SUMMARY
The molecular architecture of centromere-specific nucleosomes containing histone variant CenH3 is controversial. We have biochemically reconstituted two distinct populations of nucleosomes containing Saccharomyces cerevisiae CenH3 (Cse4). Reconstitution of octameric nucleosomes containing histones Cse4/H4/H2A/H2B is robust on noncentromere DNA, but inefficient on AT-rich centromere DNA. However, nonhistone Scm3, which is required for Cse4 deposition in vivo, facilitates in vitro reconstitution of Cse4/H4/Scm3 complexes on AT-rich centromere sequences. Scm3 has a nonspecific DNA binding domain that shows preference for AT-rich DNA and a histone chaperone domain that promotes specific loading of Cse4/H4. In live cells, Scm3-GFP is enriched at centromeres in all cell cycle phases. Chromatin immunoprecipitation confirms that Scm3 occupies centromere DNA throughout the cell cycle, even when Cse4 and H4 are temporarily dislodged in S phase. These findings suggest a model in which centromere-bound Scm3 aids recruitment of Cse4/H4 to assemble and maintain an H2A/H2B-deficient centromeric nucleosome.
INTRODUCTION
Eukaryotic genomes are packaged with histone proteins in chromatin fibers, whose fundamental unit of organization is the nucleosome. The canonical nucleosome core particle consists of ~147 bp of DNA wrapped around a histone octamer—a heterotetramer of histones H3 and H4, and two heterodimers of histones H2A and H2B—each histone associating with its partner via a common, α-helical “handshake” or histone fold motif (Arents et al., 1991; Luger et al., 1997). DNA accessibility in the nucleosome is substantially blocked and its topology tightly constrained in nearly two left-handed supercoils (Luger et al., 1997). All aspects of DNA metabolism, including gene transcription, DNA replication, recombination, and repair occur on chromatin substrates. Accordingly, nucleosomes are subject to dynamic alterations in composition, positioning, and covalent modification—processes mediated by a vast array of macromolecular enzymes and protein factors (Clapier and Cairns, 2009; Goldberg et al., 2007).
In addition, centromeres of eukaryotic chromosomes contain highly specialized nucleosomes that provide a foundation for the assembly of the kinetochore, a multiprotein superstructure that connects centromere DNA to microtubules of the spindle apparatus, thereby enabling the proper segregation of daughter chromosomes during mitotic and meiotic cell divisions (Cleveland et al., 2003). Centromeric nucleosomes are universally marked by CenH3, a conserved, nonallelic variant of histone H3 (Black and Bassett, 2008; Henikoff and Dalal, 2005; Smith, 2002). CenH3 is the focus of intensive studies, not only because of its importance early in the hierarchical assembly of the kinetochore, but also for its role as a potential epigenetic factor for the inheritance of centromeres and kinetochores (Buscaino et al., 2010; Karpen and Allshire, 1997). The histone fold motif of CenH3 (CENP-A in human) contains key amino acids responsible for its centromere-specific character (Sullivan et al., 1994). Fine mapping revealed a subdomain named CATD (CENP-A targeting domain) comprising loop 1 and the α2 helix of the CENP-A histone fold that is responsible for specifying the biological functions of CenH3 (Black et al., 2004; Black et al., 2007).
The molecular architecture of the centromeric nucleosome is the subject of much debate. Biochemical studies using α-satellite DNA of human centromeres and recombinant CENP-A protein originally demonstrated reconstitution of CenH3CENP-A nucleosomes containing all four histone subtypes (CENP-A, H4, H2A, and H2B) (Yoda et al., 2000). While the biochemical properties of these nucleosomes are generally similar to those of the canonical nucleosome, interesting differences exist, such as a reduction in nucleosomal DNA length for CenH3CENP-A nucleosomes (Yoda et al., 2000). Subsequent reconstitution studies have further confirmed both the octameric nature of the core histones and the left-handed supercoiling of DNA on the CenH3CENP-A nucleosome (Black et al., 2007; Conde e Silva et al., 2007; Sekulic et al., 2010; Tanaka et al., 2005). The crystal structure of the human (CenH3CENP-A/H4)2 tetramer has been solved (Sekulic et al., 2010).
In contrast, studies of CenH3 in Drosophila and fungi suggest strikingly different views of the organization of centromeric nucleosomes. Chemical crosslinking, atomic force microscopy, and biochemical reconstitution of Drosophila CenH3 (Cid) nucleosomes have led to the proposal of a CenH3 “hemisome” containing only one, rather than two, copies of each histone subtype—CenH3Cid, H4, H2A, and H2B (Dalal et al., 2007; Dimitriadis et al., 2010; Furuyama and Henikoff, 2009). Our studies of Saccharomyces cerevisiae (budding yeast) Scm3, a nonhistone protein required for CenH3 deposition in vivo, suggested yet another model in which Scm3 and only two of the four histone subtypes—Cse4, the budding yeast CenH3, and histone H4—are contained in the core of centromeric nucleosomes, while H2A and H2B are absent (Mizuguchi et al., 2007).
Scm3 was originally identified as a high copy suppressor of mutations in the histone fold domain of Cse4 (Stoler et al., 2007), and was independently isolated in an unbiased biochemical assay for Cse4-interacting proteins (Mizuguchi et al., 2007). Based on the genetic data, biochemical interactions between Scm3 and Cse4 were also demonstrated (Camahort et al., 2007; Stoler et al., 2007). There is broad concurrence on the main biological and biochemical properties of Scm3 (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007). In particular, conditional depletion of Scm3 was shown to cause the loss of centromere localization of Cse4 and inner kinetochore proteins, leading to cell cycle arrest in mitosis and impaired chromosome segregation. Scm3 colocalizes with Cse4 at centromeric foci as visualized by immunofluorescence; furthermore, chromatin immunoprecipitation (ChIP) experiments show that Scm3 and Cse4 occupy centromere DNA sequences that are organized in the singular centromeric nucleosome for each budding yeast chromosome (Furuyama and Biggins, 2007). These common findings provide compelling evidence to support the view that Scm3 is a bona fide inner kinetochore protein residing in close proximity to Cse4, having a critical role in the assembly and maintenance of the kinetochore.
Additional studies from our laboratory using bacterially expressed proteins found direct, specific, and stoichiometric binding of Scm3 to the Cse4/H4 tetramer in 2M NaCl to the exclusion of H2A/H2B dimers. Together with a striking depletion of H2A/H2B crosslinking to centromeres in vivo, these findings led to a hypothesis for an atypical centromeric nucleosome in which Cse4 replaces H3 and Scm3 substitutes for H2A/H2B (Mizuguchi et al., 2007). However, this model has been challenged by in vitro reconstitution of an octameric Cse4 nucleosome containing only the four histone subtypes on the 601 nucleosome positioning sequence (Camahort et al., 2009), and by observation of a transient displacement of the Schizosaccharomyces pombe Scm3 homolog from centromeres during anaphase of mitosis (Pidoux et al., 2009; Williams et al., 2009).
In this report, we have tested our hypothesis by examining biochemical interactions between Scm3, Cse4/H4, and centromere DNA in reconstitution experiments. We find that the dA,dT (AT)-rich nature of yeast centromere DNA hinders in vitro reconstitution of octameric Cse4 nucleosomes. We also show that Scm3 facilitates reconstitution of centromere DNA-protein complexes containing both Scm3 and Cse4/H4, by virtue of its distinct DNA binding domain (DBD) and Cse4/H4 binding domain (CBD). Live-cell imaging and ChIP experiments show that Scm3 occupies centromeres in vivo throughout the budding yeast cell cycle. These findings provide evidence for atypical nucleosome architecture at yeast centromeres.
RESULTS
Centromere AT-Rich DNA Hinders Reconstitution of Octameric Cse4 Nucleosomes
To investigate the ability of Cse4, the budding yeast CenH3 variant, to assemble nucleosomes, we carried out biochemical reconstitution experiments using DNA fragments the length of a nucleosome core particle, and refolded histone octamers prepared from bacterially expressed Cse4 and histones H2A, H2B, and H4 (see Figure S1A available online). Nucleosomes were reconstituted by standard salt gradient dialysis from 2M NaCl at 4°C and examined by electrophoretic mobility shift analysis (EMSA). With increasing histone concentration, we observe the formation of a major mobility shift on two noncentromere DNA sequences: the INO1 gene promoter and the 601 high affinity nucleosome positioning sequence (Lowary and Widom, 1998) (Figure 1A). The discrete nucleoprotein complex dominating at the highest protein:DNA molar ratio of 1.2 contains all four histone subtypes—Cse4, H4, H2A, and H2B (Figures S2A and S2B). It also exhibits resistance to MNase digestion typical for the canonical nucleosome core particle (Figure S2D). In concurrence with findings of Camahort et al. (2009), these results demonstrate reconstitution of Cse4-containing nucleosomes carrying two each of Cse4, H4, H2A, and H2B on noncentromere DNA templates. However, when we employed the natural centromere sequence from S. cerevisiae chromosome III (CEN3), we observe little reconstitution of a discrete Cse4-containing nucleosome (Figure 1A). Another centromere sequence from chromosome IV (CEN4) also shows inefficient nucleosome reconstitution (Figure 1B). To confirm these findings, we carried out competitive reconstitutions using Cy3-labeled CEN3 DNA and Cy5-labeled 601 DNA in the same reaction mixture (Figure 1C). This further reveals poor nucleosome formation on CEN3 DNA (Cy3) while reconstitution is efficient on 601 DNA (Cy5). Similar results are obtained when the fluorescence label is swapped between the two DNAs.
Figure 1. Reconstitution of Cse4 Nucleosomes.
