Version Changes
Revised. Amendments from Version 1
We have updated the manuscript based on reviewer comments. We have added information to the introduction about how the tools ClinEpiDB provides differ from data repositories. In the implementation section, we have added more details on data security, the types of study designs ClinEpiDB currently handles, data processing and ontology development, production of exploration applications, and the impact of date obfuscation on analysis. In the operation section, we have attempted to make it clearer why we support use of the tools in data exploration but encourage further analysis offline. While most reviewer comments and questions have been addressed directly in the manuscript, some, such as concerns over participant re-identification, have been addressed in responses to reviewers and through changes to the website. We thank the reviewers for their insightful questions and suggestions.
Abstract
The concept of open data has been gaining traction as a mechanism to increase data use, ensure that data are preserved over time, and accelerate discovery. While epidemiology data sets are increasingly deposited in databases and repositories, barriers to access still remain. ClinEpiDB was constructed as an open-access online resource for clinical and epidemiologic studies by leveraging the extensive web toolkit and infrastructure of the Eukaryotic Pathogen Database Resources (EuPathDB; a collection of databases covering 170+ eukaryotic pathogens, relevant related species, and select hosts) combined with a unified semantic web framework. Here we present an intuitive point-and-click website that allows users to visualize and subset data directly in the ClinEpiDB browser and immediately explore potential associations. Supporting study documentation aids contextualization, and data can be downloaded for advanced analyses. By facilitating access and interrogation of high-quality, large-scale data sets, ClinEpiDB aims to spur collaboration and discovery that improves global health.
Keywords: ClinEpiDB, Epidemiology database, FAIR data, Data visualization, Infectious diseases, Malaria, Enteric disease
Introduction
Large-scale epidemiological data sets offer immense potential for secondary data discovery and translational research provided the data are Findable, Accessible, Interoperable, and Reusable (FAIR) ( Wilkinson et al., 2016). Data repositories such as Dryad, dbGaP, and to a more limited extent ICPSR support the deposition of epidemiology data and metadata for download and secondary use by other researchers. A few recent studies such as Child Health and Mortality Prevention Surveillance (CHAMPS) have taken data sharing a step further and allow open access to aggregate data and online data visualization tools even as the study continues and the database is regularly updated with new data. The Clinical and Epidemiology Database (ClinEpiDB) resource was developed within this landscape as an open-access online tool to help investigators quickly and easily explore data from complex epidemiological studies and distinguishes itself from repositories and study-specific websites in two key ways: 1) ClinEpiDB maps data to common ontologies, creating a unified semantic framework that applies to all integrated studies, even those with different disease foci. 2) That framework underpins the website, where investigators are encouraged to explore data online through interactive tables, graphs, and an intuitive visual query interface, reducing the time and effort required to determine if specific data are available within one or multiple studies and worth further analysis. While some repositories have integrated tools like Survey Documentation and Analysis (SDA, Institute of Scientific Analysis) for online analysis, these tools may only be available for particular data sets and access to data may be restricted. A distinguishing feature of ClinEpiDB is that tools and visualizations are available for all studies, and aggregate data is generally publicly accessible.
For the initial prototype of ClinEpiDB, socioeconomic, demographic, clinical, and other data from the Program for Resistance, Immunology, Surveillance and Modeling of Malaria in Uganda (PRISM) ( Kamya et al., 2015), an International Center of Excellence for Malaria Research (ICEMR) ( Rao, 2015), was loaded into a relational database, leveraging infrastructure from EuPathDB (now VEuPathDB, reflecting a merger with VectorBase ( Giraldo-Calderón et al., 2015)), a collection of databases supporting multi-omics research on eukaryotic microbial pathogens, relevant non-pathogenic species, and selected hosts ( Aurrecoechea et al., 2017). Private release of the prototype to PRISM data providers prompted web tool optimization for settings with limited internet connectivity and led to rapid appreciation of the potential to facilitate data exploration by the full investigation team and raise study awareness. As a result, the PRISM study was publicly released in February 2018, even as primary publications on the data were still in preparation. The PRISM study was followed by release of ten additional studies ( Table 1), including the Global Enteric Multicenter Study (GEMS) ( Kotloff et al., 2013) and the Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health study (MAL-ED) ( Acosta et al., 2014). Additional releases containing data on malaria, enteric, respiratory, and other major global health priorities are scheduled for 2019–2020 and beyond.
Table 1. Studies publicly available via ClinEpiDB as of October 2019.
