Abstract
Seminal fluid proteins (SFPs) mediate an array of postmating reproductive processes that influence fertilization and fertility. As such, it is widely held that SFPs may contribute to postmating, prezygotic reproductive barriers between closely related taxa. We investigated seminal fluid (SF) diversification in a recently diverged passerine species pair (Passer domesticus and Passer hispaniolensis) using a combination of proteomic and comparative evolutionary genomic approaches. First, we characterized and compared the SF proteome of the two species, revealing consistencies with known aspects of SFP biology and function in other taxa, including the presence and diversification of proteins involved in immunity and sperm maturation. Second, using whole-genome resequencing data, we assessed patterns of genomic differentiation between house and Spanish sparrows. These analyses detected divergent selection on immunity-related SF genes and positive selective sweeps in regions containing a number of SF genes that also exhibited protein abundance diversification between species. Finally, we analyzed the molecular evolution of SFPs across 11 passerine species and found a significantly higher rate of positive selection in SFPs compared with the rest of the genome, as well as significant enrichments for functional pathways related to immunity in the set of positively selected SF genes. Our results suggest that selection on immunity pathways is an important determinant of passerine SF composition and evolution. Assessing the role of immunity genes in speciation in other recently diverged taxa should be prioritized given the potential role for immunity-related proteins in reproductive incompatibilities in Passer sparrows.
Keywords: cryptic female choice, immunity, fertility, positive selection, reproduction, selective sweep, sperm competition
Introduction
Understanding the contribution of trait diversification to the establishment and maintenance of reproductive isolation, as well as identifying loci underlying reproductive incompatibilities between members of divergent populations and species, are major goals in the study of biodiversity. Speciation research has traditionally focused on the role of natural selection and ecological differentiation in the evolution of reproductive barriers (Coyne and Orr 2004; Nosil 2012). The consequences of sexual selection for the evolution and maintenance of reproductive isolation and speciation are now also well established (Panhuis et al. 2001; Ritchie 2007; Kraaijeveld et al. 2011; Servedio 2012). This body of work, however, has predominately investigated precopulatory sexual selection, whereas the consequences of postcopulatory sexual selection for speciation has received relatively less attention, and thus our understanding of postmating, prezygotic (PMPZ) remains limited (Coyne and Orr 2004; Ritchie 2007; Howard et al. 2009). Although this gap in our knowledge regarding PMPZ barriers is largely attributable to the cryptic and complex nature of ejaculate–female and sperm–egg interactions (Howard et al. 2009; Pitnick and Wolfner 2009), rapid advances in the molecular characterization of reproductive systems have stimulated new research efforts into PMPZ phenotypes (McDonough et al. 2016).
Detailed genetic analyses offer the most direct means of identifying genetic changes underlying PMPZ isolation (see e.g., Sweigart 2010; Larson et al. 2013). However, such approaches are intractable or limited in their resolution in many nonmodel systems. As such, complementary genomic, transcriptomic, and proteomic analyses of reproductive systems have emerged as a powerful means of identifying loci that are rapidly diversifying and may contribute to PMPZ barriers (Andrés et al. 2008, 2013). Yet despite the expansion of omic approaches to a wide range of ecological model species, including numerous avian and mammalian speciation models (Janoušek et al. 2012; Ellegren 2014; Wolf and Ellegren 2016; Hooper et al. 2018), the majority of studies concerning the molecular diversification of reproductive systems of relevance to PMPZ barriers have been conducted in insects (McDonough et al. 2016) or externally fertilizing taxa (Vacquier and Swanson 2011; Wilburn and Swanson 2016).
Successful fertilization is dependent upon a range of male–female interactions, including ejaculate–female and sperm–egg interactions, that are mediated, at least in part, by reproductive proteins in sperm, seminal fluid (SF), the female reproductive tract, and the egg (Pitnick and Wolfner 2009; Pitnick et al. 2019). Indeed, recent investigations in fruit flies and crickets have strongly implicated the contribution of such mechanisms to PMPZ isolation (Manier, Belote, et al. 2013; Manier, Lüpold, Belote, et al. 2013; Manier, Lüpold, Pitnick, et al. 2013; Tyler et al. 2013). Additionally, interacting gamete proteins have been shown to mediate reduced fertilization rates of heterospecific relative to conspecific sperm in fish (Yeates et al. 2013) and a range of broadcast spawning marine invertebrates (e.g., sea urchin, abalone, and oysters; Palumbi 1999; Swanson and Vacquier 2002; Vacquier and Swanson 2011; Wilburn and Swanson 2016). In contrast, our understanding of the role of specific reproductive proteins in PMPZ barriers in internally fertilizing vertebrates is currently limited.
Among reproductive proteins, considerable attention has been paid to seminal fluid proteins (SFPs) because they are known to mediate a range of postmating processes that influence fertility and paternity, including decreasing female receptivity to remating, increasing rates of oogenesis, ovulation and egg laying, promoting sperm storage, and decreasing female lifespan (Wolfner 2002; Poiani 2006; Avila et al. 2011; McGraw et al. 2015). SFPs also have antimicrobial functions or are known to induce the expression of antimicrobial peptides, thus protecting the reproductive tract from infection or gametes from microbial attack (Lung et al. 2001; Rowe et al. 2013). Moreover, many SFPs are rapidly evolving and exhibit signatures of positive selection, putatively due to sexual selection (Swanson et al. 2001; Haerty et al. 2007; Ramm et al. 2007; Karn et al. 2008; Claw et al. 2018). Although the functional significance of this rapid divergence of SFPs is not well understood, it is widely held that SFPs may be involved in barriers to gene flow between diverging lineages (Civetta and Singh 1998; Turner and Hoekstra 2008; McDonough et al. 2016). As such SFPs have been the focus of studies of PMPZ genetics in several insect speciation models, including Drosophila (Wagstaff and Begun 2005), moths (Al-Wathiqui et al. 2014), and crickets (Andrés et al. 2008, 2013; Marshall et al. 2011; Larson et al. 2013).
Birds have long been at the center of speciation research (Price 2008), as well as the focus of studies on postcopulatory sexual selection. For example, numerous studies have investigated the role of postcopulatory sexual selection in driving evolutionary diversification in male reproductive biology, including sperm morphology and quality (Kleven et al. 2009; Lüpold et al. 2009; Rowe and Pruett-Jones 2011; Rowe et al. 2015). Further, experimental evidence that ejaculate–female/sperm–egg interactions can impact heterospecific fertilization success comes from a range of studies in both passerine (Pryke et al. 2010; Cramer et al. 2016) and nonpasserine (e.g., Galliformes and Anseriformes; reviewed by Birkhead and Brillard [2007]) species. Despite this, molecular investigations of PMPZ mechanisms in birds, and of reproductive proteins more generally, remain limited (see Calkins et al. 2007; Borziak et al. 2016; Álvarez-Fernández et al. 2019; Rowe et al. 2019 for exceptions). Nonetheless, such studies have the potential to reveal insights into the evolution of reproductive proteins and speciation processes (Edwards et al. 2005).
Whereas surveys for genes involved in PMPZ phenotypes were initially more reliant on comparative transcriptomic approaches, proteomics offers an unambiguous identification of genes encoding proteins that are transferred from male to female during copulation and are thus more likely to be involved in postcopulatory processes (Rowe et al. 2019). Moreover, the recent application of proteomics to SFPs in Galliformes (Borziak et al. 2016; Álvarez-Fernández et al. 2019) has resulted in the first insights into avian SFPs and paved the way for broad comparative studies of avian SF. Importantly, passerine birds appear to lack a specialized organ or tissue that contributes accessory reproductive fluids to the ejaculate (cf., tumescent lymphatic folds in fowl [Fujihara 1992], accessory glands and ejaculatory bulb of insects, and the epididymis, seminal vesicles, prostate, and bulbourethral glands of mammals [McGraw et al. 2015]). Thus, investigation of SF in passerines is likely to provide novel insight into avian SF biology and expand our knowledge of SFP function more broadly.
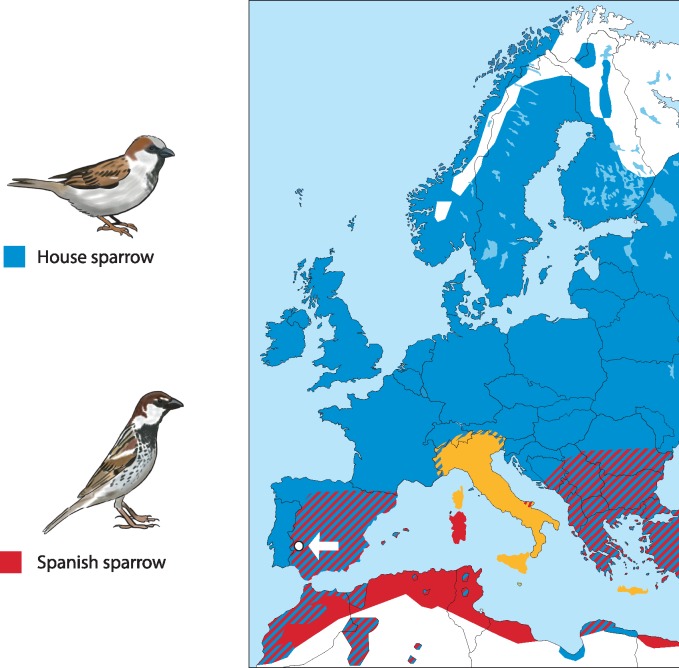
Here, we investigate SF diversification in passerine birds. We used high-throughput tandem mass spectrometry (MS/MS), which provides direct evidence of a protein’s presence and abundance in SF, in conjunction with population genetic and molecular evolutionary tests of positive selection, to characterize the diversification of SF proteomes from two closely related, ecologically similar passerine species: the house (Passer domesticus) and Spanish (Passer hispaniolensis) sparrow. These species are estimated to have diverged 0.83 Mya, although genomic evidence supports subsequent admixture in European populations (Ravinet et al. 2018). Indeed, past episodes of hybridization between house and Spanish sparrow have resulted in the formation of a homoploid hybrid species, the Italian (Passer italiae) sparrow (Elgvin et al. 2011, 2017; Hermansen et al. 2011). Present day distributions of the house and Spanish sparrow show that the two species occur in sympatry across large parts of the Spanish sparrow range, whereas the house sparrow is frequently found in allopatry (Summers-Smith 1988). Although the species are known to hybridize in a number of locations (Summers-Smith 1988; Hermansen et al. 2014; Ait Belkacem et al. 2016), current levels of gene flow are thought to be relatively low and molecular evidence suggests that genetic incompatibilities contribute to reproductive isolation between them (Hermansen et al. 2014). At the phenotypic level, differences in habitat and timing of breeding likely constitute premating barriers to gene flow (Summers-Smith 1988). Additionally, experimental crosses support the presence of postzygotic barriers between the species; female F1 house-Spanish hybrids exhibit underdeveloped ovaries and symptoms of ovarian hypofunction (Eroukhmanoff et al. 2016). Although there is currently no evidence for PMPZ barriers in this system (Cramer et al. 2014), detailed investigations are lacking. Therefore, using samples collected from a location where the house and Spanish sparrow occur sympatrically (fig. 1) and are known to interbreed (Hermansen et al. 2014), we investigated the molecular diversification of the SF proteome. As such, the aims of this study were to 1) characterize the first SF proteome of a passerine, 2) compare and contrast the SF proteome between two closely related Passer sparrow species, and 3) investigate the potential role of selection in patterns of SFP diversification.
Fig. 1.
Distribution and sampling site of the house and Spanish sparrow. Blue indicates the distribution of the house sparrow and red indicates the distribution of the Spanish sparrow. Hatched areas indicate sympatric regions where the house and Spanish sparrow distribution overlap. The yellow color indicates the distribution of the hybrid homoploid species, the Italian sparrow (not investigated in this study). White dot and arrow indicate the sampling location for seminal fluid samples used in this study.
Results
Sparrow SF Proteome Characterization
Tandem mass spectrometry (MS/MS) analysis of 2 biological replicates of SF in 2 sparrow species (16 protein fractions per replicate, 64 in total) resulted in 1,020,658 peptide spectral matches, with a comparable number of peptide spectral matches across replicates (per replicate mean ± S.D.: 255,164 ± 1,467). In total this yielded 867 high confidence protein identifications (supplementary table S1, Supplementary Material online). Analysis at the level of biological replicate (supplementary tables S2–S5, Supplementary Material online) found a consistently higher number of proteins in house sparrow samples compared with Spanish sparrow samples (i.e., 737 and 698 vs. 550 and 555; house and Spanish sparrow, respectively) and revealed substantial protein overlap between SF samples in both the house (73%) and Spanish (79%) sparrow. In light of this and to maximize protein identification per species (see below), MS/MS data were merged across replicates to generate a single SF proteome for each species. These data sets served as the foundation for our investigation of the molecular diversification of SF in this recently diverged passerine species pair.
