of network organization, regardless of genetic similarity. One possible reflection of this common change can be seen in distribution of classifier sensitivity scores (Figure 2C).
Sensitivity scores were derived using area under the receiver operating characteristic curve (AUC), which were calculated from each scan pair's true-positive and false-positive classifications. Both adult twin groups showed higher average group accuracy scores than the pediatric twin groups, whereas the adult MZ twins showed significantly higher AUC scores than the adult DZ twins (p < 0.0001). However, the pediatric MZ twins did not show significantly different AUC scores than the pediatric DZ twins. Qualitatively, the larger spread of AUC scores within both DZ groups is more pronounced in the adult DZ group and highlights a higher rate of classifier confusion for adult DZ twin pairs. This increase in classifier confusion between adult scan pairs may be due to a reduction of shared developmental changes in functional network organization and increased environmental discrepancy between co-twins that is amplified as they age.
Top Predictive Features Reveal Similar Key Network Representation
Mapping the top features repeatedly chosen across subgroups revealed that connections in the DMN and FP networks most consistently differentiated individuals, facilitating correct SVM classifier identification. It is important to note that the DMN is the most highly represented in our ROI set (57 out of a total 255). However, we find that the high similarity in percentage and type of connection between the pediatric and adult groups (Figure 3B) is informative in regard to the stability of individually unique connections. Indeed, using the features from one age group (e.g., top features from the adult sample) to predict the other age group (e.g., the pediatric subgroups) was significantly greater than chance in all but the adult DZ group, although less optimal than features derived from one's same age group (Figure 2A). Furthermore, feature set simulations choosing the same number of connection types at random highlighted the unique nature of the network representation and type of connections within both age groups' feature sets. The similarity of network representation of chosen functional connections across pediatric and adult groups contradicted our hypothesis that children and adolescents would have a different pattern of chosen connections relative to adults. Higher-order cognitive functions such as problem solving (Fedorenko et al., 2013, Hearne et al., 2016, Price et al., 2018), task switching (Church et al., 2017, Cole et al., 2013, Loose et al., 2017), and sustained attention (Crittenden et al., 2016, Lawrence et al., 2003, Marek and Dosenbach, 2018) rely on putative control networks such as the FP and CO networks and behaviorally show substantial differences between children and adults. The essential nature of these networks for human functioning is likely highly individualized in their specific instantiation and therefore likely to be included in the functional fingerprint. This study demonstrates potential age invariance in these networks' contributions.
Previous work exploring functional fingerprints also implicate the FP and DMN networks as highly predictive of an individual (Finn et al., 2015, Horien et al., 2019, Miranda-Dominguez et al., 2018), although consistency of specific connections across age groups was not fully explored. Our results expand and clarify these previous findings, suggesting that although these systems are still maturing and integrating throughout adolescence, the individualized aspects of these core connections may be established early on. However, some age-unique connectivity effects are likely embedded within these connections, as swapping the feature sets between age groups produced lower, although still significant, accuracy across nearly all groups. Based on the successful classification performance after swapping feature sets, one critical interpretation is that the inclusion of specific types of connections, at a specific ratio or count, may hold a high level of importance in identifying individuals from their functional fingerprint. More work exploring the developmental trajectory of functional connections that form a generalizable functional fingerprint is necessary, but our initial findings suggest that a unique pattern is established, stable, and useful for identifying individuals even before whole-brain functional network organization is fully solidified in early adulthood.
Motion Artifacts Unlikely Contributor to Classifier Accuracy
It is well established that motion contributes substantially to noise and rs-fcMRI results (Laumann et al., 2016, Power et al., 2012, Power et al., 2014, Van Dijk et al., 2012). In-scanner motion is linked to distance-dependent artifacts that especially affect measurement of long-range connections (Satterthwaite et al., 2012, Van Dijk et al., 2012). Our fold-rank matrices (Figures S2 and S3) revealed that the majority of chosen features for both age groups came from longer, ipsilateral connections between anterior and posterior ROIs, as well as short-range, contralateral connections within posterior parietal DMN and VIS network ROIs. Critically, these results were consistent across age groups, suggesting that age-related differences in scanner motion did not drive the contribution of long-range connections to classifier accuracy. In an effort to directly test the ability of motion to drive our classifier accuracies, the classifiers were trained on FD values in place of functional connectivity values. However, FD motion calculations provide only one value per repetition time (TR), in contrast to the 32,385 possible unique functional connections, and therefore the feature selection step could not be realistically performed. The reduced amount of data is underpowered for this type of analysis. Although testing classifier accuracies with the single vector of FD values is not an accurate comparison to our main methods, classifier accuracies using FD values did not approach significance for any group.
