Abstract
Crohn’s disease and ulcerative colitis are increasingly prevalent, relapsing and remitting inflammatory bowel diseases (IBDs) with variable disease courses and complications. Their aetiology remains unclear but current evidence shows an increasingly complex pathophysiology broadly centring on the genome, exposome, microbiome and immunome. Our increased understanding of disease pathogenesis is providing an ever-expanding arsenal of therapeutic options, but these can be expensive and patients can lose response or never respond to certain therapies. Therefore, there is now a growing need to personalise therapies on the basis of the underlying disease biology and a desire to shift our approach from “reactive” management driven by disease complications to “proactive” care with an aim to prevent disease sequelae. Precision medicine is the tailoring of medical treatment to the individual patient, encompassing a multitude of data-driven (and multi-omic) approaches to foster accurate clinical decision-making. In IBD, precision medicine would have significant benefits, enabling timely therapy that is both effective and appropriate for the individual. In this review, we summarise some of the key areas of progress towards precision medicine, including predicting disease susceptibility and its course, personalising therapies in IBD and monitoring response to therapy. We also highlight some of the challenges to be overcome in order to deliver this approach.
Keywords: inflammatory bowel disease, precision medicine, biomarkers, genomics, prognosis, therapeutics, Crohn's disease, ulcerative colitis, microbiome
Core tip
Our current understanding of inflammatory bowel disease (IBD), whilst incomplete, suggests an increasingly complex pathophysiology. Increasing biologic and small-molecule therapeutic options are available, but loss of response is common. Precision medicine in IBD, with the aim of tailoring the right therapy to the right patient at the right time on the basis of an individual patient’s biology, is an aspiration. We summarise recent progress in the pursuit of precision medicine in IBD and highlight some of the challenges that remain. In the future, precision medicine in IBD has the potential to enable delivery of truly individualised IBD care.
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are forms of idiopathic IBD that follow a relapsing and remitting course 1, 2. The incidence and prevalence of IBD are increasing worldwide, particularly in newly industrialised countries 3– 5. The pathogenesis of IBD remains unclear, although it is increasingly evident that the pathophysiological factors leading to heterogenous IBD phenotypes are increasingly complex and interwoven 6. Broadly, it appears that aetiological factors concentrate around the genome 7, exposome 8, microbiome 9 and immunome 10. Whilst many factors have been identified by studying each of these disciplines in isolation, a true understanding of an individual patient’s aetiological and pathogenic mechanisms, and thus therapeutic options, will arise only from an integrated approach 11.
A growing understanding of the immunopathophysiological mechanisms underlying the development of IBD has led to significant advances in the ability to treat IBD effectively 12– 15. In a relatively short time, we have progressed from “conventional treatment” using corticosteroids with or without immunomodulators to having an increasing array of biologic and small-molecule therapies in our medical arsenal 16. However, the associated costs of treating IBD are also increasing 17– 20. It is also becoming clear that the first biologic therapy (that is, the initial biologic therapy commenced in an individual patient) seems to be the most effective with a “dampened” response rate using second-line biologics 21. Although IBD clinicians embrace increasing therapeutic options for our patients, we also have a responsibility to ensure optimal and efficient use for each individual patient. The ability to predict response, relapse and side effects as well as optimise drug levels and early identification of loss of response is highly desirable 22. The development of biomarkers to enable targeted and effective individualised treatment is therefore necessary to allow selection of the right drug at the right time in the right patient 23.
So it is clear that a change of perspective is required to advance our understanding of IBD. IBD is a stochastic, heterogenous disease with complex aetiology and increasingly complex treatment options. Traditional research approaches focusing on isolated aspects of IBD aetiology or treatment, whilst necessary and highly informative, run the risk of oversimplification without an appreciation of complex aetiological and pharmacological associations 11. There is a growing need for a “multi-omic” approach where data from different disciplines regarding the same subject are integrated to advance our understanding of IBD immunopathogenesis, to identify new drug targets and to identify biomarkers to enable better detection and treatment of disease 24. By understanding the complex pathogenesis of IBD, we will be better able to practice precision medicine in which we can tailor medical therapy to the individual patient 25, 26.
Pathogenesis of inflammatory bowel disease
The exact pathogenesis of IBD remains enigmatic. Whilst it is understood that complex interactions between host and environment are pivotal, it is not yet clear exactly how these lead to the various disease phenotypes. It is beyond the scope of this article to provide an in-depth review of IBD pathogenesis, but we will summarise some of the key points.
Genetics
The observation that the risk of developing IBD is greater in people who have relatives with IBD suggests a genetic component 27– 29. Genome-wide association studies (GWASs) have facilitated the discovery of a large number of significant genetic risk loci, some of which are discussed later in this article 6, 30. Many loci are shared between UC and CD 31 and may overlap with loci associated with other inflammatory diseases 32. Functional gene polymorphisms are known to impact on innate immunity, adaptive immunity and regulation of the intestinal barrier 33. Despite the advances in genetic understanding, only about 25% of IBD heritability can currently be explained 30.
Environment
The incidence and prevalence of IBD are highest in the Western world, but newly industrialised countries are also experiencing increases in the incidence and prevalence of IBD, and migrants to the Western world acquire the same risk of IBD as that of the native population 34. This occurs independently of ethnicity and seems to mirror the industrialisation and westernisation of populations 4. Early life events, pollution, and diet have all been implicated in the development of IBD 8.
Microbiome
The dysbiosis seen in IBD has been well described as being characterised by a reduced microbial diversity, expansion of facultative anaerobes and decreased numbers of obligate anaerobes 35. In the most comprehensive analysis carried out to date on the microbiome in IBD, longitudinal profiles of 132 subjects were developed, demonstrating disruptions in microbial transcription and metabolite pools as well as increases in temporal variability associated with disease activity 36. It remains unknown which of these observed changes are the cause or consequence of IBD and this remains an area of intense investigation.
Immunome
The gut is a crucial immunological interface that maintains immunological balance by appropriately recognising and tolerating commensal bacteria, food antigens and self-antigens (tolerogenic response) whilst identifying and acting against pathogenic organisms (immunogenic response) 6. Dysregulated immunological responses within the gut leading to imbalances in pro- and anti-inflammatory pathways involved in innate and adaptive immunity are thought to be pivotal in the development and persistence of inflammation in IBD 37. Impaired barrier function of the epithelium coupled with epithelial neutrophil accumulation, defective antigen clearance by macrophages and impaired conditioning of dendritic cells are all innate immune factors thought to contribute to the persistence of inflammation 10. Dendritic cells bridge the gap between innate and adaptive immunity by inducing tolerance or immunogenicity amongst T and B cells and therefore can be influential in mucosal homeostasis versus mucosal inflammation 38. The balance between effector T cells (T H1, T H2, T H17 and T H9) and regulatory T (T reg) cells is crucial to maintaining tolerance or promoting chronic inflammation 39.
Concept of precision medicine and its relevance to inflammatory bowel disease
Precision medicine refers to the “tailoring of medical treatment to the individual characteristics of each patient” 40. Whilst the concepts of personalised medicine and precision medicine are very similar, precision medicine also encompasses a multidisciplinary data-driven approach to foster better clinical decision-making through a clear understanding of the molecular basis of an individual’s disease 41. In 2015, the national Precision Medicine Initiative, a novel and ambitious plan to collect multi-omic data on over 1 million patients in a new national cohort in an attempt to accelerate understanding of diseases and their treatments, was announced in the US 42. This high-profile endorsement of precision medicine has served to ignite interest in and raise awareness of the concept.
Precision medicine in IBD is conceptually attractive. We know that patients with CD or UC can have a markedly variable disease course. In Norwegian cohorts followed up over 10 years, the IBSEN study group found that up to 53% of patients with CD developed stricturing or penetrating disease and that up to 19% of patients with UC required colectomy by 10 years 43, 44. Traditional “step-up” therapy risks undertreating those patients who are destined to develop complications, but “top-down” therapy risks overtreating patients who may have remained stable and complication-free on less expensive therapies and exposes those patients to unnecessary side effects. Many clinical parameters, such as serological markers, disease location, disease behaviour, age and lifestyle, have been found to be associated with disease severity 45, 46. However, none is reliable or specific enough to alter early IBD management. Therefore, the ability to predict aggressive disease behaviour, high risk of disease complications, or response to certain treatments at or near the time of diagnosis for individual patients currently remains elusive. Nonetheless, there are encouraging signs of progress. This review aims to summarise some of the key progress made to date in the pursuit of precision medicine in IBD and aims to outline some of the challenges that have yet to be overcome.
Predicting disease susceptibility and clinical phenotype
GWASs have provided important insights into the aetiology and development of IBD 30. To date, over 240 IBD susceptibility loci have been identified through this approach 47– 50. Some of the key biological processes uncovered by GWASs include epithelial barrier dysfunction, disruption of antimicrobial defences and immune dysregulation. Whilst GWASs have provided numerous avenues to explore, these have yet to filter through as clinically useful biomarkers to help predict disease and its phenotype.
