Abstract
Proteins in saliva are needed for preprocessing food in the mouth, maintenance of tooth mineralization, and protection from microbial pathogens. Novel insights into human lineage-specific functions of salivary proteins and clues to their involvement in human disease can be gained through evolutionary studies, as recently shown for salivary amylase AMY1 and salivary agglutinin DMBT1/gp340. However, the entirety of proteins in saliva, the salivary proteome, has not yet been investigated from an evolutionary perspective. Here, we compared the proteomes of human saliva and the saliva of our closest extant evolutionary relatives, chimpanzees and gorillas, using macaques as an outgroup, with the aim to uncover features in saliva protein composition that are unique to each species. We found that humans produce a waterier saliva, containing less than half total protein than great apes and Old World monkeys. For all major salivary proteins in humans, we could identify counterparts in chimpanzee and gorilla saliva. However, we discovered unique protein profiles in saliva of humans that were distinct from those of nonhuman primates. These findings open up the possibility that dietary differences and pathogenic pressures may have shaped a distinct salivary proteome in the human lineage.
Keywords: saliva, salivary proteins, salivary proteome, great apes, Old World monkeys, evolution
Introduction
Saliva aids in the preprocessing of food, protects and remineralizes tooth enamel, and also moistens and guards the epithelial surfaces in the mouth (Mandel 1987; Dawes et al. 2015). In addition, salivary components modulate the resident oral microbiome and form a first line of defense against systemic pathogens occasionally traversing the mouth environment (Scannapieco 1994; van’t Hof et al. 2014; Marsh et al. 2016; Cross and Ruhl 2018). Many of these functions are attributed to the major abundant groups of proteins specifically secreted by the major salivary glands that is, by the bilateral parotid, submandibular, and sublingual glands (Oppenheim et al. 2007). Recent proteomic surveys have identified more than 2,000 proteins in human saliva, but only a much smaller proportion of them occur in higher abundance in saliva (i.e., each comprising >1% of total salivary protein concentration) and are intrinsically expressed by the salivary glands (Ruhl 2012). For a number of those, including salivary proline-rich proteins, amylase, and mucins, genetic polymorphisms in human populations have been found, but functional or disease associations still remain elusive (Azen and Oppenheim 1973; Biesbrock et al. 1997; Perry et al. 2007; Mandel et al. 2010; Manconi et al. 2016). What is known to-date about functions of salivary proteins, has largely been deduced from conventional in vitro experiments or by analogy to rather distantly related animal models that can only insufficiently represent the human condition with respect to saliva (Gutierrez et al. 2014; Blanchard et al. 2015). This gap in knowledge also poses an impediment to harness saliva as a diagnostic fluid for dental and systemic diseases (Baum et al. 2011; Ruhl 2012).
An alternative possibility for gaining insights into functions and disease associations of salivary proteins is through comparative studies of globally distinct human populations or through comparisons with closely related primate species (Herzberg et al. 1979; Mau et al. 2011). Recent studies using an evolutionary approach have provided major novel insights. One example is the human gene for salivary amylase (AMY1) which underwent several rounds of duplications in early hunter–gatherer ancestors and again later in traditional agricultural societies presumably driven by the sudden rise in consumption of starch (Perry et al. 2007). This evolutionary adaptation to diet led to increased levels of salivary amylase in humans with implications for taste perception and the potential to correlate with biomedically relevant phenotypes (Mandel et al. 2010; Falchi et al. 2014; Usher et al. 2015; Arredouani et al. 2016; Pajic et al. 2019). Similarly, genetic variation affecting the salivary agglutinin [deleted in malignant brain tumors 1 (DMBT1)] gene has been shown to segregate significantly between human populations, with pathogenic pressure being a likely driver (Polley et al. 2015). Our group demonstrated high polymorphism of the salivary mucin 7 (MUC7) gene among nonhuman primate species and geographically distinct human populations with possible associations to the human oral microbiome (Xu et al. 2016, 2017). Hence, the question arises whether other proteins in saliva also show functional variation among humans and nonhuman primates. Discovering such variations will allow for future studies to elucidate adaptations to human-specific dietary changes, or to new pathogen challenges that emerged with increased meat consumption beginning over 2 million years ago, and more recently with agriculture, animal husbandry, and sedentarism (Wolfe et al. 2007; Ungar and Sponheimer 2011; Smith et al. 2015; Carmody et al. 2016). To begin addressing this question, we compared the salivary proteome of humans to that of our closest extant evolutionary relatives, the great apes and the more distantly related Old World monkeys.