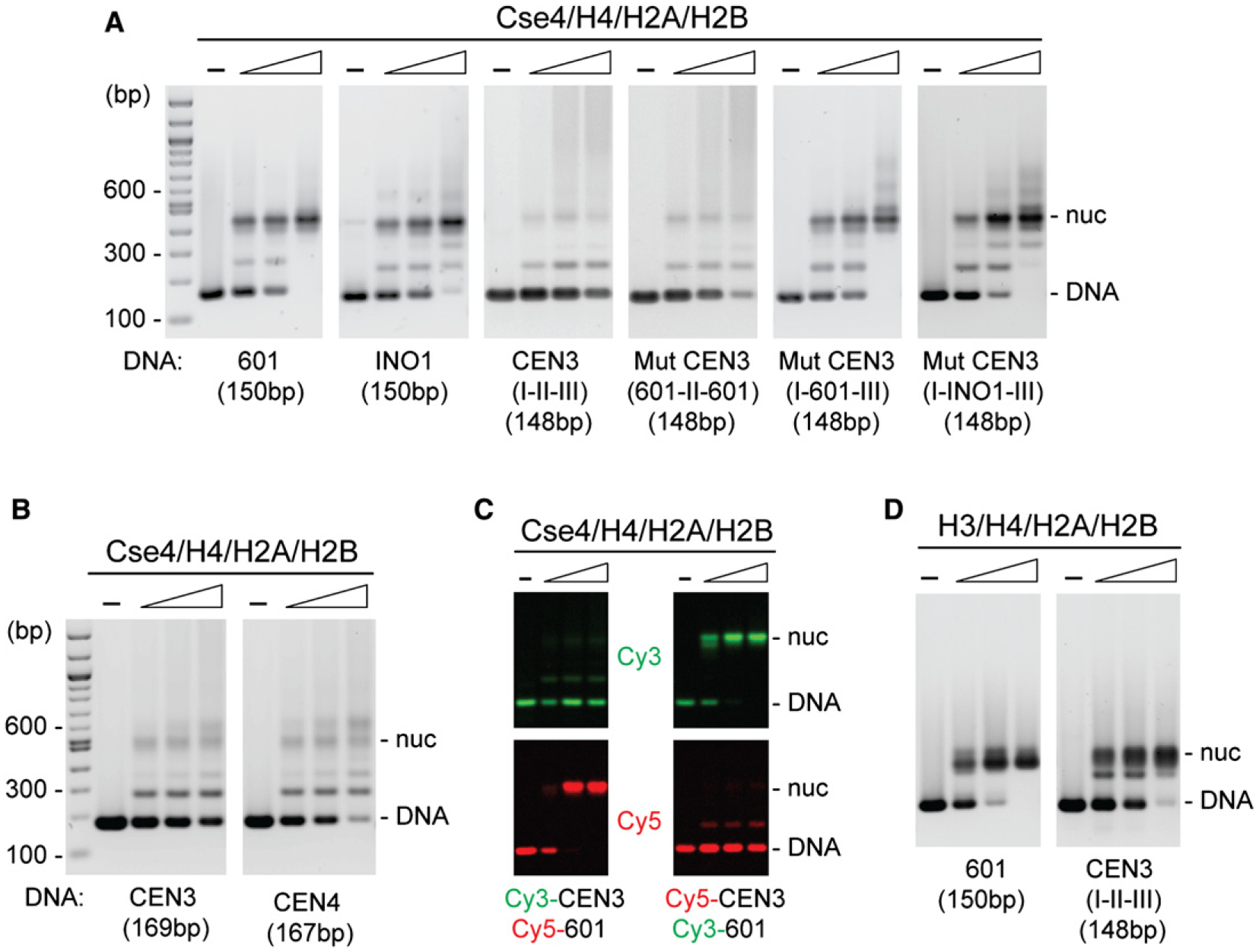
Octamers were mixed with DNA in 2 M NaCl, followed by overnight salt gradient dialysis at 4°C. Reconstituted nucleosomes were analyzed by EMSA and SYBR Green staining.
(A) Cse4 mononucleosomes were reconstituted using [Cse4/H4/H2A/H2B]2 octamers and the indicated DNAs: 150 bp 601, 150 bp INO1, 148 bp CEN3 (CDE I-II-III), and 148 bp mutant CEN3 (601-CDEII-601, CDE I-601-CDEIII, and CDE I-INO1-CDEIII) (protein:DNA molar ratio rm in ascending order, 0.3, 0.6, 1.2).
(B) CEN3 and CEN4 DNAs reconstitute Cse4 nucleosomes inefficiently.
(C) Competitive reconstitution of Cse4 nucleosomes with Cy3- and Cy5-labeled DNAs in the same reaction.
(D) Nucleosomes were reconstituted using [H3/H4/H2A/H2B]2 octamers and indicated DNAs (protein:DNA molar ratio rm in ascending order, 0.3, 0.6, 1.2). Samples were treated at 65°C overnight and analyzed by EMSA (see additional data in Figure S2).
Genetic and molecular studies have established that the centromeres of all 16 budding yeast chromosomes are composed of a contiguous arrangement of three consensus elements: CDEI, CDEII, and CDEIII, encompassing a total of ~125 bp of DNA (Clarke and Carbon, 1985). CDEI and CDEIII are short DNA sequences that bind to the CBF1 and CBF3 sequence-specific DNA binding factors, while the extended, 78–86 bp long CDEII element is distinguished by unusually high AT content (91%–95% A+T) (Clarke and Carbon, 1985; Baker and Rogers, 2005). Swapping the CDEII element of CEN3 (93% A+T) with either 601 (38% A+T) or INO1 (58% A+T) sequences of the same length substantially improves reconstitution of Cse4-containing nucleosomes (Figure 1A, I-601-III, I-INO1-III). A reciprocal swap of CDEI and CDEIII for 601 sequences leaving CDEII intact failed to improve nucleosome reconstitution (Figure 1A, 601-II-601). We conclude that the intrinsic properties of the AT-rich CDEII element disfavor reconstitution of Cse4-containing nucleosomes. It is possible to reconstitute conventional H3-containing nucleosomes on the AT-rich CEN3, albeit at a lower efficiency than for 601 DNA (Figure 2D, Figures S2C and S2D).
Figure 2. Reconstitution of Complexes Containing Cse4/H4 and Scm3 on Centromere DNA.
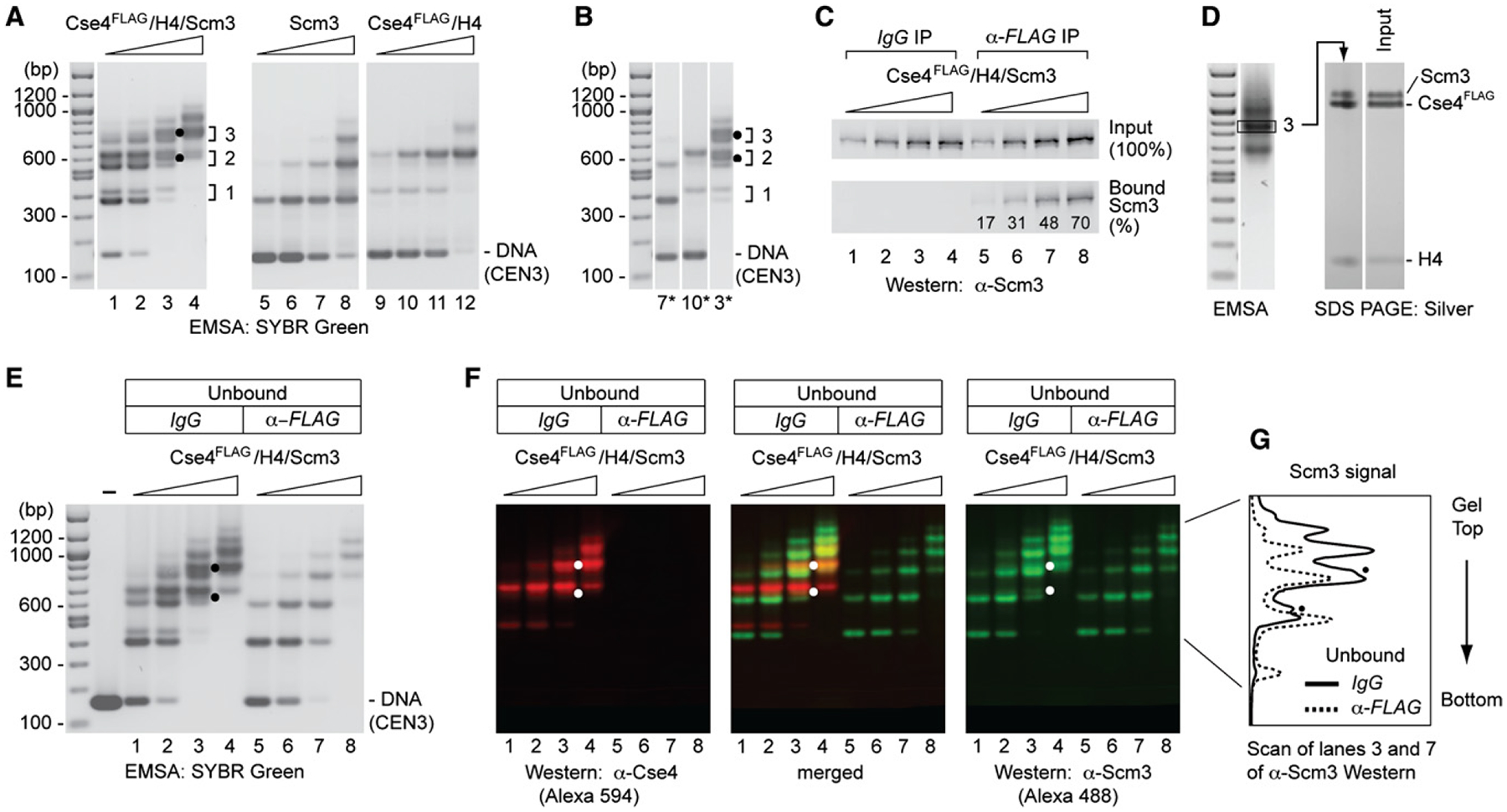
(A) Complexes were reconstituted by salt dialysis using CEN3 DNA (148 bp) and increasing amounts of Cse4FLAG/H4/Scm3 hexamers, and analyzed by EMSA and SYBR Green staining: lanes 1–4, Cse4FLAG/H4/Scm3 (protein:DNA molar ratio, rm = 0.3, 0.6, 0.9, 1.2); lanes 5–8, Scm3 (rm, = 0.125, 0.25, 0.5, 1.0); lanes 9–12, Cse4FLAG/H4 (rm = 0.27, 0.55, 1.1, and 2.2). Dots mark distinctly resolved complexes that may contain both Scm3 and Cse4FLAG (see also D–F). Brackets and numbers indicate rungs of the doublet ladders of band-shift species.