Some GWAS associations have been associated with distinct clinical phenotypes. NOD2/CARD15 is involved in pattern recognition receptor signalling in response to microbial stimuli and has been associated with an ileal fibrostenosing disease phenotype 51– 54. NOD2/CARD15 has also been associated with the need for surgery and complicated disease course 55. ATG16L1, which implicates defective autophagy as an important part of CD development 56, has been associated with ileal disease 57, and another autophagy gene, IGRM, has been associated with penetrating disease 58. IL23R, which has become a successful therapeutic target in CD 59, 60, has also been previously linked to ileal CD 61. However, most of these individual genes have limited effect sizes, meaning that applicability on their own to the general population is limited. Recent insights into the genetics of IBD reveal distinct clinical phenotypes. Exploring the associations between genetic risk scores and IBD sub-phenotypes in 29,838 patients, the UK IBD Genetics Consortium (IBDGC) was able to redefine disease subtypes into ileal CD, colonic CD and UC 62.
Similarly, the microbiome may have a role to play in helping identify “high risk” individuals. In a study of new-onset IBD, Gevers et al. found ileal microbiome signatures to be predictive of CD, even in the absence of overt inflammation 63. It is likely that the microbial signature of IBD will vary depending on the clinical phenotype 64 as well as environmental factors such as smoking and medication use. Longitudinal studies have suggested an ability to predict CD and UC phenotypes on the basis of the microbiome and an ability to distinguish patients with IBD from those with similar symptoms 65. In the future, there is the enticing possibility of generating “risk scores” for patients on the basis of their microbiome 66.
Despite this tremendous progress in genetics and the microbiome, screening for IBD in the clinic continues to rely on a combination of clinical symptoms and blood-based biomarkers. More recently, faecal calprotectin (FC) has emerged as a robust screening tool for those with suspected IBD. Studies have shown that FC has a pooled sensitivity and specificity of 0.93 (0.85–0.97) and 0.96 (0.79–0.99) respectively and a high negative predictive value (NPV) (0.96–0.98) 67, 68. FC now represents one of the core tests in clinical practice in the UK (and elsewhere) as a screening tool to identify those who require further tertiary-care investigations for suspected IBD 69. Despite its value, several practical issues reported in the literature appear to hinder its widespread clinical uptake. First, patient compliance with FC is often poor. A recent study showed that only one third of patients were compliant with FC testing, forgetfulness being the main reason for non-compliance 70. Two recent studies investigating the acceptability of FC in IBD found that sample collection was a major barrier to testing; blood testing was preferred over stool testing 71, 72. These “patient factors” pose a real challenge for clinicians in implementing FC testing in clinical care and hamper its clinical utility. FC also shows within-day variations, and the ideal time of sample collection is unclear 73, 74.
Researchers are beginning to explore novel blood-based diagnostic markers across multiple -omics platforms, including epigenetics, metabolomics and proteomics, and the results have been promising. Multicentre consortia such as IBD-Character, IBD-BIOM and Biocycle have been funded by the European Commission to develop novel biomarkers that can be transitioned into the clinic; each focuses on specific unmet needs in clinical practice. Whereas IBD-Character and IBD-BIOM have focussed on developing diagnostic and prognostic multi-omic markers in IBD, Biocycle explores innovative regimes for maintenance treatment in moderate to severe CD 75. These have yet to be translated to clinically useful biomarkers but offer a glimpse into the potential for an expanded repertoire to help diagnose IBD wherever it is suspected. However, multiple challenges exist: biomarkers should have key qualities such as being simple, easy to perform, minimally invasive, rapid and reproducible, and should consistently show high accuracy in predicting disease across several populations 76. These characteristics have yet to be ascertained amongst the biomarkers studied so far.
Predicting disease course
Historically, gastroenterologists have relied on the established clinical predictors of disease course ( Table 1) to help tailor management. Over time, we have progressed from relying on clinical predictors to using biomarkers to predict outcomes and tailor treatment algorithms based on disease course. Markers such as FC have been shown to be associated with the need for colectomy in UC 77. Similarly, in a recent retrospective study, FC was shown to predict progression in CD behaviour, hospitalisation for a flare and surgery 78. There are, however, conflicting reports on its prognostic utility 79. Furthermore, the reported prognostic performance of FC is similar to that of other non-specific blood markers such as C-reactive protein (CRP). It is clear that, despite the available predictors, accurate prognostication at the time of IBD diagnosis has yet to be achieved.
Table 1. Clinical parameters that predict unfavourable inflammatory bowel disease course.
| Disease | Time frame | Predictor of unfavourable course | References |
|---|---|---|---|
| Crohn’s disease | Within 5 years of diagnosis | Age < 40 | 88 |
| Need for steroids in first flare | 88, 89 | ||
| Perianal disease | 88, 89 | ||
| Upper gastrointestinal lesions | 90 | ||
| Ileocolonic lesions | 91 | ||
| Within 10 years of diagnosis | Age < 40 | 44, 92 | |
| Upper gastrointestinal lesions | 92 | ||
| Stricturing and penetrating behaviour | 44 | ||
| Terminal ileal lesions | 44 | ||
| Ulcerative colitis | Within 5 years of diagnosis | Younger age | 90 |
| Female gender | 90 | ||
| Within 10 years of diagnosis | Younger age | 93 | |
| Female gender | 93 | ||
| Fewer systemic symptoms | 94 | ||
| Extensive colitis | 43, 94, 95 | ||
| Non-smoking status | 93 |
We are, however, beginning to unearth the molecular profiles that associate with disease progression in IBD. The seminal study by Lee et al. 80, who explored the CD8 T-cell transcriptomic profiles in newly diagnosed IBD, identified unique RNA signatures that were associated with the need for treatment escalation or surgery (or both) over time in both CD and UC. In their follow-on study, Biasci et al. developed a multi-gene signature predicting the need for escalation using original criteria in UC (hazard ratio [HR] 3.1, 95% confidence interval [CI] 1.25–7.72, P = 0.02) and CD (HR 2.7, 95% CI 1.32–5.34, P = 0.01) 81, although this profile differs from the original T-cell profile signature. Using the same criteria for escalation, the UK IBDGC identified four prognostic genetic loci: FOXO3, XACT, a region upstream of IGFBP1 and the MHC region 82. These genes were distinct from those that predict CD susceptibility. The molecular architecture of disease course has been further defined beyond genetics at a methylome, glycome and proteome level. Studies have shown that patients with an aggressive disease course display unique circulating methylome and proteome signatures 83– 85, including markers such as serum calprotectin 79, that predict treatment escalation or surgery (or both) over time. Glycomic markers have previously been shown to be associated with IBD 86 and more recently have shown the ability to predict treatment escalation 87. All of these studies have similar clinical criteria for escalation, based on step-up approach treatment algorithms. In clinical practice, tailoring early “top-down” therapies in those with disease progression while avoiding potent therapies in those with a benign disease course at diagnosis is a real unmet need. It has yet to be ascertained whether this approach will improve clinical outcomes.
Other equally relevant definitions of disease course are being studied by IBD consortia across populations. One such consortium is the Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn’s Disease (RISK) study 96. Defining aggressive disease course as a progression in CD behaviour to either penetrating or stricturing complications over time, this prospective inception cohort study identified unique multi-omic profiles that associate with disease progression. Ileal transcriptomic data showed that expression of inflammatory response to microbe signatures versus extracellular matrix upregulation signatures discriminated between later-penetrating versus stricturing complication development. The addition of ileal transcriptomic data to a clinical and serologically based competing-risk score improved the sensitivity and specificity of the score 96. The RISK study group has also shown that by integrating summary-level GWAS and expression quantitative trait loci with RNA-seq data, transcriptional risk scores can be generated which outperform genetic risk scores in identifying CD and are able to predict CD disease course over time 97. Randomised controlled trials (RCTs) are needed to determine whether early characterisation and therapy based on these profiles have the ability to alter disease course over time in paediatric CD.
Microbial populations may have a role in helping predict disease course, as illustrated by a study in post-operative recurrence in CD 98. Here, the authors demonstrated that a decreased population of Faecalibacterium prausnitzii in the resected ileum correlated with a higher rate of recurrence 98. In a study of paediatric CD, gut microbial signatures at the time of diagnosis were found to help predict 6-month steroid-free remission 99. These studies demonstrate conceptually the potential for microbiome signatures to provide clinicians with prognostic information to help inform treatment decisions, although longitudinal studies and further validation are required.
Understanding the progression of IBD at diagnosis using several distinct yet clinically important criteria at a multi-omic level will help personalise treatment algorithms based on biology rather than symptomatology with an aim to improve clinical outcomes over time. Empowering patients with this information at diagnosis may aid progress towards personalising care in IBD.