One-dimensional protein fractionation suggested that total protein complexity (as reflected by protein banding patterns) was higher in the house sparrow relative to the Spanish sparrow, despite the standardized quantity of protein present in all samples (fig. 2A), though we cannot entirely rule out the possibility that protein quantification error contributed to this difference. Nonetheless, consistent with the idea that proteome complexity differed between the species, we identified a total of 827 proteins in house sparrow SF and 617 proteins in Spanish sparrow SF (supplementary table S1, Supplementary Material online). Thus, the SF proteome of the house sparrow is estimated to be ∼34% more complex than that of the Spanish sparrow. There was considerable overlap in the SF proteome of the 2 species, including 577 proteins comprising the “core” SF proteome shared between species (66.4% of the 867 proteins identified in total). To further support the sensitivity and consistency of proteome characterization across species, a comparison of protein abundance estimates of the core SF proteome in the house and Spanish sparrow demonstrated that they were significantly correlated (rs = 0.85, P < 0.0001, fig. 2B). As the vast majority of sparrow SFPs (95%) were identified in the SF of the house sparrow, we focused on this proteome for our initial characterization of the functional composition of the SF proteome (analysis of all 867 identified SFPs provided similar results).
Fig. 2.
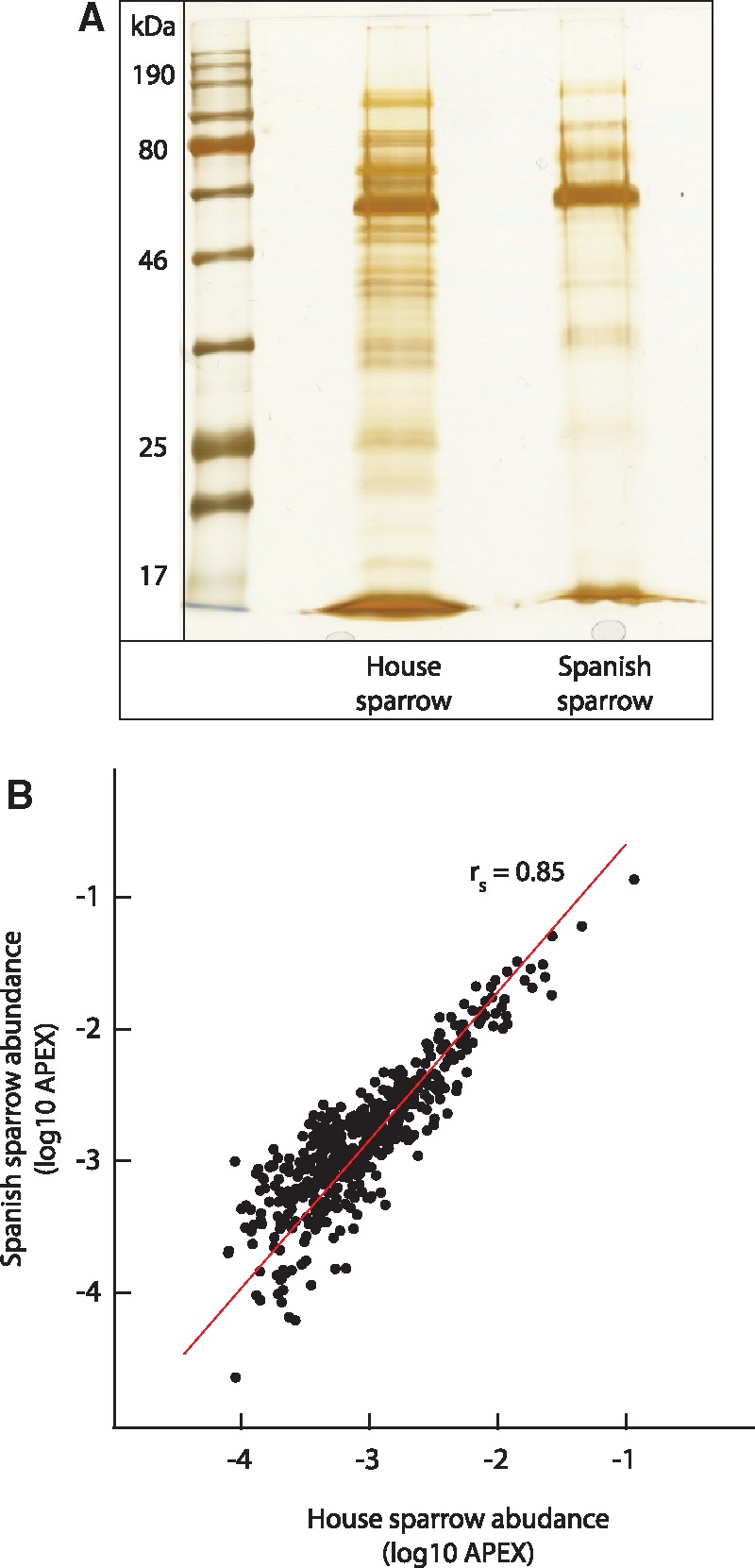
Comparison of the house sparrow and Spanish sparrow seminal fluid proteomes. (A) One-dimensional (1D) gels of house sparrow (left) and Spanish sparrow (right) seminal fluid samples. Gels were loaded with 9 μg/lane of protein and stained with silver stain. NB, gel banding patterns were consistent across biological replicates in both species (replicate gels not shown). (B) Correlation in the abundance of proteins identified in both the house sparrow and Spanish sparrow proteomes. Protein abundance is plotted as log 10 transformed APEX values, which represent normalized estimates of protein abundance for 467 shared proteins. Correlation coefficient is Spearman rank correlation.
House Sparrow SF Proteome Composition and Function
Among the 827 proteins present in house sparrow SF, several notable proteins were identified as outliers in terms of abundance (e.g., albumin [ALB], ovoinhibitor [OIH], superoxide dismutase 3 [SOD3], complement factor I [CFI], supplementary table S6, Supplementary Material online). This also included the predominant SFP, Regenerating islet-derived protein 4 (REG4), a calcium-independent lectin, which composed 8.1% of SFPs by mass and exceeded the abundance of the next most abundant protein by more than 2-fold. Lectins are sugar-binding proteins that play a number of potential roles in reproduction via sugar–protein interactions, including sperm competition and female sperm storage (Wong et al. 2008), as well as contributing to innate immune system functionality (Elliott et al. 2014). Gene Ontology (GO) analysis identified 215 enriched categories after multiple test correction (table 1 and supplementary table S7, Supplementary Material online). Among these, we highlight categories of relevance to SF biology or reproduction. First, the house sparrow SF proteome was significantly enriched in proteolytic activity, including lysosomes (P = 1.25e-9), endopeptidase activity (P = 1.89e-5), and proteasome complex (P = 0.049). Proteolysis regulators are common constituents of SF and are believed to contribute to sperm viability and a range of reproductive and postmating processes (Wolfner 2002; LaFlamme and Wolfner 2013).
Table 1.
Gene Ontology Enrichment for Proteins in the House Sparrow Seminal Fluid Proteome.
| GO ID | GO Term Description | P Value |
|---|---|---|
| GO:0043312 | Neutrophil degranulation | 3.10e-27 |
| GO:0043209 | Myelin sheath | 1.62e-26 |
| GO:0043230 | Extracellular organelle | 5.94e-21 |
| GO:1903561 | Extracellular vesicle | 5.07e-19 |
| GO:0010951 | Negative regulation of endopeptidase activity | 2.39e-15 |
| GO:0051287 | NAD binding | 9.66e-15 |
| GO:0005975 | Carbohydrate metabolic process | 1.11e-14 |
| GO:0005775 | Vacuolar lumen | 2.81e-14 |
| GO:0006521 | Regulation of cellular amino acid metabolic process | 4.34e-14 |
| GO:0031983 | Vesicle lumen | 1.29e-13 |
| GO:0038061 | NIK/NF-kappaB signaling | 1.46e-13 |
| GO:0006735 | NADH regeneration | 3.78e-13 |
| GO:1902036 | Regulation of hematopoietic stem cell differentiation | 1.51e-12 |
| GO:0061418 | Regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2.78e-12 |
| GO:0010972 | Negative regulation of G2/M transition of mitotic cell cycle | 4.11e-12 |
| GO:0051436 | Negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.11e-12 |
| GO:0051437 | Positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition | 6.74e-12 |
| GO:0033209 | Tumor necrosis factor-mediated signaling pathway | 6.74e-12 |
| GO:0060071 | Wnt signaling pathway–planar cell polarity pathway | 6.74e-12 |
| GO:0006091 | Generation of precursor metabolites and energy | 6.88e-12 |
| GO:0002479 | Antigen processing and presentation of exogenous peptide antigen via MHC class I TAP dependent | 3.40e-11 |
| GO:0006096 | Glycolytic process | 3.40e-11 |
| GO:0061718 | Glucose catabolic process to pyruvate | 5.69e-11 |
| GO:0000209 | Protein polyubiquitination | 1.42e-10 |
| GO:0051603 | Proteolysis involved in cellular protein catabolic process | 2.15e-10 |
Note.—Top 25 significantly enriched terms (for full results see supplementary table S2, Supplementary Material online).
Second, there was a significant enrichment in a wide range of membranous extracellular vesicle terms, including extracellular vesicle (P = 5.07e-19) and acrosomal vesicle (P = 0.008). Extracellular vesicles, such as exosomes, are predicted to be major contributors to the process of post-testicular sperm maturation (Sullivan and Saez 2013; Corrigan et al. 2014; Borziak et al. 2016) and are involved in the delivery of small RNAs to maturing sperm (Sharma et al. 2018). We therefore investigated the occurrence of the top 100 exosome protein markers (Keerthikumar et al. 2016) within the passerine SF proteome. This revealed the presence of 41 out of 64 (64%) of these markers (supplementary table S6, Supplementary Material online), a significant overrepresentation relative to the genome as a whole (χ2 = 209.43, P < 0.0001). However, we note that this is significantly lower than that reported in the red jungle fowl (Gallus gallus) (85.6%; two-sided binomial test, P < 0.0001). Overall, exosome protein markers represented 9.2% of SFPs by mass in the house sparrow. Although abundances of exosome protein markers were higher than nonexosome proteins on average, the difference was not statistically significant (two-tailed permutation test, Z = 1.80, P = 0.07).
Metabolic and glycolytic proteins were also enriched, including those involved in the generation of precursor metabolites and energy (P = 6.88e-12), glycolytic process (P = 3.40e-11), glutathione metabolic process (P = 7.73e-9), and glucose metabolic process (P = 0.01). We note that several of these proteins were among the most abundant in the SF proteome (e.g., alpha-enolase [ENO1], pyruvate kinase [PKM], triosephosphate isomerase [TPI1], phosphoglycerate kinase [PGK1], brain-type creatine kinase [CKB], supplementary table S6, Supplementary Material online). Widespread presence of metabolic proteins has been demonstrated in SF (Baer, Heazlewood, et al. 2009; Boes et al. 2014; Borziak et al. 2016), sperm (Dorus et al. 2006; Skerget et al. 2013; Paynter et al. 2017; Degner et al. 2019), and female sperm storage organs (Baer, Eubel, et al. 2009; Prokupek et al. 2009) and are suggested to play a fundamental role in maintaining sperm viability (den Boer et al. 2009; King et al. 2011; Paynter et al. 2017).
SFPs also have an important role in antimicrobial and immunity-related functions (e.g., Avila et al. 2011) and a number of proteins with a role in innate and adaptive immunity, as well as antimicrobial proteins, were identified in sparrow SF, including several members of the complement system (e.g., C5, C7, C9, CFD, CF1, C1R, CFH), OIH, lysozyme (LYZ), and β2-microglobulin (β2M). GO analysis also revealed a significant enrichment in terms linked to immunity, such as neutrophil degranulation (P = 3.10e-27), TAP-dependent exogenous peptide antigen processing and presentation via MHC class I (P = 3.40e-11), neutrophil mediated immunity (P = 4.42e-8), humoral immune response (P = 0.003), and lymphocyte mediated immunity (P = 0.02). Interestingly, however, we did not identify the avian antimicrobial proteins gallinacin-9 or gallinacin-10, both of which are highly abundant in the SF proteome of red jungle fowl (Borziak et al. 2016).
Finally, blood plasma proteins have been identified as a major component of red jungle fowl SF (Borziak et al. 2016). We therefore investigated the presence of blood plasma proteins in the sparrow SF proteome. Of the 120 blood plasma proteins with orthologs in the house sparrow, 55% (66/120) were present in the SF proteome (supplementary table S6, Supplementary Material online), which represents a significant enrichment relative to the entire genome (χ2 = 298.95, P < 0.0001). Blood plasma proteins composed 19.2% of SFPs by mass, which is significantly lower than the 29% (two-sided binomial test; P < 0.001) observed in the red jungle fowl (Borziak et al. 2016). Nonetheless, blood plasma proteins were, on average, significantly greater in abundance than the remainder of SFPs (two-tailed permutation test, Z = 2.89, P = 0.004).