| Study abbreviation (reference) | Study design (time
frame) |
Research focus | Record types
(# records) |
Search types | Release date/
access level |
|---|---|---|---|---|---|
| PRISM (
Dorsey
et al., 2018;
Kamya
et al., 2015) |
Longitudinal cohort
(2011–2017) |
Incidence of acute malaria and parasite
prevalence at three sites in Uganda with differing exposure to mosquito vectors |
Household (331)
Participant (1421) Observation (48,722) Entomology (17,081) |
Household
Participant Observation Entomology |
Feb 2018/Public |
| GEMS (
Gates Enterics Project
et al.,
2018; Kotloff et al., 2013) |
Case-control with
60-day follow-up (2007–2011) |
Cause, incidence, and impact of moderate-to-
severe diarrhea in children from Bangladesh, the Gambia, India, Kenya, Mali, Mozambique, and Pakistan |
Household (43,573)
Participant (22,567) Observation (60,958) |
Participant | Dec 2018/Protected |
| GEMS1A (
Gates Enterics Project
et al., 2019b; Kotloff et al., 2019) |
Case-control with
60-day follow-up (2011–2013) |
Cause, incidence, and impact of less severe
diarrhea in children from Bangladesh, the Gambia, India, Kenya, Mali, Mozambique, and Pakistan |
Household (22,770)
Participant (14,242) Observation (36,009) |
Participant | Mar 2019/ Protected |
| India ICEMR longitudinal (
Carlton
et al., 2019a; Das et al., 2012) |
Longitudinal cohort
(2013–2015) |
Prevalence and incidence of malaria at two sites
in India with varied transmission settings |
Household (110)
Participant (397) Observation (1249) |
Household
Participant Observation |
Mar 2019/Public |
| MAL-ED (
Acosta
et al., 2014;
Spiro
et al.,
2019) |
Longitudinal cohort
(2009–2014) |
Etiology, risk factors and interactions of enteric
infections and malnutrition in children from Bangladesh, Brazil, India, Nepal, Pakistan, Peru, South Africa and Tanzania |
Household (12,233)
Participant (2145) Observation (1,384,323) |
Participant
Observation |
Mar 2019/Protected |
| GEMS1 HUAS/HUAS Lite (
Gates
Enterics Project et al., 2019a; Nasrin et al., 2013) |
Household survey
(2007–2010) |
Utilization of and attitudes towards healthcare
services. Survey conducted in conjunction with GEMS1 |
Household (133,659)
Participant (133,659) Observation (133,659) |
Participant | Apr 2019/Protected |
| GEMS1A HUAS Lite ( Gates Enterics Project et al., 2019c) | Household survey
(2010–2011) |
Utilization of and attitudes towards healthcare
services. Survey conducted in conjunction with GEMS1A |
Household (62,193)
Participant (62,193) Observation (62,193) |
Participant | Apr 2019/ Protected |
| India ICEMR cross-sectional
( Carlton et al., 2019b; van Eijk et al., 2016) |
Cross-sectional
survey (2012–2014) |
Prevalence of malaria at three sites in India with
varied transmission settings |
Household (1393)
Participant (3267) Observation (3442) |
Household
Participant Observation |
Apr 2019/ Public |
| India ICEMR fever surveillance
( Carlton et al., 2019c; Rao et al., 2019) |
Health center
surveillance (2016–2017) |
Etiology of acute febrile illness in patients without
malaria |
Participant (954)
Observation (962) |
Participant | Apr 2019/ Public |
| Amazonia ICEMR Peru
( Rosas-Aguirre et al., 2017; VinetzAmazonia ICEMR Peru et al., 2019) |
Longitudinal cohort
(2012–2015) |
Prevalence and incidence of malaria in disparate
transmission settings |
Household (487)
Participant (2445) Observation (2,050,603) |
Household
Participant Observation |
Jul 2019/ Protected |
| South Asia ICEMR (
Chery
et al.,
2016; Rathod et al., 2019) |
Health center
surveillance (2012–2017) |
Correlates of clinical malaria severity and parasite
phenotypes and genotypes |
Participant (1546)
Observation (4995) |
Participant | Jul 2019/ Protected |
[i] PRISM, Program for Resistance, Immunology, Surveillance and Modeling of Malaria; GEMS, Global Enteric Multicenter Study; HUAS, Healthcare Utilization and Attitudes Survey; ICEMR, International Centers of Excellence for Malaria Research; MAL-ED, Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health.
The resulting ClinEpiDB resource facilitates easy access and exploration of epidemiologic study design details and data for each study that is loaded. Study methodology, supporting documentation, and attribution are accessible through study pages. The ClinEpiDB user interface enables point-and-click interrogation of diverse data types where variables are displayed as interactive tables and histograms, allowing users to contextualize and identify subsets of data and visualize and analyze the results. For example, users can explore the impact of geographic location, mosquito exposure, and housing design on the frequency of acute malaria versus asymptomatic Plasmodium infection in the PRISM study. Entire data sets or filtered subsets of data can be downloaded for more advanced analyses. For data sets that require advanced security, ClinEpiDB offers a tiered data access system. All ClinEpiDB data sets released to date allow complete access to aggregate data and visualization tools, but some studies require that data access requests must be approved in order to view and download disaggregate data.
Methods
Ethical statement
The ClinEpiDB platform has received approval from the University of Pennsylvania under IRB#7, Protocol #828806. All studies included in ClinEpiDB have undergone ethical approval at applicable institutions prior to data collection (ClinEpiDB is generally not involved in this process). Data providers also obtain approval from their institutions to have their data hosted on ClinEpiDB. Community engagement programs have not yet been undertaken to assess study participants’ attitudes towards data sharing via the ClinEpiDB platform.
Implementation
ClinEpiDB integrates studies conducted by various primary research groups and can accommodate a variety of study designs including observational studies (surveillance, cross-sectional, longitudinal cohort, and case-control) and randomized control trials. Researchers supply flat data files along with data dictionaries, data collection forms, and protocols to help contextualize the data. Variables within the data set may contain categorical, continuous, discrete, or free text data.