Personalising therapies in inflammatory bowel disease
The array of treatment options in IBD has grown dramatically over recent years, and a large number of therapies are in the pipeline 13. Ultimately, the goal of precision medicine is to enable preferential selection of a specific therapy based on an individual patient’s biology whilst individualising dosing to ensure that therapeutic effects are maintained and side effect risk minimised. In current clinical practice, use of therapeutic drug monitoring (TDM) “biomarkers” is well established in order to increase the chances of response and reduce risk of side effects. Despite this, it is clear that currently available TDM strategies do not completely mitigate against the risk of adverse events. Large multicentre trials such as CALM have reported an adverse event rate of up to 24 to 26% using current anti-tumour necrosis factor (anti-TNF) therapies 100.
In the case of thiopurines, we are able to predict the risk of life-threatening myelosuppression by measurement of thiopurine methyltransferase (TPMT) enabling dose reduction with intermediate and low levels at the point of treatment commencement 101. Until recently, NUDT15 polymorphisms were known to be associated with the onset of thiopurine-induced myelosuppression and were thought to be most relevant in Asian populations 102, 103. However, a recent multicentre, case control study of patients of European ancestry either affected or unaffected by thiopurine-induced myelosuppression has altered this perception 104. In that study, a GWAS and exome-wide association study (EWAS) approach identified an association between a NUDT15 variant and myelosuppression 104. Carriage of any three coding NUDT15 variants was associated with an increased risk of myelosuppression. The number needed to genotype was 95, at par with TPMT 104. These data suggest that there may be a role for NUDT15 sequencing in addition to TPMT sequencing prior to commencing thiopurines. HLA type has also been demonstrated to be a predictor of side effects in thiopurine therapy. In a multicentre study, 433 patients who had developed thiopurine-induced pancreatitis within 3 months of starting thiopurine therapy and case controls were identified. Using a GWAS approach, an association with pancreatitis development was found at rs2647047 which was found to link to HLA-DQA1 and HLA-DRB1 alleles. Compared with a baseline risk of about 4% for thiopurine-induced pancreatitis in all patients, those heterozygous for rs2647047 had a 9% risk and those who were homozygous had a 17% risk 105. During thiopurine therapy, thiopurine metabolites can be measured to determine whether a particular drug dose is optimised to produce a beneficial therapeutic effect whilst identifying poor compliance where it exists and minimising risks of side effects in an individual patient 106, 107.
Similarly, TDM for biologic therapies is increasingly embedded in clinical practice. Anti-TNF therapy monitoring with measurement of trough drug levels and detection of anti-drug antibodies allows optimisation of therapeutic effect and directs decisions to switch biologic therapy in the case of primary and secondary non-response 108. In their RCT, Steenholdt et al. compared reactive TDM for anti-TNF therapy with empiric dose escalation in the setting of secondary loss of response 109. In that RCT, TDM was associated with significant cost reductions. In the TAXIT 110 and Tailorix 111 RCTs, anti-TNF dosing based on proactive TDM was compared with dosing based on clinical features. In the TAXIT trial, there was a significantly reduced relapse rate and modest cost saving in the proactive TDM group. By contrast, the Tailorix trial found that there was no significant benefit of proactive TDM over-dosing based on clinical features. Overall, anti-TNF TDM is associated with reduced cost and improved durability of response, but there is no conclusive evidence that a proactive or reactive approach is superior 112.
Accurate pre-treatment prediction of response to biologic therapy would enable better drug selection for patients. Currently, there are no clinically available biomarkers that accurately predict treatment response, although a large number of potential candidates have been studied and extensively reviewed recently 26. Here, we report a number of new and noteworthy examples. West et al. demonstrated reproducibly in a number of data sets that high expression of oncostatin M (OSM) (a cytokine) is associated with reduced anti-TNF response 113. Verstockt et al. showed that low serum expression of Triggering Receptor Expressed on Myeloid cells 1 (TREM1) measured prior to commencing therapy is an accurate anti-TNF–specific marker for future anti-TNF response with endoscopic remission in both CD and UC 114. Telesco et al. identified and validated a 13-gene signature that predicted week 6 mucosal healing response (area under the curve receiver operating characteristics (AUC ROC) = 0.688, P = 0.002) to golimumab in the PURSUIT and PROgECT studies. The low specificity of this 13-gene signature may impact on the utility of this multi-gene panel 115. In a novel approach, Morilla et al. identified and validated a nine-microRNA signature in colonic biopsies in acute severe UC, successfully classifying responders and non-responders to corticosteroids, infliximab and cyclosporine in 90%, 84% and 80% of patients respectively 116.
Prediction of response to biologic therapy using the microbial signatures of patients with IBD has also been a focus of recent investigation. Faecal microbial diversity resembled controls in paediatric patients who responded to anti-TNF therapy compared with non-responders 117. Similarly, Ananthakrishnan et al. found differences in the distal gut microbiome of CD patients who responded to anti-integrin therapy compared with non-responders 118. The authors reported higher abundance of Roseburia inulinivorans and a Burkholderiales species as well as enrichment of 13 microbial pathways in those achieving remission. Furthermore, a neural network algorithm was able to predict drug response, although, interestingly, these findings were not apparent in UC. For ustekinumab, using 16S rRNA gene sequencing, Doherty et al. reported that at week 6 into therapy, patients in remission could be distinguished from those with active disease by characterisation of their microbiome 119. Although these studies collectively show promise as proof of concept, large-scale validation is required before these tools can become part of clinical practice.
Predicting treatment failure is equally important to potentially facilitate early switching of therapy and increase the likelihood and cost-effectiveness of recapturing response. Recently, results from the Personalised Anti-TNF Therapy in Crohn’s Disease Study (PANTS) were published 120. This multicentre, prospective, observational cohort study included 1610 patients from the point of first anti-TNF therapy exposure and followed them up for 12 months or until withdrawal of anti-TNF therapy. Low drug concentrations at week 14 were associated with primary non-response and non-remission at week 54. Obesity at baseline was associated with non-remission at week 54 for adalimumab only. Immunogenicity by week 54 was seen more commonly in patients on infliximab than on adalimumab (62.8% versus 28.5% respectively). Drug concentrations at week 14 were an independent risk factor for immunogenicity for both drugs. Obesity in the case of adalimumab and smoking in the case of infliximab were also associated with immunogenicity. Immunomodulator use was the main protective factor, and thiopurine-related reduction in immunogenicity occurred in a dose-dependent fashion. The PANTS study group has also presented data showing that HLA variants determine anti-TNF therapy immunogenicity and HLA-DQ1*05 genotype was a key factor 121. Attention has also focussed on the role of glycosylation. This complex post-translational modification of proteins affects their function and can be significantly altered in inflammation 91. Pereira et al. recently demonstrated that lower levels of branched N-glycans in colonic biopsies from a cohort of 131 patients with UC were predictive of failure to respond to standard therapy (5-ASA, corticosteroids and immunomodulators) 122. The authors also demonstrated that the sensitivity and specificity of the predictive effect were different at time of diagnosis, within 5 months of diagnosis, and at time of diagnosis in patients presenting with severe disease. The only other independent predictor of failure of standard therapy was demonstrated to be high CRP. When low levels of branched N-glycans and high CRP levels were combined in all UC versus severe UC patients at the time of diagnosis, the probability of failure of standard therapy was 46.6% versus 76.5% respectively, sensitivity was 69% versus 88% respectively, and specificity was 80% versus 75% 122.
More recent studies have used single-cell gene expression technologies in order to better understand the molecular pathophysiology of drug response. In active UC, there appears to be a differential reduction in epithelial mitochondrial genes in active UC, including the known master regulator of mitochondria biogenesis PPARGC1A ( PGC1α) and epithelial mitochondrial membrane potential ( MMP) 123. Intestinal epithelial depletion of PGC1α has been shown to negatively impact on mitochondrial function and subsequent epithelial barrier; barrier dysfunction is a feature of UC pathogenesis 124. This study also identified a gene signature that associates with anti-TNF and anti-integrin therapy response 123. In CD, a recent study investigating cellular subsets in ileal CD identified a unique cellular module in inflamed tissue that includes IgG plasma cells, mononuclear phagocytes, activated T cells and stromal cells (GIMATS module) 125. The presence of this module in patients at diagnosis was associated with a failure to achieve durable corticosteroid remission with anti-TNF therapy 125.
Monitoring response to therapy
Disease monitoring and “treat to target” (that is, using predefined outcome measures, such as clinical or endoscopic remission, as goals for the optimisation of therapy) are increasingly being recognised as a “gold standard” in IBD care. Mucosal healing is now considered a robust target and a clinical end-point for more recent drug trials 126, 127. In CD, the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) programme set out guidelines for the “treat to target” approach in order to obtain clinical and endoscopic remission 128. STRIDE recommended the use of FC and CRP to monitor response; however, until recently, there was no consensus on how to use these markers to adjust therapies. Following this, the CALM trial tested the efficacy of tight control management using 3-monthly biomarkers versus conventional management using Crohn’s Disease Activity Index on clinical and endoscopic outcomes in moderate to severe CD 100. Significantly higher numbers of patients achieved mucosal healing in the “tight control” arm compared with the conventional group. Furthermore, follow-up data from this study showed that patients with disease monitoring using biomarkers and a goal of achieving mucosal healing in the first year of treatment were less likely to have disease progression over a median of 3 years 129.
