Summary
Inbreeding and low genetic diversity can cause reductions in individual fitness and increase extinction risk in animal populations. Intentional introgression, achieved by releasing genetically diverse individuals into inbred populations, has been used as a conservation tool to improve demographic performance in endangered populations.
By the 1980s, Florida panthers (Puma concolor coryi) had been reduced to a small, inbred population that appeared to be on the brink of extinction. In 1995, female pumas from Texas (P. c. stanleyana) were released in occupied panther range as part of an intentional introgression programme to restore genetic variability and improve demographic performance of panthers.
We used 25 years (1981–2006) of continuous radiotelemetry and genetic data to estimate and model subadult and adult panther survival and cause-specific mortality to provide rigorous sex and age class-specific survival estimates and evaluate the effect of the introgression programme on these parameters.
Genetic ancestry influenced annual survival of subadults and adults after introgression, as F1 generation admixed panthers (ŝ = 0·98) survived better than pre-introgression type panthers (ŝ = 0·77) and other admixed individuals (ŝ = 0·82). Furthermore, heterozygosity was higher for admixed panthers relative to pre-introgression type panthers and positively influenced survival.
Our results are consistent with hybrid vigour; however, extrinsic factors such as low density of males in some areas of panther range may also have contributed to higher survival of F1 panthers. Regardless, improved survival of F1 subadults and adults likely contributed to the numerical increase in panthers following introgression, and our results indicate that intentional admixture, achieved here by releasing individuals from another population, appears to have been successful in improving demographic performance in this highly endangered population.
Keywords: admixture, AIC, Cox proportional hazard, demographic parameter, heterosis, hybridization, model averaging
Introduction
Hybridization, particularly between species, can have serious conservation implications if one of the parental types is rare or endangered because such populations may decline or become extinct because of hybridization (Levin, Francisco-Ortega & Jansen 1996; Rhymer & Simberloff 1996; Allendorf et al. 2001; Wolf, Takebayashi & Rieseberg 2001). However, intentional introgression, achieved by releasing individuals from different subspecies or populations into small, inbred populations, has been used as a conservation tool to restore genetic variability and improve demographic performance of endangered populations (Westemeier et al. 1998; Madsen et al. 1999). Despite the apparent success of some of these genetic manipulations (e.g. Westemeier et al. 1998), they remain controversial because of concerns regarding outbreeding depression (Greig 1979; Edmands 2007), potential loss of genetic integrity of endangered populations (Allendorf et al. 2001), and because few studies have convincingly quantified the effects of intentional introgression on demographic parameters (Beier et al. 2006).
When estimating the effect of hybridization on survival for populations that have experienced introgression, it is important to determine genetic ancestry of individuals because hybrid fitness may differ relative to parental types (Arnold & Hodges 1995; Burke & Arnold 2001). Hybrid superiority can be primarily due to exogenous selection, when hybrids are favoured under specific environmental conditions that vary across time (Grant & Grant 1992) or space (Good et al. 2000). Alternatively, hybrid superiority can be attributed to intrinsic qualities of outbred individuals as predicted by hybrid vigour theory (Shull 1908; Crow 1948). Hybrid vigour, or heterosis, is a phenomenon whereby hybrids exhibit higher fitness relative to inbred parental types due to increased heterozygosity in the F1 generation (Burke & Arnold 2001).
Florida panthers (Puma concolor coryi; Nelson & Goldman 1929) were once distributed across much of the southeastern United States, but currently exist only in one small (c. 100 individuals) breeding population in south Florida (McBride et al. 2008). This population represents the only breeding population of pumas east of the Mississippi River and is listed as endangered under the United States Endangered Species Act (Sullivan 2004). By the late 1980s and early 1990s, the population appeared to be at imminent risk of extinction as the population had declined to ≤30 individuals during this period (McBride et al. 2008) and was severely inbred and lacking in genetic diversity (Roelke, Martenson & O’Brien 1993). In 1995, eight female pumas from Texas (P. c. stanleyana; Goldman 1936) were released into south Florida as part of an intentional introgression programme to restore genetic diversity (Johnson et al. 2010). The population has increased since the initiation of this programme (McBride et al. 2008), but the effects of genetic introgression on specific demographic parameters, such as survival and sources of mortality, remain unclear. Rigorous evaluation of the effects of genetic admixture on survival rates following intentional introgression would have broad implications for the relevance of applied genetic management to the conservation of small, isolated populations (Beier et al. 2006).
We estimated survival and cause-specific mortality of radiocollared subadult and adult panthers from 1981 to 2006 and investigated the influence of multiple intrinsic variables with two main objectives. First, we sought to provide sex- and age class-specific estimates of annual survival and cause-specific mortality for panthers from 1981 to 2006 in order to provide a basic understanding of panther survival within which we could explore the influence of intentional introgression on these parameters. Second, we investigated the influence of (i) genetic ancestry, with respect to hybridization with Texas pumas released during the introgression programme and (ii) genetic diversity, as quantified by individual heterozygosity, on survival and cause-specific mortality of subadult and adult panthers from 1997 to 2006. We hypothesized that survival rates of admixed panthers would be higher than for pre-introgression type panthers and that increased survival would be most evident in the F1 generation, as predicted by hybrid vigour theory (Burke & Arnold 2001).
Materials and methods
FIELD METHODS
Florida panthers and Texas pumas were captured and then monitored by radiotelemetry from a fixed-wing aircraft ≥3 times per week from 1981 to 2006 across the range of the subspecies in south Florida, USA (Kautz et al. 2006) by biologists from the Florida Fish and Wildlife Conservation Commission (FWC) and National Park Service (NPS) using methods described by Belden et al. (1988) and Land et al. (2008). Kittens (<35 days old) were marked at natal dens with transponder chips (Land, Garman & Holt 1998; Benson, Lotz & Jansen 2008), which allowed us to more accurately estimate ages of marked kittens that were subsequently recaptured and radiocollared. Beginning in 1985, captured panthers were vaccinated against several diseases (for details, see Cunningham et al. 2008), which potentially improved their survival.
When radiocollared panthers died, a cursory examination of the carcass and surrounding area was conducted in the field, and a suspected cause of death was assigned if evidence allowed. Eighty-four of 92 (91·3%) carcasses of radiocollared panthers were also necropsied by experienced veterinarians or pathologists who attempted to determine the cause of death. We estimated the date of death from telemetry data and by assessing the condition of the carcasses found in the field.
THE INTROGRESSION PROGRAMME
Eight female pumas from west Texas were released into south Florida at five sites between 29 March and 26 July 1995 (Johnson, Land & Lotz 1997). Two were released in Everglades National Park (ENP) at one site, four in Big Cypress National Preserve (BCNP) at three different sites and two in Fakahatchee Strand Preserve State Park (FSSP) at one site. The introgression programme was undertaken to restore genetic variability to the panther population by mimicking natural gene flow that occurred between Florida panthers and other puma populations before the extirpation of pumas from most of the south-eastern United States. Five of the released Texas pumas reproduced and produced a total of 12 litters and ≥20 kittens.
