Abstract
The goal of mitosis is to form two daughter cells each containing one copy of each mother cell chromosome, replicated in the previous S phase. To achieve this, sister chromatids held together back-to-back at their primary constriction, the centromere, have to interact with microtubules of the mitotic spindle so that each chromatid takes connections with microtubules emanating from opposite spindle poles (we will refer to this condition as bipolar attachment). Only once all replicated chromosomes have reached bipolar attachments can sister chromatids lose cohesion with each other, at the onset of anaphase, and move toward opposite spindle poles, being segregated into what will soon become the daughter cell nucleus. Prevention of errors in chromosome segregation is granted by a safeguard mechanism called Spindle Assembly Checkpoint (SAC). Until all chromosomes are bipolarly oriented at the equator of the mitotic spindle, the SAC prevents loss of sister chromatid cohesion, thus anaphase onset, and maintains the mitotic state by inhibiting inactivation of the major M phase promoting kinase, the cyclin B-cdk1 complex (Cdk1). Here, we review recent mechanistic insights about the circuitry that links Cdk1 to the SAC to ensure correct achievement of the goal of mitosis.
Keywords: Cdk1, APC/C, MCC, Cdc20, CCAN, Mps1, Mad1, Mad2, Bub1, spindle assembly checkpoint, SAC.
Introduction
Maintenance of genome stability through cell generations is a crucial feature that grants health to cells, organs and organisms. In humans, genome instability is causally linked to pathological outcomes such as cancer, degenerative disorders and physical and mental retardation 1– 3. Cells have developed several mechanisms to surveil that each step required for cell division is healthy and thoroughly completed before passing to the next one. This is achieved through mechanisms called cell cycle checkpoints 4– 8. If cells experience DNA damage or sense that DNA replication or assembly of the mitotic spindle is incomplete, checkpoint mechanisms halt cell cycle progression to repair damage or complete previous cell cycle stages before moving forward in their division process. If repair or completion is frustrated, then healthy checkpoints promote cell death 9– 13. This short review will be focused on recent advancements in the mechanistic understanding of the Spindle Assembly Checkpoint (SAC), the checkpoint that prevents formation of cells with an abnormal chromosome number by delaying mitosis exit until bipolar attachment of all replicated chromosomes 14.
Progression through mitosis: a cycle of Cdk1 activation/inactivation
Progression through mitosis is granted by a wave of cyclin B-cdk1 complex (Cdk1) activity 15, 16. Cdk1 is activated at the onset of mitosis by reversal of inhibitory phosphorylations of the cdk1 moiety at threonine 14 and tyrosine 15. These phosphorylations, operated by the Myt1 and Wee1 kinases, allow accumulation of enough inactive Cdk1, during S phase and G 2, to rapidly induce mitosis upon their reversal 17, 18. Dephosphorylation and activation of Cdk1 are granted by the dual-specificity phosphatase Cdc25 19. Upon initial activation, Cdk1 phosphorylates and inhibits Myt1 and Wee1 while it phosphorylates and further activates Cdc25; this way, Cdk1 promotes positive feedback loops for its own activation 20– 22. For mitosis onset, Cdk1 activity also represses major phosphatase activities (like that of PP1 and PP2A) that otherwise would antagonize Cdk1 action. The catalytic activity of PP1 is directly inhibited by Cdk1-dependent phosphorylation, while the activity of PP2A in which B55 is the holoenzyme regulatory subunit, PP2A-B55, is kept inhibited in mitosis by the aid of Greatwall kinase (Gwl). Gwl is stimulated by Cdk1 and phosphorylates Ensa/Arpp19, two small molecules, transforming them into potent PP2A-B55 inhibitors 22.
