SUMMARY
53BP1 activity drives genome instability and lethality in BRCA1-deficient mice by inhibiting homologous recombination (HR). 53BP1’s anti-recombinogenic functions require phosphorylation-dependent interactions with PTIP and RIF1/Shieldin effector complexes. While RIF1/Shieldin blocks 5’–3’ nucleolytic processing of DNA ends, it remains unclear how PTIP antagonizes HR. Here we show that mutation of the PTIP interaction site in 53BP1 (S25A) allows sufficient DNA2-dependent end resection to rescue the lethality of BRCA1Δ11 mice, despite increasing RIF1 “end-blocking” at DNA damage sites. However, double mutant cells fail to complete HR, as excessive Shieldin activity also inhibits RNF168-mediated loading of PALB2/RAD51. As a result, BRCA1Δ1153BP1S25A mice exhibit hallmark features of HR insufficiency, including premature aging and hypersensitivity to PARPi. Disruption of Shieldin or forced targeting of PALB2 to ssDNA in BRCA1D1153BP1S25A cells restores RNF168 recruitment, RAD51 nucleofilament formation, and PARPi resistance. Our study therefore reveals a critical function of Shieldin post-resection that limits the loading of RAD51.
Graphical Abstract
Etoc blurb:
BRCA1 promotes 5’–3’ end resection and subsequently loads RAD51 onto 3’ single strand DNA to initiate homologous recombination. Callen, Zong et al. demonstrate that 53BP1 antagonizes both key steps in homologous recombination-end resection and RAD51 loading- that are coordinated by BRCA1.
INTRODUCTION
The tumor suppressor BRCA1 coordinates two key steps during DNA double strand break (DSB) repair by homologous recombination (HR) (Chen et al., 2018). Initially, BRCA1 promotes 5’–3’ end resection and subsequently, it recruits the PALB2/ BRCA2 mediator complex to load the RAD51 recombinase onto 3’ single stranded DNA (ssDNA). In BRCA1-deficient cells, HR is severely compromised, leading to genome instability during development that precipitates tumorigenesis (Chen et al., 2018). However, 53BP1 loss largely restores HR in BRCA1-deficient mice, resulting in the rescue of the embryonic lethality, and the suppression of cancer predisposition in pre-clinical mouse tumor models (Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009). The dramatic rescue of HR in BRCA1-deficient mice is in part due to 53BP1’s role in blocking end resection (Bunting et al., 2010). However, in some cell lines, BRCA1 depletion had minimal effects on end-processing under conditions where RAD51 filament formation was compromised (Zhou et al., 2014). RAD51 filament formation in BRCA1-deficient cells requires alternative recruitment of PALB2/BRCA2 through RNF168-mediated chromatin ubiquitylation (Zong et al., 2019). This raises the question of whether 53BP1, in addition to blocking resection, also antagonizes RNF168-driven RAD51 filament formation.
The pro-end-joining and anti-recombination functions of 53BP1 require phospho-dependent interactions with downstream effectors such as RIF1 and PTIP (Setiaputra and Durocher, 2019). Recently, independent laboratories have discovered components of a “Shieldin complex”, which acts downstream of RIF1 to block resection (Dev et al., 2018; Findlay et al., 2018; Gao et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018). Loss of RIF1 or Shieldin leads to defective immunoglobulin class switch recombination (CSR), blocks the fusion of dysfunctional telomeres and promotes PARPi resistance in BRCA1-deficient cells (Dev et al., 2018; Findlay et al., 2018; Gao et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018; Setiaputra and Durocher, 2019). However, RIF1 and PTIP also have essential functions in replication and transcription (Alver et al., 2017; Buonomo et al., 2009; Cho et al., 2003; Daniel et al., 2010), making their roles in 53BP1-dependent DSB repair ambiguous.
To determine the contribution of PTIP binding to 53BP1-dependent suppression of end resection and HR, we generated 53BP1-knockin mice harboring a single point mutation (S25A) in 53BP1. The 53BP1S25A mutation selectively impairs PTIP while enhances RIF1 association with DSBs. Surprisingly, the mutant 53BP1 protein is permissive for end resection sufficient to rescue the lethality of BRCA1Δ11 mice, but nevertheless blocks RAD51 loading. Our study thereby uncovers a new role of 53BP1 in antagonizing HR at the post-resection step.
Finally, we report that distinct domains of BRCA1 are capable of counteracting anti-recombinogenic functions of 53BP1 pre- and post-resection. Thus, BRCA1 mutant cells lacking exon 11 are severely impaired in DSB resection, while loss of the BRCA1 RING domain maintains resection capability but fails to support RAD51 foci formation. Since pathogenic germline mutations in the RING domain and exon 11 are frequently found in familial breast and ovarian cancers, and may predict poor responses to chemotherapy (Drost et al., 2016; Wang et al., 2016), these results have implications for our understanding of cellular mechanisms leading to PARPi and platinum resistance.
RESULTS
53BP1S25A Mutation Increases RIF1 Association with DNA Breaks
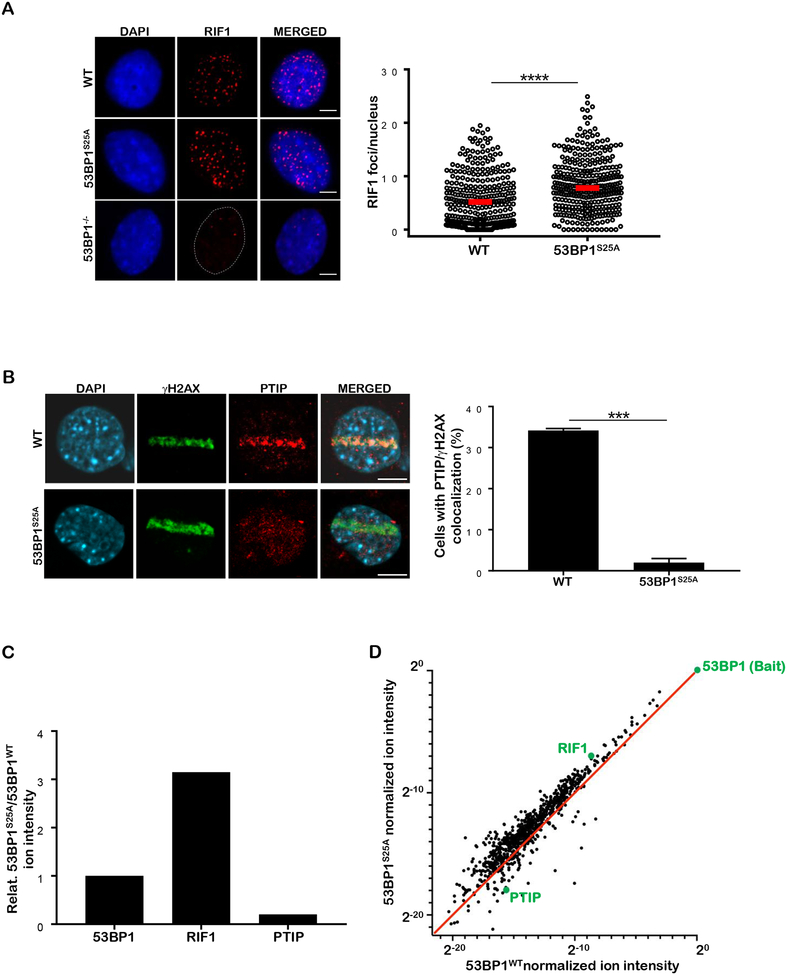
To dissect the contribution of phospho-dependent 53BP1-PTIP interaction to DSB repair in a physiological setting that does not compromise replication, MLL3/4 methyltransferase activity (Gong et al., 2009; Ray Chaudhuri et al., 2016) or 53BP1-dependent p53 activation (Cuella-Martin et al., 2016), we replaced serine 25 with alanine in embryonic stem cells (Figure S1A), which were used to generate 53BP1S25A mice. As expected, homozygous mutant 53BP1S25A mice were born at normal Mendelian frequencies and did not exhibit any overt phenotype (Figure S1A and data not shown). 53BP1 protein was present at normal levels but irradiation-induced S25 phosphorylation, which is largely ATM-dependent, was absent in 53BP1S25A mice (Figures S1B and S1C). Notably, irradiation-induced foci (IRIF) for RIF1 were significantly elevated in 53BP1S25A MEFs (Figure 1A), while PTIP recruitment to DSBs was undetectable in mutant cells (Figure 1B). Consistent with these findings, analysis by mass spectrometry revealed that RIF1 peptides were enriched, and PTIP peptides were depleted, in 53BP1 immunoprecipitates from cells expressing 53BP1S25A compared to 53BP1WT (Figures 1C, D and Table S1). Thus, the 53BP1S25A mutation specifically impairs PTIP while enhancing RIF1 association with DNA breaks. Conversely, it was shown that mutation of seven N-terminal phosphorylation sites in 53BP1 that decreased RIF1–53BP1 association led to a concomitant increase in PTIP-53BP1 interaction (Callen et al., 2013).
Figure 1. Serine 25 to Alanine mutation in 53BP1 compromises PTIP binding while increasing RIF1 association at DSBs.
(A) Left: WT, 53BP1−/− and 53BP1S25A MEFs were assayed for RIF1 (red) IRIF (10 Gy, 1 hour recovery). Cells were counterstained with DAPI (blue). Scale bar represents 10 μm. Right: quantification of RIF1 foci per nucleus in WT and 53BP1S25A MEFs, normalized by nuclear area (per 100 μm2). A minimum of 300 nuclei were quantified per condition using the Gen5 spot analysis software. A representative experiment (n=3) is shown. Statistical significance was determined by Mann-Whitney t-test. (B) Left: PTIP (red) recruitment to laser microirradiation damage in WT and 53BP1S25A MEFs. Damaged cells are indicated by γ-H2AX tracks (green). Scale bar represents 10 μm. Right: quantification of cells with PTIP/γ-H2AX colocalization. Statistical significance was determined by Mann-Whitney t-test. (C, D) Graphs depicting the relative ion intensities for 53BP1, RIF1 and PTIP peptides associated with the 53BP1 complex in HeLa-S cells expressing FLAG-HA-53BP1S25A or FLAG-HA-53BP1WT, as determined by mass spectrometry.