ESTIMATING AGE CLASSES
We knew the ages (within a few days) for panthers that were handled as kittens at natal dens and subsequently recaptured and radiocollared (n = 54, 36·7% of total). For panthers not marked as kittens, their age was estimated in the field at the time of first capture using tooth wear and pelage characteristics (Ashman & Greer 1976). We examined potential error in survival estimates because of error in age estimates and found it to be small (Appendix S1, Supporting Information). Using both types of estimates, we separated panthers into four age classes: kittens (0–1 years old), subadults (1–2·5 and 1–3·5 years old for females and males, respectively), prime adults (2·5–10 and 3·5–10 years old for females and males, respectively) and older adults (≥10 years old for both sexes). We also combined prime adults and older adults into a single adult category for some of our analyses. As panther kittens were not radiocollared until at least 4–6 months of age, and most mortality of puma kittens may occur ≤3 months after birth (Logan & Sweanor 2001), a different analytical approach was employed to estimate survival of kittens (Hostetler et al. 2010). Thus, we limited our current analyses to subadults and adults. The subadult age class was the period of a panther’s life after independence from its mother until the approximate ages when females and males establish home ranges. We estimated age of independence at just over 1 year (mean = 397 days, SD = 75 days, n = 35; FWC & NPS, unpublished data) from known-age kittens that were radiotracked simultaneously with their mothers (before and after independence). We standardized the beginning of the subadult age class to 1 year for simplicity.
GENETIC VARIABLES
We extracted total genomic DNA from blood and tissue samples obtained from wild-caught panthers and captive pumas from south Florida and west Texas during 1981–2006. We amplified and scored 23 microsatellite loci (F37, F42, FCA43, FCA57, FCA75, FCA90, FCA91, FCA94, FCA95, FCA98, FCA124, FCA133, FCA161, FCA193, FCA 243, FCA249, FCA293, FCA310, FCA369, FCA441, FCA559, FCA566, FCA668) following previously described PCR amplification conditions (Menotti-Raymond et al. 1997, 1999).
A Bayesian procedure, implemented in the program structure (Pritchard, Stephens & Donnelly 2000), was used to identify populations or genetic clusters and to estimate the genetic origin of individuals based on microsatellite allele frequencies. The structure approach assumes departures from both Hardy–Weinberg and complete linkage equilibriums to be indications of population substructure (Pritchard, Stephens & Donnelly 2000). In addition to assigning individuals to various lineages based upon composite microsatellite genotype, the analysis also allows for the estimation of proportion of genetic contribution from each group for individuals of mixed origin. We used results from the structure analysis, along with pedigree results and field evidence, to assign panthers to three groups that reflected the genetic make-up of the south Florida population (pre-introgression type panthers, F1 admixed panthers and other admixed panthers). Pre-introgression type panthers represent the genotypes present on the landscape prior to the introgression programme, and this group is composed mostly of non-admixed Florida panthers, which had no direct non-Florida relatives or < 10% non-Florida genetic contribution based on structure analyses. Additionally, one pre-introgression type panther in the sample of individuals used to investigate the influence of genetic ancestry and heterozygosity on survival (n = 98) was a descendent of a panther from the Everglades genetic population with documented genetic links to Central America (O’Brien et al. 1990; Roelke, Martenson & O’Brien 1993; Culver et al. 2000). Admixed panthers were mostly the descendents of the introduced female Texas pumas released in 1995, except for five radiocollared individuals that shared genotypic similarities with pumas maintained in a large enclosure on the Seminole Indian Reservation (SIR) adjacent to BCNP. Radiocollared panthers were documented entering the enclosure, and captive pumas were documented to have left the enclosure during our study. This captive population contained animals of unknown origins that had a genetic affinity with North American pumas (FWC, NPS, National Cancer Institute, unpublished data).
Differences in fitness between classes of hybrids are possible, and hybrid vigour is predicted to be strongest in the F1 generation (Arnold & Hodges 1995; Burke & Arnold 2001), so we included two classes of admixed panthers in our analyses: F1 and other admixed. We defined F1 admixed panthers as any offspring produced by matings between a Texas female and a pre-introgression type male. As such, F1 panthers in our sample were products of matings between Texas females and males that were either non-admixed Florida panthers or Everglades panthers. Of the 20 known kittens produced by Texas pumas, 15 were F1 admixed panthers (the remaining five were sired by F1 males). Ten of the 12 natal dens of the Texas pumas were visited shortly (< 35 days) after parturition, and all kittens were marked at these dens with transponder chips. Two F1 kittens (one from each litter) were captured from litters produced at the two dens not visited while they were still dependent offspring. Of the 15 known F1 kittens, eight were captured or recaptured and radiocollared, whereas the fates of the other 7 F1 kittens are unknown. These 8 F1 kittens came from six litters, which were produced by mating between four Texas females and four pre-introgression type males.
In summary, all individuals in our radiocollared sample were placed into one of three categories for our survival analyses: pre-introgression type panthers (n = 41), F1 admixed panthers (n = 8) and other admixed panthers (n = 49). We were also able to further classify most individuals from the other admixed category into three finer categories: backcrossed to Florida admixed panthers (n = 16), backcrossed to Everglades admixed panthers (n = 4) and backcrossed to Texas admixed panthers (n = 14). We considered potential differences in survival between pre-introgression type, F1 admixed and these finer admixture distinctions (Appendix S2, Supporting Information), but because admixed panthers beyond the F1 generation did not differ substantially in their survival, we combined all non-F1 admixed panthers into the other admixed category for our main analyses.
We estimated microsatellite-based average individual heterozygosity with the program microsat (Minch, Ruiz-Linares & Goldstein 1995) to investigate the influence of heterozygosity on survival during the same time period as the ancestry analysis. We also compared the distribution of heterozygosity values between panthers of our three ancestry categories using permutation tests (Efron & Tibshirani 1998).
SURVIVAL ANALYSES
We estimated annual survival and examined the effects of covariates using a daily time scale and Cox proportional hazard regression (Cox 1972; Therneau & Grambsch 2000). We right-censored panthers that lost their collars or whose collars failed on the last day that an active signal was heard. An important assumption of survival analyses is that radiocollar failures are independent of mortality (Therneau & Grambsch 2000). We are confident that this assumption was met in our study as we were able to confirm radiocollar failures by subsequent recapture or recovery in 22 of 32 cases.
We organized the data with records for each panther-year combination (Fieberg & DelGiudice 2009). Time in the Cox model (the baseline) was defined as day within the year. When a panther changed age class within a year, we created two records: one record with the younger age class right-censored on the transition day and another record with the older age class left-truncated (staggered entry) on the same day. To account for multiple records for the same animal existing at the same ‘time,’ we estimated robust (‘sandwich’) standard errors clustered by individual (except when including random effects; Fieberg & DelGiudice 2009; Therneau & Grambsch 2000), which were generally extremely close to uncorrected standard errors. We used the Fleming–Harrington method to generate survival estimates from the Cox analysis (Therneau & Grambsch 2000). All survival analyses were performed in r version 2.8.1 (R Development Core Team 2008), using the survival (version 2.34–1) and kinship (version 1.1.0–22) packages and additional code that we developed for our analyses (available on request).
