Abstract
TLR8 (Toll-like receptor 8) is an intracellular pattern recognition receptor that senses RNA in endosomes to initiate innate immune signaling through NF-κB, and mechanisms regulating TLR8 protein abundance are not completely understood. Protein degradation is a cellular process controlling protein concentrations, accomplished largely through ubiquitin transfer directed by E3 ligase proteins to substrates. In the present study, we show that TLR8 has a short half-life in THP-1 monocytes (∼1 h) and that TLR8 is ubiquitinated and degraded in the proteasome. Treatment with the TLR8 agonist R848 causes rapid depletion of TLR8 concentrations at early time points, an effect blocked by proteasomal inhibition. We show a novel role for RNF216 (ring finger protein 216), an E3 ligase that targets TLR8 for ubiquitination and degradation. RNF216 overexpression reduces TLR8 concentrations, whereas RNF216 knockdown stabilizes TLR8. We describe a potential role for TLR8 activation by circulating RNA ligands in humans with acute respiratory distress syndrome (ARDS): Plasma and extracted RNA fractions from subjects with ARDS activated TLR8 in vitro. MicroRNA (miRNA) expression profiling revealed several circulating miRNAs from subjects with ARDS. miRNA mimics promoted TLR8 proteasomal degradation in THP-1 cells. These data show that TLR8 proteasomal disposal through RNF216 in response to RNA ligands regulates TLR8 cellular concentrations and may have implications for innate immune signaling. In addition, TLR8 activation by circulating RNA ligands may be a previously underrecognized stimulus contributing to excessive innate immune signaling characteristic of ARDS.
Keywords: Toll-like receptor, RING finger protein, microRNA
Clinical Relevance
Toll-like receptor 8 activation may be a previously underrecognized stimulus for excessive innate immune activation in acute lung injury.
Toll-like receptor 8 (TLR8) is an endosomal pattern recognition receptor that senses single-stranded RNA and RNA degradation products, leading to activation of proinflammatory and IFN signaling pathways through NF-κB and IRF7 (IFN regulatory factor 7) (1, 2). Although classically regarded as a sensor of RNA viruses (3), TLR8 signaling has also been identified in innate immune responses to RNA derived from invading bacteria, including Streptococcus pyogenes (4), Staphylococcus aureus (5), and Enterococcus faecalis (6). Furthermore, TLR8 activation has recently been identified as a signal to discriminate dead from live bacteria (7, 8), directing adaptive immune responses. In addition to sensing viral and bacterial RNA moieties, TLR8 can be activated by circulating host-derived microRNA (miRNA, miR) (9). Thus, TLR8 activation is increasingly recognized as a key step in the innate immune host response to circulating RNA ligands.
Acute respiratory distress syndrome (ARDS) is a severe form of hypoxemic respiratory failure, with an estimated 190,000 cases in the United States annually responsible for 3.6 million hospital days and a mortality rate near 40% (10, 11). Although ARDS is a clinical diagnosis, a subset of patients with ARDS characterized by the presence of excessive inflammation has poorer outcomes, including longer ICU stays and increased mortality (12). Development of a fulminant inflammatory response is complex, but a large proximal component is activation of the innate immune system (13), with elevated concentrations of IL-6 and IL-8 being key discriminatory characteristics of patients with ARDS with poor outcomes (12). TLR8 signaling is a previously underrecognized source of innate immune activation in response to RNA ligands (1, 4–6, 14), and although there is strong evidence that TLR8 activation drives cytokine secretion in monocytes in vitro, it is unknown if TLR8 signaling contributes to inflammatory signaling in humans with ARDS.
Protein ubiquitination is a universal post-translational modification that targets proteins for degradation, largely directed by the family of E3 ligases that link the ubiquitin-transferring machinery to specific substrates (15–17). Protein degradation is a cellular regulatory mechanism implicated in diverse cell functions, including regulation of the host innate immune response to lung injury (18–25). In the present study, we show that TLR8 degradation involves the E3 ligase RNF216 (ring finger protein 216), which also ubiquitinates other members of the TLR family, including TLR4 and TLR9, among others (26–28).
In this study, we show that miRNA engagement of TLR8 promotes its disposal in the proteasome. Furthermore, we show TLR8 is ubiquitinated in response to TLR8 agonists and miRNAs at early time points. We identify the E3 ligase RNF216 as a critical regulator of TLR8 protein stability. TLR8 and RNF216 coassociate in pulldown experiments, and RNF216 knockdown increases TLR8 concentrations and TLR8 half-life. Furthermore, we show that RNA extracted from plasma of human subjects with ARDS activates TLR8 in vitro. These studies provide a novel mechanism regulating TLR8 protein abundance through RNF216-mediated ubiquitination and degradation; furthermore, these studies provide a translational link in humans to further study the TLR8–RNF216 signaling axis in severe lung injury.
Methods
Cell Culture
THP-1 and HEK293 cells (American Type Culture Collection) were cultured according to the manufacturer’s instructions.
Reagents
V5 antibody (R960) was obtained from Invitrogen. MG-132 (F1100) was purchased from UBPBio. TLR8 antibody was obtained from Novus Biologicals (NBP2-24917). RNF216 antibody was purchased from Abcam (ab25961). R848 was obtained InvivoGen (tlrl-r848). miRNA mimics were purchased from Sigma-Aldrich.
IB
IB was performed as previously described (18).
Cloning
TLR8, RNF216, and RNF219 plasmid constructs were generated using PCR-based approaches and subcloned into a pcDNA3.1D/V5-His vector (19, 29).
His Pulldown
Full-length TLR8-His-V5 plasmid was overexpressed in HEK293 cells. After 16 hours of expression, cell lysates were incubated with HisPur nickel-charged nitrilotriacetic acid agarose resin for 1 hour at 25°C. Resin pulldowns were washed before elution at 70°C in 1× denaturing loading buffer.
Sequences
RNF216 Dicer-substrate siRNAs (dsiRNAs; Integrated DNA Technologies) were generated as described in previously published literature (26). RNF216 dsiRNA 1: 5′-GGACACTATGCAATCACCCG-3′; RNF216 dsiRNA 2: 5′-GAGCAGGAGTTCTATGAGCA-3′. Cells were treated with RNF216 dsiRNA or with scrambled nontargeting universal negative control dsiRNA DS NC1 (Integrated DNA Technologies).
