Abstract
The Lhc (light-harvesting chlorophyll a/b-binding protein) superfamily represents a class of antennae proteins that play indispensable roles in capture of solar energy as well as photoprotection under stress conditions. Despite their importance, little information has been available beyond model plants. In this study, we presents a first genome-wide analysis of Lhc superfamily genes in jatropha (Jatropha curcas L., Euphorbiaceae), an oil-bearing plant for biodiesel purpose. A total of 27 members were identified from the jatropha genome, which were shown to distribute over nine out of the 11 chromosomes. The superfamily number is comparable to 28 present in castor (Ricinus communis, Euphorbiaceae), but relatively less than 35 in cassava (Manihot esculenta, Euphorbiaceae) and 34 in arabidopsis (Arabidopsis thaliana) that experienced one or two recent whole-genome duplications (WGDs), respectively. In contrast to a high number of paralogs present in cassava and arabidopsis, few duplicates were found in jatropha as observed in castor, corresponding to no recent WGD occurred in these two species. Nevertheless, 26 orthologous groups representing four defined families were found in jatropha, and nearly one-to-one orthologous relationship was observed between jatropha and castor. By contrast, a novel group named SEP6 was shown to have been lost in arabidopsis. Global transcriptome profiling revealed a predominant expression pattern of most JcLhc superfamily genes in green tissues, reflecting their key roles in photosynthesis. Moreover, their expression profiles upon hormones, drought, and salt stresses were also investigated. These findings not only improve our knowledge on species-specific evolution of the Lhc supergene family, but also provide valuable information for further studies in jatropha.
Keywords: Jatropha curcas, Ricinus communis, Manihot esculenta, Arabidopsis thaliana, Harvesting chlorophyll a/b-binding protein, Lhc superfamily, Evolution, Synteny analysis, Whole-genome duplication, Expression profile
Introduction
Light-harvesting chlorophyll a/b-binding protein (Lhc) superfamily, defined by the presence of a conserved chlorophyll-binding (CB) domain in the transmembrane alpha-helix, is composed of four distinct nuclear-encoded antennae protein families in green plants, i.e., Lhc, Lil (light-harvesting-like), PsbS (photosystem II subunit S), and FCII (ferrochelatase II) (Klimmek et al., 2006; Zou & Yang, 2019a). In contrast to an orphan group present in both PsbS and FCII families, the Lhc family, initially known as CAB (chlorophyll a/b-binding protein), contains two evolutionary groups named Lhca and Lhcb that are associated with photosystem I or II (PSI/II), respectively (Jansson, 1999; Klimmek et al., 2006). The Lil family includes four diverse subfamilies, i.e., OHP (one-helix protein), SEP (stress-enhanced protein), Lil1 or ELIP (early light-induced protein), and Lil8 or Psb33 (photosystem II protein 33) (Engelken, Brinkmann & Adamska, 2010; Zou, 2018). Among them, OHP and SEP can be further divided into several groups: the OHP subfamily includes two groups named OHP1/Lil2 and OHP2/Lil6, whereas the SEP subfamily contains six groups, i.e., SEP1/Lil4, SEP2/Lil5, SEP3/Lil3, SEP4, SEP5, and SEP6 (Engelken, Brinkmann & Adamska, 2010; Zou & Yang, 2019a). Investigation of their origin suggested that OHP is more primitive, which is more likely to result from the plastid-encoded HLIP (high light-induced protein) via gene transfer after the primary endosymbiosis (Koziol et al., 2007; Engelken, Brinkmann & Adamska, 2010). In addition to light harvesting and transport, growing evidence shows that Lhc superfamily members are also involved in regulation and distribution of excitation energy between PSI and PSII, maintenance of thylakoid membrane structure, photoprotection as well as response to various stresses (Pan et al., 2011; Fan et al., 2015; Fristedt et al., 2015; Hey et al., 2017; Myouga et al., 2018).
Jatropha curcas L. (2n = 22), commonly known as jatropha, physic nut, barbados nut, or purging nut, is a perennial large shrub or small tree (Mazumdar et al., 2018; Zou et al., 2016; Zou et al., 2018). Jatropha belongs to the Euphorbiaceae family, which also includes castor (also known as castor bean, Ricinus communis L.) and cassava (Manihot esculenta Crantz) and is characterized with high photosynthesis and high biomass (Zou et al., 2018; Zou & Yang, 2019a; Zou & Yang, 2019b). As a potential non-edible energy crop, jatropha produces high level of fossil fuel-like oil in its seeds, which can be easily processed into biodiesel (Fairless, 2007; Berchmans & Hirata, 2008; Kumar & Sharma, 2008; Maghuly & Laimer, 2013). Additionally, this species also has several unique characteristics like easy propagation, rapid growth, and adaptation to semiarid and barren soil environments (Montes & Melchinger, 2016). Although originated from Mesoamerica, jatropha can now be widely found in many tropical and subtropical countries of Africa and Asia (Wu et al., 2015; Li et al., 2017). Nevertheless, its commercial cultivation has failed mainly due to low productivity (Montes & Melchinger, 2016; Mazumdar et al., 2018). Thereby, uncovering the molecular mechanism underlying and characterization of genes involved in yield formation are prerequisites. In this study, we would like to present a first genome-wide analysis of the Lhc supergene family in jatropha, including gene structures, chromosome (Chr) locations, evolutionary relationships, sequence characteristics, global expression profiles as well as comprehensive comparison with arabidopsis, cassava, and castor. These results will not only improve our knowledge on species-specific evolution of the Lhc supergene family, but also provide valuable information for further functional analysis in jatropha.
Materials & Methods
Identification of Lhc superfamily genes
As shown in Table S1, 34 AtLhc superfamily genes were retrieved from TAIR (https://www.arabidopsis.org/, Araport11) according to previous literatures. To facilitate evolutionary analysis, 28 and 35 superfamily members present in castor and cassava (see Table S1), two Euphorbiaceous plants, were also obtained from Phytozome (Version 12, https://phytozome.jgi.doe.gov/pz/portal.html). Homologs present in the jatropha genome (Wu et al., 2015) were identified via the tBLASTn (Altschul et al., 1997; E-value, 1e–5) search by using above protein sequences as queries. Gene models of candidates were revised via aligning mRNA to loci-encoding scaffolds. Presence of the conserved CB domain (PF00504) was checked using MOTIF Search (https://www.genome.jp/tools/motif/), and exon-intron structures were displayed using Gene Structure Display Server (GSDS 2.0, https://gsds.cbi.pku.edu.cn/). Putative transmembrane helix (TMH) was predicted using CCTOP (http://cctop.enzim.ttk.mta.hu/) as well as sequence alignment. Chloroplast transit peptide (TP) of deduced proteins and biochemical parameters of mature peptides were determined using ChloroP (Version 1.1, https://www.cbs.dtu.dk/services/ChloroP/) and ProtParam (https://web.expasy.org/protparam/), respectively.
Chromosome location and synteny analysis
Gene distribution on chromosomes was analyzed using MAPchart 2.3 (Voorrips, 2002). For synteny analysis, the all-to-all BLASTP method was used to identify duplicate pairs, and MicroSyn (Cai et al., 2011) was used to detect microsynteny. Orthologs across different species were inferred from the best-reciprocal-hit (BRH)-based BLAST analysis as well as synteny analysis for jatropha and castor.
Sequence alignment, phylogenetic, and conserved motif analyses
Multiple sequence alignment was carried out using MUSCLE (Edgar, 2004). Phylogenetic tree construction was performed using MEGA7 (Kumar, Stecher & Tamura, 2016) with the maximum likelihood method (bootstrap: 1,000). Conserved motifs were identified using MEME (https://meme-suite.org/tools/meme): any number of repetitions; maximum number of motifs, 25; minimum sites, 2; and, the optimum width of each motif, between 6 and 100 residues.
Gene expression analysis
Transcriptome datasets used for expression profiling are shown in Table S2. Except for tissue-specific transcriptomes, other samples were performed for at least two biological replicates. Quality control of raw reads was carried out using fastQC (https://www. bioinformatics.babraham.ac.uk/projects/fastqc/). Read mapping were performed using Bowtie 2 (Langmead & Salzberg, 2012), and the relative transcript level of each gene was presented as FPKM (fragments per kilobase of exon per million fragments mapped, for pair-ended samples) or RPKM (Reads per kilobase per million mapped reads, for single-ended samples) (Mortazavi et al., 2008). RSEM (v1.2.27) (Li & Dewey, 2011) with parameters “log2Ratio ≥ 1” and “FDR <0.001” were used to determine differentially expressed genes.
