Abstract
Background.
Acinetobacter baumannii is a gram-negative, opportunistic pathogen. Its ability to form biofilm and increasing resistance to antibiotics present challenges for infection control. A better understanding of the impact of biofilm formation and antibiotic resistance on environmental persistence of A. baumannii in hospital settings is needed for more effective infection control.
Methods.
A. baumannii strains isolated from patients and the hospital environment were identified via MALDI-TOF mass spectrometry, rep-PCR genotyped, and antibiotic resistance was determined using Vitek®2. Biofilm mass was quantified via microtiter plate method and desiccation tolerance determined up to 56 days.
Results.
High biofilm forming, clinical, MDR positive strains were 50% less likely to die of desiccation than low biofilm, non-MDR strains. In contrast, environmental, MDR positive, low biofilm forming strains had a 2.7 times increase in risk of cell death due to desiccation compared to their MDR negative counterparts. MDR negative, high biofilm forming environmental strains had a 60% decrease in risk compared to their low biofilm forming counterparts.
Conclusions.
The MDR positive phenotype was deleterious for environmental strains and the high biofilm phenotype was critical for survival. This study provides evidence of the trade-off between antibiotic resistance and desiccation tolerance, driven by condition-dependent adaptation, and establishes rationale for research into the genetic basis of the variation in fitness cost between clinical and environmental isolates.
Keywords: Acinetobacter baumannii, desiccation tolerance, environmental survival, biofilm, multidrug resistance, hospital, environment
Introduction
Acinetobacter baumannii, a gram-negative bacterium, is an opportunistic pathogen capable of living in multiple environments that is an increasing problem in hospital settings. Once infection is established, risk of mortality is high: up to 26% for in-hospital patients (1) and up to 43% for intensive care unit (ICU) patients (2). Multidrug resistant (MDR) A. baumannii strains are increasingly reported worldwide (1), (3), and A. baumannii express several mechanisms which confer this resistance (4). It is also challenging to control, as A. baumannii can survive in the hospital environment for prolonged periods of time (5) and environmental contamination has been linked to hospital outbreaks (6).
The capacity of A. baumannii to persist in the environment may be due to its ability to form biofilms on both abiotic and biological surfaces (7), (8). Biofilm formation is also a mechanism of pathogenesis in device-related infections and provides a source of repeated transmission by prolonging survival on inanimate objects (9), (10). Under harsh environmental conditions, A. baumannii cells deep in the biofilm can undergo dormancy, becoming metabolically inactive and robust to environmental stress (11), (12). The multiple antibiotic resistance mechanisms found in A. baumannii also may play a role in its environmental survival. Boll, et al. demonstrated that resistance to cationic antimicrobial peptide drugs, such as colistin, also increases A. baumannii tolerance to desiccation (13). Gayoso et al. demonstrated that some antibiotic resistant associated proteins--which are also associated with increased tolerance to detergents--were overexpressed in A. baumannii under desiccation-stress (12). These findings suggest that some drug resistance mechanisms leverage the cells ability to survive in the open environment.
Antibiotic resistance in bacteria generally incurs a fitness cost, often manifested as reduced growth rates or a compromised competitive ability (14). Environmental pressures have been implicated for variations in phenotypic expressions (15). The trade-offs in fitness imposed by the ability to form biofilms, tolerate desiccation and multidrug resistance are potentially different for A. baumannii strains that live primarily in the environment compared to clinical strains adapted to living in the human host. To test whether these trade-offs occur, we compared the biofilm formation, antibiotic susceptibility profiles and desiccation tolerance in a collection of A. baumannii strains isolated from patients and the hospital environment. Our results suggest that a trade-off between antibiotic resistance and desiccation tolerance occurs, and biofilm formation contributes significantly to the survival of these isolates.
Materials and Methods
Collection of A. baumannii isolates:
The study protocol was approved by the University of Michigan Institutional Review Board (HUM00075484). We collected 132 clinical isolates from 115 patients and 54 environmental isolates from the University of Michigan Hospital (City, State) between August 2012 and January 2014. Only the first isolate obtained from any patient was included. For environmental isolates that were collected on the same day, from the same fomite, sharing the same rep-type banding pattern, only one was randomly chosen for inclusion; out of the 54 isolates collected, 30 environmental isolates met these criteria.