(B) Comparison of lanes 3, 7, and 10 in (A).
(C and D) Scm3 and Cse4 reside in the same DNA-protein complexes. Complexes were reconstituted as in (A) and analyzed for the presence of Scm3 and Cse4.
(C) shows percent Scm3 associated with Cse4 as analyzed by α-FLAG immunoprecipitation and western blotting. (D) shows SDS-PAGE and silver staining of a band-shift species excised from an EMSA gel. Density scan shows a Scm3:Cse4 ratio of 0.86 after normalization to that of input.
(E) Scm3-DNA complexes in the α-FLAG unbound fraction shown by EMSA and SYBR Green staining (lanes 5–8).
(F) Cse4- and Scm3-containing complexes confirmed by western blot analysis of a replicate gel as in (E), probing with α-Cse4 (rabbit) and α-Scm3 (guinea pig) antibodies, and visualized by fluorescent Alexa Fluor 594 goat-anti-rabbit and Alexa Fluor 488 goat-anti-guinea pig, respectively. Dots mark distinctly resolved complexes that are depleted by α-FLAG and therefore contain both Scm3 and Cse4FLAG.
(G) Signal intensity scans of gel lanes 3 and 7 of α-Scm3 western blot of (F) (α-FLAG IP exacerbates Cse4FLAG/H4/Scm3 dissociation, as revealed by the relative increase of the first and second mobility shifts for Cse4-free Scm3).
Reconstitution of Complexes Containing Cse4/H4 and Scm3 on Centromere DNA
When preformed Cse4FLAG/H4/Scm3 protein hexamers are reconstituted with CEN3 DNA under identical conditions, we observe a ladder-like series of increasingly retarded electrophoretic mobility shifts (doublets) on agarose gels (Figure 2A, lanes 1–4; ladder rungs labeled in increasing order, 1, 2, 3). We use agarose gels because polyacrylamide destabilizes Scm3-containing complexes (Figure S4A). At higher protein concentrations, the more retarded bands dominate, suggesting that higher-order protein complexes associate as stacked and/or tandemly bound nucleoprotein complexes. Reconstitution of Cse4FLAG/H4/Scm3 on 601 DNA gives a similar ladder of gel mobility shifts (Figure S3). Inspection of patterns produced separately by Scm3 and Cse4FLAG/H4 identifies the lower and upper bands of each doublet with Scm3 and Cse4FLAG/H4, respectively (Figure 2A, lanes 5–12), suggesting that the preformed hexamer dissociates into separate Scm3 and Cse4FLAG/H4 moieties during reconstitution. To facilitate comparisons, the shifted bands representing Scm3 (lane 7), Cse4FLAG/H4 (lane 10), and Cse4FLAG/H4/Scm3 (lane 3) are juxtaposed (Figure 2B). The presence of Cse4-free Scm3-DNA complexes was confirmed by immunodepletion of Cse4FLAG-containing complexes and EMSA of the unbound fraction (Figure 2E, lanes 5–8), and by native gel western blot analysis, probing with α-Scm3 antibody (Figure 2F, α-Scm3, green Alexa Fluor 488). Probing with α-Cse4 antibody confirms quantitative depletion of Cse4FLAG (Figure 2F, α-Cse4, red Alexa Fluor 594). A merger shows the overlap of Cse4 and Scm3 (Figure 2F, merged).
Notwithstanding such dissociation of Cse4FLAG/H4/Scm3, analysis of the α-Flag immunoprecipitate (IP) shows that a substantial fraction of Scm3 is retained within Cse4FLAG-containing nucleoprotein complexes, increasing from 17% to 70% of Scm3 input with increasing protein concentration (Figure 2C). Importantly, EMSA and native gel western blotting identifies a distinctly shifted, Scm3-containing species in the pull-down (IgG) control which migrates between dominant Scm3 and Cse4FLAG/H4 bands in rung #2 of the mobility shift ladder, and which is α-Flag depleted (Figure 2E, compare lanes 3 and 7; Figure 2G). This species thus contains both Cse4FLAG and Scm3, and may represent a Cse4/H4 tetramer and a Scm3 dimer bound to CEN3 (on the assumption that the second DNA mobility shift for Scm3 represents binding of two Scm3 dimers per DNA fragment, and that electrophoretic retardation correlates with particle mass). Higher-order complexes containing Cse4FLAG/H4/Scm3, such as in rung #3, are not comingled with substantial levels of Scm3 and Cse4FLAG/H4 bands; gel excision and SDS-PAGE show a Scm3:Cse4 molar ratio of 0.86, consistent with a composition of protein hexamers, or multiples thereof (Figure 2D). Taken together, our results document that complexes containing nearly stoichiometric Cse4FLAG, H4, and Scm3 can be reconstituted on centromere DNA under identical conditions that hinder formation of octameric Cse4 nucleosomes. This directly conflicts with the conclusions of Camahort et al. (2009), who, failing to define experimental conditions for detection of Cse4/H4/Scm3 complexes on centromere DNA, took the octameric Cse4 nucleosome on 601 DNA as a surrogate for the centromeric particle.
We find that the mixed population of Cse4/H4/Scm3 complexes on CEN3 DNA does not exhibit the resistance to MNase digestion typical of conventional or Cse4-containing nucleosomes reconstituted on 601 DNA (Figures S2D and S2E). Hence, while Scm3 is necessary for in vitro reconstitution of Cse4/H4/Scm3 nucleoprotein, it is not sufficient for assembly of a fully native, MNase-resistant centromeric nucleosome as found in vivo (Furuyama and Biggins, 2007). Additional kinetochore proteins are evidently required. When reconstituted on CEN3, the conventional H3 nucleosome displays increased sensitivity to MNase digestion, indicating that interactions are also less stable for the conventional histone octamer on AT-rich centromere DNA (Figure S2D).
Scm3 Has a Functional DNA Binding Domain with Preference for AT-Rich Sequences
Sequence analysis suggests that Scm3 contains two dissimilar regions, one of which is implicated in specific interaction with Cse4/H4. We have previously reported that this CBD of Scm3 (aa 90–193, defined as Scm3CBD) interacts directly, specifically, and stoichiometrically with Cse4/H4 in vitro (Mizuguchi et al., 2007). Sequences within this conserved CBD have also been shown to be essential for Scm3 function in vivo (Stoler et al., 2007). In addition, a consensus nuclear export signal (NES) at the N terminus of Scm3 is functionally required for cell viability (Stoler et al., 2007). Using agarose gel EMSA, we now demonstrate that bacterially expressed Scm3 has a distinct DNA binding domain (DBD) (referred to as Scm3DBD, aa 1–113, which also contains the NES) (Figure 3); note that these complexes are poorly detected on polyacrylamide gels (Figure S4A). Although Scm3 binds nonspecifically to 601 and INO1 DNA sequences, it has an ~3-fold preference for CEN3, and a similar preference was observed for CEN4 (Figures 3C and 3D). Replacing the CDEII element of CEN3 for 601 or INO1 sequences shows that it is AT-rich CDEII that is responsible for preferential binding by Scm3 (Figure S4). These findings document the presence of a DNA binding domain in Scm3 separate from the CBD. Our results are consistent with in vivo studies showing that N-terminal residues 1–103 of Scm3 are required for its binding to centromeres in vivo (M. Basrai, personal communication).
Figure 3. Scm3 Has a DNA Binding Domain with Preference for AT-Rich Sequences.