Moreover, our current work rigorously followed current best practices in neuroimaging data preprocessing and motion correction, in an effort to mitigate the influence of motion on classifier accuracy. The mean FD for all individuals and groups were well within the normative range for both pediatric and adult resting state analyses, no major motion outliers were included in our groups, and the chosen FD of 0.25 mm is at or below the commonly accepted motion censoring thresholds. Taking each of these motion considerations into account, we believe that the influence of motion on our classifier results is minimal. However, future approaches can possibly put this presumption to the test.
Limitations of the Study
Although this study expands upon recent findings in functional fingerprinting and genetic influences on rs-fcMRI organization, the following limitations must be considered. One limitation is the size of the pediatric twin sample. It is difficult to recruit and successfully retain pediatric neuroimaging participants. At the time of analysis, the neuroimaging branch of the Texas Twin Project was one of the largest pediatric twin resting state datasets, but did not provide a sufficient number of scans to fully explore analyses such as the effect of same-sex DZ pairs. Soon after completion of the current study's analyses, raw imaging data and genetic information necessary to calculate zygosity has been released from The Adolescent Brain Cognitive Development Study dataset (Casey et al., 2018, Volkow et al., 2018). This multi-site dataset includes a large number of MZ and DZ pediatric pairs, and inclusion of these data should be included in future analyses to better explore group differences. In addition, many unknowns are introduced in the realm of functional organization during development. Currently, the exact developmental trajectory of functional network organization is unknown. Incorporating future methods that quantify and account for age-related changes in network organization would further clarify the accuracy of the SVM classifier results in our pediatric subgroups.
Future analyses should explore other potential influences on classifier accuracy, such as same-sex versus opposite-sex sibling pairs. We indirectly investigated the impact of sex on identification accuracy within families by including a same-sex sibling group and comparing that with the MZ and DZ twin groups. However, siblings in this group necessarily differed by age, thus introducing a separate confound. Matching declared sex for sibling pairs did, in fact, result in higher SVM classifier accuracy of the same-sex sibling pairs than the pediatric DZ twin pairs (which was not sex-matched), suggesting a notable impact of sex variance on identification of individuals through this approach. There are limited rs-fcMRI studies that directly address sex differences in healthy twin populations, and the precise source of sex differences on functional network organization is still not fully understood (Hjelmervik et al., 2014, Satterthwaite et al., 2015). Future analyses on larger twin datasets can better parse out and control for sex differences between genetically related individuals.
Conclusions
The ability of rs-fcMRI pattern classifiers to identify matched pairs of participants in pediatric and adult samples decreased as genetic similarity decreased. This suggests a genetic influence on the pattern of highly individualized functional connections within a functional fingerprint. Connections within the identified functional fingerprint were dominated by regions in the DMN and FP networks, suggesting that integration of higher-order functional networks plays a key role in establishing the organization of individualized functional fingerprints. This pattern of connections appears to be established early in life and, importantly, is similar enough in children and adults to often allow for significant classification of individuals using the opposite age group's selected features.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This research project was supported by National Institutes of Health grants P50 HD052117 (overall PI: Jack M. Fletcher, subaward to J.A.C.), R21 HD081437 (J.A.C., E.M.T.-D.). Additional datapoints were contributed from the Developmental Cognitive Neuroscience Lab through a Brain and Behavior Research Foundation NARSAD Young Investigator award (J.A.C), start-up funds to J.A.C. (University of Texas), and a Pilot grant from the Biomedical Imaging Center at the University of Texas at Austin (20141031a).
The authors would like to thank Jack M. Fletcher for spearheading the Texas Center for Learning Disabilities (TCLD), Jenifer Juranek for heading the Houston TCLD neuroimaging team, as well as to the broader TCLD team in Austin and Houston, TX (the TCLD contributed data to the repeat pediatric samples); the Human Connectome Project and the Midnight Scan Club for their significant contributions to open science; Mary Abbe Roe, Mackenzie Mitchell, Annie Zheng, Leonel Olmedo, Joel Martinez, and Lauren Deschner for their contribution to scan data collection; and all the participating families for their time and contribution to research.
Author Contributions
Conceptualization, D.V.D., J.A.C., and J.A.L.-P.; Data Curation, L.E.E., T.N., and D.V.D.; Formal Analysis and Visualization, D.V.D.; Methodology, D.V.D., J.A.C., J.A.L.-P., L.E.E., R.M., and T.N.; Software, D.V.D., R.M., and E.M.G.; Consultation, K.P.H. and E.M.T.-D., Writing – Original Draft, D.V.D. and J.A.C.; Writing – Review & Editing, D.V.D., J.A.C., J.A.L.-P., L.E.E., R.M., E.M.G., T.N., K.P.H., and E.M.T.-D.; Supervision, J.A.C. and J.A.L.-P.; Funding Acquisition, J.A.C., and E.M.T.-D.
