Abstract
Glaucoma is a leading cause of blindness worldwide. In primary open angle glaucoma (POAG) patients, impaired trabecular meshwork (TM) function results in elevated intraocular pressure (IOP), which is the primary risk factor of developing optic neuropathy. Our previous studies showed that Wnt signaling pathway components are expressed in the human TM (HTM), and the Wnt inhibitor, secreted frizzled-related protein 1 (SFRP1) is elevated in the glaucomatous TM (GTM). Elevated SFRP1 increased IOP in mice eyes and in perfusion cultured anterior segments of the human eye. However, the cause of elevated SFRP1 in the GTM remains unknown. Promoter methylation plays a key role in regulating SFRP1 expression in certain cancer cells. In light of this, we studied whether promoter methylation is also involved in SFRP1 differential expression in the TM. Two normal TM (NTM) and two GTM cell strains were cultured for an additional 7 days after they were confluent. RNA and genomic DNA (gDNA) were isolated simultaneously to compare SFRP1 expression levels by quantitative PCR (qPCR) and to determine SFRP1 promoter methylation status by bisulfite conversion and methylation-sensitive high resolution melting analysis (MS-HRM). To study whether DNA methylation inhibitors affect SFRP1 expression in TM cells, the four TM cell strains were treated with or without 2 μM 5-aza-2′-deoxycytidine (AZA-dC) for 4 days. RNA was isolated to compare SFRP1 expression by qPCR. In addition, a human cancer cell line, NCI-H460, was used as a positive control. We found that the two GTM cell strains had significantly higher expression levels of SFRP1 than the two NTM cell strains. However, the SFRP1 promoter of all four TM cell strains was unmethylated. In addition, AZA-dC treatment did not affect SFRP1 expression in any of the TM cell strains (n = 3, p > 0.05). In contrast, the hypermethylated SFRP1 promoter of NCI-H460 cells was partially demethylated by the same treatment. AZA-dC treatment also elevated SFRP1 expression by approximately two fold in NCI-H460 cells (n = 3, p < 0.01). Our data suggest that the differential expression of SFRP1 in HTM cells is not due to differential promoter methylation.
Keywords: glaucoma, intraocular pressure, trabecular meshwork, SFRP1, DNA methylation, 5-aza-2′-deoxycytidine
1. Introduction
Glaucoma is a leading vision-threatening disease worldwide. Although the mechanism of this ocular disease is not clear, numerous studies have shown that elevated IOP is the primary risk factor for developing the characteristic optic neuropathy associated with this disease (AGIS, 2000; Heijl et al., 2002; Kass et al., 2002; Lichter et al., 2001).
In POAG patients, IOP elevation is due to increased aqueous humor outflow resistance, the majority of which arises from the TM (Tektas and Lutjen-Drecoll, 2009). Abnormal cell signaling pathway activities damage the TM, increase outflow resistance, and elevate IOP (Fleenor et al., 2006; Mao et al., 2010; Shepard et al., 2010; Wang et al., 2008; Wordinger and Clark, 2007; Wordinger et al., 2007, 2008).
One of the cell signaling pathways involved in glaucoma pathogenesis is the Wnt signaling pathway (Wang et al., 2008). In our previous study, elevated SFRP1 expression was observed in 6 GTM cell strains compared to 6 NTM cell strains at both mRNA and protein levels (Wang et al., 2008). The Wnt pathway is involved in proliferation, development, and homeostasis (Logan and Nusse, 2004). Because of the importance of the Wnt signaling pathway, Wnt signaling activity is under fine control. Otherwise, diseases may result from dysregulated Wnt signaling activity. For example, constitutively activated Wnt signaling is associated with many cancers, while in patients with low bone marrow mass and osteoporosis-pseudoglioma syndrome, this pathway is often suppressed (Levasseur et al., 2005). As well as cancers and bone diseases, the Wnt pathway is a causative factor for a range of ocular diseases including inherited retinal degenerations, Grave’s ophthalmopathy, keratoconus, as well as POAG (Mao et al., 2010). We also reported that the secreted Wnt signaling pathway inhibitor SFRP1 is elevated in GTM cells (Wang et al., 2008). In addition, exogenous SFRP1 induced ocular hypertension in both ex vivo perfusion cultured human anterior segments and mouse eyes in vivo. More importantly, this induction could be blocked by a Wnt pathway activator, which further supports the critical role of SFRP1 and the Wnt pathway in glaucoma. The SFRP1 protein is structurally similar to the Wnt pathway receptor Frizzled but lacks the transmembrane and intracellular domains (Finch et al., 1997; Uren et al., 2000). SFRP1 is able to bind to the pathway activator Wnt, which sequesters Wnt proteins and therefore down-regulates the Wnt signaling activity (Uren et al., 2000).
Although the mechanism(s) responsible for SFRP1 elevation in GTM cells is unknown, it is clear that DNA methylation-mediated SFRP1 repression exists in a number of tumor cells (Bovolenta et al., 2008; Rubin et al., 2006). DNA methylation-mediated gene repression functions in both prokaryotes and eukaryotes (Baylin and Ohm, 2006; Jones and Laird, 1999). The 5′ position of the cytosine pyrimidine ring can be methylated by DNA methyl-transferases (DNMTs). In mammals, DNA methylation usually occurs at the cytosine in CpG dinucleotides. Most of the CpG dinucleotides in the promoter region are in clusters close to the transcriptional start site. Therefore, they are also called CpG islands. Most mammalian promoters are not methylated, which permits gene expression. However, many tumor suppressor genes, including SFRP1, are repressed due to the methylation of their CpG islands (Fukui et al., 2005; Suzuki et al., 2002). Lack of SFRP1 leads to aberrantly high Wnt signaling activity, unlimited cell proliferation, and tumor formation. On the other hand, SFRP1 expression can be restored by CpG island demethylation (Suzuki et al., 2002).
Since epigenetic promoter methylation regulates SFRP1 expression in tumor cells, we determined whether DNA methylation is also a key regulator of SFRP1 expression in NTM and GTM cells.