Inactivation of Cdk1 at the end of mitosis instead depends on the ubiquitin-dependent degradation of cyclin B 14, 23– 25. This is initiated by the ubiquitin ligase Anaphase Promoting Complex/Cyclosome (APC/C) in association with its coactivator Cdc20. APC/C Cdc20 also promotes the degradation of securin, an inhibitor of separase, the protease that cleaves the protein bridge that holds sister chromatid centromeres together 14, 26– 28. This way, the onset of anaphase and Cdk1 inactivation are tightly coupled by this irreversible degradative mechanism. Initial evidence indicated that APC/C Cdc20 activity required Cdk1-dependent phosphorylation; recently, the APC/C members that are directly phosphorylated by Cdk1 were identified 29– 33. Thus, Cdk1 is also promoting a negative feedback for its own inactivation. Nevertheless, final APC/C Cdc20 activation is under the control of the SAC, which inhibits APC/C Cdc20 until bipolar attachment of all replicated chromosomes 14.
Mps1 and the SAC, in brief
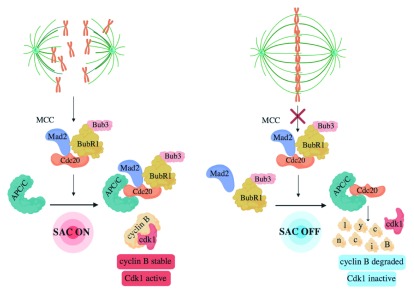
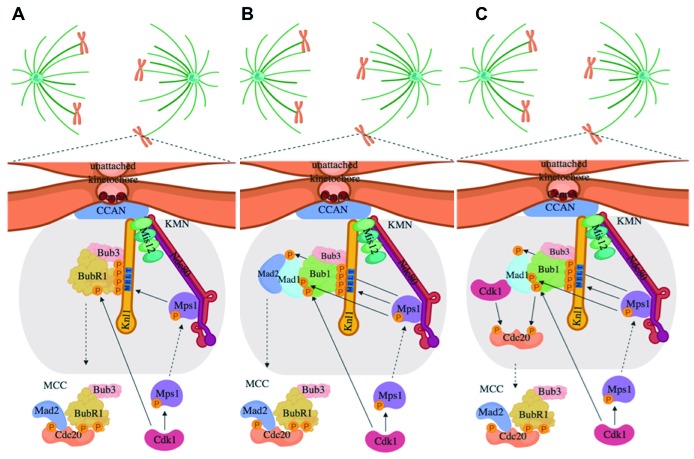
The SAC inhibits APC/C Cdc20 activation by forming a diffusible Mitotic Checkpoint Complex (MCC), composed of the proteins Mad2, Bub3, BubR1, and Cdc20 itself, in which Cdc20 is restrained from activating APC/C 14, 34– 37. MCC forms at unattached kinetochores, proteinaceous centromeric structures deputed to interact with spindle microtubules and permit chromosome segregation ( Figure 1) 14. MCC formation requires the action of crucial SAC kinases like Plk1, Aurora B, and Mps1 38– 40. These kinases also have important roles in correcting faulty chromosome–microtubule interactions to promote correct, end-on, bipolar chromosome–microtubule attachments 41. Here, however, we will primarily review recent advancements in the regulation of Mps1 in SAC control and its dependence on Cdk1 activity. Mps1 binds unattached kinetochores where it phosphorylates SAC proteins and activates them and then gets released from kinetochores upon stable microtubule binding, perhaps by competition mechanisms 42– 46. The bridge deputed to connect centromeres to microtubules is called the KMN network and is composed by the Knl1 complex, the Mis12 complex, and the Ndc80 complex 46– 50. The KMN, in the outer kinetochore, interacts with the inner kinetochore Constitutive Centromere Associated Network (CCAN), a protein network that assembles onto Cenp-A nucleosomes, a histone H3 variant found at centromeric nucleosomes 51– 53. Mps1 localizes at unattached kinetochores primarily by interacting with the Ndc80 complex 54. At kinetochores, Mps1 phosphorylates the “MELT” repeats of Knl1, promoting kinetochore recruitment of the BubR1-Bub3 and Bub1-Bub3 complexes ( Figure 2), while Knl1 dephosphorylation by PP1 appears involved in SAC silencing 43, 55– 58. Mps1 also phosphorylates Bub1, further promoting kinetochore recruitment of Mad1, another crucial SAC protein needed for the activation of Mad2 59– 61. Mad1 recruitment at kinetochores is also facilitated by the Rod-Zwilch-ZW10 (RZZ) complex 62. In addition, phosphorylation of Mad1 by Mps1 helps the Mad1-dependent conversion of Mad2 into the functional conformation required to inhibit Cdc20 in the MCC 59, 60. Thus, Mps1 is a crucial effector of the SAC mechanism by promoting MCC formation ( Figure 2).