Results
Humans Express a Waterier Saliva than Great Apes and Old World Monkeys
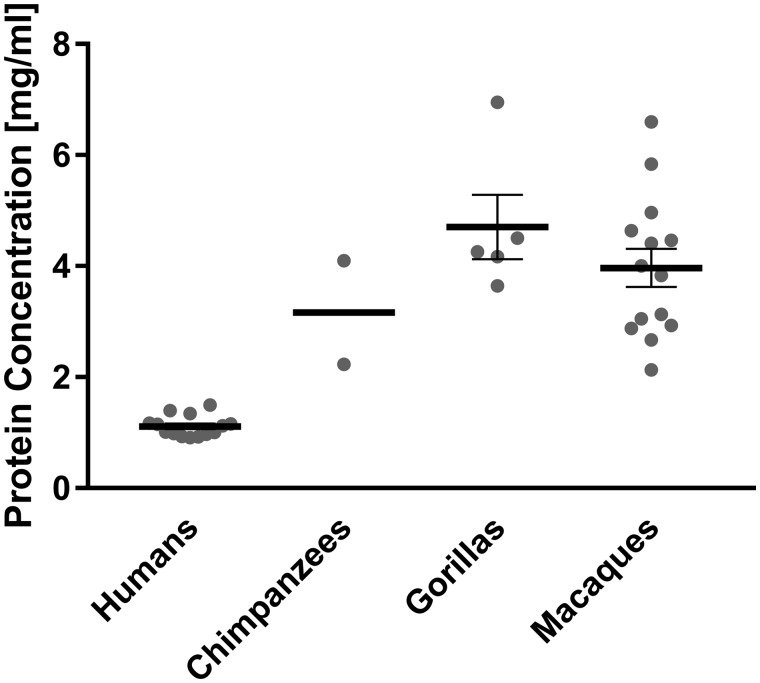
We found the mean concentration of total protein in human saliva to amount to only less than half of that measured in chimpanzee, gorilla, and macaque saliva (fig. 1). This observation remains valid, even when considering multiple samples taken from two individuals of each great ape species at different time points over a period of 2 years (supplementary fig. S1, Supplementary Material online). In those samples, protein concentrations varied less in humans than in great apes. If differences in the sampling technique were the reason for the observed interspecies differences in protein concentration, one would expect to find the most significant difference between chimpanzee and gorilla saliva, because gorilla saliva was collected by suction from the mouth with a pipet, whereas saliva from chimpanzees was obtained in the same fashion as from humans, namely by expectoration in a receptacle. However, the difference in salivary protein concentration between chimpanzees and gorillas was insignificant. Instead, we found the most significant difference between saliva of humans and nonhuman primates collectively (P < 0.0001), while differences in salivary protein concentration among chimpanzees, gorillas, and macaques were insignificant, despite the fact that humans and chimpanzees are more closely related than either are to gorillas or macaques. Overall, a higher water content of saliva would be expected to alter the physical properties in the oral cavity in humans.
Fig. 1.
Total protein concentrations of saliva from humans, chimpanzees, gorillas, and macaques. Whole saliva was collected from different individuals of each species (humans, n = 14; chimpanzees, n = 2; gorillas, n = 5; and macaques, n = 14). Gray dots represent the total salivary protein concentration of each given individual. Bold horizontal bars represent the mean total protein concentrations for each species with error bars indicating the standard error of the mean. From two human, two chimpanzee, and two gorilla individuals included in the above figure, repetitive samples were taken on different days. For those individuals the gray dot in the figure above represents the mean value of all samples that were taken from each (for detailed data see Supplementary fig. S1, Supplementary Material online).
