Abstract
FOXE1 is a thyroid-specific transcription factor essential for thyroid gland development and maintenance of the differentiated state. Interestingly, a strong association has been recently described between FOXE1 expression and susceptibility to thyroid cancer, but little is known about the mechanisms underlying FOXE1-induced thyroid tumorigenesis. Here, we used a panel of human thyroid cancer-derived cell lines covering the spectrum of thyroid cancer phenotypes to examine FOXE1 expression and to test for correlations between FOXE1 expression, the allele frequency of two SNPs and a length polymorphism in or near the FOXE1 locus associated with cancer susceptibility, and the migration ability of thyroid cancer cell lines. Results showed that FOXE1 expression correlated with differentiation status according to histological sub-type, but not with SNP genotype or cell migration ability. However, loss-and-gain-of-function experiments revealed that FOXE1 modulates cell migration, suggesting a role in epithelial-to-mesenchymal transition (EMT). Our previous genome-wide expression analysis identified Zeb1, a major EMT inducer, as a putative Foxe1 target gene. Indeed, gene silencing of FOXE1 decreased ZEB1 expression, whereas its overexpression increased ZEB1 transcriptional activity. FOXE1 was found to directly interact with the ZEB1 promoter. Lastly, ZEB1 silencing decreased the ability of thyroid tumoral cells to migrate and invade, pointing to its importance in thyroid tumor mestastases. In conclusion, we have identified ZEB1 as a bona fide target of FOXE1 in thyroid cancer cells, which provides new insights into the role of FOXE1 in regulating cell migration and invasion in thyroid cancer.
Keywords: thyroid cancer, FOXE1, ZEB1, SNP, EMT
Introduction
Thyroid cancer is the most commonly occurring endocrine malignancy and its incidence has steadily increased over the last four decades, accounting for 1% of all annual cancer diagnoses (Davies & Welch 2006, Lim et al. 2017). Papillary thyroid carcinoma (PTC), a carcinoma of follicular cell origin, is the most frequent form of differentiated thyroid carcinoma and represents 80–85% of all thyroid malignancies (Zaballos & Santisteban 2017).
Initiation and progression of thyroid cancer results from the acquisition of multiple genetic alterations. PTC is mostly driven by mutations that activate the MAPK (mitogen-activated protein kinase) signaling pathway (Zaballos & Santisteban 2017), which includes mutations in the intracellular transducer RAS and the serine/threonine kinase BRAF, and rearrangements in the cell membrane receptor tyrosine kinase RET (DeLellis 2006, Riesco-Eizaguirre & Santisteban 2016). Beyond these somatic alterations, PTC displays a strong hereditary component, since it shows the highest familial relative risk (8.60–10.30) in first-degree relatives of probands among cancers not displaying Mendelian inheritance (Goldgar et al. 1994, Pal et al. 2001).
Genome-wide association studies (GWAS) have identified SNPs associated with PTC risk (Gudmundsson et al. 2009, Matsuse et al. 2011, Mancikova et al. 2015). These allelic variations include rs965513, found in the proximal region of the FOXE1 (Forkhead Box E1) gene (approximately 57 kb from the FOXE1 locus) and rs1867277, within its promoter (NM_004473.3:c. −283G>A), and both are strongly associated with an increased risk of PTC (Landa et al. 2009, Gudmundsson et al. 2012, Jones et al. 2012).
FOXE1, formerly known as thyroid transcription factor-2, is located on chromosome 9q22 in humans and encodes a DNA-binding protein belonging to the forkhead/winged-helix family, a superfamily of evolutionarily conserved transcriptional regulators that share a highly conserved forkhead box or winged helix DNA-binding domain (Chadwick et al. 1997, Cuesta et al. 2007). This transcription factor possesses a polymorphic polyalanine (poly-A) tract just distal to its DNA-binding domain (rs71369530), which varies between 11 and 22 alanine residues, although FOXE114Ala and FOXE116Ala account for greater than 98% of reported alleles (Macchia et al. 1999, Kallel et al. 2010).
FOXE1 is a thyroid-specific transcription factor that, together with PAX8 and NKX2-1, coordinately maintains the differentiated state of the thyroid gland and is also essential for its correct development (Zannini et al. 1997, Fernandez et al. 2015). Foxe1 is also a key player in thyroid organogenesis, as its expression during early thyroid development is required for thyrocyte precursor migration (De Felice et al. 1998, De Felice & Di Lauro 2004, Parlato et al. 2004, Fernandez et al. 2015). In the differentiated thyroid, Foxe1 is a transcriptional activator of the thyroperoxidase and thyroglobulin genes and mediates the ability of cells to respond to external stimuli including thyroid stimulating hormone, insulin-like growth factor-1, and transforming growth factor-β (Santisteban et al. 1992, Ortiz et al. 1999, Lopez-Marquez et al. 2019). A previous genomic study by our group in a rat thyroid follicular cell line identified two thyroid-specific genes (Duox2 and Slc5a5) among other genes (including Cdh1 and Nr4a2) as novel Foxe1 targets (Fernandez et al. 2013).
Increasing evidence from genetic studies associates FOXE1 with PTC, implicating it as a susceptibility gene in thyroid cancer; however, its involvement in the initiation and progression of these tumors is unknown. Likewise, several studies have reported the likely contribution of FOXE1 to carcinogenesis in breast cancer (Park et al. 2012), pancreatic cancer (Sato et al. 2003), and basal and squamous cell carcinomas of the skin (Eichberger et al. 2004, Venza et al. 2010). In the context of thyroid carcinoma, several studies have focused on relating FOXE1 expression and localization in cancer cells to tumor development. For instance, FOXE1 overexpression has been described in PTC (Nonaka et al. 2008, Bychkov et al. 2013), as well as a gradual decrease in its nuclear expression according to the degree of tumor dedifferentiation, along with cytoplasmic accumulation (Zhang et al. 2006). This abnormal localization of FOXE1 could be related to thyroid tumorigenesis. In addition, an association has been described between the FOXE1 poly-A repeat region and PTC (Bullock et al. 2012).
These observations, together with the genetic studies associating SNPs in and near the FOXE1 locus with thyroid cancer risk, and the altered expression of FOXE1 (Landa et al. 2009, He et al. 2015a , Wang et al. 2017), have motivated us to further investigate the role of FOXE1 in thyroid cancer, to try to better understand the dual role of this transcription factor as both a differentiation and a tumoral factor. Accordingly, in the present study we analyzed FOXE1 expression levels in different thyroid cancer cell lines and looked for potential correlations with the genotypes of SNPs rs965513 and rs1867277 and with the length polymorphism rs71369530. Furthermore, we explored FOXE1-mediated regulation of epithelial-to-mesenchymal transition (EMT) by analyzing the expression of new genes regulated by FOXE1 in thyroid cells. Among them, ZEB1, a major EMT inducer, emerged as a putative FOXE1 target gene. Lastly, we studied the mechanism by which FOXE1 controls ZEB1 in thyroid cancer cell lines and demonstrated the involvement of ZEB1 in the regulation of EMT in thyroid cancer cells.
Methods and materials
Cell culture
Human thyroid cancer cell lines were obtained from the following sources: BCPAP and NIM from Dr M Santoro (University of Federico II, Naples, Italy); C643, Hth7, Hth83, and SW1736 from Dr N E Heldin (University of Uppsala, Uppsala, Sweden); FTC-133, K1 and Nthy-Ori-3.1 from the European Collection of Authenticated Cell Cultures (ECACC; Salisbury, Wiltshire, UK); WRO-82-1 from Dr G J F Juillard (University of California-Los Angeles School of Medicine, Los Angeles, CA, USA); TPC1 from Dr A P Dackiw (Johns Hopkins University, Baltimore, MD, USA); KTC-1 and KTC-2 from Dr Junichi Kurebayashi (Kawasaki Medical School, Japan); Cal62, ML-1, TT206-C09, and 8505c from the Leibniz-Institut DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany); T235 and T238 from Dr Lucia Roque (Portuguese Cancer Institute, Lisbon, Portugal); and OCUT2 from Dr James A Fagin (Memorial Sloan Kettering Cancer Center, New York, NY, USA). All thyroid cancer cell lines and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM). The human thyroid cell line Nthy-Ori-3.1 (ECACC #90011609) was grown in Roswell Park Memorial Institue 1640 medium. All growth media were supplemented with 10% fetal bovine serum (FBS), 50 U/mL penicillin, 50 μg/mL streptomycin, and 2 mmol/l-glutamine.
PCCl3 thyroid follicular cells, a continuous rat differentiated cell line, were cultured in Coon’s modified Ham’s F-12 medium supplemented with 5% donor calf serum (Thermo Fisher Scientific), and a six-hormone medium mixture: 1 nmol/L bovine thyroid-stimulating hormone, 10 μg/mL insulin, 10 ng/mL somatostatin, 5 μg/mL transferrin, 10 nmol/L hydrocortisone, and 10 ng/mL glycyl-l-histidyl-l-lysine acetate; all from Sigma-Aldrich.
All cell lines were used up to ten passages, maintained in 5% v/v CO2 at 37°C, and authenticated every 6 months by short tandem repeat profiling using the Applied Biosystems Identifier kit, at the Genomic Facility, Institute of Biomedical Research (IIBm; Madrid, Spain).
