Abstract
Background
Vitamin D receptor (VDR) is involved in multiple immune-mediated disorders including oral lichen planus (OLP). This study investigated the association between VDR gene polymorphisms and the risk of OLP.
Methods
In total, 177 OLP patients and 207 healthy participants were recruited from the Affiliated Hospital of Stomatology, Nanjing Medical University. Eight single nucleotide polymorphisms (SNPs: rs731236, rs739837, rs757343, rs2107301, rs2239185, rs7975232, rs11574129 and rs11568820) in the VDR gene were selected and genotyped.
Results
The results showed that OLP risk was increased in subjects with the rs2239185 TT genotype (Recessive model: adjusted Odd ratio(OR) = 2.68, 95% Confidence interval(CI) = 1.28–5.62, P = 0.009) and rs7975232 CC genotype (Recessive model: adjusted OR = 2.25, 95% CI = 1.10–4.58, P = 0.026). Moreover, rs2239185 and rs7975232 (P < 0.01) showed significant cumulative effects on OLP risk.Haplotype analysis showed that the CC haplotype (rs2239185-rs7975232) was associated with an increased risk of OLP (OR = 3.11, 95% CI = 1.42–6.83, P = 0.005), compared with the AC haplotype.
Conclusion: The rs2239185 and rs7975232 variants of VDR may influence OLP susceptibility, and VDR gene polymorphisms may be candidate susceptibility regions for OLP in a Chinese Han population.
Keywords: Oral lichen planus, Vitamin D receptors, Risk, Single nucleotide polymorphisms
Background
Oral lichen planus (OLP) is a chronic inflammatory disease of the oral mucosa mediated by T cells, whose etiology remains unknown. OLP is characterized as dense lymphocyte infiltration and basal keratinocyte degeneration observable under a microscope [1]. OLP, the typical clinical feature of which include white stripes, can manifest as reticular, papular, plaque-like, erosive, atrophic and bullous [2, 3]. Erosive-like lesions are considered to be the most threatening condition and are characterized by pain, ranging from mild discomfort to severe onset [4].The pain seriously affects the patient’s eating experience and food digestion, reducing the patients quality of life.
Previous studies have suggested that vitamin D (VD) deficiency may be associated with an increased risk of some inflammatory diseases, such as OLP and inflammatory bowel disease [5, 6]. OLP patients presented a nearly 50% reduction in mucosal VD levels, which may be caused by immunoreactions [6]. As a ligand-induced transcription factor, vitamin D receptors (VDRs) encoded by the VDR gene (chromosomal location 12q12–14) play an important role in regulating the role of vitamin D [7.8]. Increasing evidence suggests that single nucleotide polymorphisms (SNPs) in vitamin D-related genes could affect the properties of vitamin D, such as its anti-carcinogenic effects [7]. Thus, we speculated that VDR gene polymorphisms may be related to OLP. Since OLP is considered as a potential precancerous lesion, specific SNPs in VDR or vitamin D pathway genes may also play an important role in oral cancer.
Based on the above, we conducted this study in a Chinese Han population to investigate the association between key polymorphisms in VDR genes and OLP susceptibility.
Material and methods
Study groups and samples
Assuming the control group with a mutation frequency of 0.3, OR value of 2, statistical power of 0.8 and Type I error probability of 0.05, the current study may need to recruit 141 case patients and 141 control patients to be able to reject the null hypothesis that the probability is equal to 1. A total of 177 patients with OLP from the Affiliated Hospital of Stomatology, Nanjing Medical University, Jiangsu Province, China were enrolled in this study between January 2017 and June 2018.The inclusion criteria for OLP patients were as follows: (1) > 18 years old; (2) diagnosed as OLP by an oral pathologist using a biopsy specimen; and (3) treatment-naïve (the patient did not receive relevant treatment prior to inclusion in the study). Pregnant women, patients with other oral diseases (for example, lichenoid reaction lesions), patients receiving systemic or topical steroids over the past three months, and patients with autoimmune diseases were excluded. The control group included 207 healthy subjects who underwent a physical examination at the physical examination center and had no oral mucosal lesions, inflammation, infection, or autoimmune diseases such as systemic lupus erythematosus (SLE) or rheumatoid arthritis (RA). The diagnostic criteria used in this study were diagnostic guidelines developed by Van der Meij et al. according to the World Health Organization (WHO) definition of OLP [8].