53BP1S25A Rescues the Lethality of BRCA1Δ11 mice but Leads to Premature Aging
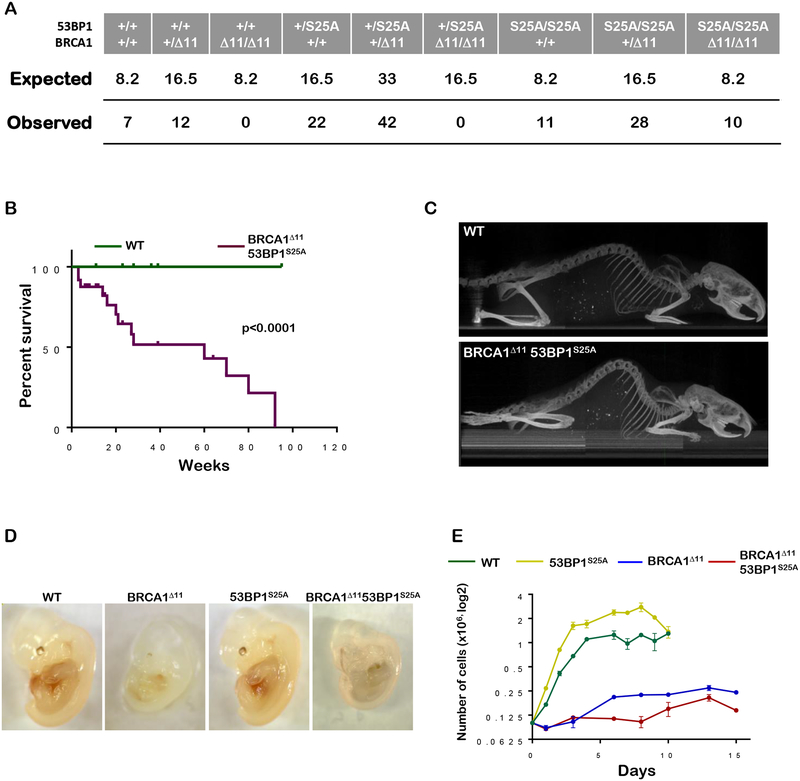
Complete loss of 53BP1 is thought to rescue the embryonic lethality of BRCA1-deficient mice by increasing DSB resection (Bunting et al., 2010) as well as by permitting RNF168-mediated loading of RAD51 onto ssDNA (Zong et al., 2019). To examine the relative contribution of each of these processes to embryonic viability, we crossed 53BP1S25A mice with mice expressing a mutant form of BRCA1 where exon 11 is deleted. While homozygous BRCA1Δ11 mutation is incompatible with viability, we found that BRCA1Δ1153BP1S25A mice were born at expected Mendelian frequencies and reached adulthood (Figures 2A and 2B). Similar to BRCA1Δ1153BP1−/−mice (Bunting et al., 2010), mutant males were sterile and showed reduced testes size (fig. S2A), most likely reflecting BRCA1’s role in meiotic sex chromosome inactivation (Turner et al., 2005). However, in striking contrast to BRCA1Δ1153BP1−/− mice which exhibit a normal lifespan (Bunting et al., 2010), BRCA1Δ1153BP1S25A mice died prematurely with a median survival of 6 months and maximum lifespan of 92 weeks (Figure 2B). BRCA1Δ1153BP1S25A mice developed pathologies consistent with accelerated aging, including kyphosis, blindness, inflammation, hair graying, and overall reduced activity with only a modest rate of cancer formation (<10%) (Figures 2C and S2B). Thus, a single point mutation in 53BP1 disrupting PTIP binding is sufficient to fully rescue the prenatal lethality of BRCA1Δ11 mice, but it is insufficient to allow normal organismal aging.
Figure 2. 53BP1S25A mutation reverts lethality of BRCA1Δ11 mice but promotes accelerated aging and an elevated rate of senescence.
(A) Table showing the expected and observed frequency of breeding outcomes from BRCA1+/Δ1153BP1+/S25A x BRCA1+/Δ1153BP1+/S25A intercrosses. (B) Kaplan-Meier survival analysis of WT and BRCA1Δ1153BP1S25A mice (n=14). A significantly shorter lifespan was observed in BRCA1Δ1153BP1S25A mice compared to WT (p < 0.0001). Log-rank (Mantel-Cox) test was used to determine statistical significance. (C) Representative X-ray CT projection image showing increased kyphosis in a 4 month old BRCA1Δ1153BP1S25A mouse compared with a WT littermate. (D) Representative image of E13.5 WT, BRCA1Δ11, 53BP1S25A and BRCA1Δ1153BP1S25A embryos showing the decrease in size of BRCA1Δ11 and BRCA1Δ1153BP1S25A animals. (E) In vitro growth of primary WT, BRCA1Δ11, 53BP1S25A and BRCA1Δ1153BP1S25ä MEFs at passage 2.
Premature aging could be due to elevated DNA damage signaling leading to senescence and apoptosis (Niedernhofer et al., 2018). Already at e13.5, BRCA1Δ1153BP1S25A embryos appear smaller than WT (Figure 2D), and mutant primary MEFs prematurely senesced at the same rate as BRCA1Δ11 cells (Figure 2E). Various tissues from mutant embryos including cerebellum, liver and limb showed TUNEL- and Caspase 3- positive staining indicative of apoptosis (Figure S2C); and increased apoptosis was associated with enhanced levels of DNA damage signaling as measured by Kap1 phosphorylation (Figure S2D). Consistent with the increased neural apoptosis, mutant mice had a smaller brain, although its overall structure was undisturbed (Figure S2E). Thus, despite loss of embryonic cells in vivo and poor growth in vitro, the rescue of viability in BRCA1Δ1153BP1S25A mice was associated with normal developmental processes but accelerated postnatal aging.
Cells from BRCA1Δ1153BP1S25A Mice Are Severely Comprised for RAD51 Loading
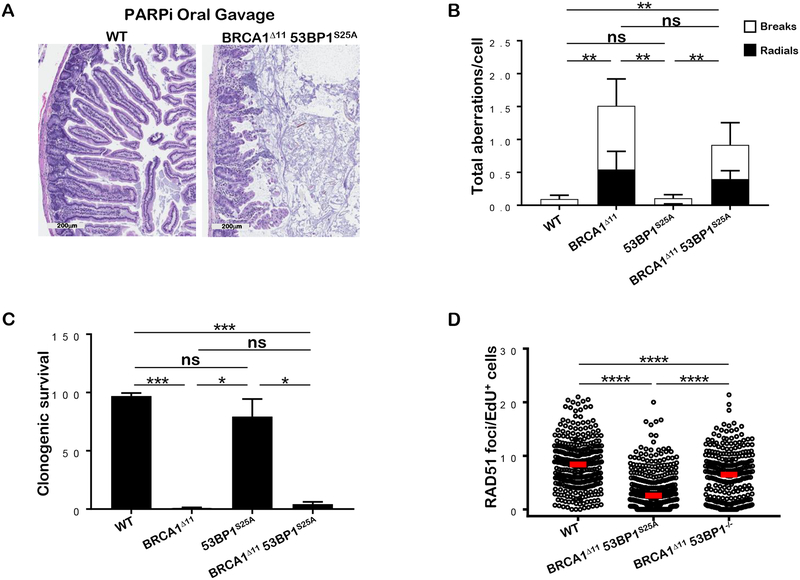
Because RAD51 -dependent HR is largely rescued in BRCA1Δ11 mice lacking 53BP1, mutant cells are resistant to PARPi and show robust RAD51 foci formation (Bunting et al., 2010). In striking contrast, BRCA1Δ1153BP1S25A mice (n=3) died within 9 days after treatment with PARPi. Histological analysis revealed severe injuries to the gastrointestinal tract, including blunting of villi and almost complete obliteration of stem cell crypts (Figure 3A). Consistent with the in vivo response, mutant B cells and MEFs showed hypersensitivity to PARPi as indicated by elevated induction of chromosomal aberrations and increased formation of radials (Figures 3B and S3A). Mutant MEFs also showed marked reduction in colony growth after PARPi treatment (Figure 3C). Moreover, we observed a severe impairment in IR-induced RAD51 foci formation in BRCA1Δ1153BP1S25A MEFs and B cells (Figures 3D and S3B). Notably, the HR defect is more severe in BRCA1Δ1153BP1S25A cells compared to that observed in BRCA1/PTIP-deficient cells (Callen et al., 2013), suggesting that PTIP has additional roles within the MLL3/4 complex independent of its interaction with 53BP1 (Ray Chaudhuri et al., 2016). Moreover, in contrast to 53BP1 ablation (Bunting et al., 2010), the 53BP1S25A mutation rescues the lethality of BRCA1Δ11 mice without restoring HR. Our results therefore indicate that severely reduced HR is compatible with viability but leads to premature aging.
Figure 3. BRCA1Δ1153BP1S25A mice are sensitive to PARPi and defective in RAD51 filament assembly.
(A) Representative histological images of the small intestine of BRCA1Δ1153BP1S25A and WT littermate controls upon treatment with PARPi by daily oral gavage. Image was taken 9 days after treatment. In BRCA1Δ1153BP1S25A mice, small intestinal villi are blunted and fused and the lamina propria is expanded by increased inflammatory cells; intestinal crypts are unorganized and cells display atypia. (B) Genomic instability (chromosome breaks and radials) measured in metaphase spreads from B lymphocytes derived from WT, BRCA1Δ11, 53BP1S25A and BRCA1Δ1153BP1S25A mice after PARPi treatment. Cells were stimulated for 2 days and then, treated for 16 hours with 1 μM PARPi. At least 50 cells were scored per condition. Experiment was repeated 5 times. Statistical significance was determined by Mann-Whitney t-test. (C) Colony formation in WT, BRCA1Δ11, 53BP1S25A and BRCA1Δ1153BP1S25A MEFs measured 9 days after continual treatment with 1 μM PARPi. Data are plotted relative to the plating efficiency of untreated controls of the same genotype. Statistical significance was determined by Mann-Whitney t-test. (D) RAD51 foci per EdU+ nucleus in WT, BRCA1Δ1153BP1S25A and BRCA1Δ1153BP1−/− MEFs measured 4 hours after 10 Gy IR, normalized by nuclear area (per 100 μm2). Statistical significance was determined by Mann-Whitney t-test. A minimum of 300 nuclei were quantified using the Gen5 spot analysis software. A representative experiment (n=2) is shown. Statistical significance was determined by Mann-Whitney t-test.
53BP1S25A Mutation Increases End Resection in BRCA1Δ11 cells
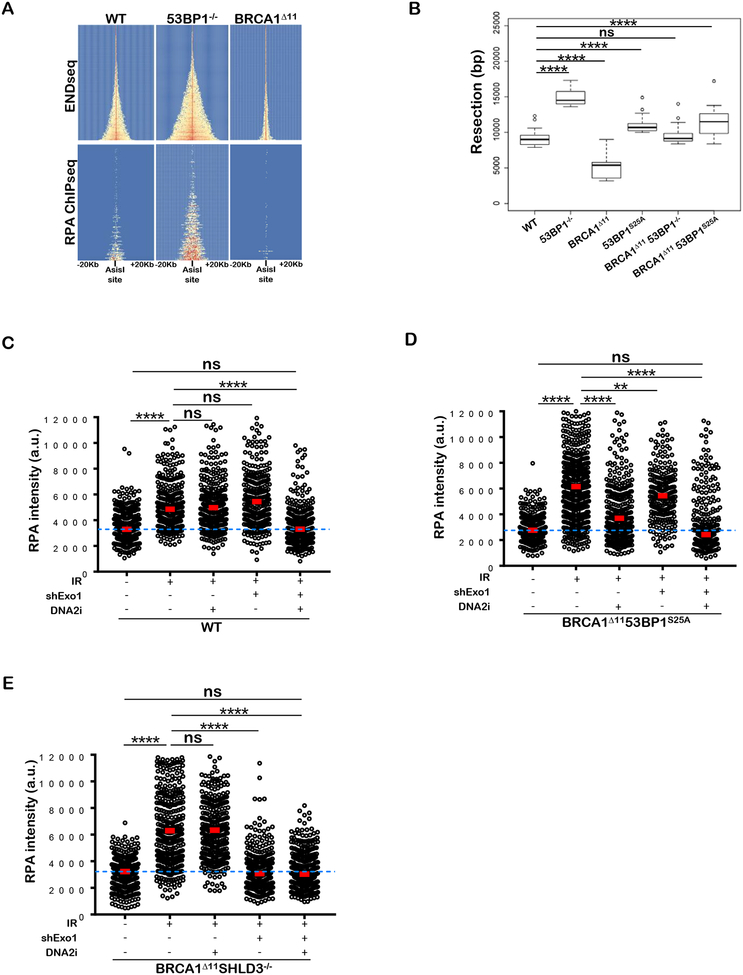
We speculated that the mutant 53BP1S25A protein might allow end resection sufficient to rescue lethality of BRCA1Δ11 mice, but inadequate for robust RAD51 foci formation or resistance to PARPi. To quantitate the level of end resection, we generated site specific DSBs by inducible expression of the restriction enzyme AsiSI and performed END-seq, which maps the dsDNA/ssDNA junctions at DSB sites (Canela et al., 2016). We observed considerable variation in resection across individual sites targeted by AsiSI (Figures 4A and S4A). Levels of end resection increased with breakage, which in turn correlated with active chromatin marks (Figures S4B and S4C) (Aymard et al., 2014). By visual inspection, it was clear that MEFs derived from 53BP1−/− mice exhibited increased end resection relative to WT, whereas BRCA1Δ11 MEFs showed reduced nucleolytic processing at all DSB sites (Figures 4A and S4A). Measurement of RPA-bound ssDNA by ChIP-seq (Brick et al., 2018), although less sensitive than END-seq, confirmed these findings (Figure 4A and S4D).