RT-qPCR
RNA was extracted with TRIzol (Thermo Fisher Scientific), and 1 µg was added to a reverse-transcriptase reaction mix (M-MLV; Promega Co.,). Quantitative PCR (qPCR) was conducted on the Mx3000P QPCR platform (Agilent Technologies). Reactions were performed in triplicate with the indicated primers and templates using the KAPA SYBR FAST qPCR Master Mix (Merck KGaA) for 40 cycles. Relative gene expression levels were quantified using the comparative threshold cycle 2−ΔΔCt method (Livak & Schmittgen 2001) by normalizing transcript levels to the expression of a housekeeping gene. The sequences of the specific primers purchased from Sigma-Aldrich are listed in Supplementary Table 1 (see section on supplementary materials given at the end of this article).
Western blotting
Total protein extracts were obtained by scraping cells in RIPA buffer containing a protease inhibitor cocktail (Roche). Equal amounts of protein (30 µg) were separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked, and incubated overnight with primary antibodies diluted in phosphate buffered saline (PBS) 0.1% v/v Tween 20 containing 5% w/v nonfat dry milk. Horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated for 1 h at room temperature and binding was detected using enhanced chemiluminescence reagents (Thermo Fisher Scientific). The following antibodies were used in this study: anti-rat Foxe1 (#PA0200, Biopat Milan, Italy); anti-human FOXE1 (#ab5080, Abcam); anti-E-cadherin (#BD 610182; BD Biosciences, Bedford, MA); anti-ZEB1 (D80D3 #3396; Cell Signaling Technology), and anti-β-actin (#sc-1616; Santa Cruz Biotechnology).
SNP genotyping
Genomic DNA from thyroid cell lines was extracted using the traditional saline method. DNA regions containing the two thyroid cancer-associated SNPs and the poly-A tract were amplified by PCR using specific primer pairs (Supplementary Table 2). PCR amplification was performed using KAPA Taq DNA polymerase (Merck KGaA) in a total volume of 15 μL containing 50 ng of DNA. PCR products were purified using the PureLink PCR Purification Kit (Invitrogen). Sequence analysis was performed on the ABI 3700 automated DNA sequencer (Applied Biosystems) using BigDye Terminator chemistry.
RNA interference, plasmids, and transfection
For FOXE1 gene silencing studies, cells were transfected with 25 nmol/L of FOXE1 siRNA (RatFoxe1 ON-TARGET Plus SMART pool or HumanFOXE1 ON-TARGET Plus SMART pool) or with scrambled siRNA (ON-TARGET Plus Non-targeting pool) using Dharma-FECT 1 Transfection Reagent (Dharmacon). For ZEB1 gene silencing studies, human ZEB1 silencer® select siRNAs n269441 (siZEB1-1) and n269443 (siZEB1-2) were used (Thermo Fisher Scientific) following the same procedure as described previously. For FOXE1 overexpressing studies, 1.5 µg of human FOXE-Flag or empty-Flag expression vectors (Clifton-Bligh et al. 1998, Carre et al. 2007) were transiently transfected. The day before transfection, cells were seeded in a six-well culture plate at a density of 2 × 105 cells per well. Samples were harvested in duplicate at different time points (24, 48, and 72 h) after transfection, and total RNA and protein was extracted.
Chromatin immunoprecipitation and electrophoretic mobility shift assay
Chromatin immunoprecipitation (ChIP) was performed as previously described (Fernandez et al. 2013) using the HighCell ChIP Kit (Diagenode Inc., Denville, NJ, USA). Cross-linked PCCl3 chromatin was immunoprecipitated using a polyclonal antibody against Foxe1 (Biopat, Milan, Italy). Two independent ChIP experiments were carried out using two different batches of Foxe1 antibody. Immunoprecipitated Foxe1 and input samples were assayed by qPCR using specific primers for the analyzed region on the Zeb1 promoter (Supplementary Table 3). The known Foxe1 target Tpo was used as a positive control for immunoprecipitation, whereas two regions located −2 kb (cis 2 kb) and −2.5 kb (cis 2.5 kb) upstream of the transcription start site were used as negative controls. PCR reactions were performed in triplicate using the SYBR Green PCR Kit (Kapa Biosystems, Woburn, MA, USA). The enrichment of target sequences in ChIP experiments was calculated relative to the negative controls and normalized to their relative amplification in the input sample (Ruiz-Llorente et al. 2012).
Electrophoretic mobility shift assays were performed using an oligonucleotide probe derived from the in silico-identified FOXE1-binding site within the human ZEB1 promoter (Oligo FOXE1: 5′-ATTCAAATAAACACTTGCATTTTA-3′). As a control, the Foxe1-binding site oligonucleotide derived from the rat Tpo promoter was used (Aza-Blanc et al. 1993). Probes were labeled with [γ32P]-ATP using T4 polynucleotide kinase (Promega) and purified using Quick Spin G-25 Sephadex columns (Roche Life Sciences). Recombinant FOXE1 was produced by in vitro transcription-translation using the TNT-coupled reticulocyte lysate system (Promega) and incubated with the labeled probe. Binding reactions were performed in a buffer containing 40 mmol/L Hepes, pH 7.9, 75 mmol/L KCl, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol, 150 ng/μL poly(dI-dC), and 5% w/v Ficoll at room temperature for 30 min. Samples were electrophoresed on a 5% w/v polyacrylamide gel in 0.5× Tris borate-EDTA. For competition, a 100-fold excess of the same (‘related’) or different (‘unrelated’) unlabeled oligonucleotides were used, as indicated in each experiment.
Luciferase assay
HeLa cells were seeded at a density of 2 × 105 cells per well in six-well tissue culture plates 24 h before transfection. Transfections were performed using the calcium phosphate co-precipitation method (Chen & Okayama 1988). The human ZEB1 gene promoter (Dave et al. 2011) was transiently transfected alone (1.5 µg) or in combination with 0.5 µg of a human FOXE-Flag (Clifton-Bligh et al. 1998, Carre et al. 2007) expression vector. One hundred nanograms of the CMV Renilla vector were cotransfected to assess transfection efficiency. After 48 h, cells were harvested, lysed, and analyzed for Luciferase and Renilla activities using the Dual-Luciferase Reporter Assay System (Promega).
Migration assay
Cell migration was evaluated using scratch wound healing assays. Cells were seeded on a six-well plate and allowed to reach confluence. Twenty-four hours after transfection, cells were treated for 2 h with 10 µg/mL mitomycin C in medium with 10% FBS to inhibit proliferation. After treatment, a single wound in the center of cell monolayer was made with a 10 μL pipette tip and cell debris was removed by washing with PBS. After 4, 8, 24, and 48 h of incubation in serum-free medium, the wound closure areas were visualized under an inverted microscope and imaged. Each experiment was performed in triplicate.
Invasion assay
Cell invasion/migration was analyzed in Transwell assays using BD BioCoat Matrigel Invasion Chambers (BD Biosciences). In total, 2 × 105 cells were suspended in 500 µL of serum-free medium and seeded into the upper chamber. The lower chamber of the Transwell was filled with 500 µL DMEM containing 20% FBS as a chemoattractant. After 18 h of incubation, cells on the surface of upper chamber were removed by scraping with a cotton swab. The invaded/migrated cells on the lower surface of the filter were fixed with 4% v/v paraformaldehyde, stained with 0.1% w/v crystal violet, imaged, and quantified by counting cells in five random fields. Experiments were performed three times in triplicate.
Bioinformatics predictions
The TCGA database was queried to assess the correlations between mRNA levels and clinical features. Firebrowse (http://firebrowse.org) was used to analyze FOXE1 mRNA levels in different tumor types. The cBioPortal (http://www.cbioportal.org) and the Cancer Regulome Explorer (http://explorer.cancerregulome.org) data portals were used to obtain the correlations using the thyroid carcinoma dataset (THCA).
Microarray data of FOXE1 levels in anaplastic thyroid carcinomas (ATCs) comparing with normal thyroid tissues were searched in The Gene Expression Omnibus (GEO) database. Two microarray datasets GSE33630 (Tomas et al. 2012) and GSE65144 (von Roemeling et al. 2015) were analyzed.
Statistical analysis
All data are reported as mean ± s.e.m. or mean ± s.d. Comparisons between two groups were made using two-tailed Student’s unpaired t-test. Statistical analysis was performed with GraphPad Prism software (GraphPad Software Inc.). Differences were considered statistically significant at P < 0.05. Associations between TCGA data and differentiation score and between FOXE1 expression and migration were assessed using Pearson’s (r) test.
Results
FOXE1 expression levels correlate with differentiation status in human thyroid cancer cell lines
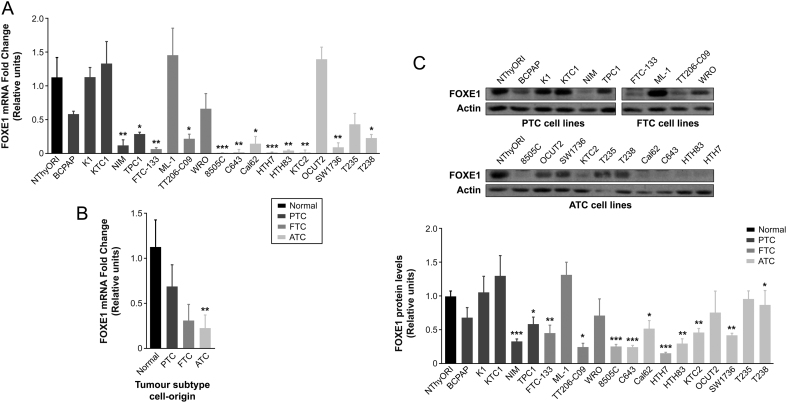
Given that FOXE1 has been described as a susceptibility gene in thyroid cancer, we first compared its level of expression in a panel of human thyroid cancer cell lines with its expression in the normal (immortalized) thyroid follicular cell line NThy-ori-3-1 (NThyOri). The cell lines used covered the spectrum of thyroid neoplasms, from well-differentiated PTC and follicular thyroid carcinomas (FTC) to anaplastic (undifferentiated) thyroid carcinoma (ATC), which is the most aggressive malignancy. Although there were some exceptions, overall FOXE1 mRNA levels were significantly higher in control and in PTC-derived cells than in FTC-derived cells, with the lowest expression observed in ATC-derived cell lines (Fig. 1A and B). Western blotting analysis of FOXE1 protein confirmed the RNA expression data, showing significantly higher expression in PTC cells than in FTC and ATC cells, with some ATC lines having almost undetectable levels of FOXE1 (Fig. 1C). These results indicate that FOXE1 expression positively correlates with the differentiation status according to the histological sub-type origin of cell lines.