The oral mucosa of all participants was assessed by two experienced oral medicine specialists. If there was a disagreement between the two examiners, a third clinically experienced oral medicine specialist would make the judgment. The main clinical features, including clinical subtype, affected sites, the number of sites, the presence of cutaneous lesions, and the types of oral lesions and symptoms, were collected for further analyses. All subjects were informed of the purpose of the study and signed the informed consents. Information, such as demographic data, alcohol consumption habits and oral hygiene, were collected by one-to-one surveys using a questionnaire designed according to our research content. Prior to OLP diagnosis, participants who drank more than 20 alcoholic drinks per week were classified as heavy drinkers [9]. The periodontal status of all subjects including the gingival index (GI), periodontal index (PI) and bleeding on probing (BOP) was evaluated in both groups. The oral hygiene of subjects was defined as poor when both GI and PI were ≥ 2, and the BOP was 1. Additionally, 10 ml of venous blood was collected from each subject for biochemical tests and SNP determination.
DNA isolation and genotyping
Genomic DNA was extracted from peripheral blood samples using protease K digestion and phenol/chloroform purification according to a standard protocol. The TaqMan allelic discrimination technology via an ABI 7900HT Sequence Detection system (Applied Biosystems, San Diego, California, USA) was used to explore polymorphisms at the chosen SNPs. Polymerase chain reaction (PCR) was executed with the following thermal profile: 50 twarer 2 min to preheat, 95 °C for 10 min for preincubate, followed by 40 cycles at 95 e with the 50 twarer 2 min to anneal thermal profile. The genotyping results were detected by LightCycler480 real-time PCR (Roche Diagnostics, Mannheim, Germany), with a 100% success rate. Two blank controls were specified to a 384-well format for quality control with 10% of the samples randomly selected as repeat samples, producing 100% concordance.
Information regarding VDR SNPs was acquired from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) and the Chinese Han population database in HapMap (http://www.hapmap.org). All SNPs were selected according to the following criteria: (1) minor allele frequency (MAF) ≥0.05 in the Chinese population and (2) a Hardy-Weinberg equilibrium test P-value ≥0.05. Tag SNPs were chosen to represent a set of variants with strong linkage disequilibrium (LD) [9]. A total of eight SNPs in the VDR gene (rs731236, rs739837, rs757343, rs2107301, rs2239185, rs7975232, rs11574129 and rs11568820) were selected according to the above steps.
Statistical analysis
All analyses were performed in Stata/SE (V.12.0 for Windows). Differences in the demographic characteristics between the case and control groups were analyzed by the Student-t test (for continuous variables) or chi-square (χ2) test (for categorical variables). The relationship between a candidate SNP and OLP risk was estimated by multivariate logistic regression analysis, and the results were expressed as OR and its 95% CI. The heterogeneity between the corresponding subgroups was examined by the Q test. The Cochran-Armitage test was used for trend analysis. Haplotype analysis was performed to explore the relationship between two significant SNPs and OLP risks. The PHASE software (v2.1) was used to estimate the haplotype frequency based on the observed genotypes. The Single-fold view software (version 4.2) was used to analyze LD parameters (i.e., D and r2) [10] and the Thesias software (version 3.1) was used to analyze associations of identified haplotypes in the VDR gene with OLP [11].
Results
Demographic information on 177 OLP patients and 207 healthy subjects is shown in Table 1. There were similar age and gender distributions between the two groups (P = 0.155 and 0.091, respectively). However, compared with the control group, OLP patients had more alcohol consumption and better oral hygiene (P < 0.05).
Table 1.