2. Material and methods
2.1. Cell culture
HTM cell strains were a kind gift from Alcon Research, Ltd. (Fort Worth, TX). Two NTM cell strains and two GTM cell strains were cultured in DMEM-low glucose medium supplemented with 2 mM glutamine, streptomycin, penicillin (Thermo-scientific, Worcester, MA), and 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Cells were passaged with Cytodex-3 microcarrier beads (Sigma–Aldrich, St. Louis, MO) as previously described (Steely et al., 1992). Confluent HTM cells in 35 mm dishes or 12-well plates or equivalent were cultured for an additional 7 days before any manipulation. Confluent TM cultures, instead of subconfluent ones were used because a) we tried to mimic in vivo conditions, under which the TM cells are confluent; and b) we found that the expression of SFRP1 in TM cultures is significantly affected by their confluence (Wang et al., 2008), and inconsistent cell confluence may dramatically affect the results. HTM cell donor information is listed in Table 1. The human lung cancer cell line, NCI-H460, was purchased from ATCC (Manassas, VA). NCI-H460 cells were cultured with the same medium as HTM cells.
Table 1.
HTM cell donor information.
| Cell strain | Age | Sex | Passage |
|---|---|---|---|
| NTM1054–05 | 58 | F | 6 |
| NTM186–06 | 80 | F | 6 |
| GTM198–04 | 80 | M | 9 |
| GTM304–04 | 75 | F | 7 |
2.2. AZA-dC treatment
Confluent HTM cells cultured for an additional 7 days in 35 mm or 12-well plates or equivalent were treated with or without the DNA methylation inhibitor, AZA-dC (Sigma) at 2 μM for 4 days.
Alternatively, HTM cells cultured to 80% confluent in 6-well plates were treated with or without AZA-dC at 2 μM for 4 days. At the end of treatment, cells in all the wells appeared to be confluent.
2.3. gDNA and RNA isolation
gDNA and RNA were extracted from HTM cells cultured in 35 mm dishes with the Illustra TriplePrep Kit (GE HealthSciences, Piscataway, NJ) according to manufacturer’s instructions. Alternatively, the RNAqueous Kit (Ambion, Austin, TX) or RNeasy Micro Kit (Qiagen, Valencia, CA) was used to extract RNA only.
2.4. MS-HRM analysis
100–500 ng gDNA from HTM cells as well as methylated and unmethylated control gDNA (Millipore, Billerica, MA) were bisulfite-converted with the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) or Cell-to-CpG Bisulfite conversion kit (Invitrogen). MS-HRM was performed with a reaction mixture (25 μl) containing 2.5 μl 10× PCR buffer (Sigma), 1.25 μl 25 mM MgCl2, 0.4 μl JumpStart Taq (Sigma), 0.5 μl 10 mM dNTPs (Promega, Madison, WI), 1.25 μl DMSO, 5 μl 5M betaine (Fisher Scientific, Pittsburgh, PA), 0.1 μl 0.1 mM forward/reverse primer, 1.25 μl 20× EvaGreen (Biotium, Hayward, CA), and 0.37 μl of the reference dye Rox. The thermoprofile was 95 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s, consisting of 40 cycles followed by a dissociation curve. SFRP1 primers were designed by Dr. Jim Herman at Johns Hopkins University (Maryland, MD). The MS-HRM study was performed in the MX-3000p thermocycler (Stratagene, Santa Clara, CA). The primers amplify a gDNA region from −25 to +161 relative to the transcriptional start site.
Primer sequences were:
Forward primer 5′ GAGGGTTYGGTYGTAGGAGTTT 3′
Reverse primer 5′ CCCCRACCAATAACRACCCT 3′ (Licchesi et al., 2008)
2.5. qPCR
After DNase I treatment, 2 μg total RNA was used for cDNA synthesis with the High Capacity cDNA Reverse Transcription Kit (AB Applied Biosystems, Carlsbad, California) in 20 μl reaction mix. qPCR was performed with 1 μl cDNA. Protocols were the same as for DNA methylation determination. The expression of SFRP1 was normalized to GAPDH with the ΔΔCt method. All primers were designed so that they either flank an intron or span an exon–exon junction. The thermoprofile was 95 °C for 15 s, 55 °C for 60 s, and 72 °C for 30 s, for a total of 40 cycles. PCR primer sequences were:
SFRP1 forward: 5′ TCTGAGGCCATCATTGAACA 3′
SFRP1 reverse: 5′ TCAGGGGCTTCTTCTTCTTG 3′, giving an amplicon of 118 bp.
GAPDH forward: 5′ GGTGAAGGTCGGAGTCAAC 3′
GAPDH reverse: 5′ CCATGGGTGGAATCATATTG 3′ (Martens et al., 2003), giving an amplicon of 153 bp.
Because the ΔΔCt method compares the relative expression level of one sample to the other, a “reference” sample is required for analysis. To compare SFRP1 expression levels among the 4 HTM cell strains (n = 3 for each cell strain), one of the three GTM198–04 samples was chosen as “reference” and its SFRP1 level was set at 100%. Then, every other sample (2 + 3 + 3 + 3) was compared to this one to obtain their relative SFRP1 expression levels for statistical analysis. To compare SFRP1 expression levels with or without AZA-dC treatment, for each cell strain, an untreated sample was used as “reference” and its SFRP1 level was set at 100%. Then, every other sample (2 untreated and 3 treated of the same strain) was compared to this one to obtain their relative SFRP1 expression levels for statistical analysis.
2.6. Data analysis
Data were analyzed using Student’s t-test, with p < 0.05 considered statistically significant.
3. Results
3.1. Differential expression of SFRP1 in HTM cells
Our previous studies showed that SFRP1 expression is elevated in GTM cells (Wang et al., 2008). To validate the TM cell strains used in this study, we compared SFRP1 expression by qPCR. Four representative cell strains, including two NTM cell strains and two GTM cell strains whose SFRP1 expression levels were significantly higher than the NTM cells (Fig. 1), were used for further analysis.
Fig. 1.