Figure 1.
FOXE1 expression in thyroid cancer cell lines. (A) FOXE1 relative mRNA expression in thyroid cell lines. Gene expression was normalized to expression of GAPDH. Values represent mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 vs NThyORI control cells. (B) FOXE1 relative mRNA expression according to tumor subtype-cell origin. Values represent mean ± s.e.m. **P < 0.01 vs NThyORI control cells. (C) Upper panel: Representative Western blot of three independent experiments showing FOXE1 protein levels in thyroid cell lines. Actin was used as a loading control. Lower panel: FOXE1 protein expression in thyroid cell lines normalized to expression of actin. Values represent mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.001, and ***P < 0.001 vs NThyORI control cells.
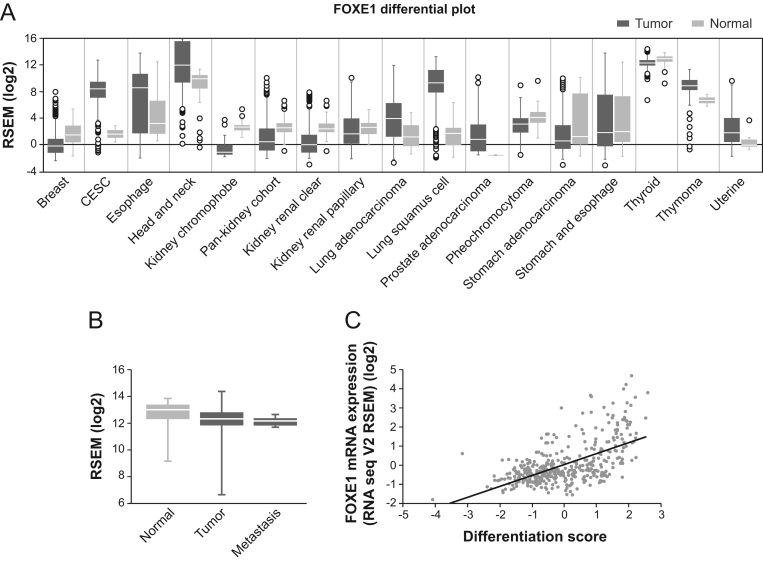
To extend these observations to human tumors in general and in particular to thyroid tumors, we analyzed the pattern of FOXE1 expression in a range of tumor samples and normal tissues using data acquired from The Cancer Genome Atlas (TCGA) (The Cancer Genome Atlas 2014) using the FireBrowse and Morpheus tools. We observed that FOXE1 levels were elevated in several cancer types, with the most striking changes seen in esophagus and lung squamous cell carcinomas (Fig. 2A). By contrast, the expression levels of FOXE1 in PTC and normal thyroid tissue were very similar (normal tissue n = 59, tumor samples n = 501) (Fig. 2B). Of note, the levels of FOXE1 in thyroid metastases (n = 8) were similar to those of the primary tumor samples and normal tissue (Fig. 2B), suggesting a role for FOXE1 in tumor progression. To further explore the implications of FOXE1 in thyroid cancer, we examined for correlations with aggressive clinical features. TCGA data from the public databases cBioportal and Cancer Regulome (Cerami et al. 2012, Gao et al. 2013) indicated that FOXE1 levels did not correlate with the risk of recurrence or with extrathyroidal extension (Supplementary Fig. 1A). However, we observed a clear correlation between FOXE1 expression and the differentiation state in PTC tumors (Fig. 2C), which is consistent with the data acquired in the thyroid tumor cells lines. This was also confirmed after analyzing FOXE1 expression levels in a genome array of human tumors that encompass all histological variants (data not shown) (Oncomine Data Set (www.oncomine.org)) (Giordano et al. 2006). In addition, microarray studies have shown that FOXE1 is among the 15 most downregulated genes in ATC (Tomas et al. 2012, von Roemeling et al. 2015) (Supplementary Fig. 1B).
Figure 2.
FOXE1 expression in human tumors. (A) FOXE1 relative mRNA expression in the indicated cancer types (CESC, cervical squamous cell carcinoma) obtained by Firebrowse analysis of TCGA database. (B) Box plot of FOXE1 mRNA expression levels in thyroid normal tissue, papillary thyroid carcinoma (PTC), and metastases: data were obtained from TCGA database. (C) Correlation between FOXE1 mRNA expression levels in thyroid tumors and differentiation score. Data were obtained from TCGA database (Pearson r = 0.5938).
Genotyping of cancer risk FOXE1 SNPs rs965513 rs1867277 and the FOXE1 poly-alanine repeat region (rs71369530) in thyroid cancer cell lines
Association studies have identified two SNPs that are clearly linked to PTC and FTC in multiple populations (Wang et al. 2018). Both rs1867277 and rs965513 SNPs are located in the same disequilibrium block; that is, their alleles co-occur on the same haplotype more often than is expected by chance and are located 238 bp and 57 kb upstream of the FOXE1 transcription start site, respectively (Gudmundsson et al. 2009, Landa et al. 2009). In addition, an association has been found between the SNP genotypes and expression levels of FOXE1. Thus, it was reported that in unaffected thyroid tissue, FOXE1 expression was significantly lower in patients with the rs965513 AA genotype when compared with those with the GG genotype, but there was no significant correlation in PTC tumors (He et al. 2015a ). Furthermore, the polymorphism in the length of the poly-A tract of FOXE1 (rs71369530), which has been associated with thyroid cancer (Kallel et al. 2010), has been demonstrated to be in tight linkage disequilibrium with rs1867277, in addition to being associated (FOXE1 16Ala) with PTC (Bullock et al. 2012).
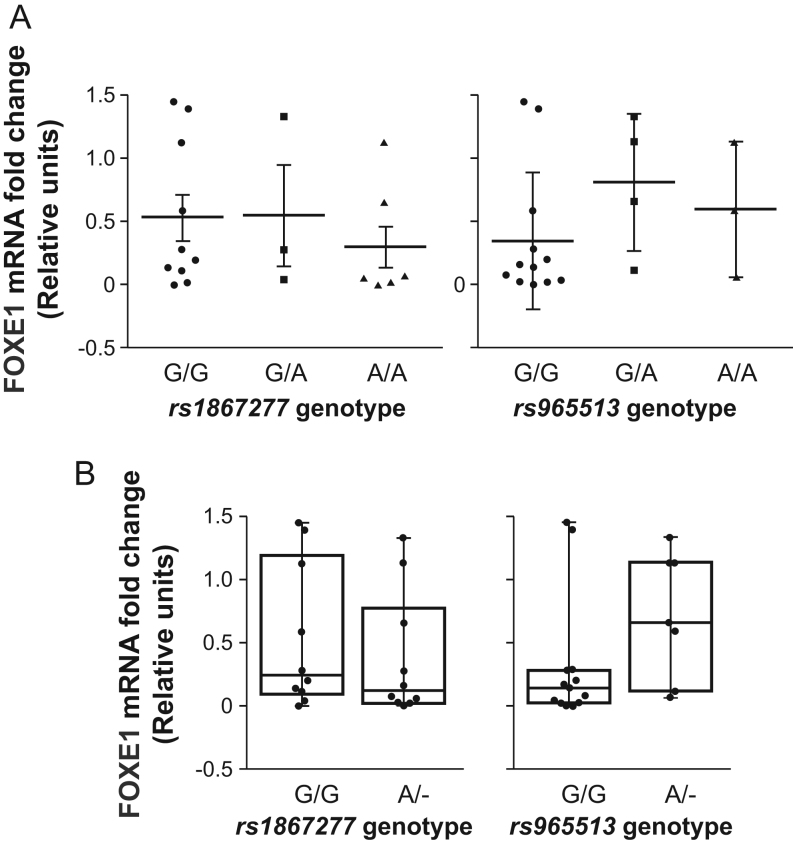
To ascertain whether a correlation exists between SNP haplotypes and FOXE1 expression in cancer, rs1867277, rs965513, and rs71369530 were genotyped by direct automated sequencing in the panel of human thyroid cancer cell lines. Allele variability was found in the analyzed cell lines, reflecting common variability in the general population, with minor allele frequency [A] of 0.5 for rs1867277 and 0.4 for rs965513 (Table 1). No significant correlation between the rs1867277 and rs965513 genotypes and the expression levels of FOXE1 were observed in the thyroid cancer cell lines analyzed (Fig. 3A). Moreover, we failed to find a correlation between FOXE1 expression and the G (G/G) or A (G/A or A/A genotype) allele (Fig. 3B). Finally, we confirmed that the most frequent FOXE1 poly-A variant in our panel of thyroid cancer cells is FOXE1 14Ala followed by FOXE1 16Ala. Other repeat lengths were rare in the thyroid cancer cell lines (Table 1). In addition, we failed to observe any correlation between the FOXE1 poly-A variant and the expression levels of FOXE1 in the thyroid cancer cell lines analyzed (data not shown).