All Major Abundant Salivary Proteins Detected in Humans are also Detectable in Chimpanzee and Gorilla Saliva
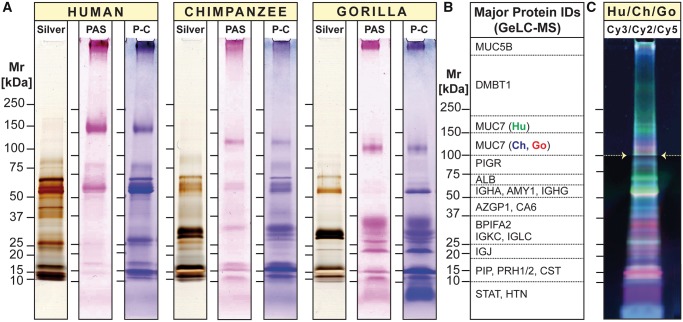
To obtain a first global comparison of the salivary proteomes, we separated human, chimpanzee, and gorilla saliva by 1D PAGE (fig. 2). We further performed gel-enhanced liquid chromatography mass spectrometry (GeLC-MS) for identification of proteins by liquid chromatography nanoelectrospray-tandem mass spectrometry (LC-ESI-MS/MS) analysis (fig. 2B, supplementary fig. S2, and supplementary table S1, Supplementary Material online). Mass spectrometric analysis of gel sections established that for all major abundant proteins that were detectable in human saliva, counterparts existed in chimpanzee and gorilla saliva (supplementary fig. S2, and supplementary table S1, Supplementary Material online). However, major differences among human saliva and that of great apes became apparent in the overall electrophoretic banding profiles (fig. 2A).
Fig. 2.
1D proteome profiles of human, chimpanzee, and gorilla saliva. (A) Equal amounts of human, chimpanzee, and gorilla saliva taken from one individual of each species were separated by 1D PAGE under reducing conditions. Protein bands were visualized by silver stain (Silver). Glycosylated protein bands were revealed by periodic acid-Schiff (PAS) staining, followed by Coomassie Blue (P-C). Note that substantial deviations of the apparent molecular size of certain protein IDs from the size expected based on amino acid composition can be explained by posttranslational modifications, most importantly extensive glycosylation that either retards the electrophoretic mobility because of the added mass of attached glycans, or increases it because of negative charge added by terminal sialic acid molecules. (B) Tracks of parallel run gel replicates for each species were longitudinally divided into consecutive slices according to molecular weight (see Supplementary fig. S2, Supplementary Material online for details). Proteins contained in each slice were identified by GeLC-MS (Supplementary table S1, Supplementary Material online for details). Listed in panel B are the major abundant proteins found in each section. (C) Differences in band intensities of differently fluorochrome-labeled salivary proteins from human (Hu: Cy3, green), chimpanzee (Ch: Cy2, blue), and gorilla (Go: Cy5, red) were visualized by 1D DIGE. Note that the image in panel C was brightened from 100 kDa (white arrows) upward for better visibility of faintly stained higher molecular weight glycoprotein bands.
A component that stood out in this regard was salivary mucin MUC7. In humans, the MUC7 glycoprotein band migrated at a higher molecular weight in electrophoresis than its counterparts in chimpanzee and gorilla saliva. This suggests that the great ape homologs of MUC7 must have either a lower molecular weight or carry a higher negative charge, thereby enhancing their mobility in electrophoresis. Indeed, we recently found that the MUC7 reference genes in these great ape species lacked one of the six proline, threonine, and serine-rich mucin repeat domains (PTS repeats) present in most human individuals (Xu et al. 2016, 2017) (for sequence alignment, see supplementary fig. S5, Supplementary Material online). Nevertheless, the lack of one PTS repeat domain consisting of only 23 amino acids cannot explain the extensive shift in electrophoretic mobility observed (Kirkbride et al. 2001). Likely, additional posttranscriptional or posttranslational features, possibly including differences in the number of negatively charged sialic acid termini, might be responsible for the larger-than-expected shift in MUC7 electrophoretic mobility.