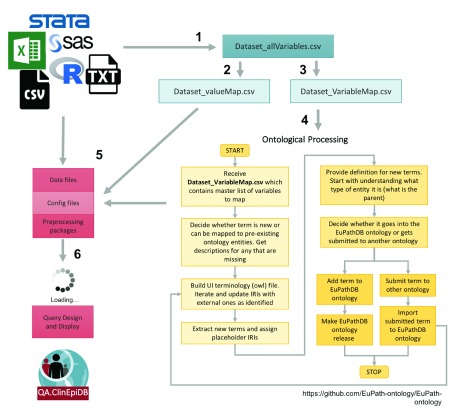
Once the data are received, a series of files are constructed according to a standard operating procedure to process the variables, map them to ontology terms, and map coded categorical values to the descriptive terms displayed on the website ( Figure 1). In rare cases where personally identifying variables – such as participant names and addresses – are included in the files received, those variables are removed before files are stored on a private file system only accessible to the data loading team to ensure participant confidentiality. All systems used to store and process the data are Federal Information Security Modernization Act (FISMA) compliant and undergo regular security review. Variables used solely for data cleaning purposes are also excluded. During initial processing, some standard checks are performed to ensure data are relatively clean, including ensuring that unique identifiers match across files, dropping variables with no data, identifying variables with unexpected values ( i.e. character values for an expected numeric variable), and identifying free-text variables that might benefit from standardization and/or translation. The study team is asked for clarification, revision, and input as needed. Creating ISA-based (Investigation, Study, Assay) files for loading often requires merging data files, and data conflicts can also be identified and fixed at that point.
Figure 1. Pipeline for processing studies.
(1) The ClinEpiDB team generates an “allVariables” file from the raw data files, data dictionaries, and data collection forms that contains all variables collected as part of the study and indicates whether each variable will be displayed on the website or not. This file is used to make (2) a “valueMap” file that maps coded categorical values to descriptive terms to be displayed on the website and (3) a “variableMap” file that maps variables to existing ontology terms and labels for display on the website. (4) The “variableMap” file is further processed by the ontology team and new ontology terms are created as needed. (5) All files are passed to the data loading team to pre-process the data, shift dates based on a random number algorithm, and create ISA files to load into the GUS4 database. (6) Once files are loaded, the data appear on an access-restricted website. Any additional searches required by a study are designed and implemented.
To deal with the challenges of integrating distinct studies with highly heterogeneous data while providing user-friendly mechanisms to identify similar variables, we employ an ontology-based approach to generate a unified semantic framework as described in Zheng et al. (2016). Wherever possible, variables are mapped to existing terms drawn from Open Biological and Biomedical Ontologies (OBO) Foundry, which supports interoperable ontologies ( Smith et al., 2007) through adhering to published principles. Necessary ontology terms are imported into the OBO Foundry registered VEuPathDB ontology, which is used as a single resource for all terms used in VEuPathDB resources. New ontology terms with VEuPathDB Internationalized Resource Identifiers (IRIs) are created as required. The use of ontologies to represent variables from different studies guides how data are loaded into relational databases and supports presentation of variables on the website to facilitate searching and analysis.
Once the data, ontology, and value mapping files are prepared, the data undergo processing to obfuscate dates to protect participant confidentiality. All dates for a given participant are consistently shifted forward or backward by 0–7 days according to a random number algorithm. While this obfuscation may introduce noise into longitudinal analysis—two events occurring on the same day for different participants will appear to have occurred 0–2 weeks apart—dates will still cluster within epidemiologically relevant timeframes for most analyses. All data are then transformed into an ISA-based format ( Sansone et al., 2012) and loaded into a relational database based on the Genomics Unified Schema, version 4 (GUS4) ( Davidson et al., 2001) running in an Oracle database management system (DBMS). Build database servers are located at the University of Pennsylvania and production instances are mirrored at the Universities of Pennsylvania and Georgia for redundancy purposes and to ensure uptime. All servers are housed in FISMA-compliant computational facilities, and industry standard backups of all data are performed.
Searches for each study are made available to users in an intuitive user interface (the “Search Wizard”), driven by a series of SQL queries against the GUS4 database (code available on GitHub, see Software availability). These searches vary depending on study design and record types. For example, in the longitudinal PRISM study, users can specifically retain or exclude observations occurring within a specified time relative to another observation through the “Related Observations” step in the Search Wizard (e.g. identify children diagnosed with febrile malaria at least twice within a six-month period). In the GEMS case-control study, users can compare cases to matching controls and choose whether to return data from the selected participants, matching cases/controls, or selected participants plus their matching cases/controls. Implementation of the strategies web development kit (WDK) ( Fischer et al., 2011) allows users to construct even more complex queries using logical operators (union, intersection, subtraction) and to save and share search strategies.
Exploration applications for additional data visualization are created with Shiny, an open-source R package for building interactive web applications ( RStudio Inc, 2019). The applications are hosted on the website via the Shiny Server Open Source software. SQL queries against the Oracle database identify all variables in the study and their format, which informs which variables appear as options to plot, how to build a custom dichotomous variable ( i.e. hemoglobin <=10 mg/dL vs. hemoglobin >10 mg/dL), and how the data are plotted within the applications.
Studies are reviewed by ClinEpiDB staff for quality control and made accessible to primary data providers using a protected internal website to ensure data accuracy and query functionality. Data are only scheduled for public release following data provider approval. Updates to the database are released every two months and can include new studies, features, and/or software updates.