Clinical isolates were obtained from patients presenting with Acinetobacter baumannii infection, and were cultured and identified using MALDI-TOF mass spectrometer (Bruker Daltonics, Bellerica, MA) by the hospital microbiology laboratory. Environmental isolates were obtained by swabbing nonporous, high and low touch areas within and outside of the infected patient’s room using CultureSwab™ (Becton, Dickinson and Co., Sparks, MD) swabs previously moistened in brain-heart infusion broth (Becton, Dickinson & Co., Sparks, MD). Sampling was performed in accordance with Healthcare Infection Control Practices Advisory Committee (HICPAC) recommendations for environmental surface sampling (16). Bacteria were recovered by homogenizing the swab with corresponding liquid using Omni-Tip™ disposable rotor stator generator probes (OMNI International, Kennesaw, GA) for 30 sec to remove cells from the swab. The homogenate was incubated at 37°C for 18 h on a rotating shaker table (150–180 rpm) during which time a 1 mL aliquot was removed at 2 hours and 18 hours, serial diluted to 10−3, plated onto CHROMagar™ plates and grown for 24 hours at 37°C. Suspected Acinetobacter colonies were subcultured onto blood agar plates (Thermal Fisher Scientific, Waltham, MA) and sent to the University of Michigan Hospital Microbiology lab for VITEK®2 and MALDI-TOF identification.
Preparation of initial inoculums:
For each experiment, initial inoculums were prepared by transferring a frozen aliquot into 2.5 mL of MHII broth and incubating at 37°C for 18±2 h on a rotating shaker table (150–180 rpm). The culture was inoculated onto BBL™ MHII agar (Becton, Dickinson and Co., Sparks, MD) and grown at 37°C. An isolated colony was transferred to MHII broth and incubated at 37°C with shaking at 150–180 rpm for 15–18 hours. From this, a starting culture with an optical density at a wavelength of 600 nm (OD600) of 0.200 ± 0.01 (Synergy™ HT Multi-Mode Microplate Reader, BioTek® Instruments, Inc.), which approximates 108 CFU/mL, was used.
Repetitive extragenic palindromic polymerase chain reaction (rep-PCR) genotyping:
Genomic DNA was extracted using a commercially available whole genome extraction kit (QIAamp® DNA Mini Kit by Qiagen, Valencia, CA). DNA quantification was performed by nanodrop (NS-1000, Thermo Scientific, Wilmington, DE) and DNA purity was evaluated by the absorbance ratio at 260 and 280 nm (A260/A280). Rep-PCR oligonucleotide primer sets published by Vila et al., (17) were prepared by Invitrogen (Carlsbad, CA). Rep-PCR conditions were followed as previously described (15), (18). Amplified products were stained using EZ-Vision® Three, DNA Dye and Buffer 6X (AMRESCO®, Solon, OH) and aliquots (10μL) of each sample were subjected to elecrophoresis in 1.2% agarose gel. A 1 Kb GeneRuler DNA Ladder Mix (#SM0333, Thermo Scientific, City, ST) was used in the first and every 4th lane of the gel. Gels were imaged using a UV trans-illuminator and imager. BioNumerics® Version 7.5 Software (Applied Maths, Inc., Austin, TX) was used to detect lanes and bands and to build phylogenetic trees using the Neighbor Joining clustering method with the Jaccard similarity coefficient (19).
Antibiotic susceptibility testing:
Antibiotic susceptibility profiles were determined using VITEK®2 (bioMérieux, Inc., Durham NC). Isolates were considered to be multidrug resistant (MDR) if they were resistant to the following three drug classes, or resistant to two and intermediate to one (20): cephalosporins, fluoroquinolones, and aminoglycosides. This definition is based on the CDC definition of MDR; bacteria resistant to at least one class of antimicrobial agents and usually resistant to all but one or two antimicrobial agents are MDROs (21). The antimicrobial agents chosen for this definition were taken from Manchada, et al (2010) who use these drug classes to define MDR Acinetobacter spp.’ in consideration of known resistance mechanisms and currently used treatments for Acinetobacter spp. infections (20).
Quantification of biofilm formation:
Each isolate was grown as described above and diluted 1:100 in MHII broth. Biofilm forming capacity was quantified in triplicate using the microtiter plate method as previously described (22), (23) using 0.1% crystal violet Gram stain solution (CAS no. 10114-58-6, Fisher Science Education, Nazareth, PA). The OD600 was measured using a microplate reader to obtain relative biofilm biomass measurements. The average of each triplicate was calculated and A. baumannii ATCC 17978 (American Type Culture Collection, Manassas, VA) was used to normalize data.
