Abstract
A quiet revolution is under way in technologies used for nanoscale cellular imaging. Focused ion beams, previously restricted to the materials sciences and semiconductor fields, are rapidly becoming powerful tools for ultrastructural imaging of biological samples. Cell and tissue architecture, as preserved in plastic-embedded resin or in plunge-frozen form, can be investigated in three dimensions by scanning electron microscopy imaging of freshly created surfaces that result from the progressive removal of material using a focused ion beam. The focused ion beam can also be used as a sculpting tool to create specific specimen shapes such as lamellae or needles that can be analyzed further by transmission electron microscopy or by methods that probe chemical composition. Here we provide an in-depth primer to the application of focused ion beams in biology, including a guide to the practical aspects of using the technology, as well as selected examples of its contribution to the generation of new insights into subcellular architecture and mechanisms underlying host-pathogen interactions.
Bridging the 3D imaging gap
Biological imaging is a mature discipline, with technologies ranging from X-ray crystallography for exploring 3D protein structures at near-atomic resolution to the creation of 3D maps of the entire human body in the Visible Human Project1. In between these extremes, higher resolution ultrastructural studies have primarily made use of transmission electron microscopy (TEM), traditionally a 2D technique. The practical limit for the thickness of a sample that can be interrogated by TEM is less than half a micrometer; in thicker samples the incident electrons in the imaging beam undergo multiple inelastic scattering events, leading to a decrease in the information content obtained. Thus large samples with thicknesses on the order of micrometers, rather than nanometers, sit in a ‘3D imaging gap’; specific imaging technologies are required to visualize their ultrastructure in three dimensions and at high resolution2.
Historically, investigations into the 3D ultrastructure of relatively large biological samples such as whole cells and tissues have used serial-section TEM (ssTEM), in which sequential sections of resin-embedded samples resembling long ribbons of material are placed on electron microscopy (EM) grids and imaged3,4. ssTEM does allow high-resolution imaging in x- and y-planes, but its z-resolution is limited by the slice thickness, as each slice is represented by just one projection image. Obtaining robust ribbons of ultrathin sections is challenging, and a lower limit of 50 nm for the slice thickness is generally accepted. ssTEM has been used with considerable success in neuroanatomy, perhaps most famously resulting in a wiring diagram of an entire nematode5, but features such as thin dendritic spine necks that are smaller than the section thickness in a direction parallel to the electron beam (the z-axis by convention) cannot be visualized with simple ssTEM.
One approach for obtaining information along the z-axis is TEM tomography, where the section is tilted along one or two axes and a series of images are acquired at various tilt angles. These 2D images are then algorithmically combined to generate a ‘tomogram’, or a 3D volume6. Tomography can yield 3D information for every slice, but the reconstruction suffers from poor resolution in the axis parallel to the imaging beam because of the ‘missing wedge’ of data arising from the restricted tilt range7, as well as from distortion and shrinkage of the tissue due to the larger electron dose8. Still, TEM tomography, especially under cryogenic conditions, has produced high-resolution 3D maps of small objects such as virions, bacteria and sections of cells, as has been reviewed elsewhere9–12. Attempts have been made to combine serial sectioning and TEM tomography for imaging larger samples13,14, so that instead of a series of 2D images, a series of tomographic reconstructions can be obtained, but this adds a layer of complexity to an already tedious and manual technique.
Methods based on the use of scanning electron microscopes (SEMs) provide exciting new opportunities for increased throughput in 3D imaging. A recently developed technique, array tomography15,16, enables coverage of very large volumes at high lateral resolution by means of automated serial sectioning. Here, a rotary microtome (i.e., an automated tape-collecting ultramicrotome) generates ribbons of sequential sections that are continuously collected on an adhesive strip, attached to a large wafer and imaged in the scanning electron microscope. In addition to its high-throughput nature, array tomography allows the user to return to the region of interest (ROI) for further investigation, as the sections can be stored long term.
Serial block face SEMs (SBF-SEMs) are also increasingly used to slice and image large resin-embedded samples17. The user records a traditional SEM image by recording scattered electrons originating from and a few nanometers below the surface of a sample. Ordinarily the electron yield from a substrate is dependent on its surface topology: highly exposed areas yield a brighter signal than recessed regions, giving SEM images a sense of depth. In the SBF-SEM, a microtome is adapted to fit inside a chamber in the microscope, and biological samples stained with heavy metals and embedded in a hard resin are iteratively sliced at user-defined thicknesses (usually 25–100 nm). Electrons scattered from the subsequently exposed faces of the sample (as opposed to from a section) are imaged by the microscope, allowing the generation of a stack of up to thousands of 2D images containing ultrastructural information for the bulk of the volume. SBF-SEMs have recently been applied to neuronal tissue18 to reconstruct neuronal maps of retinas, yielding mechanistic insight into aspects of vision and bringing big data into the field of neuronal connectomics19–21. One drawback is that a single run of an SBF-SEM destroys the entire sample, regardless of the subvolume imaged. The method can be prone to charging artifacts, but these can be mitigated to some extent by careful choice of acquisition parameters, heavy metal staining protocols22,23 and resin formulations24.
Although both array tomography and the use of SBF-SEMs are high-throughput methods, they do have some limitations. The use of a mechanical device to repeatedly slice the sample results in a predictable set of artifacts such as knife marks, holes, folds, compression and/or stretching. Poor control over the thickness of each slice can also generate artifacts in the 3D volume, potentially resulting in inaccuracies in the high-resolution 3D reconstruction of features in the sample.
The 3D imaging artifacts arising from the use of a blade to mechanically slice the sample can be reduced or eliminated by milling of the sample at a glancing angle using a focused ion beam (FIB). FIBs have been used in the semiconductor industry and materials sciences for several decades, and more recently their use has extended to biology. The effective use of FIB microscopes in 3D biological imaging is the main focus of this review.
A primer to FIB instrumentation
An FIB instrument can have many possible ion sources, including gas field ionization sources25 and inductively coupled plasma26. But for most applications, a liquid metal ion source is favored. Gallium is preferred because of its low melting point, low volatility and low vapor pressure27,28. In a typical FIB setup, when a small reservoir of Ga is heated, the liquid metal flows down a needle tip; the application of an extractor voltage to the tip results in emission, with an effective source size of ~5 nm. The resulting beam is accelerated, directed through several apertures to define the diameter and current, focused through a series of lenses and directionally controlled by deflector plates before it interacts with the sample. Ga+ ions are heavy, and when they encounter atomic nuclei in a sample they cause efficient ‘sputtering’ (i.e., removal of the substrate) at a rate that is dependent on the material itself, as well as on the beam parameters. Additionally, Ga is relatively easily ionizable and provides a well-defined correlation between beam diameter and beam current, allowing the beam to be easily controlled electronically. For example, the Ga+ beam diameter can be tuned to ~10 nm or less at low currents (<100 pA) for high-resolution work, whereas a Ga+ beam operated at 45 nA results in a beam ~1 mm thick, which is well suited to ablate large areas on a sample. The FIB can also efficiently mill through hard materials, and with careful calibration of the milling parameters, heterogeneous samples can be milled with few artifacts27,29–31. Finally, when used at a glancing angle (i.e., approximately parallel to the surface being milled), the FIB can sputter a large number of atoms from the sample with negligible implantation of Ga+ ions into the sample’s surface32. This geometry allows for very efficient sample milling, and the minimal contamination and charging from Ga+ ions embedded in the sample result in better signal-to-noise ratios and less warping in the recorded electronic image.