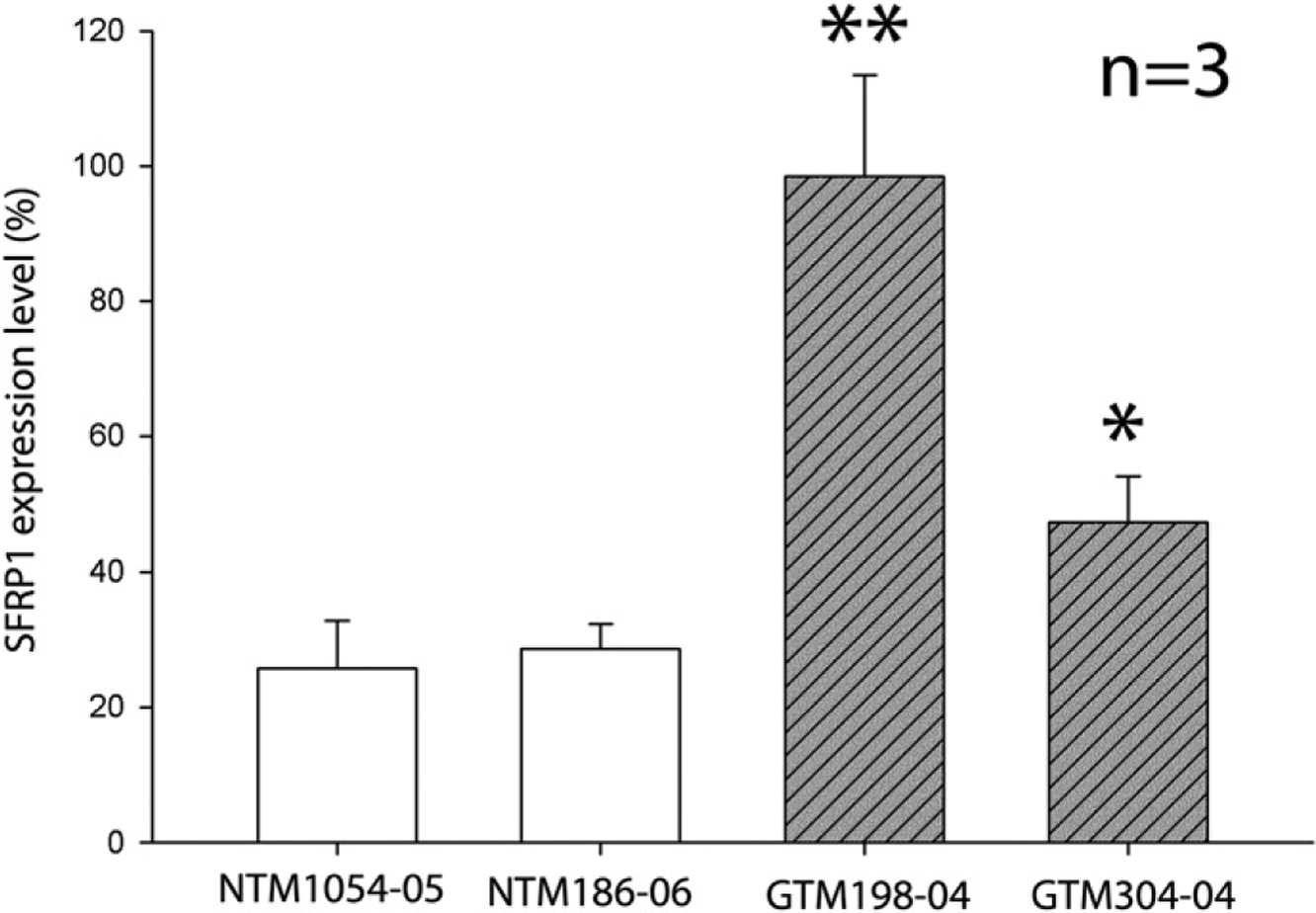
Differential expression of SFRP1 in HTM cells. Four different HTM cell strains were cultured in 35 mm dishes for an additional 7 days after they were confluent. The expression level of SFRP1 of each cell strain was studied by qPCR. The expression level of SFRP1 was first normalized to GAPDH, and then to a GTM198–04 sample, whose SFRP1 expression level was set at 100%. All experiments were performed in biological triplicates (n = 3). Bars = means ± standard deviation (SD). *: p < 0.05 in comparison with either NTM cell strain. **: p < 0.01 in comparison with either NTM cell strain.
3.2. The SFRP1 promoter of HTM cells was unmethylated
To study whether the differential expression of SFRP1 in NTM and GTM cells is due to SFRP1 promoter hypermethylation, we used the MS-HRM method (Lorente et al., 2008; Smith et al., 2009; Wojdacz and Dobrovic, 2007). Briefly, after bisulfite treatment, the CG pairs in unmethylated gDNA are converted to AT pairing, while those in methylated gDNA are preserved. If primers are designed to amplify a region containing CpG islands, PCR products will have different temperature of melting (Tm): methylated gDNA will have higher Tm; unmethylated gDNA will have lower Tm; and a mixture of methylated and unmethylated gDNA will show both Tms. This method is reported to be sensitive enough to detect methylation difference by as few as three CpG sites (Smith et al., 2009).
We chose the CpG islands that have been well-documented and whose methylation status is involved in regulating SFRP1 expression (Fig. 2A) (Fukui et al., 2005; Jost et al., 2008; Licchesi et al., 2008; Mao et al., 2010; Nojima et al., 2007). We found that PCR products from fully methylated or unmethylated control human SFRP1 promoter had their Tm at approximately 81 °C and 76 °C, respectively, while all four HTM cell strains showed Tm at 76 °C (Fig. 2B–E, and Table 2). MS-HRM studies also demonstrated high specificity because primer dimers had a small Tm peak at much lower temperature (~73 °C) (Fig. 2F), and gDNA without bisulfite conversion, whether it is unmethylated or methylated, could not be amplified (Fig. 2G).
Fig. 2.