Environmental survivability:
Environmental survivability is defined as desiccation tolerance over time and was tested over a period of 56 days as described previously (24). Each isolate was grown as described above, subcultured onto MHII agar and grown at 37°C overnight. An isolated colony was transferred to 2 mL MHII broth and incubated at 37°C with shaking at 150–180 rpm for 15–18 hours. From this, a 1 mL aliquot, with an OD600 of 0.200 ± 0.01, which approximates 108 CFU/mL, was used. Cells from each culture were pelleted using a mini centrifuge (5415R, Eppendorf, Hauppauge, NY) at 8600 × g for 5 min and the pellet was re-suspended in 1 mL of 1× PBS buffer. This was repeated twice to thoroughly wash the cells. Samples were then serially diluted in 1× PBS buffer to obtain an OD600 of 0.001, which approximates 106 CFU/mL. In duplicate, 10 μl of each culture was inoculated into the wells of a 96-well plate, preparing one for each time period of 1, 3, 7, 14, 28, and 56 days. The plates were covered with a semi-permeable membrane and incubated at approximately 72°F (22°C) with a relative humidity level of approximately 40% for the designated time period. At each time interval, the cells were revived by adding 100 uL of MHII broth to wells with gentle pipetting up and down. Samples were serial diluted 1000 fold, spread plated onto MHII agar and incubated overnight at 37°C for colony enumeration.
Statistical Analysis:
Cox proportional hazards regression analysis was performed using R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria). All other statistical analyses were performed using GraphPad Prism 6 for Windows (Version 6.01, Graph Pad Software, Inc., La Jolla, CA). Statistical significance was assessed using the paired and unpaired t tests (as appropriate), the Holm-Šídák test and one-way ANOVA, Log-rank and log-likelihood ratio with a significance level of α ≤ 0.05.
Results
Epidemiology/Isolate collection: We obtained 132 clinical isolates from 115 different patients over an 18-month period. Only the first isolate obtained from any one patient was included. Clinical isolates were equally likely to be obtained from ICU, non-ICU and outpatient locations (Table 1). A. baumannii was successfully recovered from 54 of the 314 environmental samples for an overall recovery rate of 17%. Of these 54 isolates, 30 independent isolates were included (see methods). Eighty percent (24/30) of the environmental isolates were collected from patient rooms and the remaining 20% (6/30) were obtained from non-patient areas such as nurses’ stations and medical supply areas. Clinical isolates were recovered primarily from urinary tract and respiratory tract specimens. Environmental isolates were mostly recovered from sink areas. Although some of the environmental isolates were collected from the room of an infected patient, only 10% (3/30) of the environmental isolates shared the exact same REP-type as the corresponding patient occupying that room.
Table 1:
Study isolate characteristics stratified by rep-type of 115 clinical and 30 environmental Acinetobacter baumannii isolates obtained from a University hospital between Aug 2012 and Jan 2014.