The FIB can also be used to deposit material (applications usually involve platinum or carbon) on various surfaces. When a stream of a gaseous organometallic platinum or polymeric carbon compound is flowed atop a sample, the Ga+ beam, operated at a current too low to cause a high rate of sputtering, causes the cleavage of the platinum or carbon from the volatile components of the precursor compounds. Nonvolatile elements, such as some carbon impurities and Ga itself, are then deposited on the sample surface in the immediate vicinity of the beam spot. With the ability to accurately place and modify the beam, the user can thus use Ga+ ions to either remove or add material with nanoscale spatial control, and FIBs have been used for the construction of many prototype devices on micro- to nanoscales and for other applications33,34. FIBs have also been used extensively for preparation of specimens to be analyzed by scanning transmission electron microscopy and other analytical methods, or for ‘lift-out’ protocols where the final analysis is performed outside the FIB instrument35,36. For a more in-depth look at these aspects of the FIB, we direct readers to the April 2014 edition of MRS Bulletin, and specifically to reviews by Bassim et al.37 and Cantoni et al.38
FIB-SEM dual-beam microscopy
When paired with an SEM, the FIB, in and of itself a powerful tool, becomes a useful practical instrument for 3D imaging (Fig. 1 and Supplementary Video 1). Commercial instruments have both an SEM column (usually oriented vertically) and an FIB column oriented 45°–55° with respect to the microscope column (Fig. 1a), although customized variants have been used successfully39,40. The FIB and SEM columns have their own systems of lenses, apertures and electronics, and generally operate independently of each other. A sample placed in the evacuated FIB-SEM instrument chamber can be interrogated by either of the beams, or by both beams simultaneously when placed at the coincidence point’, where the ion and electron beams coincide in space, usually at a working distance of 4–5 mm. This allows the user to mill (with the FIB) and image (with the SEM) a specific location on a sample without tilting or moving the stage. Most commonly, the sample is tilted so that its surface is orthogonal to the incident ion beam. The FIB is then used to mill a ‘trench’ in front of the ROI, revealing a polished face on the substrate that the microscope can image. This geometry allows for extremely efficient and stable milling and imaging of large volumes in exchange for a slight reduction in resolution and signal, due to the low beam currents and large working distances.
Figure 1 |.
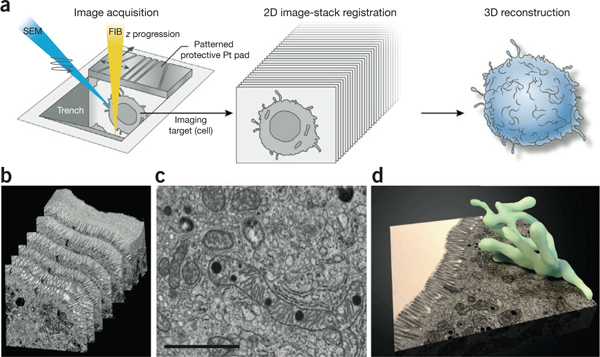
3D imaging of large biological samples by FIB-SEM. (a) Large biological samples that have been fixed either conventionally (by aldehydes) or cryogenically (by high-pressure freezing), stained by heavy metals, resin embedded and mounted are introduced into the FIB-SEM chamber. Here, chosen areas of the sample are ‘trenched’ to reveal the region of interest and then subjected to an iterative cycle of resin milling by the FIB (yellow beam) followed by SEM (blue beam) imaging of the newly revealed face to produce a 2D image stack. The patterned protective platinum (Pt) pad atop the sample to be imaged allows automatic beam tuning and slice-thickness control. The 2D image stack is then computationally converted to a 3D volume, aligned and segmented to reveal the 3D structure of interest, (b–d) A representative example of 3D tissue imaging using a mouse intestinal sample109. Shown are an image stack (b), a selected slice through the stack (c) and a segmented representation of an extensively branched mitochondrion present in the imaged volume (d). Scale bar, 1 μm. Panels b–d reprinted from Encyclopedia of Cell Biology, Vol. 2, Hartnell, L.M. et al., “Imaging cellular architecture with 3D SEM,” 44–50, Copyright 2016, with permission from Elsevier.
FIB-SEM instruments can be fitted with a wide variety of detectors depending on the imaging needs, but they almost always include an Everhart-Thornley secondary-electron detector to record electrons that are elastically or inelastically scattered from the surface and subsurface of the sample. A detector for back-scattered electrons (BSEs), which captures elastically scattered electrons, can also be useful in FIB-SEM imaging. FIB milling yields a flat surface with little topology; however, efficient elastic scattering of incident electrons by heavy elements compared with that by lighter atoms results in contrast in the BSE image41, allowing the detection of chemical differences in the sample. For biological imaging where the sample is fixed, stained and embedded with protocols similar to those for conventional TEM, the heavy metal nuclei in the stain can efficiently scatter incident electrons and generate robust secondary electron and BSE signals. Although the overall yield of BSEs is lower than that of secondary electrons, the BSE signal from the stain is detected at a better signal-to-noise ratio (SNR) and has minimal topological contributions, such as those that may arise from uneven FIB milling. The resulting image is thus similar in appearance to a conventional TEM micrograph, at slightly lower resolution and with the contrast reversed41,42. Other detectors, such as X-ray detectors for energy-dispersive X-ray spectroscopy, can also be used in conjunction with an FIB43,44, although the use of these signals in biology has yet to gain wide application. FIB-SEM instruments can also be fitted with cryostages and transfer devices for cryogenic applications45–47, as FIB processes work with remarkable fidelity at cryogenic temperatures.
In the past few years, FIB technology has been applied to biology primarily in two ways: to repetitively mill (and image by SEM) a prepared sample, and to sculpt biological samples to render them amenable to imaging by TEM or other modalities. In both cases, FIB technology has revealed biology that was previously inaccessible with other 2D imaging methods.
FIB-SEM methods for biological applications
A schematic overview of the experimental protocol used to generate 3D images in FIB-SEM imaging is provided in Figure 1a48,49. Briefly, the region to be imaged is first protected by a layer, or ‘pad’ of platinum or carbon deposited via a Ga+ beam, usually operated at 0.15–1.5 nA. An area in front of this protective pad is then milled by a FIB operated at a high current, typically 15–45 nA, exposing a face of the sample. The operator polishes this face by sweeping the FIB across it repeatedly with small advancements until the beam mills into the pad, which protects the sample face from uneven milling that could result from the FIB ‘beam tail’. At the end of the polishing, an unblemished, flat sample face is exposed. Thus the FIB operated at a glancing angle acts much as an extremely precise microtome blade, creating a sample free of artifacts associated with mechanical sectioning, but with the difference that the section abraded away is destroyed, leaving the newly exposed surface of the remainder of the sample ready for imaging. This face, and all subsequent faces of the sample exposed by iterative milling, is orthogonal to the top surface of the sample. Thus the principal axes of an FIB-SEM image stack are orthogonal to the axes described in a typical light microscopy experiment.
Typically, the FIB, operated at 0.15–1.5 nA, is advanced by a small user-defined increment, resulting in the abrasion of a certain amount of resin-embedded material and revealing a new polished face. The SEM then images this surface, often at low landing energies of 1.5–2 keV. Under these imaging conditions, the recorded back-scatter signals emerge primarily from an area <5 nm across and <10 nm thick, setting an empirical lower limit for pixel size and slice thickness in order for images to contain nonoverlapping information. The definition of resolution with an FIB-SEM is very different than in other modes of EM imaging. Although the ultimate ability to resolve features is determined by the SEM, it is difficult to quantitate the degradation of resolution due to surface damage resulting from the FIB. Nevertheless, a useful way to assess resolution is to evaluate features visibly. For example, in a best-case scenario, the two leaflets of a single lipid bilayer (~6 nm wide) can be resolved50. For typical cellular-imaging applications, sample preparation takes 2–6 h, and data collection lasts up to 60 h and results in the iterative milling and imaging of several thousand slices totaling tens of micrometers in thickness. Images in this stack are then aligned computationally and converted to an image volume, which can then be subjected to a variety of segmentation methods to reveal, in three dimensions, the features of interest in the sample.