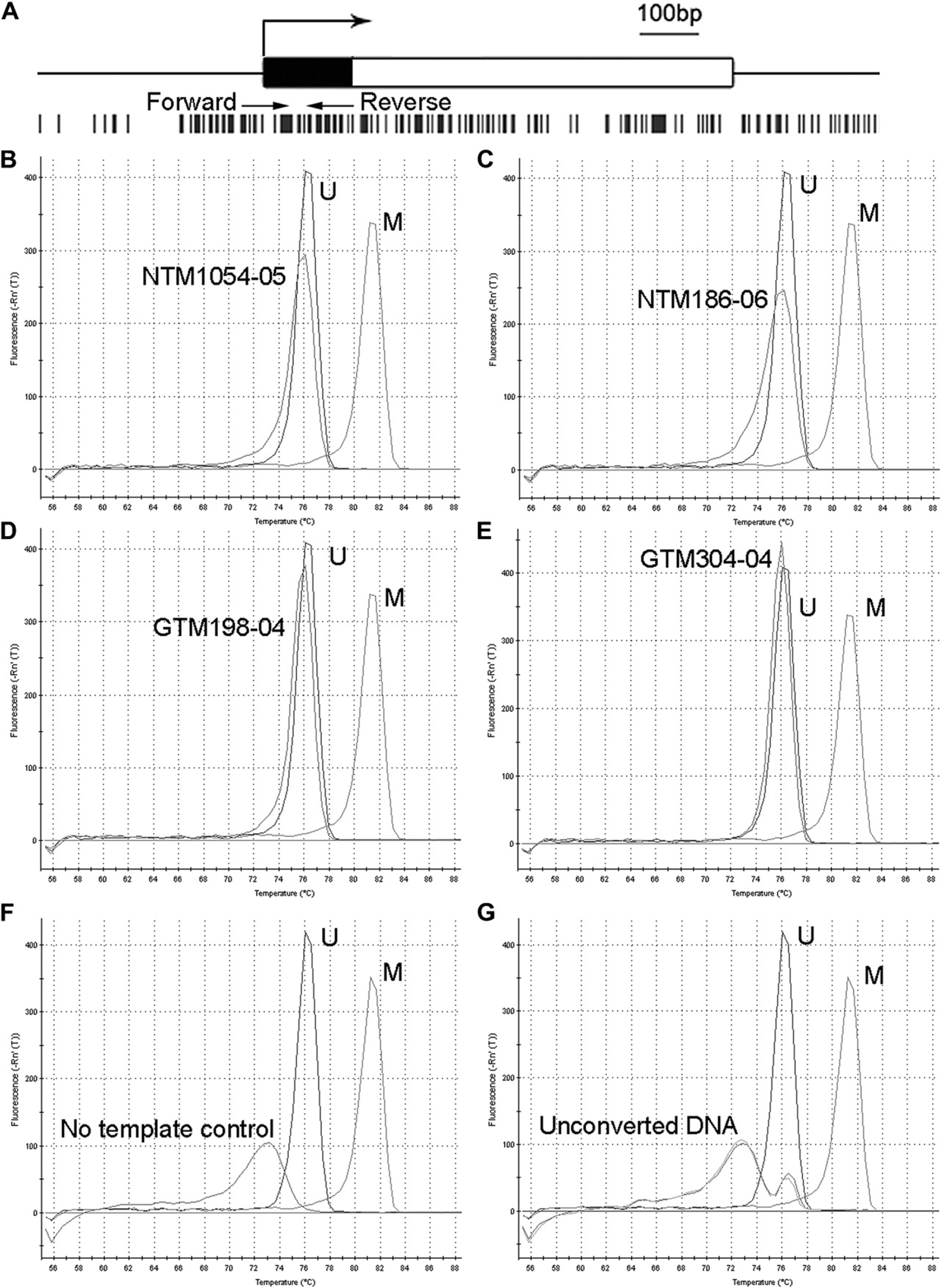
The methylation status of the SFRP1 promoter of HTM cells. The CpG islands in the SFRP1 promoter region are shown in (A). Black bar: the 5′ untranslated region. White bar: the coding region. Large arrow: transcriptional start site. Vertical black bars: CpG sites. Small arrows: the primers for MS-HRM. (B–E): MS-HRM analysis of the SFRP1 promoter of HTM cells. gDNA from the same cells as described in Fig. 1 was used for MS-HRM after bisulfite conversion. The first order derivative of fluorescence intensity was plotted against temperature. Each peak represents the Tm of the PCR product. The difference in the Tm between completely unmethylated control DNA (U) and methylated control DNA (M) was approximately 5 °C. The Tm of all four HTM cell strains was almost the same as that of the unmethylated control DNA. Negative control studies included PCR reactions without DNA template (No template control) (F), or with unmethylated or methylated DNA that were not subjected to bisulfite conversion (Unconverted DNA) (G). All experiments were performed in biological triplicates, and representative data are shown.
Table 2.
The Tm of the HTM cell strains.
| Cell strains | Tm (°C) | N | |
|---|---|---|---|
| Mean | SD | ||
| NTM1054–05 | 76.02 | 0.24 | 3a |
| NTM186–06 | 76.02 | 0.24 | 3a |
| GTM198–04 | 76.18 | 0.24 | 3a |
| GTM304–04 | 76.35 | 0.00 | 3a |
| Methylated DNA control | 81.50 | 0.25 | 3b |
| Unmethylated DNA control | 76.18 | 0.24 | 3b |
Biological replicates.
Technical replicates.
Besides these four HTM cell strains, we screened additional seven HTM cell strains (5 NTM and 2 GTM) with MS-HRM. Similarly, the seven HTM cell strains all had a Tm peak close to the unmethylated gDNA control (data not shown). Therefore, our results suggest that the CpG islands of the SFRP1 promoter in HTM cells were all unmethylated.
3.3. Methylation inhibitor treatment did not elevate SFRP1 expression in HTM cells
Although the CpG islands that we studied were unmethylated, it is possible that other CpG islands within the SFRP1 promoter are differentially methylated, which could account for differential SFRP1 expression between NTM and GTM cells. We tested this possibility by treating HTM cells with the methylation inhibitor AZA-dC, to determine whether SFRP1 promoter demethylation would elevate SFRP1 expression.