Characteristics of clinical data between OLP and control group
| Variables | OLP | Control | P-value |
|---|---|---|---|
| (n = 177) | (n = 207) | ||
| Mean age, year | 49.20 ± 14.14 | 48.47 ± 10.88 | 0.569 |
| < 45 | 61 (34.46) | 86 (41.55) | 0.155 |
| ≥ 45 | 116 (65.54) | 121 (58.45) | |
| Gender | |||
| Male (%) | 49 (27.68) | 74 (35.75) | 0.091 |
| Female (%) | 128 (72.32) | 133 (64.25) | |
| Alcohol | |||
| Heavy | 11 (6.21) | 2 (0.97) | 0.005 |
| Non Heavy | 166 (93.79) | 205 (99.03) | |
| Oral hygiene | |||
| Good | 131 (74.01) | 119 (57.49) | 0.001 |
| Poor | 46 (25.99) | 88 (42.51) | |
Abbreviation: OLP oral lichen planus
The genotype distribution of the eight SNPs in the two groups was described using dominant, recessive and additive genetic models in Table 2. The recessive genetic model computed by logistic regression analyses showed that rs2239185 and rs7975232 were significantly associated with OLP susceptibility. Patients carrying the rs2239185-TT genotype (adjusted OR = 2.39, 95%CI = 1.10–5.18, P = 0.027) and rs7975232-CC genotype (adjusted OR = 2.65, 95%CI = 1.24–5.66, P = 0.012) tended to have a higher risk of OLP.
Table 2.
Distribution of VDR genotypes between OLP and control group
| Genotype | OLP | Control | OR (95% CI) | P-value |
|---|---|---|---|---|
| rs731236 | ||||
| CC | 154 (87.01) | 180 (86.96) | 1.00 | – |
| CT | 21 (11.86) | 24 (11.59) | 0.99 (0.51–1.91) | 0.979 |
| TT | 2 (1.13) | 3 (1.45) | 0.93 (0.15–6.06) | 0.946 |
| Dominant | 0.99 (0.52–1.85) | 0.965 | ||
| Recessive | 0.94 (0.15–6.05) | 0.947 | ||
| Additive | 0.98 (0.57–1.71) | 0.954 | ||
| rs739837 | ||||
| CC | 98 (55.37) | 112 (54.11) | 1.00 | – |
| CA | 61 (34.46) | 82 (39.61) | 0.73 (0.45–1.20) | 0.218 |
| AA | 18 (10.17) | 13 (6.28) | 1.36 (0.60–3.09) | 0.468 |
| Dominant | 0.82 (0.51–1.31) | 0.404 | ||
| Recessive | 1.60 (0.73–3.50) | 0.242 | ||
| Additive | 0.98 (0.69–1.41) | 0.927 | ||
| rs757343 | ||||
| AA | 113 (63.84) | 128 (61.84) | 1.00 | – |
| AG | 53 (29.94) | 73 (35.27) | 0.70 (0.43–1.15) | 0.163 |
| GG | 11 (6.21) | 6 (2.90) | 2.06 (0.70–6.13) | 0.180 |
| Dominant | 0.81(0.50–1.29) | 0.375 | ||
| Recessive | 2.36 (0.81–6.89) | 0.117 | ||
| Additive | 0.97 (0.66–1.44) | 0.893 | ||
| rs2107301 | ||||
| CC | 83 (46.89) | 96 (46.38) | 1.00 | – |
| CT | 79 (44.63) | 93 (44.93) | 1.03 (0.67–1.60) | 0.883 |
| TT | 15 (8.47) | 18 (8.70) | 1.19 (0.54–2.58) | 0.668 |
| Dominant | 1.06 (0.69–1.61) | 0.800 | ||
| Recessive | 1.17 (0.55–2.45) | 0.686 | ||
| Additive | 1.07 (0.77–1.48) | 0.706 | ||
| rs2239185 | ||||
| CC | 90 (50.85) | 111 (53.62) | 1.00 | – |
| CT | 62 (35.03) | 83 (40.10) | 0.78 (0.47–1.27) | 0.315 |
| TT | 25 (14.12) | 13 (6.28) | 2.39 (1.10–5.18) | 0.027 |
| Dominant | 1.00 (0.63–1.59) | 0.995 | ||
| Recessive | 2.68 (1.28–5.62) | 0.009 | ||
| Additive | 1.24 (0.88–1.74) | 0.206 | ||
| rs7975232 | ||||
| AA | 81 (45.76) | 118 (57.00) | 1.00 | – |
| AC | 70 (39.55) | 74 (35.75) | 1.36 (0.83–2.23) | 0.225 |
| CC | 26 (14.69) | 15 (7.25) | 2.65 (1.24–5.66) | 0.012 |
| Dominant | 1.57 (0.97–2.50) | 0.061 | ||
| Recessive | 2.25 (1.10–4.58) | 0.026 | ||
| Additive | 1.54 (1.09–2.18) | 0.014 | ||
| rs11574129 | ||||
| CC | 119 (67.23) | 146 (70.53) | 1.00 | – |
| CT | 50 (28.25) | 57 (27.54) | 0.97 (0.59–1.61) | 0.921 |