Figure 1. Unattached or incorrectly attached chromosomes promote formation of the Mitotic Checkpoint Complex (MCC).
Until bipolar spindle assembly, the MCC, composed of Mad2, BubR1-Bub3, and Cdc20, forms, binds, and blocks APC/C action (SAC ON). Upon bipolar spindle assembly, MCC is dismantled and MCC-free Cdc20 activates APC/C (SAC OFF).
Figure 2. Paths to Mitotic Checkpoint Complex (MCC) formation.
Cdk1 phosphorylation of Mps1 helps kinetochore recruitment of Mps1 to ( A) recruit BubR1-Bub3 complex for its incorporation into MCC, ( B) recruit Bub1-Bub3 for Mad1-Mad2 docking and Mad2 incorporation into MCC, and ( C) recruit Bub1-Bub3 for Mad1-Cdk1 docking for Bub1- and Cdk1-dependent phosphorylation of Cdc20 and incorporation into MCC.
Cdk1 and the SAC
The observation that APC/C activity was promoted by Cdk1-dependent phosphorylation, while APC/C activation was inhibited by the SAC until spindle assembly, reinforced the idea that checkpoint mechanisms would oppose the forward trend of the basic cell cycle engine 4. However, in 2003, a few independent observations, from yeast and vertebrates, changed this view by showing that Cdk1 was instrumental to the SAC action 63– 65. Indeed, in yeast, SAC-defective cdk1 mutants were described and Bub1 was shown to be phosphorylated by Cdk1 for SAC proficiency 63, 64. In the Xenopus egg extract system and in human somatic cells, Cdk1 activity was revealed to be required to sustain SAC-dependent arrest and the ability of MCC members to block the APC/C 65, 66. Cdk1-dependent phosphorylation of Cdc20 appeared to have a role in reducing Cdc20 affinity for APC/C while increasing that for other MCC proteins 65– 68. Thus, Cdk1, the cell cycle engine, though paving the way for its own inactivation by phosphorylating APC/C, was instrumental for the checkpoint SAC that would block APC/C activation until correct spindle assembly 66. These observations also helped to explain why the SAC does not get reactivated at the onset of anaphase, when loss of chromatid cohesion causes loss of kinetochore tension, a condition that would have activated the SAC at earlier stages 69, 70. This was shown to be due to the concomitant reduction of Cdk1 because of the mentioned coupling of anaphase onset with degradation of cyclin B 69, 70. A few years later, the notion that Cdk1 was required for the SAC function was reinforced by the findings that, in the Xenopus egg extract system, Mps1 was phosphorylated by Cdk1 and that this phosphorylation substantially helped Mps1 activity in its fundamental role for the SAC 71.