Major differences in salivary proteins among humans and great apes were further substantiated by 1D differential in-gel electrophoresis (DIGE; Tonge et al. 2001; fig. 2C). Labeling the salivary proteins of each species with a different fluorescent Cy-dye (human, green; chimpanzee, blue; and gorilla, red) allowed to visualize differences in protein amounts and mobility by combining equal parts of salivary proteins of all three species in one gel electrophoresis. Green fluorescent protein bands indicated highest levels in humans. Pink colored bands (i.e., mixed color of blue [chimpanzee] and red [gorilla]) indicated highest levels in great apes. The low resolution of 1D gel-based analysis did not allow to identify these proteins. In sum, our results suggest that, although all abundant salivary proteins appear to be shared by humans and great apes, there exist differences among their salivary proteins in structure and quantity.
Human and Great Ape-Specific Differences of Abundant Salivary Proteins
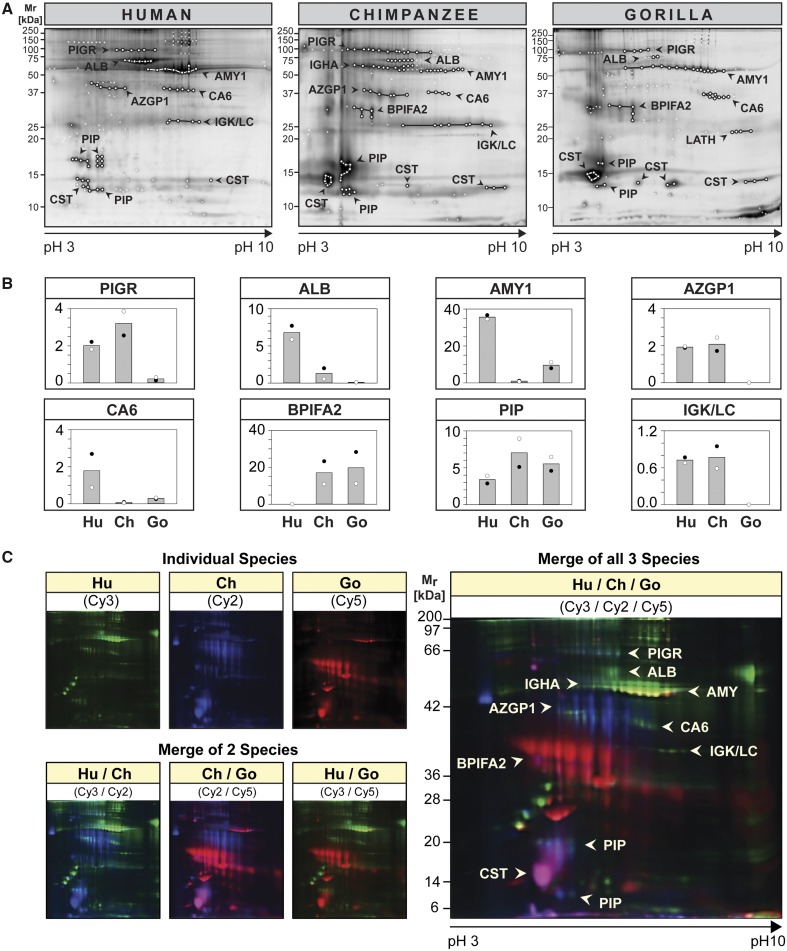
To identify salivary proteins, we used the higher resolving power of 2D gel electrophoresis followed by matrix-assisted laser desorption/ionization-time of flight MS (MALDI-TOF MS) analysis of protein spots (fig. 3). First, we examined differences in 2D spot patterns between species (fig. 3A) as compared with their variation within species (supplementary fig. S3A, Supplementary Material online). Differences in spot patterns were apparent between species, whereas differences between individuals of the same species appeared minor in comparison (supplementary fig. S3A, Supplementary Material online). This was further confirmed by interspecies comparison of salivary protein spot patterns using 2D DIGE analysis (fig. 3C) as well as by combined intra- and interspecies comparisons (supplementary fig. S4, Supplementary Material online). Although protein spots with analogous identities in saliva of humans, chimpanzees, and gorillas were detected in similar regions of the gels (fig. 3A), clear differences in their protein levels between species became apparent.