In the first reported example of FIB-SEM technology applied to 3D imaging of biological specimens, Heymann et al.51 demonstrated application of this iterative milling and imaging approach to a variety of biological specimens, including critical-point dried, plastic-embedded and high-pressure-frozen yeast cells, as well as plastic-embedded tissue specimens. Starting from these early studies, where we demonstrated 3D imaging at a milling interval of ~200 nm, we extended this technique to various cell types and tissue specimens and established workflows for imaging using traditional aldehydefixation protocols followed by staining with heavy metals and resin embedding. With these methods we were able to demonstrate higher resolutions than previously possible (6 nm lateral, with a z-slice thickness of 20 nm), enabling the description of novel subcellular features in melanoma cells such as 3D contacts between the endoplasmic reticulum and mitochondria50. Other biological examples followed; FIB-SEM imaging of diatoms revealed new architectural features associated with cell division43, and imaging of HIV-infected macrophages captured the existence of long virion-filled channels physically that connected endocytic compartments to the cell surface52, offering a possible explanation for how chronically infected cells continuously and rapidly shed infectious virions into the extracellular milieu.
More recently FIB-SEM methods have been used to describe a variety of complex cell-cell contacts and subcellular architectures (Fig. 2). Dendritic cells are antigen-presenting cells that, in conventional 2D TEM images and light microscopy (LM) phase contrast images, appear to display spaghetti-like filopodial membrane extensions53. FIB-SEM imaging of these cells in their entirety, conjugated to uninfected T cells, showed that these extensions are actually veils’ or sheets that envelop target cells and in cross-section indeed do look like thin filopodia54. The virological synapse—the interface between the cells—wass shown to be a highly convoluted membrane structure that maximizes the contact area between the dendritic cell and the T cell and might explain the high efficiency of cell-to-cell transfer of HIV. In imaging of similar cell-cell contacts between HIV-infected astrocytes55, the FIB-SEM provided the high resolution in all axes required to capture the varying angles, lengths and degrees of convolution of these features, revealing aspects of HIV biology that could not be captured by 2D imaging.
Figure 2 |.
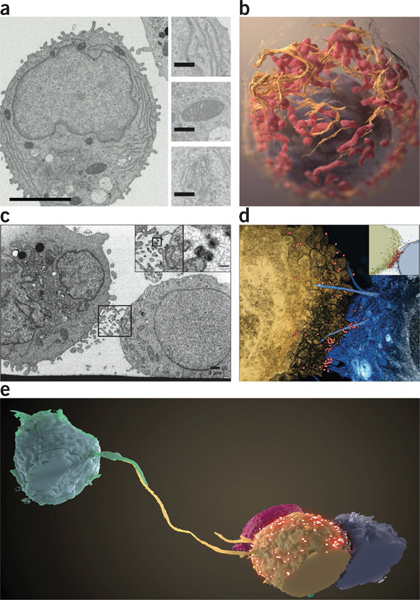
Visualizing cells and cell-cell contacts in three dimensions. (a,b) Stacks of FIB-SEM images rich in ultrastructural detail (a; inset: top, endoplasmic reticulum; middle, mitochondria; bottom, Golgi) can be segmented to reveal cellular structures in three dimensions (b; nucleus, purple; endoplasmic reticulum, orange; mitochondria, red)60. Scale bars, 3 mm (left), 400 nm (insets). (c,d) Slice from an FIB-SEM image stack (c) and segmentation of 3D volume (d) of a T cell-astrocyte virological synapse showing HIV virions (boxed and insets) appearing to surf ‘bridges’ between the cells. Scale bars, 1 μm (inset, 100 nm). (e) Long filopodia connecting distant T cells and potentially serving as conduits for HIV transmission, as captured by FIB-SEM55. Panel a reprinted from Journal of Structural Biology, Vol. 185, Narayan, K. et al, “Multi-resolution correlative focused ion beam scanning electron microscopy; applications to cell biology,” 278–284, Copyright 2014, with permission from Elsevier. Panel b reprinted from Journal of Structural Biology, Vol. 166, Heymann, J.A. et al, “3D imaging of mammalian cells with ion-abrasion scanning electron microscopy,” 1–7, Copyright 2009, with permission from Elsevier. Panels c–e copyright © American Society for Microbiology (Journal of Virology, Vol. 88, 2014, 10327–10339, doi:10.1128/JVI.00788-14).
The total volume that can be imaged in a single FIB-SEM run is limited by the maximum deflection and milling depth achievable by the FIB; runs typically yield reconstructed volumes of 104–106 μm3, often at higher 3D resolutions than achieved with an SBF-SEM. Additionally, the speed with which the required volume can be abraded by the FIB may set an upper limit on how much data can realistically be acquired in a single run. Hayworth et al.39 reported a way to circumvent this obstacle, using ‘hot knife microtomy’ to physically cut a very large resin-embedded sample into sections ~20 μm thick for imaging by FIB-SEM. The volumes were then computationally stitched back together with minimal loss to generate large 3D reconstructions of Drosophila melanogaster and Mus musculus neuronal tissue, adding to an extensive body of studies using SBF-SEMs19,20. SBF-SEMs and FIB-SEMs have also been used in parallel to capture high-resolution images of events at an organismal level, such as blood vessel fusion in zebrafish embryos56. Subtle changes in tissue architecture can also be captured by FIB-SEM on account of the high resolution in all three axes; for example, liver samples from mice suffering from a disease that causes mitochondrial dysfunction57 showed an increase in the Gaussian curvature and surface-area-to-volume ratios of diseased versus healthy mitochondria. Other researchers took advantage of the FIB’s ability to mill heterogeneous samples to create murine bone samples that allowed them to generate a detailed 3D analysis of the lacuna-canalicular network58, paving the way for detailed studies of bone mechanobiology. We recently reported high-resolution FIB-SEM imaging of skeletal muscle, which revealed complex 3D mitochondrial reticular structures that were shown to provide a conductive pathway for metabolites for efficient energy distribution in these tissues59.
Advances in FIB-SEM technology
Over the past few years, significant developments have improved the scope, resolution, robustness and speed of the FIB-SEM imaging process42,60. The use of an ‘in-column’ energy-selective back-scatter (EsB) detector has dramatically increased the SNR of samples stained with heavy metals. The most common FIB artifact is curtaining’, topological undulations on the block face parallel to the beam direction, which results in spurious signals at the secondary-electron detector. However, a grid voltage applied at the EsB detector filters out all but the highest-energy scattered electrons, so that signals are recorded almost exclusively by elastic scattering from the heavy metal-stained regions in the sample. Importantly, the EsB detector also filters out all secondary electrons generated from FIB milling, allowing for simultaneous operation of the FIB and SEM. Thus, with carefully chosen milling and imaging parameters, the ‘down time’ of the idle microscope during FIB milling and of the FIB during SEM imaging can be eliminated, dramatically speeding up data collection. Additionally, when both the ion and electron beams are kept constantly on the imaging face, the resin block, which has poor mechanical and electrical properties, is not subjected to a highly variable flux of oppositely charged ions and electrons as it would be during switching between milling and imaging modes. In our hands, this results in a measurable reduction of warping and whole-scale movements of the sample block with respect to the beams. Finally, the FIB constantly mills away material exposed to the imaging beam, solving the problem of charge buildup on the image face and thereby improving image quality.
In any 3D imaging mode involving physical slicing of the sample, controlling the slice thickness is difficult. Although groups using SBF methods have reported ~25-nm slices19,20 and some groups using FIBs have reported slices as thin as 3 or 5 nm (refs. 42,61,62), the ability to mill 3-nm slices (or slices of any particular thickness) consistently throughout the sample volume has obvious ramifications for the fidelity of the resulting 3D reconstruction. It is also important to note that reducing the pixel size below the width of the electron beam (in x and y) or the penetration depth (z) will result in oversampling and blurring of the image; we have found that a pixel size of 3–5 nm and a slice thickness of 5–15 nm are appropriate for most room-temperature resin-based applications.