TLR8 HEK Cells
TLR8 HEK-Blue reporter cells (InvivoGen) were cultured according to the manufacturer’s instructions. Cells (3 × 105/well) were seeded in 96-well plates overnight before stimulation. Cell culture supernatants were assayed for absorbance at 650 nm using a microplate reader.
miRNA Isolation and Expression Analysis
Studies were performed in conjunction with the University of Pittsburgh Genomics Research Core. miRNA was extracted from 200 μl of plasma using the miRNeasy Serum/Plasma Kit (Qiagen), followed by DNA cleanup with a 3 kD Amicon column (EMD Millipore). Extracted RNA (3 μl) was subjected to nCounter miRNA Expression Panel (NanoString Technologies) profiling to detect individual miRNAs. Data were analyzed using the nSolver software platform (NanoString Technologies). Counts of miR-451a were excluded from final data analysis because miR-451a is associated with hemolysis (30).
Human Studies
De-identified human plasma samples were obtained from the University of Pittsburgh Acute Lung Injury Biospecimen Repository (IRB number PRO10110387). Enrollment took place within 48 hours of the initiation of mechanical ventilation. Patients were retrospectively subclassified by an expert clinical panel into the following categories: 1) ARDS, as defined by Berlin criteria and agreed on by a minimum of three members of an expert clinical panel, or 2) ventilated control, as defined by intubation and mechanical ventilation for nonpulmonary critical illness without risk factors for ARDS. We performed a preliminary sample size calculation to detect a difference in two means (100% difference) in NF-κB activation (optical density 650 nm, 1.0 vs. 0.5) in patients with lung injury compared with control subjects with a power of 0.8, α of 0.05, estimated SD of 0.33, and sampling ratio of 0.33 control subjects/subjects with lung injury (given far fewer control subjects enrolled in our study). The predicted sample size included 5 control subjects and 14 subjects with ARDS. We selected 5 ventilated control subjects and 16 subjects with ARDS randomly from our database. For RNase studies, Plasma from subjects with ARDS (n = 11) were aliquoted into two 20-μl samples, and one 20-μl aliquot was treated with RNase I (100 μg/ml) for 30 minutes.
Statistical Analysis
Data were statistically analyzed by unpaired two-sample t test, Mann-Whitney U test, one-way ANOVA, or two-way ANOVA. All analyses were performed using Prism 7 software (GraphPad Software).
Results
miRNA Stimulation Reduces TLR8 Protein Levels at Early Time Points
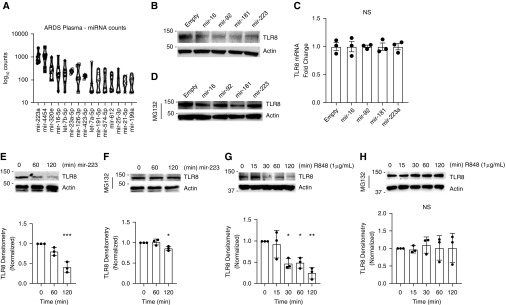
The objective of this study was to determine if circulating miRNAs in ARDS interact with TLR8. First, plasma from six subjects with ARDS (Table E1 in the data supplement) were assayed using the NanoString Technologies miRNA Expression Panel profiling array. We identified 77 circulating miRNAs from six subjects with ARDS (Figure 1A and Tables E1 and E3) with counts above baseline. The most highly expressed miRNAs in this cohort were miR-223, miR-4454, miR-320e, and miR-16a-5p (Figure 1A). miRNAs have been shown to interact with TLR8 as potential ligands (9, 31–33). We treated THP-1 cells with miRNA mimics for miR-16, miR-92, miR-181, and miR-223; miR-16 and miR-223 were identified in our plasma samples, and miR-92 and miR-181 have been shown to be associated with ARDS in whole-blood samples (34). We assayed for TLR8 protein concentrations by IB after 2 hours of miRNA stimulation and observed a decrease in TLR8 protein concentrations. The same conditions did not affect TLR2 concentrations (Figure E1A). Typically, miRNAs affect protein expression levels by binding to complementary mRNA sequences in cells and reducing transcription levels (35). Using a bioinformatics approach with TargetScan (36), we confirmed that TLR8 is not a predicted target for miR-16, miR-92, miR-181, or miR-223. We also queried a gene expression dataset of miR-223–overexpressing pulmonary epithelial cells from Neudecker and colleagues (37) (Gene Expression Omnibus accession number GSE47625) and observed no difference in TLR8 gene expression levels in miR-223–overexpressing cells (Figure E1B). We also performed reverse transcriptase–PCR for TLR8 mRNA in THP-1 cells after treatment with the same miR mimics, and we observed that miR mimic treatment did not affect TLR8 mRNA concentrations (Figure 1C). We hypothesized that this effect could be due to TLR8 protein degradation and pretreated THP-1 cells with MG-132, a proteasome inhibitor that blocks proteasomal protein degradation. MG-132 pretreatment prevented TLR8 protein concentration decreases after stimulation with most miR mimics (Figure 1D). This effect was also time dependent for miR-223 (Figures 1E and 1F), a miRNA elevated in the BAL fluid from individuals with ARDS and identified as antiinflammatory in murine acute lung injury (37). Last, we stimulated THP-1 cells with the small-molecule TLR8 agonist R848 and observed a similar effect. R848 concentrations depleted TLR8 protein concentrations after 1–2 hours stimulation, an effect blocked by MG-132 pretreatment (Figures 1G and 1H). We observed a similar effect in human peripheral blood mononuclear cells treated with R848 for 2 hours (Figure E1C). These observed effects are consistent with rapid receptor-mediated degradation, an effect observed with several other TLRs in response to ligand stimulation (19, 26).
Figure 1.
MicroRNAs (miRNA) detected in subjects with acute respiratory distress syndrome (ARDS) decrease Toll-like receptor 8 (TLR8) concentrations in vitro in monocytes, an effect mitigated by proteasome inhibition. (A) RNA was extracted from plasma of subjects with ARDS (n = 6) and subjected to NanoString Technologies miRNA Expression Panel array analysis. Figure depicts the top 15 highly detected plasma miRNAs. (B) THP-1 monocytes were transfected with miRNA mimics for miR-16, -92, -181, or -223 complexed with GenMute transfection reagent (SignaGen Laboratories) for 2 hours, and IB was performed for TLR8 or actin. (C) RT-PCR for TLR8 in THP-1 monocytes after transfection with miRNA mimics for miR-16, -92, -181, or -223 complexed with GenMute transfection reagent for 2 hours. (D) THP-1 monocytes were pretreated for 30 minutes with MG-132 (40 μmol), followed by transfection with miRNA mimics for miR-16, -92, -181, or -223 complexed with GenMute transfection reagent for 2 hours, and IB was performed for TLR8 or actin. (E and F) THP-1 monocytes were transfected with an miRNA mimic for miR-223 and assayed for TLR8 at 60 and 120 minutes after transfection, with (E) or without (F) 30 minutes of MG-132 pretreatment. ***Adjusted P = 0.006 by Dunnett multiple comparisons test, one-way ANOVA compared with 0. *Adjusted P = 0.03 by Dunnett multiple comparisons test, one-way ANOVA compared with 0. (G and H) THP-1 monocytes were stimulated with TLR8 agonist R848 (1 μg/μl), and IB was performed for TLR8 after 15, 30, 60, or 120 minutes, with (G) or without (H) 30 minutes of MG-132 pretreatment. (G) *Adjusted P = 0.01 (0 vs. 30 min), 0.02 (0 vs. 60 min). **Adjusted P = 0.001 (0 vs. 120 min) by Dunnett multiple comparisons test, one-way ANOVA. NS = nonsignificant by one-way ANOVA. THP-1 = a human monocytic cell line derived from a patient with acute monocytic leukemia.