Operation
ClinEpiDB can be accessed via any web browser at https://clinepidb.org. User support is available via the “Contact Us” link and tutorials are accessible via the “Community” drop-down menu at the top of each web page.
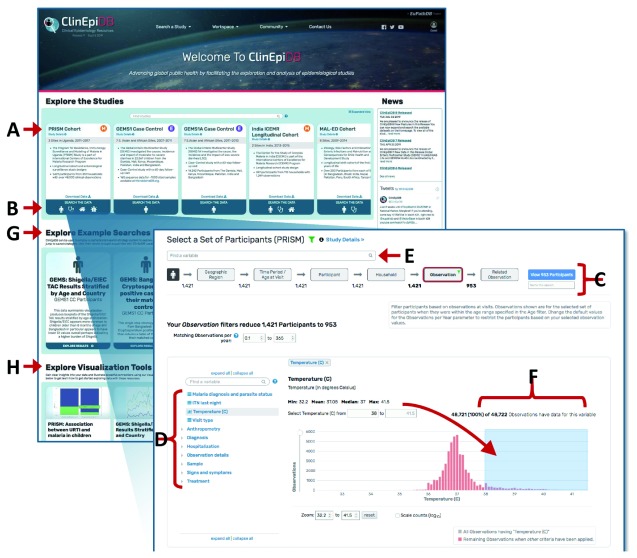
Study pages. Clicking a study name on a card on the ClinEpiDB homepage ( Figure 2A) or under the “Search a Study” drop-down menu brings up the study page, which provides a description of that particular study’s goals and objectives, methodology, investigators, and links to associated publications. This page also provides links to data collection forms and data dictionaries, which detail variable definitions, allowed values, skip patterns, etc. For studies that require permission to download data, the study page also incorporates a table listing individuals who have been granted access to the data and their brief stated purpose of use.
Figure 2. Using the Search Wizard to explore variables.
(A) Clicking a card study name opens a study page. (B) Clicking on a card search icon initiates a search. (C) The Search Wizard categorizes the variables into discrete steps. The grey buttons let users move between steps. (D) The variable tree contains all variables within that step of the Search Wizard. To subset the data, users can open a variable from this tree. (E) The “Find a variable” search bar searches for variables based on variable names and values across all Search Wizard steps. (F) Continuous data are displayed as a histogram and can be constrained by typing the exact range of values or clicking and dragging the mouse across the range of interest. (G) Clicking cards underneath “Explore Example Searches” opens up examples of searches conducted using the datasets indicated. These searches can be edited. (H) Clicking cards underneath “Explore Visualization Tools” opens up examples of how the exploration applications can be used.
Search strategies. ClinEpiDB permits users to execute searches on epidemiological data sets. Depending on available study data, up to four search types are currently supported that identify Households, Participants, Observations, or Entomology Collections of interest ( Table 2). For example, surveillance studies with just a single observation per participant offer only a participant search (e.g. “How many participants presented with both a fever and cough”). Studies with multiple participants from the same household will have household searches as well (e.g. “Which households contained children with asymptomatic parasitemia”). Longitudinal studies permit observation-level searches (e.g. “Identify all observations of children with malaria from houses with unscreened windows”). Within each search, users can subset the data based on any of the variables available through the “Search Wizard”, explore associations between variables, and return an interactive table of selected data.
Table 2. Search types currently available in ClinEpiDB.
| Search type | Default steps
available via the Search Wizard |
Results Table format | Results Table variables |
|---|---|---|---|
|
|
Household
Participant Observation |
One row per household observation
(multiple rows per household if household data was collected longitudinally) |
Household-level variables relating to geographic
location, dwelling characteristics, socioeconomic status, etc. |
|
|
Household
Participant Observation |
One row per participant | Participant-level variables relating to demographics,
enrollment, data summaries, etc. May also include upstream (household-level) variables. |
|
|
Household
Participant Observation |
One row per observation (multiple
rows per participant if data was collected longitudinally) |
Observation-level variables relating to
anthropometry, symptoms, laboratory test results, treatment, etc. May also include upstream (household- and participant-level) variables. |
|
|
Household
Entomology collection |
One row per entomology collection
(multiple rows per household if collections were done in multiple rooms or longitudinally) |
Entomology variables relating to mosquito
counts and species. May also include upstream (household-level) variables. |
From the home page, clicking a search icon on a study card initiates a search ( Figure 2B). The “Search Wizard” at the top of the page ( Figure 2C) categorizes the data, providing a step-wise approach to selecting data. On the left-hand side of the page, the variable tree presents all variables within that step of the Search Wizard ( Figure 2D), while the search bar at the top of the page allows users to search for variables across all Search Wizard steps ( Figure 2E). To subset data, users click on a variable of interest (e.g. “Temperature (C)”) and specify desired values. Continuous data are displayed as a histogram and can be selected by typing in a specific range or by clicking and dragging the cursor across the range of interest ( Figure 2F). Categorical data (e.g. “Malaria diagnosis and parasite status”) are displayed in a table and can be selected via the adjacent check boxes ( Figure 3A).