Declaration of Interests
The authors declare no competing interests.
Published: January 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.100801.
Data and Code Availability
The publicly available adult datasets are available in raw and preprocessed form. The Midnight Scan Club data can be accessed at https://openneuro.org/datasets/ds000224/versions/00001. The Human Connectome Project data can be acquired from https://www.humanconnectome.org/.
The scripts used to perform the main SVM classifier analysis can be found on the main author's github page https://github.com/iamdamion.
Supplemental Information
References
- Balsters J.H., Mantini D., Wenderoth N. Connectivity-based parcellation reveals distinct cortico-striatal connectivity fingerprints in autism spectrum disorder. NeuroImage. 2018;170:412–423. doi: 10.1016/j.neuroimage.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., Andrews-Hanna J.R., Schacter D.L. The brain’s default network: Anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Byrge L., Kennedy D.P. High-accuracy individual identification using a “thin slice” of the functional connectome. Netw. Neurosci. 2019;3:363–383. doi: 10.1162/netn_a_00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Dong Q., Niu H. The development of functional network organization in early childhood and early adolescence: a resting-state fNIRS study. Dev. Cogn. Neurosci. 2018;30:223–235. doi: 10.1016/j.dcn.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey B.J., Cannonier T., Conley M.I., Cohen A.O., Barch D.M., Heitzeg M.M., Soules M.E., Teslovich T., Dellarco D.V., Garavan H. The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church J.A., Bunge S.A., Petersen S.E., Schlaggar B.L. Preparatory engagement of cognitive control networks increases late in childhood. Cereb. Cortex. 2017;27:2139–2153. doi: 10.1093/cercor/bhw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.L., Fair D.A., Dosenbach N.U.F., Miezin F.M., Dierker D., Van Essen D.C., Schlaggar B.L., Petersen S.E. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.W., Reynolds J.R., Power J.D., Repovs G., Anticevic A., Braver T.S. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 2013;16:1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden B.M., Mitchell D.J., Duncan J. Task encoding across the multiple demand cortex is consistent with a frontoparietal and cingulo-opercular dual networks distinction. J. Neurosci. 2016;36:6147–6155. doi: 10.1523/JNEUROSCI.4590-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martino F., Valente G., Staeren N., Ashburner J., Goebel R., Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. NeuroImage. 2008;43:44–58. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- Dixon M.L., De La Vega A., Mills C., Andrews-Hanna J., Spreng R.N., Cole M.W., Christoff K. Heterogeneity within the frontoparietal control network and its relationship to the default and dorsal attention networks. Proc. Natl. Acad. Sci. U S A. 2018;115:E1598–E1607. doi: 10.1073/pnas.1715766115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., Nelson S.M., Wig G.S., Vogel A.C., Lessov-Schlaggar C.N. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A., Dosenbach N.U.F., Church J.A., Cohen A.L., Brahmbhatt S., Miezin F.M., Barch D.M., Raichle M.E., Petersen S.E., Schlaggar B.L. Development of distinct control networks through segregation and integration. Proc. Natl. Acad. Sci. U S A. 2007;104:13507–13512. [Google Scholar]
- Fedorenko E., Duncan J., Kanwisher N. Broad domain generality in focal regions of frontal and parietal cortex. Proc. Natl. Acad. Sci. U S A. 2013;110:16616–16621. doi: 10.1073/pnas.1315235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn E.S., Shen X., Scheinost D., Rosenberg M.D., Huang J., Chun M.M., Papademetris X., Constable R.T. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund J., Brandmaier A.M., Lewejohann L., Kirste I., Kritzler M., Krüger A., Sachser N., Lindenberger U., Kempermann G. Emergence of individuality in genetically identical mice. Science. 2013;340:756–759. doi: 10.1126/science.1235294. [DOI] [PubMed] [Google Scholar]