Monitoring response to therapy has been studied using conventional biomarkers such as FC and CRP. Heida et al., in a systematic analysis of six studies, showed that two consecutive elevated FC predicted disease relapse within the following 2 to 3 months 130. However, the study was not able to define an optimal FC cutoff for monitoring. An FC cutoff of not more than 250 μg/g was shown to predict endoscopic remission (positive predictive value 48.5% and NPV 96.6%) 131. Similarly, CRP has been shown to correlate with clinical disease activity and endoscopic inflammation. However, CRP is a non-specific inflammatory marker and can often be raised in other clinical scenarios such as infections. Furthermore, some patients with active disease can have normal CRP levels 132.
An area of intense interest is how best to monitor response with therapeutic de-escalation 133. The most robust study of anti-TNF de-escalation was the STORI trial, in which Louis et al. prospectively followed 115 patients with CD on combination therapy who discontinued the anti-TNF after at least 6 months of corticosteroid-free clinical remission 134. The investigators found that those who experienced a relapse and those who did not had different evolution of CRP and FC with a marked and sudden rise in these biomarkers 4 months prior to relapse 135. Several studies have explored other biomarkers of response and, in particular, mucosal healing. In a study looking at blood transcriptional biomarkers, initial whole blood microarray analysis was performed by using Affymetrix GeneChip technology to identify changes in transcriptional gene expression in endoscopically active UC compared with normal controls 136. After quantitative reverse transcription polymerase chain reaction (RT-PCR) validation, transcription of neutrophil-related genes, including CD177, haptoglobin, G protein–coupled receptor 84, and S100 calcium-binding protein A12, were demonstrated to be most associated with endoscopic disease activity. The above factors were also shown to decrease in association with a reduction in endoscopic disease activity after 14 weeks of anti-TNF treatment in a further group of patients with UC.
Glycoprotein acetylation (GlycA) is another example of a novel biomarker. In a prospective pilot study of patients with IBD, serum GlycA levels were measured via high-throughput nuclear magnetic resonance spectroscopy to investigate its role as a disease activity biomarker. GlycA was shown to normalise to healthy control levels in patients achieving mucosal healing and to significantly decrease in patients who had evidence of improvements in endoscopic appearances regardless of the treatment used. GlycA was also shown to be a useful marker of response in patients with normal CRP levels at baseline. When compared with FC and CRP, GlycA was the only marker to show consistent significant differences between responders and non-responders regardless of the treatment given. GlycA may have advantages over CRP as it is a composite marker originating from a number of acute-phase proteins and so is more stable than CRP. The cost of GlycA measurement was reported to be comparable to that of FC 137.
A number of studies have also investigated the applicability of faecal immunochemical testing (FIT) to assess disease activity and mucosal healing in UC. In a recent systematic review, the pooled sensitivity and specificity of FIT to predict mucosal healing were found to be 0.77 (95% CI 0.72–0.81) and 0.81 (95% CI 0.76–0.85) respectively, which were similar to previously published sensitivities and specificities of FC in predicting mucosal healing 138. In a study comparing sequential FIT and FC measurements in 84 UC patients having more than one colonoscopy over a 33-month period, the accuracy of assessment of disease activity and mucosal healing was studied 139. Where the previous colonoscopy and faecal markers had previously demonstrated mucosal healing, there was increased accuracy of FIT versus FC in predicting mucosal healing at the next colonoscopy. Where the previous colonoscopy and faecal markers had previously shown active inflammation, there was again increased accuracy of FIT versus FC in predicting mucosal healing at the next colonoscopy. However, where there was inflammation seen on both colonoscopies, FC better reflected changes in endoscopic activity than FIT. This interesting study suggests that FIT and FC provide slightly different information in disease monitoring in UC and that measuring both may be of benefit.
Given the well-characterised changes in the microbiome of patients with IBD, restoration of the microbial diversity of patients has been explored as a potential biomarker to assess response to therapy. Restoration of gut microbial diversity has been seen in response to anti-TNF therapy 140. In a study of paediatric IBD, Shaw et al. found that dysbiosis was correlated with inflammatory burden and that in UC, but not in CD, improvements in faecal diversity correlated with clinical response 141.
When taken together, the above studies demonstrate the potential utility of traditional and novel biomarkers as surrogates for endoscopic healing and indicators of pre-clinical relapse. Currently, it seems clear that a combination of clinically available biomarkers should be used to monitor disease activity and response to therapy. There is, however, a need for novel specific biomarkers to translate to clinical practice.
Future challenges and horizon of precision medicine in inflammatory bowel disease
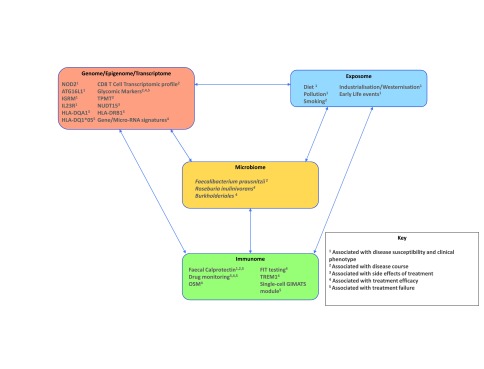
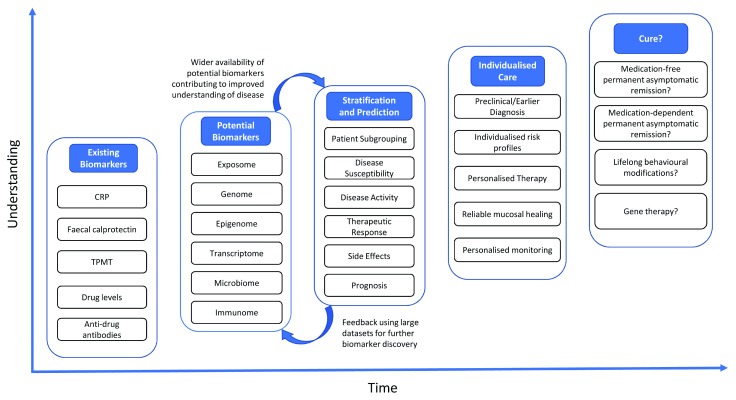
Substantial progress has been made in our ability to treat CD and UC. With the increasing array of treatment options available, the need for precision medicine in IBD as a whole is paramount. Currently available biomarkers have improved our ability to assess and monitor disease activity and therapeutic efficacy, but each has its own limitations. The aim of precision medicine in IBD is to deliver truly individualised care so that the entire patient journey from diagnosis to treatment is based on the specific biology underlying IBD in the individual patient. The current progress towards precision medicine and likely complex inter-relationships of these multi-omic data in IBD are depicted in Figure 1. Whilst there has been progress, there is still a significant path ahead before precision medicine is achieved in IBD. Our proposed pathway to precision medicine in IBD is shown in Figure 2. As our understanding improves, through using current and novel biomarkers, we will be able to better predict and stratify disease which will positively feedback into an improved ability to identify areas for further biomarker and therapeutic development. This iterative approach will increase our ability to precisely target an individual patient’s inflammation on the basis of their molecular profile, allowing matching of the patient to the most appropriate therapy and monitoring. The overall aim will be to eventually achieve cure rather than simply mucosal or histological healing.
Figure 1. Current progress towards precision medicine in inflammatory bowel disease.
This figure summarises some of the progress that has been made towards precision medicine in inflammatory bowel disease and the likely complex inter-relationship of multi-omic data.
Figure 2. The evolution of precision medicine in inflammatory bowel disease.
CRP, C-reactive protein; TPMT, thiopurine methyltransferase.
There remain many challenges in order to deliver effective precision medicine in IBD. With the need to look towards big data approaches to make progress in biomarker discovery and improving outcomes in IBD, new approaches to high-quality data sharing are required 24. This will require infrastructural changes involving sharing of data between research groups and eventually involving the use of electronic health records. The eventual goal would be to have electronic health records that are able to integrate seamlessly with large clinical and research databases to facilitate early adoption of advances in knowledge whilst providing real-time, up-to-date support to clinicians in making decisions regarding their patients 142. There will also need to be new approaches to how data are analysed to maximise their effect. With large amounts of data, there will be an increasing reliance on complex algorithms, including the use of machine learning to make sense of big data coming from multiple sources 143, 144. However, there must be stringent checks to ensure that the quality of the data is maintained and that the hypotheses generated are subjected to rigorous validation through pre-clinical and real-world trials prior to adoption in wider medical practice 24. One of the biggest barriers to precision medicine in IBD is the lack of a clearly defined mechanism (or mechanisms) of disease 145. As we discover new biomarkers and learn new ways of practicing medicine in IBD more precisely, we must use these to interrogate the existing understanding and challenge existing pathophysiological models.