Results
Identification and chromosome locations of 27 Lhc superfamily genes in jatropha
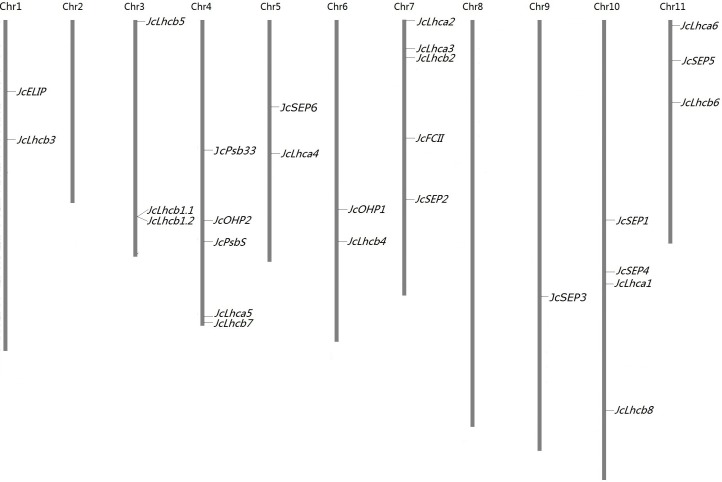
The BLAST search resulted in 27 JcLhc superfamily genes from the jatropha genome (Wu et al., 2015), which represent four previously defined families (i.e., Lhc, Lil, PsbS, and FCII) or eight subfamilies (i.e., Lhca, Lhcb, PsbS, OHP, SEP, ELIP, Psb33, and FCII). Each subfamily contains one to nine members that were named after their orthologs in castor (see below), i.e., JcLhca1–6, JcLhcb1.1–1.2, JcLhcb2–8, JcPsbS, JcELIP, JcOHP1–2, JcSEP1–6, JcPsb33, and JcFCII, respectively. These genes were shown to distribute over 25 scaffolds. Although most scaffolds contain a single member, two of them harbor two, i.e., scaffold160 (i.e., JcLhcb1.1 and JcLhcb1.2) and scaffold211 (i.e., JcLhca6 and JcSEP5) (Table 1). With the help of 1,208 available genetic markers, these genes were further anchored to nine chromosomes, and the gene number of each chromosome varies from one to five (Fig. 1).
Table 1. 27 Lhc superfamily genes identified in jatropha.
| Subfamily | Gene name | Locus ID | Scaffold position | Nucleotide length (bp, from start to stop codons) | Intron no. | EST no. | AA | TP length | TMH | Ortholog | OG | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDS | Gene | Rc | Me | At | ||||||||||
| Lhca | JcLhca1 | JCGZ_23938 | scaffold794:74368-75803(−) | 738 | 1,076 | 3 | 0 | 245 | 45 | 3 | RcLhca1 | MeLhca1.1 MeLhca1.2 |
AtLhca1 | Lhca1 |
| JcLhca2 | JCGZ_17961 | scaffold502:3176588-3178724(+) | 813 | 1,491 | 4 | 1 | 270 | 58 | 3 | RcLhca2 | MeLhca2.1 MeLhca2.2 |
AtLhca2 | Lhca2 | |
| JcLhca3 | JCGZ_15032 | scaffold42:178242-179765(−) | 816 | 1,171 | 2 | 8 | 271 | 38 | 3 | RcLhca3 | MeLhca3 | AtLhca3 | Lhca3 | |
| JcLhca4 | JCGZ_11643 | scaffold328:2197266-2195814(−) | 750 | 917 | 2 | 2 | 249 | 48 | 3 | RcLhca4 | MeLhca4.1 MeLhca4.2 |
AtLhca4 | Lhca4 | |
| JcLhca5 | JCGZ_04265 | scaffold159:84206-85704(+) | 795 | 1,255 | 5 | 0 | 264 | 57 | 3 | RcLhca5 | MeLhca5 | AtLhca5 | Lhca5 | |
| JcLhca6 | JCGZ_07509 | scaffold211:3089530-3091440(−) | 792 | 1,612 | 4 | 0 | 263 | 42 | 3 | RcLhca6 | MeLhca6 | AtLhca6 | Lhca6 | |
| Lhcb | JcLhcb1.1 | JCGZ_04588 | scaffold160:101030-101541(+) | 798 | 798 | 0 | 28 | 265 | 35 | 3 | RcLhcb1.1 RcLhcb1.2 RcLhcb1.3 |
MeLhcb1.1 MeLhcb1.2 MeLhcb1.3 |
AtLhcb1.1 AtLhcb1.2 AtLhcb1.3 AtLhcb1.4 AtLhcb1.5 |
Lhcb1 |
| JcLhcb1.2 | JCGZ_04587 | scaffold160:97668-99902(−) | 798 | 798 | 0 | 17 | 265 | 35 | 3 | RcLhcb1.1 RcLhcb1.2 RcLhcb1.3 |
MeLhcb1.1 MeLhcb1.2 MeLhcb1.3 |
AtLhcb1.1 AtLhcb1.2 AtLhcb1.3 AtLhcb1.4 AtLhcb1.5 |
Lhcb1 | |
| JcLhcb2 | JCGZ_18481 | scaffold529:239615-242657(−) | 798 | 2,225 | 1 | 10 | 265 | 37 | 3 | RcLhcb2 | MeLhcb2.1 MeLhcb2.2 |
AtLhcb2.1 AtLhcb2.2 AtLhcb2.3 |
Lhcb2 | |
| JcLhcb3 | JCGZ_00703 | scaffold108:360030-361580(−) | 804 | 1,202 | 2 | 5 | 267 | 44 | 3 | RcLhcb3 | MeLhcb3 | AtLhcb3 | Lhcb3 | |
| JcLhcb4 | JCGZ_25025 | scaffold843:295347-296708(+) | 858 | 957 | 1 | 8 | 285 | 31 | 3 | RcLhcb4 | MeLhcb4 | AtLhcb4.1 AtLhcb4.2 |
Lhcb4 | |
| JcLhcb8 | JCGZ_01281 | scaffold11:738853-741122(+) | 840 | 1,786 | 1 | 0 | 279 | 31 | 3 | RcLhcb8 | MeLhcb8 | AtLhcb8 | Lhcb8 | |
| JcLhcb5 | JCGZ_06701 | scaffold200:160436-162429(−) | 876 | 1,573 | 5 | 1 | 291 | 41 | 3 | RcLhcb5 | MeLhcb5 | AtLhcb5 | Lhcb5 | |
| JcLhcb6 | JCGZ_20203 | scaffold645:375633-376811(+) | 765 | 844 | 1 | 1 | 254 | 49 | 3 | RcLhcb6 | MeLhcb6 | AtLhcb6 | Lhcb6 | |
| JcLhcb7 | JCGZ_08016 | scaffold221:188978-192485(−) | 1,014 | 3,050 | 5 | 0 | 337 | 52 | 3 | RcLhcb7 | MeLhcb7 | AtLhcb7 | Lhcb7 | |
| PsbS | JcPsbS | JCGZ_12094 | scaffold339:857142-859344(−) | 825 | 1,726 | 3 | 2 | 274 | 62 | 4 | RcPsbS | MePsbS | AtPsbS | PsbS |
| ELIP | JcELIP | JCGZ_02231 | scaffold119:1376949-1378080(−) | 585 | 808 | 2 | 6 | 194 | 86 | 3 | RcELIP | MeELIP | AtELIP1 AtELIP2 |
ELIP |
| OHP | JcOHP1 | JCGZ_09105 | scaffold250:2426033-2426931(−) | 357 | 552 | 2 | 2 | 118 | 48 | 1 | RcOHP1 | MeOHP1 | AtOHP1 | OHP1 |
| JcOHP2 | JCGZ_08332 | scaffold224:432321-435041(+) | 552 | 2,339 | 1 | 0 | 183 | 47 | 1 | RcOHP2 | MeOHP2.1 MeOHP2.2 |
AtOHP2 | OHP2 | |
| SEP | JcSEP1 | JCGZ_23193 | scaffold7:432788-436707(−) | 438 | 3,498 | 3 | 3 | 145 | 71 | 2 | RcSEP1 | MeSEP1 | AtSEP1 | SEP1 |
| JcSEP2 | JCGZ_09398 | scaffold255:592063-593949(+) | 582 | 1,322 | 1 | 0 | 193 | 46 | 2 | RcSEP2 | MeSEP2 | AtSEP2 | SEP2 | |
| JcSEP3 | JCGZ_03488 | scaffold137:779011-781250(+) | 780 | 1,919 | 2 | 4 | 259 | 100 | 2 | RcSEP3 | MeSEP3.1 MeSEP3.2 |
AtSEP3.1 AtSEP3.2 |
SEP3 | |
| JcSEP6 | JCGZ_26324 | scaffold906:2486216-2487759(−) | 759 | 932 | 2 | 0 | 252 | 88 | 2 | RcSEP6 | MeSEP6 | – | SEP6 | |
| JcSEP4 | JCGZ_06634 | scaffold20:991705-992601(+) | 570 | 570 | 0 | 0 | 189 | 56 | 2 | RcSEP4 | MeSEP4 | AtSEP4 | SEP4 | |
| JcSEP5 | JCGZ_07816 | scaffold211:5206555-5210867(−) | 447 | 3,909 | 4 | 0 | 148 | 75 | 1 | RcSEP5 | MeSEP5 | AtSEP5 | SEP5 | |
| Psb33 | JcPsb33 | JCGZ_17235 | scaffold5:830004-833169(+) | 867 | 2,803 | 2 | 1 | 288 | 62 | 1 | RcPsb33 | MePsb33.1 MePsb33.2 |
AtPsb33 | Psb33 |
| FCII | JcFCII | JCGZ_24260 | scaffold813:61000-71385(−) | 1,500 | 9,600 | 9 | 0 | 499 | 86 | 1 | RcFCII | MeFCII | AtFCII | FCII |
Notes.