| TT | 8 (4.52) | 4 (1.93) | 2.49 (0.70–8.84) | 0.159 |
| Dominant | 1.08 (0.67–1.75) | 0.740 | ||
| Recessive | 2.50 (0.71–8.84) | 0.154 | ||
| Additive | 1.17 (0.78–1.76) | 0.439 | ||
| rs11568820 | ||||
| AA | 61 (34.46) | 74 (35.75) | 1.00 | – |
| AG | 81 (45.76) | 96 (46.38) | 1.10 (0.69–1.76) | 0.695 |
| GG | 35 (19.77) | 37 (17.87) | 1.26 (0.69–2.28) | 0.454 |
| Dominant | 1.14 (0.74–1.78) | 0.551 | ||
| Recessive | 1.19 (0.70–2.03) | 0.520 | ||
| Additive | 1.12 (0.83–1.50) | 0.455 | ||
Logistic regression analyses adjusted for age, gender, alcohol, oral hygiene
Abbreviation:VDR vitamin D receptor, OLP oral lichen planus, OR odds ratio, CI confident interval
The cumulative effects of the two SNPs on OLP were evaluated by the Cochran-Armitage trend test. The results showed that the risk of OLP increased as the number of mutations increased (Table 3). The OLP risk increased in subjects carrying one or both of the rs2239185 and rs7975232 alleles (OR = 2.33, 95% CI = 1.22–4.43). In stratified analyses on the combined variant alleles (rs2239185 and rs7975232) and OLP risk susceptibility, no heterogeneity was observed (Table 4).
Table 3.
Cumulative Effects of rs2239185 and rs7975232 on OLP risk
| Variables | OLP n(%) | Control n(%) | OR(95%CI) | P-value |
|---|---|---|---|---|
| 0 | 145 (81.92) | 188 (90.82) | 1.00 | – |
| 1 | 13 (7.34) | 10 (4.83) | 1.82 (0.74–4.48) | 0.190 |
| 2 | 19 (10.73) | 9 (4.35) | 2.87 (1.21–6.81) | 0.017* |
| Trend | P a = 0.007 | |||
| 0 | 145 (81.92) | 188 (90.82) | 1.00 | – |
| 1–2 | 32 (18.08) | 19 (9.18) | 2.33 (1.22–4.43) | 0.010 |
Variables are numbers of combined unfavorable alleles (rs2239185-TT and rs7975232-CC)
Logistic regression analyses adjusted for age, gender, alcohol consumption and oral hygiene. Pa-value was analyzed by Cochran-Armitage trend test
Abbreviation: OLP oral lichen planus, OR odds ratio, CI confident interval
Table 4.
Stratified analyses on combined variant alleles (rs2239185 and rs7975232) and OLP risk
| Variables | OLP risk (0 vs.1–2) | OR (95%CI) | P a | P b | |
|---|---|---|---|---|---|
| OLP | Control | ||||
| Age, year | |||||
| ≥ 45 | 50/11 | 75/11 | 1.69 (0.60–4.71) | 0.318 | 0.457 |
| < 45 | 95/21 | 133/8 | 3.13 (1.28–7.65) | 0.012 | |
| Gender | |||||
| Male | 40/9 | 58/16 | 0.61 (0.21–1.72) | 0.347 | 0.267 |
| Female | 105/23 | 130/3 | 10.11 (2.82–36.30) | < 0.001 | |
| Alcohol | |||||
| Heavy | 8/3 | 1/1 | – | – | – |
| Non Heavy | 137/29 | 187/18 | 2.42 (1.25–4.67) | 0.008 | |
| Oral hygiene | |||||
| Good | 109/22 | 103/16 | 1.59 (0.75–3.37) | 0.231 | 0.453 |
| Poor | 36/10 | 85/3 | 6.47 (1.56–26.90) | 0.010 | |
Logistic regression was used in the implicit model to determine the adjusted P a value according to age, gender, alcohol consumption and oral hygiene, heterogeneity was used to test
P b-value
Abbreviation: OLP oral lichen planus, OR odds ratio, CI confident interval
LD information for the two SNPs was shown in Table 5. We performed haplotype analysis to assess the effects of the rs2239185 and rs7975232 haplotype variant alleles on the OLP risk (Table 5). When compared with the most frequent AC haplotype, the CC haplotype was significantly associated with OLP susceptibility (OR = 3.11, 95%CI = 1.42–6.83), which was consistent with the single SNP analysis.