TLR8 Is Ubiquitinated and Degraded in the Proteasome
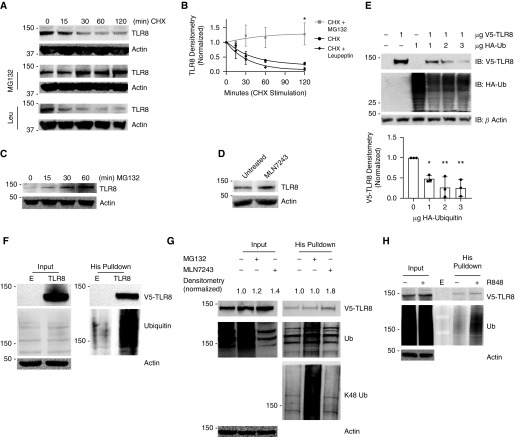
TLR8 protein half-life was determined in THP-1 cells by treating them with cycloheximide (CHX), a compound that inhibits new protein translation but permits protein degradation to occur unchecked. TLR8 concentrations decreased in a time-dependent manner after CHX treatment, with a half-life of approximately 60 minutes (Figures 2A and 2B). Protein degradation occurs in both the proteasome and the lysosome (16). We pretreated THP-1 cells with either the proteasome inhibitor MG-132 or the lysosomal hydrolase inhibitor leupeptin (Leu) before CHX stimulation and observed that MG-132 treatment, but not leupeptin treatment, stabilized TLR8 expression (Figures 2A and 2B), suggesting that TLR8 degradation occurs in the proteasome. Furthermore, TLR8 accumulated with MG-132 treatment alone (Figure 1C). Thus, TLR8 is processed via the proteasome for its disposal in THP-1 cells. Ubiquitination is a common post-translational modification that tags proteins for degradation (17), and we found that TLR8 accumulated in cells after treatment with an upstream E1-activating enzyme inhibitor, MLM7243, which prevents ubiquitin transfer to downstream substrates (Figure 2D). Furthermore, in HEK293 cells, ectopically expressed V5-TLR8 concentrations decreased with increased coexpression of hemagglutinin (HA)-tagged ubiquitin (Figure 2E). To determine if TLR8 is a substrate for ubiquitination, a plasmid containing full-length V5-His-TLR8 was transfected into HEK293 cells, followed by treatment with MG-132 for 4 hours before His pulldown. A polyubiquitin band was observed in V5-His-TLR8–transfected cells but not in empty vector control cells (Figure 2F), suggesting that the His-TLR8–enriched cellular fraction was polyubiquitinated. Furthermore, we transfected cells with V5-His-TLR8 and treated them with the E1-activating enzyme inhibitor MLN7243 or MG-132, followed by His pulldown and IB for V5, ubiquitin, or the more specific K48-linked ubiquitin. As expected, treatment with MG-132 or MLN7243 increased input concentrations of V5-TLR8. After enrichment for TLR8 by His pulldown, a polyubiquitin smear was present in MG-132–treated cells; furthermore, IB of the same TLR8-enriched cellular fractions with the more specific K48-polyubiquitin antibody revealed that V5-His-TLR8 is K48 polyubiquitinated (Figure 2G), and K48 polyubiquitination is the most common tag for disposal in the proteasome (15, 38). Last, we observed an increase in ectopically expressed TLR8 polyubiquitination after R848 stimulation (Figure 2H). Taken together, these results suggest that TLR8 is K48 polyubiquitinated and disposed of in the proteasome and that ligand stimulation promotes rapid TLR8 polyubiquitination.
Figure 2.
TLR8 has a short half-life and undergoes proteasomal degradation. (A) TLR8 concentrations decrease with cycloheximide (CHX) treatment and are stabilized with MG-132. THP-1 cells were treated with CHX (40 μg/μl) or CHX + MG-132 (40 μmol) or leupeptin (40 μmol) for 0, 15, 30, 60, or 120 minutes, and IB was performed for TLR8 or β-actin. IB is representative of three independent experiments. (B) Densitometric quantification of three independent experiments. The data represent mean ± SEM (n = 3). *P < 0.001, extra sum-of-squares F test. (C) THP-1 cells were treated with MG-132 (40 μmol) for indicated times, and IB was performed for TLR8 or β-actin. (D) THP-1 cells were treated with the E1-activating enzyme inhibitor MLN7243 for 2 hours, and IB was performed for TLR8 or β-actin. (E) HEK293 cells were transfected with TLR8 (1 μg) or cotransfected with increasing amounts of hemagglutinin-tagged ubiquitin (HA-Ub) plasmid, and IB was performed for V5-TLR8, HA-Ub, or β-actin 6 hours after transfection. IB is representative of three independent experiments. Figure shows densitometric quantification of three independent experiments. *Adjusted P = 0.01 by Dunnett multiple comparisons test, one-way ANOVA compared with 0. **Adjusted P = 0.002 by Dunnett multiple comparisons test, one-way ANOVA compared with 0. (F) HEK293 cells were transfected with an empty vector or TLR8, treated with MG-132 (40 μmol) for 4 hours, and subjected to His pulldown, and IB was performed for V5, Ub, or β-actin. (G) HEK293 cells were transfected with TLR8 and were treated with MG-132 (40 μmol) or MLN7243 or were untreated. Cell lysates were subjected to His pulldown, and IB was performed for V5, Ub, K48-linked Ub, or β-actin. (H) TLR8-transfected HEK cells were pretreated with MG-132 (40 μmol) for 2 hours, followed by stimulation with R848 (1 μg/μl) for 2 hours, and subjected to His pulldown followed by IB for V5-TLR8, Ub, or actin. His = histidine; Leu = leucine.