Compositional Divergence of Sparrow SF Proteomes
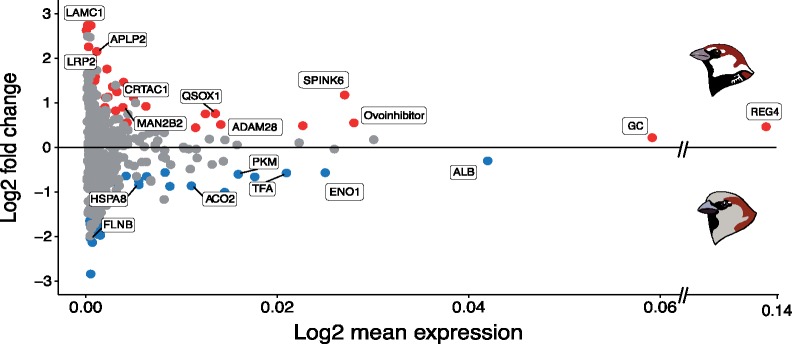
The core SF proteome abundance profile was relatively consistent across the two sparrow species (fig. 2B). Most notably, as for house sparrow, REG4 was the most abundant protein in Spanish sparrow SF, in which it composed 11.4% of SFPs by mass. Despite this consistency, 61 proteins (ca., 11% of the core proteome) showed significantly different abundance estimates between the species, including 34 that were more abundant in the Spanish sparrow and 27 more abundant in the house sparrow (P < 0.05 after BH correction for multiple testing; fig. 3 and supplementary table S8, Supplementary Material online). Proteins that were significantly more abundant in the Spanish sparrow included a number of proteins linked to sperm maturation (e.g., LRP2, MAN2B2, and MAN2A2; Fisher and Howie 2006; Skerget et al. 2015), immunity and antioxidant defense (e.g., REG4, OIH, and SOD3), and sperm–egg interactions (e.g., MAN2B2 and PLG), as well as several proteins that appear to be the target of selection (e.g., F2, GC, APLP2, and see below). House sparrow SF also exhibited a greater abundance of proteins linked to immunity, although, by definition, these were a distinct set of proteins from those of higher abundance in Spanish sparrow (e.g., ALB and ANXA1). A number of glycolytic proteins (i.e., PKM and ENO1) and several members of the heat shock protein family HSP70 (i.e., HSPA8, HSPA9, HSPA2, and HSPA4) were also in greater abundance in house sparrow SF.
Fig. 3.
MA plot comparing protein abundances between the house and Spanish sparrow seminal fluid proteomes. Differential abundance analysis for proteins identified in both species revealed 61 significant proteins after Benjamini–Hochberg multiple testing correction. Proteins of significantly greater abundance in the house sparrow are shown in blue, whereas proteins of significantly greater abundance in the Spanish sparrow are shown in red, and nonsignificant proteins are in gray. Proteins with P values <0.001 have been labeled with protein names.
Although the vast majority of the SFPs identified were shared between the two species, we also found a number of proteins unique to each; there were 250 SFPs unique to the house sparrow and 40 unique to the Spanish sparrow. Although many species-specific proteins were present in relatively high abundance (i.e., protein abundance greater than median protein abundance for the combined SF proteome), proteins unique to one or the other species showed lower abundances on average than those identified as shared proteins (two-sided permutation test: Z = 2.49, P = 0.013). Given this, and the potential for false negative error due to difficulty in identifying proteins of low abundance by MS/MS (Chandramouli and Qian 2009), we refrain from drawing strong conclusions regarding the absence of proteins from a given species. Nonetheless, the bias toward the house sparrow in terms of numbers of unique proteins does not appear to be accounted for by low protein abundance alone; that is, when excluding the 25% of proteins with the lowest abundance values, we find no difference between the ratio of house to Spanish unique protein in this restricted protein set and the ratio obtained using all protein data (χ2 = 2.28, P = 0.131). Among the 250 proteins unique to the house sparrow, there were 23 GO terms that were significantly overrepresented (supplementary table S9, Supplementary Material online). Notable among these enriched terms were the following: neutrophil degranulation (P = 4.45e-5), negative regulation of endopeptidase activity (P = 3.21e-4), endopeptidase inhibitor activity (P = 2.72e-3), and zona pellucida receptor complex (P = 0.005). Furthermore, in some instances, these species-specific proteins appear to be the target of positive selection and are discussed where relevant below.
Population Genomics of the SF Proteome
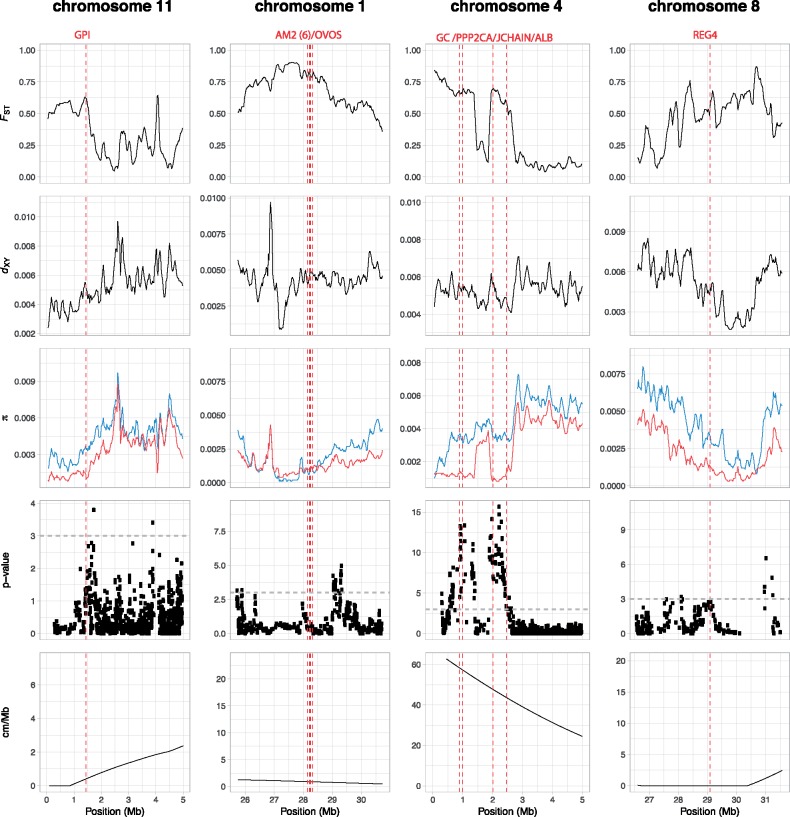
We used whole-genome resequencing data of house (n = 46) and Spanish (n = 43) sparrows to investigate patterns of genetic diversity, differentiation, and divergence in SF proteome genes between the two species. First, to identify highly differentiated genomic regions, we estimated FST, a relative measure of differentiation, for each SF gene. The mean FST of SF genes was 0.125, and the FST distribution was not significantly different from the distribution for all other genes in the genome (two-tailed permutation test, Z = 1.11, P = 0.27). We identified 35 highly differentiated SF genes (i.e., genes situated above the 95th percentile of the genome-wide gene FST distribution), including 27 on autosomes, 7 on the Z chromosome, and a single gene located on an unplaced scaffold (supplementary table S10, Supplementary Material online). Next, given that there are a number of modifying factors (e.g., recombination rate, selection, and gene flow) that can complicate the interpretation of patterns of genomic differentiation between lineages (Ravinet et al. 2017), we further characterized the regions surrounding the SF genes by investigating absolute nucleotide divergence (dXY), nucleotide diversity (π, a measure of within-species genetic variation), and recombination rate. This approach enabled us to gain a more refined understanding of the types of evolutionary processes (e.g., diversifying selection vs. background selection) acting on these SF genes. In our data set, gene density is low in regions of low recombination (supplementary fig. S1, Supplementary Material online), suggesting that Hill–Robertson effects are less likely to explain the patterns we observe. Taken together, these analyses identified a number of candidate genes showing genomic patterns suggestive of signatures of selection. These included Glucose-6-phosphate isomerase (GPI) on chromosome 11, which appears to show a strong signature of divergent selection; with corresponding peaks in FST and dXY, as well as reduced nucleotide diversity in both species, but especially prominent in the Spanish sparrow (fig. 4). GPI has been identified as a dual functioning protein: intracellularly, GPI functions as a glycolytic enzyme, whereas extracellularly, the protein functions as a lymphokine and induces immunoglobulin secretion, and has been identified as a sperm antigen in mice (Yakirevich and Naot 2000). Additionally, a cluster of immunity-related genes, including Alpha-2-macroglobulin (A2M) and Ovostatin (OVOS), on chromosome 1 fall in a region of high FST with a corresponding reduction in nucleotide diversity in both species, and moderate recombination rates. In this case, however, the pattern of dXY is less clear. Although the immune genes reside upon a local peak in dXY, the region immediately surrounding these genes exhibits low levels of absolute divergence (dXY) relative to the remainder of the chromosome (fig. 4). Thus, in contrast to GPI, this suggests a potential role for purifying selection conserving this cluster of SF immune genes in both species.
Fig. 4.
Patterns of genomic differentiation and divergence and signatures of selection in SF proteome genes between house and Spanish sparrows. Red dashed lines represent midpoint of genes of interest on chromosomes 11, 1, 4, and 8, respectively. Differentiation and divergence was measured as FST and dXY, and nucleotide diversity measured as π (blue for house sparrow and red for Spanish sparrow). Extended haplotype homozygosity (xpEHH) statistics show evidence of recent selective sweeps. Horizontal dashed line represents threshold of significance equivalent to P value of 0.001.
We also tested for signatures of selection on SF genes between house and Spanish sparrow using long-range haplotype selection statistics, which are designed to detect the increase in haplotype homozygosity around a target of selection during a recent selective sweep (Vitti et al. 2013). Specifically, we used the cross-population extended haplotype homozygosity (xpEHH) statistic to identify SF genes where a selective sweep occurred in at least one lineage (Vitti et al. 2013). This identified 1,006 outlier single-nucleotide polymorphisms (SNPs) where the log 10 P value of xpEHH between house and Spanish sparrow was >3, representing a cutoff of P = 0.001. We then identified all SF genes occurring within 100 kb of these outlier SNPs, and identified 55 SF genes falling within these regions (supplementary table S11, Supplementary Material online). GO analysis identified three gene pathways with evidence of enrichment among this outlier gene set (supplementary table S12, Supplementary Material online), including glutathione metabolic process (P = 0.01) neutrophil degranulation (P = 0.01), and extracellular vesicle (P = 0.04). A gene of interest in the extracellular vesicle pathway is Serum albumin (ALB) (fig. 4), encoding a blood plasma protein with a suggested role in innate immunity (Giles and Czuprynski 2003). Furthermore, in domestic turkeys, addition of serum albumin under in vitro conditions appears to increase the proportion of motile sperm and sperm velocity (Bakst and Cecil 1992; Atikuzzaman et al. 2017). Additionally, Group-specific component (GC), also on chromosome 4, encodes the major vitamin D binding protein, a member of the albumin family of proteins (fig. 4). Importantly, the protein products of both of these genes exhibit significant differences in SF abundance between the two species. Additionally, xpEHH statistics also suggest recent positive selective sweeps have occurred in the region of both REG4 and Plasminogen, two proteins exhibiting differential abundance between the house and Spanish sparrow SF proteome. In the case of REG4, the region is characterized by a peak in FST and decrease in nucleotide diversity, particularly in the Spanish sparrow (fig. 4).
Some of the highest peaks in xpEHH occurred on chromosome 4 and these outlier SNPs were associated with three SF genes, PPP2CA, NAAA, and JCHAIN. These SF genes were also identified as outlier genes in terms of FST, showed a small local peak in dXY, and a decrease in nucleotide diversity in the Spanish sparrow (fig. 4). PPP2CA—serine/threonine-protein phosphatase—is involved in signaling events required for sperm motility acquisition in the mammalian epididymis (Freitas et al. 2017). JCHAIN is a small, highly conserved protein associated with immunoglobulin (IgA and IgM) polymerization and secretion. Notably, these secretory antibodies have a high antigen avidity, making them particularly well suited to agglutinating bacteria and viruses, and, in some cases, play a role in the activation of complement (Johansen et al. 2000).