Table 5.
Haplotypes analysis of rs2239185 and rs7975232 with OLP risk
| Haplotype | OLP n(%) | Control n(%) | OR | P |
|---|---|---|---|---|
| AC | 218 (61.59) | 295 (71.26) | 1.00 | 1.00 |
| CT | 98 (27.68) | 94 (22.70) | 1.38 (0.96–1.97) | 0.084 |
| CC | 24 (6.78) | 10 (2.42) | 3.11 (1.42–6.83) | 0.005 |
| AT | 14 (3.95) | 15 (3.62) | 1.21 (0.55–2.66) | 0.633 |
Logistic regression analyses adjusted for age, gender,alcohol consumption and oral hygiene
SNPs order: rs2239185 and rs797523
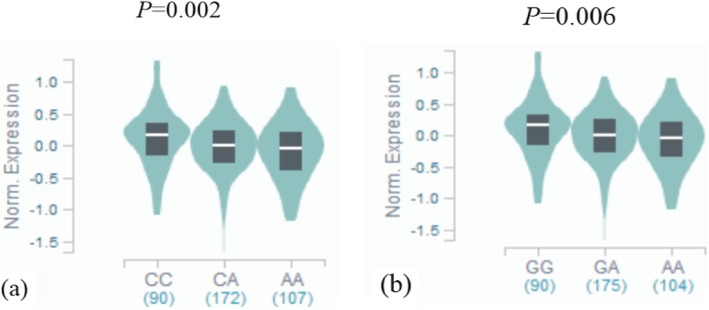
To further explore the biological significance of rs2239185 and rs7975232 in VDR, we also searched for expression quantitative trait loci (eQTL) evidence based on the public Genotype-Tissue Expression(GTEx) database (https://gtexportal.org/). We found that the VDR rs2239185 and rs7975232 genotypes were significantly associated with the expression of VDR in whole blood. Mutations associated with VDR rs7975232 and rs2239185 down-regulated of VDR gene expression in whole blood (P = 0.002 and 0.006, respectively, Fig. 1).
Fig. 1.
a Results of eQTL analysis on VDR rs7975232 loci. in whole blood. b. Results of eQTL analysis on VDR rs2239185 loci. in whole blood
Discussion
OLP is one of the main causes of oral cancer and approximately 0.5 to 2.0% of patients with OLP can progress to malignant transformation [12]. Finding new markers that can effectively identify people at a high risk for OLP can help prevent and control OLP, thereby reducing the progression of malignant transformation. Previous studies have confirmed the role of multiple factors including genetic polymorphisms in OLP susceptibility [13]. The current study aimed to further explore the effects of genetic variation in the VDR gene and related environmental factors on OLP susceptibility in Chinese Han population and the results showed that VDR rs2239185-TT and rs7975232-CC increased the susceptibility to OLP, after adjusting for factors such as drinking and oral hygiene [14, 15].