Very recently, through careful biochemical dissection, important observations have described in closer detail how Cdk1 is an integral part of the SAC mechanisms 72, 73. Indeed, it has been shown that kinetochore localization of Mps1, in human cells, greatly depends on direct phosphorylation by Cdk1; thus, Cdk1 controls activity and localization of Mps1 72. Mps1, in turn, helps kinetochore localization of Cdk1 73– 76. As mentioned earlier, by phosphorylating Knl1, Mps1 creates a docking site for kinetochore localization of Bub1, and cooperative Cdk1- and Mps1-dependent phosphorylations of Bub1 are required to recruit Mad1 at kinetochores 42, 43, 50, 59, 77. Kinetochore localization of Mad1 is crucial for its ability to convert Mad2 in the effective form that incorporates into the MCC 61. However, it has also recently been shown that Mad1 stably interacts with Cdk1 and that Mps1, through kinetochore recruitment of Mad1, in turn, promotes kinetochore localization of Cdk1 ( Figure 2) 72, 73. At kinetochores, Cdk1 may further phosphorylate other substrates to sustain the SAC like Cdc20 or BubR1 and possibly also help error correction and SAC resolution by favoring BubR1 interaction with the protein phosphatase PP2A-B56 65, 78– 80. Recent evidence also indicated how the indirect downregulation of the protein phosphatase PP2A-B55 activity by Cdk1 is instrumental for the SAC-promoting action of Cdk1 itself 72, 81. In addition, it should be noted that kinetochore localization of Mps1 is favored by the activity of Aurora B, perhaps by phosphorylating members of the Ndc80 complex 42. However, centromere localization of Aurora B depends on other components of the Chromosomal Passenger Complex (CPC), composed of survivin, borealin, INCENP, and Aurora B itself, and Cdk1 activity is required, directly and indirectly, for CPC centromeric localization 82– 84. Thus, even by mastering CPC localization, Cdk1 affects Mps1 and is fundamental for SAC action.
Concluding remarks and further questions
The recent advancements, reviewed here, in the mechanisms of mitotic exit and in particular in how Cdk1 mechanistically serves the SAC, suggest that Cdk1 is an integral part of the SAC system. Thus, perhaps the cell cycle engine, Cdk1, and the checkpoint, SAC, are not to be viewed any longer as separate mechanisms but rather as integrated systems that ensure correct execution of complex biological tasks. Important hints have also been recently provided on how the SAC can be silenced, such as on priming mechanisms for protein phosphatases that would reverse SAC-activating phosphorylations upon bipolar chromosome attachments, in addition to the notion that the MCC itself undergoes proteasome-dependent turnover for rapid SAC silencing 79, 84– 88. Nevertheless, major phosphatases like PP1 and PP2A are directly or indirectly inhibited by Cdk1 activity 22. Thus, it is still unclear whether chromosome attachment and kinetochore tension are sufficient to dislodge kinases and let phosphatases take the upper hand for SAC silencing or whether these conditions also affect the activity of crucial SAC kinases 40. Based on our previous observations, we hypothesize in this regard that Cdk1 activity could be locally downregulated by non-proteolytic means upon bipolar chromosome attachment and that this would lead to SAC silencing 89– 92. If this were true, a proteolysis-independent negative control of Cdk1 would be required for SAC silencing, ahead of and for final, proteolysis-dependent, Cdk1 inactivation and mitotic exit.