However, there are grounds for significant optimism. A number of ongoing studies will advance our understanding of precision medicine in IBD. The Predicting outcomes For Crohn’s disease using a molecular biomarker (PROFILE) trial (ISRCTN11808228) is the first biomarker-stratified trial in IBD 146. It is currently recruiting and aims to recruit 400 patients from about 50 centres across the UK. In that trial, a gene expression signature previously found to determine more aggressive disease 80 will be tested for by means of a whole blood quantitative polymerase chain reaction assay that has been previously validated 147. Trial participants in each biomarker group will be randomly assigned 1:1 to receive accelerated step-up therapy or top-down therapy. The primary outcome measure for that trial will be sustained surgery and steroid-free remission, and one of the secondary outcome measures will be mucosal healing. The PRognostic effect of Environmental factors in Crohn’s and Colitis (PREdiCCt) Study (ISRCTN67248113) is also currently recruiting and aims to recruit a total of 3100 patients with CD or UC in remission. The study aims to follow these patients over two years and will collect information on diet, lifestyle and gut bacteria to see how these are associated with IBD flare and recovery.
Following the efforts of several consortia to generate multi-omic datasets, there is now a need to define the IBD “interactome”: a disease network where perturbations in individual -omes cause intestinal inflammation, mediated by dysfunctional molecular modules 11. There are now several systems biology software tools that facilitate multi-level data integration such as iCluster 148 that identifies patient subtypes within IBD on the basis of multi-omic high-throughput data while other tools allow visualisation of the interactome such as Cytoscape 149. Methodologies that facilitate data integration are comprehensively summarised in a recent review 11. Future drug innovations based primarily on IBD network interactions will transform the field of therapeutics in IBD 11.
Conclusions
Significant progress has been made towards the goal of precision medicine in IBD, but clearly there is still a substantial unmet need for further biomarkers with which to stratify patients and their treatment. It is vital that we continue to develop existing practice by assimilating advances in knowledge whilst optimising novel and existing biomarkers to continually make advances towards truly individualised care. Precision medicine in IBD will require not only the knowledge but the tools and the infrastructure with which to implement it effectively. This will require coordinated investment in technology to enable seamless sharing of big data and the ability to perform high-throughput analysis to create a continually learning IBD community. The recent announcement of the HDRUK G.I. Know Health Data Research Hub for IBD across the UK means that positive steps are already being taken towards integrated big data discovery to advance the field of precision medicine in IBD ( https://www.hdruk.ac.uk/infrastructure/the-hubs/g-i-know/). Precision medicine has huge potential to impact on outcomes in IBD. We all have a responsibility to play our part in practicing high-quality biomarker-driven IBD care whilst contributing to big data in IBD so that in the future we will be able to offer truly individualised IBD care.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Claudio Fiocchi, Department of Inflammation and Immunity, Lerner Research Institute, Department of Gastroenterology and Hepatology, Cleveland Clinic, Cleveland, OH, USA
Sunanda Kane, Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; peer review: 2 approved]
References
- 1. Torres J, Mehandru S, Colombel JF, et al. : Crohn's disease. Lancet. 2017;389(10080):1741–55. 10.1016/S0140-6736(16)31711-1 [DOI] [PubMed] [Google Scholar]
- 2. Ungaro R, Mehandru S, Allen PB, et al. : Ulcerative colitis. Lancet. 2017;389(10080):1756–70. 10.1016/S0140-6736(16)32126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Molodecky NA, Soon IS, Rabi DM, et al. : Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42; quiz e30. 10.1053/j.gastro.2011.10.001 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Ng SC, Shi HY, Hamidi N, et al. : Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390(10114):2769–78. 10.1016/S0140-6736(17)32448-0 [DOI] [PubMed] [Google Scholar]
- 5. Sýkora J, Pomahačová R, Kreslová M, et al. : Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol. 2018;24(25):2741–63. 10.3748/wjg.v24.i25.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xavier RJ, Podolsky DK: Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–34. 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Verstockt B, Smith KG, Lee JC: Genome-wide association studies in Crohn's disease: Past, present and future. Clin Transl Immunology. 2018;7(1):e1001. 10.1002/cti2.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, et al. : Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15(1):39–49. 10.1038/nrgastro.2017.136 [DOI] [PubMed] [Google Scholar]
- 9. McIlroy J, Ianiro G, Mukhopadhya I, et al. : Review article: the gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment Pharmacol Ther. 2018;47(1):26–42. 10.1111/apt.14384 [DOI] [PubMed] [Google Scholar]
- 10. de Souza HS, Fiocchi C: Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. 10.1038/nrgastro.2015.186 [DOI] [PubMed] [Google Scholar]
- 11. de Souza HSP, Fiocchi C, Iliopoulos D: The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. 2017;14(12):739–49. 10.1038/nrgastro.2017.110 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Paramsothy S, Rosenstein AK, Mehandru S, et al. : The current state of the art for biological therapies and new small molecules in inflammatory bowel disease. Mucosal Immunol. 2018;11(6):1558–70. 10.1038/s41385-018-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weisshof R, El Jurdi K, Zmeter N, et al. : Emerging Therapies for Inflammatory Bowel Disease. Adv Ther. 2018;35(11):1746–62. 10.1007/s12325-018-0795-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verstockt B, Ferrante M, Vermeire S, et al. : New treatment options for inflammatory bowel diseases. J Gastroenterol. 2018;53(5):585–90. 10.1007/s00535-018-1449-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Odes S: How expensive is inflammatory bowel disease? A critical analysis. World J Gastroenterol. 2008;14(43):6641–7. 10.3748/wjg.14.6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reinglas J, Gonczi L, Kurt Z, et al. : Positioning of old and new biologicals and small molecules in the treatment of inflammatory bowel diseases. World J Gastroenterol. 2018;24(32):3567–82. 10.3748/wjg.v24.i32.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burisch J, Jess T, Martinato M, et al. : The burden of inflammatory bowel disease in Europe. J Crohns Colitis. 2013;7(4):322–37. 10.1016/j.crohns.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 18. Bähler C, Vavricka SR, Schoepfer AM, et al. : Trends in prevalence, mortality, health care utilization and health care costs of Swiss IBD patients: a claims data based study of the years 2010, 2012 and 2014. BMC Gastroenterol. 2017;17(1):138. 10.1186/s12876-017-0681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peery AF, Crockett SD, Murphy CC, et al. : Burden and Cost of Gastrointestinal, Liver, and Pancreatic Diseases in the United States: Update 2018. Gastroenterology. 2019;156(1):254–272.e11. 10.1053/j.gastro.2018.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coward S, Clement F, Benchimol EI, et al. : Past and Future Burden of Inflammatory Bowel Diseases Based on Modeling of Population-Based Data. Gastroenterology. 2019;156(5):1345–1353.e4. 10.1053/j.gastro.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 21. Singh S, George J, Boland BS, et al. : Primary Non-Response to Tumor Necrosis Factor Antagonists is Associated with Inferior Response to Second-line Biologics in Patients with Inflammatory Bowel Diseases: A Systematic Review and Meta-analysis. J Crohns Colitis. 2018;12(6):635–43. 10.1093/ecco-jcc/jjy004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Lee JC: Predicting the course of IBD: light at the end of the tunnel? Dig Dis. 2012;30 Suppl 1:95–9. 10.1159/000341132 [DOI] [PubMed] [Google Scholar]
- 23. Lewis JD: The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140(6):1817–1826.e2. 10.1053/j.gastro.2010.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olivera P, Danese S, Jay N, et al. : Big data in IBD: a look into the future. Nat Rev Gastroenterol Hepatol. 2019;16(5):312–21. 10.1038/s41575-019-0102-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Viennois E, Zhao Y, Merlin D: Biomarkers of Inflammatory Bowel Disease: From Classical Laboratory Tools to Personalized Medicine. Inflamm Bowel Dis. 2015;21(10):2467–74. 10.1097/MIB.0000000000000444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stevens TW, Matheeuwsen M, Lönnkvist MH, et al. : Systematic review: predictive biomarkers of therapeutic response in inflammatory bowel disease-personalised medicine in its infancy. Aliment Pharmacol Ther. 2018;48(11–12):1213–31. 10.1111/apt.15033 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Orholm M, Fonager K, Sørensen HT: Risk of ulcerative colitis and Crohn's disease among offspring of patients with chronic inflammatory bowel disease. Am J Gastroenterol. 1999;94(11):3236–8. 10.1111/j.1572-0241.1999.01526.x [DOI] [PubMed] [Google Scholar]