Table 1.
Genotypes of FOXE1 SNPs in thyroid cancer cell lines.
| Cancer subtype | Cell line | Mutation | SNPs genotype | Other described mutations | ||
|---|---|---|---|---|---|---|
| rs1867277 | rs965513 | Poly Alanine | ||||
| PTC | BCPAP | BRAF V600E | G/G | A/A | 14/14 | p53 D259Y/AKT1 copy gain |
| PTC | K1 | BRAF V600E | A/A | A/A | 16/16 | PI3KCA E542K |
| PTC | KTC1 | BRAF V600E | G/A | G/A | 14/14 | – |
| PTC | NIM | BRAF V600E | G/G | G/A | 14/14 | – |
| PTC | TPC1 | RET/PTC1 | G/G | G/G | 14/14 | – |
| FTC | FTC-133 | PTEN R130* | A/A | A/A | 16/16 | p53 R273H |
| FTC | ML-1 | – | G/G | G/G | 14/14 | p53 frameshift |
| FTC | TT206-C09 | NRAS | G/A | G/G | 14/14 | p53 R273C |
| FTC | WRO | BRAF V600E | A/A | G/A | 14/16 | – |
| ATC | 8505c | BRAF V600E | G/G | G/G | 14/14 | p53 R248G |
| ATC | C643 | HRAS | A/A | G/G | 14/14 | p53 R248Q and K286E |
| ATC | Cal62 | KRAS | G/G | G/G | 14/14 | p53 A161D |
| ATC | HTH7 | NRAS | A/A | G/G | 14/16 | p53 G245S |
| ATC | HTH83 | HRAS | G/A | G/G | 12/14 | – |
| ATC | KTC2 | BRAF V600E | G/G | G/G | 14/14 | – |
| ATC | OCUT2 | BRAF V600E | G/G | G/G | 14/14 | PI3KCA H1047R |
| ATC | SW1736 | BRAF V600E | A/A | G/G | 14/16 | p53 Null |
| ATC | T235 | BRAF V600E | G/G | G/G | 14/14 | – |
| ATC | T238 | BRAF V600E | A/A | G/G | 16/16 | p53 S183*/PI3KCA E542K |
| – | NThyORI | – | G/G | G/A | 14/14 | – |
The genotypes of the rs1867277, rs965513 SNPs and the length of the poly-alanine tract are shown in the central columns. The cancer subtype from which the cell lines originated, as well as the driver mutations, is also shown.
Figure 3.
No correlation between single nucleotide polymorphism genotypes and FOXE1 expression levels. (A) Representation of correlation of FOXE1 mRNA expression and genotypes (GG, AG, or AA) of rs1867277 and rs965513. Values represent mean ± s.e.m. (B) Box plot of FOXE1 mRNA expression in thyroid cancer cell lines vs genotypes with or without risk allele A of rs1867277 and rs965513. Values represent mean ± max to min.
These data suggest that FOXE1 expression in tumoral thyroid cell lines is independent of the allele/genotype.
Analysis of migration in thyroid cancer cell lines
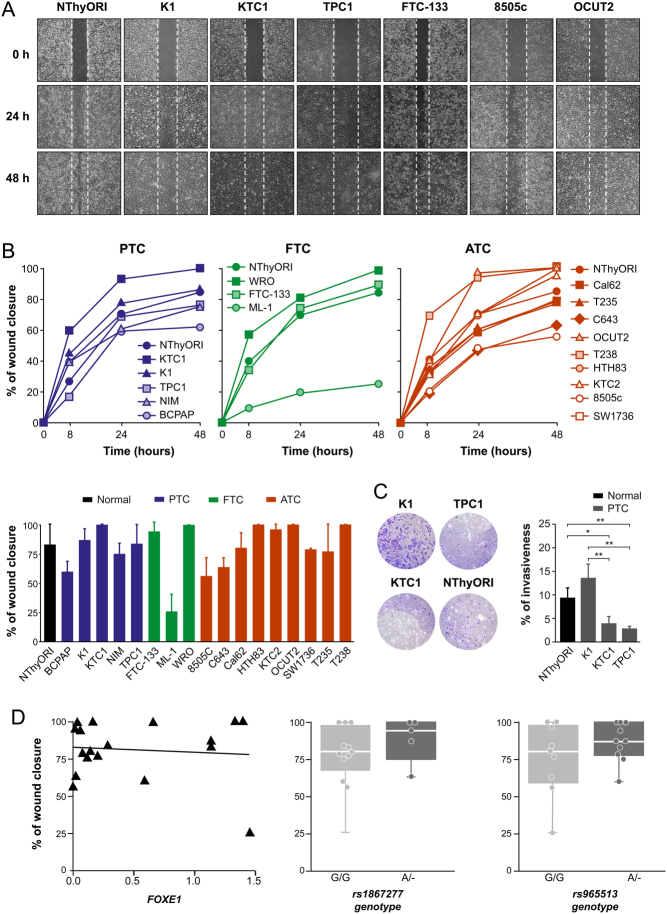
In an attempt to discern the role of FOXE1 in thyroid tumorigenesis, and considering that this gene is involved in the migration of thyroid cells from the pharyngeal floor to the trachea during development (De Felice et al. 1998), we hypothesized that FOXE1 could be involved in thyroid tumor cell migration. To study intrinsic cellular motility, we first performed scratch wound-healing assays in thyroid cancer cell lines. Results showed that among the PTC cell lines, K1 and KTC1 showed the highest cellular migration, which was greater than that of control NThy-ori-3-1 cells (Fig. 4A and B). Likewise, the FTC cell lines WRO and FTC-133 and the ATC cell lines T238 and OCUT2 showed the greatest extent of wound closure after 24 and 48 h (Fig. 4B). By contrast, the 8505c and C643 cell lines exhibited the lowest percentages of wound closure among ATC-derived cells (Fig. 4A and B).
Figure 4.
Analysis of thyroid cancer cell migration and invasion. (A) Representative images of wound-healing assay of PTC cell lines: K1, KTC1, and TPC1; FTC cell line: FTC-133; ATC cell lines: 8505c, OCUT2 and NThyORI cells. (B) Time course of wound closure in PTC-, FTC-, and ATC-derived cells. Cells were photographed at 0, 8, 24 and 48 h, and wound closure area was quantified using ImageJ software. Bar graph shows wound closure after 48 h of migration. Values represent mean ± s.e.m. of the percentage of the closure of original wound from three independent experiments performed in triplicate. (C) NThyORI, K1, KTC1, and TPC1 cells were seeded in the upper chambers of Transwells, allowed to migrate for 18 h, and photographed. Left: representative images of the lower chamber (invading cells). Right: percentage of invasiveness by direct measurement with ImageJ. Values represent mean ± s.e.m. from three independent experiments performed in triplicate. *P < 0.05, **P < 0.01. (D) Correlation analysis between migration, SNP genotype, and FOXE1 expression levels. Left: representation of correlation between wound closure and levels of FOXE1 (Pearson r = −0.091). Center and Right: representation of the correlation between wound closure and genotype in rs1867277 and rs965513 (G/G; –/A).
We also performed Matrigel assays to evaluate the invasive capacity of the three PTC cell lines showing the greatest migration capacity compared with NThy-ori-3-1 cells. We found that K1 cells had a significantly higher invasive capacity through Matrigel than KTC1, TPC1, and NThy-ori-3-1 cells (Fig. 4C). In turn, KTC1 and TPC1 cells had a significantly lower invasive capacity than control NThy-ori-3-1 cells (Fig. 4C). We then evaluated whether a correlation exists between SNP genotype, FOXE1 levels, and cellular migration. We found that the accelerated wound closure did not correlate with expression levels of FOXE1 (Pearson r = −0.091) or with the presence of one copy of the [A] allele in rs1867277 and rs965513 (Fig. 4D).