Figure 4. BRCA 1Δ1153BP1S25A cells exhibit normal levels of end resection catalyzed mainly by DNA2.
(A) Top panel: Heat map of END-seq signals across individual AsiSI sites in WT, 53BP1−/− and BRCA1Δ11 MEFs measured 5 hours after AsiSI induction. Lower panel: ChIP-seq for ssDNA bound by RPA in the same cells. Heat maps are ordered by END-seq signal intensity in WT cells. (B) Box plots showing quantification of resection end points in the top 10% resected breaks in WT, 53BP1−/−, BRCA1Δ11, 53BP1S25A, BRCA1An53BP1−/− and BRCA1Δ1153BP1S25A MEFs at AsiSI cleaved DSB sites. Welch’s t-test was used to determine statistical significance. (C) Quantification of the intensity of chromatin bound RPA in individual EdU-positive nuclei from WT and EXO1-depleted MEFs, either pre-treated or not with 1 μM DNA2i prior to 10 Gy IR. Cells were analyzed 4 hours post-IR. (D) Quantification of the intensity of chromatin bound RPA in individual EdU-positive nuclei from EXO1-proficient and EXO1-depleted BRCA1Δ1153BP1S25A MEFs, either pre-treated or not with 1 μM DNA2i prior to 10 Gy IR. Cells were analyzed 4 hours post-IR. (E) Quantification of the intensity of chromatin bound RPA in individual EdU-positive nuclei from EXO1-proficient and EXO1-depleted BRCA1Δ11SHLD3−/− MEFs, either pre-treated or not with 1 μM DNA2i prior to 10 Gy IR. Cells were analyzed 4 hours post-IR. In panels C, D, and E a minimum of 300 nuclei per condition were quantified using the Gen5 spot analysis software. A representative experiment (n=2) is shown. Statistical significance was determined by Mann-Whitney t-test.
53BP1S25A mutant MEFs showed slightly longer resection tracts than WT (Figure 4B and S4E). Consistently, we observed mildly increased levels of chromatin bound RPA in mutant cells (Figure S4F). Moreover, single strand annealing (SSA), a mutagenic RAD51-independent homology-mediated repair pathway that requires end resection (Stark et al., 2004), was slightly elevated (1.4 fold) in 53BP1S25A cells (Figure S4G). The average resection length in BRCA1Δ1153BP1−/− MEFs was similar to WT and intermediate between 53BP1- and BRCA1-deficiency (Figure 4B). Surprisingly, despite severely compromised IR-induced RAD51 foci (Figure 3D), BRCA1Δ1153BP1S25A MEFs accumulated elevated levels of ssDNA at AsiSI sites relative to BRCA1Δ11 or WT (Figure 4B). Moreover, IR-induced chromatin binding of RPA was increased in BRCA1Δ1153BP1S25A cells (Figure S4F). Thus, disrupting the 53BP1-PTIP interaction largely rescues the end resection defect associated with BRCA1-deficiency, but BRCA1Δ1153BP1S25A cells are nevertheless defective in loading RAD51.
End Resection is predominantly catalyzed by DNA2 in BRCA1Δ1153BP1S25A cells
The nucleases EXO1 and DNA2 are thought to act redundantly to promote end resection (Symington, 2016). Consistently, whereas depletion of EXO1 alone or treatment with the chemical inhibitor of DNA2 (NSC-105808) by itself did not decrease IR-induced RPA chromatin binding in replicating WT cells, end resection was significantly decreased when both nucleases were inactivated simultaneously (Figure 4C). However, in BRCA1Δ1153BP1S25A cells, treatment with DNA2i alone significantly reduced IR-induced RPA recruitment to damaged sites while EXO1 inhibition had a smaller impact (Figure 4D). Similarly BLM inhibition (BLMi) significantly reduced RPA foci formation in BRCA1Δ1153BP1S25A cells relative to WT (Figure S4H). These data implied that DNA2 activity is normally blocked by 53BP1-PTIP interaction. To determine which machinery catalyzes end resection in the absence of Shieldin, we examined nucleolytic processing in BRCA1Δ11SHLD3−/− cells as well as in BRCA1Δ11 cells depleted for RIF1. In contrast to BRCA1Δ1153BP1S25A cells, DNA damage induced RPA chromatin binding in BRCA1Δ11SHLD3−/− and BRCA1Δ11/RIF1-depleted cells was largely dependent on EXO1 (Figures 4E and S4I). Thus, distinct domains in 53BP1 recruit effectors to suppress resection by different nucleases.
Shieldin Interferes with RAD51 loading in BRCA1Δ1153BP1S25A Cells
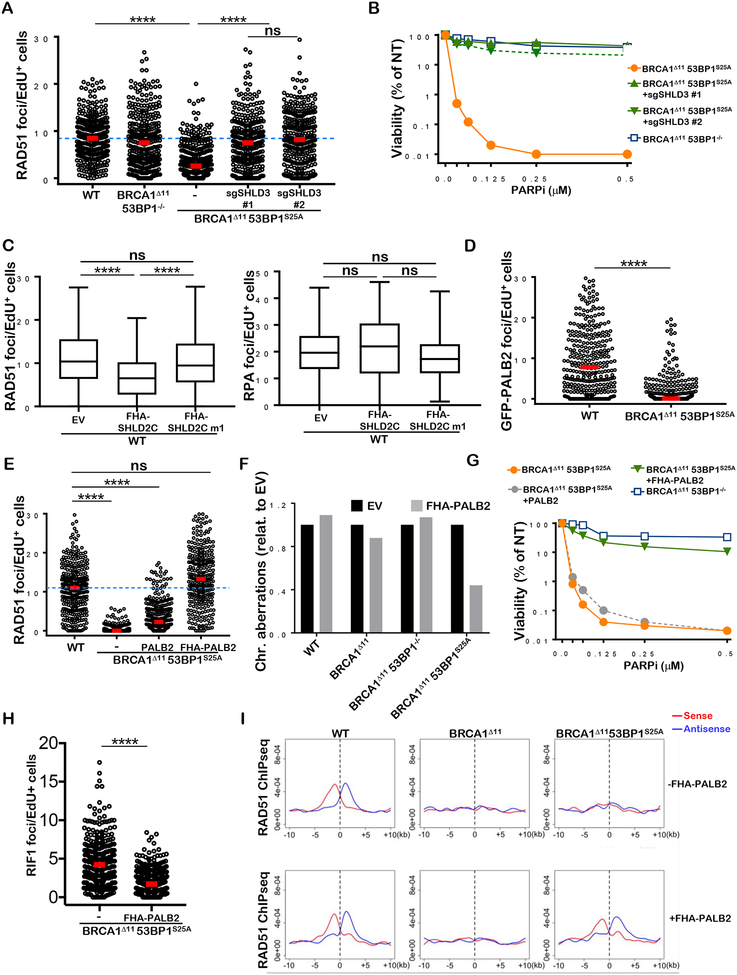
To explain the apparent inability of BRCA1Δ1153BP1S25A cells to load RAD51 despite abundant generation of ssDNA, we speculated that the RIF1/Shieldin complex might directly interfere with RAD51 filament formation in BRCA1Δ1153BP1S25A cells. Indeed, similar to the 53BP1S25A single mutant (Figure 1A), BRCA1Δ1153BP1S25A MEFs showed significantly elevated levels of RIF1 foci after IR (Figure S5A). To test the role of Shieldin in limiting RAD51 foci formation on resected DNA ends, we depleted Shld3 in BRCA1Δ1153BP1S25A MEFs both by siRNA- and CRISPR/CAS9-mediated deletion. Notably, IR-induced RAD51 foci were restored when Shld3 was depleted from BRCA1Δ1153BP1S25A MEFs by either method (Figures 5A and S5B). Moreover, Shld3 depletion suppressed PARPi hypersensitivity in BRCA1Δ1153BP1S25A MEFs (Figures 5B and S5C). We conclude that Shieldin blocks RAD51 filament formation post-resection in BRCA1Δ1153BP1S25A cells.
Figure 5. RAD51 loading defect in BRCA1Δ1153BP1S25A cells is corrected by Shld3 depletion or forced PALB2 chromatin binding.
(A) RAD51 foci per EdU+ nucleus in WT, BRCA1Δ1153BP1−/−, BRCA1Δ1153BP1S25A and BRCA1Δ1153BP1S25ä MEFs deficient in SHLD3 measured 4 hours after 5 Gy IR. Two independent BRCA1Δ1153BP1S25ASHLD3−/− clones (#1 and #2) were used. Foci numbers are normalized by nuclear area (per 100 μm2). (B) Viability of WT, BRCA1Δ1153BP1S25A and BRCA1Δ1153BPIS25ASHLD3−/− MEFs measured by CellTiter-Glo 10 days after PARPi treatment. (C) RAD51 (left panel) and RPA (right panel) foci formation 4 hours after 10 Gy IR in WT cells expressing FHA-SHLD2 containing (SHLD2C) or lacking (SHLD2Cm1) the OB fold (Noordermeer et al., 2018). (D) GFP-PALB2 foci per EdU+ nucleus in WT and BRCA1Δ1153BP1S25A MEFs overexpressing PALB2 measured 4 hours after 5 Gy IR. (E) RAD51 foci per EdU+ nucleus in WT, BRCA1Δ1153BP1S25A, and BRCA1Δ1153BP1S25A MEFs overexpressing PALB2 or PALB2 fused with the FHA domain of RNF8, measured 4 hours after 10 Gy IR. Foci numbers are normalized by nuclear area (per 100 μm). (F) Chromosomal aberrations in WT, BRCA1Δ11, BRCA1Δ1153BP1−/− and BRCA1Δ1153BP1S25A expressing empty vector or FHA-PALB2. (G) Viability of BRCA1Δ1153BP1S25A and BRCA1Δ1153BPΓ MEFs with or without FHA-PALB2 protein measured by CellTiter-Glo 10 days after PARPi treatment. (H) RIF1 foci per EdU+-nucleus in BRCA1Δ1153BP1S25A and BRCA1Δ1153BP1S25A MEFs expressing FHA-PALB2 measured 4 hours after 10 Gy IR. (I) Aggregate plot for ssDNA bound by RAD51 as measured by ChIP-seq at AsiSI sites, separated into sense and antisense strands. WT, BRCA1Δ11 and BRCA1Δ1153BP1S25A MEFs (top panels) are compared with their respective counterparts expressing FHA-PALB2 (bottom panels). In panels A, D, E and H, a minimum of 300 nuclei per condition were quantified using the Gen5 spot analysis software. In panel C, a minimum of 130 nuclei per condition were quantified. A representative experiment (n=2) is shown. Statistical significance was determined by Mann-Whitney t-test in all the indicated panels.