Vitamin D and its receptors are mainly involved in immune regulation and calcium and phosphorus metabolism regulation. Evidence from population studies suggests that vitamin D deficiency is highly prevalent among the general population in China, which may be related to genetic variation in VDR [16]. Previous studies have shown that VDR genetic variants (such as rs1544410, rs7975232 and rs731236) can participate in the development of various tumors (such as prostate cancer and hepatocellular carcinoma) by affecting the biological effects of vitamin D [17, 18]. Shortly thereafter, vitamin D deficiency was found to be associated to immune-related diseases (systemic lupus erythematosus) and infectious diseases (tuberculosis) [19]. On this basis, our study found the association between the genetic variation of VDR and OLP susceptibility, which further confirmed the findings of Zhao Bin et al. And their study found that the VD/VDR signaling pathway could prevent OLP development by mediating the NF-κB pathway [20]. The current results revealed that mutations in VDR rs2239185 and rs7975232 increased the risk of OLP, and OLP risk increased with increasing adverse alleles. The heterogeneity test suggested that our final results were robust and unaffected by other factors. Additionally, due to the strong LD between VDR rs7975232 and rs2239185, haplotype analysis was further conducted with the results suggested that SNP-SNP interactions in the VDR gene increased susceptibility to OLP, which further validated the single-gene analysis results [21–23].
The gene encoding VDR, which is located on chromosome 12q13, consists of 9 exons and 8 introns, and is approximately 75 kb in length [24]. Both rs2239185 and rs7975232 are located in a VDR gene intron, and their polymorphism can affect messenger RNA transcription and protein translation by regulating gene transcription to change the expression and function of VDR [25]. Moreover, the GTEx database showed that VDR expression in the blood was indeed affected by genetic variation associated with rs2239185 and rs7975232. Therefore, we hypothesized that the rs2239185 and rs7975232 variants, as well as the CC haplotypes between the two, may affect VDR expression in whole blood by interfering with transcription. The binding of VD to VDR in whole blood and oral keratinocytes was reduced, preventing the activation of the vitamin D/VDR signaling pathway [20], thereby inhibiting functions related to the VDR signaling pathway, such as anti-inflammatory effects, and eventually led to the occurrence of OLP.
It is worth noting that our findings differ from those of Bojan Kujundzic et al., which found that the genetic polymorphism associated with OLP risk was not VDR rs7975232 but VDR rs2228570 (rs2239185 was not included in the study) [9]. And the haplotype analysis involved rs7975232 and rs731236 suggested that the AT haplotype decreased OLP risk (compared with the most common haplotype CT), further confirming that OLP susceptibility was not caused by a single SNP. Many previous epidemiological studies have shown that the incidence of OLP, clinical symptoms and the mutation rate of VDR genes are different in different ethnic populations, so we speculate that this may also be the main reason for the difference between the two studies [26, 27]. The sample size for this study was smaller than our study (only 65 patients with oral lichen planus and 100 healthy blood donors), and all of study subjects were Caucasian and Serbian. Nonetheless, the results needs to be further confirmed in larger sample cohort studies and functional experimental studies.
We acknowledge that there are limitations in this study. Although various analyses were used to reveal the potential impact of VDR gene polymorphisms on OLP susceptibility, the existing epidemiological evidence presented in this study was not sufficient to speculate on the role of the VDR gene, indicating that our findings require confirmation by functional studies. However, our study also has advantages. First, this is the first study to find genetic evidence that the VDR gene is associated with OLP susceptibility in a Chinese Han population. Second, the sample size of this study is larger than the existing related research [9].
Conclusion
This is the first study to show that genetic mutations in VDR are associated with OLP susceptibility, and that rs2239185 and rs7975232 may be the genetic markers for OLP susceptibility in a Chinese Han population. However, larger scale prospective studies and functional experiments are required to validate our findings.
Acknowledgements
The authors thank Haozhi Fan,Feng Zang,Mei Liu,Lingyun Zhuo and Jingjing Wu for their help with laboratory work.
Abbreviations
- BOP
Bleeding on probing
- CI
Confidence interval
- eQTL
Expression quantitative trait loci
- GI
Gingival index
- GTEx
Genotype-Tissue Expression
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- NJMU
Nanjing Medical University
- OLP
Oral lichen planus
- OR
Odd ratio
- PCR
Polymerase chain reaction
- PI
Periodontal index
- RA
Rheumatoid arthritis
- SLE
Systemic lupus erythematosus
- SNPs
Single nucleotide polymorphisms
- VD
Vitamin D
- VDR
Vitamin D receptor
- VDRs
Vitamin D receptors
- WHO
World Health Organization
Author contributions
HS wrote the paper. HS, PH, RY and JY participated in the design of the study. HS, QL and GW carried out the surveys and experiments. HS, FZ and LZ performed the statistical analysis.ML, LZ, JW and HF contributed materials and analysis tools. All authors read and approved the final manuscript.