For our survival analyses, we used an information-theoretic approach (Akaike’s Information Criterion; AIC) for model selection and statistical inference (Burnham & Anderson 2002). Information-theoretic approaches allow comparison of non-nested models, selection of models that should best predict future data (from the same statistical population) and model-averaged estimates to address model selection uncertainty (Burnham & Anderson 2002). We calculated AIC values, Akaike differences (ΔAICi, difference between AIC value of the ith model and the top-ranked model) and Akaike weights (wi, the weight of evidence that the ith model is actually the best model of the models being considered given the data) as in Burnham & Anderson (2002). Generally, we considered models with ΔAIC <2 to have substantial empirical support, models with ΔAIC of 2–4 to be plausible models with less empirical support and models with ΔAIC >4 to have much less empirical support. In addition, the sum of the weights of models including a given variable can be interpreted as a measure of its importance, relative to other variables (Burnham & Anderson 2002). We calculated model-averaged estimates of annual survival and unconditional variance for each level of categorical variables and across continuous covariates (Burnham & Anderson 2002; Tinker et al. 2006). To generate these model-averaged estimates of survival, we used all models included in the model selection process for a given analysis, weighted by their Akaike weights. Models with no effects of the covariates being presented were included in the averages (as having the same survival for all values of the covariate); therefore, the model averages represent unconditional estimates of survival (Burnham & Anderson 2002). The unconditional standard errors estimated around the model averages are generally larger than standard errors estimated for parameters of single models and should not be interpreted as tests of statistical significance. We conducted two separate survival analyses: a main analysis to estimate survival from 1981 to 2006 with the entire data set and a subset analysis using data from subadults and prime adults from 1997 to 2006 to investigate the effects of the introgression programme on survival.
In the first set of analyses, we investigated the influence of sex, age class and year, and included 144 radiocollared subadults and adults from 1981 to 2006 (one panther whose death was capture related was excluded). Previous studies have documented differences in survival between sexes and ages of pumas (Logan & Sweanor 2001; Lambert et al. 2006; Robinson et al. 2008) making these logical variables to include in our models. We started by selecting the best model or set of models for sex and age (all models with ΔAIC <2). Because the subadult and prime adult age classes were defined differently for males and females and because we had a priori reason to believe that the patterns of mortality would be different across ages for the two sexes, we only considered the interactive effect of sex and age for the difference between subadults and adults. We considered differences in survival between prime adult and older adult age classes both additively and interactively with sex, which resulted in five sex and age class models.
We used the best model or models (i.e. lowest AIC) from the preceding analysis as base models to investigate potential temporal variation in survival using Gaussian mixed effects models (sometimes referred to within survival analysis as shared frailty models) with year as a random effect (Therneau & Grambsch 2000). Each year was treated as a separate category, except for 1981–1986, which were combined as one category because of small sample sizes. We estimated temporal variance (variance of the random effect) for each model using a Laplace approximation under a maximum likelihood approach (Pankratz, de Andrade & Therneau 2005). Because our intent was to estimate the temporal variance rather than produce an estimate of survival for each year, we counted the random effect as a single parameter for purposes of model comparison (Vaida & Blanchard 2005).
In a second set of analyses, we investigated the potential effects of genetic introgression, in terms of genetic ancestry and genetic diversity, on survival rates. Given that the first offspring produced by a Texas female did not appear in our sample of subadults and adults until 1997, we conducted this subset analysis using data only from 1997 until the end of this study on December 31, 2006. Only two admixed panthers (born during 1995 and 1996) reached 10 years of age by the end of 2006, and both for periods of <1 year; thus, we excluded older adult panthers from these analyses to account for possible differences in survival for older animals. In addition to including an ancestry model where the three ancestry classes were separated (Ancest1), we included ancestry models where other admixed panthers were combined with either pre-introgression type (to test specifically for F1 hybrid vigour; Ancest2) or F1 panthers (to test for general hybrid superiority; Ancest3). We combined these three models with the effects of sex and age class (subadult and prime adult; Age3), and the effect of heterozygosity (Het).
Some radiocollared panthers were removed from the wild and held in captivity for various reasons, and some received medical treatment. We right-censored panthers that were permanently removed from the wild as a result of livestock depredation incidents (n = 2; including 1 F1 panther) on the date of removal. We also right-censored panthers that were temporarily removed for treatment and rehabilitation of injuries judged not to have been fatal (n = 3), capture-related injuries (n = 3; including 1 F1 panther) or for reproductive evaluation (n = 1) on date of removal and then re-entered them into the analyses upon release to the wild. Panthers removed for various reasons as kittens (<1 year old) and later released into the wild (n = 7; including 1 F1 panther) were entered into our analyses either on the day of release (if adults or subadults) or upon reaching the subadult age class after being released as kittens. Panthers removed from the wild because of injuries or illness judged to be fatal (n = 4) were treated as mortalities, and the injury or illness was assigned as cause of death.
To estimate and model the importance of different mortality agents on rates and patterns of mortality for subadult and adult Florida panthers, we performed cause-specific mortality analyses. We attributed mortality of radiocollared panthers to 1 of 4 causes: (1) hit by vehicle, (2) intraspecific aggression, (3) other (included known causes of death such as disease, heart failure and infections unrelated to intraspecific aggression) and (4) unknown (mortalities for which evidence from field and necropsy examinations was insufficient to assign cause of death). We estimated cause-specific annual mortality rates for Florida panthers overall and within categories (sex, age class within sex and ancestry within sex), using the nonparametric cumulative incidence function estimator (NPCIFE; Heisey & Patterson 2006). The NPCIFE is a generalization of the staggered-entry Kaplan–Meier method of survival estimation (Pollock et al. 1989). All cause-specific mortality analyses were performed using SPLUS code from Heisey & Patterson (2006) which we modified for use in R. Additional details of the methods used for these analyses can be found in Appendix S3 (Supporting Information).
Results
AGE, SEX AND YEAR
We recorded 93 deaths among the 144 radiotracked panthers, but these deaths were not distributed evenly by sex or age class. Models that incorporated the interactive effects of sex and age class (either 2 or 3 age classes per sex) had substantially higher empirical support than models with neither or only the effect of sex (Table 1). Survival rates were higher for females than for males, but these varied among age classes (Table 2). For males, prime adults had the highest survival, whereas for females, subadults had the highest survival (Table 2). The highest ranked model included an additive effect of sex and old age, indicating that survival for this age class differed from other age classes, but there was no support for an interactive effect of sex and old age (Table 1). Survival estimates were lower for older adults of both sexes (Table 2). The model with a random effect of year had marginally lower support than the equivalent model with no random effect (Table 1B), and the estimated temporal variance of the hazard rate for model 2 was small (0·115), suggesting that the temporal variance in survival rates was also small.
Table 1.