Abbreviations
APC/C, Anaphase Promoting Complex/Cyclosome; Cdk1, cyclin B-cdk1 complex; CPC, Chromosomal Passenger Complex; Gwl, Greatwall kinase; CCAN, Constitutive Centromere Associated Network; KNM, Knl1 complex, Ndc80 complex, Mis12 complex; MCC, Mitotic Checkpoint Complex; SAC, Spindle Assembly Checkpoint
Acknowledgments
The authors acknowledge Associazione Italiana per la Ricerca sul Cancro (AIRC) for support and all the relevant works on this topic apologizing for not having cited them all.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Jonne Raaijmakers, Oncode Institute, Division of Cell Biology, The Netherlands Cancer Institute, Amsterdam, The Netherlands
Jakob Nilsson, The Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, Copenhagen, Denmark
Funding Statement
This work was supported by a grant from AIRC (IG grant 2017; Id. 19851 to DG).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- 1. Taylor AMR, Rothblum-Oviatt C, Ellis NA, et al. : Chromosome instability syndromes. Nat Rev Dis Primers. 2019;5(1):64. 10.1038/s41572-019-0113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Bach DH, Zhang W, Sood AK: Chromosomal Instability in Tumor Initiation and Development. Cancer Res. 2019;79(16):3995–4002. 10.1158/0008-5472.CAN-18-3235 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 3. de Wolf B, Kops GJPL: Kinetochore Malfunction in Human Pathologies. Adv Exp Med Biol. 2017;1002:69–91. 10.1007/978-3-319-57127-0_4 [DOI] [PubMed] [Google Scholar]
- 4. Hartwell LH, Weinert TA: Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–34. 10.1126/science.2683079 [DOI] [PubMed] [Google Scholar]
- 5. Murray AW: Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992;359(6396):599–604. 10.1038/359599a0 [DOI] [PubMed] [Google Scholar]
- 6. Elledge SJ: Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274(5293):1664–72. 10.1126/science.274.5293.1664 [DOI] [PubMed] [Google Scholar]
- 7. Nurse P: Checkpoint pathways come of age. Cell. 1997;91(7):865–7. 10.1016/s0092-8674(00)80476-6 [DOI] [PubMed] [Google Scholar]
- 8. Musacchio A: Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3595–604. 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartwell LH, Kastan MB: Cell cycle control and cancer. Science. 1994;266(5192):1821–8. 10.1126/science.7997877 [DOI] [PubMed] [Google Scholar]
- 10. Sorger PK, Dobles M, Tournebize R, et al. : Coupling cell division and cell death to microtubule dynamics. Curr Opin Cell Biol. 1997;9(6):807–14. 10.1016/s0955-0674(97)80081-6 [DOI] [PubMed] [Google Scholar]
- 11. Jacotot E, Ferri KF, Kroemer G: Apoptosis and cell cycle: distinct checkpoints with overlapping upstream control. Pathol Biol (Paris). 2000;48(3):271–9. [PubMed] [Google Scholar]
- 12. Clarke PR, Allan LA: Cell-cycle control in the face of damage--a matter of life or death. Trends Cell Biol. 2009;19(3):89–98. 10.1016/j.tcb.2008.12.003 [DOI] [PubMed] [Google Scholar]
- 13. Krenning L, van den Berg J, Medema RH: Life or Death after a Break: What Determines the Choice? Mol Cell. 2019;76(2):346–58. 10.1016/j.molcel.2019.08.023 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Musacchio A, Salmon ED: The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–93. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- 15. Nurse P: Universal control mechanism regulating onset of M-phase. Nature. 1990;344(6266):503–8. 10.1038/344503a0 [DOI] [PubMed] [Google Scholar]
- 16. Minshull J, Pines J, Golsteyn R, et al. : The role of cyclin synthesis, modification and destruction in the control of cell division. J Cell Sci. 1989;12:77–97. 10.1242/jcs.1989.supplement_12.8 [DOI] [PubMed] [Google Scholar]
- 17. Atherton-Fessler S, Hannig G, Piwnica-Worms H: Reversible tyrosine phosphorylation and cell cycle control. Semin Cell Biol. 1993;4(6):433–42. 10.1006/scel.1993.1051 [DOI] [PubMed] [Google Scholar]
- 18. Deibler RW, Kirschner MW: Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37(6):753–67. 10.1016/j.molcel.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Perdiguero E, Nebreda AR: Regulation of Cdc25C activity during the meiotic G2/M transition. Cell Cycle. 2014;3(6):733–7. 10.4161/cc.3.6.906 [DOI] [PubMed] [Google Scholar]