| Clinical | Environmental | Population | |||
|---|---|---|---|---|---|
| Dominant† | Sporadic‡ | Dominant† | Sporadic‡ | Total | |
| n (%) | n (%) | n (%) | n (%) | n (%) | |
| Total (n) | 52 (100) | 63 (100) | 21 (100) | 9 (100) | 145 (100) |
| Patient characteristics | |||||
| Mean Age (years) | 52.7 | 41.5 | n/a | n/a | 47.1 |
| Males | 24 (46) | 38 (60) | n/a | n/a | 62 (54) |
| Females | 28 (53) | 25 (40) | n/a | n/a | 53 (46) |
| Hospital Location | |||||
| ICU Unit | 24 (46) | 4 (7) | 17 (81) | 4 (44) | 49 (34) |
| Non-ICU Unit | 11 (21) | 26 (41) | 4 (19) | 5 (56) | 46 (32) |
| Outpatient | 17 (33) | 33 (52) | n/a | n/a | 50 (34) |
| Site of Isolation | |||||
| Urinary | 19 (36) | 29 (46) | n/a | n/a | 48 (33) |
| Respiratory | 17 (33) | 17 (27) | n/a | n/a | 34 (23) |
| Soft Tissue | 10 (19) | 9 (14) | n/a | n/a | 19 (13) |
| Blood | 4 (8) | 3 (5) | n/a | n/a | 7 (5) |
| *Other Body Site | 2 (4) | 5 (8) | n/a | n/a | 7 (5) |
| Keypad | n/a | n/a | 4 (19) | 2 (22) | 6 (4) |
| Sink Area | n/a | n/a | 7 (33) | 3 (33) | 10 (7) |
| Floor | n/a | n/a | 4 (19) | 1 (11) | 5 (3) |
| Computer Area | n/a | n/a | 2 (9.5) | 1 (11) | 3 (2) |
| Bed Rail | n/a | n/a | 2 (9.5) | 0 (0) | 2 (1) |
| **Other Fomite | n/a | n/a | 2 (9.5) | 2 (22) | 4 (3) |
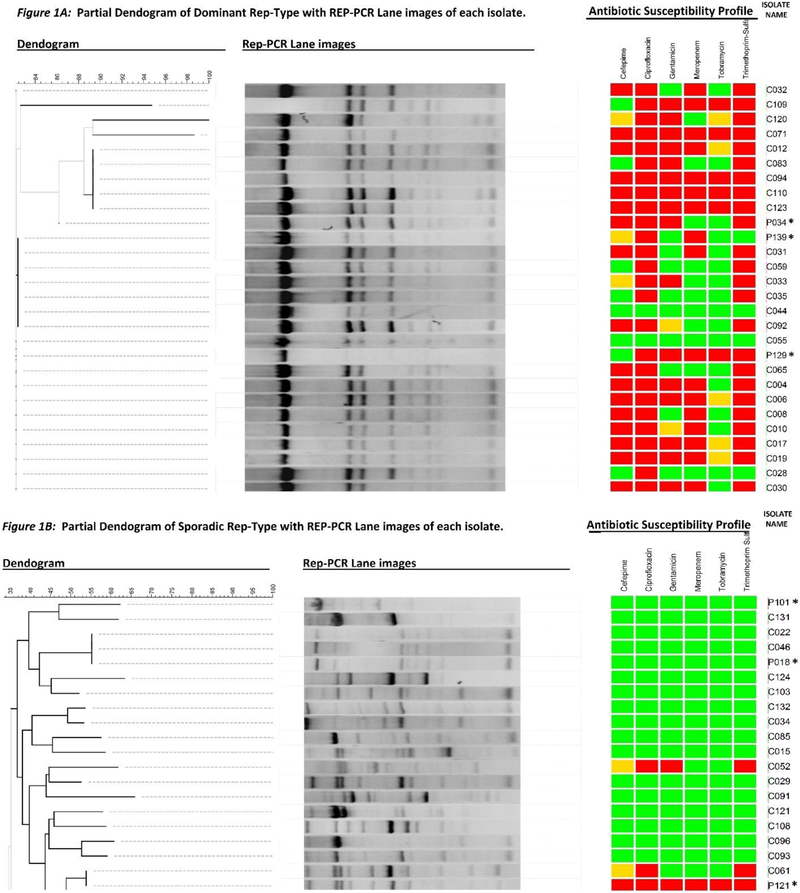
Dominant rep-type strains are those with a common banding pattern (Figs 1A).
Sporadic rep-type strains are those with banding patterns highly dissimilar from the dominant type (Figs 1B).
Other body sites include CSF, drainage, para fluid, and bone.
Other fomites include hallway ledge, phone, bed-side table, and counter top.
Rep-PCR genotyping:
We identified a common banding pattern that had 8 distinct bands, with a prominent band at approximately 4000 base-pairs. Of the145 isolates, 64 displayed the common banding pattern exactly and 9 differed only by 1 band; we will refer to these 73 strains as the “dominant rep-type” for the remainder of this paper (Figure 1A). The remaining 72 isolates displayed banding patterns highly dissimilar from the dominant type (Figure 1B). We will refer to this set of strains as “sporadic rep-type”. We identified 59 unique banding patterns among the sporadic rep-types. The distribution of dominant and sporadic rep-types varied by isolate source. Urinary isolates were predominantly of the sporadic rep-type (29/48, 60%) but half of respiratory isolates were comprised of the dominant-type. One-third of the environmental isolates (10/30) were isolated from the sink area and these were predominantly of the dominant type (7/10, 70%). Full dendograms of the dominant and sporadic rep-types are available upon request from the authors.
Figure 1:
Dendogram of 73 dominant rep-type strains consisting of 52 clinical and 21 environmental isolates (1A) and dendogram of 72 sporadic rep-type strains consisting of 63 clinical and 9 environmental isolates (1B) with corresponding gel lanes and antibiotic susceptibility profiles for each isolate. Antibiotic susceptibility is indicated as green=susceptible, yellow=intermediate, red=resistant. *Environmental isolates.