Model comparison results for the effects of sex, age class (A) and year (B) on Florida panther survival. Year was included as a random effect using the top-ranked sex and age model as a base. For each model, we present the number of parameters, the difference in Akaike’s Information Criterion (ΔAIC) and the Akaike weight (wi)
| Model | Parameters | ΔAIC | wi |
|---|---|---|---|
| A. Sex and age class models | |||
| Sex × Age1a + Olderb | 4 | 0 | 0·658 |
| Sex × Age2c | 5 | 1·98 | 0·245 |
| Sex × Age1 | 3 | 4·69 | 0·063 |
| Sex | 1 | 5·95 | 0·034 |
| Constantd | 0 | 15·25 | 0·000 |
| B. Additive effect of year (random) | |||
| Sex × Age1 + Older | 4 | 0·00 | 0·522 |
| Sex × Age1 + Older + rand(Year)e | 5 | 0·18 | 0·478 |
Age1 divides panthers into subadults (1–2·5 and 1–3·5 years for females and males, respectively) and adults (≥2·5 and ≥3·5 years for females and males, respectively).
Older refers to older adults (≥10 years); the first model therefore has the same older adult effect for both sexes, whereas the second model allows for different older adult effects between sexes.
Age2 divides the panthers into subadults (same as Age1), prime adults (2·5–10 and 3·5–10 years for females andmales, respectively) and older adults (≥10 years).
No predictor variables.
rand(Year) refers to a random effect of year as a categorical variable, with 1981–1986 grouped together (temporal variance).
Table 2.
Model-averaged annual survival rates (ŝ), standard errors (SÊ) and number of Florida panthers tracked (n) in sex and age class categories. All models from Table 1A were used for model averages
| Category | Females | Males | ||||
|---|---|---|---|---|---|---|
| ŝ | SÊ | n | ŝ | SÊ | n | |
| Subadulta | 0·951 | 0·034 | 40 | 0·713 | 0·049 | 54 |
| Prime adultb | 0·872 | 0·023 | 64 | 0·799 | 0·036 | 44 |
| Older adultc | 0·760 | 0·056 | 12 | 0·635 | 0·083 | 11 |
1–2·5 and 1–3·5 years old (estimated) for males and females, respectively.
2·5–10 and 3·5–10 years old (estimated) for males and females, respectively.
≥10 years old (estimated) for both males and females.
GENETIC ANCESTRY AND HETEROZYGOSITY
We recorded 47 deaths among the 98 panthers included in the genetic survival analysis; these deaths were not distributed evenly by genetic ancestry or heterozygosity. There was evidence that genetic ancestry influenced survival as all models with substantial empirical support (ΔAIC ≤2) included an ancestry variable, the ΔAIC of the top-ranked model containing neither ancestry nor heterozygosity was 7·61, and the sum of the weights of models including ancestry variables was 0·928 (Table 3). Ancestry variables included in the top-ranked models were all in agreement that the survival of F1 admixed panthers differed from that of other ancestry categories (sum of weights of such models was 0·890); however, evidence for a difference between other admixed panthers and pre-introgression type panthers was weaker (sum of weights = 0·315; Table 3). Model-specific and model-averaged survival was higher for F1 admixed panthers than for other ancestry classes (Table 4; Fig. 1).
Table 3.
Model comparison results showing top-ranked models (difference in Akaike’s Information Criterion [ΔAIC] <4) for the effects of ancestry, heterozygosity, sex and age class (subadults and prime adults only) on Florida panther survival during 1997–2006. For each model, we present the number of parameters, ΔAIC and the Akaike weight (wi). The full table is presented in Appendix S4 (Supporting Information)
| Model | Parameters | ΔAIC | wi |
|---|---|---|---|
| Sex × Age3a + Ancest2b + Hetc | 5 | 0·00 | 0·244 |
| Sex × Age3 + Ancest2 | 4 | 0·23 | 0·217 |
| Sex × Age3 + Ancest1d | 5 | 1·35 | 0·124 |
| Sex × Age3 + Ancest1 + Het | 6 | 1·98 | 0·090 |
| Sex + Ancest2 + Het | 3 | 2·77 | 0·061 |
| Sex + Ancest2 | 2 | 2·98 | 0·055 |
| Sex × Age3 + Het | 4 | 3·09 | 0·052 |
Age3 differentiates subadult (age 1–2·5 for females and 1–3·5 for males) and prime adult (ages 2·5–10 for females and 3·5–10 for males) panthers.
Ancest2 divides panthers into two ancestry categories: F1 admixed, and other admixed and pre-introgression type combined.
Het refers to individual average heterozygosity.
Ancest1 divides panthers into three ancestry categories: F1 admixed, other admixed and pre-introgression type.
Table 4.
Model-specific estimated annual survival rates (with SÊ and number of panthers) by ancestry, sex and age class, with the estimating models and their Akaike weights (wi)
| Group | Pre-introgression type | Other admixed | F1 admixed | Model | wi |
|---|---|---|---|---|---|
| All | 0·775 (0·039; 41) | 0·821 (0·036; 49) | 0·978 (0·021; 8) | Ancest1 | 0·007 |
| Females | 0·837 (0·041; 20) | 0·864 (0·035; 22) | 0·982 (0·018; 6) | Sex + Ancest1 | 0·025 |
| Males | 0·713 (0·055; 21) | 0·758 (0·056; 27) | 0·967 (0·032; 2) | Sex + Ancest1 | 0·025 |
| Female subadultsa | 0·953 (0·049; 9) | 0·964 (0·036; 14) | 0·995 (0·007; 4) | Sex × Age3 + Ancest1 | 0·124 |
| Female prime adultsb | 0·803 (0·047; 19) | 0·848 (0·041; 22) | 0·977 (0·022; 6) | Sex × Age3 + Ancest1 | 0·124 |
| Male subadultsc | 0·606 (0·088; 14) | 0·686 (0·073; 22) | 0·949 (0·050; 2) | Sex × Age3 + Ancest1 | 0·124 |
| Male prime adultsd | 0·789 (0·059; 12) | 0·836 (0·056; 13) | 0·976 (0·024; 2) | Sex × Age3 + Ancest1 | 0·124 |
Female subadults were 1–2·5 years old.
Female prime adults were 2·5–10 years old.
Male subadults were 1–3·5 years old.
Male prime adults were 3·5–10 years old.
Fig. 1.