- AA
- amino acid
- At
- Arabidopsis thaliana
- bp
- base pair
- CDS
- coding sequence
- EST
- expressed sequence tag
- Me
- Manihot esculenta
- OG
- orthologous group
- Rc
- Ricinus communis
- TMH
- transmembrane helix
- TP
- transit peptide)
Figure 1. Chromosomal locations of JcLhc superfamily genes.
Chromosome serial numbers are indicated at the top of each chromosome. Chr: chromosome.
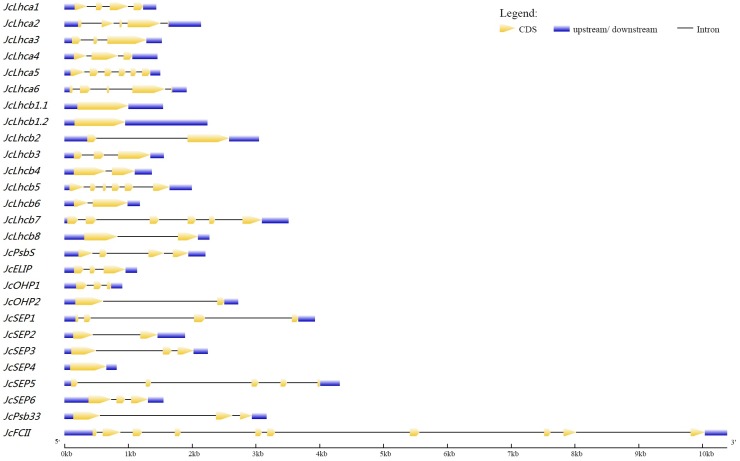
The expression of all JcLhc genes was supported by available Sanger sequencing-derived expressed sequence tags (ESTs) and/or RNA sequencing (RNA-seq), where JcLhcb1.1 harbors the maximum of 28 EST hits. The intron number of these genes varies from zero to nine: approximately 11.11% of genes are intronless, and 29.63%, 22.22%, 11.11%, 11.11%, 11.11% or 3.70% contain two, one, three, four, five and nine introns, respectively (Table 1 and Fig. 2). Similar exon-intron structure was also observed in castor, cassava, and arabidopsis (see Table S1), implying a conserved evolution between these species. The average length of coding sequences (CDS) is about 760 bp, varying from 357 bp of JcOHP1 to 1,500 bp of JcFCII. Compared with CDS, the intron length is relatively more variable, ranging from 79 bp of JcLhcb6 to 8,100 bp of JcFCII and with the average length of 1,259 bp (Table 1 and Fig. 2). The CDS of JcLhcb1.1 and JcLhcb1.2, which are reversely clustered on scaffold160, was shown to exhibit 96.7% identity. Thereby, they are more likely to result from tandem duplication (Zou, Yang & Zhang, 2019; Zou & Yang, 2019a; Zou & Yang, 2019c).
Figure 2. Exon-intron structures of JcLhc superfamily genes.
The graphic representation of the gene models is displayed using GSDS. GSDS: gene structure display server.
Synteny analysis and determination of orthologous groups
Orthologs of JcLhc superfamily genes in castor, cassava, and arabidopsis were further identified by using the BRH method, resulting in 26 orthologous groups (OGs) when the definition was confined to at least one member present in more than two species examined (Table 1). The result is highly consistent with phylogenetic analysis (see below) as well as synteny analysis performed between jatropha and castor, where one-to-one orthologous relationship was observed with exception of the Lhcb1 group with two-to-three (Table 1). Interestingly, RcLhcb1.2, which may originate by dispersed duplication, is located on scaffold30005 together with RcLhca3. However, no collinear gene was found for RcLhcb1.2 in jatropha (see Fig. S1). By contrast, orthologous relationships between jatropha and cassava/arabidopsis are relatively complex, which include one-to-one, one-to-two, one-to-three, and two-to-five, corresponding to one or more recent WGDs occurred in these two species. It is worth noting that, SEP6, a recently identified group that is present in jatropha, castor, and cassava, is absent from arabidopsis (Table 1), implying species/lineage-specific gene loss. Additionally, species-specific gene expansion was also observed: duplicates identified in jatropha (one) and castor (two) were shown to result from tandem or dispersed duplication, respectively; nine duplicates identified in cassava were derived from tandem duplication (three) and whole-genome duplication (WGD) (six); in arabidopsis, four or five duplicates were derived from tandem duplication and WGD, respectively (Table 1 and Table S1).
Although not exactly the same within a family, the exon-intron structure is highly conserved within a certain OG: Lhcb1 and SEP4 are intronless; one-intron-containing groups include Lhcb2/-4/-6/-8, OHP2, and SEP2, whereas two-intron groups include Lhca3/-4, Lhcb3, ELIP, OHP1, SEP3/-6, and Psb33; Lhca1 and SEP1 feature three introns, whereas Lhca2/-6, and SEP5 feature four introns; three groups (i.e., Lhcb5/-7 and Lhca5) contain five introns, whereas only one group (i.e., FCII) harbors nine introns (Fig. 2, Table 1, and Table S1).
Phylogenetic analysis, sequence features, and conserved motifs
As shown in Table 1, the deduced JcLhc superfamily proteins consist of 118–499 amino acids (AA) with one to four TMHs (Fig. S2), and the predicted length of transit peptide ranges from 31 to 100 residues (Table 1). Several physical and chemical parameters of mature peptides were further calculated: the molecular weight (MW) and isoelectric point (pI) values of mature proteins in jatropha range from 7.55 to 46.76 kilodalton (kDa) or from 4.56 to 9.99, respectively; about 81.48% of JcLhc superfamily proteins harbor a pI value of less than 7, which is relatively less than 85.71% in castor, 91.43% in cassava, or 91.18% in arabidopsis; and, about 59.26% of JcLhc superfamily proteins harbor a grand average of hydropathicity (GRAVY) value of less than 0, which is relatively less than 64.29% in castor, 62.86% in cassava, or 73.53% in arabidopsis (Table 1 and Table S1).