Molecular Evolution of the Sparrow SF Proteome
Genome-wide molecular evolutionary rate analyses (including 7,670 genes) were used to determine dN and dS for the house sparrow-specific lineage. We found no significant difference between SF genes and genomic background distributions for either dN (two-sided permutation test, Z = 0.44, P = 0.66, mean: 0.0083 vs. 0.0081, SF and genomic background respectively, supplementary fig. S2a, Supplementary Material online) or dN/dS (two-sided permutation test, Z = −0.41, P = 0.68, mean: 0.1631 vs. 0.1696, SF and genomic background respectively, supplementary fig. S2b, Supplementary Material online). Similarly, we found no association between SFP abundance divergence and values of either dN (Kruskal–Wallis χ2 = 3.99, P = 0.41, supplementary fig. S3a, Supplementary Material online) or dN/dS (Kruskal–Wallis χ2 = 0.87, P = 0.93, supplementary fig. S3b, Supplementary Material online). We also examined variation in dN/dS values with respect to estimates of tissue specificity (i.e., τ; Yanai et al. 2005) generated through the integration of our SF semiquantitative protein abundance data and gene expression data from the collared flycatcher (Ficedula albicolis) (Uebbing et al. 2016), one of the species used in our molecular evolutionary analysis. This revealed an elevated value of average dN/dS in SFPs exhibiting higher values of τ (supplementary fig. S4, Supplementary Material online). This finding suggests that SF-specific proteins evolve more rapidly than SF proteins that exhibit broader expression patterns. Although we acknowledge that these analyses should be interpreted with caution due to the use of expression data from a different passerine species, our finding that tissue-specific proteins evolve more rapidly in SF is consistent with patterns observed in reproductive tissues in the flycatcher (e.g., ovary, testis, supplementary fig. S4, Supplementary Material online) and previous studies (e.g., Duret and Mouchiroud 2000).
Outlier analyses identified 31 SF encoding genes with high dN values (i.e., dN > 0.027, supplementary table S6, Supplementary Material online), including a number of immune genes (β2M, C7, C9, and F2), genes linked to spermatogenesis (SPATA18), zona pellucida binding (ZPBP2), and the REG4 gene encoding the most abundant SFP. Similarly, this analysis identified 26 genes with elevated dN/dS ratios (i.e., dN/dS > 0.57, supplementary table S6, Supplementary Material online), suggesting a number of SF genes are rapidly evolving. These included a number of genes linked to immune function, such as complement component 7 (C7), Avidin, and Annexin (ANXA1), as well as genes linked to proteolysis (SERPINF2) and sperm–egg binding (ZPBP2). A single gene showed strong evidence of rapid evolution across the full length of the protein-coding region (as inferred by a dN/dS ratio > 1): CD81, encoding a protein (CD81) in the tetraspanin family. Taken together, this suggests that SFPs linked to immunity, antimicrobial defense, and sperm–oocyte interactions have been likely targets of selection on the house sparrow lineage.
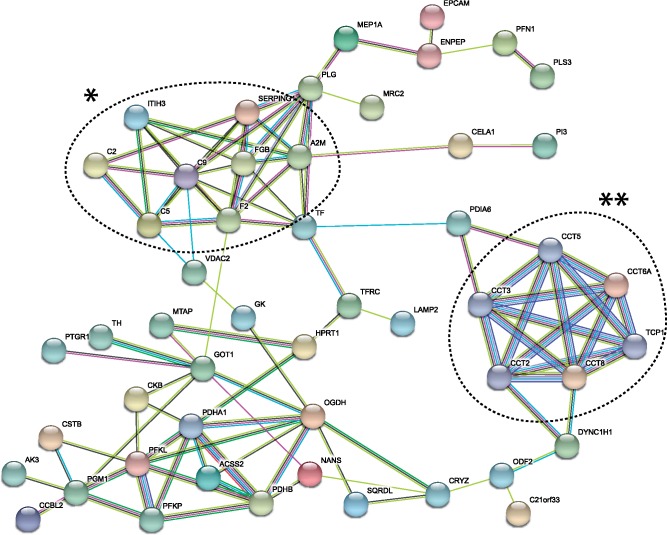
Genome-wide likelihood ratio comparisons of neutral (M8a) and selection models (M8) using Benjamini–Hochberg multiple test correction and a false discovery rate (FDR) of <0.01 were used to assess positive selection acting on genes of the SF proteome across passerine birds. Evidence of positive selection was observed for 84 of the 572 SF genes (14.7%; supplementary table S6, Supplementary Material online), which was significantly higher than the remainder of the genome (i.e., 10.2%; one-tailed binomial test; P = 0.0004). GO analysis of these genes revealed significant enrichments in 18 terms (supplementary table S13, Supplementary Material online), including several terms linked to immunity (e.g., protein activation cascade: P = 1.01e-7, humoral immune response: P = 0.003). Complement system genes (e.g., CFH, F2, C9, C5, and CFI) were notable members of these enriched GO categories. Additionally, the enriched term zona pellucida receptor complex (P = 0.005) included genes coding for two subunits of the chaperonin TCP1 complex (CCT8, CCT6A) and ZPBP2, all of which have been reported to mediate mammalian sperm–oocyte interactions (Dun et al. 2011). The interaction network of proteins encoded by positively selected genes (Szklarczyk et al. 2015) was highly consistent with these findings, supporting the putative involvement of these genes into similar biological processes and pathways. Following the elimination of isolated nodes, the protein–protein interaction network consisted of 51 nodes and was significantly larger and exhibited greater connectivity than expected (103 edges identified; PPI enrichment P < 1.0e-16). Prominent clusters within the network included proteins linked to immunity (e.g., complement pathway) and proteins involved in sperm–egg interactions (fig. 5).
Fig. 5.
Protein–protein interaction network for genes showing evidence of positive selection. Genes exhibiting signature of positive selection (i.e., comparison of M8 and M8a site models) were analyzed using the STRING protein–protein interaction database, allowing for up to five additional protein interactors. Network nodes represent proteins and edges represent protein–protein associations. Different colored edges represent complementary types of empirical evidence supporting interactions, including evidence from curated databases, experimentally determined interactions, and coexpression studies. The protein network exhibited significantly greater connectivity that expected (PPI enrichment P < 1.0e-16). Proteins linked to immunity form a cluster (*) including several complement proteins (e.g., C5 and C2), whereas the cluster on the right (**) includes several proteins involved in sperm–egg interactions (e.g., TCP1 and CCT5).
Male reproductive systems are often characterized by rapid gene evolution and gene gain/loss, including de novo gene gain (Swanson and Vacquier 2002; Hahn et al. 2007; Findlay et al. 2008). Such processes can contribute to orthology loss in reproductive proteins, especially among more distantly related species (Whittington et al. 2017). As such, we compared the proportion of SFPs with orthologs to the proportion observed for the entire genome. No significant differences were observed for the 11 species in our data set (two-sided binomial test, P = 0.081). Next, although no difference in abundance was observed between SFPs under positive selection and other SFPs (two-sided permutation test, Z = −0.34, P = 0.73), we note that six were among the most abundant proteins in house sparrow SF. This included brain-subtype creatine kinase (CKB), phosphotransferase, CFI, Plasminogen, aspartate aminotransferase (GOT1), and serum iron transport protein transferrin (TFA). Two of these proteins (Plasminogen and TFA) were also among the proteins found to be differentially abundant between the sparrow species, as were a further four proteins under positive selection (transferrin receptor [TFRC], galactocerebrosidase precursor [GALC], coagulation factor II [F2], Annexin, and chymotrypsin like elastase family member 1). Finally, SF genes under selection also included 23 genes encoding proteins that were unique to the house sparrow SF. These included several genes linked to sperm maturation and function (VDAC2 and SPATA18; Bansal et al. 2015) and zona pellucida adhesion and sperm–egg interactions (CCT8, CCT6A, and ZPBP2; Dun et al. 2011).
Discussion
This study provides the first proteomic characterization of SF in passerine birds, as well as evolutionary genomic analyses of SF proteome diversification in the recently diverged species pair, the house and Spanish sparrow. Our data revealed a diversity of SFPs, with a total of 827 unique proteins identified in the SF of the house sparrow (our representative passerine SF proteome). As such, the proteomic complexity of passerine SF is comparable in size to the SF proteome of humans (Pilch and Mann 2006) and the red jungle fowl (Borziak et al. 2016), but considerably higher than that observed in insects (e.g., Drosophila [Findlay et al. 2008; Sepil et al. 2019] and mosquito [Sirot et al. 2011; Boes et al. 2014; Degner et al. 2019]) and some nonhuman primates (Claw et al. 2018). The overall functional composition of house sparrow SF, however, was generally consistent with SF proteomes in a broad range of taxa (e.g., mammals [Claw et al. 2018], red jungle fowl [Borziak et al. 2016], and insects [Findlay et al. 2008; Kelleher et al. 2009; Boes et al. 2014; Degner et al. 2019]), demonstrating that passerine SF is a complex mix of proteins enriched in functional categories, such as proteolysis regulators, immunity, antioxidant defense, and metabolism. The functional coherence of the passerine SF proteome with those of other species highlights the importance of these functional groups for SF biology and reproductive processes more generally. Additionally, evolutionary analyses indicate that the SF proteome appears to be an enhanced target for selection. Of particular importance in the current study is the fact that several differentially abundant proteins also exhibited evidence of either positive selection or recent selective sweeps; though we note that we found no significant associations between protein abundance divergence and estimates of evolutionary change (i.e., dN or dN/dS). Nonetheless, at least in some cases, there may be a relationship between proteome composition diversification and the evolution of the genes encoding these proteins, with one possibility being that selective sweeps in regulatory regions have led to protein abundance differences. Previous molecular studies have identified genetic incompatibilities between the house and Spanish sparrow (Hermansen et al. 2014), though it is unclear if they are related to PMPZ or to postzygotic barriers. Our finding of SF proteome diversification at both the phenotypic and genomic level, however, is consistent with a possible role for SFPs in these incompatibilities.
One notable protein that exhibits significant differences in protein abundance between the species, as well as signatures of putative divergent selection and genomic differentiation, is REG4. Interestingly, REG4 is also the predominant SFP in both species, comprising between 8.1% and 11.4% of SFPs by mass (house and Spanish sparrow, respectively). REG4 is a lectin, a class of sugar-binding protein that can play several potential roles in reproduction. For example, in Drosophila, lectins are important for female sperm storage, influencing both sperm retention in (i.e., Acp29AB; Wong et al. 2008) and release from storage organs (i.e., lectin-46Cb, lectin-46Ca; Ravi Ram and Wolfner 2007), and the outcome of sperm competition (Wong et al. 2008). In birds, sperm are stored in specialized sperm storage tubules (SSTs) (Bobr et al. 1964), a process considered to be a basic requirement of avian reproductive physiology. Lectins bind to the epithelium and microvilli of SSTs, which may be indicative of sperm–epithelial SST cell interactions, although the importance of this binding for sperm storage is unclear (Bakst and Bauchan 2016). Nonetheless, one possibility is that REG4 may play a role in avian sperm storage. Interestingly, lectins are also well represented in the SF of red jungle fowl, although the REG protein family is not. As such, it is possible that different lectins are important for sperm storage in different avian taxa. Additionally, members of the REG protein family exhibit bactericidal activity through targeting of bacterial peptidoglycans (Elliott et al. 2014; Mukherjee et al. 2014) and regulate gut microbiota (Vaishnava et al. 2011; Cao et al. 2016), whereas mannose-binding lectins (e.g., REG4) activate the complement system via the lectin pathway. Thus, REG4 could influence reproductive outcomes via a range of mechanisms.
A diversity of antimicrobial and innate and adaptive immunity proteins were identified in sparrow SF, including several that either were rapidly evolving or exhibit signatures of positive selection. Furthermore, positively selected SF genes appeared to be, as a class, enriched in immune-related pathways. We note that the rapid evolution of immunity genes may not be the result of selection associated with their function in ejaculates. Indeed, immune genes, more broadly, are hotspots of positive selection across a wide taxonomic range of birds (Shultz and Sackton 2019). Nonetheless, even if nonreproductive selection drives SF immunity protein evolution, this may have important consequences for SF function and, given that PMPZ barriers have been suggested to be mediated by the immune system (Haley and Abplanalp 1970; Ghaderi et al. 2011; also reviewed in Birkhead and Brillard 2007; Wigby et al. 2019), reproductive isolation. Although the widespread presence of immunity pathways in male and female reproductive systems is well established (Wira et al. 2005; Dorus et al. 2012), a full understanding of the significance of this association to reproductive outcomes has yet to be achieved. Here, we discuss some of the more prominent SF immunity proteins and explore their potential functional role in resisting infection or the mediation of other processes relevant to reproductive outcomes.
Antimicrobial proteins were prominent in the sparrow SF proteome and have been identified as abundant components of SF in other avian systems (Borziak et al. 2016). Furthermore, functional assays have demonstrated antimicrobial activity of ejaculates in a range of taxa (Poiani 2006), including birds (Rowe et al. 2011, 2013). Bacteria have been shown to negatively impact sperm function (e.g., Diemer et al. 1996; Zan Bar et al. 2008; Kaur et al. 2009; Haines et al. 2013), and bacteria in ejaculates can have negative consequences for male fertility (Maroto Martín et al. 2010; Haines et al. 2015). The ejaculates of both house and Spanish sparrows harbor complex bacterial communities that include species that reduce sperm motility (Rowe, M. and Czirják, G. Á unpublished data). As such, antimicrobial SF proteins may serve to protect sperm from microbial attack, provide antimicrobial defense to the female’s reproductive tract or eggs (Lung et al. 2001), or limit the transmission of sexually transmitted infections to females.