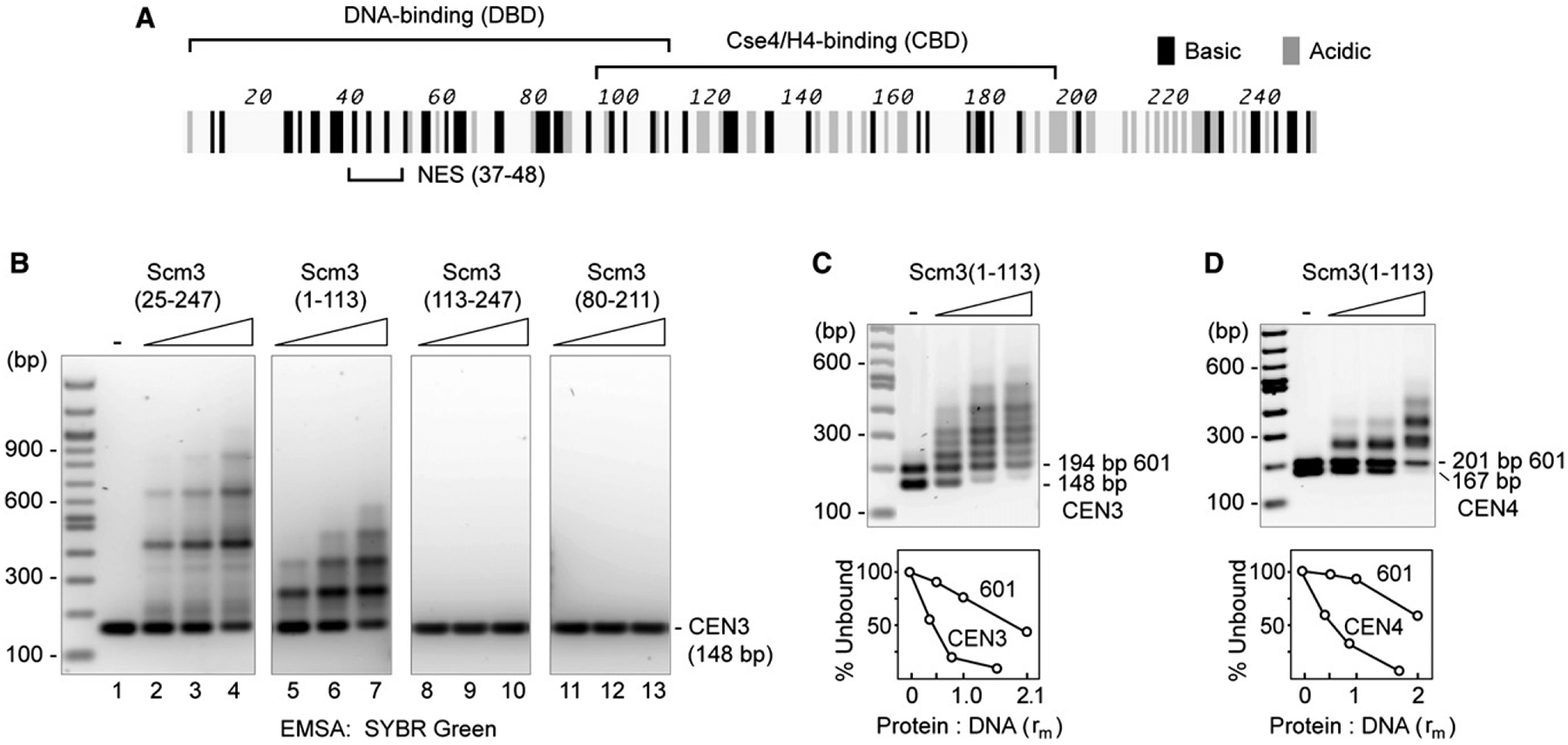
(A) Map of domains on the Scm3 open reading frame.
(B) Binding of CEN3 DNA to Scm3(25–247), Scm3(1–113), Scm3(113–247), and Scm3(80–211), shown by EMSA and SYBR Green staining. Essentially full-length, His-tagged Scm3(25–247) or Scm3 subfragments (Figure S1) were mixed with CEN3 DNA (protein:DNA ratio rm = 0.26, 0.52, and 1.04) in 2M NaCl and diluted to 0.2M NaCl in a final volume of 50 μl. After incubation at room temperature for 30 min, samples were analyzed by EMSA and SYBR Green staining.
(C and D) Scm3(1–113) has preference for CEN over 601 DNA (see also Figure S3). Binding of a mixture of CEN and 601 DNA (1 μg each) to Scm3(1–113) (protein:DNA ratio rm = 0.52, 1.04, 2.08) was analyzed as in (A). Graph shows percent unbound DNA to Scm3(1–113):DNA ratios.
Scm3 Exhibits Histone Chaperone Activity for Cse4/H4
To further characterize biochemical activities of Scm3, we examined its effects on the renaturation and folding of the Cse4/H4/Scm3 hexamer in solution. Gel filtration chromatography shows a dramatic loss of Cse4/H4 in the absence of Scm3 when denatured proteins are refolded in 2M NaCl at 4°C, indicating that the nonhistone protein helps prevent aggregation of Cse4/H4 (Figure 4A). Moreover, purified Scm3CBD strikingly improves loading of soluble Cse4/H4 on CEN3 DNA in vitro. The presence of an equimolar amount of Scm3CBD with Cse4FLAG/H4 before abrupt dilution from 2M NaCl provides an ~5-fold enhancement of two discrete complexes observed with EMSA that likely represent single and double Cse4FLAG/H4 tetrasomes (Figure 4B, lane 2). The enhancement of Cse4/H4 loading is not a trivial consequence of preventing Cse4FLAG/H4 from aggregation, as histones in the absence of Scm3 remain in the soluble supernatant after centrifugation (Figure 4C). This histone chaperone activity of Scm3CBD mimics the deposition of Cse4/H4 by salt gradient dialysis (Figure 4B, lanes 8, 9). The chaperone activity of Scm3CBD is specific for Cse4, having no discernable effect on the loading of H3/H4 tetramers on CEN3 DNA (Figure 4D). Also consistent with the behavior of a histone chaperone (Akey and Luger, 2003), Scm3CBD is not retained on the reconstituted nucleoprotein complex, even when introduced in stoichiometric excess to Cse4FLAG/H4 (Figure 4E). Addition of full-length Scm3 to Cse4/H4 enhances the formation of not only Cse4/H4 tetrasomes but also Cse4/H4/Scm3-CEN3 complexes, indicating that preformation of protein hexamers is not obligatory for reconstitution (Figure 4B, lanes 5–7).
Figure 4. Scm3 Shows Chaperone-like Activity for Cse4/H4.
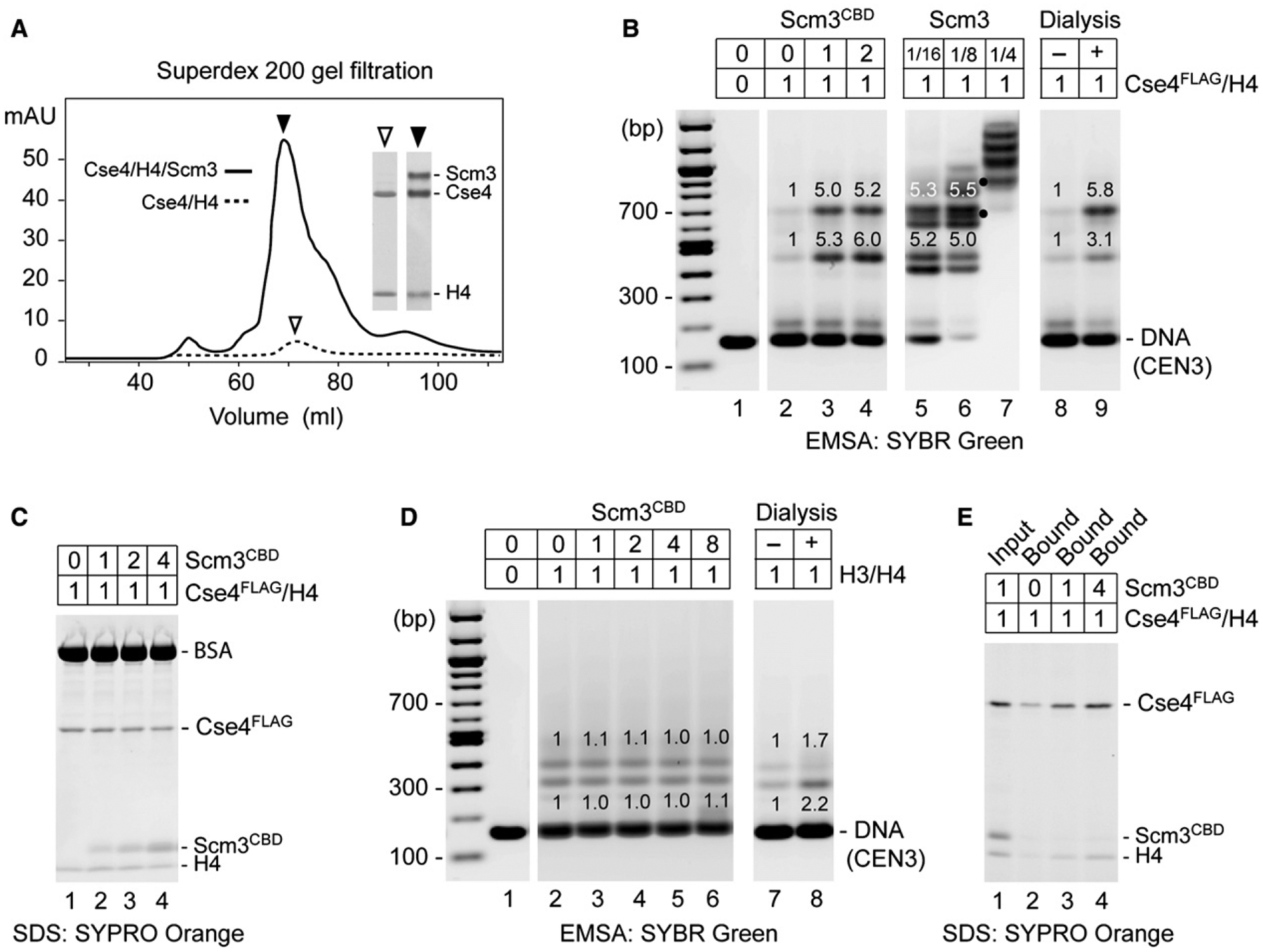
(A) Scm3 prevents loss of Cse4 and H4 during protein refolding and chromatography. Shown is Superdex 200 gel filtration of refolded Cse4/H4/Scm3 and Cse4/H4 complexes. Insets show SDS-PAGE and Coomassie blue staining of protein peak fractions (2 μl for Cse4/H4/Scm3 fraction and 20 μl for Cse4/H4).