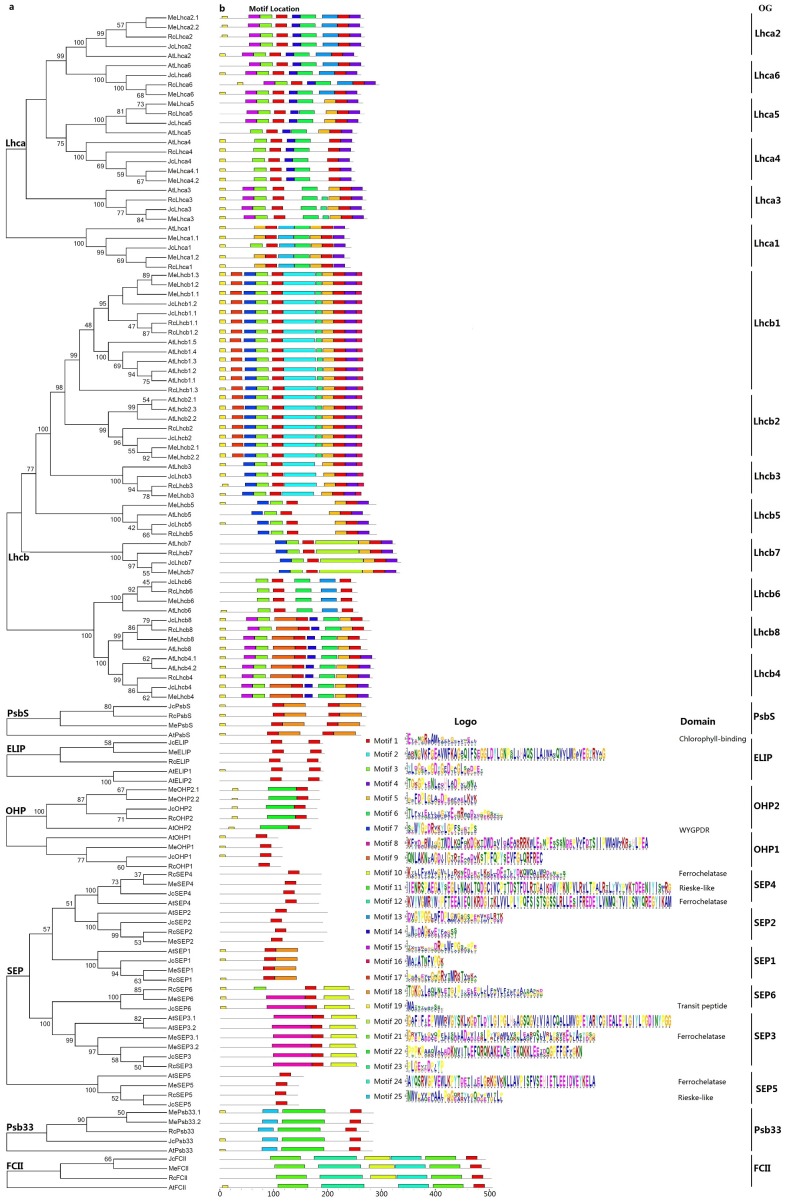
Except for JcPsb33 that contains a CB-like domain (see Fig. S3), all other JcLhc superfamily proteins include the core CB domain of approximately 20 AA (see Table S1). Nevertheless, their overall sequence similarity was shown to be considerably low, even within the conserved Lhc family (ranging from 27.2% to 98.5%, see Table S3). To keep the analysis reliable, an independent phylogenetic tree was constructed for each subfamily by using full-length proteins from jatropha, castor, cassava, and arabidopsis. As shown in Fig. 3A, subfamilies Lhca, Lhcb, OHP, and SEP are clearly clustered into six, eight, two or six phylogenetic groups respectively, corresponding to 22 OGs as described above, i.e., Lhca1–6, Lhcb1–8, OHP1, OHP2, and SEP1–6. Among them, Lhcb8 and SEP6 exhibit a closer relationship with Lhcb4 and SEP3, with a similarity of 79.5% or 55.5%, respectively (Table S3), where JcLhcb8 harbors a relative shorter C-terminal in relation to JcLhcb4 (Fig. S2).
Figure 3. Phylogenetic and conserved motif analyses of jatropha, castor, cassava, and arabidopsis Lhc superfamily proteins.
(A) Phylogenetic analysis of Lhca, Lhcb, PsbS, ELIP, OHP, SEP, Psb33, and FCII subfamilies; (B) Distribution of conserved motifs. Sequence alignment was performed using MUSCLE and unrooted phylogenetic trees were constructed using MEGA7 (maximum likelihood method; bootstrap, 1,000 replicates). Only bootstrap values at nodes supported by a posterior probability of ≥50% are given. The distance scale denotes the number of amino acid substitutions per site. The name of each OG is indicated next to the corresponding group. OG: orthologous group.
To reveal possible divergence of members within a certain OG and between different OGs/(sub)families, conserved motifs were analyzed using MEME. As shown in Fig. 3B and Table 2, motifs are highly variable between subfamilies or even between different evolutionary groups, and considerably more motifs were identified in the Lhc family as compared with PsbS, FCII, and Lil families. Among 25 motifs identified, Motif 1, which is characterized as the CB domain, is widely distributed, including Psb33s. Motif 19, which is characterized as chloroplast transit peptide, is also widely found. Motifs 25 and 11 are characterized as part of the Rieske-like domain (PF13806), where Motif 11 is Psb33-specific and Motif 25 is also present in Lhca1s. The Ferrochelatase domain (PF00762), which is FCII-specific, was shown to include Motifs 21, 12, 10, and 24. Among them, Motif 10 is also present in SEP3s and SEP6s. Motif 7, which includes the WYGPDR/WYGEER domain, is widely found in Lhcb1, −2, − 3, − 5, and −7 groups. WYGPDR has been proven to be essential for trimerization (Hobe et al., 1995; Rogl & Kühlbrandt, 1999), however, experimental evidence is still needed for WYGPD and WYGEER varieties. By contrast, little information is available for other motifs, including several group-specific motifs such as Motifs 8, 9, 18, 20, and 22.
Table 2. Detailed information of 25 motifs identified in this study.
Motifs were identified using MEME.
| Motif | E-value | Sites | Width | Best match |
|---|---|---|---|---|
| Motif 1 | 2.5e−1,988 | 205 | 21 | EJINGRLAMLGFLGFLVQEIL |
| Motif 2 | 6.2e−1,170 | 24 | 60 | ARNGVKFGEAVWFKAGAQIFSEGGLDYLGNPSLIHAQSILAIWACQVVLMGAVEGYRVAG |
| Motif 3 | 9.4e−992 | 69 | 23 | YLDGELPGDYGFDPAGLSADPET |
| Motif 4 | 8.6e−894 | 64 | 21 | TGKGPJENLADHLADPVHNNI |
| Motif 5 | 2.6e−582 | 58 | 21 | GSFDPLGLADDPEAFAELKVK |
| Motif 6 | 3.0e−546 | 40 | 29 | TLFVIELJLIGYVEFRRWADLDNPGSVYP |
| Motif 7 | 1.9e−374 | 32 | 21 | SPWYGPDRVKYLGPFSGETPS |
| Motif 8 | 4.2e−331 | 8 | 73 | KFVDPRWIGGTWDLKQFZKDGKTDWDAVIDAEAKRRKWLEENPESSSNDEPVVFDTSIIPWWAWIKRYHLPEA |
| Motif 9 | 1.4E−216 | 9 | 41 | QNLAKNVAGDIIGTRTEAADVKSTPFQPYSEVFGLQRFREC |
| Motif 10 | 3.5E−205 | 12 | 48 | KTLLFVAVAGVLLIRKNEDIETLKKLLDETTLYDKQWQATWKDZNPSS |
| Motif 11 | 9E−200 | 5 | 80 | IENRSPAEGAYSEGLJNAKLTQDGCIVCPTTDSTFDLRTGAIKDWYPKNPVLRVLTPALRTLYVYPVKTDEENIYISLRG |
| Motif 12 | 1E−170 | 4 | 80 | KVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAM |
| Motif 13 | 8.1E–166 | 13 | 29 | DVGYPGGLWFDPLGWGSGSPEKVKELRTK |
| Motif 14 | 3.6E–159 | 27 | 15 | SWYDAGKVEYFAGSS |
| Motif 15 | 1.8E–154 | 25 | 21 | TVCVKADPDRPLWFPGSTPPE |
| Motif 16 | 5.4E–153 | 24 | 11 | WAYATNFVPGK |
| Motif 17 | 7.6E–149 | 20 | 21 | PSAPEVMGNGRVTMRKTVKKA |
| Motif 18 | 8.6E–142 | 12 | 41 | TGKGLLAQLNJETGJPIYELEPLVLFNVLFALFAAINASKD |
| Motif 19 | 3.6E–140 | 80 | 11 | MATSTLAASSS |
| Motif 20 | 6.1E–140 | 4 | 80 | GAFHFIEPVWWRVGYSKLKGDTLDYLGIPGLHLAGSQGVIVIAICQALLMVGPEYARYCGIEALEPLGIYLPGDINYPGG |
| Motif 21 | 3.4E–142 | 8 | 57 | GPEPLLGVZPFLINLLADPVIERLPYVGAFLVKPLEAFISLVPLPKVEEGLASYGGG |
| Motif 22 | 2E–98 | 5 | 53 | PPPKPAAQVALDDKNVITLEFQRQKAKELQEYFKQKKLEETDQGPFFGFLGKN |
| Motif 23 | 4.3E–98 | 23 | 11 | PLGEVTDPJYP |
| Motif 24 | 4.6E–92 | 4 | 57 | AYQSRVGPVEWLKPYTDETIIELGRKGVKNLLAVPISFVSEHIETLEEIDVEYKELA |
| Motif 25 | 1.4E–106 | 10 | 29 | NWVPAVPLAALPGGZATYJGQPVPTGLLP |
Although these motifs are usually conserved within a certain OG, species-specific gain or loss was also observed. For example, the conserved Motifs 15, 23, and 10 are absent from AtLhca5, AtLhca3 or AtFCII, respectively, whereas the widely present Motif 8 in SEP3s and SEP6s is replaced by Motif 3 in MeSEP6 (Fig. 3).