Of particular note were proteins of the complement system, which were prominent in the sparrow SF proteome. Complement activity has been demonstrated in avian SF (Rowe et al. 2011), thus supporting the functional relevance of SF proteome composition. The complement system is an integral part of the innate immune system, and complement activation results in the rapid elimination of invading bacteria via direct killing of Gram-negative bacteria, bacterial labeling of complement products to stimulate phagocytic killing, and the stimulation of adaptive immune cells (Heesterbeek et al. 2018). Complement is also a major element of innate immunity in the mammalian female reproductive tract, where it can negatively impact sperm survival and function (Harris et al. 2006). Sperm appear to be able to evade complement attack, however, via the presence of complement regulators in SF and on sperm (Harris et al. 2006; Sakaue et al. 2010), and diminished SF complement inhibitory activity is associated with reduced sperm quality (Chowdhury et al. 1996). Our results show that complement regulators are a feature of passerine SF (e.g., CFI and CFH), including CFI, which is among the most abundant proteins in the SF proteome, and exhibit signatures of positive selection. However, complement regulators in SF are postulated to protect sperm in the lower female reproductive tract, whereas sperm reaching the upper portion of the female tract are thought to rely on complement regulators on the sperm surface (Harris et al. 2006). This, combined with a putative role for complement and complement regulators in sperm–oocyte interactions (Harris et al. 2006), suggests that the complement system has complex, and potentially overlapping roles, in bacterial defense and ejaculate–female/sperm–egg interactions.
Also of note, is the cluster of five Alpha-2-macroglobulin (A2M) genes, which showed evidence of being under selection in both sparrow species, and the identification of beta-2-microglobulin (β2M) as a rapidly evolving protein. A2M is an innate immunity protein that has been shown to function as a scavenger of proteases introduced by parasites and pathogens, with functional consequences documented for antiviral activity (Chen et al. 2010) and disease resistance (Freedman 1991; Araujo-Jorge et al. 1992). Given that proteases are abundant in both SF and the female reproductive tract (male- and female-derived proteases, respectively), it is not unreasonable to speculate that the canonical function of A2M in immunity could be co-opted for similar functions related to endogenous, reproductive proteases. In contrast, β2M is a component of MHC class I complex that functions to present antigens to cells of the adaptive immune system and is involved in self-recognition and microbial defense. MHC-dependent gamete fusion has been demonstrated in a range of vertebrate taxa, though the mechanisms underlying female choice of sperm from either MHC dissimilar males or males with “optimal” MHC similarity remains unclear (reviewed by Firman et al. [2017]). The presence of MHC proteins in red jungle fowl SF has been suggested as a molecular mechanism underlying cryptic female choice (Borziak et al. 2016). Moreover, MHC genes are suggested to play a crucial role in species divergence (Blais et al. 2007; Eizaguirre et al. 2009; Malmstrøm et al. 2016), though more frequently in the context of mate choice and selection against hybrids (Eizaguirre et al. 2009). The occurrence of rapidly evolving MHC proteins in avian SF emphasizes the potential importance of immunity genes to PMPZ mechanisms that bias fertilization toward conspecific sperm.
In addition to rapidly evolving immunity proteins, our analyses also revealed proteins linked to sperm–egg interactions that were rapidly evolving (e.g. ZPBP2), under positive selection (e.g., CCT8 and CCT6A), or exhibit signatures of divergent selection between the sparrow lineages (e.g., GPI, Plasminogen). Furthermore, in some cases, these proteins are differentially abundant between the species (e.g., Plasminogen) or unique to one of the sparrow species (e.g., CCT8 and CCT6A in house sparrow). Proteins involved in gametic interactions evolve rapidly, and, in the case of some externally fertilizing taxa, have been shown to mediate fertilization success and underlie conspecific sperm precedence (Palumbi 1999; Swanson and Vacquier 2002; Vacquier and Swanson 2011; Wilburn and Swanson 2016). Whereas the functional consequences of rapid evolution of proteins with sperm–egg interaction annotations in our system are unclear, it is plausible that they may contribute to PMPZ barriers that restrict gene flow between lineages, as has been suggested for reproductive proteins more generally (Civetta and Singh 1998; Turner and Hoekstra 2008; McDonough et al. 2016).
In the past, extensive hybridization between the house and Spanish sparrow resulted in the formation of the hybrid species, the Italian sparrow (Elgvin et al. 2011; Hermansen et al. 2011; Elgvin et al. 2017). Current levels of hybridization, however, appear to be relatively low (Hermansen et al. 2014) and, despite extensive geographic distribution overlap (fig. 1), there are no known contemporary hybrid zones. There is evidence of strong postmating isolation via mito-nuclear and Z-linked genetic incompatibilities between the species (Hermansen et al. 2014) and previous work suggests that this is due, at least in part, to postzygotic barriers (Eroukhmanoff et al. 2016). The current study builds on this body of work and identifies rapidly diversifying aspects of the SF proteome that could potentially contribute to PMPZ barriers between the species, though it remains unclear whether SFPs could have been responsible for incipient reproductive isolation or simply accumulated after species divergence. Nonetheless, evidence of selection in the regions of immunity-related genes, raises the possibility that immunological mechanisms may contribute to reproductive incompatibilities in this system. Of particular note is the strong pattern of differentiation we observed in GPI, which encodes a sperm-binding protein (GPI) identified as a sperm antigen in mice (Yakirevich and Naot 2000). Sperm surface antigenicity has been implicated in the reduced ability of turkey sperm to traverse the vagina of chickens following artificial insemination, suggesting a localized immunological mechanism of conspecific sperm selection (Steele and Wishart 1992). Thus, divergent selection on the GPI gene may result in sperm antigenicity that acts as an immunological barrier to heterospecific fertilization in the sparrow system. Similarly, the rapid evolution of interacting complement and complement regulatory proteins might lead to a scenario in which sperm suffer from reduced complement inhibitory activity in a heterospecific female reproductive tract leading to reduced survival. These findings suggest that differential effects of female immunological responses could form the basis of cryptic female choice mechanisms which favor conspecific sperm. Lastly, our comparative genomic approach detected signatures of divergent selection on genes that exhibited interspecific protein abundance, including ALB, GC, REG4, and Plasminogen. Thus, consistent patterns of differential abundance, rapid evolution, or signatures of selection may provide a means for prioritizing candidates that may influence reproductive incompatibilities in this system.
In conclusion, through the use of proteomic and evolutionary genomic analyses, we have been able to gain considerable insight into the functional evolution of passerine SF. Of particular note is the diversity of immunity molecules in sparrow SF, and our finding that several are rapidly evolving and appear to be under the influence of positive selection. These observations are consistent with selection associated with microbes, which are common in the reproductive tracts of both male and female birds (Stewart and Rambo 2000; Hupton et al. 2003), but may also be attributable to more complex functionalities in reproductive processes that govern sperm survival, storage, usage, and fertilization competency. These nonmutually exclusive scenarios are further supported by the diversity and abundance of immunity proteins in red jungle fowl SF (Borziak et al. 2016). Finally, the identification of interspecific differences in immunity-related SFPs at the phenotypic (i.e., protein abundances) and genomic level point toward immunological barriers that may contribute to reproductive isolation in these species. Our results build on a body of literature supporting a role for immune-related genes in species diversification (Eizaguirre et al. 2009; Karvonen and Seehausen 2012) and highlight the need for future studies regarding the contributions of immunity incompatibilities to PMPZ mechanisms of reproductive isolation.
Materials and Methods
Ejaculate Collection and SF Isolation
Wild, free-living house and Spanish sparrows were studied near Olivenza, Spain (38°40′56″N, 7°11′17″W), where the species occur in sympatry, including breeding in the same stork nests, during the Spring of 2015 (8–12 April). Birds were trapped using mist nets and males were transported to the University of Extremadura (Badajoz) for further processing. After sampling, all birds were released back at the site of initial capture. All trapping and sampling of birds was conducted in accordance with Spanish Animal Protection Regulation RD53/2013, and all methods were approved by the Institutional Commission of Bioethics at the University of Extremadura (CBUE 49/2011).
Whole ejaculate samples were collected from a total of 44 males (22 house and 22 Spanish sparrow) using cloacal massage (Wolfson 1952). Prior to sampling, the exterior of the cloaca was thoroughly cleaned with alcohol, and, for each individual, the collector (Rowe) wore new nitrile gloves. Exuded semen (ca., 0.5–4 µl) was immediately collected in a sterile, calibrated microcapillary tube, and then transferred to a sterile eppendorf tube, diluted in 15 µl of sterile phosphate buffered saline and centrifuged at 3,500 × g for 30–60 s to separate sperm from SF. The SF supernatant was then transferred to a new, sterile tube and stored at −80 °C. Prior to protein quantification, samples were subjected to a second round of centrifugation (3,500 × g for 60 s), after which a small aliquot of the supernatant was examined using phase contrast microscopy to confirm it was free of any contaminating sperm and sperm parts. After initial protein quantification of six house sparrow and six Spanish sparrow samples showed there was insufficient protein in a single sample for analysis, we pooled SF samples from eight males to create two biological replicates for each species. Protein quantitation of these pooled samples showed that total protein content ranged from 9.28 to 24.31 µg (mean ± S.D.: 16.56 ± 6.9) per pooled sample.
Protein Preparation and 1-Dimensional SDS-PAGE
Solubilization buffer (4% Chaps, 7 M urea, and 2 M thiourea) was added to each SF sample. Next, 9 μg of total protein from each pooled replicate was separated in parallel on 4–12% Nupage Bis-Tris gels running on an XCell SureLock Mini-Cell PowerEase 200 system (Life-Technologies). Gels were then fixed in 45% methanol and 1.0% acetic acid for 1 h, washed with Milli-Q water, pretreated for 1 min with 0.02% Na2S2O3, and then stained with silver (0.2% AgNO3) for 20 min. Gels were transferred to a gel slicer where each lane was cut horizontally into 16 slices, resulting in total of 64 samples for analysis. Gel slices were subjected to proteolytic digestion using an Automated Preparation Station (Perkin Elmer). Proteins were first reduced and alkylated followed by digestion with trypsin at 37 °C for 16 h.
Tandem Mass Spectrometry (MS/MS)
Tandem mass spectrometry (MS/MS) experiments were performed using a Dionex Ultimate 3000 RSLC nanoUPLC (Thermo Fisher Scientific Inc., Waltham, MA) system and a QExactive Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.). Separation of peptides was performed by reverse-phase chromatography at a flow rate of 300 nl/min and a Thermo Scientific reverse-phase nano Easy-spray column (Thermo Scientific PepMap C18, 2-μm particle size, 100-A pore size, 75-μm i.d. × 50-cm length). Peptides were loaded onto a precolumn (Thermo Scientific PepMap 100 C18, 5-μm particle size, 100-A pore size, 300-μm i.d. × 5-mm length) from the Ultimate 3000 autosampler with 0.1% formic acid for 3 min at a flow rate of 10 μl/min. After this period, the column valve was switched to allow elution of peptides from the precolumn onto the analytical column. Solvent A was water + 0.1% formic acid and solvent B was 80% acetonitrile, 20% water + 0.1% formic acid. The linear gradient employed was 2–40% B in 30 min. The LC eluant was sprayed into the mass spectrometer by means of an Easy-spray source (Thermo Fisher Scientific Inc.). All m/z values of eluting ions were measured in an Orbitrap mass analyzer, set at a resolution of 70,000. Data-dependent scans (Top 20) were employed to automatically isolate and generate fragment ions by higher energy collisional dissociation in the quadrupole mass analyzer and measurement of the resulting fragment ions was performed in the Orbitrap analyzer, set at a resolution of 17,500. Peptide ions with charge states of 2+ and above were selected for fragmentation. Postrun, the data were processed and peak-lists generated using Proteome Discoverer (version 1.3, ThermoFisher). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org; last accessed May 4, 2019) via the PRIDE partner repository (Vizcaíno et al. 2016) with the data set identifier PXD013748.