SHLD2 features OB-fold domains that bind to ssDNA (Dev et al., 2018; Findlay et al., 2018; Gao et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018). Forced targeting of SHLD2 to DSBs (by fusing SHLD2 to the RNF8 FHA domain) was found to suppress RAD51 foci formation in BRCA1/53BP1 deficient cells in a manner dependent on the SHLD2 OB-fold (Noordermeer et al., 2018). Similarly, we found that expression of FHA-SHLD2 was sufficient to reduce IR-induced RAD51 foci in WT cells only when the OB fold of SHLD2 was intact (Fig. 5C). However, FHA-SHLD2 did not block IR-induced RPA foci formation (Fig. 5C). Thus, in the presence of RPA-bound ssDNA, the loading of RAD51 is inhibited when SHLD2 is targeted to chromatin.
Since RNF168-dependent PALB2 recruitment is essential for loading RAD51 in BRCA1/53BP1 deficient cells (Zong et al., 2019), we examined PALB2 foci formation in BRCA1Δ1153BP1S25A cells by stable overexpression of GFP-PALB2. While IR-induced PALB2 foci were readily detectable in WT cells, PALB2 foci formation was severely compromised in BRCA1Δ1153BP1S25A cells (Figure 5D and S5D). Moreover, PALB2 overexpression did not overcome the defect in RAD51 foci formation (Figure 5E). Based on these findings, we speculated that RIF1/Shieldin interferes with PALB2/BRCA2 loading on ssDNA which prevents efficient RAD51 filament formation in BRCA1Δ1153BP1S25A cells.
To test this, we targeted PALB2 to chromatin by fusing PALB2 to the FHA domain of RNF8 (FHA-PALB2) (Zong et al., 2019). Previously, we have shown that forced PALB2 targeting to resected DSBs overcomes the severe HR defect in BRCA1Δ11/+ RNF168−/− cells (Zong et al., 2019). While FHA-PALB2 failed to rescue IR-induced RAD51 foci in resection-deficient BRCA1Δ11 cells, it restored RAD51 foci formation to WT levels in BRCA1Δ1153BP1S25A cells that are proficient in end resection (Figures 5E and S5E). Similarly, whereas FHA-PALB2 failed to mitigate PARPi sensitivity in BRCA1Δ11 cells or enhance PARPi resistance in BRCA1Δ1153BP1−/− cells, it markedly reduced the sensitivity of BRCA1Δ1153BP1S25A cells to PARPi (Figures 5F and 5G). PALB2 chromatin targeting also significantly reduced RIF1 foci formation (Figure 5H), consistent with the idea that elevated RIF1/Shieldin activity blocks PALB2 function in BRCA1Δ1153BP1S25A cells. Finally, the ability of FHA-PALB2 to rescue PARPi resistance and RAD51 foci formation in BRCA1Δ1153BP1S25A was dependent on its BRCA2-interaction domain (WD40) but not its chromatin association motif (ChAM) (Figure S5F, S5G), consistent with the idea that PALB2 recruits and is reliant on BRCA2 to assemble RAD51 (Kowalczykowski, 2015).
To directly examine RAD51 loading on ssDNA, we performed chromatin immunoprecipitation of ssDNA-bound RAD51 (Brick et al., 2018) at AsiSI cleaved sites. Consistent with immunofluorescence analyses, forced targeting of PALB2 to chromatin permitted RAD51 loading onto 3’ ssDNA in BRCA1Δ1153BP1S25A cells, while it did not nucleate RAD51 filaments at DSB sites in BRCA1Δ11 cells (Figure 5I and S5H), presumably because of the lack of sufficient ssDNA. Thus, the post-resection HR defect in BRCA1Δ1153BP1S25A cells is caused by an inability to recruit PALB2/BRCA2 to damaged chromatin.
Shieldin Interferes with the RNF168 Ubiquitylation Pathway that Loads RAD51
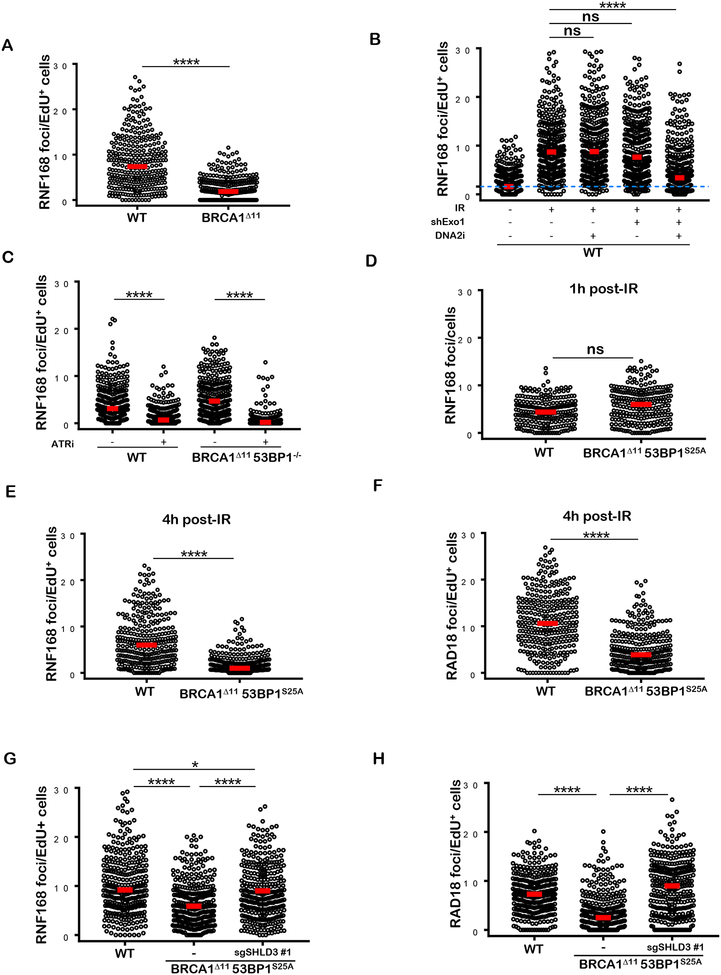
We have recently shown that in addition to its requirement for initiating 53BP1 recruitment to DSBs early after DNA damage, the E3 ubiquitin ligase RNF168 acts redundantly with BRCA1 to load PALB2/RAD51 on resected DNA ends (Zong et al., 2019). Indeed, we found that the accumulation of RNF168 at DSB sites during S-phase is dependent on end-resection as it is abrogated in BRCA1 -deficient cells, and in WT cells deficient in EXO1/DNA2 (Figures 6A and 6B). Moreover, RNF168 loading in S phase is dependent on ATR signaling as it is severely compromised when cells are pretreated with ATRi (Fig. 6C). Because 53BP1 associates with Shieldin on H2A-K15-Ub (Setiaputra and Durocher, 2019), which is also recognized by RNF168 (Doil et al., 2009; Stewart et al., 2009), we hypothesized that the block in PALB2 loading in BRCA1Δ1153BP125A cells might be due to defective association of RNF168 with its own mark post-resection. Indeed, the accumulation of RNF168 foci observed at 4 hours after IR was severely compromised in S-phase BRCA1Δ1153BP1S25A cells whereas the initial RNF168 recruitment, necessary for 53BP1/RIF1/Shieldin deposition was not altered (Figures 6D and 6E). RAD 18, which binds H2A-K15-Ub produced by RNF8/RNF168 (Hu et al 2017; Huang et al., 2009; Panier et al., 2012), also showed defective IRIF in S phase BRCA1Δ1153BP1S25A cells 4 hours post IR (Figure 6F and 6H). Importantly, deletion of Shld3 in BRCA1Δ1153BP1S25A rescued both RNF168 and RAD18 recruitment (Figures 6G and 6H). Thus, Shieldin blocks the RNF168-mediated chromatin ubiquitylation pathway in BRCA1Δ1153BP1S25A cells post resection, thereby compromising BRCA1-independent HR.
Figure 6. Shieldin blocks RNF168 recruitment post-resection in BRCA1Δ1153BP1S25A cells.
(A) Quantification of RNF168 foci in individual EdU+ nuclei from WT and BRCA1Δ11 MEFs. Cells were irradiated with 5 Gy and analyzed 4 hours post-IR. (B) Quantification of RNF168 foci in individual EdU+ nuclei from WT and EXO1-depleted MEFs, pretreated or not with DNA2i (1 μM). Cells were either un-irradiated or irradiated with 10 Gy and analyzed 4 hours post-IR. (C) Quantification of RNF168 foci in individual EdU+ nuclei from WT and BRCA1Δ1153BP1−/− MEFs pretreated with ATRi (AZ20, 10 μM). Cells were either un-irradiated or irradiated with 10 Gy and analyzed 4 hours post-IR. (D) Quantification of RNF168 foci in WT and BRCA1Δ1153BP1S25A MEFs 1 hour after 10 Gy IR. Statistical significance was determined by Wilcoxon Rank Sum test. (E) Quantification of RNF168 foci in individual EdU+ nuclei from WT and BRCA1Δ1153BP1S25A MEFs 4 hours post IR (5 Gy). Statistical significance was determined by Wilcoxon Rank Sum test. (F) Quantification of RAD 18 foci in individual EdU+ nuclei from WT and BRCA1Δ1153BP1S25A MEFs. Cells were irradiated with 5 Gy and analyzed 4 hours post-IR. (G) Quantification of RNF168 foci in individual EdU+ nuclei from WT, BRCA1Δ1153BP1S25A and BRCA1Δ1153BP1S25ASHLD3−/− MEFs. Cells were irradiated with 5 Gy and analyzed 4 hours post IR. (H) Quantification of RAD 18 foci in individual EdU+ nuclei from WT, BRCA1Δ1153BP1S25A MEFs and BRCA1Δ1153BP1S25ASHLD3−/− MEFs. Cells were irradiated with 5 Gy and analyzed 4 hours post-IR. In panels A-H, a minimum of 300 nuclei per condition were quantified using the Gen5 spot analysis software. A representative experiment (n=2) is shown. Unless otherwise noted, statistical significance was determined by Mann-Whitney t-test in all the panels.
RING-less BRCA1 Supports End Resection but not RAD51 Loading
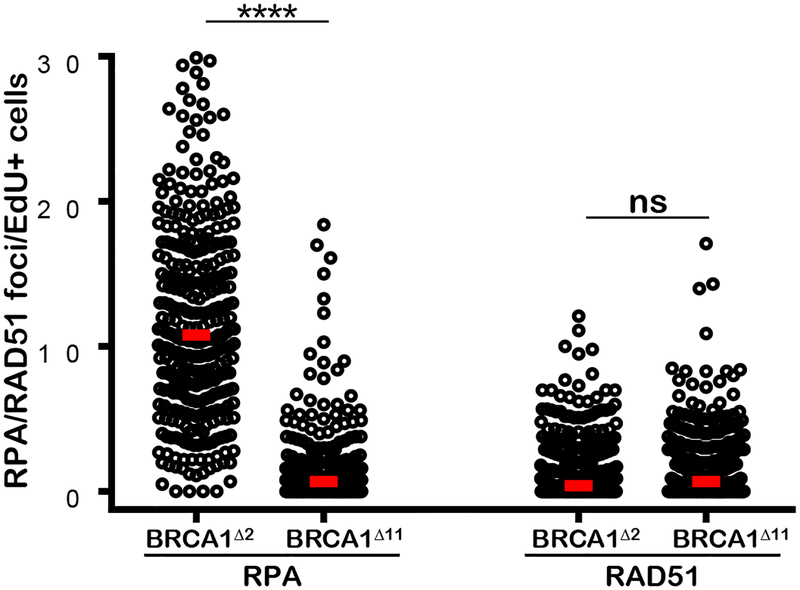
The RING (really interesting new gene) domain of BRCA1 posesses ubiquitin ligase activity and is required for stable interaction with BARD1 (Baer and Ludwig, 2002). Deletion of BRCA1 exon 2 (BRCA1Δ2/Δ2) results in an alternative translation initiation producing an N-terminally truncated BRCA1 protein lacking the RING domain (RING-less BRCA1) (Drost et al., 2016; Li et al., 2016; Wang et al., 2016). RING-less BRCA1 fails to stabilize BARD1 and is defective for HR (Drost et al., 2016; Li et al., 2016; Wang et al., 2016; Zong et al., 2019). In contrast to BRCA1Δ11/Δ11, we found that BRCA1Δ2/Δ2 MEFs exhibited robust levels of DSB resection measured by RPA foci, but failed to form RAD51 foci (Figure 7). Since loss of 53BP1 or RNF168 rescues the RAD51 loading defect in BRCA1Δ2/Δ2 cells (Zong et al., 2019), this indicates that 53BP1 primarily blocks RAD51 loading post-resection in cells expressing mutant BRCA1 lacking the RING domain.