Expression profiles of JcLhc superfamily genes
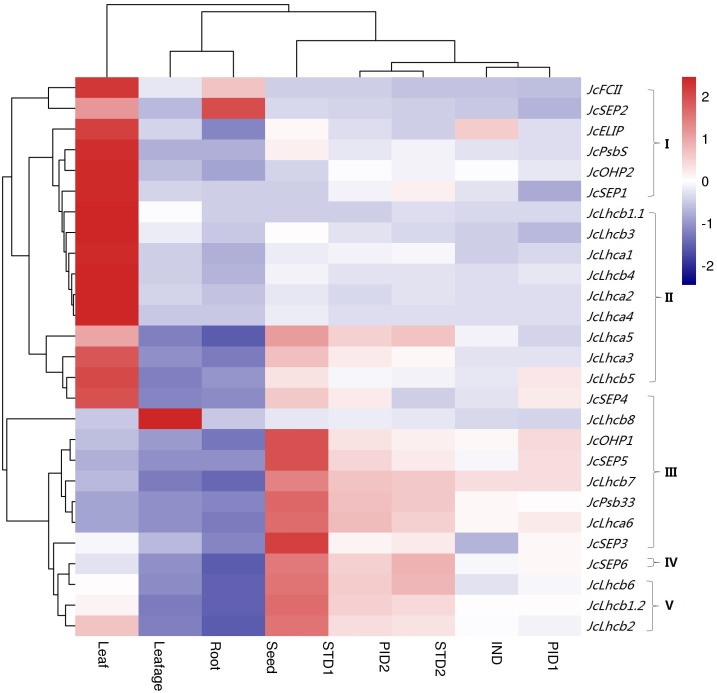
Global gene expression profiles were investigated in various tissues, i.e., root (from 15-day-old seedlings), leafage (from 4-year-old plants, half expanded), leaf (mature leaf, fully expanded), IND (undifferentiated inflorescence of 0.5 cm diameter), PID1 (female flower with carpel primordia beginning to differentiate), PID2 (female flower with three distinct carpels formed), STD1 (male flower with stamen primordia beginning to differentiate), STD2 (male flower with ten complete stamens formed), and developing seed (19–28 days after pollination). Despite the expression of all identified genes, their transcript levels are highly variable over different tissues. As shown in Fig. 4, the majority of JcLhc superfamily genes are predominantly expressed in leaf, and the total transcript level of the whole superfamily in STD1, PID2, STD2, PID1, leafage, root, and seed accounts for 42.82%, 24.26%, 23.71%, 21.41%, 21.16%, 18.01%, 5.83%, or 1.75% of that in leaf, respectively. According to tissue-specific expression patterns, JcLhc superfamily genes can be divided into five main clusters: Cluster I is mostly expressed in leafage, including JcPsbS, JcFCII, and four Lils (i.e., JcOHP2, JcSEP1, JcSEP2, and JcELIP); Clusters II and III that contain the vast majority of Lhc family members and include approximately 63.00% of the whole superfamily are predominantly expressed in mature leaf, where Cluster III is also highly abundant in leafage (i.e., JcLhca6, JcLhcb7, JcLhcb8, JcOHP1, JcSEP3, JcSEP4, JcSEP5, and JcPsb33); Cluster IV that only includes JcSEP6 is preferentially expressed in root, whereas Cluster V is typically expressed in STD1 (i.e., JcLhcb1.2, JcLhcb2, and JcLhcb6) (Fig. 4). It is worth noting that, JcLhcb1.1 represents the most expressed gene in most tissues examined, implying its key roles.
Figure 4. Tissue-specific expression profiles of JcLhc superfamily genes.
Color scale represents FPKM normalized log10 transformed counts where navy indicates low expression and firebrick3 indicates high expression. FPKM: fragments per kilobase of exon per million fragments mapped; IND, undifferentiated inflorescence of 0.5 cm diameter; PID1, female flower with carpel primordia beginning to differentiate; PID2, female flower with three distinct carpels formed; STD1, male flower with stamen primordia beginning to differentiate; STD2, male flower with ten complete stamens formed.
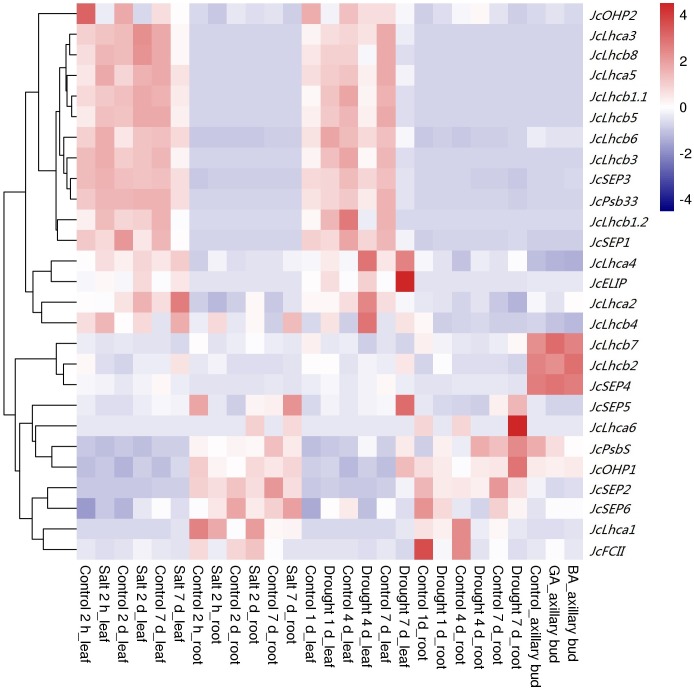
Given drought and salt are two of the most important abiotic stresses affecting plant growth and development, photosynthesis, and crop yield, we thereby investigated the response patterns of JcLhc superfamily genes post drought or salt treatment in leaves and roots of eight-week-old seedlings. After withholding irrigation for 1, 4 or 7 d, the total superfamily transcripts in roots were not significantly changed, by contrast, initial increase followed by significant decrease were observed in leaves. The result is consistent with the fact that the net photosynthesis rate (Pn) and stomatal conductance had decreased to 80% or 20% of those in the control after the start of the stress treatment for 2 and 7 d, respectively (Zhang et al., 2015). For 1 d, seven or three genes were significantly regulated in leaves and roots, respectively. Among them, JcLhcb1.1, JcLhcb1.2, JcLhcb3, JcELIP, JcSEP2, and JcSEP5 were upregulated in leaves, whereas JcOHP2 was downregulated; JcLhcb3 and JcFCII were downregulated in roots, whereas JcPsbS was upregulated. For 4 d, ten or seven genes were significantly regulated in leaves and roots, respectively. Among them, JcLhca4, JcLhcb2, JcLhcb4, and JcELIP were upregulated in leaves, whereas JcLhcb1.1, JcLhcb1.2, JcLhcb3, JcLhcb5, JcLhcb7, and JcLhcb8 were downregulated; JcLhca3, JcLhca4, JcLhcb3, JcLhcb8, and JcPsbS were upregulated in roots, whereas JcLhca1 and JcFCII were downregulated. For 7 d, 19 or seven genes were significantly regulated in leaves and roots, respectively. Among them, JcLhca4, JcLhcb2, JcPsbS, JcELIP, JcOHP1, JcSEP2, and JcSEP5 were upregulated in leaves, whereas JcLhca3, JcLhca5, JcLhcb1.1, JcLhcb1.2, JcLhcb3, JcLhcb5, JcLhcb6, JcLhcb8, JcSEP1, JcSEP3, JcSEP4, and JcPsb33 were downregulated; JcLhca1, JcLhcb1.1, JcLhcb1.2, JcLhcb3, JcLhcb5, and JcFCII were downregulated in roots, whereas JcOHP1 was upregulated (Fig. 5).
Figure 5. Expression profiles of JcLhc superfamily genes upon drought, salt, BA, or GA treatments.
Color scale represents RPKM normalized log10 transformed counts where navy indicates low expression and firebrick3 indicates high expression. RPKM, Reads per kilobase per million mapped reads.
Similar to drought treatment, after applying 100 mM NaCl for 2 h, 2 d or 7 d, gradual downregulation of total transcripts was only observed in leaves. For 2 h, five or three genes were significantly regulated in leaves and roots, respectively. Among them, JcLhcb2, JcOHP2, JcSEP2, and JcFCII were downregulated in leaves, whereas JcLhcb1.2 was upregulated; in roots, JcLhcb8, JcSEP5, and JcFCII were downregulated. For 2 d, six genes were significantly regulated in both leaves and roots, respectively. Among them, JcELIP, JcSEP2, JcSEP4, and JcFCII were upregulated in leaves, whereas JcOHP2 and JcSEP1 were downregulated; JcLhca1, JcLhca2, JcLhcb8, and JcSEP5 were upregulated in roots, whereas JcLhcb1.1 and JcLhcb1.2 were downregulated. For 7 d, nine or seven genes were significantly regulated in both leaves and roots, respectively. Among them, JcLhca3, JcLhcb1.1, JcLhcb1.2, JcLhcb3, JcLhcb5, JcLhcb8, JcSEP1, and JcSEP4 were downregulated in leaves, whereas JcLhcb2 was upregulated; JcLhca3, JcLhcb1.2, JcLhcb3, JcLhcb5, and JcSEP5 were downregulated in roots, whereas JcELIP and JcFCII were upregulated (Fig. 5). Downregulation of most regulated genes is highly consistent with gradual decrease of Pn, where the Pn values of 2 and 7 d after stress treatment accounted for 83% or 50% of the control, respectively (Zhang et al., 2014).