(B) Scm3CBD promotes loading of Cse4FLAG/H4 on CEN3 DNA. Proteins were abruptly diluted from 2.0M to 0.2M NaCl and incubated with DNA. EMSA shows binding of CEN3 DNA (0.5 μg) by Cse4FLAG/H4 (1.2 μg) in the presence of 0-, 1-, and 2-fold molar excess of Scm3CBD (lanes 2–4) and in the presence of full-length Scm3 (lanes 5–7 where dots mark distinctly resolved complexes that may contain both Scm3 and Cse4FLAG), as compared to salt dialysis (lanes 8 and 9). Relative fluorescence intensity is indicated above gel bands.
(C) SDS-PAGE of proteins that remained in solution after DNA binding reaction. Protein binding to CEN3 was performed as in (B).
(D) Scm3CBD has no effect on H3/H4 loading. Reactions with increasing molar ratios of Scm3CBD to H3/H4 were analyzed as in (B).
(E) Scm3CBD fails to stably associate with Cse4FLAG/H4 on DNA. Protein binding to biotin-labeled CEN3 was performed as in (B), in the presence of 0-, 1-, and 4-fold molar excess of Scm3CBD. DNA-bound proteins were pulled down with streptavidin-coated beads and analyzed by SDS-PAGE.
Scm3-GFP Is Enriched at Centromeres in All Phases of the Cell Cycle
On fixed mitotic chromosome spreads, Scm3 colocalizes with Cse4 as a single focus of immunofluorescence that corresponds to the cluster of 16 haploid yeast centromeres (Mizuguchi et al., 2007). To address whether the focal distribution of Scm3 persists through the cell cycle, we engineered a strain in which the SCM3 coding sequence was replaced with a SCM3-GFP fusion under control of the endogenous Scm3 promoter. The strain also carries a kinetochore marker, Nuf2-mCherry. In haploid living cells, Scm3-GFP colocalizes with Nuf2-mCherry at either one or two centromere clusters, depending on cell cycle stage (Figures 5A–5C). However, there is substantial S. cerevisiae Scm3-GFP distributed diffusely throughout cell nuclei (Figure 5C). Despite this global nuclear localization, Scm3-GFP appears enriched at centromeres throughout the cell cycle in a manner similar to Cse4-GFP (Joglekar et al., 2006), and in particular throughout mitotic division, including anaphase (Figure 5D). This continuous presence of centromeric Scm3-GFP during rapid separation of daughter chromosomes in anaphase is evident in Movie S1 (Movie S2 shows Cse4-GFP). This persistence is in contrast to the transient depletion of S. pombe Scm3-GFP during anaphase (Pidoux et al., 2009; Williams et al., 2009; J.W., unpublished data).
Figure 5. Scm3 Is Enriched at Centromere Clusters throughout Cell Cycle.
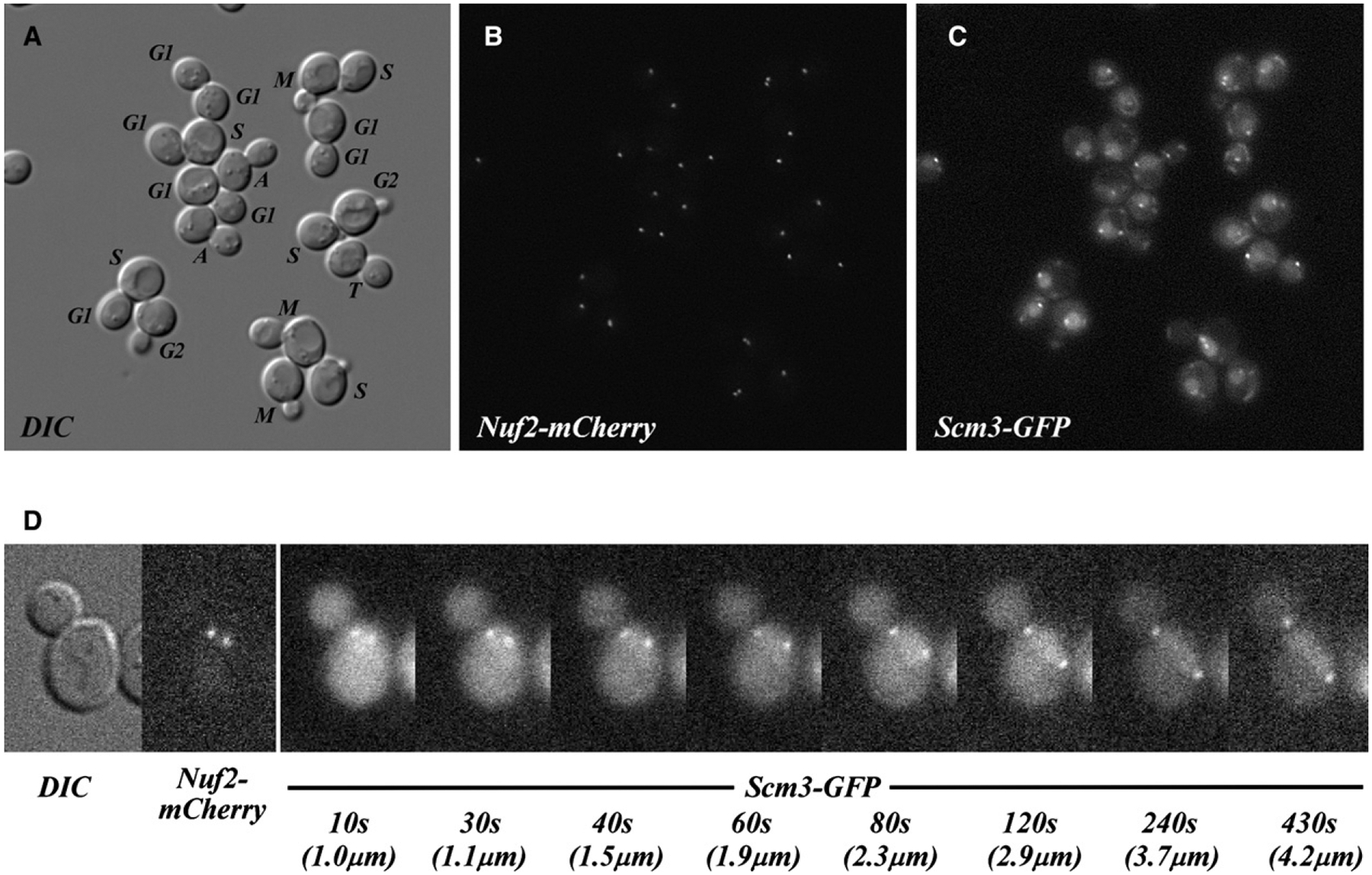
(A) DIC image of live asynchronous cells coexpressing Scm3-GFP and Nuf2-mCherry (as sole gene copies). The central plane of the Z stack is depicted. Cell cycle stages were assigned by inspection of all image planes, the position of centromeres, and the orientation of nuclei, as revealed in (B) and (C) (M, metaphase; A, anaphase; T, telophase). In a separate analysis of live, asynchronous cells, all 2069 cells examined showed centromeric enrichment of Scm3-GFP in addition to diffuse nuclear fluorescence.
(B) Compressed Z stack showing localization of the Nuf2-mCherry centromere marker.
(C) Compressed Z stack showing localization of Scm3-GFP in the same cells. To allow comparison between images, the brightness of (B) and (C) was linearly scaled within the same range of the original 16-bit signal registered by the camera. Slight differences in the positions of individual Nuf2 and Scm3 foci are consistent with observed movements of centromeres during sequential imaging.
(D) Selected frames from time-lapse series of a Scm3-GFP- and Nuf2-mCherry-expressing cell undergoing anaphase (enlarged 2×). DIC and Nuf2-mCherry images at the start are shown, followed by Scm3-GFP images at the indicated elapsed times (Scm3-GFP is enriched on both centromere clusters in all consecutive frames taken 10 s apart, for 650 s). Images (0.5 s exposure) were obtained in EM-CCD mode (EM gain = 100). Elapsed time (seconds) and the separation between centromere clusters (μm) are indicated.
Persistent Scm3 Occupancy at CEN3 and CEN4 throughout the Cell Cycle
To confirm the binding of Scm3 to centromere DNA sequences in vivo, we examined Scm3 occupancy at two yeast centromeres, CEN3 and CEN4, in synchronized cell populations after release from α factor arrest. Formaldehyde crosslinking, ChIP, and qPCR analysis show that Scm3 is enriched at the CEN3 and the CEN4 DNA loci relative to a pericentric region for all analyzed cell cycle stages (Figures 6A and 6B). Cse4 is similarly enriched (Figures 6A and 6B), except for a temporary depletion of Cse4 and histone H4 at 20 min after release from α factor arrest (Figures 6A–6D). FACS analysis reveals the onset of S phase in this time period (Figure 6E), suggesting that the depletion of Cse4 and H4 may be related to DNA replication. Such depletion of Cse4 at centromeres in S phase was previously reported (Hajra et al., 2006), but the codepletion of H4 was not known until this study. Importantly, histones H2B and H3 also appear to be depleted at CEN3 and CEN4 throughout the cell cycle (Figures 6C and 6D; see quantitative analysis for all ChIP-qPCR results in Figure S5). Hence, for a short time during S phase, both Cse4 and H4 are apparently dislodged from centromeres, while Scm3 persists. However, western blotting shows that the drop in Cse4 ChIP signal is not due to loss of Cse4 protein (Figure S5C).
Figure 6. Scm3 Persists on Centromeres throughout the Cell Cycle.