- Fu Y., Ma Z., Hamilton C., Liang Z., Hou X., Ma X., Hu X., He Q., Deng W., Wang Y. Genetic influences on resting-state functional networks: a twin study. Hum. Brain Mapp. 2015;36:3959–3972. doi: 10.1002/hbm.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Gilmore J.H., Shen D., Smith J.K., Zhu H., Lin W. The synchronization within and interaction between the default and dorsal attention networks in early infancy. Cereb. Cortex. 2013;23:594–603. doi: 10.1093/cercor/bhs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D.C., Winkler A.M., Kochunov P., Almasy L., Duggirala R., Carless M.A., Curran J.C., Olvera R.L., Laird A.R., Smith S.M. Genetic control over the resting brain. Proc. Natl. Acad. Sci. U S A. 2010;107:1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E.M., Laumann T.O., Adeyemo B., Petersen S.E. Individual variability of the system-level organization of the human brain. Cereb. Cortex. 2017;27:386–399. doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E.M., Lynch C.J., Gratton C., Laumann T.O., Gilmore A.W., Greene D.J., Ortega M., Nguyen A.L., Schlaggar B.L., Petersen S.E. Three distinct sets of connector hubs integrate human brain function. Cell Rep. 2018;24:1687–1695.e4. doi: 10.1016/j.celrep.2018.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C., Laumann T.O., Nielsen A.N., Greene D.J., Gordon E.M., Gilmore A.W., Greene D.J., Ortega M., Nguyen A.L., Schlaggar B.L. Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron. 2018;98:439–452.e5. doi: 10.1016/j.neuron.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.S., Fair D.A. Development of large-scale functional networks from birth to adulthood: a guide to the neuroimaging literature. NeuroImage. 2017;160:15–31. doi: 10.1016/j.neuroimage.2017.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene D.J., Church J.A., Dosenbach N.U.F., Nielsen A.N., Adeyemo B., Nardos B., Petersen S.E., Black K.J., Schlaggar B.L. Multivariate pattern classification of pediatric Tourette syndrome using functional connectivity MRI. Dev. Sci. 2016;19:581–598. doi: 10.1111/desc.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak K.V., Marquand A.F., Beckmann C.F. Connectopic mapping with resting-state fMRI. NeuroImage. 2018;170:83–94. doi: 10.1016/j.neuroimage.2017.06.075. [DOI] [PubMed] [Google Scholar]
- Hearne L.J., Mattingley J.B., Cocchi L. Functional brain networks related to individual differences in human intelligence at rest. Sci. Rep. 2016;6:32328. doi: 10.1038/srep32328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmervik H., Hausmann M., Osnes B., Westerhausen R., Specht K. Resting states are resting traits – an fMRI study of sex differences and menstrual cycle effects in resting state cognitive control networks. PLoS One. 2014;9:e103492. doi: 10.1371/journal.pone.0103492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C., Shen X., Scheinost D., Constable R.T. The individual functional connectome is unique and stable over months to years. NeuroImage. 2019;189:676–687. doi: 10.1016/j.neuroimage.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T., Alnæs D., Doan N.T., Brandt C.L., Andreassen O.A., Westlye L.T. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat. Neurosci. 2017;20:513–515. doi: 10.1038/nn.4511. [DOI] [PubMed] [Google Scholar]
- LaConte S., Strother S., Cherkassky V., Anderson J., Hu X. Support vector machines for temporal classification of block design fMRI data. NeuroImage. 2005;26:317–329. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Laumann T.O., Snyder A.Z., Mitra A., Gordon E.M., Gratton C., Adeyemo B., Gilmore A.W., Nelson S.M., Berg J.J., Greene D.J. On the stability of BOLD fMRI correlations. Cereb. Cortex. 2016;27:4719–4732. doi: 10.1093/cercor/bhw265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N.S., Ross T.J., Hoffmann R., Garavan H., Stein E.A. Multiple neuronal networks mediate sustained attention. J. Cogn. Neurosci. 2003;15:1028–1038. doi: 10.1162/089892903770007416. [DOI] [PubMed] [Google Scholar]
- Lin W., Zhu Q., Gao W., Chen Y., Toh C.H., Styner M., Gerig G., Smith J.K., Biswal B., Gilmore J.H. Functional connectivity MR imaging reveals cortical functional connectivity in the developing brain. Am. J. Neuroradiol. 2008;29:1883–1889. doi: 10.3174/ajnr.A1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose L.S., Wisniewski D., Rusconi M., Goschke T., Haynes J.-D. Switch-independent task representations in frontal and parietal cortex. J. Neurosci. 2017;37:8033–8042. doi: 10.1523/JNEUROSCI.3656-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Dosenbach N.U.F. The frontoparietal network: function, electrophysiology, and importance of individual precision mapping. Dialogues Clin. Neurosci. 2018;20:133–140. doi: 10.31887/DCNS.2018.20.2/smarek. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Hwang K., Foran W., Hallquist M.N., Luna B. The contribution of network organization and integration to the development of cognitive control. PLoS Biol. 2015;13:1–25. doi: 10.1371/journal.pbio.