Responses to gibberellin acid (GA) and 6-benzylaminopurine (BA) treatments were also examined in young axillary buds. Application of 10 µM GA for 12 h resulted in one upregulated (i.e., JcFCII) and two downregulated (i.e., JcSEP2 and JcSEP5) genes, in contrast, no evident effect was observed for the same concentration of BA (Fig. 5).
Discussion
In green plants, the Lhc superfamily is consisted of four antennae protein families that play essential roles in light-harvesting and photoprotection (Jansson, 1999; Klimmek et al., 2006; Zou, 2018). Despite their importance, extensive research is still limited to the model plant arabidopsis and few other species such as Chlamydomonas reinhardtii, Physcomitrella patens, castor, and cassava (Elrad & Grossman, 2004; Klimmek et al., 2006; Alboresi et al., 2008; Engelken, Brinkmann & Adamska, 2010; Zou, Huang & An, 2013; Zou & Yang, 2019a). Among them, it’s well established that cassava and arabidopsis experienced one or two additional WGDs after the so-called γ hexaploidization event shared by all core eudicots: the recent WGD occurred in cassava is called ρ, which was estimated to occur within a window of 39–47 million years ago (Mya) (Bredeson et al., 2016; Zou, Yang & Zhang, 2019; Zou & Yang, 2019a; Zou & Yang, 2019b; Zou & Yang, 2019c), whereas two recent WGDs occurred in arabidopsis are known as β and α, which were estimated to occur within a window of 61–65 or 23–50 Mya, respectively (Bowers et al., 2003). From this point of view, analysis of species without recent WGDs may improve our knowledge on species-specific evolution of this special gene family. Jatropha, another economically Euphorbiaceous plant for potential biodiesel purpose, is a good candidate for such study. According to comparative genomics analysis, jatropha may share a common ancestor with cassava and castor at approximately 65 Mya (Bredeson et al., 2016), and no additional WGD occurred after their divergence.
In the present study, a first genome-wide identification and global analysis of Lhc superfamily genes were performed in jatropha, and the superfamily number of 27 members identified in this species is comparable to 28 reported in castor but relatively less than 35 or 34 present in cassava and arabidopsis, respectively (Klimmek et al., 2006; Engelken, Brinkmann & Adamska, 2010; Zou, Huang & An, 2013; Zou & Yang, 2019a). Nevertheless, 26 OGs representing four families (i.e., Lhc, Lil, PsbS, and FCII) were found in jatropha as observed in castor and cassava (Zou, Huang & An, 2013; Zou & Yang, 2019a). Few recent duplicates were identified in jatropha as well as castor, corresponding to no recent WGD occurred in these two species (Chan et al., 2010; Wu et al., 2015). Compared with castor that contains two dispersed duplicates, only one duplicate derived from tandem duplication was found in jatropha. By contrast, considerably more duplicates, i.e., nine, were identified in both cassava and arabidopsis (Table S1), reflecting the occurrence of one or two recent WGDs (Bowers et al., 2003; Bredeson et al., 2016). Despite having the same number of duplicates, cassava contains relatively more WGD duplicates (6 vs 5) but less local duplicates (3 vs 4), implying species-specific evolution pattern. Interestingly, duplicates in both jatropha and castor are confined to the Lhcb1 group, however, in cassava and arabidopsis, local duplicates were also found in the Lhcb2 group (Table S1).
Among four families identified, both PsbS and FCII include a single member in four species examined in this study. By contrast, Lhc and Lil families are relatively complex. The Lhc family contains 14 OGs representing two subfamilies (i.e., Lhca and Lhcb), whereas the Lil family includes ten OGs representing four subfamilies (i.e., ELIP, OHP, SEP, and Psb33). According to crystal analyses, the Lhc family members usually contain three alpha-helixes, whereas the PsbS family features four (Kühlbrandt, Wang & Fujiyoshi, 1994; Pan et al., 2011; Fan et al., 2015). By contrast, one to three helixes were shown to be present in Lil proteins, i.e., one for OHPs and Psb33s, two for SEPs, and three for ELIPs (Jansson, 1999; Engelken, Brinkmann & Adamska, 2010; Fristedt et al., 2015; Beck et al., 2017). Similar results were also observed in this study, however, both JcSEP5 and JcFCII were shown to contain a single helix.
It is noteworthy that, among 26 OGs identified, SEP6 is absent from arabidopsis. SEP6 exhibits about 41.7%, 40.5% or 39.0–40.3% sequence identity with SEP3 in jatropha, castor and cassava respectively, implying their early divergence and species-specific gene loss. Indeed, SEP6 orthologs were broadly found in dicots, including Carica papaya and Aquilegia coerulea (Zou & Yang, 2019a). Additionally, Lhcb8 shows approximately 72.0%, 69.9%, 72.6% or 64.2–65.3% identity with Lhcb4 in jatropha, castor, cassava, and arabidopsis, respectively. Lhcb8 is widely present in core eudicots but not in A. coerulea and monocots, suggesting its recent origin. According to synteny analysis performed in arabidopsis, Lhcb8 is more likely to be a duplicate of Lhcb4 generated along the γ event (Bowers et al., 2003; Wang, Tan & Paterson, 2013).
Potential roles of JcLhc superfamily genes could be inferred from their expression patterns and function-characterized orthologs in arabidopsis and other species. According to GO annotation, they belong to thylakoid membrane proteins that have activity of chlorophyll binding, pigment binding, xanthophyll binding, lipid binding, protein binding, iron-sulfur cluster binding, oxidoreductase, ferrochelatase as summarized in Table S4. Our transcriptional profiling not only supports the expression of all 27 JcLhc superfamily genes identified in this study, but also reveals key genes in a certain tissue, development stage or environment condition. Similar to that reported in arabidopsis (Jansson, 1999; Klimmek et al., 2006), genes encoding JcLhca1 to −4 and JcLhcb1 to −6, which are characterized as abundant Lhc proteins, were highly expressed in most examined jatropha tissues, especially in mature leaf. By contrast, four genes encoding so-called rare Lhc proteins (i.e., JcLhca5, JcLhca6, JcLhcb7, and JcLhcb8) are lowly expressed, exhibiting a similar expression pattern to members of Lil, PsbS, and FCII families. The result is consistent with our cluster analysis, which divides JcLhc superfamily genes into five clusters named I, II, III, IV, and V. These genes encoding abundant Lhc proteins belong to Clusters II and V, whereas rare Lhc genes were divided into Cluster III with the exception of JcLhca5. Clustering JcLhca5 into Cluster II is due to its leaf-preferential expression pattern, but not the transcript level. Compared with mature leaf, leafage is considerably more sensitive to high light and other stresses. This is not surprising that the majority of members in Lil, PsbS, and FCII families are highly expressed in this special tissue, comprising Clusters I and III. It is worth noting that, JcSEP6, the unique member in Clusters IV, is lowly expressed in most examined tissues with the exception of root. As a recently identified superfamily member, the detailed function of SEP6 still needs to be investigated. Furthermore, most JcLhc superfamily genes were regulated by drought and/or salt, two most important abiotic stresses affecting crop growth and yield (Zhang et al., 2014; Zhang et al., 2015). As expected, more genes are downregulated, especially for the leaf tissue, corresponding to decrease of transpiration rate and stomatal conductance (Zhang et al., 2015). Nevertheless, frequent upregulation of certain members was also observed, i.e., JcLhca4, JcELIP, JcPsbS, JcSEP2, JcSEP5, and JcOHP1. Interestingly, JcLhca2 and JcLhca5 exhibit distinct responses upon drought or salt stress, whereas JcLhca2 was specifically regulated by salt and JcLhca4, JcLhcb4, JcLhcb6, JcLhcb7, JcPsbS, JcOHP1, JcSEP2 and JcPsb33 were only regulated by drought. The involvement of Lhc superfamily genes in stress response has been well documented in arabidopsis and other species, including high light, chloroplast retrograde signal, oxidative stress, abscisic acid, etc. (Montané & Kloppstech, 2000; Staneloni, Rodriguez-Batiller & Casal, 2008; Gerotto et al., 2011; Tibiletti et al., 2016). For example, ELIPs, as the name suggests, are induced by early and high light, as well as other stresses such as UV-B, cold, heat, drought, salt, hypoxia, and anoxia described in this study and elsewhere (Heddad et al., 2006; Hayami et al., 2015). In arabidopsis, knockout and overexpression of ELIPs resulted in decreased chlorophyll levels (Casazza et al., 2005; Tzvetkova-Chevolleau et al., 2007). Another high-light induced gene, PsbS, acts as the main sensor of the low pH in plants and plays an essential role in nonphotochemical quenching (Bergantino et al., 2003; Li et al., 2004; Gerotto et al., 2011; Fan et al., 2015; Tibiletti et al., 2016). OHPs, which are related to HLIPs in cyanobacteria, are essential for the formation of the PSII reaction center. In arabidopsis, mutations in AtOHP1 or AtOHP2 caused severe growth deficits, reduced pigmentation, and disturbed thylakoid architecture (Beck et al., 2017; Hey & Grimm, 2018; Myouga et al., 2018; Li et al., 2019). By contrast, two plant hormones (i.e., BA and GA), which can improve shoot branching after application to young axillary buds (Ni et al., 2017), had little effect on transcriptional regulation of JcLhc superfamily genes.