We included a human lung cancer cell line, NCI-H460, as a control since the SFRP1 promoter of NCI-H460 cells is hypermethylated (Fukui et al., 2005; Licchesi et al., 2008). Furthermore, these hypermethylated CpG islands can be partially demethylated by AZA-dC, which results in elevated SFRP1 expression (Fukui et al., 2005; Licchesi et al., 2008). We applied the same AZA-dC regimen to HTM and NCI-460 cells. After AZA-dC treatment, the expression of SFRP1 was not significantly elevated in any of the four HTM cell strains (each sample in biological triplicates, p > 0.05) (Fig. 3). Since DNA methylation may be significantly affected by DNA replication and cell division, it is likely that AZA-dC treatment will have different effects on proliferating HTM cells. We tested this possibility with NTM1054–05 and GTM198–04 cells, the two cell strains with the lowest and highest SFRP1 expression, respectively (Fig. 1). However, the expression of SFRP1 was not changed in either cell strain (each sample in biological triplicates, p > 0.05) (Fig. 4).
Fig. 3.
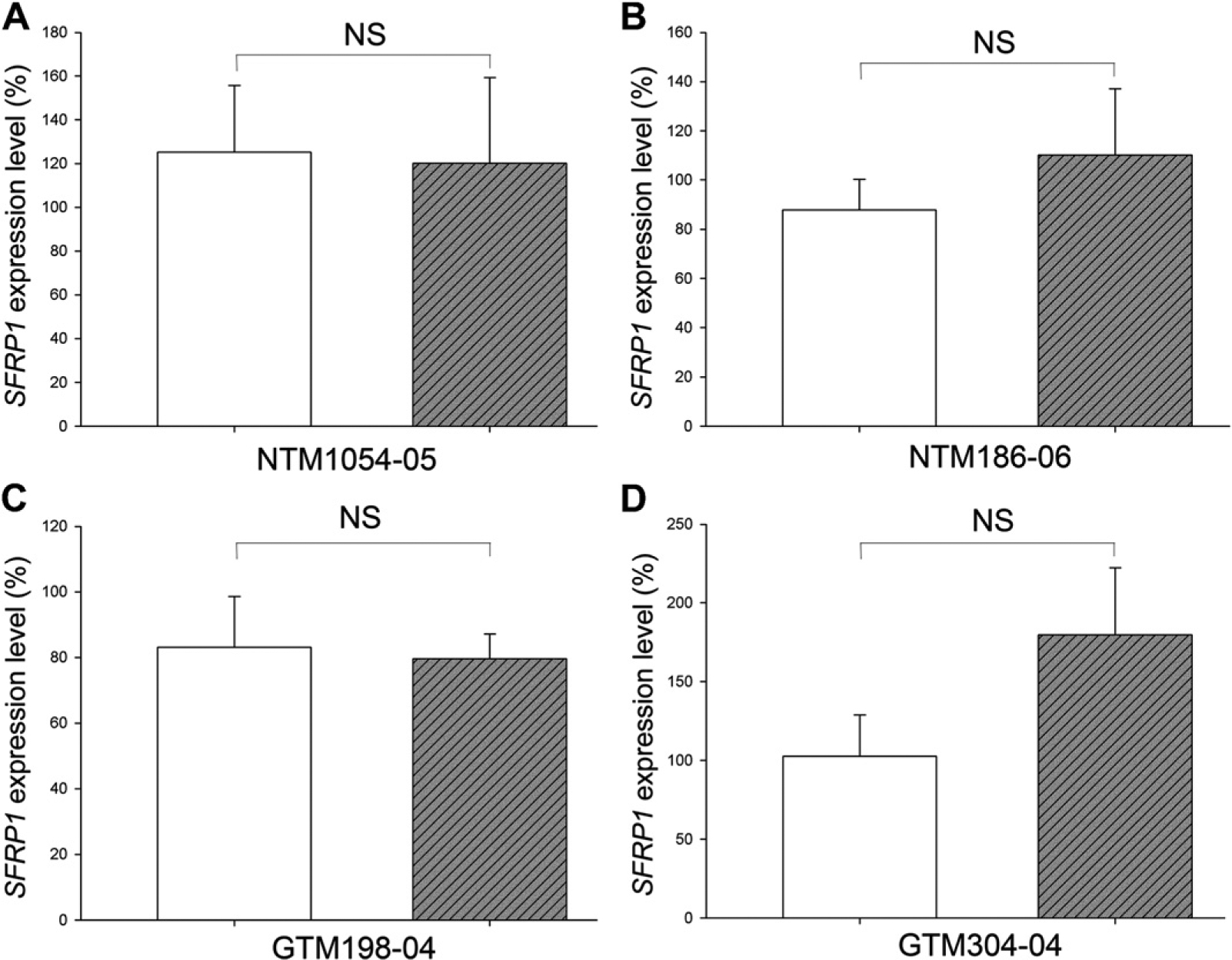
AZA-dC treatment did not affect SFRP1 expression in confluent HTM cells. Confluent HTM cells cultured for additional 7 days were subjected to treatment with (hatched bars) or without (open bars) 2 μM AZA-dC for 4 days. The expression levels of SFRP1 were studied by qPCR. The expression level of SFRP1 was first normalized to GAPDH, and then to a control sample, whose SFRP1 expression level was set at 100%. All experiments were performed in biological triplicates (n = 3). Bars = means ± standard deviation (SD). NS: not significant (p > 0.05).
Fig. 4.