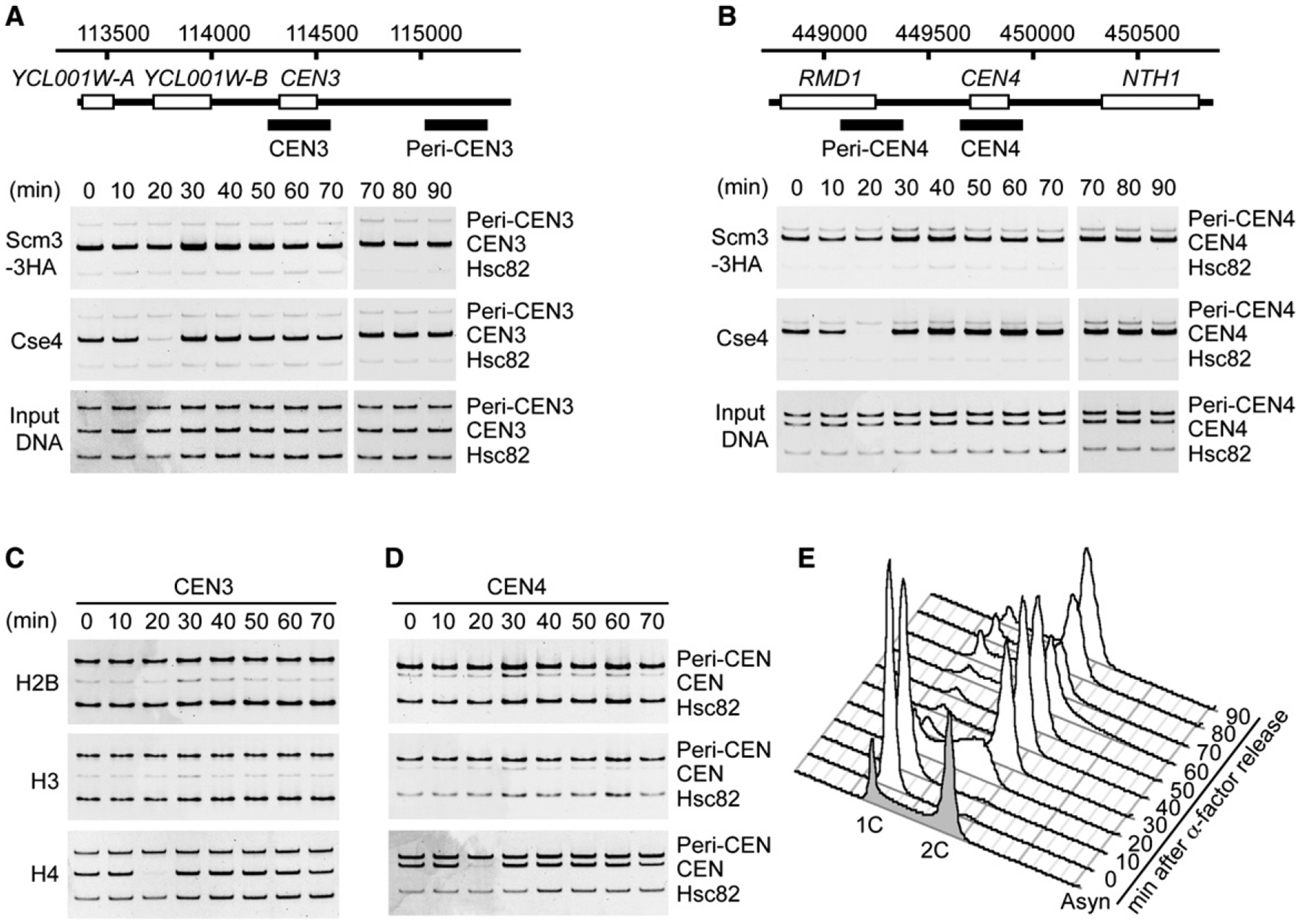
(A and B) ChIP shows Scm3 occupancy of CEN3 and CEN4 at indicated times after release from α factor arrest. Occupancy by Cse4 and H4 is diminished at 20 min and restored at 30 min. CEN and Peri-CEN sequences analyzed by PCR in the ChIP assay are indicated by solid bars. A segment of the HSC82 proximal promoter, containing a positioned canonical nucleosome, was assayed as an internal normalization control (Hsc82).
(C and D) Persistent depletion of H2B and H3 and temporary dislocation of H4 at CEN3 and CEN4 through the cell cycle. Input DNA samples are the same as in (A) and (B). ChIP quantification is given as bar graphs in Figure S6.
(E) DNA content of the synchronized cell population upon α factor release, shown by FACS analysis. The number of cells in mitosis was determined by microscopy after DAPI staining (10 min, 0%; 20 min, 0%; 30 min, 26%; 40 min, 69%; 50 min, 83%; 60 min, 48%; 70 min, 13%).
DISCUSSION
The CDEII elements of all 16 budding yeast centromeres are extremely AT rich (Clarke and Carbon, 1985). Moreover, CDEII sequences are unique among AT-rich sequences in the yeast genome by their highly nonrandom arrangement of homopolymer runs of five to seven nucleotides extending over ~80 bp (Baker and Rogers, 2005). The physical properties of AT-rich sequences—such as intrinsic curvature and twist and the ability to bend and twist in response to external forces—deviate from those of mixed sequence B-form DNA and influence the stability of reconstituted nucleosomes in vitro (Haran and Mohanty, 2009; Widom, 2001). In vivo, poly(A) stretches destabilize nucleosomes in a length-dependent manner in yeast cells (Shimizu et al., 2000; Struhl, 1985), and short A tracts are preferentially excluded from isolated yeast nucleosome core particles (Rando and Chang, 2009). The physical properties of CDEIIs are unknown, except for modest sequence-directed bending of DNA (Bechert et al., 1999; Ng et al., 1986). Based on available predictions, all 16 CDEIIs score at or near the bottom of the nucleosome occupancy scale (Kaplan et al., 2008).
CDEII elements are responsible for the observed resistance to biochemical reconstitution of octameric Cse4 nucleosomes under standard in vitro conditions, as their replacement with 601 and INO1 sequences of the same length, but lower AT content, abolishes resistance. While the exact mechanism of this inhibition is unknown, it seems likely that the intrinsic structure of CDEII resists DNA wrapping over the octameric histone core (or might affect interactions with H2A/H2B by changing Cse4/H4 conformation). By contrast, inclusion of Scm3 with Cse4/H4 enables reconstitution of complexes containing all three proteins on centromere (and 601) DNA. However, Cse4/H4/Scm3 complexes tend to dissociate into Cse4/H4 and Scm3 moieties during salt dialysis from 2M NaCl, especially at low protein concentrations, yielding a mixed population of protein-DNA complexes. Such dissociation is probably a consequence of a lack of stabilizing interactions with the CDEI and CDEIII DNA binding factors CBF1 and CBF3. Nonetheless, our in vitro experiments demonstrate that at near-stoichiometric ratios of Cse4/H4/Scm3 to CEN3 DNA, a substantial fraction of Scm3 coexists with Cse4/H4 in a nucleoprotein complex, as protein hexamers or multiples thereof. These findings are consistent with results of a single-molecule study showing that the center of the λ phage genome enriched for poly(A·T) tracts excludes octameric Cse4 nucleosomes, but not Cse4/H4/Scm3-λDNA complexes (Visnapuu and Greene, 2009). In the future, it will be important to examine complexes reconstituted in the presence of inner kinetochore proteins such as CBF3.
Reconstitution of Cse4/H4/Scm3 complexes on AT-rich centromere DNA requires both the DBD and the conserved CBD of Scm3. Purified Scm3DBD binds preferentially to AT-rich CDEII DNA, but this DBD alone does not promote in vitro reconstitution of Cse4/H4 on CEN3 DNA (our unpublished data). In contrast, Scm3CBD (without the DBD) is able to substantially enhance reconstitution of soluble Cse4/H4 on centromere DNA. Under conditions of abrupt dilution from 2M NaCl to physiological salt, Scm3CBD is not retained in the Cse4/H4-CEN3 nucleoprotein complex but released into solution, thus demonstrating the Cse4 histone chaperone function of Scm3 as previously proposed (Stoler et al., 2007). In this regard, Scm3 functions similarly to human HJURP, which is also required for deposition of CenH3CENP-A on human centromeres. HJURP shares conserved residues with Scm3CBD and facilitates in vitro loading of CenH3CENP-A/H4 tetramers on human centromere DNA (Dunleavy et al., 2009; Foltz et al., 2009; Sanchez-Pulido et al., 2009; Shuaib et al., 2010).
The NMR structure of the Scm3/Cse4/H4 protein complex reveals specific helix-helix contacts between conserved and functionally important residues of Scm3CBD and Cse4 (Zhou et al., 2011). Importantly, Scm3CBD binding is structurally incompatible with DNA binding by Cse4/H4, suggesting a mutual exclusion mechanism for its chaperone activity. Thus, retention of full-length Scm3 within the atypical centromeric nucleosome is dependent on its distinct DBD, which is apparently unaffected by the loss of Scm3-Cse4 contacts. In this context, we note that there are no inconsistencies between our results and experimental data of Camahort et al. (2009) and Pidoux et al. (2009), who observed a separation of Scm3 and Cse4 into distinct subcellular fractions upon extensive MNase digestion. This MNase-induced separation—which may have misled investigators to conclude that Scm3 is not within the nucleosomal particle—would be expected if Scm3 and Cse4 were bound adjacently on CDEII with little, if any mutual contact, and thus separable by nuclease cleavage. The MNase sensitivity of reconstituted Cse4/H4/Scm3 nucleoprotein complexes is consistent with this view.