Conclusion
This study presents a first genomics analysis of the Lhc supergene family in jatropha, resulting in 27 members that are distributed across nine out of 11 chromosomes. Despite a relatively smaller number of members, 26 orthologous groups representing four families were found, where SEP6 represents a novel group that has been lost in the model plant arabidopsis. Nearly one-to-one orthologous relationship was observed between jatropha and castor, however, species-specific gene expansion was observed in these two species as well as cassava and arabidopsis. Exon-intron structures, protein motifs, and expression profiles of JcLhc superfamily genes were also analyzed and discussed. These findings provide valuable information for further studies in jatropha and species beyond.
Supplemental Information
The chromosome serial numbers are indicated at the top of each chromosome, and 27 RcLhc superfamily genes are shown just behind their collinear genes in jatropha (where RcLhcb1.2 with no collinear gene in jatropha). (Chr: chromosome; Jc: Jatropha curcas; Rc: Ricinus communis).
(A) Sequence alignment of the JcLhc family; (B) Sequence alignment of the JcLil family; (C) Sequence alignment of JcPsbS with RcPsbS, MePsbS, and AtPsbS; (D) Sequence alignment of JcFCII with RcFCII, MeFCII, and AtFCII. Sequence alignment was performed using MUSCLE and predicted TMHs were underlined. (TMH: transmembrane helix).
Sequence alignment was performed using MUSCLE. The predicted chloroplast transit peptide and Lhc motif-bearing TMH region were boxed. (TMH: transmembrane helix).
1 Conserved domains were predicted using MOTIF (CB, Chloroa_b-bind, PF00504; Rieske-like, PF00355; Ferrochelatase, PF00762). 2 Chloroplast transit peptide was predicted using ChloroP. 3 Duplicated modes in arabidopsis were determined based on studies of Wang, Tan & Paterson (2013) and related references therein. (AA, amino acid; CB, chlorophyll a/b-binding; GRAVY, grand average of hydropathicity; p I, isoelectric point; kDa, kilodalton; MW, molecular weight; TP, transit peptide; TMH, transmembrane helix; WGD, whole-genome duplication).
Acknowledgments
The authors appreciate those contributors who make the related genome and transcriptome data accessible in public databases. The authors also thank the editor and two reviewers for their helpful suggestions.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31700580 and 31971688), the Natural Science Foundation of Hainan province (319MS093), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630022019017 and 1630052017011), and the Research Fund of Guangdong University of Petrochemical Technology (2018rc55). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Yongguo Zhao performed the experiments, analyzed the data, and approved the final draft.
Hua Kong and Yunling Guo performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Zhi Zou conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The data is available in the Supplementary Files.
References
- Alboresi et al. (2008).Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLOS ONE. 2008;3(4):e2033. doi: 10.1371/journal.pone.0002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul et al. (1997).Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck et al. (2017).Beck J, Lohscheider JN, Albert S, Andersson U, Mendgen KW, Rojas-Stütz MC, Adamska I, Funck D. Small one-helix proteins are essential for photosynthesis in Arabidopsis. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.00007. Article 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchmans & Hirata (2008).Berchmans HJ, Hirata S. Biodiesel production from crude Jatropha curcas L. seed oil with a high content of free fatty acids. Bioresource Technology. 2008;99(6):1716–1721. doi: 10.1016/j.biortech.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Bergantino et al. (2003).Bergantino E, Segalla A, Brunetta A, Teardo E, Rigoni F, Giacometti GM, Szabò I. Light- and pH-dependent structural changes in the PsbS subunit of photosystem II. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):15265–15270. doi: 10.1073/pnas.2533072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers et al. (2003).Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422(6930):433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Bredeson et al. (2016).Bredeson JV, Lyons JB, Prochnik SE, Wu GA, Ha CM, Edsinger-Gonzales E, Grimwood J, Schmutz J, Rabbi IY, Egesi C, Nauluvula P, Lebot V, Ndunguru J, Mkamilo G, Bart RS, Setter TL, Gleadow RM, Kulakow P, Ferguson ME, Rounsley S, Rokhsar DS. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology. 2016;34(5):562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- Cai et al. (2011).Cai B, Yang X, Tuskan GA, Cheng ZM. MicroSyn: a user friendly tool for detection of microsynteny in a gene family. BMC Bioinformatics. 2011;12:79. doi: 10.1186/1471-2105-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casazza et al. (2005).Casazza AP, Rossini S, Rosso MG, Soave C. Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Molecular Biology. 2005;58(1):41–51. doi: 10.1007/s11103-005-4090-1. [DOI] [PubMed] [Google Scholar]
- Chan et al. (2010).Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, Cahoon EB, Gedil M, Stanke M, Haas BJ, Wortman JR, Fraser-Liggett CM, Ravel J, Rabinowicz PD. Draft genome sequence of the oilseed species Ricinus communis. Nature Biotechnology. 2010;28(9):951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar (2004).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrad & Grossman (2004).Elrad D, Grossman AR. A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Current Genetics. 2004;45(2):61–75. doi: 10.1007/s00294-003-0460-x. [DOI] [PubMed] [Google Scholar]
- Engelken, Brinkmann & Adamska (2010).Engelken J, Brinkmann H, Adamska I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evolution Biology. 2010;10:233. doi: 10.1186/1471-2148-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairless (2007).Fairless D. Biofuel: the little shrub that could—maybe. Nature. 2007;449(7163):652–655. doi: 10.1038/449652a. [DOI] [PubMed] [Google Scholar]
- Fan et al. (2015).Fan M, Li M, Liu Z, Cao P, Pan X, Zhang H, Zhao X, Zhang J, Chang W. Crystal structures of the PsbS protein essential for photoprotection in plants. Nature Structural & Molecular Biology. 2015;22(9):729–735. doi: 10.1038/nsmb.3068. [DOI] [PubMed] [Google Scholar]
- Fristedt et al. (2015).Fristedt R, Herdean A, Blaby-Haas CE, Mamedov F, Merchant SS, Last RL, Lundin B. PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiology. 2015;167(2):481–492. doi: 10.1104/pp.114.253336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto et al. (2011).Gerotto C, Alboresi A, Giacometti GM, Bassi R, Morosinotto T. Role of PSBS and LHCSR in Physcomitrella patens acclimation to high light and low temperature. Plant Cell & Environment. 2011;34:922–932. doi: 10.1111/j.1365-3040.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- Hayami et al. (2015).Hayami N, Sakai Y, Kimura M, Saito T, Tokizawa M, Iuchi S, Kurihara Y, Matsui M, Nomoto M, Tada Y, Yamamoto YY. The responses of Arabidopsis early light-induced protein2 to ultraviolet B, high light, and cold stress are regulated by a transcriptional regulatory unit composed of two elements. Plant Physiology. 2015;169(1):840–855. doi: 10.1104/pp.15.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad et al. (2006).Heddad M, Norén H, Reiser V, Dunaeva M, Andersson B, Adamska I. Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiology. 2006;142(1):75–87. doi: 10.1104/pp.106.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey & Grimm (2018).Hey D, Grimm B. Requirement of ONE-HELIX PROTEIN 1 (OHP1) in early Arabidopsis seedling development and under high light intensity. Plant Signaling & Behavior. 2018;29:1–3. doi: 10.1080/15592324.2018.1550317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey et al. (2017).Hey D, Rothbart M, Herbst J, Wang P, Müller J, Wittmann D, Gruhl K, Grimm B. LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiology. 2017;174(2):1037–1050. doi: 10.1104/pp.17.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe et al. (1995).Hobe S, Förster R, Klingler J, Paulsen H. N-proximal sequence motif in light-harvesting chlorophyll a/b-binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry. 1995;34(32):10224–10228. doi: 10.1021/bi00032a016. [DOI] [PubMed] [Google Scholar]