Funding
Scientific research Fund of National Health Commission of the People’s republic of China (201502002)
Science and technology development fund project of Nanjing Medical University(2016NJMU056)
Availability of data and materials
The data used and analysis during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the research and ethical committee of Nanjing Medical University, (ethical review 2017(445), Nanjing, China), and written consent was acquired from the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hong Shen and Qinglan Liu contributed equally to this work.
Contributor Information
Rongbin Yu, Email: rongbinyu@njmu.edu.cn.
Jianrong Yang, Email: jrjskq@163.com.
References
- 1.Gupta S, Jawanda MK. Oral lichen planus:An update on etiology,pathogenesis,clinical presentation,diagnosis and management. Ind J Dermatol. 2015;60(3):222–229. doi: 10.4103/0019-5154.156315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van der Meij EH, Schepman KP, van der Waal I. The possible premalignant character of oral lichen planus and oral lichenoid lesions:a prospective study. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2003;96(2):164–171. doi: 10.1016/S1079-2104(03)00305-6. [DOI] [PubMed] [Google Scholar]
- 3.Ismail SB, Kumar SKS, Zain RB. Oral lichen planus and Lichenoid reactions; etiopathogenesis, diagnosis, management and malignant transformation. J Oral Sci. 2007;49(2):89–106. doi: 10.2334/josnusd.49.89. [DOI] [PubMed] [Google Scholar]
- 4.Payeras MR, Cherubini K, Figueiredo MA, Salum FG. Oral lichen planus:Focus on etiopathogenesis. Arch Oral Biol. 2013;58(9):1057–1069. doi: 10.1016/j.archoralbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Du J, Chen Y, Shi Y, Liu T, Cao Y, Tang Y, Ge X, Nie H, Zheng C, Li YC. 1,25-Dihydroxyvitamin D Protects Intestinal Epithelial Barrier by Regulating the Myosin Light Chain Kinase Signaling Pathway. Inflamm Bowel Dis. 2015;21(11):2495–2506. doi: 10.1097/MIB.0000000000000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J, Li R, Yu FY, Yang F, Wang JH, Chen QY, Wang XY, Zhao B, Zhang F. Experimental study on 1,25(OH)2 D3 amelioration of oral lichen planus through regulating NF--kB signaling pathway. Oral Dis. 2017;23:770–778. doi: 10.1111/odi.12659. [DOI] [PubMed] [Google Scholar]
- 7.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 8.Van der Meij EH, van der Waal I. Lack of clinicopathologic correlation in the diagnosis of oral lichen planus based on the presently available diagnostic criteria and suggestions for modifications. J Oral Pathol Med. 2003;32:507–512. doi: 10.1034/j.1600-0714.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 9.Kujundzic B, Zeljic K, Magic GSM, Stanimirovic D, Ilic V. Association of vdr, cyp27b1, cyp24a1 and mthfr gene polymorphisms with oral lichen planus risk. Clin Oral Invest. 2016;20:781–789. doi: 10.1007/s00784-015-1572-7. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Tregouet DA, Garelle V. A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics. 2007;23(8):1038–1039. doi: 10.1093/bioinformatics/btm058. [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick SG, Hirsch SA, Gordon SC. The malignant transformation of oral lichen planus and oral lichen oid lesions:a systematic review. J Am Dent Assoc. 2014;145(1):45–56. doi: 10.14219/jada.2013.10. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Yao H, Cui B, Ning G, Tang GY. Genetic linkage analysis of oral lichen planus in a Chinese family. Genet Mol Res. 2011;10(3):1427–1433. doi: 10.4238/vol10-3gmr1137. [DOI] [PubMed] [Google Scholar]