We reported recently60 that depositing a pad consisting of a carbon layer atop a patterned platinum layer results in a high-contrast set of notches appearing at predictable (x, y) locations in a milled image face. When the location of the notches is tracked in real time with the SEM, the FIB can be advanced or retracted just enough to maintain a near-constant slice thickness for the length of the FIB-SEM run, allowing for high-fidelity 3D reconstructions of small moieties in the sample volume. These notches can also be used to correct for thermal and mechanical drifts in x and y that occur during a long data run. Additionally, they serve as high-resolution markers for automated beam-tuning protocols that can be executed at pre-specified intervals during a long acquisition run; the focus and stigmation of the electron beam must be continuously changed and checked as the FIB continues to mill into a sample and the imaging face recedes. With these advances, extremely stable and high-resolution 2D image stacks can be generated, allowing for the location and accurate 3D reconstruction of features as small as individual HIV cores (~60 nm in length) present in the cytoplasm of a mammalian cell60 (~10,000 mm3) (Fig. 3).
Figure 3 |.
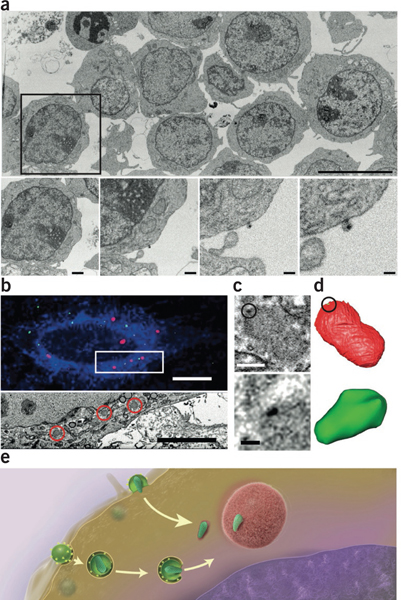
Imaging the large and small by FIB-SEM. (a) Very large fields of view including many HIV-infected cells (top) can be imaged at resolutions high enough to visualize individual virions in the same image (bottom; panels show increasing magnifications of the boxed region above)55. (b) Using fiducial markers, confocal (top) and FIB-SEM image volumes (bottom, corresponding to the boxed area above) can be registered; correlative fiducials are denoted by red channels (top) and areas circled in red (bottom). (c,d) FIB-SEM image (c) and segmentation (d) of individual HIV cores (dark signal in c (top and bottom), segmented green region in d) associated with TRIM5-α bodies (fuzzy gray body in c (top and bottom), segmented red region in d) in the cytoplasm of infected cells. With CLEM one can locate these cores and visualize them in three dimensions, achieving imaging across a volume scale of 109 in a single FIB-SEM imaging experiment60. (e) A model for HIV transport to the nucleus; the postfusion intermediate of the viral core is stabilized by association with the surface of TRIM5-α bodies. Scale bars, 9 μm (a, top); 1.2 μm, 600 nm, 300 nm and 150 nm (a, bottom, from left to right); 10 μm (b, top); 5 μm (b, bottom); 400 nm (c, top); and 100 nm (c, bottom). Panel a copyright © American Society for Microbiology (Journal of Virology, Vol. 88, 2014, 10327–10339, doi:10.1128/JVI.00788-14). Panels b–d reprinted from Journal of Structural Biology, Vol. 185, Narayan, K. et al., “Multi-resolution correlative focused ion beam scanning electron microscopy; applications to cell biology,” 278–284, Copyright 2014, with permission from Elsevier.
The age-old problem of image resolution versus image size may also be solved with the ability to image only certain ROIs on an image face. In most modes of imaging, one must choose between imaging large areas at poor resolution and imaging just a fraction of the field of view at high resolution. SEM images as large as 1 gigapixel can nowbe generated, with any shape or aspect ratio, allowing the imaging of very large areas at high resolution (Fig. 3). Alternatively, the user can apply ‘keyframe’ imaging to increase the efficiency of an FIB-SEM run, as we have recently reported60. In this approach, the entire field of view is imaged at lower resolution in x and y, and at sparser z-slice intervals, and only subareas of the entire field that are ROIs are imaged at higher resolution in x, y and z. Thus each sample yields two data sets: the keyframes give an overall ultrastructural context, which may be useful for correlation with light imaging (described below), and the ROI images can be used to create high-resolution subvolume reconstructions.
Correlative imaging with the FIB-SEM
Correlation of light and electron microscopy (CLEM) is by no means a new concept; the combination was first used more than 50 years ago to image the ultrastructure of goldfish retinas63, as was the use of heavy metal tissue stains that are both electron and light opaque64,65. Given that fluorescence allows the imaging of specific moieties with extremely high SNRs, as well as of live cells, combining these techniques with EM to obtain ultrastructural context is appealing. Several groups have reported advances recently, primarily involving the combination of fluorescence microscopy with TEM (at either room or cryogenic temperatures) or scanning electron microscopy, and these have been reviewed elsewhere66–71. Combining fluorescence microscopy with use of the FIB-SEM allows the user to investigate larger volumes; however, there are several technical challenges related to sample preparation and post-processing of images. There have been no reports yet of CLEM combined with cryo–FIB-SEM imaging, so here we restrict our CLEM discussion to room-temperature imaging and discuss cryo-FIB applications separately.
There are several ways to combine CLEM with FIB-SEM imaging. In one approach, the fluorescence image is acquired first via either live-cell or fixed-cell microscopy. The sample is then prepared by high-pressure freezing and freeze substitution or standard conventional methods and imaged by FIB-SEM (Fig. 4). Alternatively, resin-embedded samples can be imaged by both LM and EM. Pre-embedding LM has a major advantage in that it can follow live fluorescence microscopy. In such a workflow, cells on an alphanumerically coded gridded glass coverslip can be imaged live, fixed and embedded; transfer of the grid pattern to the resin block allows the cells to be located in the SEM and imaged by FIB-SEM. For robust and unbiased correlation, the 3D EM volume generated must be registered to the 3D LM data by the use of fiducials (markers that are fixed in space and visible in both imaging modalities) or by aligning the membranes and running an affine transform to correct for minor deformations caused by the fixing and embedding procedures (Fig. 4). Various groups have used fluorescent tags or dyes that retain their fluorescence after staining with altered heavy metal concentrations and resin embedding72–74, and recently a combination of soluble NSF-attachment proteins and Halo tags coupled with synthetic fluorophores was shown to retain signal even at high uranyl acetate concentrations, allowing for superior TEM images75. However, to our knowledge, the retention of fluorescence upon interaction with the FIB has not been demonstrated in FIB-SEM experiments.
Figure 4 |.
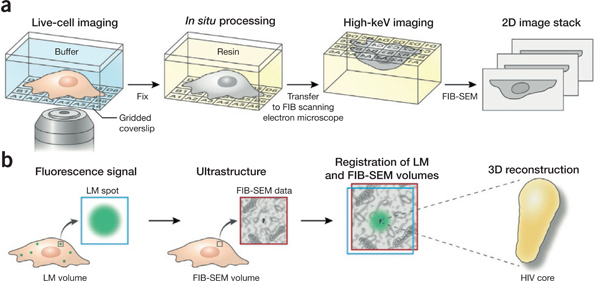
3D imaging of specific targets with correlative LM and FIB-SEM. (a) LM of a biological sample grown on or attached to an alphanumerically coded gridded coverslip produces a ‘coordinate map’ whose fidelity is maintained after resin embedding in situ, allowing location of the ROI for FIB-SEM imaging, (b) Fluorescence microscopy of tagged targets in the biological sample yields a 2D or 3D ‘target map’. FIB-SEM images subsequently acquired from the same sample can then be registered to the fluorescence image, allowing the user to reliably obtain the nanoscale 3D structures of specific fluorescent targets in a sample.