- 28. Spehlmann ME, Begun AZ, Burghardt J, et al. : Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14(7):968–76. 10.1002/ibd.20380 [DOI] [PubMed] [Google Scholar]
- 29. Moller FT, Andersen V, Wohlfahrt J, et al. : Familial risk of inflammatory bowel disease: a population-based cohort study 1977-2011. Am J Gastroenterol. 2015;110(4):564–71. 10.1038/ajg.2015.50 [DOI] [PubMed] [Google Scholar]
- 30. McGovern DP, Kugathasan S, Cho JH: Genetics of Inflammatory Bowel Diseases. Gastroenterology. 2015;149(5):1163–1176.e2. 10.1053/j.gastro.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos GP, Papadakis KA: Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin Proc. 2019;94(1):155–65. 10.1016/j.mayocp.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhernakova A, van Diemen CC, Wijmenga C: Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10(1):43–55. 10.1038/nrg2489 [DOI] [PubMed] [Google Scholar]
- 33. Zhao M, Burisch J: Impact of Genes and the Environment on the Pathogenesis and Disease Course of Inflammatory Bowel Disease. Dig Dis Sci. 2019;64(7):1759–69. 10.1007/s10620-019-05648-w [DOI] [PubMed] [Google Scholar]
- 34. Kaplan GG: The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–7. 10.1038/nrgastro.2015.150 [DOI] [PubMed] [Google Scholar]
- 35. Rigottier-Gois L: Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 2013;7(7):1256–61. 10.1038/ismej.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. : Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–62. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park JH, Peyrin-Biroulet L, Eisenhut M, et al. : IBD immunopathogenesis: A comprehensive review of inflammatory molecules. Autoimmun Rev. 2017;16(4):416–26. 10.1016/j.autrev.2017.02.013 [DOI] [PubMed] [Google Scholar]
- 38. Rescigno M, Di Sabatino A: Dendritic cells in intestinal homeostasis and disease. J Clin Invest. 2009;119(9):2441–50. 10.1172/JCI39134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahluwalia B, Moraes L, Magnusson MK, et al. : Immunopathogenesis of inflammatory bowel disease and mechanisms of biological therapies. Scand J Gastroenterol. 2018;53(4):379–89. 10.1080/00365521.2018.1447597 [DOI] [PubMed] [Google Scholar]
- 40. National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease: Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. The National Academies Collection: Reports funded by National Institutes of Health. Washington (DC): National Academies Press (US) National Academy of Sciences.2011. [PubMed] [Google Scholar]
- 41. Ginsburg GS, Phillips KA: Precision Medicine: From Science To Value. Health Aff (Millwood). 2018;37(5):694–701. 10.1377/hlthaff.2017.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Collins FS, Varmus H: A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. 10.1056/NEJMp1500523 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Solberg IC, Lygren I, Jahnsen J, et al. : Clinical course during the first 10 years of ulcerative colitis: results from a population-based inception cohort (IBSEN Study). Scand J Gastroenterol. 2009;44(4):431–40. 10.1080/00365520802600961 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Solberg IC, Vatn MH, Høie O, et al. : Clinical course in Crohn's disease: results of a Norwegian population-based ten-year follow-up study. Clin Gastroenterol Hepatol. 2007;5(12):1430–8. 10.1016/j.cgh.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 45. Beaugerie L, Sokol H: Clinical, serological and genetic predictors of inflammatory bowel disease course. World J Gastroenterol. 2012;18(29):3806–13. 10.3748/wjg.v18.i29.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peyrin-Biroulet L, Panés J, Sandborn WJ, et al. : Defining Disease Severity in Inflammatory Bowel Diseases: Current and Future Directions. Clin Gastroenterol Hepatol. 2016;14(3):348–354.e17. 10.1016/j.cgh.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 47. de Lange KM, Moutsianas L, Lee JC, et al. : Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–61. 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hong M, Ye BD, Yang SK, et al. : Immunochip Meta-Analysis of Inflammatory Bowel Disease Identifies Three Novel Loci and Four Novel Associations in Previously Reported Loci. J Crohns Colitis. 2018;12(6):730–41. 10.1093/ecco-jcc/jjy002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Liu JZ, van Sommeren S, Huang H, et al. : Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Jostins L, Ripke S, Weersma RK, et al. : Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–24. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Girardin SE, Hugot JP, Sansonetti PJ: Lessons from Nod2 studies: towards a link between Crohn's disease and bacterial sensing. Trends Immunol. 2003;24(12):652–8. 10.1016/j.it.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 52. Abreu MT, Taylor KD, Lin YC, et al. : Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123(3):679–88. 10.1053/gast.2002.35393 [DOI] [PubMed] [Google Scholar]
- 53. Lesage S, Zouali H, Cézard JP, et al. : CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70(4):845–57. 10.1086/339432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cuthbert AP, Fisher SA, Mirza MM, et al. : The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122(4):867–74. 10.1053/gast.2002.32415 [DOI] [PubMed] [Google Scholar]
- 55. Cleynen I, González JR, Figueroa C, et al. : Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut. 2013;62(11):1556–65. 10.1136/gutjnl-2011-300777 [DOI] [PubMed] [Google Scholar]
- 56. Salem M, Ammitzboell M, Nys K, et al. : ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy. 2015;11(4):585–94. 10.1080/15548627.2015.1017187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fowler EV, Doecke J, Simms LA, et al. : ATG16L1 T300A shows strong associations with disease subgroups in a large Australian IBD population: further support for significant disease heterogeneity. Am J Gastroenterol. 2008;103(10):2519–26. [DOI] [PubMed] [Google Scholar]
- 58. Latiano A, Palmieri O, Cucchiara S, et al. : Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am J Gastroenterol. 2009;104(1):110–6. [DOI] [PubMed] [Google Scholar]
- 59. Deepak P, Sandborn WJ: Ustekinumab and Anti-Interleukin-23 Agents in Crohn's Disease. Gastroenterol Clin North Am. 2017;46(3):603–26. 10.1016/j.gtc.2017.05.013 [DOI] [PubMed] [Google Scholar]
- 60. Duerr RH, Taylor KD, Brant SR, et al. : A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–3. 10.1126/science.1135245 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Okazaki T, Wang MH, Rawsthorne P, et al. : Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn's disease risk in a population-based case-control study: evidence of gene-gene interactions. Inflamm Bowel Dis. 2008;14(11):1528–41. 10.1002/ibd.20512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cleynen I, Boucher G, Jostins L, et al. : Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387(10014):156–67. 10.1016/S0140-6736(15)00465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Gevers D, Kugathasan S, Denson LA, et al. : The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Pascal V, Pozuelo M, Borruel N, et al. : A microbial signature for Crohn's disease. Gut. 2017;66(5):813–22. 10.1136/gutjnl-2016-313235 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Papa E, Docktor M, Smillie C, et al. : Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7(6):e39242. 10.1371/journal.pone.0039242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Somineni HK, Kugathasan S: The Microbiome in Patients With Inflammatory Diseases. Clin Gastroenterol Hepatol. 2019;17(2):243–55. 10.1016/j.cgh.2018.08.078 [DOI] [PubMed] [Google Scholar]
- 67. van Rheenen PF, Van de Vijver E, Fidler V: Faecal calprotectin for screening of patients with suspected inflammatory bowel disease: diagnostic meta-analysis. BMJ. 2010;341:c3369. 10.1136/bmj.c3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kennedy NA, Clark A, Walkden A, et al. : Clinical utility and diagnostic accuracy of faecal calprotectin for IBD at first presentation to gastroenterology services in adults aged 16–50 years. J Crohns Colitis. 2015;9(1):41–9. 10.1016/j.crohns.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. NICE: DG11: Faecal calprotectin diagnostic tests for inflammatory diseases of the bowel. London: NICE;2013. Reference Source [Google Scholar]
- 70. Maréchal C, Aimone-Gastin I, Baumann C, et al. : Compliance with the faecal calprotectin test in patients with inflammatory bowel disease. United European Gastroenterol J. 2017;5(5):702–7. 10.1177/2050640616686517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kalla R, Boyapati R, Vatn S, et al. : Patients’ perceptions of faecal calprotectin testing in inflammatory bowel disease: Results from a prospective multicentre patient-based survey. Scand J Gastroenterol. 2018;53(12):1437–42. 10.1080/00365521.2018.1527394 [DOI] [PubMed] [Google Scholar]