- Jansson (1999).Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends in Plant Science. 1999;4(6):236–240. doi: 10.1016/S1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Klimmek et al. (2006).Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiology. 2006;140(3):793–804. doi: 10.1104/pp.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol et al. (2007).Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiology. 2007;143(4):1802–1816. doi: 10.1104/pp.106.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt, Wang & Fujiyoshi (1994).Kühlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367(6464):614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- Kumar & Sharma (2008).Kumar A, Sharma S. An evaluation of a multipurpose oil seed crop for industrial uses (Jatropha curcas L): a review. Industrial Crops & Products. 2008;28:1–10. doi: 10.1016/j.indcrop.2008.01.001. [DOI] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead & Salzberg (2012).Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Dewey (2011).Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2017).Li H, Tsuchimoto S, Harada K, Yamasaki M, Sakai H, Wada N, Alipour A, Sasai T, Tsunekawa A, Tsujimoto H, Ando T, Tomemori H, Sato S, Hirakawa H, Quintero VP, Zamarripa A, Santos P, Hegazy A, Ali AM, Fukui K. Genetic tracing of Jatropha curcas L. from its mesoamerican origin to the world. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.01539. Article 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2004).Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. Journal of Biological Chemistry. 2004;279:22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- Li et al. (2019).Li Y, Liu B, Zhang J, Kong F, Zhang L, Meng H, Li W, Rochaix JD, Li D, Peng L. OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiology. 2019;179(1):195–208. doi: 10.1104/pp.18.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghuly & Laimer (2013).Maghuly F, Laimer M. Jatropha curcas, a biofuel crop: functional genomics for understanding metabolic pathways and genetic improvement. Biotechnology Journal. 2013;8:1172–1182. doi: 10.1002/biot.201300231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar et al. (2018).Mazumdar P, Singh P, Babu S, Siva R, Harikrishna JA. An update on biological advancement of Jatropha curcas L.: new insight and challenges. Renewable & Sustainable Energy Reviews. 2018;91:903–917. doi: 10.1016/j.rser.2018.04.082. [DOI] [Google Scholar]
- Montané & Kloppstech (2000).Montané MH, Kloppstech K. The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene. 2000;258(1–2):1–8. doi: 10.1016/S0378-1119(00)00413-3. [DOI] [PubMed] [Google Scholar]
- Montes & Melchinger (2016).Montes JM, Melchinger AE. Domestication and breeding of Jatropha curcas L. Trends in Plant Science. 2016;21:1045–1057. doi: 10.1016/j.tplants.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Mortazavi et al. (2008).Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Myouga et al. (2018).Myouga F, Takahashi K, Tanaka R, Nagata N, Kiss AZ, Funk C, Nomura Y, Nakagami H, Jansson S, Shinozaki K. Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiology. 2018;176(3):2277–2291. doi: 10.1104/pp.17.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni et al. (2017).Ni J, Zhao ML, Chen MS, Pan BZ, Tao YB, Xu ZF. Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6-benzyladenine in Jatropha curcas. Scientific Reports. 2017;7(1):11417. doi: 10.1038/s41598-017-11588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan et al. (2011).Pan X, Li M, Wan T, Wang L, Jia C, Hou Z, Zhao X, Zhang J, Chang W. Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nature Structural & Molecular Biology. 2011;18(3):309–315. doi: 10.1038/nsmb.2008. [DOI] [PubMed] [Google Scholar]
- Rogl & Kühlbrandt (1999).Rogl H, Kühlbrandt W. Mutant trimers of light-harvesting complex II exhibit altered pigment content and spectroscopic features. Biochemistry. 1999;38(49):16214–16222. doi: 10.1021/bi990739p. [DOI] [PubMed] [Google Scholar]
- Staneloni, Rodriguez-Batiller & Casal (2008).Staneloni RJ, Rodriguez-Batiller MJ, Casal JJ. Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Molecular Plant. 2008;1(1):75–83. doi: 10.1093/mp/ssm007. [DOI] [PubMed] [Google Scholar]
- Tibiletti et al. (2016).Tibiletti T, Auroy P, Peltier G, Caffarri S. Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiology. 2016;171(4):2717–2730. doi: 10.1104/pp.16.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau et al. (2007).Tzvetkova-Chevolleau T, Franck F, Alawady AE, Dall’Osto L, Carrière F, Bassi R, Grimm B, Nussaume L, Havaux M. The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant Journal. 2007;50(5):795–809. doi: 10.1111/j.1365-313X.2007.03090.x. [DOI] [PubMed] [Google Scholar]
- Voorrips (2002).Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang, Tan & Paterson (2013).Wang Y, Tan X, Paterson AH. Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics. 2013;14:652. doi: 10.1186/1471-2164-14-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2015).Wu P, Zhou C, Cheng S, Wu Z, Lu W, Han J, Chen Y, Chen Y, Ni P, Wang Y, Xu X, Huang Y, Song C, Wang Z, Shi N, Zhang X, Fang X, Yang Q, Jiang H, Chen Y, Li M, Wang Y, Chen F, Wang J, Wu G. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant Journal. 2015;81(5):810–821. doi: 10.1111/tpj.12761. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang C, Zhang L, Zhang S, Zhu S, Wu P, Chen Y, Li M, Jiang H, Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to drought stress. BMC Plant Biology. 2015;15:17. doi: 10.1186/s12870-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang L, Zhang C, Wu P, Chen Y, Li M, Jiang H, Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLOS ONE. 2014;9(5):e97878. doi: 10.1371/journal.pone.0097878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou (2018).Zou Z. Mining gene families in the castor bean genome. In: Rabinowicz P, Kole C, editors. The castor bean genome. Springer; Switzerland: 2018. pp. 135–173. [Google Scholar]
- Zou, Huang & An (2013).Zou Z, Huang QX, An F. Genome-wide identification, classification and expression analysis of Lhc supergene family in castor bean (Ricinus communis L.) Agriculture Biotechnology. 2013;2:44–48, 51. [Google Scholar]
- Zou et al. (2018).Zou Z, Huang QX, Xie GS, Yang LF. Genome-wide comparative analysis of papain-like cysteine protease family genes in castor bean and physic nut. Scientific Reports. 2018;8:331. doi: 10.1038/s41598-017-18760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou & Yang (2019a).Zou Z, Yang JH. Genomics analysis of the light-harvesting chlorophyll a/b-binding (Lhc) superfamily in cassava (Manihot esculenta Crantz) Gene. 2019a;702:171–181. doi: 10.1016/j.gene.2019.03.071. [DOI] [PubMed] [Google Scholar]
- Zou & Yang (2019b).Zou Z, Yang JH. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genomics. 2019b;20:380. doi: 10.1186/s12864-019-5780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou & Yang (2019c).Zou Z, Yang JH. Genomic analysis of Dof transcription factors in Hevea brasiliensis, a rubber-producing tree. Industrial Crops & Products. 2019c;134:271–283. doi: 10.1016/j.indcrop.2019.04.013. [DOI] [Google Scholar]
- Zou, Yang & Zhang (2019).Zou Z, Yang JH, Zhang XC. Insights into genes encoding respiratory burst oxidase homologs (RBOHs) in rubber tree (Hevea brasiliensis Muell. Arg.) Industrial Crops & Products. 2019;128:126–139. doi: 10.1016/j.indcrop.2018.11.005. [DOI] [Google Scholar]
- Zou et al. (2016).Zou Z, Yang LF, Gong J, Mo YY, Wang JK, Cao JH, An F, Xie GS. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Frontiers in Plant Science. 2016;7 doi: 10.3389/fpls.2016.00395. Article 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The chromosome serial numbers are indicated at the top of each chromosome, and 27 RcLhc superfamily genes are shown just behind their collinear genes in jatropha (where RcLhcb1.2 with no collinear gene in jatropha). (Chr: chromosome; Jc: Jatropha curcas; Rc: Ricinus communis).
(A) Sequence alignment of the JcLhc family; (B) Sequence alignment of the JcLil family; (C) Sequence alignment of JcPsbS with RcPsbS, MePsbS, and AtPsbS; (D) Sequence alignment of JcFCII with RcFCII, MeFCII, and AtFCII. Sequence alignment was performed using MUSCLE and predicted TMHs were underlined. (TMH: transmembrane helix).
Sequence alignment was performed using MUSCLE. The predicted chloroplast transit peptide and Lhc motif-bearing TMH region were boxed. (TMH: transmembrane helix).
1 Conserved domains were predicted using MOTIF (CB, Chloroa_b-bind, PF00504; Rieske-like, PF00355; Ferrochelatase, PF00762). 2 Chloroplast transit peptide was predicted using ChloroP. 3 Duplicated modes in arabidopsis were determined based on studies of Wang, Tan & Paterson (2013) and related references therein. (AA, amino acid; CB, chlorophyll a/b-binding; GRAVY, grand average of hydropathicity; p I, isoelectric point; kDa, kilodalton; MW, molecular weight; TP, transit peptide; TMH, transmembrane helix; WGD, whole-genome duplication).
Data Availability Statement
The following information was supplied regarding data availability:
The data is available in the Supplementary Files.