Figure 7. RING-less BRCA1 supports end resection but not RAD51 filament assembly.
Left: IR-induced RPA foci per EdU+nuclei in BRCA1F2/F2 MEFs with adenoviral Cre infection which deletes BRCA1 exon 2 (BRCA2Δ2/Δ2) and in BRCA1Δ11 cells. Right: IR-induced RAD51 foci per EdU+ nucleus in BRCA2Δ2/Δ2 and BRCA1Δ11 cells. A minimum of 300 nuclei per condition were quantified using the Gen5 spot analysis software. A representative experiment (n=2) is shown. Statistical significance was determined by Mann-Whitney t-test.
DISCUSSION
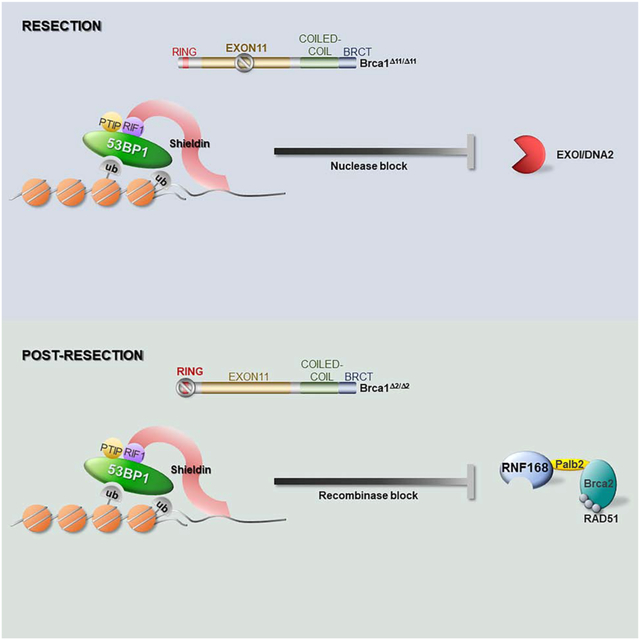
Our study supports a model in which RIF1/Shieldin and PTIP associate independently with 53BP1 to regulate distinct end resection pathways. Although they interact with different phosphorylation sites on 53BP1, RIF1 and PTIP appear to partially occlude one another. This results in the observed increase in IRIF for RIF1 in 53BP1S25A mutant cells that are incapable of binding PTIP. Nevertheless, elevated RIF1/Shieldin activity at DSBs is insufficient to inhibit end resection on its own. Interestingly, long-range resection is normally mediated by a combination of EXO1 and DNA2, but is catalyzed predominantly by DNA2 in PTIP-defective BRCA1Δ1153BP1S25A cells (Figures 4D and S6) and by EXO1 in Shieldin-defective BRCA1Δ11SHLD3−/− or BRCA1Δ11/RIF1-depleted cells (Figures 4E and S6). Thus, mutually exclusive phosphorylation-mediated interactions with PTIP and RIF1/Shieldin enable 53BP1 to independently suppress end resection by distinct nucleases.
Although loss of either 53BP1 effectors restores end resection to normal levels in BRCA1-deficient cells, they differentially impact downstream steps of HR. In BRCA1Δ1153BP1S25A cells, the continued presence of Shieldin on chromatin post-resection greatly reduces the loading of RAD51 on ssDNA (Figure S6). Defective RAD51 loading may in turn promote further hyperresection in BRCA1Δ1153BP1S25A cells. Inefficient RAD51 filament assembly is apparently sufficient to rescue the prenatal lethality associated with BRCA1Δ11 mutation, but is inadequate to support normal lifespan in BRCA1Δ1153BP1S25ä mice even though the cancer rate is not markedly increased. These phenotypes are reminiscent of dominant negative RAD51 mutations found in Fanconi anemia, which do not produce typical features of bone marrow failure or cancer predisposition, but instead cause neuronal defects, microcephaly and sensitivity to DNA damaging agents (Ameziane et al., 2015; Takenaka et al., 2019; Wang et al., 2015).
How does 53BP1/Shieldin block RAD51 filament assembly? RAD51 recombinase activity is facilitated by numerous proteins including BRCA2, RAD54 and the family of RAD51 paralogs which act at different stages of HR (Kowalczykowski, 2015). BRCA2 must first displace RPA from resected DNA ends to enable subsequent deposition of RAD51 on ssDNA (Kowalczykowski, 2015). Shieldin, RPA and BRCA2/PALB2 are all OB-fold containing complexes with ssDNA binding activity (Setiaputra and Durocher, 2019). An interesting possibility is that RPA and Shieldin may form mixed polymers on ssDNA. Although BRCA2 has higher affinity for ssDNA than RPA, it is possible that BRCA2 cannot efficiently displace Shieldin to allow nucléation of RAD51 filament. Indeed, we show that forced targeting of SHLD2 to DSBs is inhibitory to the replacement of RPA by RAD51 (Fig. 5C). Nevertheless, the block in HR in BRCA1Δ1153BP1S25A cells, associated with elevated of RIF1/Shieldin activity, can be overcome by forced PALB2 loading to chromatin (Figures 5E–5H) in a manner dependent on PALB2’s interaction with BRCA2 (Fig. S5F and S5G). In BRCA1 -deficient cells, the recruitment of PALB2/BRCA2 to ssDNA is dependent on RNF168-driven H2A-K15 ubiquitylation (Zong et al., 2019). Our data thus suggest that Shieldin binding to resected ends can directly compete with RNF168, leading to defective PALB2 accrual which prevents the nucléation of RAD51 filaments (Fig. S6). However, a more direct role of Shieldin in antagonizing BRCA2 activity cannot be discounted.
The post-resection anti-recombination functions of 53BP1 described herein may be relevant to the mechanism of chemotherapy resistance in a subset of BRCA1-mutated tumors. Germline pathogenic mutations in the RING domain, exemplified by the 185delAG founder mutation commonly found in the Ashkenazi Jewish population, are associated with breast and ovarian cancers, and predict poor responses to chemotherapy (Drost et al., 2016; Wang et al., 2016). Therapy resistance in this setting has been linked to overexpression of the hypomorphic RING-less BRCA1 protein, and is associated with partially recovered RAD51 loading and HR (Drost et al., 2016; Wang et al., 2016). By contrast, we found that non-cancerous cells expressing RING-less BRCA1 at physiological levels, despite robust end resection, are highly defective in RAD51 loading (Figures 7), which can be corrected by loss of 53BP1(Zong et al., 2019). We therefore propose that when the RING-less BRCA1 is overexpressed in cancers, its residual hypomorphic activity is able to partially overcome 53BP1/Shieldin binding to resected DNA ends, leading to partial restoration of HR and development of chemotherapy resistance.
In summary, expression of 53BP1S25A on a BRCA1-mutant background produces a striking uncoupling of end resection from downstream RAD51 loading and suggests that embryonic viability requires end resection but not necessarily efficient HR. These data support a model in which 53BP1 antagonizes two key steps in HR that are normally mediated by BRCA1.
STAR Methods
Contact for reagents and resource sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Andre Nussenzweig (andre_nussenzweig@nih.gov).
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental Model
A targeting construct containing the S25A mutation in murine 53BP1 was constructed by amplification of sequences from BAC clone RP23–179G19 (CHORI, Oakland, CA). Briefly, the left homology arm consisting of 5.1 kB of sequence prior to exon 2 of 53BP1 was inserted into an Entry clone using Gateway recombinational cloning (ThermoFisher, Carlsbad, CA) and flanked by Gateway attB4 and attB1 sites. The right homology arm consisting of exon 2 with an embedded S25A mutation and 3.1 kB of downstream homologous DNA in the subsequent intron was constructed using overlap extension PCR to introduce the appropriate mutation (TCT to GCT) and flanked by Gateway attB2 and attB3 sequences. Homology arm clones were sequenced in their entirety to ensure that no PCR mutations were introduced during amplification. Validated clones were used in a Multisite Gateway recombination reaction along with a selection cassette consisting of an attB1-attB2 flanked reverse orientation PGK promoter-neomycin resistance-BgH polyA fragment and introduced into a Gateway pUC19 destination vector with flanking attR4 and attR3 sites. The final targeting clone consists of the 9.1 kB sequence: left homology arm-(pA-neo-PGK)-exon 2 S25A-right homology arm and is flanked by NotI sites for release of the transgene from the vector backbone. Two derivative targeted ES clones were injected into C57BL/6 blastocysts. The resultant chimeric offspring was backcrossed with wild-type C57BL/6 mice, producing 53BP1+/S25A animals. Germline transmission of the targeted allele was confirmed by PCR (forward primer: 5’ ggagatggctgagaaagtgc 3’; reverse primer: 5’ tcccctggaatggaataaca 3’). The PGK-neo cassette was removed by crossing to β-actin-Cre transgenic mice (Jackson Laboratory). BRCA1+/Δ11 mice were obtained from the NCI mouse repository. 53BP1−/− mice were a gift from Junjie Chen. p53−/−mice were obtained from Taconic Biosciences. All mouse breeding and experimentation followed protocols approved by the National Institutes of Health Institutional Animal Care and Use Committee.
Method details
MEFs generation
Mouse embryonic fibroblasts (MEFs) were generated from E13.5 embryos and grown in Dulbecco’s Modified Eagle’s Medium (DMEM, GIBCO) supplemented with 15% heat-inactivated fet al bovine serum (FBS, Gemini Bio-Products) and 1% penicillin + streptomycin (GIBCO). To establish immortalized MEF cell lines, primary MEFs between passages 2–4 were transiently transfected with a vector encoding SV40 T-antigen (pCMV-SV40T). SV40-immortalized MEFs were routinely cultured in DMEM supplemented with 10 or 15% FBS.
For knockdown of Shieldin-3 in MEFs, CRISPR/Cas9 technology was used by cloning the gene-specific gRNAs into pX459 vector (Sense 5’ CACCGGGAAGTTTGGACTCATCGTA 3’ and Antisense 5’ AAACTACGATGAGTCCAAACTTCCC 3’) as described (Gupta et al., 2018). Cells were transfected with Lipofectamine following manufacturer’s procedure, selected with puromycin for 48 hours and subjected to single clone isolation. Confirmation of the mutated loci was done through PCR amplification and sequencing of the targeted region.