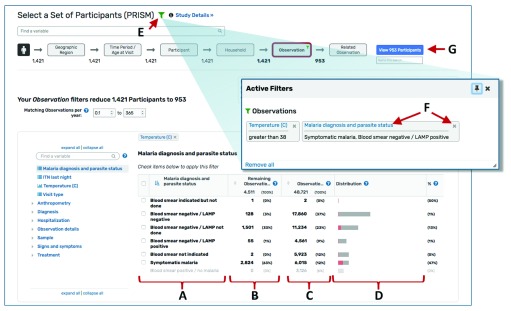
Figure 3. Adding, editing, and removing filters.
Categorical data are displayed in a table and (A) can be selected via check boxes next to the values. (B) The “Remaining” column indicates the data remaining given all other data selections (including selections in upstream steps), while the (C) Observations column indicates the total counts for all data. (D) For both continuous and categorical variables, data that meet the filter criteria (“remain”) are shown in red on the distribution graph while data that do not meet the filter criteria are shown in grey. (E) Clicking the green filter icon brings up a box that lists all applied filters. (F) Users can click the blue link to edit a filter or the “x” to remove it. (G) The blue button takes the user to the results page.
As data are selected, the data available for other variables in that Search Wizard step and any downstream steps are dynamically updated so the user can visualize the impact of their selection(s) on other variables. The “Remaining” column in the variable tables indicates the data remaining given all upstream filters ( Figure 3B), while the column to the immediate right indicates the total counts ( Figure 3C). For both continuous and categorical variables, data meeting upstream selection criteria are shown in red on the distribution graph while data excluded by the selection criteria are shown in grey ( Figure 3D). Selections can be reviewed, edited, or removed by clicking the green filter icon ( Figure 3E) and then clicking the blue link to edit selections for a variable or the “X” to remove it ( Figure 3F). Combined with data visualization through bar charts and histograms, this ability to conveniently add, edit, and remove filters makes it possible to rapidly assess the structure of the data and potential associations between variables of interest.
New users wanting to get a sense of what types of searches are possible can choose to view and edit publicly available searches under the “Explore Example Searches” section of the homepage ( Figure 2G).
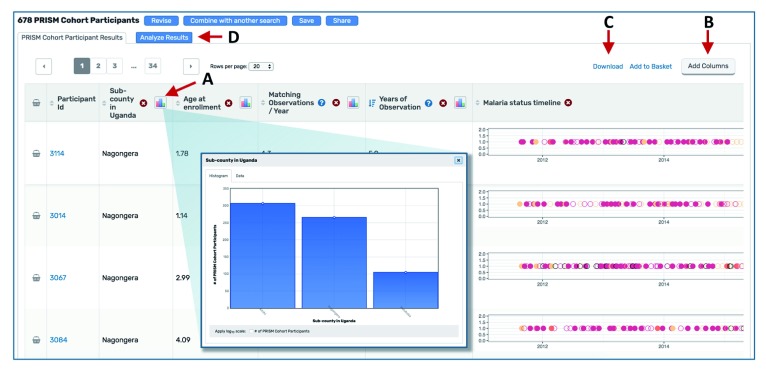
Results page and exploration apps. Data selected as described above are displayed on the Results Page ( Figure 4) when the user clicks the blue button at the right-hand terminus of the Search Wizard ( Figure 3G). The selected data are displayed as a table ( Figure 4) and may be passed to a suite of web applications for additional visualization and analysis. Variables available as columns are based on the type of search performed ( Table 2). Histogram icons in the column headers allow users to assess the distribution of the subset of data for that variable ( Figure 4A); links in the top right corner allow users to add additional variable columns ( Figure 4B) or download the selected data ( Figure 4C).
Figure 4. Using the Results Page.
(A) Clicking a histogram icon opens a pop up showing the distribution of data for that variable. (B) The “Add columns” button allows users to change which variables are shown in the table. (C) The “Download” link directs users to a page where they can choose which variables to download. The data subset is based on the selections applied in the Search Wizard. (D) The “Analyze Results” tab leads to a suite of applications for further data visualization.
The “Analyze Results” tab ( Figure 4D) leads to a suite of web applications. Three applications are currently available in ClinEpiDB: Distributions, Contingency Tables, and Data Summaries (also accessible under “Explore Visualization Tools” on the homepage; Figure 2H). The Distributions application shows the distribution of any variable in the data set and allows stratification based on other variables. The Contingency Table application generates a 2 × 2 contingency table for two selected variables and calculates a p-value, odds ratio, and relative risk, enabling assessment of associations (note that these statistics should be interpreted with caution as they do not control for confounding or other forms of bias). The Data Summaries application plots a variable of interest over time for longitudinal studies or two variables of interest against each other for non-longitudinal studies. For each app, users can toggle between tabs to define plot parameters, view summary statistics, display a plot grid or individual plots, and obtain help. Drop-down menus allow users to specify which variables to graph and whether to stratify data based on additional variables. Updating parameters automatically regenerates all statistics and plots. By default, the applications utilize the entire study data set, but users may choose to examine data selected in the Search Wizard by stratifying based on search results. The plots may be downloaded, but we encourage users to use the applications primarily for initial data exploration and to make their own graphs for presentations and publications after careful execution of a statistically robust, thorough data analysis, considering any potential biases and statistical assumptions.