Another CLEM approach is to use true correlative tags, and much work has been done recently to discover and use such markers in biology, mostly with TEM75,76. Importantly, although many of these tags can be and are being used with FIB-SEM imaging, the automated iterative milling and imaging steps preclude any protocol that involves post-sectioning staining steps. Fluoronanogold and quantum dots77,78 are fluorescent, and their electron density allows their visualization in TEM, but the lower resolution of FIB-SEM imaging has made it challenging to locate these particles unambiguously in the volume. Alternatively, tetracysteine-tagged proteins can be combined with biarsenical labeling reagents such as TrAsH or ReAsH for visualization, and singlet oxygen–generating protein systems such as miniSOG or flavin mononucleotide79,80, or indeed GFP itself81, can serve as genetically encoded tags that generate singlet oxygen species upon illumination; the local photoconversion of diaminobenzidine to an osmophilic polymer allows a preferential deposition of osmium tetroxide that appear as foci of contrast in the EM image volume.
Regardless of the correlative method used, if there is a time gap between live-cell imaging and EM, deformations and shrinkage caused by the conventional EM preparation protocols will result in some displacement of the signal in 3D images. Although shrinkage artifacts can be reduced to some extent by high-pressure freezing protocols82, this displacement combined with the lower resolution of LM, especially along the z-axis, creates a Volume of uncertainty’ in correlation that could extend up to several hundred nanometers in any direction. Accurate alignment algorithms have significantly reduced this error73, but a final manual search is still often required. Ultimately, although there may be no silver bullet that yields perfect CLEM results, a combination of advances in imaging, sample preparation and the discovery of new tags is likely to produce higher correlative fidelity and better images in both modalities in future experiments.
Cryo-FIB operations
It has been known for several decades that fixing a biological sample in vitreous ice preserves it in a near-native state83. Still, there are limits on the thickness of a sample to be imaged by TEM84,85, and this has restricted microscopy at cryogenic temperatures to studies of small bacteria, viruses and proteins, or the very edges of cells. Thicker samples have been studied by vitreous sectioning86, but these samples are affected by the same artifacts as regular microtomy samples, as discussed above, as well as the inherent issues of contamination and fragility of samples maintained at approximately −135 °C. FIB operations, however, perform well under cryogenic conditions, and different groups have exploited this to generate TEM-ready lamellae from thick biological samples using various approaches.
Marko et al.46 demonstrated that plunge-frozen bacteria located on an exposed area of a TEM grid could be thinned using a tangential FIB milling protocol of gradually decreasing currents to mill away the material around the sample without causing local heating. The final electron-transparent lamella still encased in vitreous ice could then be imaged in scanning transmission electron microscopy mode in the FIB itself, or it could be transferred to the transmission electron microscope for high-resolution imaging. Other groups have grown cells directly on a modified TEM grid reinforced by a modified specimen support or cartridge45,47,87 and, after plunge-freezing, ablated parts of the cell at a glancing angle with an FIB mill protocol executed at low current. Depending on the sample, one can use either a single milling pattern, which generates a thin wedge-shaped volume, or two parallel milling patterns above and below a certain height, cutting through part of the cell and leaving a thin ‘shelf’ attached to the cell on either side (Fig. 5). Annular FIB milling of cellular samples to generate a pillar shape allows complete rotation of the specimen during tomography88, and other approaches have included FIB milling of cells that have been high-pressure frozen89 or drawn in a capillary90, allowing cryo-TEM imaging of various structures located in areas of the sample too thick to be electron transparent.
Figure 5 |.
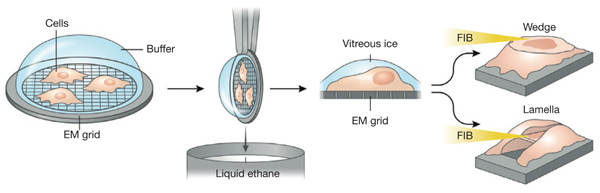
FIB thinning and lift-out of specimens for cryo-imaging. Cells grown or pipetted on TEM grids are plunge-frozen in liquid ethane and transferred to an FIB-SEM under cryogenic temperatures. A chosen area is FIB-milled tangentially either from one direction to produce a wedge or from above and below to produce a lamella, revealing the region to be imaged. This region, still encased in vitreous ice, is now thin enough to be imaged in the FIB-SEM in scanning transmission electron microscopy mode, or it can be transferred to a transmission electron microscope under cryogenic conditions and imaged at high resolution.
Although in these examples the FIB was used as a preparatory tool, recent studies have shown that FIB-SEM imaging, in the manner described above, can be done under cryogenic conditions. Schertel et al.61 recently published a protocol by which high-pressure frozen mammalian cells can be sequentially milled and imaged by FIB-SEM. Although the contrast may be generated from some level of local sublimation, this approach has the potential to yield further improvements in resolution and image quality, and it might be possible to extend it to cover mammalian cells in their entirety.
Chemical imaging
In addition to the architectural imaging described so far, the FIB has also been used in conjunction with unrelated imaging techniques to probe the spatial distributions of chemical species in biological samples (Fig. 6). Whereas the information contained in each pixel in microscopy is typically a single grayscale value, various technologies can be used to record a chemical signal or spectrum in each pixel. These techniques maybe loosely clumped under the appellation “chemical imaging” and are discussed in detail elsewhere91,92; here we discuss a few techniques whose scope has been expanded by the use of the FIB.
Figure 6 |.
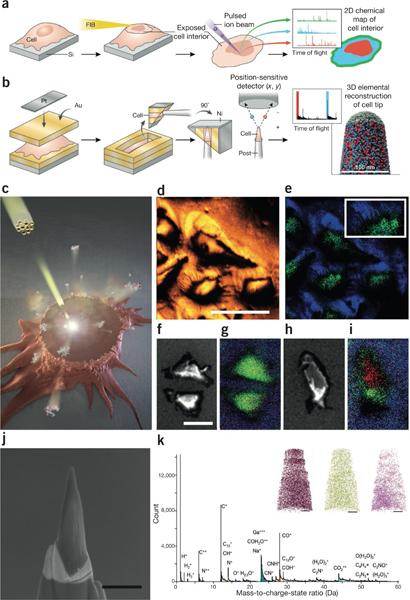
FIBs in chemical imaging, (a) Cells grown on a silicon nitride substrate and then plunge-frozen and freeze-dried can be FIB-milled almost parallel to the substrate surface; this clean cross-sectional cut of the cell exposes its interior, which can then be probed by SIMS to produce a spatial map of the chemical composition of the inside of the cell, (b) Using a circular FIB milling pattern with decreasing radii, portions of freeze-dried cells are sculpted into a thin needle shape ~100 nm wide. The resulting sample can be probed by APT to yield an information-rich 3D atomic-level chemical map. (c) Principle of approach to chemical imaging using SIMS in which cellular components are sputtered by the incident beam (yellow). (d,e) SIMS images from cells grown on a silicon substrate revealing all ions (d) and plasma membrane markers (e; phosphatidylcholine headgroups, green; silicon substrate, blue) resolve some filopodial extensions (inset). (f–i) Low-dose SEM (f,h) and SIMS chemical images (g,i) of cells before (f,g) and after (h,i) tangential FIB milling reveal the structural and chemical signatures of the cells midway through the cell volume. Green, phosphatidylcholine; red, DAPI (cell nucleus)96. (j,k) In APT, an annular pattern of milling by the FIB can be used to generate ‘tips’ from cells (j), from which low-mass ions can be extracted to obtain a mass spectrum (k) and 3D nanoscale elemental maps can be reconstructed (k, inset; C+ m/z = 12, brown; K+ m/z = 39, green; Na+ m/z = 23, pink)102. Scale bars, (d) 50 μm, (f) 30 μm, (j) 1 μm and (k, insets) 50 nm. Panels b, j and k reprinted from Journal of Structural Biology, Vol. 178, Narayan, K. et al., “Chemical mapping of mammalian cells by atom probe tomography,” 98–107, Copyright 2012, with permission from Elsevier. Panels c–i from Szakal, C. et al., “Compositional mapping of the surface and interior of mammalian cells at submicrometer resolution,” Anal. Chem. 83, 1207–1213 (2011), American Chemical Society.