For transient depletion of endogenous Shieldin-3, siRNA technology (Invitrogen) was used (Sense 5’ GACUGCACAGUAGAUCUCUUGGAGU 3’ and Antisense 5’ ACUCCAAGAGAUCUACUGUGCAGUC 3’). siRNA was transfected with Lipofectamine RNAiMAX (Invitrogen) as per manufacturer’s instructions.
For knockdown of EXO1, a pool of TRC lentiviral short hairpins were used (Dharmacon. 71124 antisense 5’ ATAGAACTAGACCTACAGAGC 3’, 71126 antisense 5’ TTATTCCTCATCTTAGACGGG 3’ and 71127 antisense 5’ ATCCGTCAAATATGAGAATCG 3’). Lentiviral particles were produced by transfecting 293T cells with Lipofectamine-3000 as per manufacturer’s instructions. Forty-eight hours later, viral supernatant was harvested and used to transduce MEFs. Cells were selected with puromycin and used two days later. EXO1 knockdown was confirmed by RT-PCR using the following primers: Forward: 5’ AGGGGAACAGAACTCCAAGC 3’, Reverse2: 5’ CCAGGAACCTTGTTCCGTCT 3’. Samples were run and analyzed on a BioRad CFX96 Real-Time PCR detection system. For microirradiation, MEFs were presensitized in DMEM media containing 0.1 μg/ml of Hoechst 33342 for 60 min before replacing it with phenol red free media containing 5 mM HEPES, and then irradiated with the 364-nm laser line on a LSM510 confocal microscope (Carl Zeiss, Inc.) equipped with a heated stage. Cells were allowed to recover for 1 hour before processing for immunofluorescence.
Plasmids and transfection
Retroviral pMX-PIE-based vectors encoding fusion protein of EGFP-FHA-PALB2(del)s were produced as follows: cDNAs corresponding to truncated human PALB2 proteins were produced by PCR amplification from either pDEST-FRT-TO-GFP-PALB2 1–103, -PALB2-deltaCHAM, -PALB2-deltaMRG15, or -PALB2-deltaWD40(gifts from Dr. Daniel Durocher) then ligated with cDNA corresponding to FHA domain of human RNF8 fused with EGFP (Zong et al., 2019). The resulting EGFP-FHA-PALB2(del) fragments were subcloned into pMX-PIE vector at the multi-cloning site (MluI/PacI). Infection-competent retroviral particles were assembled in BOSC23 cells co-transfected with the pCL-ECO helper virus.
Retroviral supernatant was collected 40–48 h later to transduce MEFs
Immunoblotting and Immunofluorescence
Western blotting was performed as described previously (Zong et al., 2019). Briefly, cells were collected and lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5% Tween-20, 0.5% NP-40, 2 mM PMSF, 2.5 mM β-glycerophosphate (all from Sigma-Aldrich) and protease inhibitor cocktail tablet (complete Mini, Roche Diagnostics). Equal amounts of protein were loaded into precast mini-gels (Invitrogen) and resolved by SDS-PAGE. Proteins were blotted onto a nitrocellulose membrane, blocked with 5% membrane blocking agent (GE Healthcare) in TBS and incubated with the corresponding primary antibody. Primary antibodies were used at the following dilutions: anti-53BP1 (1:1000, Novus Biologicals), anti-phospho53BP1-S25 (1:500, Cell Signaling), anti-Tubulin (1:10,000, Sigma- Aldrich), antiphospho-53(S15) (1:500, Cell Signaling), anti-Chk2 (1:1500, Upstate Biotechnology). Fluorescent secondary antibodies were used at a dilution of 1:15,000 (Li-Cor). Detection of protein bands was performed by fluorescence imaging using a Li-Cor Odyssey CLx imaging system (Li-Cor Biosciences).
For immunofluorescence staining, MEFs were grown on 18 mm × 18 mm glass coverslips, and B lymphocytes were attached to slides coated with CellTak (BD Biosciences). Prior to γ-irradiation (137Cs Mark 1 irradiator, JL Shepherd), cells were incubated with 10 μM EdU (Invitrogen) for 20 min. Where indicated, cells were additionally pretreated with 1 μM NSC-105808 (DNA2i) or 10 μM AZ20 (ATRi) for 1 hour. Following irradiation, cells were allowed to recover for 1 hour or 4 hours. Pre-extraction and fixation of samples were carried out as previously described (Zong et al., 2019). The antibodies used for standard immunofluorescence experiments were anti-RIF1 (1:5,000, gift of Davide Robbiani, Rockefeller University), anti-53BP1 (1:1000, Novus), anti-RAD51 (1:250, Abcam), anti-RPA (1:5,000, Abcam), anti-RNF168 (1:100, R&D Systems), anti-RAD18 (1:5000, Millipore), anti-GFP (1:500, Roche). For laser microirradiation experiments, primary antibodies were anti-γH2AX (1:5000, Upstate Biotechnology) and anti-PTIP (1 μg/ml, gift of Kai Ge). Detection was achieved using fluorochrome-conjugated secondary antibodies (Invitrogen). Where indicated, Click-IT chemistry was performed as per manufacturer’s instructions. Finally, DNA was counterstained with DAPI (Thermo Fisher Scientific). Images were captured at 63× magnification with an AxioCam MRC5 attached to an Axio Observer Z1 epifluorescence microscope (Zeiss) or at 40× magnification on a Lionheart LX automated microscope (BioTek Instruments, Inc.). Quantification of nuclear foci and total nuclear intensity was performed using the Gen5 spot analysis software (BioTek). ZEN Blue (Zeiss) was used to quantify fluorescence intensities of laser stripes.
Metaphase spreads, clonogenic survival and viability assays
Activated asynchronous B cells and MEFs were treated with 1 μM PARPi (AZD2281, Selleckchem) for 16 hours, subsequently arrested at mitosis with 0.1 μg/ml colcemid (Roche) and metaphase chromosome spreads were prepared as previously described (Zong et al., 2019). Images were acquired using a Metafer automated scanning and imaging platform (MetaSystems).
To assay for clonogenic survival, MEFs were seeded in 6 cm dishes and treated continuously with 1 μM PARPi (Selleckchem) or exposed to the indicated doses of IR. After 9 days, culture dishes were stained with 0.5% crystal violet. Colonies containing >50 cells were counted. Clonogenic survival for a given treatment was calculated relative to the plating efficiency in non-treated controls.
To determine cell growth and viability, MEFs were plated in 6-well plates (10,000 per well) and treated continuously with different doses of PARPi for 10 days. The drug-containing medium was replenished every three days and cells were subcultured when they approach confluency. On day 10, cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) as per manufacturer’s instructions.
Single Strand Annealing (SSA) assay
To measure SSA, MEFs resuspended in 800 μl of OptiMEM were elecroporated with 15 μg of linearized SA-GFP reporter (Stark et al., 2004). Cells were selected in puromycin, and puromycin-resistant clones were pooled for analysis. The pools of clones were seeded onto a 24 well dish at 5–7 × 105 cells per well, transfected the next day with 0.5 μg of I-SceI expression plasmid (pCBASce) or GFP expression plasmid with 1.8 μl Lipofectamine 2000 (Thermofisher) overnight. GFP+ cells were analyzed three days after the start of transfection, using a CyAN ADP cytometer (Beckman Coulter). Repair frequencies were normalized to transfection efficiency, using the parallel transfections with the GFP expression vector.
Purification of TAP-53BP1 complex and MS/MS analysis
Purification of 53BP1 was performed using an established tandem immunoaffinity method (Nakatani and Ogryzko, 2003). Flag-HA-53BP1 was stably expressed after transduction of pOZ-Flag-HA-53BP1 retrovirus into HeLa-S cells. Cells were harvested and resuspendend in 5X volume of hypotonic buffer (10 mM Tris, pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors (Pierce)). Cells were pelleted by spinning at 2,500 rpm, resuspended in 1X pellet volume of hypotonic buffer and homogenized using a dounce tissue grinder (Wheaton). Nuclear material was pelleted, resuspended in 0.5X pellet volume of low salt buffer (20 mM Tris pH 7.3, 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors), and dounced again. 0.5X pellet volume of high salt buffer (20 mM Tris pH 7.3, 1.2M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 10 mM β-mercaptoethanol, and protease inhibitors) was slowly added to nuclear extract, which was subsequently stirred for 30–45 minutes. Extract was spun down at 14,000 rpm for 30 minutes and the soluble material was dialyzed in BC100 buffer (20 mM Tris pH 7.3, 100 mM KCl, 0.2 mM EDTA, 20% Glycerol, 0.2 mM PMSF, 1 mM β-mercaptoethanol). 53BP1 complex was purified from the nuclear extract using anti-Flag (M2) resin (Sigma), followed by purification using anti-HA (F-7) resin (Santa Cruz) in TAP buffer (50 mM Tris-HCl pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, and protease inhibitors). For both Flag and HA purifications, nuclear extract was rotated with resin for 4 hours, washed extensively with TAP buffer, and eluted with 0.4 mg/mL of Flag or HA peptide (Sigma). After elution, the complex was TCA precipitated and associated proteins were identified by LC-MS/MS using an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (ThermoFisher) and Sequest software.
In vivo PARP inhibitor treatment and histopathological analyses
BRCA1Δ1153BP1S25A and WT littermate control mice were administered PARPi (AZD2281) prepared in 10%DMSO/90% Captisol via oral gavage. Mice were treated with 40 mg/kg of PARPi daily in a volume of approximately 100 μ1 and monitored daily for changes in health status following PARPi treatment. H&E staining was routinely performed on formalin-fixed, paraffin-embedded tissues. Gut rolls of the small intestine, cecum, and large intestine were digitized with an Aperio ScanScope XT (Leica) at 200× in a single z-plane. Aperio whole-slide images were evaluated by a board-certified veterinary pathologist.
Sections were photographed at 10× magnification. Mice were perfused transcardially with 4% paraformaldehyde (PFA), while embryos were drop-fixed in 4% PFA and tissues were cryoprotected in 25% PBS-buffered sucrose solution, embedded in O.C.T. and sectioned sagittally at 10 μm using an HM500M cryostat (Microm). Immunohistochemistry was performed after antigen retrieval. Antibodies used were: anti-Tbr1 (1:250; Synaptic Systems #328005) and anti-phospho KAP1(Ser-824) (1:500; Bethyl Labs, #A300–767A). Immunostaining of active caspase-3 was visualized with a VIP substrate kit (Vector Laboratories) and biotinylated secondary antibody and avidin-biotin complex (Vectastain Elite kit; Vector Laboratories). Sections were counterstained with 0.1% methyl green (Vector Laboratories), dehydrated and mounted with DPX (Fluka). For fluorescence detection, FITC or Cy3 conjugated secondary antibodies (Jackson Immunologicals) were used and nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) or propidium iodide (Vector Laboratories). TUNEL staining was done using the ApopTag system (Chemicon).
Magnetic Resonance and X-Ray imaging
Magnetic resonance imaging (MRI) was performed using a 7 T Bruker ClinScan system (Bruker BioSpin MRI GmbH, Germany) equipped with 12S gradient coil. A two-channel surface coil was used for MR imaging. Animals were anesthetized and maintained with 1.5–2% isoflurane during MRI sessions. Data analysis was done by manually segmenting the regions and computing volumes using OsiriX (v5.7, Pixmeo, Switzerland).