- 72. Buisson A, Gonzalez F, Poullenot F, et al. : Comparative Acceptability and Perceived Clinical Utility of Monitoring Tools: A Nationwide Survey of Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23(8):1425–33. 10.1097/MIB.0000000000001140 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Calafat M, Cabré E, Mañosa M, et al. : High within-day variability of fecal calprotectin levels in patients with active ulcerative colitis: what is the best timing for stool sampling? Inflamm Bowel Dis. 2015;21(5):1072–6. 10.1097/MIB.0000000000000349 [DOI] [PubMed] [Google Scholar]
- 74. Du L, Foshaug R, Huang VW, et al. : Within-Stool and Within-Day Sample Variability of Fecal Calprotectin in Patients With Inflammatory Bowel Disease: A Prospective Observational Study. J Clin Gastroenterol. 2018;52(3):235–40. 10.1097/MCG.0000000000000776 [DOI] [PubMed] [Google Scholar]
- 75. Satsangi J, Kitten O, Chavez M, et al. : How to Apply for and Secure EU Funding for Collaborative IBD Research Projects. J Crohns Colitis. 2016;10(3):363–70. 10.1093/ecco-jcc/jjv237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vermeire S, Van Assche G, Rutgeerts P: Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut. 2006;55(3):426–31. 10.1136/gut.2005.069476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ho GT, Lee HM, Brydon G, et al. : Fecal calprotectin predicts the clinical course of acute severe ulcerative colitis. Am J Gastroenterol. 2009;104(3):673–8. [DOI] [PubMed] [Google Scholar]
- 78. Kennedy NA, Jones GR, Plevris N, et al. : Association Between Level of Fecal Calprotectin and Progression of Crohn's Disease. Clin Gastroenterol Hepatol. 2019;17(11):2269–2276.e4. 10.1016/j.cgh.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kalla R, Kennedy NA, Ventham NT, et al. : Serum Calprotectin: A Novel Diagnostic and Prognostic Marker in Inflammatory Bowel Diseases. Am J Gastroenterol. 2016;111(12):1796–805. 10.1038/ajg.2016.342 [DOI] [PubMed] [Google Scholar]
- 80. Lee JC, Lyons PA, McKinney EF, et al. : Gene expression profiling of CD8 + T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest. 2011;121(10):4170–9. 10.1172/JCI59255 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Biasci D, Lee JC, Noor NM, et al. : A blood-based prognostic biomarker in IBD. Gut. 2019;68(8):1386–95. 10.1136/gutjnl-2019-318343 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Lee JC, Biasci D, Roberts R, et al. : Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn's disease. Nat Genet. 2017;49(2):262–8. 10.1038/ng.3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kalla R, Adams A, Vatn S, et al. : Proximity Extension Assay based Proteins Show Immune Cell Specificity and can Diagnose and Predict Outcomes in Inflammatory Bowel Diseases: IBD Character Study. Gastroenterology. 2017;152:S606–S607. 10.1016/S0016-5085(17)32161-3 [DOI] [Google Scholar]
- 84. Kalla R, Adams A, Vatn S, et al. : OC-047 Epigenetic alterations at diagnosis predict susceptibility, prognosis and treatment escalation in inflammatory bowel disease – ibd character. Gut. 2017;66(Suppl 2):A24–A5. 10.1136/gutjnl-2017-314472.47 [DOI] [Google Scholar]
- 85. Ventham NT, Kennedy NA, Adams AT, et al. : Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun. 2016;7:13507. 10.1038/ncomms13507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Trbojević Akmačić I, Ventham NT, Theodoratou E, et al. : Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm Bowel Dis. 2015;21(6):1237–47. 10.1097/MIB.0000000000000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shubhakar A, Jansen B, Adams A, et al. : DOP10 Serum N-glycomic biomarkers predict treatment escalation in inflammatory bowel disease. J Crohns Colitis. 2019;13:S032–S033. 10.1093/ecco-jcc/jjy222.045 [DOI] [PubMed] [Google Scholar]
- 88. Beaugerie L, Seksik P, Nion-Larmurier I, et al. : Predictors of Crohn's disease. Gastroenterology. 2006;130(3):650–6. 10.1053/j.gastro.2005.12.019 [DOI] [PubMed] [Google Scholar]
- 89. Loly C, Belaiche J, Louis E: Predictors of severe Crohn's disease. Scand J Gastroenterol. 2009;43(8):948–54. 10.1080/00365520801957149 [DOI] [PubMed] [Google Scholar]
- 90. Henriksen M, Jahnsen J, Lygren I, et al. : Are there any differences in phenotype or disease course between familial and sporadic cases of inflammatory bowel disease? Results of a population-based follow-up study. Am J Gastroenterol. 2007;102(9):1955–63. [DOI] [PubMed] [Google Scholar]
- 91. Clerc F, Novokmet M, Dotz V, et al. : Plasma N-Glycan Signatures Are Associated With Features of Inflammatory Bowel Diseases. Gastroenterology. 2018;155(3):829–43. 10.1053/j.gastro.2018.05.030 [DOI] [PubMed] [Google Scholar]
- 92. Wolters FL, Russel MG, Sijbrandij J, et al. : Phenotype at diagnosis predicts recurrence rates in Crohn's disease. Gut. 2005;55(8):1124–30. 10.1136/gut.2005.084061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Höie O, Wolters F, Riis L, et al. : Ulcerative colitis: patient characteristics may predict 10-yr disease recurrence in a European-wide population-based cohort. Am J Gastroenterol. 2007;102(8):1692–701. [DOI] [PubMed] [Google Scholar]
- 94. Langholz E, Munkholm P, Davidsen M, et al. : Course of ulcerative colitis: analysis of changes in disease activity over years. Gastroenterology. 1994;107(1):3–11. 10.1016/0016-5085(94)90054-x [DOI] [PubMed] [Google Scholar]
- 95. Hoie O, Wolters FL, Riis L, et al. : Low colectomy rates in ulcerative colitis in an unselected European cohort followed for 10 years. Gastroenterology. 2007;132(2):507–5. 10.1053/j.gastro.2006.11.015 [DOI] [PubMed] [Google Scholar]
- 96. Kugathasan S, Denson LA, Walters TD, et al. : Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017;389:1710–8. 10.1016/S0140-6736(17)30317-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Marigorta UM, Denson LA, Hyams JS, et al. : Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn's disease. Nat Genet. 2017;49(10):1517–21. 10.1038/ng.3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sokol H, Pigneur B, Watterlot L, et al. : Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 99. Haberman Y, Tickle TL, Dexheimer PJ, et al. : Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124(8):3617–33. 10.1172/JCI75436 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Colombel JF, Panaccione R, Bossuyt P, et al. : Effect of tight control management on Crohn's disease (CALM): a multicentre, randomised, controlled phase 3 trial. Lancet. 2018;390(10114):2779–89. 10.1016/S0140-6736(17)32641-7 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Dean L: Azathioprine Therapy and TPMT Genotype. In: Pratt V, McLeod H, Rubinstein W, Dean L, Kattman B, Malheiro A, editors. Medical Genetics Summaries.Bethesda (MD): National Center for Biotechnology Information (US).2012. [PubMed] [Google Scholar]
- 102. Yang SK, Hong M, Baek J, et al. : A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46(9):1017–20. 10.1038/ng.3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Moriyama T, Nishii R, Perez-Andreu V, et al. : NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48(4):367–73. 10.1038/ng.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Walker GJ, Harrison JW, Heap GA, et al. : Association of Genetic Variants in NUDT15 With Thiopurine-Induced Myelosuppression in Patients With Inflammatory Bowel Disease. JAMA. 2019;321(8):773–785. 10.1001/jama.2019.0709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Heap GA, Weedon MN, Bewshea CM, et al. : HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat Genet. 2014;46(10):1131–4. 10.1038/ng.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Warner B, Johnston E, Arenas-Hernandez M, et al. : A practical guide to thiopurine prescribing and monitoring in IBD. Frontline Gastroenterol. 2018;9(1):10–5. 10.1136/flgastro-2016-100738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. de Boer NKH, Peyrin-Biroulet L, Jharap B, et al. : Thiopurines in Inflammatory Bowel Disease: New Findings and Perspectives. J Crohns Colitis. 2018;12(5):610–20. 10.1093/ecco-jcc/jjx181 [DOI] [PubMed] [Google Scholar]
- 108. Mitrev N, Vande Casteele N, Seow CH, et al. : Review article: consensus statements on therapeutic drug monitoring of anti-tumour necrosis factor therapy in inflammatory bowel diseases. Aliment Pharmacol Ther. 2017;46(11–12):1037–53. 10.1111/apt.14368 [DOI] [PubMed] [Google Scholar]
- 109. Steenholdt C, Brynskov J, Thomsen OØ, et al. : Individualised therapy is more cost-effective than dose intensification in patients with Crohn's disease who lose response to anti-TNF treatment: a randomised, controlled trial. Gut. 2014;63(6):919–27. 10.1136/gutjnl-2013-305279 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 110. Vande Casteele N, Ferrante M, Van Assche G, et al. : Trough concentrations of infliximab guide dosing for patients with inflammatory bowel disease. Gastroenterology. 2015;148(7):1320–1329.e3. 10.1053/j.gastro.2015.02.031 [DOI] [PubMed] [Google Scholar]