- 20. Kapuy O, He E, López-Avilés S, et al. : System-level feedbacks control cell cycle progression. FEBS Lett. 2009;583(24):3992–8. 10.1016/j.febslet.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Domingo-Sananes MR, Kapuy O, Hunt T, et al. : Switches and latches: A biochemical tug-of-war between the kinases and phosphatases that control mitosis. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3584–94. 10.1098/rstb.2011.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunt T: On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv Biol Regul. 2013;53(2):173–8. 10.1016/j.jbior.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 23. Evans T, Rosenthal ET, Youngblom J, et al. : Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33(2):389–96. 10.1016/0092-8674(83)90420-8 [DOI] [PubMed] [Google Scholar]
- 24. Murray AW, Solomon MJ, Kirschner MW: The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339(6222):280–6. 10.1038/339280a0 [DOI] [PubMed] [Google Scholar]
- 25. King RW, Peters JM, Tugendreich S, et al. : A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81(2):279–88. 10.1016/0092-8674(95)90338-0 [DOI] [PubMed] [Google Scholar]
- 26. Michaelis C, Ciosk R, Nasmyth K: Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. 10.1016/s0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- 27. Ciosk R, Zachariae W, Michaelis C, et al. : An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93(6):1067–76. 10.1016/s0092-8674(00)81211-8 [DOI] [PubMed] [Google Scholar]
- 28. Lu D, Girard JR, Li W, et al. : Quantitative framework for ordered degradation of APC/C substrates. BMC Biol. 2015;13:96. 10.1186/s12915-015-0205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shteinberg M, Protopopov Y, Listovsky T, et al. : Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem Biophys Res Commun. 1999;260(1):193–8. 10.1006/bbrc.1999.0884 [DOI] [PubMed] [Google Scholar]
- 30. Rudner AD, Murray AW: Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149(7):1377–90. 10.1083/jcb.149.7.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang S, Chang L, Alfieri C, et al. : Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533(7602):260–4. 10.1038/nature17973 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Fujimitsu K, Grimaldi M, Yamano H: Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science. 2016;352(6289):1121–4. 10.1126/science.aad3925 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Qiao R, Weissmann F, Yamaguchi M, et al. : Mechanism of APC/C CDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci U S A. 2016;113(19):E2570–E2578. 10.1073/pnas.1604929113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Sudakin V, Chan GK, Yen TJ: Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–36. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Díaz-Martínez LA, Yu H: Running on a treadmill: dynamic inhibition of APC/C by the spindle checkpoint. Cell Div. 2007;2: 23. 10.1186/1747-1028-2-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Izawa D, Pines J: The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517(7536):631–4. 10.1038/nature13911 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Alfieri C, Chang L, Zhang Z, et al. : Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536(7617):431–6. 10.1038/nature19083 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Nigg EA: Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2(1):21–32. 10.1038/35048096 [DOI] [PubMed] [Google Scholar]
- 39. Suijkerbuijk SJ, Kops GJ: Preventing aneuploidy: the contribution of mitotic checkpoint proteins. Biochim Biophys Acta. 2008;1786(1):24–31. 10.1016/j.bbcan.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 40. Saurin AT: Kinase and Phosphatase Cross-Talk at the Kinetochore. Front Cell Dev Biol. 2018;6:62. 10.3389/fcell.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Manic G, Corradi F, Sistigu A, et al. : Molecular Regulation of the Spindle Assembly Checkpoint by Kinases and Phosphatases. Int Rev Cell Mol Biol. 2017;328:105–61. 10.1016/bs.ircmb.2016.08.004 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Santaguida S, Tighe A, D'Alise AM, et al. : Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190(1):73–87. 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamagishi Y, Yang CH, Tanno Y, et al. : MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14(7):746–52. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- 44. Hiruma Y, Sacristan C, Pachis ST, et al. : CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348(6240):1264–7. 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Ji Z, Gao H, Yu H: CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 2015;348(6240):1260–4. 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- 46. Aravamudhan P, Goldfarb AA, Joglekar AP: The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol. 2015;17(7):868–79. 