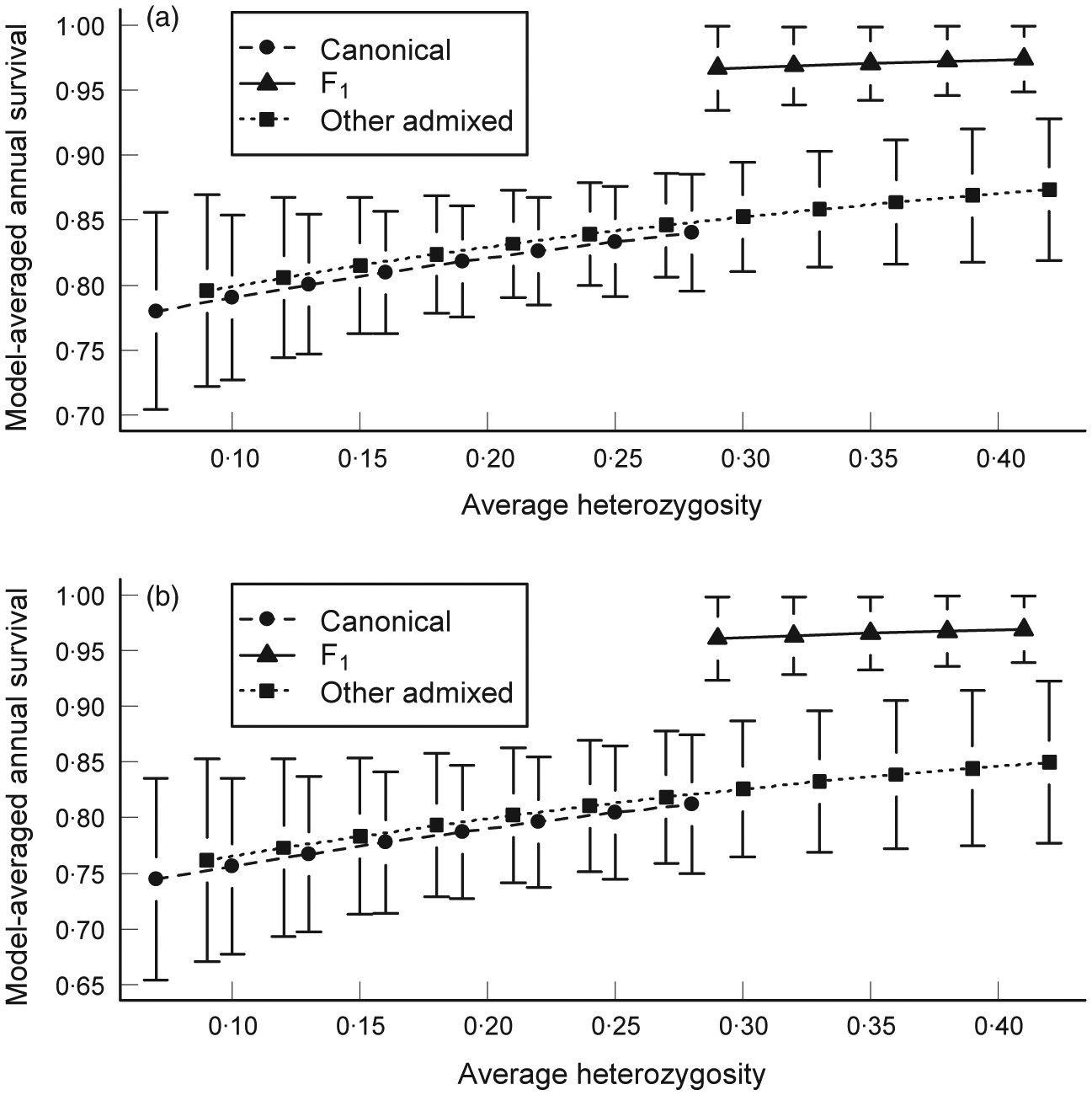
Model-averaged effects of ancestry and heterozygosity on annual survival of (a) female and (b) male prime adult Florida panthers from 1997 to 2006. Genetic factors were additive to sex and age class in all models. The range of heterozygosity values presented for each ancestry category approximately corresponds to those found in the data set (all subadult and prime adult panthers from 1997 to 2006 with that ancestry), except for one outlier with high heterozygosity excluded from the range for the pre-introgression type. Error bars represent unconditional standard errors.
There was also evidence that average heterozygosity influenced survival of subadult and prime adult panthers after genetic introgression (1997–2006), as the sum of the weights of models including heterozygosity was 0·537 (Table 3) and model-averaged annual survival probability increased with heterozygosity (Fig. 1). Average heterozygosity was highest for F1 (mean = 0·337, SD = 0·035, n = 8), intermediate for other admixed (mean = 0·261, SD = 0·065, n = 49) and lowest for pre-introgression type panthers (mean = 0·161, SD = 0·063, n = 41). Each ancestry group differed significantly in average heterozygosity from both other groups in pairwise comparisons (all P ≤ 0·002; permutation tests). There was also evidence that heterozygosity positively influenced survival even within ancestry groups, as the top-ranked model included both factors (Table 3). Although model-averaged survival was similar for pre-introgression type and other admixed panthers at a given level of heterozygosity, heterozygosity ranged higher for other admixed individuals, leading to an increased probability of survival (Fig. 1; Table 4).
The full AIC table and selected Cox model coefficients for the genetic comparisons are presented in Appendix S4 (Supporting Information). Robust z-tests on individual coefficients (in the context of sex and age class) also indicated that F1 admixed panthers survived better than panthers in other ancestry classes and that heterozygosity positively influenced survival (Appendix S4, Supporting Information).
CAUSE-SPECIFIC MORTALITY
The greatest cause of mortality for radiocollared Florida panthers was intraspecific aggression, followed by mortality from unknown causes, vehicles and other (Appendix S3, Supporting Information). When sexes were combined, pre-introgression type panthers had a higher level of mortality because of intraspecific aggression than admixed panthers (z = 2·404, P = 0·016, risk ratio = 3·06). Increasing heterozygosity also significantly decreased the risk because of intraspecific aggression (z = −2·943, P = 0·003, risk ratio = 0·480). Additional details of the results of these analyses are available in Appendix S3 (Supporting Information).
Discussion
We provide evidence of hybrid vigour in the panther population following intentional introgression, as genetic ancestry and heterozygosity influenced survival of subadult and prime adult panthers. Our findings correspond very closely to the outcomes of hybridization on an inbred population predicted by theory in that survival and heterozygosity were higher for F1 admixed individuals compared with pre-introgression type Florida panthers, but evidence for higher survival in admixed generations beyond the F1 was far weaker (Crow 1948; Burke & Arnold 2001). It should be noted that we did not compare survival between F1 panthers and the released Texas females; thus, we investigated hybrid vigour by comparing survival between F1 panthers and only one of the parental populations.
Hybrid vigour was first recognized by crossing divergent lines of agricultural plants and extensive research in agricultural genetics has confirmed the phenomenon (Crow 1948; Whitlock, Ingvarsson & Hatfield 2000; Birchler, Auger & Riddle 2003). Much less is known about the fitness consequences of hybridization for plants and animals in nature (Grant & Grant 1992; Campbell & Waser 2001). For animals, hybrid vigour has been invoked by previous studies for a variety of species including insects (Ebert et al. 2002), fish (Rosenfield et al. 2004), salamanders (Fitzpatrick & Shaffer 2007) and corals (Slattery et al. 2008). We are unaware of previous studies of free-ranging mammals demonstrating superior fitness of admixed individuals (or superiority of a component of fitness, as in our example) that was clearly attributed to intrinsic hybrid vigour rather than hybrids being favoured under specific environmental conditions. Broadly speaking, hybrid fitness may be influenced by endogenous or exogenous selection, and hybrid superiority is often assumed to be primarily due to the latter (Burke & Arnold 2001) as hybrids may be favoured under environmental conditions that vary across time (Grant & Grant 1992) or space (Moore 1977; Good et al. 2000). Our demonstration of higher survival in F1 admixed panthers relative to an inbred parental type is consistent with an intrinsic hybrid vigour effect (Burke & Arnold 2001), and temporal variation in environmental conditions were minimized in our study by limiting the analysis to years during which admixed and pre-introgression type panthers coexisted on the south Florida landscape. However, environmental conditions could have varied spatially across the range of the panther and potentially contributed to the higher survival of F1 admixed panthers in some instances. We suggest that variation in at least two extrinsic factors, panther density and habitat conditions, could have influenced panther survival across ancestry categories.