Antibiotic Susceptibility:
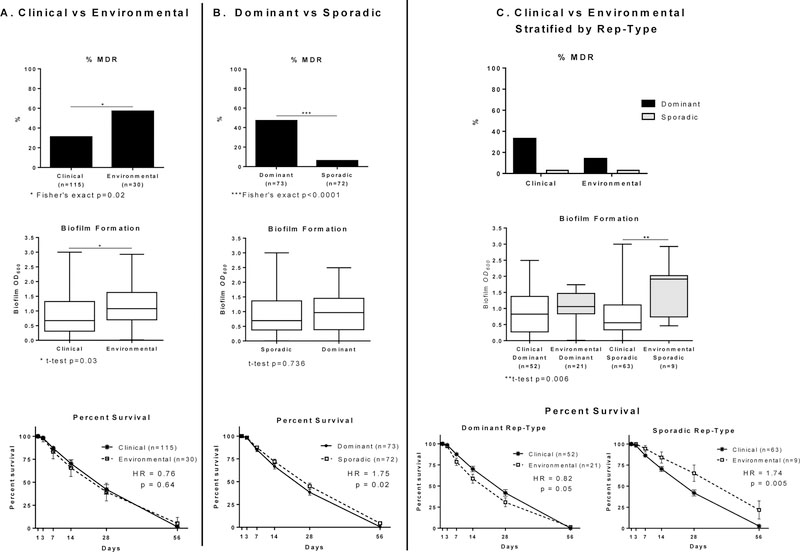
Environmental isolates were almost 3 times more likely to be MDR than clinical isolates (57% (17/30) vs. 31% (36/115), odds ratio 2.87, p=0.02), (Figure 2A). Figure 2C shows the percentage of clinical and environmental isolates that were MDR, stratified by rep-type. The higher percentages of resistance seen among both the clinical and environmental isolates can be attributed to the dominant rep-type. Overall, isolates of the dominant rep-type were almost 20 times more likely to be MDR than sporadic rep-type strains (64% (47/73) vs. 8% (6/72), odds ratio=19.88, Fisher’s exact test p<0.0001).
Figure 2:
Percent Multidrug Resistant (MDR), Box plots of biofilm OD600 and Survival Curves with standard error bars, hazard ratio (HR) and p-values of A. baumannii isolates collected from a University Hospital between Aug 2012 and Jan 2014, comparing (A) 115 clinical and 30 environmental isolates (B) 73 dominant and 72 sporadic rep-type isolates and (C) clinical and environmental isolates stratified by rep-type. Multidrug resistance was defined as being resistant to the following three drug classes (20): cephalosporins, fluoroquinolones, and aminoglycosides or resistant to two and intermediate to one.
Clinical isolates collected from patients in the ICU were also more likely to be MDR than those collected from patients outside the ICU: (67.5% (19/28) vs. 16.2% (6/37), odds ratio= 9.05, Fisher’s exact test p=0.0001). Although the numbers of environmental isolates were small they show a similar trend (ICU: 62% (13/21) were MDR vs. 44% (4/9) non-ICU). Clinical and environmental isolates collected from the ICU were more likely to be of the dominant rep-type (chi-square p-value <0.0001) compared to those collected outside the ICU but there was no difference in MDR prevalence by isolate source (t-test p=0.99).
Biofilm Formation:
Mean OD600 values for environmental and clinical isolates were 1.17 and 0.88 respectively (t-test p=0.03) (Figure 2). Environmental isolates produced more biofilm biomass than clinical isolates, regardless of rep-type. For the dominant rep-type, the mean OD600 values for clinical and environmental strains were 0.92 and 1.10 respectively (t-test p=0.30) and for the sporadic-type, clinical and environmental strains mean OD600 values were 0.84 and 1.55 respectively (t-test p=0.006). The mean biofilm OD600 values of non-ICU environmental isolates (n=9) and ICU derived environmental isolates (n=21) were 1.35 and 1.18 respectively, (t-test p=0.50).