10.1038/ncb3179 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Varma D, Wan X, Cheerambathur D, et al. : Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol. 2013;202(5):735–46. 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petrovic A, Keller J, Liu Y, et al. : Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell. 2016;167(4):1028–1040.e15. 10.1016/j.cell.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Umbreit NT, Gestaut DR, Tien JF, et al. : The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci U S A. 2012;109(40):16113–8. 10.1073/pnas.1209615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varma D, Salmon ED: The KMN protein network--chief conductors of the kinetochore orchestra. J Cell Sci. 2012;125(Pt 24):5927–36. 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hori T, Amano M, Suzuki A, et al. : CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135(6):1039–52. 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- 52. Screpanti E, de Antoni A, Alushin GM, et al. : Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21(5):391–8. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Hori T, Shang WH, Takeuchi K, et al. : The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200(1):45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stucke VM, Baumann C, Nigg EA: Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113(1):1–15. 10.1007/s00412-004-0288-2 [DOI] [PubMed] [Google Scholar]
- 55. Shepperd LA, Meadows JC, Sochaj AM, et al. : Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22(10):891–9. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Primorac I, Weir JR, Chiroli E, et al. : Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2013;2:e01030. 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang G, Mendez BL, Sedgwick GG, et al. : Two functionally distinct kinetochore pools of BubR1 ensure accurate chromosome segregation. Nat Commun. 2016;7: 12256. 10.1038/ncomms12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rosenberg JS, Cross FR, Funabiki H: KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21(11):942–7. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ji Z, Gao H, Jia L, et al. : A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife. 2017;6: pii: e22513. 10.7554/eLife.22513 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Faesen AC, Thanasoula M, Maffini S, et al. : Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017;542(7642):498–502. 10.1038/nature21384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sironi L, Melixetian M, Faretta M, et al. : Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20(22):6371–82. 10.1093/emboj/20.22.6371 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Caldas GV, Lynch TR, Anderson R, et al. : The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 2015;5(11): pii: 150160. 10.1098/rsob.150160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kitazono AA, Garza DA, Kron SJ: Mutations in the yeast cyclin-dependent kinase Cdc28 reveal a role in the spindle assembly checkpoint. Mol Genet Genomics. 2003;269(5):672–84. 10.1007/s00438-003-0870-y [DOI] [PubMed] [Google Scholar]
- 64. Yamaguchi S, Decottignies A, Nurse P: Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 2003;22(5):1075–87. 10.1093/emboj/cdg100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D'Angiolella V, Mari C, Nocera D, et al. : The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 2003;17(20):2520–5. 10.1101/gad.267603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. D'Angiolella V, Grieco D: Attach first, then detach: a role for cyclin B-dependent kinase 1 in coordinating proteolysis with spindle assembly. Cell Cycle. 2004;3(2):132–3. 10.4161/cc.3.2.664 [DOI] [PubMed] [Google Scholar]
- 67. Hein JB, Hertz EPT, Garvanska DH, et al. : Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat Cell Biol. 2017;19(12):1433–40. 10.1038/ncb3634 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Labit H, Fujimitsu K, Bayin NS, et al. : Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 2012;31(15):3351–62. 10.1038/emboj.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rattani A, Vinod PK, Godwin J, et al. : Dependency of the spindle assembly checkpoint on Cdk1 renders the anaphase transition irreversible. Curr Biol. 2014;24(6):630–7. 10.1016/j.cub.2014.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vázquez-Novelle MD, Sansregret L, Dick AE, et al. : Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr Biol. 2014;24(6):638–45. 10.1016/j.cub.2014.