The resistance to reconstitution of an octameric Cse4 nucleosome on centromere DNA indicates that the ~80 bp of AT-rich CDEII resists wrapping along the left-handed superhelical ramp provided by the histone octamer. However, the Scm3-facilitated association of CDEII with Cse4/H4 tetramers suggests that a shorter wrap—primarily ~30 bp on each side of the dyad axis, or ~60 bp in total—is tolerated over histone fold domains of the Cse4/H4 tetramer. Wrapping of AT-rich sequences on the histone core of a nucleosome is influenced not only by the intrinsic properties of DNA but also by environmental temperature. A poly(A·T) stretch that is inhibitory to nucleosome reconstitution under standard conditions of salt gradient dialysis at 4°C is nonetheless accommodated by reconstitution at higher temperatures (Puhl and Behe, 1995). A nucleosome containing an A16 stretch has been crystallized and atomic structure solved (Bao et al., 2006). Accordingly, we examined the temperature dependence of Cse4 nucleosome reconstitution on CEN3 and CEN4 DNA and found substantial increases in reconstitution efficiency of an octameric Cse4 nucleosome at temperatures of 23°C and 37°C (Figures S7A and S7B), consistent with a recent report (Kingston et al., 2011). Such permissive conditions allow the simultaneous formation on centromere DNA of two mutually exclusive Cse4 nucleosome populations: octameric Cse4 nucleosomes and atypical Cse4/H4/Scm3-containing complexes (Figure S7C).
Given that yeast grow at 30°C, rather than at 4°C, the inherent instability of an octameric Cse4 nucleosome should be partially mitigated. However, binding of Scm3DBD to CDEII would sterically occlude DNA sites for H2A/H2B binding. Although Scm3DBD has a modest sequence preference that should help direct it to the AT-rich element, it is CBF3, the CDEIII-specific DNA binding complex, that is responsible for centromere-specific recruitment of Scm3 (Camahort et al., 2007; our unpublished data). This is consistent with genetic studies showing that Scm3 and the Ndc10 component of CBF3 are mutually required for binding to centromeres in vivo (Camahort et al., 2007; Mizuguchi et al., 2007).
A Model of the Centromeric Nucleosome
We propose that there are two populations of Cse4 nucleosomes in yeast chromatin. One, indisputably octameric, occurs on noncentromeric DNAs and is the likely form of Cse4 that is incorporated at noncentromeric loci in the budding yeast genome (Camahort et al., 2009; Lefrancois et al., 2009). The second population—at centromeres—contains predominantly Scm3 and Cse4/H4 (and inner kinetochore proteins) and is deficient for H2A/H2B. We document simple experimental conditions that allow partial reconstitution of such atypical nucleosomes.
In the proposed model (Figure 7), we place a Cse4/H4 tetramer on CDEII, based on genetic interactions between Cse4 and CDEII (Keith and Fitzgerald-Hayes, 2000), high-resolution ChIP analysis (Camahort et al., 2009), and the likely exclusion of Cse4/H4 from CDEI and CDEIII by CBF1 and CBF3. Our in vivo evidence shows that at least two copies of Cse4 exist in the centromeric nucleosome (Figure S6), in agreement with Camahort et al. (2009) and Chen et al. (2000). A Scm3 dimer binds to the periphery of CDEII without stable contact with Cse4/H4. If the path of DNA in the Cse4/H4 tetrasome follows canonical structure, ~60 bp of CDEII wraps over histone fold domains of the Cse4/H4 tetramer in a left-handed supercoil for ~3/4 of one turn (Luger et al., 1997). In the absence of H2A/H2B, the trajectory of DNA emerging from the tetrasome and from Scm3 is unknown. On both sides of CDEII, inner kinetochore proteins CBF1 and CBF3 induce bending on CDEI and CDEIII by ~70° and ~55°, respectively (Niedenthal et al., 1993; Pietrasanta et al., 1999), but it is unknown whether DNA follows a left- or right-handed solenoidal supercoil, a plectonemic (interwound) supercoil (Cozarelli et al., 1990), or no supercoil. Thus, there are alternative configurations to a proposed switch from left- to right-handed DNA wrap over centromeric histones that could account for the 1.33 linking number change of a CEN3-containing yeast plasmid upon conditional mutation of the Ndc10 component of CBF3 (Furuyama and Henikoff, 2009). This mutation causes not only loss of CBF3 and Cse4, but also of Scm3, and thus of CBF1 as well (Camahort et al., 2007; Mizuguchi et al., 2007). Irrespective of handedness, the model we propose involves essentially one wrap of DNA.
Figure 7. Model of Budding Yeast Centromeric Nucleosome with Associated Inner Kinetochore Proteins.
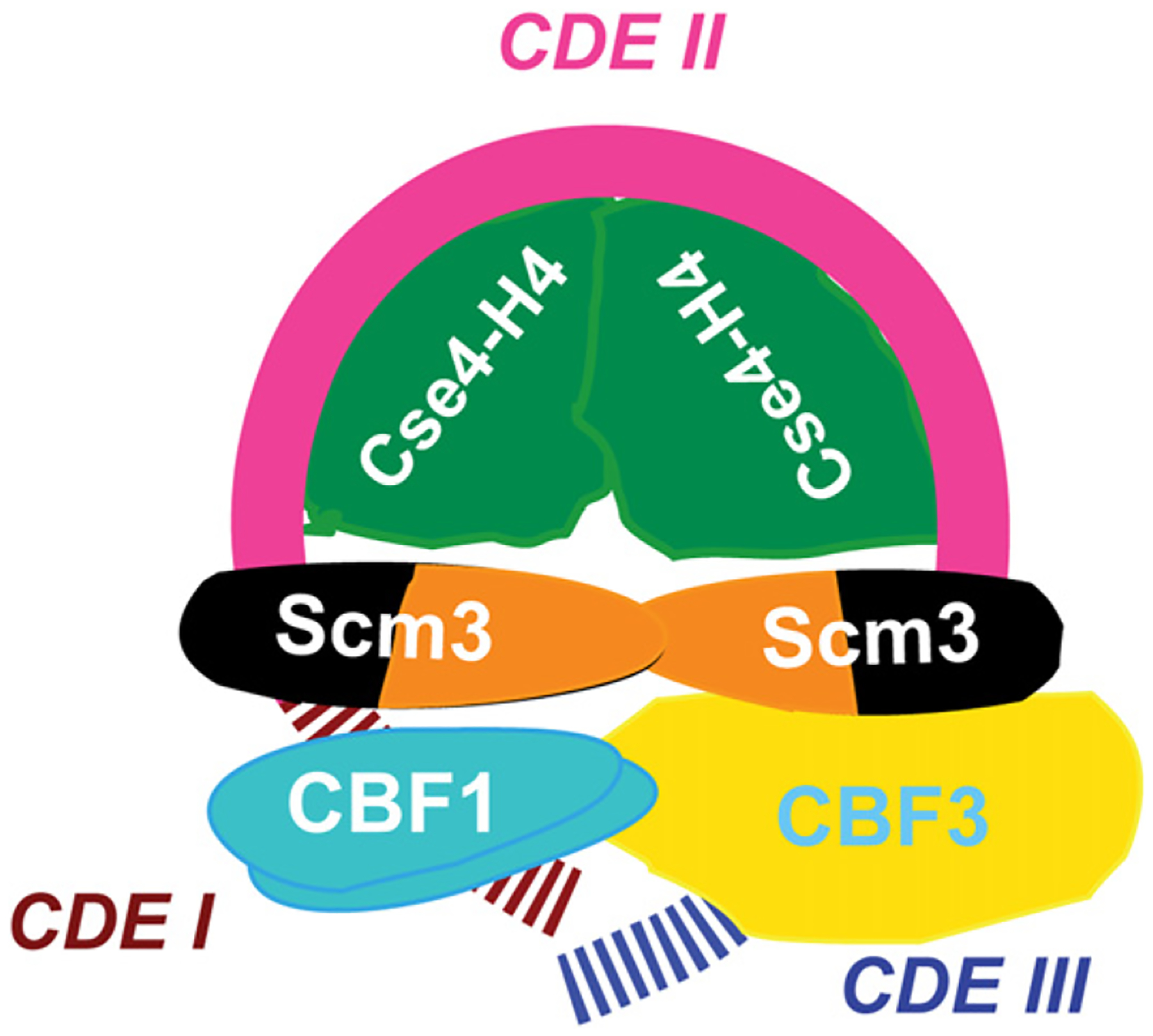
The model involves essentially one wrap of DNA. Cse4/H4 centers on CDEII, occupying ~60 bp, and Scm3 is at the remaining ~20 bp of CDEII adjacent to CDEI and CDEIII, consistent with high-resolution ChIP (Camahort et al., 2009). Scm3 is no longer in stable contact with Cse4 but contacts CBF3, which contacts CBF1 (Camahort et al., 2009; Hemmerich et al., 2000). The trajectory of DNA on Scm3, Cbf1, CBF3, and beyond is unknown. Scm3DBD is in dark green, and Scm3CBD is in orange.
As revealed by ChIP-qPCR and Scm3-GFP fluorescence in live cells, the steady-state persistence of Scm3 on centromere DNA throughout the cell cycle is strikingly correlated with a continued deficiency of H2B (and probably H2A) from centromere sequences. Thus, our model represents the prevalent composition for the Cse4 nucleosome at centromeres. (A conjecture that Scm3 occupancy may be restricted to a specific cell-cycle stage [Black and Cleveland, 2011] is not supported by our experimental data.) It is of interest that while Scm3 occupancy is persistent, Cse4/H4 is temporarily dislodged from centromere DNA in early S phase, as demonstrated by ChIP-qPCR. Such loss of Cse4 was previously reported in cells synchronized either by release from α factor arrest or by use of a temperature-sensitive mutation in a cell cycle regulator (cdc15–2). This indicates that the temporary dislocation of Cse4 is unlikely to be an artifact of cell synchronization method, but is related to DNA replication (Hajra et al., 2006).