Desiccation tolerance:
Overall, there was no statistical difference in survival between clinical and environmental isolates with respect to desiccation tolerance (Figure 2). When stratified by rep-type, dominant-type clinical strains survived better than dominant-type environmental strains. By contrast, sporadic-type environmental strains survived better than sporadic-type clinical strains. We used the Cox proportional hazards model to examine the effect of MDR and biofilm phenotypes of clinical and environmental isolates as explanatory variables, after accounting for rep-type, for risk of cell death due to desiccation after 56 days of follow-up (Cox proportional hazards model results are available upon request from the authors). Biofilm formation capacity and MDR phenotype were statistically significant and each had a statistically significant interaction terms with clinical/environmental status. However, the source of isolation (clinical versus environmental) was not statistically significant and therefore does not affect survival without the added effect of biofilm formation or MDR phenotype.
Discussion
Biofilms increase desiccation tolerance (24), (25), (7) and may confer antibiotic resistance (26). We described and compared the prevalence and interactions among biofilm formation, antibiotic resistance and desiccation tolerance in a collection of 115 clinical and 30 environmental A. baumannii isolates. Our results suggest a fitness trade-off for the MDR positive phenotype in A. baumannii that is dependent upon environmental conditions; the MDR positive environmental isolates had significantly decreased survival whereas the MDR positive clinical isolates had significantly increased survival. In addition, while the high biofilm phenotype was important for both clinical and environmental isolates to tolerate desiccation, it was critically important for the environmental isolates. We also identified a highly antibiotic resistant, dominant strain with a distinct rep-PCR banding pattern that was endemic in this hospital among clinical and environmental isolates.
Among clinical isolates, the MDR phenotype confers desiccation tolerance and the high biofilm phenotype works synergistically to improve tolerance.
Our finding that the MDR positive phenotype among clinical isolates increased desiccation tolerance is consistent with previous studies demonstrating concordance between antibiotic resistances with increased desiccation tolerance among clinical strains. Boll, et al. demonstrated that resistance to cationic antimicrobial peptide drugs also increases A. baumannii tolerance to desiccation by fortifying the fatty acid lipid content of the lipid A in the outer membrane via the production of hepta-acylated lipid A (13). Further, Gayoso et al., found that some proteins such as AmpC and Oxa51 that are associated with antibiotic resistance and increased tolerance to detergents like sodium dodecyl sulfate were overexpressed in A. baumannii clinical strain AbH12O-A2 when subjected to desiccation-stress (12). We defined MDR as resistance to cephalosporins, fluoroquinolones, and aminoglycosides, or resistant to two and intermediate to one (20). Studies using other definitions might have slightly different outcomes, but our study provides evidence that clinical isolates with the MDR positive phenotype can have increased tolerance to desiccation.
Among environmental isolates, the MDR phenotype carries a fitness cost of decreased desiccation tolerance, and the high biofilm phenotype buffers this cost.
Previous studies have demonstrated a genetic fitness cost for the MDR phenotype in bacteria and the potential cost of antibiotic resistance for clinical strains in vivo is well documented (14), (27). However, to our knowledge, no other studies have evaluated the fitness cost of drug resistance in environmental strains. We provide evidence of variation in desiccation tolerance between clinical and environmental isolates of the same phenotypes suggesting a different set of fitness-costs under different environmental conditions. A possible explanation for this may be the effects of epistasis. Epistatic outcomes can be influenced by a number of factors including the genotype in which the mutation occurs, growth environment and the level of stress or other selective pressures imposed upon the cell (27). For example, rifampicin resistance can be beneficial or deleterious for the microorganism, depending upon the environmental conditions (28), (29), (30). However, we cannot rule out that environmental sampling selected for more desiccant tolerant strains.
The high biofilm forming phenotype provided increased tolerance to desiccation for both clinical and environmental isolates, but was critical for environmental isolate survival.
Biofilm formation is suspected of being one of the key pathogenic features of A. baumannii, particularly with device-related infections (31), (32). We show a trend of increased survival for high biofilm forming clinical isolates with additional tolerance when coupled with the MDR positive phenotype. By contrast, biofilm formation had a significant impact on desiccation tolerance for environmental isolates, which likely comes at a cost of reduced drug resistance and may be driven by condition-dependent survival responses. Biofilm genes may vary in expression in response to environmental conditions. Longo et al. report different pili-like structures mediating adhesion among clinical isolates of A. baumannii, resulting in wide variability in the ability for different strains to adhere biotic or abiotic surfaces, suggesting that the genes involved in biofilm development on abiotic surfaces are not correlated with those for biofilm development on biological surfaces (26). This may offer some clues to help explain why we observed variation in the effect of high biofilm formation on desiccation tolerance between environmental and clinical isolates. For example, the clinical isolates in this study that were catheter associated could have a different expression of biofilm in vitro compared to non-catheter associated isolates. Unfortunately, we did not have access to this information and therefore were only able to consider biofilm in terms of an isolate’s ability to form biofilm. Further studies are needed to identify if the variation observed between clinical and environmental isolates resulted from different expressions of the same set of genes.