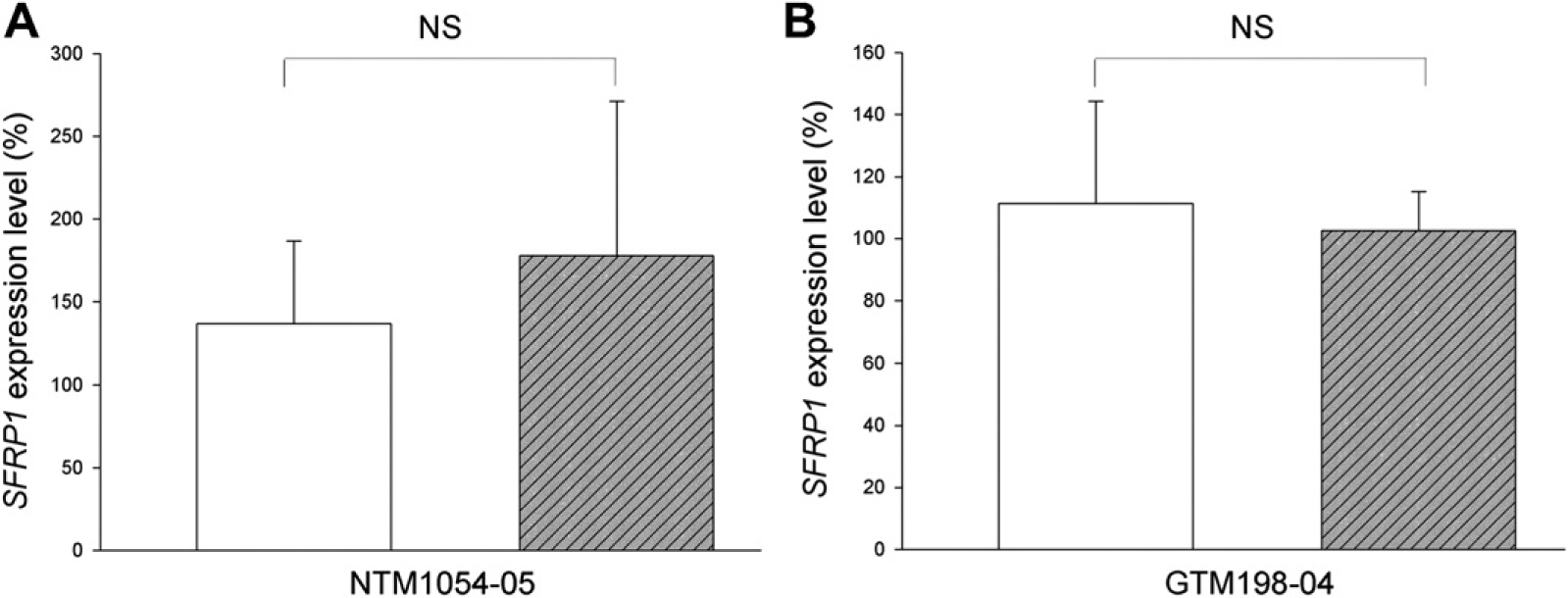
AZA-dC treatment did not affect SFRP1 expression in non-confluent HTM cells. Eighty percent confluent HTM cells were subjected to treatment with (hatched bars) or without (open bars) 2 μM AZA-dC for 4 days. The expression levels of SFRP1 were studied by qPCR. The expression level of SFRP1 was first normalized to GAPDH, and then to a control sample, whose SFRP1 expression level was set at 100%. All experiments were performed in biological triplicates (n = 3). Bars = means ± SD. NS: not significant (p > 0.05).
In contrast, the same treatment increased SFRP1 expression in NCI-H460 cells by more than two fold (64 ± 32% vs. 185 ± 23%, n = 3, p < 0.01) (Fig. 5A). Meanwhile, the SFRP1 promoters became partially demethylated (Fig. 5B and C).
Fig. 5.
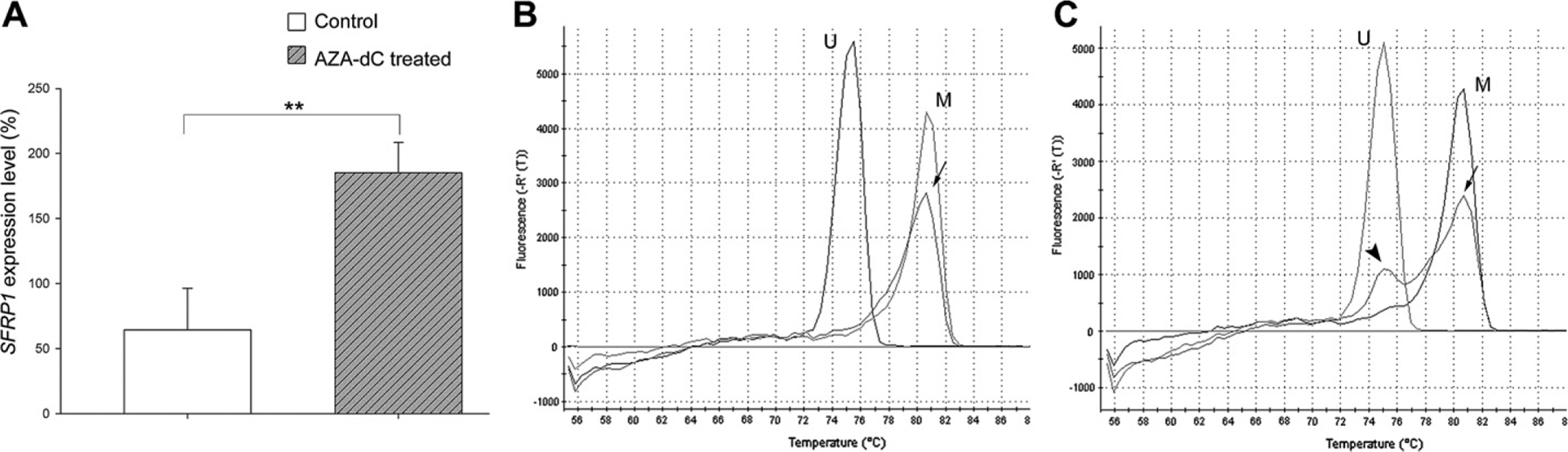
AZA-dC treatment induced SFRP1 expression and demethylation of the SFRP1 promoter in NCI-H460 cells. NCI-H460 cells were treated with or without 2 μM AZA-dC for 4 days. (A) The expression level of SFRP1 of each cell strain was studied by qPCR. The expression level of SFRP1 was first normalized to GAPDH, and then to a control sample, whose SFRP1 expression level was set at 100%. The experiment was performed in biological triplicates (n = 3). Bars = means ± SD. **: p < 0.01. (B–C) MS-HRM studies showed that NCI-H460 cells without AZA-dC treatment had methylated SFRP1 promoters (B, arrow). AZA-dC treatment caused demethylation of the SFRP1 promoter in some cells, which resulted in two peaks on the melting curve (C). The major peak (arrow) represented the methylated promoters while the minor peak (arrowhead) represented the unmethylated promoters. U: unmethylated DNA control. M: methylated DNA control.