Secondary ion mass spectrometry (SIMS) is a powerful and sensitive surface-analysis technique that has been applied to biological samples93,94. In SIMS, the top layer (1–2 nm) of a substrate is sputtered by a highly focused rastering primary ion beam, and the resulting ions are separated according to their mass-to-charge ratios in a mass analyzer to yield a mass spectral image of the sample. Here the Ga+ FIB is used not as a primary ion source but rather to mill away large portions of the sample to expose surfaces that would not be accessible otherwise, with fewer concerns regarding implantation and reconstruction accuracy95. Szakal et al.96 recently demonstrated abrasion of the top few micrometers of adherent mammalian cells grown on a silicon substrate by a tangential FIB mill parallel to the substrate, yielding a clean cross-section of the cells that exposed their interior. These cells were then probed by a cluster beam, and chemical signatures from the nucleus and cytoplasm were retrieved with ~400-nm spatial resolution (Fig. 6). In a similar study, James et al.97 used an FIB to sequentially ablate layers of adherent macrophages and used scanning X-ray microscopy to visualize and measure the uptake of ZnO nanoparticles in the cells.
A very different chemical imaging technique called atom probe tomography98 (APT) is much more dependent on FIB milling; a central requirement of APT is that the substrate be needle-shaped, with a diameter of ~100 nm. APT has been used to generate elemental maps of materials such as nanowires, as well as other materials that can be milled by the FIB until they attain the requisite shape and size99,100. In a pilot application to biological material, we showed recently that when adherent cells are encased in multiple layers of conductive material to mitigate their poor mechanical and electrical properties, portions of the cells could be carefully excised via FIB ‘wedge’ techniques101 and subjected to a circular FIB milling protocol with a continuously decreasing radius to generate an appropriately shaped substrate; this substrate was probed by APT to create an elemental map of a portion of a cell102 (Fig. 6).
Future prospects
Despite recent developments in FIB technology in biology, hurdles remain. For example, the current rate of success of generating cryo-TEM samples with targeted FIB milling is highly variable, with mechanical stability, de-vitrification and contamination accounting for a large number of failures. With FIB-SEM imaging, mechanical and electronic constraints of current instrument design, as well as the inherent speed limitations of FIB milling, may impose practical limitations on the resolution and size of imageable volumes, as well as the number of samples that can be imaged in any given study. Indeed, for large samples, FIBs may have to be supplanted by laser ablation to mill away the bulk of material to quickly get to the ROIs103. The precision with which FIB milling is used to excavate material to reach target locations also needs improvement. Mistakes in FIB milling are irreversible given the destructive nature of the FIB; however, the development of fiducials and cross-imaging platforms is leading to improvements in this area104.
Recent improvements in support systems and cryo-compatible transfer devices are making the transfer of cryogenic samples more dependable. The use of FIB-SEMs for cryo-techniques is in its infancy, but this approach holds promise and is likely to yield biological breakthroughs once current issues of charging and stability are resolved. Correlative platforms are improving the efficiency of locating and imaging ROIs in multiple modalities. The toolbox of correlative and chemical tags is set to expand, and this will aid in efforts toward complete automation of 3D correlation from multiple imaging modalities. Hybrid methodologies including physical sectioning along with FIB-SEM imaging are increasing the scope of the technique. Beneficial changes are also being made to the instruments; new column and detector technologies are improving image quality, and powerful scan generators have dramatically increased the scope of FIBs. Software solutions are allowing robust automated image acquisition with minimal manual intervention and maximum image quality. Through all of these advances, the FIB-SEM is gradually being converted from a high-end instrument into a robust imaging tool.
A major bottleneck in large-volume imaging is data segmentation. Image-processing tools can now align and process large amounts of acquired data relatively quickly, but the completely automated extraction of features of choice from a variety of 3D cellular images is far from a reality. Although some significant advances have been made with different approaches, especially in the field of connectomics19,105–107, the segmentation of these data for analysis, interpretation and presentation in a meaningful manner continues to be data set-specific, and mostly manual and slow. The creation of a centralized set of programs with widely applicable commands for FIB-SEM image processing, segmentation and visualization, perhaps similar to IMOD108, could help catalyze some of these steps. One optimistic scenario is that a convergence of sample processing, specific tags and image-acquisition protocols may result in broad categories of data set ‘types5 that could be subjected to sets of generalizable segmentation algorithms and parameters to extract features of interest. Paired with standardization of FIB-SEM metadata, these steps could potentially transform the visualization and distribution of 3D data sets and/or features of interest. FIB-SEM imaging is poised to become part of mainstream biological imaging, and when paired with other imaging modalities, it could open up new areas of biology to investigation in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the intramural program of the National Cancer Institute, US National Institutes of Health, Bethesda, Maryland, USA. The authors thank E. Tyler for artistic rendering of the figures; E. He, A. Brust and D. Bliss for help creating Supplementary Video 1 describing the FIB-SEM imaging process; and L. Earl and other members of their laboratory for many fruitful discussions. The authors apologize to those colleagues whose work has not been cited owing to space constraints.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Datafiles are available in the online version of the paper.
References
- 1.Ackerman MJ, Spitzer VM, Scherzinger AL & Whitlock DG The Visible Human data set: an image resource for anatomical visualization. Medinfo 8, 1195–1198 (1995). [PubMed] [Google Scholar]
- 2.Subramaniam S Bridging the imaging gap: visualizing subcellular architecture with electron tomography. Curr. Opin. Microbiol. 8, 316–322 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spacek J & Lieberman AR Ultrastructure and three-dimensional organization of synaptic glomeruli in rat somatosensory thalamus. J. Anat. 117, 487–516 (1974). [PMC free article] [PubMed] [Google Scholar]
- 4.Thaemert JC Ultrastructural interrelationships of nerve processes and smooth muscle cells in three dimensions. J. Cell Biol. 28, 37–49 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White JG, Southgate E, Thomson JN & Brenner S The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 6.Baumeister W, Grimm R & Walz J Electron tomography of molecules and cells. Trends Cell Biol. 9, 81–85 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Bartesaghi A et al. Classification and 3D averaging with missing wedge correction in biological electron tomography. J. Struct. Biol. 162, 436–450 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantele F, Zampighi L, Radermacher M, Zampighi G & Lanzavecchia S Local refinement: an attempt to correct for shrinkage and distortion in electron tomography. J. Struct. Biol. 158, 59–70 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan L & Jensen GJ Electron tomography of cells. Q. Rev. Biophys. 45, 27–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang J, Liu X, Rochat RH, Baker ML & Chiu W Reconstructing virus structures from nanometer to near-atomic resolutions with cryo-electron microscopy and tomography. Adv. Exp. Med. Biol. 726, 49–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lučič V, Rigort A & Baumeister W Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 202, 407–419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs J. A. Structural biology in situ—the potential of subtomogram averaging. Curr. Opin. Struct. Biol. 23, 261–267 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Noske AB, Costin AJ, Morgan GP & Marsh BJ Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J. Struct. Biol. 161, 298–313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soto GE et al. Serial section electron tomography: a method for three-dimensional reconstruction of large structures. Neuroimage 1, 230–243 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Micheva KD & Smith SJ Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55, 25–36 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayworth KJ et al. Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Front. Neural Circuits 8, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denk W & Horstmann H Serialblock-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2, e329 (2004).This paper describes the idea of putting an adapted microtome inside a scanning electron microscope to obtain automated serial sections; also see refs. 19 and 20.