The sporadic rep-types were more likely to be susceptible to antibiotics and tolerated desiccation better than the multidrug resistant, endemic, dominant rep-types.
Half of our collection of 145 isolates shared a common rep-PCR banding pattern; isolates of the dominant rep-type were 19.9 times more likely to be MDR positive than sporadic-type isolates (Figures 1–2). Luo et al. also report a higher level of antibiotic resistance among 169 endemic strains compared to 121 sporadic strains collected at a large hospital system (15). This is not to minimize the clinical significance of sporadic isolates which can cause significant disease among compromised patients, and with their increased desiccation tolerance have a higher probability of environmental spread.
In summary, we demonstrate that the MDR positive phenotype imposes a fitness cost on A. baumannii environmental isolates by significantly decreasing desiccation tolerance, even in the presence of the high biofilm phenotype. By contrast the MDR positive phenotype does not affect desiccation tolerance among clinical isolates, and the high biofilm phenotype increases desiccation tolerance. In the absence of the MDR phenotype, biofilm formation improved desiccation tolerance in both clinical and environmental isolates but the impact on survival was significantly greater for environmental isolates. This study provides data on the characteristics of both clinical and environmental isolates, such as biofilm formation and behavior in the hospital environment that is needed to improve our understanding of the environmental transmission of infectious agents and develop more effective infection control. The risk of environmental transmission of drug resistant pathogens can be reduced by inhibiting their ability to survive in the environment. Our research increases current understanding of the association of the MDR phenotype with persistence, and demonstrates that the association is mediated by environmental conditions.
Figure 3:
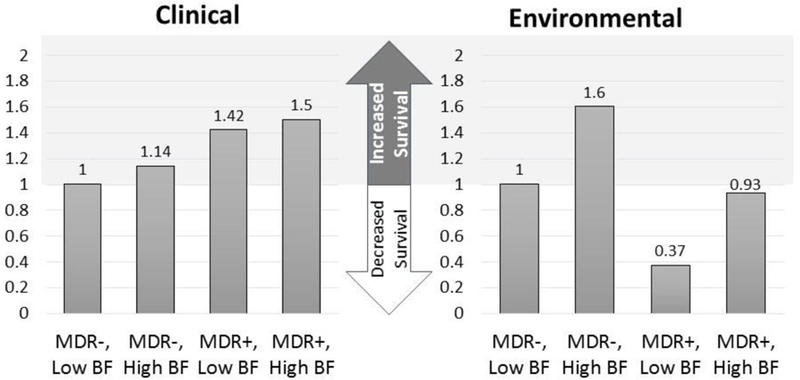
Likelihood of survival of 115 clinical and 30 environmental isolates of A. baumannii determined using a Cox proportional hazards model, accounting for rep-type, clinical/environmental status, biofilm formation capability and MDR phenotype. Clinical comparison reference group: MDR−, Low BF, clinical isolates. Environmental comparison reference group: MDR−, Low BF, environmental isolates. MDR−, multidrug negative phenotype; MDR+, multidrug positive phenotype; BF, Biofilm.
Highlights.
We identified a dominant rep-type of A. baumannii from clinical & environment sources.
The majority of isolates of the dominant rep-type were multidrug resistant.
High biofilm phenotype is critical for all A. baumannii isolates to tolerate desiccation
We show a trade-off between MDR and desiccation tolerance for environmental isolates.
Acknowledgments
The authors thank the University of Michigan Hospital Microbiology lab for providing clinical isolates and Connie Brenke and Jeana Houseman for providing technical assistance.
Funding information: This work was partially supported by an internal grant to CX at the University of Michigan, National Institutes of Health grant No. R01GM098350 to CX, a National Institutes of Health-sponsored Training Program in Infectious Disease grant (No. T32 AI049816) to BF, and the University of Michigan Risk Science Center fellowship to CG. These funding sources did not influence the design of this study, the collection and interpretation of the data, or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest. All authors report no conflicts of interest.