Data downloads. Data in ClinEpiDB may be downloaded in two ways. Clicking the “Download” link on the Results Page ( Figure 4C) allows users to customize downloads, specifying which variables to retrieve based on the search type (see Table 2). All other variables can be downloaded, and data can be linked across files via observation, participant, and household IDs. Users may specify .txt or .csv formats, both of which can be consumed by most modern data analysis tools. Users can also download the entire data set via the “Download Data” link on the homepage study card and the study page. An ontology term association file links variables to their original study labels so users can reference study data collection forms and data dictionaries to learn more about each variable. Variables are also mapped to ontology terms via Internationalized Resource Identifiers (IRIs) which are included in each column header of the download file ( Ong et al., 2017). Following OBO Foundry principles, the terms are reused or requested from existing ontologies when possible but placeholder terms are also created as needed. Once defined, the terms are made public in the VEuPathDB application ontology along with imported terms from other ontologies and are searchable on Ontobee.
Accessibility of datasets
ClinEpiDB is committed to making epidemiologic data sets accessible to global research and biomedical communities while protecting the rights of study participants and data providers. Prior to viewing the website for the first time, users are required to agree to a Data Access and Use Policy outlining expectations regarding data use, protection of participant privacy, and acknowledgement of data providers and ClinEpiDB.
Some studies require data access restrictions at the data provider’s discretion. There are five access levels data providers can choose from that differ in their requirements for users to view aggregate versus disaggregate data ( Table 3). Aggregate data are accessible in the Search Wizard and through the exploration applications, while disaggregate data can be found on the results page, individual record pages, and in the download files. Except for studies classified as private, which require approval to see any data, users can see variables and aggregate data for all studies. Note that on occasion, variables that are potential indirect identifiers are loaded into the database but not displayed on the website. These variables are available for download to individuals with access approval.
Table 3. Data access restriction levels.
| Access
level |
Description |
|---|---|
| Public | No access restrictions. Users can view and download all data as a “Guest” without logging in. |
| Controlled | Users can view data in the Search Wizard, in exploration applications, and view the results pages
and recordpages as a “Guest” without logging in, but must obtain approval from the data providers to download data. |
| Limited | Users can view data in the Search Wizard and exploration applications as a “Guest” without logging
in, but must log in with a registered account to view more than 20 rows of data on the results page or view individual record pages. Users must obtain approval from the data providers to download data. |
| Protected | Users can view data in the Search Wizard and exploration applications as a “Guest” without logging
in, but must obtain approval from the data providers to view more than 20 rows of data on the results page, view individual record pages, or download data. |
| Private | Users must request and obtain approval to access any aspect of the data. |
When a user reaches a restricted section of the website, they are automatically prompted to either log in with a ClinEpiDB account or log in and submit a data access request, depending on the access restrictions. The data access request form requires the purpose for which the requested data will be used, whether the requester has been in contact with the study team, hypotheses and/or research questions, analysis plan, and planned dissemination of results. The request is then sent to data providers for approval. Users are contacted within a few days with any conflicts that are identified or with notification of approval. Once approved, they may view and download that study’s data at any point by logging into their ClinEpiDB account. To ensure transparency and promote collaboration within the wider scientific community, the requestor’s name, organization, request date, and indicated purpose appear publicly on the corresponding study page once approved.
Use cases
ClinEpiDB provides a powerful web-based platform that enables the research community to easily access and explore clinical epidemiological data for primary and secondary use via an intuitive point-and-click interface, maximizing potential for generating new, data-driven hypotheses and promoting collaborations between researchers.
Two examples focusing on the PRISM data ( Dorsey et al., 2018), the first study released on ClinEpiDB, illustrate how the website can be used by potential collaborators looking for samples and analysts looking for data to inform modeling. In the first instance, a collaborator interested in accessing and analyzing peripheral blood mononuclear cell (PBMC) samples from timepoints close to when a participant was diagnosed with malaria was able to identify the appropriate samples themselves using ClinEpiDB and begin generating preliminary data. By initiating an observation search and setting “Sample type” to “PBMC”, they were able to determine that 5295 PBMC samples were collected during the study. Next, by going to the “Related Observation” step in the Search Wizard, opting to “ Keep Observations within 0–10 days after the Related Observation specified below” and selecting data where “Malaria diagnosis and parasite status” was “Symptomatic malaria”, they were able to identify 130 PBMC samples collected within 10 days of a malaria diagnosis (see saved strategy). In a second example, a student was able to examine the data using ClinEpiDB to determine a difference in the percent of malaria-attributable fever based on whether fever was self-reported or measured. By running an observation search and limiting “Temperature (C)” to greater than or equal to 38 then looking at where “Asexual Plasmodium parasites present, by microscopy” was positive, they found that 2824 of 4508 observations of measured fever (62.6%) could be attributed to malaria. In contrast, looking at observations where “Subjective fever” was reported and where “Asexual Plasmodium parasites present, by microscopy” was positive revealed that 6006 of 15,228 observations (39.4%) of self-reported fever could be attributed to malaria. They planned to use those statistics to adjust a model that uses data on self-reported fever.