- 18.Tapia JC et al. High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nat. Protoc. 7,193–206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmstaedter M et al. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Briggman KL, Helmstaedter M & Denk W Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Briggman KL & Denk W Towards neural circuit reconstruction with volume electron microscopy techniques. Curr. Opin. Neurobiol. 16, 562–570 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Seligman AM, Wasserkrug HL & Hanker JS A new staining method (OTO) for enhancing contrast of lipid-containing membranes and droplets in osmium tetroxide–fixed tissue with osmiophilic thiocarbohydrazide(TCH). J. Cell Biol. 30, 424–432 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams JC Heavy metal intensification of DAB-based HRP reaction product. J. Histochem. Cytochem. 29, 775 (1981). [DOI] [PubMed] [Google Scholar]
- 24.Kizilyaprak C, Longo G, Daraspe J & Humbel BM Investigation of resins suitable for the preparation of biological sample for 3-D electron microscopy. J. Struct. Biol. 189, 135–146 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Livengood RH et al. The neon gas field ion source—a first characterization of neon nano machining properties. Nucl. Instrum. Methods Phys. Res. A 645, 136–140 (2011). [Google Scholar]
- 26.Smith NS et al. High brightness inductively coupled plasma source for high current focused ion beam applications. J. Vac. Sci. TechnoL B Nanotechnol. Microelectron. 24, 2902–2906 (2006). [Google Scholar]
- 27.Holzer L, Indutnyi F, Gasser PH, Munch B & Wegmann M Three-dimensional analysis of porous BaTi03 ceramics using FIB nanotomography. J. Microsc. 216, 84–95 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Orloff J High-resolution focused ion-beams. Rev. Sci. Instrum. 64, 1105–1130 (1993). [Google Scholar]
- 29.Burdet P, Vannod J, Hessler-Wyser A, Rappaz M & Cantoni M Three-dimensional chemical analysis of laser-welded NiTi-stainless steel wires using a dual-beam FIB. Acta Mater. 61, 3090–3098 (2013). [Google Scholar]
- 30.Inkson BJ, Steer T, Mobus G & Wagner T Subsurface nanoindentation deformation of Cu-Al multilayers mapped in 3D by focused ion beam microscopy. J. Microsc. 201, 256–269 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Keller LM et al. Characterization of multi-scale microstructural features in Opalinus Clay. Microporous Mesoporous Mater. 170, 83–94 (2013). [Google Scholar]
- 32.Volkert CA & Minor AM Focused ion beam microscopy and micromachining. MRS Bull. 32, 389–395 (2007). [Google Scholar]
- 33.Fu YQ, Kok N & Bryan A Microfabrication of microlens array by focused ion beam technology. Microelectron. Eng. 54, 211–221 (2000). [Google Scholar]
- 34.Vasile MJ, Nassar R, Xie J & Guo H Microfabrication techniques using focused ion beams and emergent applications. Micron 30, 235–244 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Giannuzzi LA, Drown JL, Brown SR, Irwin RB & Stevie F Applications of the FIB lift-out technique for TEM specimen preparation. Microsc. Res. Tech. 41, 285–290 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Giannuzzi LA Routine backside FIB milling with EXpressLO (TM). In Proc. 38th International Symposium for Testing and Failure Analysis (ISTFA 2012) 388–390 (ASM International, 2012). [Google Scholar]
- 37.Bassim N, Scott K & Giannuzzi LA Recent advances in focused ion beam technology and applications. MRS Bull. 39, 317–325 (2014). [Google Scholar]
- 38.Cantoni M & Holzer L Advances in 3D focused ion beam tomography. MRS Bull. 39, 354–360 (2014).References 37 and 38 are good reviews that describe basic aspects of the physics of FIBs and their use, primarily in materials sciences.
- 39.Hayworth KJ et al. Ultrastructurally smooth thick partitioning and volume stitching for large-scale connectomics. Nat. Methods 12, 319–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kopek BG, Shtengel G, Xu CS, Clayton DA & Hess HF Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. USA 109, 6136–6141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Winter DAM et al. Tomography of insulating biological and geological materials using focused ion beam (FIB) sectioning and low-kV BSE imaging. J. Microsc. 233, 372–383 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Murphy GE et al. Correlative 3D imaging of whole mammalian cells with light and electron microscopy. J. Struct. Biol. 176, 268–278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hildebrand M, Kim S, Shi D, Scott K & Subramaniam S 3D imaging of diatoms with ion-abrasion scanning electron microscopy. J. Struct. Biol. 166, 316–328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott K & Ritchie NWM Analysis of 3D elemental mapping artefacts in biological specimens using Monte Carlo simulation. J. Microsc. 233, 331–339 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Wang K, Strunk K, Zhao GP, Gray JL & Zhang PJ 3D structure determination of native mammalian cells using cryo-FIB and cryo-electron tomography. J. Struct. Biol. 180, 318–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marko M, Hsieh C, Schalek R, Frank J & Mannella C Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods 4, 215–217 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Rigort A et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl. Acad. Sci. USA 109, 4449–4454 (2012).References 46 and 47 describe approaches for using the FIB to thin cryo-specimens for high-resolution TEM imaging.
- 48.Knott G, Rosset S & Cantoni M Focussed ion beam milling and scanning electron microscopy of brain tissue. J. Vis. Exp. 2011, e2588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bushby AJ et al. Imaging three-dimensional tissue architectures by focused ion beam scanning electron microscopy. Nat. Protoc. 6, 845–858 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Heymann JA et al. 3D imaging of mammalian cells with ion-abrasion scanning electron microscopy. J. Struct. Biol. 166, 1–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heymann JA et al. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J. Struct. Biol. 155, 63–73 (2006).This is the first report of the use of FIB-SEM imaging to obtain a 3D-volume reconstruction of a cell or tissue specimen.
- 52.Bennett AE et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 5, el000591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Voorhis WC, Hair LS, Steinman RM & Kaplan G Human dendritic cells. Enrichment and characterization from peripheral blood. J. Exp. Med. 155, 1172–1187 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felts RL et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc. Natl. Acad. Sci. USA 107, 13336–13341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Do T et al. Three-dimensional imaging of HIV-1 virological synapses reveals membrane architectures involved in virus transmission. J. Virol. 88,10327–10339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Armer HE et al. Imaging transient blood vessel fusion events in zebrafish by correlative volume electron microscopy. PLoS One 4, e7716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy GE et al. Ion-abrasion scanning electron microscopy reveals distorted liver mitochondrial morphology in murine methylmalonic acidemia. J. Struct. Biol. 171, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider P, Meier M, Wepf R & Muller R Serial FIB/S EM imaging for quantitative 3D assessment of the osteocyte lacuno-canalicular network. Bone 49, 304–311 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Glancy B et al. Mitochondrial reticulum for cellular energy distribution in muscle. Nature 523, 617–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narayan K et al. Multi-resolution correlative focused ion beam scanning electron microscopy: applications to cell biology. J. Struct. Biol. 185, 278–284 (2014).This paper describes some of the recent advances in increasing the scope and robustness of FIB-SEM imaging of biological samples.
- 61.Schertel A et al. Cryo FIB-SEM: volume imaging of cellular ultrastructure in native frozen specimens. J. Struct. Biol. 184, 355–360 (2013).This study describes cryo-FIB-SEM, in which biological samples are FIB-milled and imaged by the SEM at cryogenic temperatures.