FOXE1 regulates genes involved in epithelial-to-mesenchymal transition
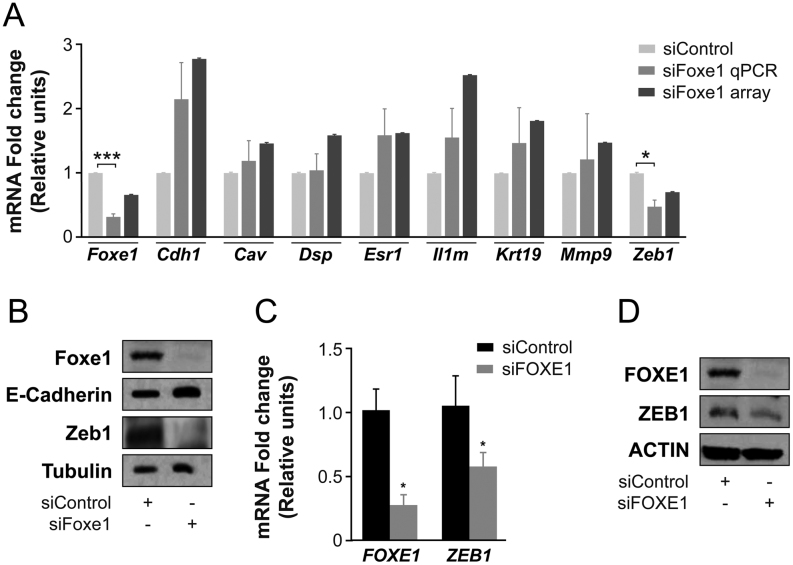
In a previous genome-wide screening analysis in Foxe1-silenced rat PCCl3 cells, we identified several novel genes potentially regulated by Foxe1 (Fernandez et al. 2013). Among the upregulated genes was Cdh1, which is widely involved in EMT (Peinado et al. 2004, Lamouille et al. 2014). However, we were unable to demonstrate direct binding of Foxe1 to the Cdh1 promoter (Fernandez et al. 2013). Using this previous analysis as a guide, and to assess the possible role of FOXE1 in regulating invasion and migration, we analyzed the mRNA levels of seven randomly chosen EMT-associated genes (Cav, Dsp, Esr1, Il1rn, Krt19, Mmp9, and Zeb1) from the list of putative Foxe1 targets (Fernandez et al. 2013). We first silenced Foxe1 in PCCl3 cells and confirmed that the mRNA level of Foxe1 was decreased and the mRNA level of Cdh1 was increased as compared with control-silenced cells (Fig. 5A). Expression qPCR analysis of the seven genes showed that six genes were moderately upregulated and one (Zeb1) was significantly downregulated in Foxe1-silenced PCC13 cells (Fig. 5A), which is in accord with our previous expression array analysis (Fernandez et al. 2013). As expected, Western blotting showed that protein levels of Zeb1 were lower in Foxe1-silenced cells than in control-silenced cells, whereas the opposite was observed for E-Cdh1 (Fig. 5B). Although on the surface these results might seem contradictory considering that Foxe1 is a differentiation-related gene, they can be reconciled if Foxe1 has a bona fide role in migration and consequently in EMT. To test whether FOXE1 regulates ZEB1 abundance in human thyroid cancer cells, we silenced its expression in K1 cells, which exhibited the greatest migration and invasion ability of all the PTC lines analyzed. In accord with the findings in rat PCCI3 cells, FOXE1 silencing resulted in a significant decrease in ZEB1 mRNA (Fig. 5C) and protein (Fig. 5D) abundance, suggesting that ZEB1 is a target of FOXE1.
Figure 5.
Experimental validation of microarray results by quantitative PCR. (A) Relative expression of seven EMT-specific genes in Foxe1-silenced PCCl3 cells assessed by qPCR. Silenced Foxe1 microarray data are from Fernandez et al. (2013). As controls, we evaluated Foxe1 and Cdh1 mRNA expression levels. Relative gene expression in silenced Foxe1 (siFoxe1) samples was calculated using the corresponding siScramble samples as a reference. Values represent mean ± s.e.m. of four independent experiments. *P < 0.05, ***P < 0.001 vs siScramble. (B) Total protein extracts from PCC13 cells were analyzed to assess the protein levels of Foxe1, E-Cadherin, and Zeb1; tubulin was used as loading control. A representative Western blot of four independent experiments is shown. (C) FOXE1 and ZEB1 expression in the K1 thyroid cancer cell line after 48 h of siFOXE1 transfection assessed by qPCR analysis using the corresponding siScramble samples as a reference. Values are mean ± s.e.m. of three independent experiments. *P < 0.01 vs siScramble. (D) Total protein extracts were analyzed to assess the protein levels of FOXE1 and ZEB1 after 72 h of siFOXE1 transfection of K1 cells. Actin was used as a loading control. A representative Western blot analysis of four independent experiments is shown.
FOXE1 binds specifically to the ZEB1 promoter and regulates its transcriptional activity
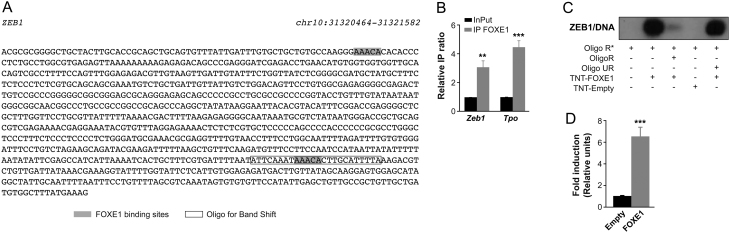
Given the aforementioned results, we next searched for canonical FOXE1-binding sites within the proximal human ZEB1 promoter (±1010 bp), finding two potential FOXE1-binding domains (Fig. 6A). We performed ChIP analysis to determine whether Foxe1 binds to the Zeb1 promoter sequence in PCCl3 cells. We used a polyclonal antibody against Foxe1 and analyzed the immunoprecipitated DNA of two independent experiments using qPCR. Foxe1 binding to the Tpo promoter was used as a positive control (Fernandez et al. 2013). ChIP analysis showed unequivocally that Foxe1 interacts with the Zeb1 promoter when normalized to the control (Fig. 6B). Also, an electrophoretic mobility shift assay showed that recombinant FOXE1 binds specifically to the probe derived from the ZEB1 promoter (Fig. 6C).
Figure 6.
FOXE1 binding to and transcriptional activation of the human ZEB1 promoter. (A) Chromosomal location of putative FOXE1-binding sites in the human ZEB1 promoter. We searched for FOXE1-binding sites in promoter regions (−1010/+26 bp relative to the transcription start site) of ZEB1. FASTA promoter sequence of ZEB1 was extracted from the Ensemble database (http://www.ensemble.org). (B) ChIP experiments for FoxE1 binding to Zeb1 promoter and qPCR analysis of immunoprecipitated chromatin in PCCl3 cells using a Foxe1 antibody. The enrichment of target sequence was calculated as the immunoprecipitation ratio (arbitrary units) relative to the negative control Cis 2.5 kb and normalized to the relative amplification in the input sample. A sequence from the Tpo promoter was used as a positive control. Values represent mean ± s.e.m. of four independent experiments each performed in triplicate. (C) EMSA assays were performed with a 32P-labeled probe containing the specific recognition sequence for FOXE1. The 32P-labeled probe (Oligo R*) was incubated alone (lane 1), with TNT-translated FOXE1 (lanes 2, 3, and 5) or with TNT-translated proteins from an empty vector (lane 4). Competition was performed with an excess of unlabeled related (Oligo R, lane 3) or unrelated oligonucleotides (Oligo UR, lane 5). (D) The human ZEB1 promoter was cotransfected into HeLa cells with 3 µg of the empty expression vector or with 3 µg of a vector harboring the cDNA for FOXE1. ZEB1 promoter activity is expressed as fold-induction relative to the activity observed with the empty expression vector. Luciferase activity was normalized to Renilla activity derived from the cotransfected pRL-SV40 to adjust for transfection efficiency. Values represent mean ± s.d. of four independent experiments. ***P < 0.001 vs control.
Lastly, we cotransfected HeLa cells with a FOXE1 expression vector and a luciferase reporter vector containing the ZEB1 promoter. Reassuringly, results showed a significant six-fold increase in ZEB1 promoter activity by FOXE1 expression (Fig. 6D). Overall, these results indicate that FOXE1 functionally transactivates the ZEB1 promoter.
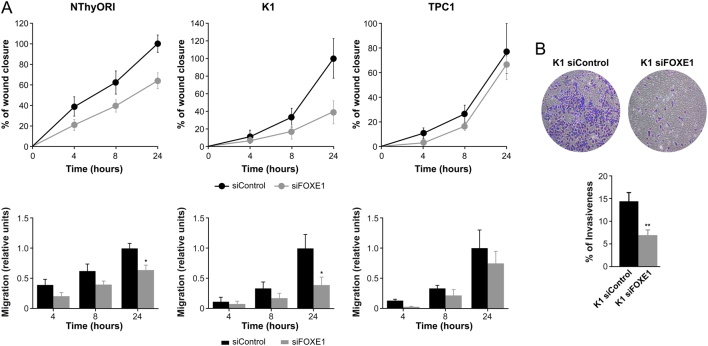
FOXE1 promotes thyroid cancer cell migration and invasion
We investigated whether FOXE1 affects the migration and invasion capability of thyroid cancer cells. We thus silenced FOXE1 expression in NThy-ori-3-1, K1, and TPC1 cells and repeated the scratch-wound and Transwell analysis. The results showed that FOXE1 silencing significantly impaired cell migration relative to control non-silenced cells (Fig. 7A). In addition, FOXE1 depletion significantly suppressed the invasion of K1 thyroid cancer cells in Transwell assays with Matrigel (Fig. 7B). Reciprocal experiments in NThy-ori-3-1, K1, and TPC1 cells showed that the over-expression of FOXE1 markedly increased, in a time-dependent manner, cell migration ability (Supplementary Fig. 2). These findings indicate that FOXE1 is closely associated with metastatic phenotypes of thyroid cancer cells.
Figure 7.
FOXE1 modulates migration and invasion in thyroid cancer cells. (A) Time course of wound closure in NThyORI, K1, and TPC1 cells silenced or not for FOXE1 expression (upper panel). Cells were photographed at 0, 4, 8, and 24 h, and wound closure area was quantified using ImageJ software. Quantification of migration rates in FOXE1-silenced cells vs control cells are shown in lower panel. Bar graph shows migration after 4, 8, and 24 h. (B) Invasiveness of K1 cells after FOXE1 silencing. Top: representative images of the lower chamber (invading cells). Bottom: percentage of invasiveness relative to siControl cells. Values represent mean ± s.e.m. from three independent experiments *P < 0.05, **P < 0.01. A full color version of this figure is available at https://doi.org/10.1530/ERC-19-0156.