- 111. D’Haens G, Vermeire S, Lambrecht G, et al. : Increasing Infliximab Dose Based on Symptoms, Biomarkers, and Serum Drug Concentrations Does Not Increase Clinical, Endoscopic, and Corticosteroid-Free Remission in Patients With Active Luminal Crohn’s Disease. Gastroenterology. 2018;154(5):1343–1351.e1. 10.1053/j.gastro.2018.01.004 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Ricciuto A, Dhaliwal J, Walters TD, et al. : Clinical Outcomes With Therapeutic Drug Monitoring in Inflammatory Bowel Disease: A Systematic Review With Meta-Analysis. J Crohns Colitis. 2018;12(11):1302–15. 10.1093/ecco-jcc/jjy109 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. West NR, Hegazy AN, Owens BMJ, et al. : Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–89. 10.1038/nm.4307 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Verstockt B, Verstockt S, Dehairs J, et al. : Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine. 2019;40:733–42. 10.1016/j.ebiom.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 115. Telesco SE, Brodmerkel C, Zhang H, et al. : Gene Expression Signature for Prediction of Golimumab Response in a Phase 2a Open-Label Trial of Patients With Ulcerative Colitis. Gastroenterology. 2018;155(4):1008–1011.e8. 10.1053/j.gastro.2018.06.077 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 116. Morilla I, Uzzan M, Laharie D, et al. : Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17(5):905–13. 10.1016/j.cgh.2018.08.068 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 117. Kolho KL, Korpela K, Jaakkola T, et al. : Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am J Gastroenterol. 2015;110(6):921–30. 10.1038/ajg.2015.149 [DOI] [PubMed] [Google Scholar]
- 118. Ananthakrishnan AN, Luo C, Yajnik V, et al. : Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017;21(5):603–610.e3. 10.1016/j.chom.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 119. Doherty MK, Ding T, Koumpouras C, et al. : Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn's Disease Patients. mBio. 2018;9(2):pii: e02120-17. 10.1128/mBio.02120-17 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Kennedy NA, Heap GA, Green HD, et al. : Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn's disease: a prospective, multicentre, cohort study. Lancet Gastroenterol Hepatol. 2019;4(5):341–53. 10.1016/S2468-1253(19)30012-3 [DOI] [PubMed] [Google Scholar]
- 121. Sazonovs A, Kennedy NA, Bewshea C, et al. : OP013 HLA-DQA1 contributes to the development of antibodies to anti-TNF therapy in Crohn’s disease. J Crohns Colitis. 2018;12(supplement_1):S009–S010. 10.1093/ecco-jcc/jjx180.012 [DOI] [Google Scholar]
- 122. Pereira MS, Maia L, Azevedo LF, et al. : A [Glyco]biomarker that Predicts Failure to Standard Therapy in Ulcerative Colitis Patients. J Crohns Colitis. 2019;13(1):39–49. 10.1093/ecco-jcc/jjy139 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 123. Haberman Y, Karns R, Dexheimer PJ, et al. : Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10(1):38. 10.1038/s41467-018-07841-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 124. Cunningham KE, Vincent G, Sodhi CP, et al. : Peroxisome Proliferator-activated Receptor-γ Coactivator 1-α (PGC1α) Protects against Experimental Murine Colitis. J Biol Chem. 2016;291(19):10184–200. 10.1074/jbc.M115.688812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Martin JC, Chang C, Boschetti G, et al. : Single-Cell Analysis of Crohn's Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell. 2019;178(6):1493–1508.e20. 10.1016/j.cell.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ma C, Hussein IM, Al-Abbar YJ, et al. : Heterogeneity in Definitions of Efficacy and Safety Endpoints for Clinical Trials of Crohn’s Disease: A Systematic Review. Clin Gastroenterol Hepatol. 2018;16(9):1407–1419.e22. 10.1016/j.cgh.2018.02.051 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 127. Ma C, Panaccione R, Fedorak RN, et al. : Heterogeneity in Definitions of Endpoints for Clinical Trials of Ulcerative Colitis: A Systematic Review for Development of a Core Outcome Set. Clin Gastroenterol Hepatol. 2018;16(5):637–647.e13. 10.1016/j.cgh.2017.08.025 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 128. Peyrin-Biroulet L, Sandborn W, Sands BE, et al. : Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE): Determining Therapeutic Goals for Treat-to-Target. Am J Gastroenterol. 2015;110(9):1324–38. 10.1038/ajg.2015.233 [DOI] [PubMed] [Google Scholar]
- 129. Yzet C, Ungaro R, Bossuyt P, et al. : OP35 Endoscopic and deep remission at 1 year prevents disease progression in early Crohn’s disease: Long-term data from CALM. J Crohns Colitis. 2019;13:S024–S025. 10.1093/ecco-jcc/jjy222.032 [DOI] [Google Scholar]
- 130. Heida A, Park KT, van Rheenen PF: Clinical Utility of Fecal Calprotectin Monitoring in Asymptomatic Patients with Inflammatory Bowel Disease: A Systematic Review and Practical Guide. Inflamm Bowel Dis. 2017;23(6):894–902. 10.1097/MIB.0000000000001082 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 131. DʼHaens G, Ferrante M, Vermeire S, et al. : Fecal calprotectin is a surrogate marker for endoscopic lesions in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(12):2218–24. 10.1002/ibd.22917 [DOI] [PubMed] [Google Scholar]
- 132. Florin TH, Paterson EW, Fowler EV, et al. : Clinically active Crohn's disease in the presence of a low C-reactive protein. Scand J Gastroenterol. 2006;41(3):306–11. 10.1080/00365520500217118 [DOI] [PubMed] [Google Scholar]
- 133. Boyapati RK, Torres J, Palmela C, et al. : Withdrawal of immunosuppressant or biologic therapy for patients with quiescent Crohn's disease. Cochrane Database Syst Rev. 2018;5:CD012540. 10.1002/14651858.CD012540.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Louis E, Mary JY, Vernier-Massouille G, et al. : Maintenance of remission among patients with Crohn's disease on antimetabolite therapy after infliximab therapy is stopped. Gastroenterology. 2012;142(1):63–70.e5. 10.1053/j.gastro.2011.09.034 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 135. de Suray N, Salleron J, Vernier-Massouille G, et al. : 864 Close Monitoring of CRP and Fecal Calprotectin is Able to Predict Clinical Relapse in Patients With Crohn's Disease in Remission After Infliximab Withdrawal. a Sub-Analysis of the Stori Study. Gastroenterology. 2012;142(5):S-149 10.1016/S0016-5085(12)60560-5 [DOI] [Google Scholar]
- 136. Planell N, Masamunt MC, Leal RF, et al. : Usefulness of Transcriptional Blood Biomarkers as a Non-invasive Surrogate Marker of Mucosal Healing and Endoscopic Response in Ulcerative Colitis. J Crohns Colitis. 2017;11(11):1335–46. 10.1093/ecco-jcc/jjx091 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 137. Dierckx T, Verstockt B, Vermeire S, et al. : GlycA, a Nuclear Magnetic Resonance Spectroscopy Measure for Protein Glycosylation, is a Viable Biomarker for Disease Activity in IBD. J Crohns Colitis. 2019;13(3):389–94. 10.1093/ecco-jcc/jjy162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Dai C, Jiang M, Sun MJ, et al. : Fecal immunochemical test for predicting mucosal healing in ulcerative colitis patients: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2018;33(5):990–7. 10.1111/jgh.14121 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 139. Hiraoka S, Inokuchi T, Nakarai A, et al. : Fecal Immunochemical Test and Fecal Calprotectin Results Show Different Profiles in Disease Monitoring for Ulcerative Colitis. Gut Liver. 2018;12(2):142–8. 10.5009/gnl17013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 140. Lewis JD, Chen EZ, Baldassano RN, et al. : Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn's Disease. Cell Host Microbe. 2015;18(4):489–500. 10.1016/j.chom.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 141. Shaw KA, Bertha M, Hofmekler T, et al. : Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8(1):75. 10.1186/s13073-016-0331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Sitapati A, Kim H, Berkovich B, et al. : Integrated precision medicine: the role of electronic health records in delivering personalized treatment. Wiley Interdiscip Rev Syst Biol Med. 2017;9(3). 10.1002/wsbm.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Love-Koh J, Peel A, Rejon-Parrilla JC, et al. : The Future of Precision Medicine: Potential Impacts for Health Technology Assessment. Pharmacoeconomics. 2018;36(12):1439–51. 10.1007/s40273-018-0686-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Waljee AK, Sauder K, Patel A, et al. : Machine Learning Algorithms for Objective Remission and Clinical Outcomes with Thiopurines. J Crohns Colitis. 2017;11(7):801–10. 10.1093/ecco-jcc/jjx014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ashton JJ, Mossotto E, Ennis S, et al. : Personalising medicine in inflammatory bowel disease-current and future perspectives. Transl Pediatr. 2019;8(1):56–69. 10.21037/tp.2018.12.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Parkes M, Noor NM, Dowling F, et al. : PRedicting Outcomes For Crohn's dIsease using a moLecular biomarkEr (PROFILE): protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open. 2018;8(12):e026767. 10.1136/bmjopen-2018-026767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Lee J, Biasci D, Noor N, et al. : OC-044 Profile trial: predicting outcomes for crohn’s disease using a molecular biomarker. Gut. 2017;66(Suppl 2):A22–A3. 10.1136/gutjnl-2017-314472.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Shen R, Olshen AB, Ladanyi M: Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25(22):2906–12. 10.1093/bioinformatics/btp543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Shannon P, Markiel A, Ozier O, et al. : Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]