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.F., Norton M.I., Van Horn J.D., Wegner D.M., Grafton S.T., Macrae C.N. Wandering minds: the default network and stimulus-independent thought. Science. 2007;315:393–395. doi: 10.1126/science.1131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O., Feczko E., Grayson D.S., Walum H., Nigg J.T., Fair D.A. Heritability of the human connectome: a connectotyping study. Netw. Neurosci. 2018;2:175–199. doi: 10.1162/netn_a_00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O., Mills B.D., Carpenter S.D., Grant K.A., Kroenke C.D., Nigg J.T., Fair D.A. Connectotyping: model based fingerprinting of the functional connectome. PLoS One. 2014;9:e111048. doi: 10.1371/journal.pone.0111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A., Snyder A.Z., Tagliazucchi E., Laufs H., Elison J., Emerson R.W., Shen M.D., Wolff J.J., Botteron K.N., Dager S. Resting-state fMRI in sleeping infants more closely resembles adult sleep than adult wakefulness. PLoS One. 2017;12:1–19. doi: 10.1371/journal.pone.0188122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack R.A., Laumann T.O., Koyejo O., Gregory B., Hover A., Chen M.-Y., Gorgolewski K.J., Luci J., Joo S.J., Boyd R.L. Long-term neural and physiological phenotyping of a single human. Nat. Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Fair D.A., Schlaggar B.L., Petersen S.E. The Development of human functional brain networks. Neuron. 2010;67:735–748. doi: 10.1016/j.neuron.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Schlaggar B.L., Lessov-Schlaggar C.N., Petersen S.E. Evidence for hubs in human functional brain networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.R., Yeo D.J., Wilkey E.D., Cutting L.E. Prospective relations between resting-state connectivity of parietal subdivisions and arithmetic competence. Dev. Cogn. Neurosci. 2018;30:280–290. doi: 10.1016/j.dcn.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle M.E. The brain’s default mode network. Annu. Rev. Neurosci. 2015;38:433–447. doi: 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- Satterthwaite T.D., Wolf D.H., Loughead J., Ruparel K., Elliott M.A., Hakonarson H., Gur R.C., Gur R.E. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. NeuroImage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite T.D., Wolf D.H., Roalf D.R., Ruparel K., Erus G., Vandekar S., Gennatas E.D., Elliott M.A., Smith A., Hakonarson H. Linked sex differences in cognition and functional connectivity in youth. Cereb. Cortex. 2015;25:2383–2394. doi: 10.1093/cercor/bhu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite T.D., Wolf D.H., Ruparel K., Erus G., Elliott M.A., Eickhoff S.B., Gennatas E.D., Jackson C., Prabhakaran K., Smith A. Heterogeneous impact of motion on fundamental patterns of developmental changes in functional connectivity during youth. NeuroImage. 2013;83:45–57. doi: 10.1016/j.neuroimage.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk K.R.A., Sabuncu M.R., Buckner R.L. The influence of head motion on intrinsic functional connectivity MRI. NeuroImage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Koob G.F., Croyle R.T., Bianchi D.W., Gordon J.A., Koroshetz W.J., Pérez-Stable E.J., Riley W.T., Bloch M.H., Conway K. The conception of the ABCD study: from substance use to a broad NIH collaboration. Dev. Cogn. Neurosci. 2018;32:4–7. doi: 10.1016/j.dcn.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yin X., Ge H., Han Y., Pang Z., Liu B., Liu S., Friston K. Heritability of the effective connectivity in the resting-state default mode network. Cereb. Cortex. 2017;27:5626–5634. doi: 10.1093/cercor/bhw332. [DOI] [PubMed] [Google Scholar]
- Xu T., Opitz A., Craddock R.C., Wright M.J., Zuo X.N., Milham M.P. Assessing variations in areal organization for the intrinsic brain: from fingerprints to reliability. Cereb. Cortex. 2016;26:4192–4211. doi: 10.1093/cercor/bhw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanto T.P., Gazzaley A. Fronto-parietal network: flexible hub of cognitive control. Trends Cogn. Sci. 2013;17:602–603. doi: 10.1016/j.tics.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The publicly available adult datasets are available in raw and preprocessed form. The Midnight Scan Club data can be accessed at https://openneuro.org/datasets/ds000224/versions/00001. The Human Connectome Project data can be acquired from https://www.humanconnectome.org/.
The scripts used to perform the main SVM classifier analysis can be found on the main author's github page https://github.com/iamdamion.