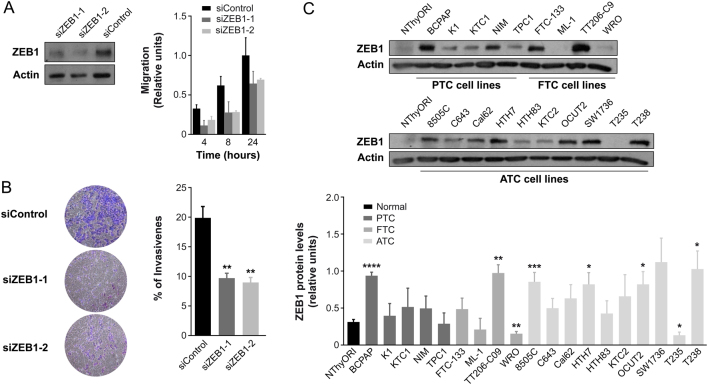
ZEB1 knockdown suppresses cell migration and invasion in thyroid cancer cells
Given the potentially crucial role of ZEB1 in EMT, we hypothesized that FOXE1 may regulate the EMT process in thyroid cancer cells, at least partly, by targeting ZEB1. Thus, we examined the effects of ZEB1 silencing on cell migration and invasion. Wound healing and Transwell analysis showed that ZEB1 is likely important for the migration and invasion of thyroid cancer cells, as both parameters were decreased in silenced cells (Fig. 8A and B). We also examined the expression of ZEB1 in a panel of human thyroid cancer cell lines, but we could not establish a clear correlation between ZEB1 and FOXE1 expression (Fig. 8C).
Figure 8.
ZEB1 silencing reduces migration and invasion in thyroid cancer cells. (A) Analysis of ZEB1 silencing in K1 cells (left panel). Total protein extracts were analyzed by Western blot to assess the protein levels of ZEB1 after 48 h of siZEB1-1 and siZEB1-2 transfection. Actin was used as a loading control. A representative Western blot analysis of three independent experiments is shown. Quantification of migration rates in ZEB1-silenced cells vs control cells is shown in the right panel. (B) Invasion ability of ZEB1-silenced K1 cells. Left: representative images of the lower chamber (invading cells). Right: percentage of invasiveness relative to siControl cells. Values represent mean ± s.e.m. from four independent experiments **P < 0.01. (C) Upper panel: Representative Western blot of three independent experiments showing ZEB1 protein levels in thyroid cell lines. Actin was used as a loading control. Lower panel: ZEB1 protein expression in thyroid cell lines normalized to expression of actin. Values represent mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.001, ****P < 0.0001 vs NThyORI control cells. A full color version of this figure is available at https://doi.org/10.1530/ERC-19-0156.
Discussion
Beyond its role in organogenesis and differentiation (De Felice & Di Lauro 2004, Fernandez et al. 2015), FOXE1 has been described as being strongly associated with susceptibility to several types of cancer (Brune et al. 2008, Venza et al. 2010, Park et al. 2012) including PTC (Gudmundsson et al. 2009, Landa et al. 2009, Bullock et al. 2012, He et al. 2015a ). These seemingly opposing roles are intriguing and have been the focus of our study. Indeed, several genes have been identified with dual roles as oncogenes and tumor suppressors (Shen et al. 2018), but few examples exist of genes related to differentiation, and therefore necessary for normal cell development, and at the same time involved in cancer. This characteristic, however, has been attributed to the Forkhead-box family of transcription factors, as they are involved in differentiation, embryogenesis, longevity, DNA repair, and carcinogenesis (Katoh et al. 2013). Accordingly, FOXE1 might also exert this dual action.
The role of Foxe1 in development and differentiation is well understood (Santisteban et al. 1992, Aza-Blanc et al. 1993, Ortiz et al. 1997, 1999, De Felice et al. 1998, De Felice & Di Lauro 2004, Fernandez et al. 2015, Lopez-Marquez et al. 2019), yet little is known about its potential role in thyroid carcinogenesis, or how different allelic variants in or near FOXE1 are associated with thyroid cancer risk. In the present study, we characterized FOXE1 expression levels in a panel of thyroid cancer cell lines and analyzed the potential role of FOXE1 in the regulation of EMT.
FOXE1 expression levels are unaltered or are upregulated in human tumors, suggesting that it might be important in the initiation and progression of these tumors (Sato et al. 2003, Eichberger et al. 2004, Venza et al. 2010, Park et al. 2012, Sugimachi et al. 2016). Loss of thyroid-specific proteins and differentiation markers is common in thyroid carcinogenesis. Indeed, several studies have reported the abnormal expression of thyroid-specific transcription factors in some thyroid carcinomas and propose that their deregulation is a pivotal event for the initiation and progression of thyroid neoplasms (Fabbro et al. 1994, Ros et al. 1999, Zhang et al. 2006). In the case of FOXE1, it has been described that nuclear expression is lost according to the degree of tumor dedifferentiation, which seems to be related to the progression of thyroid tumorigenesis (Zhang et al. 2006, Bychkov et al. 2013). Conversely, it has also been described that FOXE1 is overexpressed in PTC, and shows an aberrant cytoplasmic location in PTC cells, which again has been related to cancer cell biology (Nonaka et al. 2008, Bychkov et al. 2013). These two seemingly opposing processes makes the study of this factor in thyroid cancer challenging.
Our analysis of FOXE1 expression in a panel of thyroid cancer cell lines shows that FOXE1 levels inversely correlate with differentiation degree according to histological sub-type origin of cell lines, which is consistent with data on the expression patterns of FOXE1 in PTC and normal tissues by TCGA and fits with data of FOXE1 expression in a genome array of human tumors that include all histological variants (Giordano et al. 2006). It should be mentioned that other recent studies have reported overexpression of FOXE1 in PTC samples as well as in K1 and TPC1 cell lines (Ding et al. 2019, Ma et al. 2019), which contrasts with our data. We do not have an explanation for this apparent discrepancy other than the use of different antibodies in the studies; however, our results showing lower levels of FOXE1 in PTC cells than in control cells are in agreement with data from TCGA on more than 500 samples of patients with PTC.
We genotyped the rs1867277 and rs965513 SNPs in the same panel of thyroid cancer cell lines, as different variants in and near FOXE1 have been associated with the predisposition to PTC (Gudmundsson et al. 2009, Landa et al. 2009, Matsuse et al. 2011). In this regard, He et al. reported that the risk [A] allele of DNA variant rs965513 was associated with low expression levels of FOXE1 in unaffected thyroid tissue, but no correlation was found between rs965513 genotype and FOXE1 levels in PTC tissues (He et al. 2015a ). Further, in a functional study of rs1867277, Landa et al. revealed that the risk [A] allele led to the differential recruitment of USF1 and USF2 transcription factors, affecting the transcriptional regulation of FOXE1 and conferring a role in the pathogenesis of PTC. In our correlation studies, however, we failed to observe an association between FOXE1 levels and the risk [A] allele of rs965513 or of rs1867277 in the thyroid cancer cell lines analyzed. A possible explanation for this is the complexity of the 9q22 region, as there is evidence supporting a different regulatory model that may govern FOXE1 promoter activity (He et al. 2015b , Wang et al. 2017), together with possible epigenetic modifications due to the proximity of CpG islands (Abu-Khudir et al. 2014) and the potential involvement of other transcription factors (Landa et al. 2009, Lopez-Marquez et al. 2019). Similarly, although the existence of a relationship between the poly-A tract and PTC tumors has been described (Bullock et al. 2012), we did not observe this in the panel of cell lines analyzed herein.
FOXE1 plays a crucial role in thyroid morphogenesis by promoting thyroid precursor cell migration during gland development (De Felice et al. 1998), suggesting the involvement of FOXE1 in cell migration and EMT. We therefore analyzed cell migration in the panel of thyroid cancer cell lines in relation to FOXE1 expression and SNP genotype. Although FOXE1 levels positively correlated with migration rate in some cell lines, we could not establish a correlation between FOXE1 expression and the ability of thyroid cancer cell lines to migrate and invade. Nevertheless, silencing of FOXE1 expression resulted in impaired thyroid cancer cell migration and invasion, and the opposite was observed after FOXE1 overexpression. Similarly, the presence of one copy of the [A] allele in rs1867277 and rs965513 did not significantly correlate with accelerated wound closure; however, it seems that there is a high migration capacity in cell lines containing the [A] risk allele of SNP rs1867277.
Foxe1 binds to DNA sequences present in the promoters of thyroglobulin (Santisteban et al. 1992) and thyroperoxidase (Aza-Blanc et al. 1993), promoting their transcriptional activation. In a previous study, we identified novel Foxe1 downstream targets using expression arrays in Foxe1-silenced thyroid epithelial cells (Fernandez et al. 2013), supporting the involvement of FOXE1 in relevant biological processes and pathways. One of the hallmarks of EMT is the functional loss of E-cadherin (encoded by Cdh1), which was upregulated in Foxe1-silenced thyroid cells, suggesting that FOXE1 modulates the expression of Cdh1. In this study, we show that ZEB1, a key factor that modulates E-cadherin expression and the induction of EMT, is regulated by FOXE1 in thyroid cells. In addition, we demonstrate a direct interaction of FOXE1 with the ZEB1 promoter and an increase in ZEB1 transcriptional activity in FOXE1-transfected cells. Interestingly, loss-of-function experiments revealed that cells silenced for ZEB1 show blunted migration and invasion relative to control non-silenced cells, a behavior similar to that observed in silenced FOXE1 cells, which clearly demonstrates that FOXE1 regulates migration and invasion in thyroid cancer cells, at least in part, through ZEB1.