The E3 Ligase RNF216 Targets TLR8 for Ubiquitination and Degradation
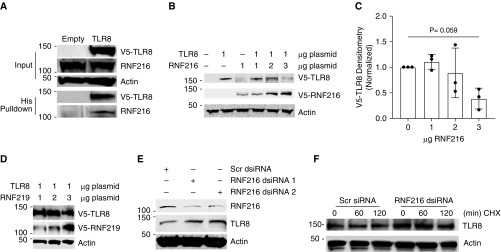
RNF216 is an E3 ligase that ubiquitinates TLR4 and TLR9, targeting them for proteasomal degradation (26). TLR8 shares a common cytoplasmic Toll/IL-1 receptor (TIR) domain (39), and we hypothesized that RNF216 would also target TLR8 for degradation. First, we detected an association between overexpressed TLR8 and endogenous RNF216 in HEK cells after TLR8 overexpression and His pulldown (Figure 3A). Increasing amounts of co-overexpressed RNF216 reduced overexpressed TLR8 concentrations in HEK cells (Figures 3B and 3C), an effect not observed with co-overexpression of the E3 ligase RNF219 (Figure 3D). In THP-1 cells, knockdown of RNF216 with dsiRNA was associated with a concomitant increase in TLR8 protein concentrations (Figure 3E). Furthermore, RNF216 knockdown stabilized TLR8 concentrations after CHX treatment in THP-1 cells (Figure 3F). Taken together, these data suggest that the E3 ligase RNF216 is a key regulator of TLR8 protein stability.
Figure 3.
E3 ligase RNF216 (ring finger protein 216) targets TLR8 for ubiquitination and degradation. (A) RNF216 associates with TLR8. HEK293 cells were transfected with an empty vector or TLR8 and subjected to His pulldown, and IB was performed for V5-TLR8, RNF216, or β-actin. (B) HEK293 cells were transfected with TLR8 (1 μg) or cotransfected with increasing amounts of RNF216, and IB was performed for V5-TLR8, RNF216, or β-actin 16 hours after transfection. IB is representative of three independent experiments. (C) Densitometric quantification of three independent experiments. The data represent mean ± SEM (n = 3). Adjusted P = 0.059 by Dunnett multiple comparisons test, one-way ANOVA compared with 0. (D) HEK293 cells were transfected with TLR8 (1 μg) or cotransfected with increasing amounts of RNF219, and IB was performed for V5-TLR8, RNF216, or β-actin 16 hours after transfection. (E) THP-1 cells were transfected with Dicer-substrate siRNA (dsiRNA) against RNF216 or a scrambled dsiRNA (scr), and IB was performed for RNF216, TLR8, or β-actin 72 hours after transfection. (F) THP-1 cells were transfected with dsiRNA against RNF216 or a scrambled dsiRNA (scr) and treated with CHX (40 μg/ml) for the indicated times, followed by IB for TLR8 or β-actin 72 hours after transfection.
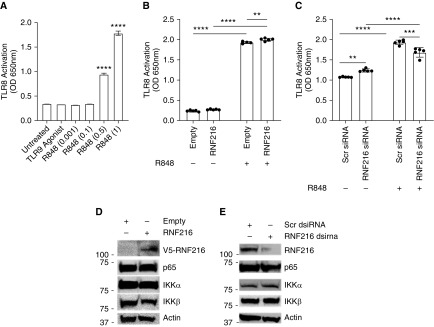
RNF216 Regulates TLR8 Stability but Not TLR8-Dependent Signaling In Vitro
To determine if RNF216 regulates TLR8-dependent downstream signaling events, we used a TLR8 reporter cell line (InvivoGen) that measures TLR8-dependent NF-κB activation. We observed TLR8 activation with increasing doses of TLR8 agonist R848 but not of the TLR9 agonist ODN 2006 (Figure 4A). RNF216 overexpression (Figure 4B) or RNF216 knockdown (Figure 4C) had little effect on TLR8-dependent signaling. RNF216 overexpression (Figure 4C) or RNF216 knockdown (Figure 4D) also did not affect protein concentrations of NF-κB signaling intermediaries, a pathway known to be heavily regulated by the ubiquitin/proteasome system (40–42). Taken together, these results suggest that RNF216 regulated TLR8 stability through the ubiquitin/proteasome system, but not necessarily TLR8-dependent NF-κB activation.
Figure 4.
TLR8 activation is not affected by RNF216 overexpression or knockdown in TLR8/NF-κB reporter cell lines. (A) TLR8-HEK-Blue (InvivoGen) reporter cell lines were stimulated with TLR9 agonist ODN 2006 (negative control) or increasing amounts of R848 for 16 hours, and NF-κB SEAP activity was measured by optical density (OD) at 650 nm. ****Adjusted P < 0.0001 by Dunnett multiple comparisons test, one-way ANOVA compared with untreated. (B) TLR8-HEK-Blue cells were transfected with an empty vector or RNF216 and treated with R848 (1 μg/μl) for 16 hours, and NF-κB SEAP activity was measured by OD at 650 nm. **Adjusted P = 0.003 by Tukey multiple comparisons test, two-way ANOVA. ****Adjusted P < 0.0001 by Tukey multiple comparisons test, two-way ANOVA. (C) TLR8-HEK-Blue cells were transfected with scrambled dsiRNA or RNF216 dsiRNA and treated with R848 (1 μg/μl) for 16 hours, and NF-κB SEAP activity was measured by OD at 650 nm. **Adjusted P = 0.006 by Tukey multiple comparisons test, two-way ANOVA. ***Adjusted P = 0.0001 by Tukey multiple comparisons test, two-way ANOVA. ****Adjusted P < 0.0001 by Tukey multiple comparisons test, two-way ANOVA. (D) HEK cells were transfected with an empty vector or RNF216, and IB was performed for RNF216, p65, IKKα, IKKβ, or actin. (E) HEK cells were transfected with scrambled dsiRNA or RNF216 dsiRNA, and IB was performed for RNF216, p65, IKKα, IKKβ, or actin. SEAP = secreted alkaline phosphatase.