4. Discussion
In this study, we used the MS-HRM method to detect the methylation status of the SFRP1 promoter instead of traditional methods like methylation sensitive PCR and bisulfite PCR followed by restriction analysis. If primers are carefully designed, this method is able to simultaneously differentiate methylated DNA, unmethylated DNA, and partially methylated DNA.
By the MS-HRM method, we found the CpG islands in the SFRP1 promoter were all unmethylated, regardless of TM cell strains or SFRP1 expression levels. Also, the DNA methylation inhibitor AZA-dC did not significantly alter SFRP1 expression. These findings suggest that promoter methylation does not play a significant role in regulating SFRP1 expression in the TM. As a positive control, we also evaluated NCI-H460 cancer cells, where SFRP1 expression was controlled by promoter methylation.
Combining the data from our previous study (Wang et al., 2008), we found totally 9 out of the 11 GTM cell strains had higher SFRP1 expression compared to the NTM cell strains with only 6 out of 17 having higher SFRP1 expression (data not shown), which suggests that the majority of the GTM cells have elevated SFRP1 expression. The NTM or GTM cell strains in both studies were selected based on the disease history of organ donors. All GTM cell strains were isolated from donors with clinically diagnosed POAG. Although the IOP data of the organ donors are not available, we believe that those POAG patients had elevated IOP because otherwise they would have been diagnosed as normal tension glaucoma. Since there was SFRP1 dose-dependent IOP elevation in mouse eyes (Wang et al., 2008), it would be very interesting to further investigate whether a similar effect also exists in POAG patients. However, for NTM cells, it is possible that the donors had not developed clinically manifest POAG before death, since age is another important risk factor of POAG. Also, this may explain why some of the NTM cell strains have higher SFRP1 expression levels.
Epigenetics is defined as a change in gene expression without changing the DNA sequence. One of the most frequently studied epigenetic gene regulation mechanism is DNA methylation. DNA methylation has been extensively studied in development and cancers. In contrast, the role of this important mechanism has not been investigated in the TM in glaucoma. Although an association between DNA methylation and SFRP1 expression was not found in this study, we cannot rule out the possibility that DNA methylation plays a role in the regulation of other glaucoma-associated molecules. For example, the expression of TGF-β2 is elevated in the aqueous humor (Inatani et al., 2001; Picht et al., 2001; Tripathi et al., 1994) and TM (Tovar-Vidales et al., 2011) of glaucoma patients. The expression of the BMP inhibitor, GREMLIN, is also elevated in the glaucomatous TM (Wordinger et al., 2007) and optic nerve head cells (Zode et al., 2009). The mechanism(s) responsible for elevated levels of these molecules is still unclear. In cancer cells, the expression of the key pathway components, TGF-β receptors and GREMLIN, are both affected by DNA methylation (Kang et al., 2010, 1999; Munoz-Antonia et al., 2009; Pinto et al., 2003). Therefore, further studies are required to find out whether other glaucoma-associated molecules are regulated by DNA methylation or other epigenetic mechanisms such as histone acetylation or microRNAs.
Acknowledgements
The authors would like to thank Alcon Research, Ltd. (Fort Worth, TX) for the HTM cell strains.
Abbreviations:
- AZA-dC
5-aza-2′-deoxycytidine
- gDNA
genomic DNA
- GTM
glaucomatous trabecular meshwork
- HTM
human trabecular meshwork
- IOP
intraocular pressure
- MS-HRM
methylation sensitive high resolution melting analysis
- NTM
normal trabecular meshwork
- POAG
primary open angle glaucoma
- qPCR
quantitative PCR
- SFRP1
secreted frizzled-related protein 1
- Tm
temperature of melting
- TM
trabecular meshwork
References
- The AGIS Investigators, 2000. The Advanced Glaucoma Intervention Study (AGIS): 7. The relationship between control of intraocular pressure and visual field deterioration. Am. J. Ophthalmol 130, 429–440. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE, 2006. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 6, 107–116. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J, 2008. Beyond Wnt inhibition: new functions of secreted frizzled-related proteins in development and disease. J. Cell. Sci 121, 737–746. [DOI] [PubMed] [Google Scholar]
- Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS, 1997. Purification and molecular cloning of a secreted, frizzled-related antagonist of Wnt action. Proc. Natl. Acad. Sci. U S A 94, 6770–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleenor DL, Shepard AR, Hellberg PE, Jacobson N, Pang IH, Clark AF, 2006. TGFbeta2-induced changes in human trabecular meshwork: implications for intraocular pressure. Invest. Ophthalmol. Vis. Sci 47, 226–234. [DOI] [PubMed] [Google Scholar]
- Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, Yokoi K, Ueda Y, Shimokata K, Sekido Y, 2005. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene 24, 6323–6327. [DOI] [PubMed] [Google Scholar]
- Heijl A, Leske MC, Bengtsson B, Hyman L, Hussein M, 2002. Reduction of intraocular pressure and glaucoma progression: results from the early manifest glaucoma trial. Arch. Ophthalmol 120, 1268–1279. [DOI] [PubMed] [Google Scholar]
- Inatani M, Tanihara H, Katsuta H, Honjo M, Kido N, Honda Y, 2001. Transforming growth factor-beta 2 levels in aqueous humor of glaucomatous eyes. Graefes Arch. Clin. Exp. Ophthalmol 239, 109–113. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW, 1999. Cancer epigenetics comes of age. Nat. Genet 21,163–167. [DOI] [PubMed] [Google Scholar]
- Jost E, Schmid J, Wilop S, Schubert C, Suzuki H, Herman JG, Osieka R, Galm O, 2008. Epigenetic inactivation of secreted frizzled-related proteins in acute myeloid leukaemia. Br. J. Haematol 142, 745–753. [DOI] [PubMed] [Google Scholar]