Taken all this together, we postulate that FOXE1 has a crucial role in thyroid tumor cell migration and invasion, as shown by the results of loss/gain-of-function of FOXE1 on migration/invasion, and with the increasing evidence of the role of forkhead box proteins in the development and progression of cancer (Myatt & Lam 2007, Katoh et al. 2013).
Our results are also consistent with other studies, demonstrating that FOXE1 can interact with other factors and transactivate key genes in cancer such as SNAIL (Xu et al. 2015), an E-cadherin transcriptional repressor, or TERT (telomerase reverse transcriptase), which is coregulated by FOXE1 and the ETS factor ELK1 (Bullock et al. 2016). Along this line, it would be interesting to search for FOXE1 interacting partners in thyroid cancer, which may reveal unique or separate signaling pathways.
In conclusion, we have identified ZEB1 as a bona fide target of FOXE1 in thyroid cancer cells, which provides new insights into the role of FOXE1 in regulating EMT in thyroid cancer.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by grants SAF2016-75531-R from Ministerio de Ciencia, Innovación y Universidades (MICIU), Spain, Fondo Europeo de Desarrollo Regional FEDER, B2017/BMD-3724 from Comunidad de Madrid, and GCB14142311CRES from Fundación Española Contra el Cáncer (AECC).
Acknowledgements
The authors thank Dr A Garcia de Herreros (IMIN, PRB, Barcelona, Spain) for ZEB1 promoter construct, Dr K McCreath for helpful comments on the manuscript and Javier Pérez (Instituto de Investigaciones Biomédicas, Madrid, Spain) for the artwork.
References
- Abu-Khudir R, Magne F, Chanoine JP, Deal C, Van Vliet G, Deladoey J. 2014. Role for tissue-dependent methylation differences in the expression of FOXE1 in nontumoral thyroid glands. Journal of Clinical Endocrinology and Metabolism E1120–E1129. ( 10.1210/jc.2013-4414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P, Di Lauro R, Santisteban P. 1993. Identification of a cis-regulatory element and a thyroid-specific nuclear factor mediating the hormonal regulation of rat thyroid peroxidase promoter activity. Molecular Endocrinology 1297–1306. ( 10.1210/mend.7.10.8264661) [DOI] [PubMed] [Google Scholar]
- Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, Yang D, Omura N, Eshleman J, Canto M, et al 2008. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiology, Biomarkers and Prevention 3536–3542. ( 10.1158/1055-9965.EPI-08-0630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock M, Duncan EL, O’Neill C, Tacon L, Sywak M, Sidhu S, Delbridge L, Learoyd D, Robinson BG, Ludgate M, et al 2012. Association of FOXE1 polyalanine repeat region with papillary thyroid cancer. Journal of Clinical Endocrinology and Metabolism E1814–E1819. ( 10.1210/jc.2012-1456) [DOI] [PubMed] [Google Scholar]
- Bullock M, Lim G, Li C, Choi IH, Kochhar S, Liddle C, Zhang L, Clifton-Bligh RJ. 2016. Thyroid transcription factor FOXE1 interacts with ETS factor ELK1 to co-regulate TERT. Oncotarget 85948–85962. ( 10.18632/oncotarget.13288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bychkov A, Saenko V, Nakashima M, Mitsutake N, Rogounovitch T, Nikitski A, Orim F, Yamashita S. 2013. Patterns of FOXE1 expression in papillary thyroid carcinoma by immunohistochemistry. Thyroid 817–828. ( 10.1089/thy.2012.0466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre A, Castanet M, Sura-Trueba S, Szinnai G, Van Vliet G, Trochet D, Amiel J, Leger J, Czernichow P, Scotet V, et al 2007. Polymorphic length of FOXE1 alanine stretch: evidence for genetic susceptibility to thyroid dysgenesis. Human Genetics 467–476. ( 10.1007/s00439-007-0420-5) [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 401–404. ( 10.1158/2159-8290.CD-12-0095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Obermayr F, Frischauf AM. 1997. FKHL15, a new human member of the forkhead gene family located on chromosome 9q22. Genomics 390–396. ( 10.1006/geno.1997.4692) [DOI] [PubMed] [Google Scholar]
- Chen CA, Okayama H. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 632–638. [PubMed] [Google Scholar]
- Clifton-Bligh RJ, Wentworth JM, Heinz P, Crisp MS, John R, Lazarus JH, Ludgate M, Chatterjee VK. 1998. Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nature Genetics 399–401. ( 10.1038/1294) [DOI] [PubMed] [Google Scholar]
- Cuesta I, Zaret KS, Santisteban P. 2007. The forkhead factor FoxE1 binds to the thyroperoxidase promoter during thyroid cell differentiation and modifies compacted chromatin structure. Molecular and Cellular Biology 7302–7314. ( 10.1128/MCB.00758-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, de Herreros AG. 2011. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. Journal of Biological Chemistry 12024–12032. ( 10.1074/jbc.M110.168625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L, Welch HG. 2006. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA 2164–2167. ( 10.1001/jama.295.18.2164) [DOI] [PubMed] [Google Scholar]
- De Felice M, Di Lauro R. 2004. Thyroid development and its disorders: genetics and molecular mechanisms. Endocrine Reviews 722–746. ( 10.1210/er.2003-0028) [DOI] [PubMed] [Google Scholar]
- De Felice M, Ovitt C, Biffali E, Rodriguez-Mallon A, Arra C, Anastassiadis K, Macchia PE, Mattei MG, Mariano A, Scholer H, et al 1998. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nature Genetics 395–398. ( 10.1038/1289) [DOI] [PubMed] [Google Scholar]
- DeLellis RA. 2006. Pathology and genetics of thyroid carcinoma. Journal of Surgical Oncology 662–669. ( 10.1002/jso.20700) [DOI] [PubMed] [Google Scholar]
- Ding Z, Ke R, Zhang Y, Fan Y, Fan J. 2019. FOXE1 inhibits cell proliferation, migration and invasion of papillary thyroid cancer by regulating PDGFA. Molecular and Cellular Endocrinology 110420 ( 10.1016/j.mce.2019.03.010) [DOI] [PubMed] [Google Scholar]
- Eichberger T, Regl G, Ikram MS, Neill GW, Philpott MP, Aberger F, Frischauf AM. 2004. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. Journal of Investigative Dermatology 1180–1187. ( 10.1111/j.0022-202X.2004.22505.x) [DOI] [PubMed] [Google Scholar]
- Fabbro D, Di Loreto C, Beltrami CA, Belfiore A, Di Lauro R, Damante G. 1994. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Research 4744–4749. ( 10.1016/0959-8049(95)00560-9) [DOI] [PubMed] [Google Scholar]
- Fernandez LP, Lopez-Marquez A, Martinez AM, Gomez-Lopez G, Santisteban P. 2013. New insights into FoxE1 functions: identification of direct FoxE1 targets in thyroid cells. PLoS One e62849 ( 10.1371/journal.pone.0062849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LP, Lopez-Marquez A, Santisteban P. 2015. Thyroid transcription factors in development, differentiation and disease. Nature Reviews Endocrinology 29–42. ( 10.1038/nrendo.2014.186) [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling pl1 ( 10.1126/scisignal.2004088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, Wilhelm KG, Jr, Vinco M, Misek DE, Sanders D, Zhu Z, et al 2006. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clinical Cancer Research 1983–1993. ( 10.1158/1078-0432.CCR-05-2039) [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. 1994. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. Journal of the National Cancer Institute 1600–1608. ( 10.1093/jnci/86.21.1600) [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, et al 2009. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nature Genetics 460–464. ( 10.1038/ng.339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, He H, Jonasdottir A, Sigurdsson A, Stacey SN, Johannsdottir H, et al 2012. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nature Genetics 319–322. ( 10.1038/ng.1046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Li W, Liyanarachchi S, Jendrzejewski J, Srinivas M, Davuluri RV, Nagy R, de la Chapelle A. 2015a. Genetic predisposition to papillary thyroid carcinoma: involvement of FOXE1, TSHR, and a novel lincRNA gene, PTCSC2. Journal of Clinical Endocrinology and Metabolism E164–E172. ( 10.1210/jc.2014-2147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Li W, Liyanarachchi S, Srinivas M, Wang Y, Akagi K, Wu D, Wang Q, Jin V, Symer DE, et al 2015b. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. PNAS 6128–6133. ( 10.1073/pnas.1506255112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Howarth KM, Martin L, Gorman M, Mihai R, Moss L, Auton A, Lemon C, Mehanna H, Mohan H, et al 2012. Thyroid cancer susceptibility polymorphisms: confirmation of loci on chromosomes 9q22 and 14q13, validation of a recessive 8q24 locus and failure to replicate a locus on 5q24. Journal of Medical Genetics 158–163. ( 10.1136/jmedgenet-2011-100586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallel R, Belguith-Maalej S, Akdi A, Mnif M, Charfeddine I, Galofre P, Ghorbel A, Abid M, Marcos R, Ayadi H, et al 2010. Genetic investigation of FOXE1 polyalanine tract in thyroid diseases: new insight on the role of FOXE1 in thyroid carcinoma. Cancer Biomarkers 43–51. ( 10.3233/DMA-2011-0824) [DOI] [PubMed] [Google Scholar]