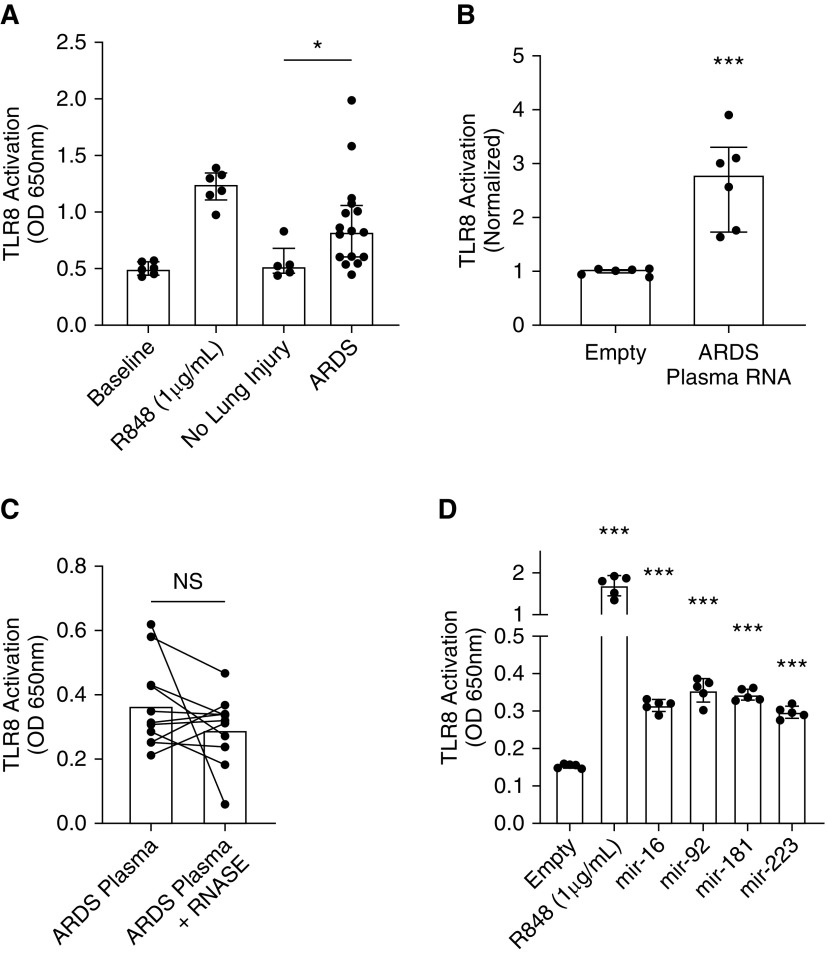
Plasma and Plasma RNA from Subjects with ARDS Activates TLR8 In Vitro
The role of TLR8 in acute lung injury is unknown, but TLR8-dependent proinflammatory signaling is activated in response to bacterial RNA and some host RNA ligands, including miRNAs and mitochondrial RNA (4–7, 14). Furthermore, bacteremia and sepsis are common risk factors for the development of ARDS (43). We asked whether plasma from subjects with ARDS could activate TLR8. We stimulated TLR8-HEK-Blue cells with plasma from subjects enrolled in the University of Pittsburgh Acute Lung Injury Registry (Table 1). Although plasma from subjects without lung injury had a minimal effect to activate TLR8, plasma from subjects with ARDS increased TLR8 activation (P = 0.014 by Mann-Whitney U test compared with subjects with no lung injury) (Figure 5A). Notably, this cohort had trends of higher median concentrations of other proinflammatory cytokines, but there were no significant correlations between proinflammatory cytokine concentrations and TLR8 activation (Table E2 and Figure E2), suggesting that the activation of NF-κB in the reporter cells was TLR8 specific. Because TLR8 is an RNA sensor, we asked whether RNA extracted from ARDS plasma was sufficient to activate TLR8. We stimulated TLR8-HEK-Blue cells with RNA from six patients from our NanoString Technologies miRNA Expression Panel analysis cohort (Figure 1A and Table E1) and observed TLR8 activation (Figure 5B). Thus, the extracted RNA fraction from plasma was sufficient to activate TLR8 in vitro. Plasma miRNAs are generally protected from RNase degradation because they are contained in exosomes or bound to carrier proteins, including AGO2 (argonaute 2) (44–46), so as an orthogonal approach, we divided 11 ARDS plasma samples into two 20-μl aliquots and treated one with RNase (100 μg/ml) for 30 minutes, and both aliquots were added to TLR8-HEK-Blue cells. RNase treatment did not significantly reduce TLR8 activation (Figure 5C), further supporting the notion that the TLR8-stimulating potential of the plasma samples derives not from free, uncomplexed RNA; rather, it derives from ligands protected from RNase degradation, which would include miRNAs and potentially other RNA ligands contained in exosomes or complexed with proteins. Last, we treated TLR8-HEK-Blue reporter cells with R848 or transfected them with miRNA mimics for miR-16, miR-92, miR-181, or miR-223. As expected, R848 strongly activated TLR8, whereas miR-16, miR-92, miR-181, and miR-223 mimics also increased TLR8 activation, but to a much lesser extent than R848 (Figure 5D). Although these same miRNA mimics promoted TLR8 degradation, they may also be weak TLR8 activators, but to a much lesser extent than a known ligand such as R848.
Table 1.
Clinical and Demographic Characteristics of Subjects with Acute Respiratory Distress Syndrome in Toll-like Receptor 8 Activity Assay
| No Lung Injury | ARDS | |
|---|---|---|
| n | 5 | 16 |
| Sex, F, n (%) | 2 (40) | 7 (44) |
| Age, yr | 49.5 | 56 |
| FiO2 (%), average | 46 | 62 |
| PEEP (mm Hg), average | 5.6 | 9.8 |
| Receiving vasopressor, n (%) | 1 (20) | 7 (44) |
| WBC (x 10−9 per liter) | 11.42 | 12.34375 |
Definition of abbreviations: ARDS = acute respiratory distress syndrome; PEEP = positive end-expiratory pressure; WBC = white blood cells.
Figure 5.
Plasma and extracted plasma RNA from subjects with ARDS activates TLR8 in vitro. (A) Plasma from subjects with ARDS activates TLR8. TLR8-HEK-Blue cells were stimulated with 40 μl of plasma from mechanically ventilated subjects with no lung injury or with ARDS, and TLR8-dependent NF-κB SEAP activity was measured by a plate reader at 650 nm after 16 hours. *P = 0.014 by Mann-Whitney U test compared with subjects with no lung injury. (B) TLR8-HEK-Blue cells were stimulated with extracted plasma RNA from subjects with ARDS, and TLR8-dependent NF-κB SEAP activity was measured by plate reader at 650 nm after 16 hours. ***P = 0.002 by Mann-Whitney U test compared with empty vector. (C) Plasma from subjects with ARDS (n = 11) was aliquoted into two 20-μl samples. One 20-μl aliquot was treated with RNase I (100 μg/ml) for 30 minutes, and both aliquots were added to TLR8-HEK-Blue cells. OD values at 650 nm were recorded after 16 hours. (D) TLR8-HEK-Blue cells were transfected with R848 or miRNA mimics for miR-16, miR-92, miR-181, or miR-223, and TLR8-dependent NF-κB SEAP activity was measured by plate reader at 650 nm after 16 hours. ***Adjusted P < 0.005 by Dunnett multiple comparisons test, one-way ANOVA.