An additional hypothetical example highlights how users might explore data in ClinEpiDB before deciding to submit a data access request to download the data for further analysis. A user might be interested in re-analyzing risk factors for rotavirus infection and disease in children based on new molecular diagnostics testing for enteropathogens in MAL-ED stool samples ( Platts-Mills et al., 2018; Spiro et al., 2019) instead of ELISAs, as done previously ( Mohan et al., 2017). To quickly determine if secondary analysis is worthwhile, the user would perform an Observation-level search of MAL-ED, choosing the Observation step from the Search Wizard, and selecting the entire range of Cycle threshold (Ct) values under “Rotavirus Ct value, by TAC result” to limit analysis to samples that underwent TAC testing for rotavirus. Setting “Stool type” to “Diarrhea” reveals that 6745 diarrheal stool samples were tested for rotavirus using TAC. By navigating back to “Rotavirus CT value, by TAC result” and setting the range of Ct values to “<31.7” (the TAC cut-off for rotavirus defined in Platts-Mills et al. (2018) and then returning to “Stool type”, the user would observe that 568 (8.4%) of 6745 diarrheal stool samples were positive for rotavirus using TAC (see saved strategy). Substituting “Rotavirus, by ELISA” for “Rotavirus CT value, by TAC result,” the user would then discover that 535 (5.7%) of 9301 diarrheal stool samples were positive for rotavirus by ELISA, consistent with the report by Mohan et al. (2017) (see saved strategy). Such study exploration enables rapid evaluation of whether or not a robust statistical reanalysis using the more sensitive molecular diagnostic data would be feasible.
Conclusions
Journals and funders increasingly require that data be made publicly available ( National Institutes of Health, 2003; The Wellcome Trust, 2011), but data hidden in supplementary data files or stored in data repositories are often difficult to locate, interpret, or use by those not actively engaged in the study. ClinEpiDB strives to follow FAIR Guiding Principles ( Wilkinson et al., 2016) by creating resources, tools, vocabularies, and infrastructure that supports third-party discovery and reuse of primary epidemiological research data. Studies loaded into ClinEpiDB are provided with stable, unique identifiers, making them “Findable.” An intuitive interface and visualization tools allow users to see and directly query the data, lowering the barrier for exploratory data analysis. While these tools are not a substitute for rigorous, controlled statistical analyses, data can be downloaded in common machine-readable formats for robust analysis, making it more “Accessible.” The implementation of standardized, publicly available ontologies makes the data more “Interoperable.” Even when similar variables in different studies map to distinct ontology terms, the display labels, definitions, and position of the variable in the variable tree provide useful information that allow users to generate similar queries for different studies. Study pages are always public and provide context that makes the data more “Reusable.”
As ClinEpiDB continues to be developed, users can expect to see the release of additional studies focusing on malaria, enteric disease, respiratory disease, and more. Additional long-term development plans include strengthening and expanding data visualization and exploration tools. Epidemiologic data loaded into ClinEpiDB is currently separate from genomic data available via other EuPathDB resources such as PlasmoDB ( Bahl et al., 2003) or MicrobiomeDB ( Oliveira et al., 2018), but the use of common infrastructure creates the possibility of queries across currently disparate resources, facilitating additional secondary data use.
In summary, the ClinEpiDB platform promotes access and interrogation of complex epidemiological studies loaded in the database through a user interface that enables visualization of and interaction with all data within a study. Regular release of additional studies along with new features is expected to further support secondary data use. Similar to what has been achieved through the EuPathDB websites, production of ClinEpiDB will help maximize the impact of the epidemiology studies that are loaded and abbreviate time to discovery while stimulating productive collaborations between research groups.
Data availability
All data underlying the results are available as part of the article and no additional source data are required.
Software availability
Infrastructure description of repositories available from: https://eupathdb.org/eupathdb/wdkCustomization/jsp/questions/XmlQuestions.Infrastructure.jsp
Source code available from: https://github.com/VEuPathDB
Archived source code at the time of publication: https://doi.org/10.5281/zenodo.3522209
License: GNU Library General Public License v2
All GitHub repositories are publicly available except for ClinEpiPresenters, since this repository may contain information on studies that are not yet ready for release. The archived source code includes a version of this repository where information has been redacted for studies that have not yet been released.
Acknowledgements
We thank the study participants and investigators for their important contributions. We thank the data providers for their time and dedication, which aided public release of data on ClinEpiDB, and we thank the EuPathDB team for their work on extending the architecture and developing the framework that allowed this project to proceed.