For the kyphosis study, mice were imaged in the prone position for a 3-bed position X-ray Computed Tomography (CT) (Inveon Multi-Modality PET/CT, Siemens Medical Solutions, Knoxville, TN). X-ray CT step and shoot acquisition parameters were: 80 kVp, 500 μA, 1000 msec per step, 180 steps covering 360-degrees. X-ray CT images were reconstructed using a Feldkamp cone beam algorithm with a Shepp-Logan smoothing filter resulting in 512×512×1170 matrix (0.08 × 0.08 × 0.08 μm pixel size). Images were linearly calibrated to Hounsfield units (air: −1000 HU, water: 0 HU). The X-ray CT 3D images (orthogonal axial, coronal, and sagittal views) were displayed using a medical image viewer (MIM v 6.6.5, MIM Software Inc, Cleveland, OH) and a pseudo-3D maximum intensity projection (MIP) was implemented to rotate and translate the mouse into perpendicular orientation for analysis
END-seq and ChIP-SSDS
A retrovirus encoding AsiSI (pTRE3G-HA-ER-AsiSI) was stably transduced into the indicated MEF cell lines as previously described (Canela et al., 2016). Exponentially growing AsiSI-expressing MEFs were treated with 1 μM doxocyclin (DOX) for 24 hours and then, for 5 hours with 4-OHT, resulting in the nuclear translocation of AsiSI. MEFs were harvested and 9 million cells were embedded in agarose plugs, after which END-seq was performed as described (Canela et al., 2017). END-seq reads were aligned to the mouse (GRCm38p2/mm10) genomes using Bowtie (version 1.1.2) (Langmead et al., 2009) with parameters -n 3 -k 1 −1 50. Break intensity was measured by integrating the RPKM values within 100 bp of each AsiSI break site. To quantify the width of maximum resection endpoint (in bp), a sliding window containing twenty 100 bp bins was used, starting from the AsiSI site out to 20 kb to the right side. When more than half of the bins within this sliding window had an RPKM value equal to or lower than the background, then the last bin within the window with a detectable signal over background was regarded as the maximum resection endpoint. Background was determined by the maximum END-seq signal for 100 bp bins more than 20 kb (within 20 kb-30 kb) away from individual AsiSI sites.
To map ssDNA bound by RAD51 and RPA at AsiSI sites (ChIP-SSDS), we first captured DNA bound by RAD51 or RPA. Twenty million cells were harvested for chromatin immunoprecipitation (ChIP) using an anti-RAD51 (Abcam #176458, 10 μg/sample) or anti-RPA antibody (Abcam #10359, 10 μg/sample). ssDNA that spontaneously forms hairpins after heat renaturation was then enriched and sequenced as described (Brick et al., 2018).
Statistical analyses
Unless indicated, all data are presented as individual replicates. The total number of replicates, mean and error bars are explained in the figure legends. The statistical tests (Mann-Whitney, Welch’s, Mantel-Cox and Wilcox Rank Sum) and resulting P values (represented by asterisks) are indicated in the figure legends and/or figure panels and were calculated using GraphPad Prism and R software (ns = p>0.05; * = p≤0.05; ** = p≤0.01; *** = p≤0.001; **** = p≤0.0001).
Data and code availability
The datasets generated during this study are available at GEO with accession GEO: GSE133808. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133808.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-53BP1 | Novus Biologicals | Cat# NB100–305; RRID: AB_10001695 |
| Rabbit polyclonal anti-53BP1 (phosho S25) | Abcam | Cat# ab70323; RRID:AB_1267592 |
| Mouse monoclonal anti-α-Tubulin | Sigma-Aldrich | Cat# T-5168; RRID: AB_477579 |
| Rabbit polyclonal anti-Phospho-p53 (Ser15) | Cell Signaling | Cat# 9284; RRID:AB_331464 |
| Mouse monoclonal anti-Chk2 | Millipore | Cat# 05–649; RRID:AB_2244941 |
| Mouse monoclonal anti-GFP | Roche Applied Science | Cat# 11814460001; RRID: AB_390913 |
| Rabbit polyclonal anti-Rad51 | Abcam | Cat# ab176458; RRID:AB_2665405 |
| Rabbit polyclonal antiRPA32/RPA2 | Abcam | Cat# ab10359; RRID:AB_297095 |
| Rabbit polyclonal antiRPA32/RPA2 | Abcam | Cat# ab76420; RRID:AB_1524336 |
| Sheep polyclonal anti-RNF168 | R&D System | Cat# AF7217; RRID:AB_10971653 |
| Mouse monoclonal anti-RAD18 | Millipore | Cat# ABE1377 |
| Mouse monoclonal anti-FLAG (M2) | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-RIF1 | Gift from Davide Robbiani | N/A |
| Mouse monoclonal anti-H2AX (pS139) | Cat# 05–636; RRID:AB_309864 | |
| Guinea pig polyclonal antibody anti-Tbr1 | Synaptic Systems | Cat# 328 005; RRID:AB_2620072 |
| Rabbit polyclonal antibody anti-Kap1 (Ser 824) | Bethyl | Cat# A300–767A; RRID:AB_669740 |
| IRDye 680RD Goat anti-Mouse IgG (H+L) | LI-COR Biosciences | Cat# 926–68070; RRID:AB_10956588 |
| IRDye 800CW Goat anti-Rabbit IgG (H+L) | LI-COR Biosciences | Cat# 925–32211; RRID:AB_2651127 |
| FITC-Goat anti-Guinea Pig IgG(H+L) | Jackson ImmunoResearch | Cat# 106–096-003; RRID:AB_2337418 |
| Cy3-Goat anti-Rabbit IgG(H+L) | Jackson ImmunoResearch | Cat# 111–166-003; RRID:AB_2338007 |
| Alexa Fluor 488 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-11001; RRID:AB_2534069 |
| Alexa Fluor 568 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-11031; RRID:AB_144696 |
| Alexa Fluor 568 Goat anti-Rat IgG (H+L) | Thermo Fisher Scientific | Cat# A-11077; RRID:AB_2534121 |
| Alexa Fluor 488 Chicken anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A21441; RRID:AB_10563745 |
| Alexa Fluor 568 Goat anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-11011; RRID:AB_143157 |
| Alexa Fluor 568 Donkey anti-Sheep IgG (H+L) | Thermo Fisher Scientific | Cat# A21099; RRID:AB_10055702 |
| Purified Rat anti-Mouse CD180 (RP/14) | BD Biosciences | Cat# 552128; RRID:AB_394343 |
| Bacterial and Virus Strains | ||
| Bacteria: TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C404006 |
| Retrovirus: pCL-ECO | Addgene | Cat# 12371 |
| Lentivirus: pSpCas9(BB)-2A-Puro (PX459) | Addgene | RRID:Addgene_48139 |
| Mammalian expression pCMV-VsVg envelope lentiviral protein | Gift from Katrin F. Chua | N/A |
| Mammalian expression lentiviral packaging pCMV-dR8.2 | Gift from Katrin F. Chua | N/A |
| Retrovirus: pMX-empty-IRES-GFP-Puro | Jiri Lukas, (Zong et al., 2015) | N/A |
| Retrovirus: pMX-empty(no IRES-GFP)-Puro | Gift from Davide Robbiani | N/A |
| Retrovirus: pMX-GFP-PALB2-Puro | (Zong et al., 2015) | N/A |
| Retrovirus: pMX-GFP-FHA(RNF8)-PALB2-Puro | (Zong et al., 2015) | N/A |
| Mammalian expression: pCBASceI | Addgene | RRID:Addgene_26477 |
| Mammalian expression: pEGFP-N1 | Jeremy M. Stark | N/A |
| Adenovirus: Ad5-CMV-eGFP | Addgene | N/A |
| Adenovirus: Ad5-CMV-Cre-eGFP | Addgene | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Olaparib | Selleckchem | Cat# AZD2281 |
| ATR inhibitor (AZ20) | Selleckchem | Cat# S7050 |
| DNA2 inhibitor | NIH Developmental Therapeutics Program | Cat# NSC-105808 |
| Bloom inhibitor (ML216) | Sigma-Aldrich | Cat# SML0661 |
| 4-Hydroxytamoxifen | Sigma-Aldrich | Cat# H7904 |
| Doxycycline hyclate | Sigma-Aldrich | Cat# D9891 |
| Lipopolysaccharide (LPS) from E. coli O111:B4 | Sigma-Aldrich | Cat# L2630 |
| Interleukin 4 (IL-4) from mouse | Sigma-Aldrich | Cat# I1020 |
| CD43 microbeads (Ly-48) | Miltenyi Biotec | Cat# 130–049-801 |
| Protein G Magnetic Beads | Active Motif | Cat# 104502 |
| X-tremeGENE™ 9 DNA Transfection Reagent | Roche Diagnostics | Cat# 6365809001 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000015 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat# 62249 |
| GeneArt Seamless Cloning Enzyme Mix | Thermo Fisher Scientific | |
| Zero Blunt™ PCR Cloning Kit | Thermo Fisher Scientific | Cat# K270020 |
| PNA probe for telomeres Cy3-(CCCTAA)3 | PNA Bio | Cat# F1002 |
| DAPI | Thermo Fisher Scientific | Cat# 62248 |
| EdU | Thermo Fisher Scientific | Cat# A10044 |
| Colcemid | Roche Diagnostics | Cat# 10295892001 |
| cOmplete, Mini Protease inhibitor cocktail | Roche Diagnostics | Cat# 11836153001 |
| Crystal Violet | Sigma-Aldrich | Cat# 0775 |
| Anti-FLAG M2 magnetic beads | Sigma | Cat# M8823; RRID:AB_2637089 |
| Trizol | Invitrogen | Cat# 15596026 |
| Puregene Proteinase K enzyme | Qiagen | Cat# 158920 |
| Puregene RNase A Solution | Qiagen | Cat# 158924 |
| T4 DNA Polymerase | NEB | Cat# M0203L |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | Cat# M0210L |
| Exonuclease T (ExoT) | NEB | Cat# M0265L |
| Exonuclease VII (ExoVII) | NEB | Cat# M0379L |
| Klenow Fragment (3′→5′ exo-) | NEB | Cat# M0212L |
| Quick Ligation Kit | NEB | Cat# M2200L |
| USER enzyme | NEB | Cat# M5505L |
| KAPA HiFi HotStart ReadyMix (2X) | KAPA Biosystems | Cat# KK2600 |
| MyOne Streptavidin C1 Beads | ThermoFisher | Cat# 650–01 |
| Agencourt AMPure XP beads | Beckman Coulter | Cat# A63881 |
| Dynabeads Protein A | Thermo Fisher Scientific | Cat# 10002D |
| Critical Commercial Assays | ||
| Click-IT EdU Alexa Fluor 488 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10425 |
| Click-IT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10634 |
| Vector VIP Substrate kit | Vector laboratories | Cat# SK-4600; RRID:AB_2336848 |
| ApopTag Fluorescein Apoptosis detection kit | Chemicon | Cat# S7110 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat# G7571 |
| KAPA Library Quantification Kit | Kapa Biosciences | Cat# KK4824 |
| CHEF Mammalian Genomic DNA plug kit | Bio-Rad | Cat# 1703591 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE133808 |
| ChIP-seq for H3K27ac, H3K79me2, H3K27me3 | (Chronis et al., 2017) | GSE90893 |
| Experimental Models: Cell Lines | ||
| MEF: Wildtype | This paper | N/A |
| MEF: BRCA1Δ11/Δ11 | This paper | N/A |
| MEF: 53BP1S25A/S25A | This paper | N/A |
| MEF: 53BP1−/− | (Bunting et al., 2010) | |
| MEF: BRCA1 Δ11/Δ11 53BP1−/− | (Bunting et al., 2010) | |
| MEF: BRCA1 Δ11/Δ11 53BP1S25A/S25A | This paper | |
| MEF: BRCA1Δ2/Δ2 | (Bunting et al., 2010) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: BRCA1+/Δ11. B6/129 | NCI mouse repository | Strain # 01XC9 |
| Mouse: 53BP1S25A/S25A. B6/129 | This paper | N/A |
| Mouse: 53BP1−/−. B6/129 | (Ward et al., 2003) | N/A |
| Mouse: p53+/−. B6/129 | Taconic Biosciences | N/A |
| Mouse: BRCA1+/Δ2. B6/129 | (Ludwig et al., 1997) | N/A |
| Oligonucleotides | ||
| Primers for genotyping 53BP1_S25A allele: Forward, 5’-ggagatggctgagaaagtgc; Reverse, 5’-tcccctggaatggaataaca | This paper | N/A |
| sgRNA targeting endogenous mouse Shieldin-3 locus: Sense 5’ CACCGGGAAGTTTGGACTCATCGTA; Sense 5’ CACCGGGAAGTTTGGACTCATCGTA | (Gupta et al., 2018) | N/A |
| siRNAs targeting endogenous mouse Shieldin-3 locus: Sense 5’ GACUGCACAGUAGAUCUCUUGGAGU and Antisense 5’ ACUCCAAGAGAUCUACUGUGCAGUC | This paper | N/A |
| shRNAs targeting endogenous mouse Exo1 locus: Pool of antisense 5’-ATAGAACTAGACCTACAGAGC, antisense 5’ TTATTCCTCATCTTAGACGGG and antisense 5’ ATCCGTCAAATATGAGAATCG | This paper | N/A |
| Primers to confirm Exo1 knockdown by RT-PCR: Forward: 5’ AGGGGAACAGAACTCCAAGC, Reverse: 5’ CCAGGAACCTTGTTCCGTCT | This paper(Sasanuma et al., 2018)(Sasanuma et al., 2018)(Sasanuma et al., 2018) | N/A |
| Recombinant DNA | ||
| Plasmid: FLAG-PALB2 | (Orthwein et al., 2015) | N/A |
| Plasmid: GFP-PALB2 | (Orthwein et al., 2015) | N/A |
| Software and Algorithms | ||
| ZEN 2 (blue edition) | Zeiss | https://www.zeiss.com/ |
| Gen5 spot analysis | BioTek | https://www.biotek.com/ |