- 62.Villinger C et al. FIB/S EM tomography with TEM-like resolution for 3D imaging of high-pressure frozen cells. Histochem. Cell Biol. 138, 549–556 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Stell WK Correlated light + electron microscope observations on Golgi preparations of goldfish retina. J. Cell Biol. 23, 89A (1964).14228521 [Google Scholar]
- 64.Hanker JS, Deb C, Wasserkrug HL & Seligman AM Staining tissue for light and electron microscopy by bridging metals with multidentate ligands. Science 152,1631–1634 (1966). [DOI] [PubMed] [Google Scholar]
- 65.Seligman AM, Ueno H, Wasserkrug HL & Hanker JS Esterase method for light and electron microscopy via the formation of osmiophilic diazothioethers (1, 2). Ann. Histochim. 11, 115–129 (1966). [PubMed] [Google Scholar]
- 66.de Boer P, Hoogenboom JP & Giepmans BN Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods 12, 503–513 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Zhang P Correlative cryo-electron tomography and optical microscopy of cells. Curr. Opin. Struct. Biol. 23, 763–770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Modla S & Czymmek KJ Correlative microscopy: a powerful tool for exploring neurological cells and tissues. Micron 42, 773–792 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Sjollema KA, Schnell U, Kuipers J, Kalicharan R & Giepmans BN Correlated light microscopy and electron microscopy. Methods Cell Biol. Ill, 157–173 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Hanein D & Volkmann N Correlative light-electron microscopy. Adv. Protein Chem. Struct. Biol. 82, 91–99 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Rigort A, Villa E, Bauerlein FJ, Engel BD & PLitzko JM Integrative approaches for cellular cryo-electron tomography: correlative imaging and focused ion beam micromachining. Methods Cell Biol. Ill, 259–281 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Biel SS, Kawaschinski K, Wittern KP, Hintze U & Wepf R From tissue to cellular ultrastructure: closing the gap between micro- and nanostructural imaging. J. Microsc. 212, 91–99 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Kukulski W et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 192,111–119 (2011).This paper describes an approach for getting extremely accurate correlation between LM and EM images of cryogenic samples.
- 74.Watanabe S et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods 8, 80–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perkovic M et al. Correlative light- and electron microscopy with chemical tags. J. Struct. Biol. 186, 205–213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sosinsky GE, Giepmans BN, Deerinck TJ, Gaietta GM & Ellisman MH Markers for correlated light and electron microscopy. Methods Cell Biol. 79, 575–591 (2007).This report provides a detailed list of markers and approaches available for CLEM.
- 77.Brown E & Verkade P The use of markers for correlative light electron microscopy. Protoplasma 244, 91–97 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ & Ellisman MH Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods 2, 743–749 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Gaietta G et al. Multicolor and electron microscopic imaging of connexin trafficking. Science 296, 503–507 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Shu X et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, el001041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grabenbauer M et al. Correlative microscopy and electron tomography of GFP through photooxidation. Nat. Methods 2, 857–862 (2005). [DOI] [PubMed] [Google Scholar]
- 82.McDonald KL A review of high-pressure freezing preparation techniques for correlative light and electron microscopy of the same cells and tissues. J. Microsc. 235, 273–281 (2009).This review provides a detailed description of high-pressure freezing protocols used to avoid ultrastructural artifacts commonly seen with conventionally fixed biological samples.
- 83.Taylor KA & Glaeser RM Retrospective on the early development of cryoelectron microscopy of macro molecules and a prospective on opportunities for the future. J. Struct Biol. 163, 214–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koster AJ et al. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120, 276–308 (1997). [DOI] [PubMed] [Google Scholar]
- 85.McIntosh R, Nicastro D & Mastronarde D New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 15, 43–51 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Al-Amoudi A et al. Cryo-electron microscopy of vitreous sections. EMB0 J. 23, 3583–3588 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harapin J et al. Structural analysis of multicellular organisms with cryoelectron tomography. Nat. Methods 12, 634–636 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Kamino T, Yaguchi T, Ohnishi T, Ishitani T & Osumi M Application of a FIB-STEM system for 3D observation of a resin-embedded yeast cell. J. Electron Microsc. (Tokyo) 53, 563–566 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Hsieh C, Schmelzer T, Kishchenko G, Wagenknecht T & Marko M Practical workflow for cryo focused-ion-beam milling of tissues and cells for cryo-TEM tomography. J. Struct. Biol. 185, 32–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hayles MF et al. The making of frozen-hydrated, vitreous lamellas from cells for cryo-electron microscopy. J. Struct. Biol. 172,180–190 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Evans CL & Xie XS Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 1, 883–909 (2008). [DOI] [PubMed] [Google Scholar]
- 92.Petibois C Imaging methods for elemental, chemical, molecular, and morphological analyses of single cells. Anal. Bioanal. Chem. 397, 2051–2065 (2010). [DOI] [PubMed] [Google Scholar]
- 93.Fletcher JS & Vickerman JC Secondary ion mass spectrometry: characterizing complex samples in two and three dimensions. Anal. Chem. 85, 610–639 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Ostrowski SG, Van Bell CT, Winograd N & Ewing AG Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science 305, 71–73 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fletcher JS, Lockyer NP, Vaidyanathan S & Vickerman JC T0F-SIMS 3D biomolecular imaging of Xenopus laevis oocytes using buckminsterfullerene (C-60) primary ions. Anal. Chem. 79, 2199–2206 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Szakal C, Narayan K, Fu J, Lefman J & Subramaniam S Compositional mapping of the surface and interior of mammalian cells at submicrometer resolution. Anal. Chem. 83,1207–1213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.James SA et al. Quantification of ZnO nanoparticle uptake, distribution, and dissolution within individual human macrophages. ACS Nano 7, 10621–10635 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Kelly TF & Miller MK Invited review article: atom probe tomography. Rev. Sci. Instrum. 78, 031101 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Allen JE et al. High-resolution detection of Au catalyst atoms in Si nanowires. Nat. Nanotechnol. 3, 168–173 (2008). [DOI] [PubMed] [Google Scholar]
- 100.Thompson K et al. In situ site-specific specimen preparation for atom probe tomography. Ultramicroscopy 107, 131–139 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Langford RM & Clinton C In situ lift-out using a FIB-SEM system. Micron 35, 607–611 (2004). [DOI] [PubMed] [Google Scholar]
- 102.Narayan K, Prosa TJ, Fu J, Kelly TF & Subramaniam S Chemical mapping of mammalian cells by atom probe tomography. J. Struct. Biol. 178, 98–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stegmann FI., Dömer H, Rosenkranz R & Zschech E Efficient target preparation by combined pulsed laser ablation and FIB milling. Microsc. Microanal. 17, 658–659 (2011). [Google Scholar]
- 104.Smith C Microscopy: two microscopes are better than one. Nature 492, 293–297 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Liu T, Jones C, Seyedhosseini M & Tasdizen T A modular hierarchical approach to 3D electron microscopy image segmentation. J. Neurosci. Methods 226, 88–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nunez-Iglesias J, Kennedy R, Plaza SM, Chakraborty A & Katz WT Graph-based active learning of agglomeration (GALA): a Python library to segment 2D and 3D neuroimages. Front. Neuroinform. 8, 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chklovskii DB, Vitaladevuni S & Scheffer LK Semi-automated reconstruction of neural circuits using electron microscopy. Curr. Opin. Neurobiol. 20, 667–675 (2010).This report describes an approach for segmenting and reconstructing specific shapes (here, neurons) in a large 3D EM image volume.
- 108.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IM0D. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 109.Hartnell LM, Earl LA, Bliss D, Moran A & Subramaniam S Imaging cellular architecture with 3D SEM In Encyclopedia of Cell Biology Vol. 2 (Eds. Bradshaw R & Stahl P) 44–50 (Academic Press, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