RNF216 Gene Expression Is Downregulated in Critically Ill Subjects at Risk for or with ARDS
RNF216 regulates the stability of several TLRs and proteins that regulate innate immune responses (26–28, 47); thus, changes in RNF216 expression might also alter the function of its substrates, including TLRs. We queried publicly available gene datasets (48–50) in patients with ARDS or in patients with risk factors for ARDS (sepsis, pneumonia), with uninjured patients or healthy volunteers serving as control subjects. We restricted our search criteria to examine cells or tissues known to express TLR8 (blood leukocytes or whole blood), and we excluded studies that examined cells or tissues in which TLR8 expression/function is unknown (lung tissue or BAL). We found three studies meeting these criteria, and results are summarized in Table E4. RNF216 gene expression was significantly downregulated in all three datasets (Table E4 and Figure E3). Because RNF216 controls TLR4- and TLR9-dependent inflammatory responses, and because we show that it also regulates TLR8 stability, decreased RNF216 expression may be an additional mechanism promoting excessive inflammation in subjects with or at risk for ARDS.
Discussion
The family of TLRs are key regulators of the innate immune response to invading pathogens by sensing pathogen-associated molecular pattern molecules as well as host-derived damage-associated molecular pattern (DAMP) signals (51–53). In the present study, we show a novel mechanism regulating TLR8 protein concentrations through the E3 ligase RNF216, as well as a potential link showing TLR8 activation in subjects with ARDS.
TLR8 is a promiscuous intracellular RNA sensor activated by RNA degradation products rich in uridine residues (1, 14). TLR8 activation occurs in response to viral RNA, bacterial RNA, host mitochondrial RNA, and host miRNAs (1, 3–6, 9, 14, 54). Its role in ARDS and other hyperinflammatory disorders is not well characterized. TLR8 activation and downstream inflammatory signaling are partially dependent on lysosomal acidification, because treatment with bafilomycin ameliorates TLR8-dependent NF-κB activation in the presence of ligands (4). Together with the proteasome, the lysosome is the other major site of protein disposal in cells. Although we did not observe an effect on TLR8 half-life with lysosomal inhibition, further studies are warranted to determine if TLR8 degradation occurs exclusively in the proteasome or whether the lysosome might be partially responsible for TLR8 disposal, especially after encountering ligands.
TLR8 is sequestered in endosomes, and TLR8 ligands are encountered in the endolysosomal compartment. Generally, extracellular ligands (viruses, circulating RNA, exosomes) can enter cells via receptor-mediated endocytosis and fuse with endosomes, delivering their cargo to TLR8-containing endosomes (1, 55, 56). Alternatively, cellular autophagy can deliver TLR-containing endosomes to ligands (2). One cellular receptor involved in the uptake of extracellular RNA is the receptor for advanced glycation end products (RAGE) (57), and we have recently shown a role for RAGE degradation by the E3 ligase subunit FBXO10 (F-box only protein 10) in response to a DNA ligand ODN 2006 (18). Future studies can investigate if RAGE/FBXO10 is involved in sensing extracellular RNA in ARDS or whether miRNA-containing exosomes can activate TLR8 independent of receptor-mediated endocytosis.
The role of TLR8 signaling and activation in murine or human lung injury remains uncharacterized. Our data suggest that 1) miRNAs may engage with TLR8, promoting its proteasomal degradation at early time points, and 2) extracted plasma RNA from subjects with ARDS is capable of driving TLR8 activation in vitro in a TLR8 reporter cell line. The mechanistic role of some miRNAs in lung injury has been reported. In fact, the most highly expressed plasma miRNA in our cohort (miR-223) has been described as an antiinflammatory miRNA in acute lung injury, and concentrations of miR-223 are elevated in the BAL fluid of patients with ARDS compared with healthy control subjects (37). Further, this study showed that neutrophil transfer of miR-223 to epithelial cells restrained inflammation through miR-223–dependent PARP1 (poly[ADP-ribose] polymerase 1) mRNA degradation. Our data might suggest that another potential antiinflammatory mechanism of miR-223 (and other circulating miRNAs) could be through TLR8 proteasomal degradation, limiting the availability of TLR8 to sense other proinflammatory RNA ligands. However, we also show that miRNA mimics were weak activators of TLR8-dependent NF-κB activation in TLR8 reporter cells. Indeed, this phenomenon has been reported previously (9), but it may be cell and tissue specific because no increase in TLR8-dependent NF-κB activation was observed with miR-21 treatment in another study (33). Thus, the effects of circulating miRNAs are pleotropic, and additional studies are warranted to further characterize their relative contributions to excessive inflammation during acute lung injury, including whether TLR8 degradation is detrimental or beneficial to the host.
We examined TLR8 protein concentrations at early time points (<2 h), and the results show a reduction in TLR8 concentrations in response to TLR8 ligands. Several other studies have reported increases in TLR8 protein concentrations after agonist stimulation, but at much later time points (24 h), with a mechanism dependent on increases in TLR8 gene transcription (58, 59). Thus, the interplay between RNF216-mediated TLR8 degradation and TLR8 transcription in response to TLR8 agonists remains to be fully characterized. This phenomenon may be particularly relevant in ARDS, in which a sustained hyperinflammatory phenotype (over days) is associated with worse outcomes (12, 60). Our data suggest that RNF216 might be important in regulating TLR8 concentrations at early time points, but its role in regulating TLR8, as well as other TLRs, at later time points remains an active area of investigation.
We show a role for the E3 ubiquitin ligase RNF216 in regulating TLR8 protein concentrations. Thus, pharmacological modulation of RNF216 activity may be a novel strategy to regulate TLR8 concentrations and TLR8-dependent signaling. RNF216 was first identified as a negative regulator of TLR4 and TLR9 signaling (26). In addition, RNF216 has also been shown to target RIP1 (peceptor-interacting protein 1), TRAF3 (TNF receptor–associated factor 3), and BECN1 (Beclin 1) (27, 28, 47), proteins involved in innate immune signaling and autophagy. Thus, although pharmacological inhibition of RNF216 would be a novel strategy to boost TLR4, TLR9, and TLR8 immune responses, it might also affect proteins involved in the activation of autophagy. In addition, RNF216 might also target other proteins for ubiquitination and degradation, especially those with TIR domains; thus, RNF216 inhibition might also have unexpected off-target effects by interfering with these interactions.