- Kang SH, Bang YJ, Im YH, Yang HK, Lee DA, Lee HY, Lee HS, Kim NK, Kim SJ, 1999. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene 18, 7280–7286. [DOI] [PubMed] [Google Scholar]
- Kang S, Dong SM, Park NH, 2010. Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol. Oncol 118, 58–63. [DOI] [PubMed] [Google Scholar]
- Kass MA, Heuer DK, Higginbotham EJ, Johnson CA, Keltner JL, Miller JP, Parrish RK 2nd, Wilson MR, Gordon MO, 2002. The ocular hypertension treatment study: a randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch. Ophthalmol 120, 701–713. [DOI] [PubMed] [Google Scholar]
- Levasseur R, Lacombe D, de Vernejoul MC, 2005. LRP5 mutations in osteoporosis-pseudoglioma syndrome and high-bone-mass disorders. Joint Bone Spine 72, 207–214. [DOI] [PubMed] [Google Scholar]
- Licchesi JD, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG, 2008. Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis 29, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter PR, Musch DC, Gillespie BW, Guire KE, Janz NK, Wren PA, Mills RP, 2001. Interim clinical outcomes in the collaborative initial glaucoma treatment study comparing initial treatment randomized to medications or surgery. Ophthalmology 108, 1943–1953. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R, 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell. Dev. Biol 20, 781–810. [DOI] [PubMed] [Google Scholar]
- Lorente A, Mueller W, Urdangarin E, Lazcoz P, von Deimling A, Castresana JS, 2008. Detection of methylation in promoter sequences by melting curve analysis-based semiquantitative real time PCR. BMC Cancer 8, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Wordinger RJ, Clark AF, 2010. Focus on molecules: SFRP1. Exp. Eye Res 91, 552–553. [DOI] [PubMed] [Google Scholar]
- Martens JW, Sieuwerts AM, Bolt-deVries J, Bosma PT, Swiggers SJ, Klijn JG, Foekens JA, 2003. Aging of stromal-derived human breast fibroblasts might contribute to breast cancer progression. Thromb. Haemost 89, 393–404. [PubMed] [Google Scholar]
- Munoz-Antonia T, Torrellas-Ruiz M, Clavell J, Mathews LA, Muro-Cacho CA, Baez A, 2009. Aberrant methylation inactivates transforming growth factor beta receptor I in head and neck squamous cell carcinoma. Int. J. Otolaryngol 2009, 848695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, Hirata K, Itoh F, Tokino T, Mori M, Imai K, Shinomura Y, 2007. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene 26, 4699–4713. [DOI] [PubMed] [Google Scholar]
- Picht G, Welge-Luessen U, Grehn F, Lutjen-Drecoll E, 2001. Transforming growth factor beta 2 levels in the aqueous humor in different types of glaucoma and the relation to filtering bleb development. Graefes Arch. Clin. Exp. Ophthalmol 239, 199–207. [DOI] [PubMed] [Google Scholar]
- Pinto M, Oliveira C, Cirnes L, Carlos Machado J, Ramires M, Nogueira A, Carneiro F, Seruca R, 2003. Promoter methylation of TGFbeta receptor I and mutation of TGFbeta receptor II are frequent events in MSI sporadic gastric carcinomas. J. Pathol 200, 32–38. [DOI] [PubMed] [Google Scholar]
- Rubin JS, Barshishat-Kupper M, Feroze-Merzoug F, Xi ZF, 2006. Secreted WNT antagonists as tumor suppressors: pro and con. Front. Biosci 11, 2093–2105. [DOI] [PubMed] [Google Scholar]
- Shepard AR, Millar JC, Pang IH, Jacobson N, Wang WH, Clark AF, 2010. Adenoviral gene transfer of active human transforming growth factor-{beta}2 elevates intraocular pressure and reduces outflow facility in rodent eyes. Invest. Ophthalmol. Vis. Sci 51, 2067–2076. [DOI] [PubMed] [Google Scholar]
- Smith E, Jones ME, Drew PA, 2009. Quantitation of DNA methylation by melt curve analysis. BMC Cancer 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steely HT, Browder SL, Julian MB, Miggans ST, Wilson KL, Clark AF, 1992. The effects of dexamethasone on fibronectin expression in cultured human trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci 33, 2242–2250. [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB, 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet 31, 141–149. [DOI] [PubMed] [Google Scholar]
- Tektas OY, Lutjen-Drecoll E, 2009. Structural changes of the trabecular meshwork in different kinds of glaucoma. Exp. Eye Res 88, 769–775. [DOI] [PubMed] [Google Scholar]
- Tovar-Vidales T, Clark AF, Wordinger RJ, 2011. Transforming growth factor-beta2 utilizes the canonical Smad-signaling pathway to regulate tissue transglutaminase expression in human trabecular meshwork cells. Exp. Eye Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi RC, Li J, Chan WF, Tripathi BJ, 1994. Aqueous humor in glaucomatous eyes contains an increased level of TGF-beta 2. Exp. Eye Res 59, 723–727. [DOI] [PubMed] [Google Scholar]
- Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS, 2000. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem 275, 4374–4382. [DOI] [PubMed] [Google Scholar]
- Wang WH, McNatt LG, Pang IH, Millar JC, Hellberg PE, Hellberg MH, Steely HT, Rubin JS, Fingert JH, Sheffield VC, Stone EM, Clark AF, 2008. Increased expression of the WNT antagonist sFRP-1 in glaucoma elevates intraocular pressure. J. Clin. Invest 118, 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdacz TK, Dobrovic A, 2007. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 35 e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordinger RJ, Clark AF, 2007. Bone morphogenetic proteins and their receptors in the eye. Exp. Biol. Med. (Maywood) 232, 979–992. [DOI] [PubMed] [Google Scholar]
- Wordinger RJ, Fleenor DL, Hellberg PE, Pang IH, Tovar TO, Zode GS, Fuller JA, Clark AF, 2007. Effects of TGF-beta2, BMP-4, and gremlin in the trabecular meshwork: implications for glaucoma. Invest. Ophthalmol. Vis. Sci 48, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Wordinger RJ, Zode G, Clark AF, 2008. Focus on molecules: gremlin. Exp. Eye Res 87, 78–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zode GS, Clark AF, Wordinger RJ, 2009. Bone morphogenetic protein 4 inhibits TGF-beta2 stimulation of extracellular matrix proteins in optic nerve head cells: role of gremlin in ECM modulation. Glia 57, 755–766. [DOI] [PubMed] [Google Scholar]