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Morin V, Prieto S, Melines S, et al. : CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr Biol. 2012;22(4):289–95. 10.1016/j.cub.2011.12.048 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Hayward D, Alfonso-Pérez T, Cundell MJ, et al. : CDK1-CCNB1 creates a spindle checkpoint-permissive state by enabling MPS1 kinetochore localization. J Cell Biol. 2019;218(4):1182–99. 10.1083/jcb.201808014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Alfonso-Pérez T, Hayward D, Holder J, et al. : MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J Cell Biol. 2019;218(4):1108–17. 10.1083/jcb.201808015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Pfaff KL, King RW: Determinants of human cyclin B1 association with mitotic chromosomes. PLoS One. 2013;8(3):e59169. 10.1371/journal.pone.0059169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Allan LA, Reis M, Liu Y, et al. : Cyclin B1 scaffolds MAD1 at the corona to activate the spindle assembly checkpoint. BioRxiv. 2019; 726224. 10.1101/726224 [DOI] [Google Scholar]
- 76. Jackman M, Marcozzi C, Pardo M, et al. : Cyclin B1-Cdk1 binding to MAD1 links nuclear pore disassembly to chromosomal stability. BioRxiv. 2019; 701474. 10.1101/701474 [DOI] [Google Scholar]
- 77. Zhang G, Kruse T, López-Méndez B, et al. : Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat Commun. 2017;8: 15822. 10.1038/ncomms15822 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Wong OK, Fang G: Cdk1 phosphorylation of BubR1 controls spindle checkpoint arrest and Plk1-mediated formation of the 3F3/2 epitope. J Cell Biol. 2007;179(4):611–7. 10.1083/jcb.200708044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kruse T, Zhang G, Larsen MS, et al. : Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci. 2013;126(Pt 5):1086–92. 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- 80. Hayward D, Bancroft J, Mangat D, et al. : Checkpoint signaling and error correction require regulation of the MPS1 T-loop by PP2A-B56. J Cell Biol. 2019;218(10):3188–99. 10.1083/jcb.201905026 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Diril MK, Bisteau X, Kitagawa M, et al. : Loss of the Greatwall Kinase Weakens the Spindle Assembly Checkpoint. PLoS Genet. 2016;12(9):e1006310. 10.1371/journal.pgen.1006310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ruchaud S, Carmena M, Earnshaw WC: Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- 83. Tsukahara T, Tanno Y, Watanabe Y: Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 2010;467(7316):719–23. 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Feng H, Raasholm M, Moosmann A, et al. : Switching of INCENP paralogs controls transitions in mitotic chromosomal passenger complex functions. Cell Cycle. 2019;18(17):2006–25. 10.1080/15384101.2019.1634954 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Nijenhuis W, Vallardi G, Teixeira A, et al. : Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol. 2014;16(12):1257–64. 10.1038/ncb3065 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Ge S, Skaar JR, Pagano M: APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle. 2014;8(1):167–71. 10.4161/cc.8.1.7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Visconti R, Palazzo L, Grieco D: Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2014;9(3):564–9. 10.4161/cc.9.3.10581 [DOI] [PubMed] [Google Scholar]
- 88. Varetti G, Guida C, Santaguida S, et al. : Homeostatic control of mitotic arrest. Mol Cell. 2011;44(5):710–20. 10.1016/j.molcel.2011.11.014 [DOI] [PubMed] [Google Scholar]
- 89. D'Angiolella V, Palazzo L, Santarpia C, et al. : Role for non-proteolytic control of M-phase-promoting factor activity at M-phase exit. PLoS One. 2007;2(2):e247. 10.1371/journal.pone.0000247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Visconti R, Palazzo L, Della Monica R, et al. : Fcp1-dependent dephosphorylation is required for M-phase-promoting factor inactivation at mitosis exit. Nat Commun. 2012;3: 894. 10.1038/ncomms1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Visconti R, Della Monica R, Palazzo L, et al. : The Fcp1-Wee1-Cdk1 axis affects spindle assembly checkpoint robustness and sensitivity to antimicrotubule cancer drugs. Cell Death Differ. 2015;22(9):1551–60. 10.1038/cdd.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Della Monica R, Visconti R, Cervone N, et al. : Fcp1 phosphatase controls Greatwall kinase to promote PP2A-B55 activation and mitotic progression. eLife. 2015;4: pii: e10399. 10.7554/eLife.10399 [DOI] [PMC free article] [PubMed] [Google Scholar]