First, survey results and capture efforts suggested that adult males were at very low density in ENP and portions of BCNP (FWC & NPS, unpublished data). Low local density of adult males could have provided a survival advantage to subadult and adult F1 panthers in these areas (n = 4) by decreasing the risk of death by intraspecific aggression. This possibility is consistent with our results as pre-introgression type panthers were more frequently killed by intraspecific aggression than admixed panthers. Furthermore, ENP was likely devoid of females when the Texas females were released, which could have contributed to higher survival of female F1 panthers in ENP (n = 3) through decreased competition and reducing the need for dispersal. However, half of the radiocollared F1 panthers inhabited areas known to be occupied by both adult male and female panthers, so even if low density of adult panthers influenced our results in some portions of the range, it would only offer a partial potential explanation for the higher survival of F1 panthers.
A second extrinsic factor that may have influenced our results is variation in habitat conditions across the range of panthers, and we recognize the potential for interactions between habitat quality, genetic ancestry and survival. Theoretical (Moore 1977; Moore & Price 1993) and empirical work (Rand & Harrison 1989; Good et al. 2000) suggest that hybrids sometimes thrive in different habitat types than parental types, leading to the production of relatively fit hybrids and to the establishment and maintenance of hybrid zones. Novel phenotypes produced through hybridization can allow for niche differentiation between hybrid and parental types when some of the available habitat is not suitable for the parental types (Lewontin & Birch 1966; Buerkle et al. 2000). It has been suggested that since introgression, panthers have moved into areas in south Florida that were not occupied during years of lower population size just prior to introgression (Pimm, Dollar & Bass 2006), meaning that admixed individuals might have occupied and survived in areas that were not used by pre-introgression type panthers. However, Creel (2006) pointed out that such an expansion into new areas could have simply been driven by demographics, as growing populations will often expand into new habitats. An intriguing possibility to consider with future analysis is whether admixed panthers (especially F1s) used different habitat types than pre-introgression type panthers and whether genotype-specific habitat selection patterns influenced survival (and other components of fitness).
An ideal test of the hybrid vigour theory would involve a carefully designed experiment with large sample sizes for robust statistical inferences. However, such experiments and sample sizes are rarely possible for elusive and highly endangered species of large carnivores that typically occur at low density. Populations small enough to experience inbreeding often result in smaller sample sizes than would be preferred for more robust statistical inferences. Nonetheless, analysing these data is critically important in terms of increasing understanding of the dynamics of highly endangered populations. Although our sample of radiocollared F1 individuals is numerically small (n = 8), it represents a substantial proportion of the total F1 offspring produced by the introgression programme (38–53%, depending on the range of possible litter sizes for the two dens not visited); therefore, the survival of the F1 individuals we studied should be representative. Although we believe that hybrid vigour is the most parsimonious explanation for the superior survival of F1 admixed panthers, we suggest our results should be interpreted cautiously because of small number of F1 admixed panthers in our analysis. The small sample leaves open the possibility that factors other than hybrid vigour may have contributed to the higher survival of the F1 individuals (e.g. density, habitat variables and other unknown factors), as well as increasing the potential for model overfitting (unreliable model predictions owing to a large number of parameters relative to number of data points; Harrell 2001).
Pimm, Dollar & Bass (2006) evaluated the effects of the introgression programme on panther demographic parameters, including survival, but their results with respect to adult survival were equivocal (Creel 2006). Our data set, analyses and results differed from those of Pimm, Dollar & Bass (2006) in several important ways. Pimm, Dollar & Bass (2006) excluded 15 panthers because they did not know the genetic ancestry of these individuals, whereas we determined ancestry for the entire data set with updated genetic analyses completed in 2009. We also addressed aspects of the analysis by Pimm, Dollar & Bass (2006) following recommendations made by Creel (2006). First, we separated adults from subadults to consider possible differences in age structure between pre-introgression type and admixed categories, whereas Pimm, Dollar & Bass (2006) categorized all panthers independent from their mothers as adults. We also excluded older panthers (≥10 years) from our analysis because these individuals, which survived poorly relative to other age classes, were underrepresented in the admixed classes. Second, Pimm, Dollar & Bass (2006) compared demographic parameters of some panthers from years prior to introgression to those of admixed individuals following introgression, thus failing to control for potential temporal variation in panther demographic variables. We limited our comparison of survival between admixed and pre-introgression type genotypes to years when both coexisted on the landscape (i.e. 1997–2006). It should be noted that this restriction by itself has little effect on inference, except to weaken the evidence for an effect of heterozygosity (J. Hostetler, unpublished data). Perhaps the most important difference between our analysis and that of Pimm, Dollar & Bass (2006) is that we separated F1 generation offspring from other admixed panthers, as recommended by previous researchers investigating fitness consequences of hybridization (reviewed by Arnold & Hodges 1995; Burke & Arnold 2001). Subadult and adult survival improved most dramatically for F1 admixed panthers after introgression, and this effect would be diluted by pooling all admixed panthers into a single category. Finally, we included data on heterozygosity to determine whether differences in survival across genotypes were associated with differences in genetic diversity. Thus, we believe our approach has provided additional insight into the effect of the introgression programme on subadult and adult panther survival and represents a well-documented example of the utility of genetic introgression in improving a demographic parameter of an inbred population.
Age and sex influenced survival of panthers as survival rates for females were higher than for males in each age class. The subadult age class is the period when male panthers are dispersing and attempting to locate and establish home ranges (Maehr et al. 2002), and our results indicate this is a dangerous period for male panthers. Conversely, female survival rates were highest for the subadult age class. Female pumas are often philopatric, dispersing less frequently and for shorter distances than males (Sweanor, Logan & Hornocker 2000; Maehr et al. 2002), consistent with most species of polygynous mammals (Greenwood 1980). Our results also strongly suggest that older panthers (≥10 years) survived poorly compared with other age classes, despite the small sample size of older adults and the potential bias against detecting survival senescence because of heterogeneity in individual survival (Cam et al. 2002).
Our results, and those of Hostetler et al. (2010) with respect to kitten survival, are important steps to determining the demographic mechanisms that led to the numerical increase in panthers after introgression and indicate that intentional introgression can be a valuable tool for conserving small, inbred populations. However, potential variation in the response of populations to admixture, the possibility of outbreeding depression through loss of co-adapted gene combinations or adaptation to local environmental conditions (Greig 1979; Templeton 1997; Edmands 2007), and the problem of losing genetically unique populations through swamping (Allendorf et al. 2001; Creel 2006) suggest that intentional admixture of wild populations should be undertaken only when extinction appears imminent, as in the panther example. Despite the apparent success of the introgression programme for panthers (at least in the short term), the problems that led to a small population size and inbreeding, habitat loss and isolation from other populations, have not been corrected and will likely be exacerbated as development and human population growth are projected to increase in south Florida (Kautz et al. 2006). Therefore, investigating the influence of introgression on other demographic parameters (e.g. fecundity) and population growth and determining the longevity of any demographic benefits associated with intentional introgression will be important next steps for evaluating whether genetic augmentation will be an effective long-term management tool for panthers.