Peptide Identification and Protein Annotation
Raw data from each MS/MS run were analyzed by X!Tandem (Craig and Beavis 2004), Comet (Eng et al. 2013), and Mascot (Perkins et al. 1999) against the P. domesticus protein annotation, including 14,260 proteins, generated as part of a high-quality de novo reference genome (Elgvin et al. 2017). Only the longest protein isoform of each gene was included in the search database. To address the absence of immunoglobulin proteins in the sparrow genome annotation, manually curated avian immunoglobulin proteins were added to our search database. A fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 10.0 PPM were used. Iodoacetamide derivative of cysteine was specified as a fixed modification, whereas oxidation of methionine was specified as a variable modification. Peptides were allowed up to two missed trypsin cleavage sites. All downstream analyses of the Comet and X!Tanden results were conducted using the Trans-Proteomic Pipeline (TPP v4.7 POLAR VORTEX rev 1; Deutsch et al. 2010). FDRs were estimated with a randomized decoy database using PeptideProphet, employing accurate mass binning model and the nonparametric negative distribution model. Mascot results were curated using Scaffold (4.5.1, Proteome Software Inc., Portland, OR), including estimation of FDRs using the Scaffold Local FDR algorithm. Peptide identifications were accepted if they could be established at >95.0% probability.
To ensure that protein assignations were robust and reproducible, protein identification was only accepted if they 1) could be established at >99.0% probability, 2) include a minimum of two peptide matches, and 3) were obtained in at least two of the three search engines used. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. To account for potential protein divergence between P. domesticus and P. hispaniolensis, X!Tandem was run using the parameters previously described, but allowing for peptide identification with a single amino acid substitution. This analysis led to the identification of only 21 additional proteins (∼3.4% of total proteome), most of which (14 of 21) were already identified in the house sparrow SF. Protein divergence therefore had a limited impact on the characterization of SF composition in these species.
Orthology Determination
ProteinOrtho was used with default settings to establish orthology relationships between P. domesticus and zebra finch (Taeniopygia guttata). For proteins that were not in ProteinOrtho, BlastP was used to identify homologous zebra finch proteins (e-value of <1e-7 and requiring homology ≥50% of the protein length) to establish GO classification (see below). In turn, orthology relationships with the domestic chicken (Gallus gallus domesticus) were obtained using precalculated orthology relationships in Ensembl Genes 91 in BioMart. Orthology relationships facilitated direct comparison to the exosome ExoCarta high-quality protein marker (exosome) data set (Keerthikumar et al. 2016), the red jungle fowl (Gallus gallus) SF proteome (Borziak et al. 2016), and the domestic chicken blood plasma proteome (Ma et al. 2014).
Functional Annotation and Enrichment Analysis
GO functional annotations and gene descriptions were obtained for protein sequences from the entire house sparrow protein set using PANNZER (Koskinen et al. 2015). This approach allowed us to generate a genomic background specific to the house sparrow for all further analyses. When specific proteins of interest lacked PANNZER annotations we used BLAST to obtain functional annotations. Functional enrichment of GO terms present in the SF proteome (i.e., house sparrow SF proteome and a number of proteome subsets) relative to the entire genomic background was performed using the clusterProfiler (Yu et al. 2012), and Benjamini–Hochberg multiple test correction was used to determine significance enrichment levels (at P < 0.05).
Semiquantitative Protein Analysis
Protein quantitation was conducted using the APEX Quantitative Proteomics Tool (Braisted et al. 2008). The 50 proteins with the highest number of spectral counts and protein identification probabilities were utilized for the training data set. The 35 physicochemical properties available in the APEX tool were used for prediction of peptide detection/nondetection in the construction of a training data set file. Protein probabilities (Oi) were computed using the Random Forest classifier algorithm trained with the data set generated in the previous step. APEX protein abundances from a single combined data set per species were calculated using the protXML file generated by InterProphet within TPP. Interspecific protein abundance variation was assessed using the Z statistic as implemented by the APEX software and significant differences identified via Benjamini–Hochberg corrected P values (P < 0.05).
Population Genomic Analysis
We complemented our proteomic analysis using whole-genome resequencing data of house (n = 46) and Spanish (n = 43) sparrows (see Elgvin et al. 2017; Ravinet et al. 2018) for detailed methods on sequencing and data processing). Briefly, following data quality filtering, variants were called at all sites (variant and invariant) using GATK (3.7) HaplotypeCaller (DePristo et al. 2011). We then established two high-quality data sets for different downstream analyses (see below). The first data set (hereafter “variant only”) included only polymorphic, biallelic SNPs occurring in at least 80% of individuals, with minimum site and genotype quality scores of 20 and a mean site depth of between 10× and 40×. In addition, we masked all genotypes with a depth below 5× and above 60×, and then filtered for a minor allele frequency (MAF) threshold of 0.05. The second data set (hereafter “all sites”) was filtered for the same criteria, with the exception of the MAF threshold, and included calls at all sites (i.e., variant and invariant positions). All filtering was conducted using vcftools 0.1.13 (Danecek et al. 2011) and bcftools 1.1(Danecek and McCarthy 2017).
To identify genes encoding SFPs putatively under selection, we estimated FST, a relative measure of differentiation, for each SF gene as the mean FST of all SNPs within the annotated gene region using vcftools (Danecek et al. 2011) from our “variant only” data set. We then tested whether genes underlying the SF proteome exhibited a greater signal of divergence relative to the overall genomic background. To do this, we compared the distribution of FST values for SF proteome genes to all other annotated genes in the genome. Next, in order to identify SF genes that were more strongly divergent than average, we identified SF genes located above the 95th percentile of the empirical distribution of FST values for the entire genome (supplementary fig. S5, Supplementary Material online). We then examined the identity of these outlier genes manually.
To gain further insight into the evolutionary processes acting on SF genes, we further examined regions surrounding the SF outlier genes of interest. Specifically, we first calculated FST for these regions using 100-kb sliding windows with a 25-kb step using vcftools 0.1.13 (Danecek et al. 2011). For these same windows, we estimated dXY, an absolute measure of divergence, and nucleotide diversity (π) for both the house and Spanish sparrow using popgenWindows.py (Martin et al. 2015) with our “all sites” data set. Finally, we used 100-kb sliding window estimates of recombination rate data for these regions taken from Elgvin et al. (2017).
Next, given that variation in FST and dXY can be partially explained by genome-wide variation in recombination rate, we calculated an additional measure of divergent selection based on extended haplotype length-based selection statistics. These statistics incorporate information on linkage disequilibrium and recombination rate variation and have high power for detecting recent selective sweeps (Vitti et al. 2013). Specifically, we calculated per-SNP estimates of xpEHH. xpEHH compares measures of haplotype lengths between populations in order to identify regions where a selective sweep may have occurred in at least one lineage. To calculate xpEHH, we first phased data using ShapeIt2 (O’Connell et al. 2014). For all autosomes, the previously published linkage map (Elgvin et al. 2017) was used to inform phasing and we did not include the Z chromosome in this analysis; see Ravinet et al (2018) for further details on phasing. xpEHH was then calculated on the phased “variant only” data set using the R package rehh (Gautier et al. 2017). We identified peaks in xpEHH using the cutoff log10 (P value) xpEHH > 3, representing a P value of 0.001.
Molecular Evolutionary Analysis
Molecular evolutionary analyses of orthology groups across Passerines were based on the orthology relationships present in OrthoDB (Passeriformes level), including the P. domesticus relationships established previously (see above). Nucleotide coding sequences from the 11 species in OrthoDB were batch downloaded from NCBI (Acanthisitta chloris, Corvus brachyrhynchos, Corvus cornix, Geospiza fortis, Manacus vitellinus, P. domesticus, Pseudopodoces humilis, Serinus canaria, and Zonotrichia albicollis) or ENSEMBL (Ficedula albicollis and T. guttata), and aligned “in-frame” using the L-INS-i algorithm of MAFFT (Katoh and Standley 2013). SFPs have a tendency for lineage-specific gene gains and losses (Hahn et al. 2007; Findlay et al. 2008), which may make identification of orthologs more difficult for SFPs compared with nonreproductive proteins, especially among more distantly related species (Whittington et al. 2017). We therefore examined orthology loss in SFPs relative to the genome as a whole. Next, a phylogeny was established for these species from the time-calibrated molecular phylogeny of all extant avian species (Jetz et al. 2012). Specifically, we downloaded 1,000 randomly selected phylogenetic trees for the 11 species from those available at www.birdtree.org using the Hackett sequenced species backbone and summarized the sample of trees onto a single maximum clade credibility tree with mean node heights using TreeAnnotator (version 1.8.0, BEAST; Drummond et al. 2012).
Using this phylogeny (supplementary fig. S6, Supplementary Material online), we estimated house sparrow lineage-specific values of dN (nonsynonymous substitution rate) and dS (synonymous substitution rate) using the free-ratio model in codeml in PAML (Yang 2007). To gain a preliminary understanding of tissue specificity of SF protein genes, we made use of average gene expression data from the collared flycatcher (Ficedula albicolis) (Uebbing et al. 2016), in conjunction with our SF semiquantitative protein abundance data, in order to examine tissue specificity of expression in SF. Orthology relationships, as determined above, were used to integrate flycatcher gene expression data with sparrow SF composition. Expression specificity (τ) was calculated following Yanai et al. (2005). We then divided values of τ for each tissue type into quartiles and compared house sparrow lineage-specific average values of dN/dS among these quartiles. Next, we implemented the M8 and M8a site models for 7,670 genes using codeml in PAML (Yang 2007), and compared the model M8 to the null model M8a in order to infer the influence of positive selection. The null distribution was the 50:50 mixture of point mass 0 and chi-square (Self and Liang 1987; Wong et al. 2004). P values were adjusted with Benjamini–Hochberg multiple test correction, and we used a conservative threshold of significance of P < 0.01. Interactions among proteins with evidence of positive selection were predicted using STRING (v10.0; Szklarczyk et al. 2015). Default settings were used to identify interactions with a minimum score of 0.4. We included a maximum of five interactions in the first shell of the PPI, and proteins without any interactions were excluded from the resultant network.
Statistical Analysis
All statistical analyses were performed using the R statistical package (v 3.5.1; R Development Core Team 2019).
Supplementary Material
Acknowledgments
The authors are extremely grateful to Scott Pitnick, Caitlin McDonough, Erin McCullough, Jane Pascar, Zeeshan Syed, and two anonymous reviewers for helpful comments on this manuscript and to Alfonso Marzal Reynolds, Sergio Magallanes Argany, and Diana Carneiro for their generous assistance with fieldwork. They also wish to thank the Cambridge Proteomics Facility, including Mike Deery, Renata Feret, and Kathryn Lilley for excellent proteomic support and Eric Sedore and Larne Pekowsky for computational advice. This research was supported by a Research Council of Norway grant (230434 to M.R.). E.W. was supported by a Marilyn Kerr Fellowship and S.D. by funding from Syracuse University.