The persistence of Scm3 in S phase indicates that Scm3 soon reoccupies centromere DNA after replication fork passage, and is therefore in position to target Cse4/H4 for reassembly. Constant, steady-state Scm3 occupancy may indicate the biological necessity for a centromere-bound, Cse4-specific chaperone to ensure stable retention of Cse4 in the single centromeric nucleosome, as demonstrated by FRAP analysis of Cse4-GFP (Pearson et al., 2004). Such a role is consistent with the finding that Scm3 is genetically required to maintain kinetochore function throughout the cell cycle (Camahort et al., 2007).
General Implications
Our analysis of budding yeast Scm3 should provide insights on the architecture of centromeric nucleosomes among the fungi, which possess clear Scm3 homologs with single or multiple DNA binding motifs linked to the conserved Scm3-CBD motif (Aravind et al., 2007; Mizuguchi et al., 2007). Thus, all fungal Scm3 proteins should at least facilitate deposition of CenH3 by a DNA binding, CenH3-specific chaperone mechanism. Whether Scm3 occupies centromeres throughout the cell cycle of other fungi is unclear. In fission yeast, Scm3 is indeed localized to centromeres through most of the cell cycle, except for a transient period in metaphase-anaphase of mitosis, when it clearly drops below detection (Pidoux et al., 2009; Williams et al., 2009).
HJURP, the human homolog of Scm3, is also critical for the centromeric deposition of human CenH3CENP-A in vivo and has CenH3-specific histone chaperone activity in vitro. However, HJURF does not persist on centromeres through most of the cell cycle (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010). Moreover, biochemical evidence that CENP-A exists in octameric nucleosomes on human centromeric α-satellite sequences is compelling. However, given the reported instability of H2A/H2B dimers within the octameric CENP-A nucleosome reconstituted from bacterially expressed proteins (Conde e Silva et al., 2007), the possibility of deviations from the norm should not be overlooked for the minute subpopulation of CENP-A nucleosomes making functional contact with spindle microtubules via the kinetochore.
EXPERIMENTAL PROCEDURES
Reconstitution of Nucleoprotein Complexes
Protein purification, reconstitution of histone tetramers, octamers and Scm3-histone complexes, and reconstitution of nucleosomes were performed using established procedures (see the Supplemental Experimental Procedures).
Abrupt Dilution Assay for Histone Chaperone Activity of Scm3
Binding reactions were carried out in 50 μl volume, typically containing 200 mM NaCl, 10 mM Tris-Cl (pH 7.5), 1 mM EDTA, 0.02% NP-40, 2 mM β-mercaptoethanol, 400 μg/ml BSA, 0.5–4.0 μg of purified DNA fragments, and up to 5 μl of purified proteins or protein complexes. Histones were first mixed with Scm3 or Scm3 fragments in a total of 5 μl in 2M NaCl. The mixture was abruptly diluted to binding buffer to give a final of 200 mM NaCl. After incubation at room temperature for 10 min, DNA was added to the reactions, incubated for 30 min at room temperature, and analyzed by EMSA and SYBR Green staining.
Immunoprecipitation
Reconstituted protein-DNA complexes were incubated for 20 min at room temperature with anti-FLAG antibodies or control IgG coupled to Dynabeads-protein G. Beads were washed with binding buffer containing 200 mM NaCl, and bound proteins analyzed by SDS PAGE and western blotting. Unbound fractions were analyzed by EMSA and SYBR Green staining. A duplicate EMSA gel was transferred to PVDF filter for western blotting to identify protein-DNA complexes containing Cse4/H4, Scm3, or both.
Cell Synchronization
Cells were grown in YPD medium at 30°C. Cells were blocked in G1 by the addition of 0.3 μg/ml mating pheromone α factor, and 0.2 and 0.1 μg/ml at 45 and 90 min, respectively. After 110 min, >95% of cells were unbudded and were released into medium lacking α factor.
ChIP
ChIP was done essentially as described previously (Mizuguchi et al., 2007), using PCR primers specific for CEN3 and CEN4 with their flanking sequence. “CEN3” and “peri-CEN3” PCR products (Figure 6 and Figure S5) represent DNA between coordinates 114,319–114,562, and 115,019–115,321. “CEN4” and “peri-CEN4” PCR products span coordinates 448,899–449,166, and 449,644–449,891, respectively. Coordinates for the “Hsc82” primer sets are CHRXIII 631551 and 631729. Percent IP value (Figure S5) was determined by normalizing to a primer set internal to Hsc82 PCR signal.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ed Luk for performing FACS analysis; members of our laboratory for helpful suggestions; and Mitch Smith, David Clark, Michael Lichten, Victor Zhurkin, Kerry Bloom, and the reviewers for helpful comments on the manuscript. We are grateful for the gift of 601 nucleosome positioning sequences from Jon Widom. This manuscript is dedicated to his memory. The work was supported by the intramural research program of the National Cancer Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, two movies, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.molcel.2011.07.009.
REFERENCES
- Akey CW, and Luger K (2003). Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol 13, 6–14. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, and Wu C (2007). Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle 6, 2511–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, and Moudrianakis EN (1991). The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88, 10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, and Rogers K (2005). Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae. Genetics 171, 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, White CL, and Luger K (2006). Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol 361, 617–624. [DOI] [PubMed] [Google Scholar]
- Bechert T, Heck S, Fleig U, Diekmann S, and Hegemann JH (1999). All 16 centromere DNAs from Saccharomyces cerevisiae show DNA curvature. Nucleic Acids Res. 27, 1444–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, and Bassett EA (2008). The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol 20, 91–100. [DOI] [PubMed] [Google Scholar]
- Black BE, and Cleveland DW (2011). Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr., and Cleveland DW (2004). Structural determinants for generating centromeric chromatin. Nature 430, 578–582. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, and Cleveland DW (2007). Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309–322. [DOI] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, and Pidoux A (2010). Building centromeres: home sweet home or a nomadic existence? Curr. Opin. Genet. Dev 20, 118–126. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, and Gerton JL (2007). Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26, 853–865. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, and Gerton JL (2009). Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, and Fitzgerald-Hayes M (2000). The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol 20, 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, and Cairns BR (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem 78, 273–304. [DOI] [PubMed] [Google Scholar]
- Clarke L, and Carbon J (1985). The structure and function of yeast centromeres. Annu. Rev. Genet 19, 29–55. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, and Sullivan KF (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, and Prunell A (2007). CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol 370, 555–573. [DOI] [PubMed] [Google Scholar]
- Cozarelli NR, Boles TC, and White JH (1990). DNA Topology and Its Biological Effects (New York: Cold Spring Laboratory Press; ). [Google Scholar]
- Dalal Y, Wang H, Lindsay S, and Henikoff S (2007). Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5, e218 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, and Dalal Y (2010). Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 107, 20317–20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, and Almouzni-Pettinotti G (2009). HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, and Cleveland DW (2009). Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, and Biggins S (2007). Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104, 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, and Henikoff S (2009). Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, and Bernstein E (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. [DOI] [PubMed] [Google Scholar]
- Hajra S, Ghosh SK, and Jayaram M (2006). The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol 174, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran TE, and Mohanty U (2009). The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys 42, 41–81. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Stoyan T, Wieland G, Koch M, Lechner J, and Diekmann S (2000). Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc. Natl. Acad. Sci. USA 97, 12583–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, and Dalal Y (2005). Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev 15, 177–184. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, and Salmon ED (2006). Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol 8, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, and Segal E (2008). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, and Allshire RC (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Keith KC, and Fitzgerald-Hayes M (2000). CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics 156, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, and Singleton MR (2011). Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem 286, 4021–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, and Snyder M (2009). Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10, 37 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol 276, 19–42. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, and Wu C (2007). Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164. [DOI] [PubMed] [Google Scholar]
- Ng R, Ness J, and Carbon J (1986). Structural studies on centromeres in the yeast Saccharomyces cerevisiae. Basic Life Sci. 40, 479–492. [DOI] [PubMed] [Google Scholar]
- Niedenthal RK, Sen-Gupta M, Wilmen A, and Hegemann JH (1993). Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21, 4726–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, and Bloom K (2004). Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol 14, 1962–1967. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, and Allshire RC (2009). Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrasanta LI, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, and Hansma H (1999). Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci. USA 96, 3757–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl HL, and Behe MJ (1995). Poly(dA).poly(dT) forms very stable nucleosomes at higher temperatures. J. Mol. Biol 245, 559–567. [DOI] [PubMed] [Google Scholar]
- Rando OJ, and Chang HY (2009). Genome-wide views of chromatin structure. Annu. Rev. Biochem 78, 245–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, and Allshire RC (2009). Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, and Black BE (2010). The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 467, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Mori T, Sakurai T, and Shindo H (2000). Destabilization of nucleosomes by an unusual DNA conformation adopted by poly(dA) small middle dotpoly(dT) tracts in vivo. EMBO J. 19, 3358–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, and Hamiche A (2010). HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA 107, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM (2002). Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol 14, 279–285. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, and Baker RE (2007). Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA 104, 10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (1985). Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. USA 82, 8419–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, and Masri K (1994). Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol 127, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, and Yokoyama S (2005). Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. J. Biol. Chem 280, 41609–41618. [DOI] [PubMed] [Google Scholar]
- Visnapuu ML, and Greene EC (2009). Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat. Struct. Mol. Biol 16, 1056–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J (2001). Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys 34, 269–324. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, and Russell P (2009). Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, and Okazaki T (2000). Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, and Bai Y (2011). Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