Supplementary Material
Acknowledgements
We thank D. Land, M. Cunningham, Roy McBride, M. Lotz, D. Shindle, S. Schulze, D. Giardina, A. Johnson, S. Bass, L. Oberhofer, M. Alvarado, H. Fitting and Rocky McBride, Rowdy McBride, C. McBride and others for assistance with fieldwork. We also thank B. Bolker, D. Valle, J. White, J. Nichols and G. White for statistical assistance or advice and D. Land, Roy McBride, B. Bolker, J. Nichols, A. Singh, M. Lotz, M. Cunningham, T. O’Meara, J. Gore, P. Mahoney and two anonymous reviewers for reviewing earlier versions of this manuscript. This work was funded through the Florida Panther Research and Management Trust Fund, National Park Service, University of Florida, and a grant from the United States Fish and Wildlife Service.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Appendix S1. Age error estimation.
Appendix S2. Fine scale ancestry testing.
Appendix S3. Cause-specific mortality analysis.
Appendix S4. Inference results for ancestry and heterozygosity.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Allendorf FW, Leary RF, Spruell P & Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution, 16, 613–622. [Google Scholar]
- Arnold ML & Hodges SA (1995) Are natural hybrids fit or unfit relative to their parents? Trends in Ecology & Evolution, 10, 67–71. [DOI] [PubMed] [Google Scholar]
- Ashman DL & Greer K (1976) Age techniques Transactions of the Mountain Lion Workshop (eds Christensen GC & Fisher RJ), pp. 199–204. U.S. Fish and Wildlife Service, Portland, OR. [Google Scholar]
- Beier P, Vaughan MR, Conroy MJ & Quigley H (2006) Evaluating scientific inferences about the Florida panther. Journal of Wildlife Management, 70, 236–245. [Google Scholar]
- Belden RC, Frankenberger WB, McBride RT & Schwikert ST (1988) Panther habitat use in southern Florida. Journal of Wildlife Management, 52, 660–663. [Google Scholar]
- Benson JF, Lotz MA & Jansen D (2008) Natal den selection by Florida panthers. Journal of Wildlife Management, 72, 405–410. [Google Scholar]
- Birchler JA, Auger DL & Riddle NC (2003) In search of the molecular basis of heterosis. Plant Cell, 15, 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA & Rieseberg LH (2000) The likelihood of homoploid hybrid speciation. Heredity, 84, 441–451. [DOI] [PubMed] [Google Scholar]
- Burke JM& Arnold ML (2001) Genetics and the fitness of hybrids. Annual Review of Genetics, 35, 31–52. [DOI] [PubMed] [Google Scholar]
- Burnham KP & Anderson DR (2002) Model Selection and Multimodel Inference, 2nd edn Springer, New York, NY. [Google Scholar]
- Cam E, Link WA, Cooch EG, Monnat JY & Danchin E (2002) Individual covariation in life-history traits: seeing the trees despite the forest. American Naturalist, 159, 96–105. [DOI] [PubMed] [Google Scholar]
- Campbell DR & Waser NM (2001) Genotype-by-environment interaction and the fitness of plant hybrids in the wild. Evolution, 55, 669–676. [DOI] [PubMed] [Google Scholar]
- Cox DR (1972) Regression models and life-tables. Journal of the Royal Statistical Society Series B-Statistical Methodology, 34, 187–220. [Google Scholar]
- Creel S (2006) Recovery of the Florida panther – genetic rescue, demographic rescue, or both? Response to Pimm et al. (2006) Animal Conservation, 9, 125–126. [Google Scholar]
- Crow JF (1948) Alternative hypotheses of hybrid vigor. Genetics, 33, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver M, Johnson WE, Pecon-Slattery J & O’Brien SJ (2000) Genomic ancestry of the American puma (Puma concolor). Journal of Heredity, 91, 186–197. [DOI] [PubMed] [Google Scholar]
- Cunningham MW, Brown MA, Shindle DB, Terrell SP, Hayes KA, Ferree BC, McBride RT, Blankenship EL, Jansen D, Citino SB, Roelke ME, Kiltie RA, Troyer JL & O’Brien SJ (2008) Epizootiology and management of feline leukemia virus in the Florida puma. Journal of Wildlife Diseases, 44, 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Haag C, Kirkpatrick M, Riek M, Hottinger JW & Pajunen VI (2002) A selective advantage to immigrant genes in a Daphnia metapopulation. Science, 295, 485–488. [DOI] [PubMed] [Google Scholar]
- Edmands S (2007) Between a rock and a hard place: evaluating the relative risk of inbreeding and outbreeding for conservation and management. Molecular Ecology, 16, 463–475. [DOI] [PubMed] [Google Scholar]
- Efron B & Tibshirani RJ (1998) An Introduction to the Bootstrap. Chapman and Hall/CRC Press, Boca Raton, FL. [Google Scholar]
- Fieberg J & DelGiudice GD (2009) What time is it? Choice of time origin and scale in extended proportional hazards models. Ecology, 90, 1687–1697. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM & Shaffer HB (2007) Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proceedings of the National Academy of Sciences of the United States of America, 104, 15793–15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman EA (1936) A new puma from Texas. Proceedings of the Biological Society of Washington, 49, 137–138. [Google Scholar]
- Good TP, Ellis JC, Annett CA & Pierotti R (2000) Bounded hybrid superiority in an avian hybrid zone: effects of mate, diet, and habitat choice. Evolution, 54, 1774–1783. [DOI] [PubMed] [Google Scholar]
- Grant PR & Grant BR (1992) Hybridization of bird species. Science, 256, 193–197. [DOI] [PubMed] [Google Scholar]
- Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Animal Behaviour, 28, 1140–1162. [Google Scholar]
- Greig JC (1979) Principles of genetic conservation in relation to wildlife management in southern Africa. South African Journal of Wildlife Research, 9, 57–78. [Google Scholar]
- Harrell FE (2001) Regression Modeling Strategies. Springer-Verlag, New York, NY. [Google Scholar]
- Heisey DM & Patterson BR (2006) A review of methods to estimate cause-specific mortality in presence of competing risks. Journal of Wildlife Management, 70, 1544–1555. [Google Scholar]
- Hostetler JA, Onorato DP, Nichols JD, Johnson WE, Roelke ME, O’Brien SJ, Jansen D & Oli MK (2010) Genetic introgression and the survival of Florida panther kittens. Biological Conservation, 143, 2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Land ED & Lotz MA (1997) Florida Panther Genetic Restoration and Management. Annual Performance Report. Florida Fish and Wildlife Conservation Commission, Tallahassee, FL. [Google Scholar]
- Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden C, McBride R, Jansen D, Lotz M, Shindle D, Howard J, Wildt DE, Penfold LM, Hostetler JA, Oli MK & O’Brien SJ (2010) Genetic restoration of the Florida panther. Science, 329, 1641–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautz R, Kawula R, Hoctor T, Comiskey J, Jansen D, Jennings D, Kasbohm J, Mazzotti F, McBride R, Richardson L & Root K (2006) How much is enough? Landscape-scale conservation for the Florida panther Biological Conservation, 130, 118–133. [Google Scholar]