References
- 1.Sunenshine RH, Wright MO, Maragakis LL, Harris AD, Song X, Hebden J, et al. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis 2007;13(1):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control 2010;38(5 Suppl 1):S25–33. [DOI] [PubMed] [Google Scholar]
- 3.Tacconelli E, Cataldo MA, De Pascale G, Manno D, Spanu T, Cambieri A, et al. Prediction models to identify hospitalized patients at risk of being colonized or infected with multidrug-resistant Acinetobacter baumannii calcoaceticus complex. J Antimicrobial Chemother 2008;62(5):1130–7. [DOI] [PubMed] [Google Scholar]
- 4.Lin MF, Lan CY. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases 2014;2(12):787–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendt C, Dietze B, Dietz E, Ruden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 1997;35(6):1394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977–2000. Infect Control Hosp Epidemiol 2003;24(4):284–95. [DOI] [PubMed] [Google Scholar]
- 7.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 2003;149(Pt 12):3473–84. [DOI] [PubMed] [Google Scholar]
- 8.McQueary CN, Actis LA. Acinetobacter baumannii biofilms: variations among strains and correlations with other cell properties. J Microbiol 2011;49(2):243–50. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K Persister cells. Annual Review of Microbiol 2010;64:357–72. [DOI] [PubMed] [Google Scholar]
- 10.Lee HW, Koh YM, Kim J, Lee JC, Lee YC, Seol SY, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 2008;14(1):49–54. [DOI] [PubMed] [Google Scholar]
- 11.Roberts ME, Stewart PS. Modelling protection from antimicrobial agents in biofilms through the formation of persister cells. Microbiology 2005;151(Pt 1):75–80. [DOI] [PubMed] [Google Scholar]
- 12.Gayoso CM, Mateos J, Mendez JA, Fernandez-Puente P, Rumbo C, Tomas M, et al. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Research 2014;13(2):460–76. [DOI] [PubMed] [Google Scholar]
- 13.Boll JM, Tucker AT, Klein DR, Beltran AM, Brodbelt JS, Davies BW, et al. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. MBio 2015;6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 2015;8(3):284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo TL, Rickard AH, Srinivasan U, Kaye KS, Foxman B. Association of blaOXA-23 and bap with the persistence of Acinetobacter baumannii within a major healthcare system. Front Microbiol 2015;6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sehulster L, Chinn RY. Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). MMWR. Recommendations and reports : Morbidity and mortality weekly report. Recommendations and reports / Centers for Disease Control 2003;52(RR-10):1–42. [PubMed] [Google Scholar]
- 17.Vila J, Marcos MA, Jimenez de Anta MT. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J Med Microbiol 1996;44(6):482–9. [DOI] [PubMed] [Google Scholar]
- 18.Snelling AM, Gerner-Smidt P, Hawkey PM, Heritage J, Parnell P, Porter C, et al. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J Clin Microbiol 1996;34(5):1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol and Evolution 1987;4(4):406–25. [DOI] [PubMed] [Google Scholar]
- 20.Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Global Infect Dis 2010;2(3):291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multidrug-resistant organisms in health care settings, 2006. American journal of infection control 2007;35(10 Suppl 2):S165–93. [DOI] [PubMed] [Google Scholar]
- 22.Badave GK, Kulkarni D. Biofilm Producing Multidrug Resistant Acinetobacter baumannii: An Emerging Challenge. J Clin Diagn Res 2015;9(1):DC08–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 1985;22(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinal P, Marti S, Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 2012;80(1):56–60. [DOI] [PubMed] [Google Scholar]
- 25.de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, Bloemberg G, et al. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 2010;5(5):e10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longo F, Vuotto C, Donelli G. Biofilm formation in Acinetobacter baumannii. The New Microbiologica 2014;37(2):119–27. [PubMed] [Google Scholar]
- 27.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evolutionary Applications 2015;8(3):273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Verdugo A, Gaut BS, Tenaillon O. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evolutionary Biol 2013;13:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi Q, Preston GM, MacLean RC. Linking system-wide impacts of RNA polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa. MBio 2014;5(6):e01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall AR, Angst DC, Schiessl KT, Ackermann M. Costs of antibiotic resistance-separating trait effects and selective effects. Evolutionary Applications 2015;8(3):261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis 2006;42(5):692–9. [DOI] [PubMed] [Google Scholar]
- 32.Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Intl J Antimicrobial Agents 2010;35(3):219–26. [DOI] [PubMed] [Google Scholar]