Funding Statement
This work was supported by the Bill and Melinda Gates Foundation [OPP1169785 to DSR]; and the National Institutes of Health (NIH) [HHSN272201400030C to DSR and JCK; U19AI089674 to GD].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- Acosta AM, Chavez CB, Flores JT, et al. : The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis. 2014;59(suppl 4):S193–S206. 10.1093/cid/ciu653 [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Barreto A, Basenko EY, et al. : EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017;45(D1):D581–D591. 10.1093/nar/gkw1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl A, Brunk B, Crabtree J, et al. : PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31(1):212–215. 10.1093/nar/gkg081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Eapen A, Sharma S, et al. : Data Set: India ICEMR Longitudinal Cohort. ClinEpiDB. 2019a. Reference Source [Google Scholar]
- Carlton JM, Eapen A, Sharma S, et al. : Data Set: India ICEMR Cross-Sectional. ClinEpiDB. 2019b. Reference Source [Google Scholar]
- Carlton JM, Mohanty S, Satpathi S: Data Set: India ICEMR Fever Surveillance. ClinEpiDB. 2019c. Reference Source [Google Scholar]
- Chery L, Maki JN, Mascarenhas A, et al. : Demographic and clinical profiles of Plasmodium falciparum and Plasmodium vivax patients at a tertiary care centre in southwestern India. Malar J. 2016;15(1):569. 10.1186/s12936-016-1619-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Anvikar AR, Cator LJ, et al. : Malaria in India: the center for the study of complex malaria in India. Acta Trop. 2012;121(3):267–273. 10.1016/j.actatropica.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SB, Crabtree J, Brunk BP, et al. : K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst J. 2001;40(2):512–531. 10.1147/sj.402.0512 [DOI] [Google Scholar]
- Dorsey G, Kamya M, Greenhouse B, et al. : Data Set: PRISM Cohort. ClinEpiDB. 2018. Reference Source [Google Scholar]
- Fischer S, Aurrecoechea C, Brunk BP, et al. : The Strategies WDK: a graphical search interface and web development kit for functional genomics databases. Database (Oxford). 2011;2011:bar027. 10.1093/database/bar027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates Enterics Project, Levine MM, Kotloff K, et al. : Data Set: GEMS1 Case Control. ClinEpiDB. 2018. Reference Source [Google Scholar]
- Gates Enterics Project, Levine MM, Kotloff K, et al. : Data Set: GEMS1 HUAS/HUAS Lite. ClinEpiDB. 2019a. Reference Source [Google Scholar]
- Gates Enterics Project, Levine MM, Kotloff K, et al. : Data Set: GEMS1A Case Control. ClinEpiDB. 2019b. Reference Source [Google Scholar]
- Gates Enterics Project, Levine MM, Kotloff K, et al. : Data Set: GEMS1A HUAS Lite. ClinEpiDB. 2019c. Reference Source [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, et al. : VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43(Database issue):D707–13. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamya MR, Arinaitwe E, Wanzira H, et al. : Malaria transmission, infection, and disease at three sites with varied transmission intensity in Uganda: implications for malaria control. Am J Trop Med Hyg. 2015;92(5):903–912. 10.4269/ajtmh.14-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nasrin D, Blackwelder WC, et al. : The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Glob Health. 2019;7(5):e568–e584. 10.1016/S2214-109X(19)30076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, et al. : Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- Mohan VR, Karthikeyan R, Babji S, et al. : Rotavirus Infection and Disease in a Multisite Birth Cohort: Results From the MAL-ED Study. J Infect Dis. 2017;216(3):305–316. 10.1093/infdis/jix199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin D, Wu Y, Blackwelder WC, et al. : Health care seeking for childhood diarrhea in developing countries: evidence from seven sites in Africa and Asia. Am J Trop Med Hyg. 2013;89(1 Suppl):3–12. 10.4269/ajtmh.12-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health: Final NIH statement on sharing research data. 2003. Reference Source [Google Scholar]
- Oliveira FS, Brestelli J, Cade S, et al. : MicrobiomeDB: a systems biology platform for integrating, mining and analyzing microbiome experiments. Nucleic Acids Res. 2018;46(D1):D684–D691. 10.1093/nar/gkx1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E, Xiang Z, Zhao B, et al. : Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Res. 2017;45(D1):D347–D352. 10.1093/nar/gkw918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts-Mills JA, Liu J, Rogawski ET, et al. : Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Health. 2018;6(12):e1309–e1318. 10.1016/S2214-109X(18)30349-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MR: Foreword: International Centers of Excellence for Malaria Research. Am J Trop Med Hyg. 2015;93(3 Suppl):1–4. 10.4269/ajtmh.15-0407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PN, van Eijk AM, Choubey S, et al. : Dengue, chikungunya, and scrub typhus are important etiologies of non-malarial febrile illness in Rourkela, Odisha, India. BMC Infect Dis. 2019;19(1):572. 10.1186/s12879-019-4161-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod PK, Kakati S, Gomes E, et al. : Data Set: South Asia ICEMR Surveillance. ClinEpiDB. 2019. Reference Source [Google Scholar]
- Rosas-Aguirre A, Guzman-Guzman M, Gamboa D, et al. : Micro-heterogeneity of malaria transmission in the Peruvian Amazon: a baseline assessment underlying a population-based cohort study. Malar J. 2017;16(1):312. 10.1186/s12936-017-1957-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Inc: shiny: web application framework for R. Retrieved May 6, 2019,2019. Reference Source [Google Scholar]
- Sansone SA, Rocca-Serra P, Field D, et al. : Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. 10.1038/ng.1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, et al. : The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. 10.1038/nbt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro D, Gottlieb M, Glass R, et al. : Data Set: MAL-ED Cohort. ClinEpiDB. 2019. Reference Source [Google Scholar]
- The Wellcome Trust: Sharing research data to improve public health: full joint statement by funders of health research. 2011. Reference Source [Google Scholar]
- van Eijk AM, Ramanathapuram L, Sutton PL, et al. : The use of mosquito repellents at three sites in India with declining malaria transmission: surveys in the community and clinic. Parasit Vectors. 2016;9(1):418. 10.1186/s13071-016-1709-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinetz JM, Llanos-Cuentas A, Chuquiyauri R, et al. : Data Set: Amazonia ICEMR Peru Cohort. ClinEpiDB. 2019. Reference Source [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. : The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Cade JS, Brunk B, et al. : Malaria study data integration and information retrieval based on OBO Foundry ontologies. CEUR Workshop Proceedings. 2016;1747 Reference Source [Google Scholar]