- Lambert CMS, Wielgus RB, Robinson HS, Katnik DD, Cruickshank HS, Clarke R & Almack J (2006) Cougar population dynamics and viability in the Pacific northwest. Journal of Wildlife Management, 70, 246–254. [Google Scholar]
- Land ED, Garman DR & Holt GA (1998) Monitoring female Florida panthers via cellular telephone. Wildlife Society Bulletin, 26, 29–31. [Google Scholar]
- Land ED, Shindle DB, Kawula RJ, Benson JF, Lotz MA & Onorato DP (2008) Florida panther habitat selection analysis of concurrent GPS and VHF telemetry data. Journal of Wildlife Management, 72, 633–639. [Google Scholar]
- Levin DA, Francisco-Ortega J & Jansen RK (1996) Hybridization and the extinction of rare plant species. Conservation Biology, 10, 10–16. [Google Scholar]
- Lewontin RC & Birch LC (1966) Hybridization as a source of variation for adaptation to new environments. Evolution, 20, 315–336. [DOI] [PubMed] [Google Scholar]
- Logan KA & Sweanor LL (2001) Desert Puma: Evolutionary Ecology and Conservation of an Enduring Carnivore. Island Press, Washington, DC. [Google Scholar]
- Madsen T, Shine R, Olsson M & Wittzell H (1999) Restoration of an inbred adder population. Nature, 402, 34–35. [Google Scholar]
- Maehr DS, Land ED, Shindle DB, Bass OL & Hoctor TS (2002) Florida panther dispersal and conservation. Biological Conservation, 106, 187–197. [Google Scholar]
- McBride RT, McBride RT, McBride RM & McBride CE (2008) Counting pumas by categorizing physical evidence. Southeastern Naturalist, 7, 381–400. [Google Scholar]
- Menotti-Raymond M, David VA, Stephens JC, Lyons LA & O’Brien SJ (1997) Genetic individualization of domestic cats using feline STR loci for forensic applications. Journal of Forensic Science, 42, 1039–1051. [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schäffer AA, Tomlin JF, Hutton MK & O’Brien SJ (1999) A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics, 57, 9–23. [DOI] [PubMed] [Google Scholar]
- Minch E, Ruiz-Linares A & Goldstein DB (1995) MICROSAT http://hpgl.stanford.edu/projects/microsat/. [Google Scholar]
- Moore WS (1977) An evaluation of narrow hybrid zones in vertebrates. Quarterly Review of Biology, 52, 263–277. [Google Scholar]
- Moore WS & Price JT (1993) Nature of selection in the northern flicker hybrid zone and its implications for speciation theory Hybrid Zones and the Evolutionary Process (ed. Harrison RG), pp. 196–225. Oxford University Press, New York, NY. [Google Scholar]
- Nelson EW & Goldman EA (1929) List of the pumas with three described as new. Journal of Mammalogy, 10, 345–350. [Google Scholar]
- O’Brien SJ, Roelke ME, Yuhki N, Richards KW, Johnson WE, Franklin WL, Anderson AE, Bass OL, Belden RC & Martenson JS (1990) Genetic introgression within the Florida panther Felis concolor coryi. National Geographic Research, 6, 485–494. [Google Scholar]
- Pankratz VS, deAndrade M & Therneau TM (2005) Random-effects Cox proportional hazards model: general variance components methods for time-to-event data. Genetic Epidemiology, 28, 97–109. [DOI] [PubMed] [Google Scholar]
- Pimm SL, Dollar L & Bass OL (2006) The genetic rescue of the Florida panther. Animal Conservation, 9, 115–122. [Google Scholar]
- Pollock KH, Winterstein SR, Bunck CM & Curtis PD (1989) Survival analysis in telemetry studies: the staggered entry design. Journal of Wildlife Management, 53, 7–15. [Google Scholar]
- Pritchard JK, Stephens M & Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rand DM & Harrison RG (1989) Ecological genetics of a mosaic hybrid zone: mitochondrial, nuclear, and reproductive differentiation of crickets by soil type. Evolution, 43, 432–449. [DOI] [PubMed] [Google Scholar]
- Rhymer JM & Simberloff D (1996) Extinction by hybridization and introgression. Annual Review of Ecology and Systematics, 27, 83–109. [Google Scholar]
- Robinson HS, Wielgus RB, Cooley HS & Cooley SW (2008) Sink populations in carnivore management: cougar demography and immigration in a hunted population. Ecological Applications, 18, 1028–1037. [DOI] [PubMed] [Google Scholar]
- Roelke ME, Martenson JS & O’Brien SJ (1993) The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Current Biology, 3, 340–350. [DOI] [PubMed] [Google Scholar]
- Rosenfield JA, Nolasco S, Lindauer S, Sandoval C & Kodric-Brown A (2004) The role of hybrid vigor in the replacement of Pecos pupfish by its hybrids with sheepshead minnow. Conservation Biology, 18, 1589–1598. [Google Scholar]
- Shull GH(1908) The composition of a field of maize. Reports of the American Breeders Association, 4, 296–301. [Google Scholar]
- Slattery M, Kamel HN, Ankisetty S, Gochfeld DJ, Hoover CA & Thacker RW (2008) Hybrid vigor in a tropical Pacific soft-coral community. Ecological Monographs, 78, 423–443. [Google Scholar]
- Sullivan JD Jr (2004) Florida’s Endangered Species, Threatened Species, and Species of Special Concern. Florida Fish and Wildlife Conservation Commission, Tallahassee, FL. [Google Scholar]
- Sweanor LL, Logan KA & Hornocker MG (2000) Cougar dispersal patterns, metapopulation dynamics, and conservation. Conservation Biology, 14, 798–808. [Google Scholar]
- Templeton AR (1997) Coadaptation, local adaptation, and outbreeding depression Principles of Conservation Biology (eds Meffe GK & Carroll CR), pp. 171–172. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Therneau TM & Grambsch PM (2000) Modeling Survival Data: Extending the Cox Model. Springer, New York, NY. [Google Scholar]
- Tinker MT, Doak DF, Estes JA, Hatfield BB, Hatfield BB, Staedler MM & Bodkin JL (2006) Incorporating diverse data and realistic complexity into demographic estimation procedures for sea otters. Ecological Applications, 16, 2293–2312. [DOI] [PubMed] [Google Scholar]
- Vaida F & Blanchard S (2005) Conditional Akaike information for mixed-effects models. Biometrika, 92, 351–370. [Google Scholar]
- Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, Bouzat JL & Paige KN (1998) Tracking the long-term decline and recovery of an isolated population. Science, 282, 1695–1698. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Ingvarsson PK & Hatfield T (2000) Local drift load and the heterosis of interconnected populations. Heredity, 84, 452–457. [DOI] [PubMed] [Google Scholar]
- Wolf DE, Takebayashi N & Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conservation Biology, 15, 1039–1053. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