We provide data suggesting that plasma RNA from subjects with ARDS can activate TLR8 in vitro. Several nucleic acids function as DAMPs, including bacterial and viral RNA (4, 6, 54, 59), cell-free mitochondrial DNA (mtDNA) (61, 62), and mitochondrial RNA (14). Furthermore, RNA modifications, including oxidation from peroxisomes or 2′-O-methylation, can alter the immunogenicity of RNA ligands (63). Mechanisms regulating nucleic acid DAMPs are only beginning to be fully characterized. For example, elevated concentrations of cell-free mtDNA is associated with mortality in critically ill humans (64), and mtDNA is a known ligand for TLR9 (65). How mtDNA interacts with the innate immune system during acute lung injury is relatively unknown, but a recent report shows a role for red blood cells as scavengers of mtDNA (66). During acute inflammation, the ability of red blood cells to scavenge mtDNA is reduced, thus increasing the available fraction of circulating cell-free mtDNA to interact with immune cells and activate proinflammatory cytokine signaling. Mechanisms regulating RNA DAMPs are relatively unexplored.
Circulating nucleic acid DAMPs interact with TLRs, generally confined to the intracellular compartment in endosomes. Ligand engagement sets off a signaling cascade mediated by recruitment of adapter proteins to TLR cytosolic domains. For example, TLR8 and TLR9 require MyD88 to signal, and MyD88 is essential for host defense in pulmonary infection (67). TRAF proteins also play an important role in TLR signal transduction, and we have recently shown a role for a ubiquitin E3 ligase, FBXO3, in regulating TRAF proteins in Pseudomonas lung injury (24). Thus, although we show a role for the E3 ligase RNF216 in regulating TLR8 at early time points, several other proteins involved in the TLR8 signaling cascade are known to be regulated by the ubiquitin proteasome system, and how RNA DAMPs affect these intermediaries has not been characterized.
In summary, we show that TLR8 is a substrate for proteasomal degradation through the E3 ligase RNF216. TLR8 is proteasomally degraded at early time points in response to TLR8 agonists in a mechanism dependent on RNF216. We show a putative, previously unreported role for TLR8 activation in ARDS, based on both total plasma and extracted RNA from subjects with ARDS activating TLR8 in vitro.
Supplementary Material
Acknowledgments
Acknowledgment
The authors offer a special acknowledgment to the patients and their families who were enrolled in the University of Pittsburgh Acute Lung Injury Biospecimen Repository. The authors acknowledge Sarah Rapport, B.S., for coordinating patient enrollment in the repository. The authors also acknowledge the University of Pittsburgh Genomics Core because this project used the University of Pittsburgh Health Sciences Core Research Facilities Genomics Research Core plasma miRNA extraction and cleanup services.
Footnotes
Supported by the National Institutes of Health under National Heart, Lung, and Blood Institute (NHLBI) grants 1F32HL137258-01 and 1K08HL144820 (J.E.); NHLBI grants HL116472 and HL132862; NHLBI R35 grant HL139860 (B.B.C.); NHLBI grants HL096376, HL097376, HL098174, 5T32HL007563-28, and P01 HL114453 (R.K.M.); by the U.S. Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development; and by a Merit Review Award from the U.S. Department of Veterans Affairs (R.K.M.).
Author Contributions: J.E. designed the studies, performed experiments, analyzed the data, and wrote the manuscript. J.E., T.L., C.B., Y.C., V.W., J.V., and J.L. performed in vitro experiments. B.M. and G.D.K. directed human sample experiments. J.E., B.M., and G.D.K. performed and analyzed human sample experiments. Y.L. designed the study and analyzed the data. R.K.M. and B.B.C. directed the study.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0373OC on August 6, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat Struct Mol Biol. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- 2.Blasius AL, Beutler B. Intracellular Toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, El-Far M, Dupuy FP, Abdel-Hakeem MS, He Z, Procopio FA, et al. HCV RNA activates APCs via TLR7/TLR8 while virus selectively stimulates macrophages without inducing antiviral responses. Sci Rep. 2016;6:29447. doi: 10.1038/srep29447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eigenbrod T, Pelka K, Latz E, Kreikemeyer B, Dalpke AH. TLR8 senses bacterial RNA in human monocytes and plays a nonredundant role for recognition of Streptococcus pyogenes. J Immunol. 2015;195:1092–1099. doi: 10.4049/jimmunol.1403173. [DOI] [PubMed] [Google Scholar]
- 5.Bergstrøm B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, et al. TLR8 senses Staphylococcus aureus RNA in human primary monocytes and macrophages and induces IFN-β production via a TAK1-IKKβ-IRF5 signaling pathway. J Immunol. 2015;195:1100–1111. doi: 10.4049/jimmunol.1403176. [DOI] [PubMed] [Google Scholar]
- 6.Nishibayashi R, Inoue R, Harada Y, Watanabe T, Makioka Y, Ushida K. RNA of Enterococcus faecalis strain EC-12 is a major component inducing interleukin-12 production from human monocytic cells. PLoS One. 2015;10:e0129806. doi: 10.1371/journal.pone.0129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ugolini M, Gerhard J, Burkert S, Jensen KJ, Georg P, Ebner F, et al. Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. Nat Immunol. 2018;19:386–396. doi: 10.1038/s41590-018-0068-4. [DOI] [PubMed] [Google Scholar]
- 8.Barbet G, Sander LE, Geswell M, Leonardi I, Cerutti A, Iliev I, et al. Sensing microbial viability through bacterial RNA augments T follicular helper cell and antibody responses. Immunity. 2018;48:584–598, e5. doi: 10.1016/j.immuni.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. LUNG SAFE Investigators; ESICM Trials Group. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 11.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, et al. Incidence and outcomes of acute lung injury. N Engl J Med. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 12.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA NHLBI ARDS Network. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Mallampalli RK. The acute respiratory distress syndrome: from mechanism to translation. J Immunol. 2015;194:855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krüger A, Oldenburg M, Chebrolu C, Beisser D, Kolter J, Sigmund AM, et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 2015;16:1656–1663. doi: 10.15252/embr.201540861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 16.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 17.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 18.Evankovich J, Lear T, McKelvey A, Dunn S, Londino J, Liu Y, et al. Receptor for advanced glycation end products is targeted by FBXO10 for ubiquitination and degradation. FASEB J. 2017;31:3894–3903. doi: 10.1096/fj.201700031R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKelvey AC, Lear TB, Dunn SR, Evankovich J, Londino JD, Bednash JS, et al. RING finger E3 ligase PPP1R11 regulates TLR2 signaling and innate immunity. Elife. 