- Katoh M, Igarashi M, Fukuda H, Nakagama H. 2013. Cancer genetics and genomics of human FOX family genes. Cancer Letters 198–206. ( 10.1016/j.canlet.2012.09.017) [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology 178–196. ( 10.1038/nrm3758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I, et al 2009. The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genetics e1000637 ( 10.1371/journal.pgen.1000637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. 2017. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA 1338–1348. ( 10.1001/jama.2017.2719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- Lopez-Marquez A, Fernandez-Mendez C, Recacha P, Santisteban P. 2019. Regulation of Foxe1 by TSH and TGFbeta depends on the interplay between thyroid-specific, CREB and SMAD transcription factors. Thyroid 714–725. ( 10.1089/thy.2018.0136) [DOI] [PubMed] [Google Scholar]
- Ma J, Huang X, Li Z, Shen Y, Lai J, Su Q, Zhao J, Xu J. 2019. FOXE1 supports the tumor promotion of Gli2 on papillary thyroid carcinoma by the Wnt/beta-catenin pathway. Journal of Cellular Physiology 17739–17748. ( 10.1002/jcp.28399) [DOI] [PubMed] [Google Scholar]
- Macchia PE, Mattei MG, Lapi P, Fenzi G, Di Lauro R. 1999. Cloning, chromosomal localization and identification of polymorphisms in the human thyroid transcription factor 2 gene (TITF2). Biochimie 433–440. ( 10.1016/S0300-9084(99)80092-3) [DOI] [PubMed] [Google Scholar]
- Mancikova V, Cruz R, Inglada-Perez L, Fernandez-Rozadilla C, Landa I, Cameselle-Teijeiro J, Celeiro C, Pastor S, Velazquez A, Marcos R, et al 2015. Thyroid cancer GWAS identifies 10q26.12 and 6q14.1 as novel susceptibility loci and reveals genetic heterogeneity among populations. International Journal of Cancer 1870–1878. ( 10.1002/ijc.29557) [DOI] [PubMed] [Google Scholar]
- Matsuse M, Takahashi M, Mitsutake N, Nishihara E, Hirokawa M, Kawaguchi T, Rogounovitch T, Saenko V, Bychkov A, Suzuki K, et al 2011. The FOXE1 and NKX2-1 loci are associated with susceptibility to papillary thyroid carcinoma in the Japanese population. Journal of Medical Genetics 645–648. ( 10.1136/jmedgenet-2011-100063) [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. 2007. The emerging roles of forkhead box (Fox) proteins in cancer. Nature Reviews Cancer 847–859. ( 10.1038/nrc2223) [DOI] [PubMed] [Google Scholar]
- Nonaka D, Tang Y, Chiriboga L, Rivera M, Ghossein R. 2008. Diagnostic utility of thyroid transcription factors Pax8 and TTF-2 (FoxE1) in thyroid epithelial neoplasms. Modern Pathology 192–200. ( 10.1038/modpathol.3801002) [DOI] [PubMed] [Google Scholar]
- Ortiz L, Zannini M, Di Lauro R, Santisteban P. 1997. Transcriptional control of the forkhead thyroid transcription factor TTF-2 by thyrotropin, insulin, and insulin-like growth factor I. Journal of Biological Chemistry 23334–23339. ( 10.1074/jbc.272.37.23334) [DOI] [PubMed] [Google Scholar]
- Ortiz L, Aza-Blanc P, Zannini M, Cato AC, Santisteban P. 1999. The interaction between the forkhead thyroid transcription factor TTF-2 and the constitutive factor CTF/NF-1 is required for efficient hormonal regulation of the thyroperoxidase gene transcription. Journal of Biological Chemistry 15213–15221. ( 10.1074/jbc.274.21.15213) [DOI] [PubMed] [Google Scholar]
- Pal T, Vogl FD, Chappuis PO, Tsang R, Brierley J, Renard H, Sanders K, Kantemiroff T, Bagha S, Goldgar DE, et al 2001. Increased risk for nonmedullary thyroid cancer in the first degree relatives of prevalent cases of nonmedullary thyroid cancer: a hospital-based study. Journal of Clinical Endocrinology and Metabolism 5307–5312. ( 10.1210/jcem.86.11.8010) [DOI] [PubMed] [Google Scholar]
- Park E, Gong EY, Romanelli MG, Lee K. 2012. Suppression of estrogen receptor-alpha transactivation by thyroid transcription factor-2 in breast cancer cells. Biochemical and Biophysical Research Communications 532–537. ( 10.1016/j.bbrc.2012.04.039) [DOI] [PubMed] [Google Scholar]
- Parlato R, Rosica A, Rodriguez-Mallon A, Affuso A, Postiglione MP, Arra C, Mansouri A, Kimura S, Di Lauro R, De Felice M. 2004. An integrated regulatory network controlling survival and migration in thyroid organogenesis. Developmental Biology 464–475. ( 10.1016/j.ydbio.2004.08.048) [DOI] [PubMed] [Google Scholar]
- Peinado H, Portillo F, Cano A. 2004. Transcriptional regulation of cadherins during development and carcinogenesis. International Journal of Developmental Biology 365–375. ( 10.1387/ijdb.041794hp) [DOI] [PubMed] [Google Scholar]
- Riesco-Eizaguirre G, Santisteban P. 2016. ENDOCRINE TUMOURS: Advances in the molecular pathogenesis of thyroid cancer: lessons from the cancer genome. European Journal of Endocrinology R203–R217. ( 10.1530/EJE-16-0202) [DOI] [PubMed] [Google Scholar]
- Ros P, Rossi DL, Acebron A, Santisteban P. 1999. Thyroid-specific gene expression in the multi-step process of thyroid carcinogenesis. Biochimie 389–396. ( 10.1016/S0300-9084(99)80086-8) [DOI] [PubMed] [Google Scholar]
- Ruiz-Llorente S, Carrillo Santa de Pau E, Sastre-Perona A, Montero-Conde C, Gomez-Lopez G, Fagin JA, Valencia A, Pisano DG, Santisteban P. 2012. Genome-wide analysis of Pax8 binding provides new insights into thyroid functions. BMC Genomics 147 ( 10.1186/1471-2164-13-147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban P, Acebron A, Polycarpou-Schwarz M, Di Lauro R. 1992. Insulin and insulin-like growth factor I regulate a thyroid-specific nuclear protein that binds to the thyroglobulin promoter. Molecular Endocrinology 1310–1317. ( 10.1210/mend.6.8.1406708) [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. 2003. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Research 3735–3742. [PubMed] [Google Scholar]
- Shen L, Shi Q, Wang W. 2018. Double agents: genes with both oncogenic and tumor-suppressor functions. Oncogenesis 25 ( 10.1038/s41389-018-0034-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimachi K, Matsumura T, Shimamura T, Hirata H, Uchi R, Ueda M, Sakimura S, Iguchi T, Eguchi H, Masuda T, et al 2016. Aberrant methylation of FOXE1 contributes to a poor prognosis for patients with colorectal cancer. Annals of Surgical Oncology 3948–3955. ( 10.1245/s10434-016-5289-x) [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 676–690. ( 10.1016/j.cell.2014.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas G, Tarabichi M, Gacquer D, Hebrant A, Dom G, Dumont JE, Keutgen X, Fahey TJ, 3rd, Maenhaut C, Detours V. 2012. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene 4490–4498. ( 10.1038/onc.2011.626) [DOI] [PubMed] [Google Scholar]
- Venza I, Visalli M, Tripodo B, De Grazia G, Loddo S, Teti D, Venza M. 2010. FOXE1 is a target for aberrant methylation in cutaneous squamous cell carcinoma. British Journal of Dermatology 1093–1097. ( 10.1111/j.1365-2133.2009.09560.x) [DOI] [PubMed] [Google Scholar]
- von Roemeling CA, Marlow LA, Pinkerton AB, Crist A, Miller J, Tun HW, Smallridge RC, Copland JA. 2015. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. Journal of Clinical Endocrinology and Metabolism E697–E709. ( 10.1210/jc.2014-2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He H, Li W, Phay J, Shen R, Yu L, Hancioglu B, de la Chapelle A. 2017. MYH9 binds to lncRNA gene PTCSC2 and regulates FOXE1 in the 9q22 thyroid cancer risk locus. PNAS 474–479. ( 10.1073/pnas.1619917114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He H, Liyanarachchi S, Genutis LK, Li W, Yu L, Phay JE, Shen R, Brock P, de la Chapelle A. 2018. The role of SMAD3 in the genetic predisposition to papillary thyroid carcinoma. Genetics in Medicine 927–935. ( 10.1038/gim.2017.224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chang R, Peng Z, Wang Y, Ji W, Guo J, Song L, Dai C, Wei W, Wu Y, et al 2015. Loss of polarity protein AF6 promotes pancreatic cancer metastasis by inducing Snail expression. Nature Communication 7184 ( 10.1038/ncomms8184) [DOI] [PubMed] [Google Scholar]
- Zaballos MA, Santisteban P. 2017. Key signaling pathways in thyroid cancer. Journal of Endocrinology R43–R61. ( 10.1530/JOE-17-0266) [DOI] [PubMed] [Google Scholar]
- Zannini M, Avantaggiato V, Biffali E, Arnone MI, Sato K, Pischetola M, Taylor BA, Phillips SJ, Simeone A, Di Lauro R. 1997. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO Journal 3185–3197. ( 10.1093/emboj/16.11.3185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zuo H, Nakamura Y, Nakamura M, Wakasa T, Kakudo K. 2006. Immunohistochemical analysis of thyroid-specific transcription factors in thyroid tumors. Pathology International 240–245. ( 10.1111/j.1440-1827.2006.01959.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a