References
- Ait Belkacem A, Gast O, Stuckas H, Canal D, LoValvo M, Giacalone G, Päckert M.. 2016. North African hybrid sparrows (Passer domesticus, P. hispaniolensis) back from oblivion—ecological segregation and asymmetric mitochondrial introgression between parental species. Ecol Evol. 6(15):5190–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Fernández A, Borziak K, McDonald GC, Dorus S, Pizzari T.. 2019. Female novelty and male status dynamically modulate ejaculate expenditure and seminal fluid proteome over successive matings in red junglefowl. Sci Rep. 9:5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Wathiqui N, Lewis SM, Dopman EB.. 2014. Using RNA sequencing to characterize female reproductive genes between Z and E Strains of European Corn Borer moth (Ostrinia nubilalis). BMC Genomics. 15(1):189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés JA, Larson EL, Bogdanowicz SM, Harrison RG.. 2013. Patterns of transcriptome divergence in the male accessory gland of two closely related species of field crickets. Genetics 193(2):501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés JA, Maroja LS, Harrison RG.. 2008. Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. Proc R Soc B 275(1646):1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo-Jorge TC, Lage MJ, Rivera MT, Carlier Y, Van Leuven F.. 1992. Trypanosoma cruzi: enhanced alpha-macroglobulin levels correlate with the resistance of BALB/cj mice to acute infection. Parasitol Res. 78(3):215–221. [DOI] [PubMed] [Google Scholar]
- Atikuzzaman M, Alvarez-Rodriguez M, Carrillo AV, Johnsson M, Wright D, Rodriguez-Martinez H.. 2017. Conserved gene expression in sperm reservoirs between birds and mammals in response to mating. BMC Genomics. 18(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF.. 2011. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 56(1):21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B, Eubel H, Taylor NL, O’Toole N, Millar AH.. 2009. Insights into female sperm storage from the spermathecal fluid proteome of the honeybee Apis mellifera. Genome Biol. 10(6):R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B, Heazlewood JL, Taylor NL, Eubel H, Millar AH.. 2009. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics 9(8):2085–2097. [DOI] [PubMed] [Google Scholar]
- Bakst MR, Bauchan G.. 2016. Lectin staining of the uterovaginal junction and sperm-storage tubule epithelia in broiler hens. Poult Sci. 95(4):948–955. [DOI] [PubMed] [Google Scholar]
- Bakst MR, Cecil HC.. 1992. Effect of bovine serum albumin on motility and fecundity of turkey spermatozoa before and after storage. J Reprod Fertil. 94(2):287–293. [DOI] [PubMed] [Google Scholar]
- Bansal SK, Gupta N, Sankhwar SN, Rajender S.. 2015. Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS One 10(5):e0127007–e0127021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead TR, Brillard JP.. 2007. Reproductive isolation in birds: postcopulatory prezygotic barriers. Trends Ecol Evol. 22(5):266–272. [DOI] [PubMed] [Google Scholar]
- Blais J, Rico C, van Oosterhout C, Cable J, Turner GF, Bernatchez L.. 2007. MHC adaptive divergence between closely related and sympatric African cichlids. PLoS One 2(8):e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobr LW, Lorenz FW, Ogasawara FX.. 1964. Distribution of spermatozoa in the oviduct and fertility in domestic birds I: residence sites of spermatozoa in fowl oviducts. J Reprod Fertil. 8(1):39–47. [DOI] [PubMed] [Google Scholar]
- Boes KE, Ribeiro JMC, Wong A, Harrington LC, Wolfner MF, Sirot LK.. 2014. Identification and characterization of seminal fluid proteins in the Asian tiger mosquito, Aedes albopictus. PLoS Negl Trop Dis. 8(6):e2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borziak K, Álvarez-Fernández A, Karr TL, Pizzari T, Dorus S.. 2016. The Seminal fluid proteome of the polyandrous Red junglefowl offers insights into the molecular basis of fertility, reproductive ageing and domestication. Sci Rep. 6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braisted JC, Kuntumalla S, Vogel C, Marcotte EM, Rodrigues AR, Wang R, Huang S-T, Ferlanti ES, Saeed AI, Fleischmann RD.. 2008. The APEX Quantitative Proteomics Tool: generating protein quantitation estimates from LC-MS/MS proteomics results. BMC Bioinformatics 9(1):529–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins JD, El-Hinn D, Swanson WJ.. 2007. Adaptive evolution in an avian reproductive protein: ZP3. J Mol Evol. 65(5):555–563. [DOI] [PubMed] [Google Scholar]
- Cao S, Su X, Zeng B, Yan H, Huang Y, Wang E, Yun H, Zhang Y, Liu F, Li W.. 2016. The gut epithelial receptor LRRC19 promotes the recruitment of immune cells and gut inflammation. Cell Rep. 14:695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K, Qian P-Y.. 2009. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics 2009:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Zhang X-Q, Lo C-W, Liu P-F, Liu Y-T, Gallo RL, Hsieh M-F, Schooley RT, Huang C-M.. 2010. The essentiality of α-2-macroglobulin in human salivary innate immunity against new H1N1 swine origin influenza A virus. Proteomics 10(12):2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury NA, Kamada M, Takikawa M, Mori H, Gima H, Aono T.. 1996. Complement-inhibiting activity of human seminal plasma and semen quality. Arch Androl. 36(2):109–118. [DOI] [PubMed] [Google Scholar]
- Civetta A, Singh RS.. 1998. Sex-related genes, directional sexual selection, and speciation. Mol Biol Evol. 15(7):901–909. [DOI] [PubMed] [Google Scholar]
- Claw KG, George RD, MacCoss MJ, Swanson WJ.. 2018. Quantitative evolutionary proteomics of seminal fluid from primates with different mating systems. BMC Genomics. 19(1):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan L, Redhai S, Leiblich A, Fan S-J, Perera SMW, Patel R, Gandy C, Wainwright SM, Morris JF, Hamdy F.. 2014. BMP-regulated exosomes from Drosophila male reproductive glands reprogram female behavior. J Cell Biol. 206(5):671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Craig R, Beavis RC.. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20(9):1466–1467. [DOI] [PubMed] [Google Scholar]
- Cramer ERA, Ålund M, McFarlane SE, Johnsen A, Qvarnström A.. 2016. Females discriminate against heterospecific sperm in a natural hybrid zone. Evolution 70(8):1844–1855. [DOI] [PubMed] [Google Scholar]
- Cramer ERA, Laskemoen T, Eroukhmanoff F, Haas F, Hermansen JS, Lifjeld JT, Rowe M, Sætre G-P, Johnsen A.. 2014. Testing a post-copulatory pre-zygotic reproductive barrier in a passerine species pair. Behav Ecol Sociobiol. 68(7):1133–1144. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST.. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, McCarthy SA.. 2017. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33(13):2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner EC, Ahmed-Braimah Y, Borziak K, Wolfner MF, Harrington LC, Dorus S. 2019. Proteins, Transcripts, and Genetic Architecture of Seminal Fluid and Sperm in the Mosquito Aedes aegypti. Molecular & Cellular Proteomics. 18:S6–S22. [DOI] [PMC free article] [PubMed]
- den Boer SPA, Boomsma JJ, Baer B.. 2009. Honey bee males and queens use glandular secretions to enhance sperm viability before and after storage. J Insect Physiol. 55:538–543. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, et al. 2010. A guided tour of the Trans-Proteomic Pipeline. Proteomics 10(6):1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer T, Weidner W, Michelmann HW, Schiefer HG, Rovan E, Mayer F.. 1996. Influence of Escherichia coli on motility parameters of human spermatozoa in vitro. Int J Androl. 19(5):271–277. [DOI] [PubMed] [Google Scholar]
- Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL.. 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 38(12):1440–1445. [DOI] [PubMed] [Google Scholar]
- Dorus S, Skerget S, Karr TL.. 2012. Proteomic discovery of diverse immunity molecules in mammalian spermatozoa. Syst Biol Reprod Med. 58(4):218–228. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun MD, Smith ND, Baker MA, Lin M, Aitken RJ, Nixon B.. 2011. The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm–oocyte interaction. J Biol Chem. 286(42):36875–36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D.. 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 17(1):68–74. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Kingan SB, Calkins JD, Balakrishnan CN, Jennings WB, Swanson WJ, Sorenson MD.. 2005. Speciation in birds: genes, geography, and sexual selection. Proc Natl Acad Sci U S A. 102(Suppl 1):6550–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizaguirre C, Lenz TL, Traulsen A, Milinski M.. 2009. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol Lett. 12(1):5–12. [DOI] [PubMed] [Google Scholar]
- Elgvin TO, Hermansen JS, Fijarczyk A, Bonnet T, Borge T, Saether SA, Voje KL, Saetre G-P.. 2011. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol Ecol. 20(18):3823–3837. [DOI] [PubMed] [Google Scholar]
- Elgvin TO, Trier CN, Tørresen OK, Hagen IJ, Lien S, Nederbragt AJ, Ravinet M, Jensen H, Sætre G-P.. 2017. The genomic mosaicism of hybrid speciation. Sci Adv. 3(6):e1602996.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51–63. [DOI] [PubMed] [Google Scholar]
- Elliott DE, Siddique SS, Weinstock JV.. 2014. Innate immunity in disease. Clin Gastroenterol Hepatol. 12(5):749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, Hoopmann MR.. 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics 13(1):22–24. [DOI] [PubMed] [Google Scholar]
- Eroukhmanoff F, Rowe M, Cramer ERA, Haas F, Hermansen JS, Runemark A, Johnsen A, Sætre G-P.. 2016. Experimental evidence for ovarian hypofunction in sparrow hybrids. Avian Res. 7(1):3. [Google Scholar]
- Findlay GD, Yi X, MacCoss MJ, Swanson WJ.. 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6(7):e178.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman RC, Gasparini C, Manier MK, Pizzari T.. 2017. Postmating female control: 20 years of cryptic female choice. Trends Ecol Evol. 32(5):368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CE, Howie S.. 2006. The role of megalin (LRP-2/Gp330) during development. Dev Biol. 296(2):279–297. [DOI] [PubMed] [Google Scholar]
- Freedman SJ. 1991. The role of alpha 2-macroglobulin in furunculosis: a comparison of rainbow trout and brook trout. Comp Biochem Physiol B 98(4):549–553. [DOI] [PubMed] [Google Scholar]
- Freitas MJ, Vijayaraghavan S, Fardilha M.. 2017. Signaling mechanisms in mammalian sperm motility. Biol Reprod. 96(1):2–12. [DOI] [PubMed] [Google Scholar]
- Fujihara N. 1992. Accessory reproductive fluids and organs in male domestic birds. Worlds Poult Sci J. 48(1):39–56. [Google Scholar]
- Gautier M, Klassmann A, Vitalis R.. 2017. rehh2.0: a reimplementation of the R package rehhto detect positive selection from haplotype structure. Mol Ecol Resour. 17(1):78–90. [DOI] [PubMed] [Google Scholar]
- Ghaderi D, Springer SA, Ma F, Cohen M, Secrest P, Taylor RE, Varki A, Gagneux P.. 2011. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc Natl Acad Sci. 108(43):17743–17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles S, Czuprynski C.. 2003. Novel role for albumin in innate immunity: serum albumin inhibits the growth of Blastomyces dermatitidis yeast form in vitro. Infect Immun. 71(11):6648–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, Wong A, Ravi Ram K, Sirot LK, Levesque L, Artieri CG, Wolfner MF, Civetta A, et al. 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177(3):1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han S-G.. 2007. Gene family evolution across 12 Drosophila Genomes. PLoS Genet. 3(11):e197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines MD, Parker HM, McDaniel CD, Kiess AS.. 2013. Impact of 6 different intestinal bacteria on broiler breeder sperm motility in vitro. Poult Sci. 92(8):2174–2181. [DOI] [PubMed] [Google Scholar]
- Haines MD, Parker HM, McDaniel CD, Kiess AS.. 2015. When rooster semen is exposed to Lactobacillus fertility is reduced. Int J Poult Sci . 14:541–547. [Google Scholar]
- Haley LE, Abplanalp H.. 1970. Possible immunological basis for a reduction of fertility in cross-mating fowl with Japanese quail. J Reprod Fertil. 23(3):375–381. [DOI] [PubMed] [Google Scholar]
- Harris CL, Mizuno M, Morgan BP.. 2006. Complement and complement regulators in the male reproductive system. Mol Immunol. 43:57–67. [DOI] [PubMed] [Google Scholar]
- Heesterbeek DAC, Angelier ML, Harrison RA, Rooijakkers S.. 2018. Complement and bacterial infections: from molecular mechanisms to therapeutic applications. J Innate Immun. 10(5–6):455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansen JS, Haas F, Trier CN, Bailey RI, Nederbragt AJ, Marzal A, Saetre G-P.. 2014. Hybrid speciation through sorting of parental incompatibilities in Italian sparrows. Mol Ecol. 23(23):5831–5842. [DOI] [PubMed] [Google Scholar]
- Hermansen JS, Saether SA, Elgvin TO, Borge T, Hjelle E, Saetre G-P.. 2011. Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol Ecol. 20(18):3812–3822. [DOI] [PubMed] [Google Scholar]
- Hooper DM, Griffith SC, Price TD.. 2018. Sex chromosome inversions enforce reproductive isolation across an avian hybrid zone. Mol Ecol. 21:610–617. [DOI] [PubMed] [Google Scholar]
- Howard DJ, Palumbi SR, Birge LM, Manier MK.. 2009. Sperm and speciation In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm biology: an evolutionary perspective. Oxford: Academic Press; p. 363–399. [Google Scholar]
- Hupton G, Portocarrero S, Newman M, Westneat DF.. 2003. Bacteria in the reproductive tracts of red-winged blackbirds. Condor 105(3):453–464. [Google Scholar]
- Janoušek V, Wang L, Luzynski KEN, Dufková P, Vyskočilová MM, Nachman MW, Munclinger P, Macholán M, Piálek J, Tucker PK.. 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol. 21(12):3032–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AØ.. 2012. The global diversity of birds in space and time. Nature 491(7424):444–448. [DOI] [PubMed] [Google Scholar]
- Johansen FE, Braathen R, Brandtzaeg P.. 2000. Role of J chain in secretory immunoglobulin formation. Scand J Immunol. 52:240–248. [DOI] [PubMed] [Google Scholar]
- Karn RC, Clark NL, Nguyen ED, Swanson WJ.. 2008. Adaptive evolution in rodent seminal vesicle secretion proteins. Mol Biol Evol. 25(11):2301–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvonen A, Seehausen O.. 2012. The role of parasitism in adaptive radiations—when might parasites promote and when might they constrain ecological speciation? Int J Ecol. 2012:1–20. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Prabha V, Shukla G, Sarwal A.. 2009. Interference of human spermatozoal motility by live Staphylococcus aureus. Am J Biomed Sci. 2:91–97. [Google Scholar]
- Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, et al. 2016. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 428(4):688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Watts TD, LaFlamme BA, Haynes PA, Markow TA.. 2009. Proteomic analysis of Drosophila mojavensis male accessory glands suggests novel classes of seminal fluid proteins. Insect Biochem Mol Biol. 39(5–6):366–371. [DOI] [PubMed] [Google Scholar]
- King M, Eubel H, Millar AH, Baer B.. 2011. Proteins within the seminal fluid are crucial to keep sperm viable in the honeybee Apis mellifera. J Insect Physiol. 57(3):409–414. [DOI] [PubMed] [Google Scholar]
- Kleven O, Fossøy F, Laskemoen T, Robertson RJ, Rudolfsen G, Lifjeld JT.. 2009. Comparative evidence for the evolution of sperm swimming speed by sperm competition and female sperm storage duration in passerine birds. Evolution 63(9):2466–2473. [DOI] [PubMed] [Google Scholar]
- Koskinen P, Törönen P, Nokso-Koivisto J, Holm L.. 2015. PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics 31(10):1544–1552. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld K, Kraaijeveld-Smit FJL, Maan ME.. 2011. Sexual selection and speciation: the comparative evidence revisited. Biol Rev. 86(2):367–377. [DOI] [PubMed] [Google Scholar]
- LaFlamme BA, Wolfner MF.. 2013. Identification and function of proteolysis regulators in seminal fluid. Mol Reprod Dev. 80(2):80–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Andrés JA, Bogdanowicz SM, Harrison RG.. 2013. Differential introgression in a mosaic hybrid zone reveals candidate barrier genes. Evolution 67(12):3653–3661. [DOI] [PubMed] [Google Scholar]
- Lung O, Kuo L, Wolfner MF.. 2001. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J Insect Physiol. 47(6):617–622. [DOI] [PubMed] [Google Scholar]
- Lüpold S, Calhim S, Immler S, Birkhead TR.. 2009. Sperm morphology and sperm velocity in passerine birds. Proc R Soc B 276(1659):1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Sun Z, de Matos R, Zhang J, Odunsi K, Lin B.. 2014. Towards an animal model of ovarian cancer: cataloging chicken blood proteins using combinatorial peptide ligand libraries coupled with shotgun proteomic analysis for translational research. OMICS 18(5):280–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W, et al. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. 48(10):1204–1210. [DOI] [PubMed] [Google Scholar]
- Manier MK, Belote JM, Berben KS, Lüpold S, Ala-Honkola O, Collins WF, Pitnick S.. 2013. Rapid diversification of sperm precedence traits and processes among three sibling Drosophila species. Evolution 67(8):2348–2362. [DOI] [PubMed] [Google Scholar]
- Manier MK, Lüpold S, Belote JM, Starmer WT, Berben KS, Ala-Honkola O, Collins WF, Pitnick S.. 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr Biol. 23(19):1853–1862. [DOI] [PubMed] [Google Scholar]
- Manier MK, Lüpold S, Pitnick S, Starmer WT.. 2013. An analytical framework for estimating fertilization bias and the fertilization set from multiple sperm-storage organs. Am Nat. 182(4):552–561. [DOI] [PubMed] [Google Scholar]
- Maroto Martín LO, Muñoz EC, De Cupere F, Van Driessche E, Echemendia-Blanco D, Rodríguez JMM, Beeckmans S.. 2010. Bacterial contamination of boar semen affects the litter size. Anim Reprod Sci. 120(1–4):95–104. [DOI] [PubMed] [Google Scholar]
- Marshall JL, Huestis DL, Garcia C, Hiromasa Y, Wheeler S, Noh S, Tomich JM, Howard DJ.. 2011. Comparative proteomics uncovers the signature of natural selection acting on the ejaculate proteomes of two cricket species isolated by postmating, prezygotic phenotypes. Mol Biol Evol. 28(1):423–435. [DOI] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough CE, Whittington E, Pitnick S, Dorus S.. 2016. Proteomics of reproductive systems: towards a molecular understanding of postmating, prezygotic reproductive barriers. J Proteomics 135:26–37. [DOI] [PubMed] [Google Scholar]
- McGraw LA, Suarez SS, Wolfner MF.. 2015. On a matter of seminal importance. BioEssays 37(2):142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang Q-X.. 2014. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature 505:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. 2012. Ecological speciation. Oxford: Oxford University Press. [Google Scholar]
- O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, Traglia M, Huang J, Huffman JE, Rudan I, et al. 2014. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10:e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR. 1999. All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci U S A. 96(22):12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis TM, Butlin R, Zuk M, Tregenza T.. 2001. Sexual selection and speciation. Trends Ecol Evol. 16(7):364–371. [DOI] [PubMed] [Google Scholar]
- Paynter E, Millar AH, Welch M, Baer-Imhoof B, Cao D, Baer B.. 2017. Insights into the molecular basis of long-term storage and survival of sperm in the honeybee (Apis mellifera). Sci Rep. 7:40236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS.. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20(18):3551–3567. [DOI] [PubMed] [Google Scholar]
- Pilch B, Mann M.. 2006. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 7(5):R40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S, Wolfner MF.. 2009. Ejaculate-female and sperm–female interactions In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: an evolutionary perspective. San Diego: Academic Press; p. 247–304. [Google Scholar]
- Pitnick S, Wolfner M, Dorus S, Forthcoming 2019. Post-ejaculate modifications to sperm (PEMS). Biol Rev. [DOI] [PMC free article] [PubMed]
- Poiani A. 2006. Complexity of seminal fluid: a review. Behav Ecol Sociobiol. 60(3):289–310. [Google Scholar]
- Price T. 2008. Speciation in Birds. Colorado: Roberts and Company. [Google Scholar]
- Prokupek AM, Kachman SD, Ladunga I, Harshman LG.. 2009. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Mol Biol. 18(4):465–475. [DOI] [PubMed] [Google Scholar]
- Pryke SR, Rollins LA, Griffith SC.. 2010. Females use multiple mating and genetically loaded sperm competition to target compatible genes. Science 329(5994):964–967. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2019. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD.. 2007. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 25(1):207–219. [DOI] [PubMed] [Google Scholar]
- Ravi Ram K, Wolfner MF.. 2007. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 47(3):427–445. [DOI] [PubMed] [Google Scholar]
- Ravinet M, Elgvin TO, Trier CN, Aliabadian M, Gavrilov A, Sætre G-P.. 2018. Signatures of human-commensalism in the house sparrow genome. Proc R Soc B 285(1884):pii: 20181246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, Noor MAF, Mehlig B, Westram AM.. 2017. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J Evol Biol. 30(8):1450–1477. [DOI] [PubMed] [Google Scholar]
- Ritchie MG. 2007. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 38(1):79–102. [Google Scholar]
- Rowe M, Albrecht T, Cramer ERA, Johnsen A, Laskemoen T, Weir JT, Lifjeld JT.. 2015. Postcopulatory sexual selection is associated with accelerated evolution of sperm morphology. Evolution 69(4):1044–1052. [DOI] [PubMed] [Google Scholar]
- Rowe M, Czirják GÁ, Lifjeld JT, Giraudeau M.. 2013. Lysozyme-associated bactericidal activity in the ejaculate of a wild passerine. Biol J Linn Soc Lond. 109(1):92–100. [Google Scholar]
- Rowe M, Czirják GÁ, McGraw KJ, Giraudeau M.. 2011. Sexual ornamentation reflects antibacterial activity of ejaculates in mallards. Biol Lett. 7(5):740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M, Skerget S, Rosenow MA, Karr TL. 2019. Identification and characterization of the zebra finch (Taeniopygia guttata) sperm proteome. Journal of Proteomics, 193:192–204. [DOI] [PubMed] [Google Scholar]
- Rowe M, Pruett-Jones S.. 2011. Sperm competition selects for sperm quantity and quality in the Australian Maluridae. PLoS One 6(1):e15720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue T, Takeuchi K, Maeda T, Yamamoto Y, Nishi K, Ohkubo I.. 2010. Factor H in porcine seminal plasma protects sperm against complement attack in genital tracts. J Biol Chem. 285(3):2184–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self SG, Liang K-Y.. 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc. 82(398):605–610. [Google Scholar]
- Sepil I, Hopkins BR, Dean R, Thézénas M-L, Charles PD, Konietzny R, Fischer R, Kessler B, Wigby S.. 2019. Quantitative proteomics identification of seminal fluid proteins in male Drosophila melanogaster. Mol Cell Proteomics 18(Suppl 1):S46–S58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servedio MR. 2012. The relationship between sexual selection and speciation. Curr Zool. 58(3):413–415. [Google Scholar]
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ.. 2018. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell 46(4):481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz AJ, Sackton TB.. 2019. Immune genes are hotspots of shared positive selection across birds and mammals. eLife 8:1703–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot LK, Hardstone MC, Helinski MEH, Ribeiro JMC, Kimura M, Deewatthanawong P, Wolfner MF, Harrington LC.. 2011. Towards a semen proteome of the dengue vector mosquito: protein identification and potential functions. PLoS Negl Trop Dis. 5(3):e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerget S, Rosenow M, Polpitiya A, Petritis K, Dorus S, Karr TL.. 2013. The rhesus macaque (Macaca mulatta) sperm proteome. Mol Cell Proteomics 12(11):3052–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerget S, Rosenow MA, Petritis K, Karr TL.. 2015. Sperm proteome maturation in the mouse epididymis. PLoS One 10(11):e0140650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele MG, Wishart GJ.. 1992. Evidence for a species-specific barrier to sperm transport within the vagina of the chicken hen. Theriogenology 38(6):1107–1114. [DOI] [PubMed] [Google Scholar]
- Stewart R, Rambo TB.. 2000. Cloacal microbes in house sparrows. Condor 102(3):679–684. [Google Scholar]
- Sullivan R, Saez F.. 2013. Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction 146(1):R21–R35. [DOI] [PubMed] [Google Scholar]
- Summers-Smith JD. 1988. The sparrows. Calton: Bloomsbury.
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF.. 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci U S A. 98(13):7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD.. 2002. The rapid evolution of reproductive proteins. Nat Rev Genet. 3(2):137–144. [DOI] [PubMed] [Google Scholar]
- Sweigart AL. 2010. The genetics of postmating, prezygotic reproductive isolation between Drosophila virilis and D. americana. Genetics 184(2):401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43(D1):D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Hoekstra HE.. 2008. Causes and consequences of the evolution of reproductive proteins. Int J Dev Biol. 52(5–6):769–780. [DOI] [PubMed] [Google Scholar]
- Tyler F, Harrison XA, Bretman A, Veen T, Rodríguez-Muñoz R, Tregenza T.. 2013. Multiple post-mating barriers to hybridization in field crickets. Mol Ecol. 22(6):1640–1649. [DOI] [PubMed] [Google Scholar]
- Uebbing S, Künstner A, Mäkinen H, Backström N, Bolivar P, Burri R, Dutoit L, Mugal CF, Nater A, Aken B, et al. 2016. Divergence in gene expression within and between two closely related flycatcher species. Mol Ecol. 25(9):2015–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ.. 2011. Selection in the rapid evolution of gamete recognition proteins in marine invertebrates. Cold Spring Harbor Perspect Biol. 3(11):a002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV.. 2011. The antibacterial lectin RegIII-gamma promotes the spatial segregation of microbiota and host in the intestine. Science 334(6053):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC.. 2013. Detecting natural selection in genomic data. Annu Rev Genet. 47(1):97–120. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44(D1):D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, Begun DJ.. 2005. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171(3):1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington E, Forsythe D, Borziak K, Karr TL, Walters JR, Dorus S.. 2017. Contrasting patterns of evolutionary constraint and novelty revealed by comparative sperm proteomic analysis in Lepidoptera. BMC Genomics. 18(1):931.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S, Suarez SS, Lazzaro BP, Pizzari T, Wolfner MF.. 2019. Sperm success and immunity. Curr Top Dev Biol. 135:287–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilburn DB, Swanson WJ.. 2016. From molecules to mating: rapid evolution and biochemical studies of reproductive proteins. J Proteomics 135:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wira CR, Fahey JV, Sentman CL, Pioli PA, Shen L.. 2005. Innate and adaptive immunity in female genital tract: cellular responses and interactions. Immunol Rev. 206(1):306–335. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Ellegren H.. 2016. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 18:87–100. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in. Heredity 88(2):85–93. [DOI] [PubMed] [Google Scholar]
- Wolfson A. 1952. The cloacal protuberance—a means for determining breeding condition in live male passerines. Bird-Banding 23(4):159–165. [Google Scholar]
- Wong A, Albright SN, Giebel JD, Ram KR, Ji S, Fiumera AC, Wolfner MF.. 2008. A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics 180(2):921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R.. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168(2):1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakirevich E, Naot Y.. 2000. Cloning of a glucose phosphate isomerase/neuroleukin-like sperm antigen involved in sperm agglutination. Biol Reprod. 62(4):1016–1023. [DOI] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yeates SE, Diamond SE, Einum S, Emerson BC, Holt WV, Gage M.. 2013. Cryptic choice of conspecific sperm controlled by the impact of ovarian fluid on sperm. Evolution 67(12):3523–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Bar T, Yehuda R, Hacham T, Krupnik S, Bartoov B.. 2008. Influence of Campylobacter fetus subsp. fetus on ram sperm cell quality. J Med Microbiol. 57(11):1405–1410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.