- 14.Georgakopoulou Eleni A., Achtari Marina D., Achtaris Michael, Foukas Periklis G., Kotsinas Athanassios. Oral Lichen Planus as a Preneoplastic Inflammatory Model. Journal of Biomedicine and Biotechnology. 2012;2012:1–8. doi: 10.1155/2012/759626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccaglini L, Thongprasom K, Carrozzo M, Bigby M. Urban legends series: lichen planus. Oral Dis. 2013;19(2):128–143. doi: 10.1111/j.1601-0825.2012.01953.x. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Wareth L, Haq A, Turner A, Khan S, Salem A, Mustafa F, Hussein N, Pallinalakam F, Grundy L, Patras G, Rajah J. Total vitamin D assay comparison of the Roche diagnostics “vitamin D total” electrochemiluminescence protein binding assay with the Chromsystems HPLC method in a population with both D2 and D3 forms of vitamin D. Nutrients. 2013;5:971–980. doi: 10.3390/nu5030971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salimi S, Farajian-Mashhadi F, Alavi-Naini R, Talebian G, Narooie-Nejad M. Association between vitamin D receptor polymorphisms and haplotypes with pulmonary tuberculosis. Biomed Rep. 2015;3:189–194. doi: 10.3892/br.2014.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao X, Zeng H, Zhang G, Zhou W, Yan Q, Dai L, Wang X. The associated ion between the VDR gene polymorphisms and susceptibility to hepatocellular carcinoma and the clinicopathological features in subjects infected with HBV. Biomed Res Int. 2013;953974. [DOI] [PMC free article] [PubMed]
- 19.Carvalho C, Marinho A, Leal B, Bettencourt A, Boleixa D, Almeida I, Farinha F, Costa PP, Vasconcelos C, Silva BM. Association between vitamin D receptor (VDR) gene polymorphisms and systemic lupus erythematosus in Portuguese patients. Lupus. 2015;24:846–853. doi: 10.1177/0961203314566636. [DOI] [PubMed] [Google Scholar]
- 20.Zhao B, Li R, Yang F, et al. LPS-induced vitamin D receptor decrease in oral keratinocytes is associated with oral lichen planus. Sci Rep. 2018;8:763. doi: 10.1038/s41598-018-19234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jingwi EY, Abbas M, Ricks-Santi L, Winchester D, Beyene D, Day A, Naab TJ, Kassim OO, Dunston GM, Copeland RL, Jr, Kanaan YM. Vitamin D receptor genetic polymorphisms are associated with PSA level, Gleason score and prostate cancer risk in African-American men. Anticancer Res. 2015;35(3):1549–1558. [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Guo L, Li H, Sun C, Cui X, Song G, Wang J, Zhang Q. Polymorphism rs2239185 in vitamin D receptor gene is associated with severe community-acquired pneumonia of children in Chinese Han population: a case-control study. Eur J Pediatr. 2015;174(5):621–629. doi: 10.1007/s00431-014-2433-0. [DOI] [PubMed] [Google Scholar]
- 23.Alves Pereira Neto T, Costa Pereira AA, Costa Hanemann JA, LFL C, LCC M. DC-SIGN and VDR polymorphisms are associated with chronic form of paracoccidioidomycosis with oral manifestations. Mycoses. 2019;62(2):186–192. doi: 10.1111/myc.12866. [DOI] [PubMed] [Google Scholar]
- 24.Wu M, Yue M, Huang P, Zhang Y, Xie C, Yu R, Li J, Wang J. Vitamin D level and vitamin D receptor genetic variations contribute to HCV infection susceptibility and chronicity in a Chinese population. Infect Genet Evol. 2016;41:146–152. doi: 10.1016/j.meegid.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Valdivielso JM, Fernandez E. Vitamin D receptor polymorphisms and diseases. Clin Chim Acta. 2006;371:1–12. doi: 10.1016/j.cca.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Papadopoulou A, Kouis P, Middleton N, Kolokotroni O, Karpathios T, Nicolaidou P, Yiallouros PK. Association of vitamin D receptorgene polymorphisms and vitamin D levels with asthma and atopy in Cypriot adolescents: a case-control study. Multidiscip Respir Med. 2015;10(1):26. doi: 10.1186/s40248-015-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thongprasom K, Mravak-Stipetić M, Luckprom P, et al. Oral lichen planus: a retrospective comparative study between Thai and Croatian patients. Acta Dermatovenerol Croat. 2009;17(1):2–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analysis during the current study are available from the corresponding author on reasonable request.