2016;5:e18496. doi: 10.7554/eLife.18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coon TA, McKelvey AC, Lear T, Rajbhandari S, Dunn SR, Connelly W, et al. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci Transl Med. 2015;7:295ra109. doi: 10.1126/scitranslmed.aab3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vadász I, Weiss CH, Sznajder JI. Ubiquitination and proteolysis in acute lung injury. Chest. 2012;141:763–771. doi: 10.1378/chest.11-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallampalli RK, Coon TA, Glasser JR, Wang C, Dunn SR, Weathington NM, et al. Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J Immunol. 2013;191:5247–5255. doi: 10.4049/jimmunol.1300456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen BB, Coon TA, Glasser JR, Zou C, Ellis B, Das T, et al. E3 ligase subunit Fbxo15 and PINK1 kinase regulate cardiolipin synthase 1 stability and mitochondrial function in pneumonia. Cell Rep. 2014;7:476–487. doi: 10.1016/j.celrep.2014.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, et al. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol. 2013;14:470–479. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, et al. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy. 2014;10:2239–2250. doi: 10.4161/15548627.2014.981792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakhaei P, Mesplede T, Solis M, Sun Q, Zhao T, Yang L, et al. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009;5:e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lear T, Dunn SR, McKelvey AC, Mir A, Evankovich J, Chen BB, et al. RING finger protein 113A regulates C-X-C chemokine receptor type 4 stability and signaling. Am J Physiol Cell Physiol. 2017;313:C584–C592. doi: 10.1152/ajpcell.00193.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cervantes JL, Weinerman B, Basole C, Salazar JC. TLR8: the forgotten relative revindicated. Cell Mol Immunol. 2012;9:434–438. doi: 10.1038/cmi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Liang H, Zhang J, Zen K, Zhang CY. microRNAs are ligands of Toll-like receptors. RNA. 2013;19:737–739. doi: 10.1261/rna.036319.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZJ, Guo JS, Li SS, Wu XB, Cao DL, Jiang BC, et al. TLR8 and its endogenous ligand miR-21 contribute to neuropathic pain in murine DRG. J Exp Med. 2018;215:3019–3037. doi: 10.1084/jem.20180800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Liang L, Zhang R, Wei Y, Su L, Tejera P, et al. Whole blood microRNA markers are associated with acute respiratory distress syndrome. Intensive Care Med Exp. 2017;5:38. doi: 10.1186/s40635-017-0155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal V, Bell GW, Nam JW, Bartel DP.Predicting effective microRNA target sites in mammalian mRNAs Elife 20154e05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med. 2017;9:eaah5360. doi: 10.1126/scitranslmed.aah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopal R, Waller AS, Mendoza JD, Wightman PD. The covalent modification and regulation of TLR8 in HEK-293 cells stimulated with imidazoquinoline agonists. Biochem J. 2008;409:275–287. doi: 10.1042/BJ20070519. [DOI] [PubMed] [Google Scholar]
- 40.Heaton SM, Borg NA, Dixit VM. Ubiquitin in the activation and attenuation of innate antiviral immunity. J Exp Med. 2016;213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Chen ZJ. Regulation of NF-κB by ubiquitination. Curr Opin Immunol. 2013;25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377:562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 44.Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 2016;7:11106. doi: 10.1038/ncomms11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fearns C, Pan Q, Mathison JC, Chuang TH. Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem. 2006;281:34592–34600. doi: 10.1074/jbc.M604019200. [DOI] [PubMed] [Google Scholar]
- 48.Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cazalis MA, Lepape A, Venet F, Frager F, Mougin B, Vallin H, et al. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive Care Med Exp. 2014;2:20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scicluna BP, Klein Klouwenberg PM, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Respir Crit Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Baral P, Batra S, Zemans RL, Downey GP, Jeyaseelan S. Divergent functions of Toll-like receptors during bacterial lung infections. Am J Respir Crit Care Med. 2014;190:722–732. doi: 10.1164/rccm.201406-1101PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balamayooran T, Balamayooran G, Jeyaseelan S. Review: Toll-like receptors and NOD-like receptors in pulmonary antibacterial immunity. Innate Immun. 2010;16:201–210. doi: 10.1177/1753425910366058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernard MA, Zhao H, Yue SC, Anandaiah A, Koziel H, Tachado SD. Novel HIV-1 miRNAs stimulate TNFα release in human macrophages via TLR8 signaling pathway. PLoS One. 2014;9:e106006. doi: 10.1371/journal.pone.0106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 56.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bertheloot D, Naumovski AL, Langhoff P, Horvath GL, Jin T, Xiao TS, et al. RAGE enhances TLR responses through binding and internalization of RNA. J Immunol. 2016;197:4118–4126. doi: 10.4049/jimmunol.1502169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zannetti C, Bonnay F, Takeshita F, Parroche P, Ménétrier-Caux C, Tommasino M, et al. C/EBPδ and STAT-1 are required for TLR8 transcriptional activity. J Biol Chem. 2010;285:34773–34780. doi: 10.1074/jbc.M110.133884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farina A, Peruzzi G, Lacconi V, Lenna S, Quarta S, Rosato E, et al. Epstein-Barr virus lytic infection promotes activation of Toll-like receptor 8 innate immune response in systemic sclerosis monocytes. Arthritis Res Ther. 2017;19:39. doi: 10.1186/s13075-017-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delucchi K, Famous KR, Ware LB, Parsons PE, Thompson BT, Calfee CS ARDS Network. Stability of ARDS subphenotypes over time in two randomised controlled trials. Thorax. 2018;73:439–445. doi: 10.1136/thoraxjnl-2017-211090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simmons JD, Lee YL, Mulekar S, Kuck JL, Brevard SB, Gonzalez RP, et al. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects Ann Surg 2013258591–596.[Discussion, pp. 596–598.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rimbach K, Kaiser S, Helm M, Dalpke AH, Eigenbrod T. 2′-O-Methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J Innate Immun. 2015;7:482–493. doi: 10.1159/000375460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumagai Y, Takeuchi O, Akira S. TLR9 as a key receptor for the recognition of DNA. Adv Drug Deliv Rev. 2008;60:795–804. doi: 10.1016/j.addr.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, et al. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med. 2018;197:470–480. doi: 10.1164/rccm.201706-1161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai S, Batra S, Shen L, Wakamatsu N, Jeyaseelan S. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J Immunol. 2009;183:6629–6638. doi: 10.4049/jimmunol.0901033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.