| Metafer 4 | MetaSystems | https://metasystems-international.com/ |
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| RStudio | RStudio Team | https://www.rstudio.com/ |
| Bowtie 1.1.2 | (Langmead et al., 2009) | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.1.2/ |
| MACS 1.4.3 | (Zhang et al., 2008) | https://pypi.org/pypi/MACS/1.4.3 |
| UCSC database | (Karolchick et al., 2004) | https://genome.ucsc.edu |
| UCSC genome browser | (Kent et al., 2002) | https://genome.ucsc.edu |
| Bedtools | (Quinlan and Hall, 2010) | https://github.com/arq5x/bedtools2 |
| R 3.3.5 | R Core Team, 2008 | https://www.r-project.org/ |
| Sequest | (Eng et al., 1994) | N/A |
| OsiriX v5.7 | Pixmeo | https://www.osirix-viewer.com/ |
| MIM v6.6.5 | MIM Software Inc. | https://www.mimsoftware.com/ |
| FlowJo (10.1) | FlowJo LLC | https://www.flowjo.com/ |
| Other | ||
| BOSC23 retrovirus packaging cells | ATCC | Cat# CRL-11270; RRID: CVCL_4401 |
| 293T lentivirus packaging cells | ATCC | Cat# CRL-11268; RRID:CVCL_1926 |
| NuPAGE™ 4–12% Bis-Tris Protein Gels | ThermoFisher Scientific | Cat# NP0321BOX |
| Glass Bottom Microwell Dishes | MatTek Corporation | Cat# P35G-1.5–14-C |
| FluoroBrite DMEM Media | Thermo Fisher Scientific | Cat# A1896701 |
| LSM510 confocal microscope | Zeiss | N/A |
| Axio Observer Z1 epifluorescence microscope | Zeiss | N/A |
| IN Cell Analyzer | GE Healthcare | N/A |
| Odyssey® CLx Imaging System | LI-COR Biosciences | N/A |
| Lion heart LX automated microscope | BioTek Instruments | N/A |
| CyAN ADP cytometer | Beckman Coulter | N/A |
| FACSCalibur | BD Biosciences | N/A |
| Cytogenetic drying chamber | Thermotron | N/A |
| LTQ Orbitrap Velos Pro ion-trap mass spectrometer | Thermo Fisher Scientific | N/A |
| Aperio ScanScope XT | Leica | N/A |
| Nano Quant Infinite M200 Pro microplate reader | Tecan | N/A |
| Mark 1 137Cs irradiator | JL Shepherd | N/A |
Highlights.
Point mutation 53BP1S25A rescues lethality of BRCA1Δ11 mice without restoring HR
53BP1S25A mutation uncouples 53BP1 end blocking activities pre- and post-resection
RIF1/Shieldin blocks BRCA1 independent loading of RAD51 onto single strand DNA
Shieldin and PTIP associate with 53BP1 to regulate distinct end-resection pathways
ACKNOWLEDGMENTS
We thank Anthony Tubbs for comments on the paper; Jennifer Mehalko and Dom Esposito (Protein Expression Laboratory, Frederick National Laboratory for Cancer Research) for transgenic constructs; Karim Baktiar, Diana Haines and Elijah Edmonson (Pathology/histotechnology Laboratory, Frederick National Laboratory for Cancer Research) for rodent necropsy, pathology analysis and imaging; Joseph Kalen and Nimit Patel (Small Animal Imaging Program, Frederick National Laboratory for Cancer Research) for X-ray CTscan imaging; Jennifer Wise and Kelly Smith for assistance with animal work; Davide Robbiani and Kai Ge for antibodies; Dan Durocher for Shieldin constructs; David Goldstein and the CCR Genomics core for sequencing support. Neil Johnson for discussions. Research in the J.M.S. laboratory is supported by the NIH grant R01CA197506. The A.N. laboratory is supported by the Intramural Research Program of the NIH, an Ellison Medical Foundation Senior Scholar in Aging Award (AG-SS- 2633–11), the Department of Defense Idea Expansion (W81XWH-15–2-006) and Breakthrough (W81XWH-16–1-599) Awards, the Alex Lemonade Stand Foundation Award, and an NIH Intramural FLEX Award.
Footnotes
Supplemental Information
Table S1. Mass Spectrometry analysis of WT and mutant 53BP1. Proteins with >1 total peptides for 53BP1+/+ are shown.
Related to Figure 1.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alver RC, Chadha GS, Gillespie PJ, and Blow JJ (2017). Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cell Rep 18, 2508–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H, et al. (2015). A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun 6, 8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. (2014). Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21, 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R, and Ludwig T (2002). The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev 12, 86–91. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. (2010). 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Pratto F, Sun CY, Camerini-Otero RD, and Petukhova G (2018). Analysis of Meiotic Double-Strand Break Initiation in Mammals. Methods Enzymol 601, 391–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. (2010). 53BP1 inhibits homologous recombination in BRCA1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, and de Lange T (2009). Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol 187, 385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. (2013). 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. (2017). Genome Organization Drives Chromosome Fragility. Cell 170, 507–521 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, and Nussenzweig A (2016). DNA Breaks and End Resection Measured Genome-wide by End Sequencing. Mol Cell 63, 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. (2009). A selective requirement for 53BP1 in the biological response to genomic instability induced by BRCA1 deficiency. Mol Cell 35, 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Feng W, Lim PX, Kass EM, and Jasin M (2018). Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu Rev Cancer Biol 2, 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EA, Prindle MJ, and Dressler GR (2003). BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol 23, 1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, and Plath K (2017). Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 168, 442–459 e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, and Chapman JR (2016). 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms. Mol Cell 64, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, et al. (2010). PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science 329, 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev H, Chiang TW, Lescale C, de Krijger I, Martin AG, Pilger D, Coates J, Sczaniecka-Clift M, Wei W, Ostermaier M, et al. (2018). Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol 20, 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. (2009). RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446. [DOI] [PubMed] [Google Scholar]
- Drost R, Dhillon KK, van der Gulden H, van der Heijden I, Brandsma I, Cruz C, Chondronasiou D, Castroviejo-Bermejo M, Boon U, Schut E, et al. (2016). BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest 126, 2903–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Findlay S, Heath J, Luo VM, Malina A, Morin T, Coulombe Y, Djerir B, Li Z, Samiei A, Simo-Cheyou E, et al. (2018). SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Feng S, Ning S, Liu J, Zhao H, Xu Y, Shang J, Li K, Li Q, Guo R, et al. (2018). An OB-fold complex controls the repair pathways for DNA double-strand breaks. Nat Commun 9, 3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Oliveira C, Becker JR, Bilham K, Moralli D, Anzilotti C, Fischer R, Deobagkar-Lele M, Sanchiz-Calvo M, Fueyo-Marcos E, et al. (2018). 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature 560, 122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, and Chen J (2009). Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem 284, 7284–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, Kremer M, Typas D, Lammers M, Mailand N, Nussenzweig A,et al. (2018). DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173, 972–988 e923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Botuyan MV, Cui G, Zhao D, and Mer G (2017). Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18. Mol Cell 66, 473–487 e479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, and Chen J (2009). RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32, D493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC (2015). An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cole F, Patel DS, Misenko SM, Her J, Malhowski A, Alhamza A, Zheng H, Baer R, Ludwig T, et al. (2016). 53BP1 ablation rescues genomic instability in mice expressing ‘RING-less’ BRCA1. EMBO Rep 17, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Chapman DL, Papaioannou VE, and Efstratiadis A (1997). Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of BRCA1, Brca2, BRCA1/Brca2, BRCA1/p53, and Brca2/p53 nullizygous embryos. Genes Dev 11, 1226–1241. [DOI] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, Bianchi A, Zimmermann M, Durocher D, and de Lange T (2018). 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature 560, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, and Ogryzko V (2003). Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 370, 430–444. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Gurkar AU, Wang Y, Vijg J, Hoeijmakers JHJ, and Robbins PD (2018). Nuclear Genomic Instability and Aging. Annu Rev Biochem 87, 295–322. [DOI] [PubMed] [Google Scholar]
- Noordermeer SM, Adam S, Setiaputra D, Barazas M, Pettitt SJ, Ling AK, Olivieri M, Alvarez-Quilon A, Moatti N, Zimmermann M, et al. (2018). The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. (2015). A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH, and Durocher D (2012). Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell 47, 383–395. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, and Durocher D (2019). Shieldin - the protector of DNA ends. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, and Jasin M (2004). Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24, 9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. (2009). The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434. [DOI] [PubMed] [Google Scholar]
- Symington LS (2016). Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51, 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka S, Kuroda Y, Ohta S, Mizuno Y, Hiwatari M, Miyatake S, Matsumoto N, and Oka A (2019). A Japanese patient with RAD51-associated Fanconi anemia. Am J Med Genet A 179, 900–902. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, and Burgoyne PS (2005). Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37, 41–47. [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. (2015). A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell 59, 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, Nicolas E, Peri S, van der Gulden H, van der Heijden I, O’Brien SW, et al. (2016). The BRCA1-Delta11q Alternative Splice Isoform Bypasses Germline Mutations and Promotes Therapeutic Resistance to PARP Inhibition and Cisplatin. Cancer Res 76, 2778–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, and Chen J (2003). p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23, 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G, and Paull TT (2014). Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res 42, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong D, Adam S, Wang Y, Sasanuma H, Callen E, Murga M, Day A, Kruhlak MJ, Wong N, Munro M, et al. (2019). BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation. Mol Cell 73, 1267–1281 e1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available at GEO with accession GEO: GSE133808. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133808.