Fig. 3.
2D proteome profiles of human, chimpanzee and gorilla saliva. (A and B) Equal amounts of protein in saliva from two individuals of each species were differently fluorochrome-labeled (Individual 1: Cy3; Individual 2: Cy5) and pooled with corresponding unlabeled saliva prior to coseparation by 2D PAGE. The 2D proteome profiles for each individual were recorded using a fluorescence scanner set at the appropriate wavelength for each respective cyanine fluorochrome (see supplementary fig. S3A, Supplementary Material online for individual scans). (A) Each gel was counter-stained with Deep Purple and scanned again to reveal all protein spots. Protein spots were excised and analyzed by MALDI-TOF and LC-ESI-MS/MS for protein identification (see Supplementary fig. S3B–D, Supplementary Material online for spot reference number and Supplementary table S2, Supplementary Material online for protein IDs). Protein spots with successful identifications are encircled and spot groups were linked by lines if they contained proteins of the same identity. (B) Fluorescence intensities of protein spots belonging to the same protein ID were quantified using the DeCyder 2D software, the values of individual spots summed up and averaged (individual 1: black circle,●; individual 2: empty circle,◯). The columns represent the mean fluorescence intensity between two individuals of the same species. (C) Differences in the 2D proteome profiles of differently fluorochrome-labeled salivary proteins from human (Hu: Cy3, green), chimpanzee (Ch: Cy2, blue), and gorilla (Go: Cy5, red), one individual each, were visualized using a fluorescence laser scanner at appropriate wavelengths for each cyanine dye. The composite salivary protein spot patterns of two or three species were derived using the ImageQuant software.
We chose eight distinctly demarcated spot groups, well supported by protein identifications, for further comparison by densitometry among the three different species (fig. 3B). Humans showed higher salivary amylase (AMY1) levels than the great apes, confirming earlier reports (McGeachin and Akin 1982; Perry et al. 2007; Behringer et al. 2013). The same trend was found for carbonic anhydrase VI (CA6) and albumin (ALB). An opposite trend was observed for parotid secretory protein [bactericidal/permeability-increasing fold containing family A member 2 (BPIFA2)/short palate, lung, and nasal epithelium clone 2 (SPLUNC2)] where humans showed lower levels than the great apes. Other proteins, including the poly-immunoglobulin receptor (PIGR, secretory component), zinc α-2 glycoprotein (AZGP1), and immunoglobulin light chains (IGK/LC), showed high levels in both humans and chimpanzees, but far lower ones in gorillas.
Two-dimensional DIGE analysis (fig. 3C) further confirmed these findings in that major spot groups including albumin, AMY1, and CA6 stood out with green fluorescence, indicating highest levels in human saliva. Spot groups containing AZGP1, BPIFA2, prolactin-inducible protein (PIP), and salivary cystatins [presumably cystatins S (CST2) and SA (CST4) according to the location of the spot in the acidic region of the gel] stood out in either blue, red, or pink, indicating higher levels in chimpanzees, gorillas, or both these species, than in humans. A unique spot group identified as latherin (LATH) was revealed only in gorilla saliva (fig. 3A). Latherin was not detectable in human saliva but was detected in chimpanzees by 1D GeLC-MS analysis (supplementary table S1, Supplementary Material online). The absence of latherin in human saliva is explainable by the previously reported frameshift mutation of the gene for latherin in the human lineage after divergence from chimpanzees, making it a pseudogene in humans (Hahn and Lee 2005; Vance et al. 2013). Taken together, the results confirmed that all abundant proteins identified in human saliva could also be detected in chimpanzees and gorillas. In addition, we could identify three salivary protein components (AMY1, CA6, and BPIFA2) that follow human-specific trends in their levels.
Estimation of the Extent of Human and Nonhuman Primate-Specific Trends in Salivary Protein Levels
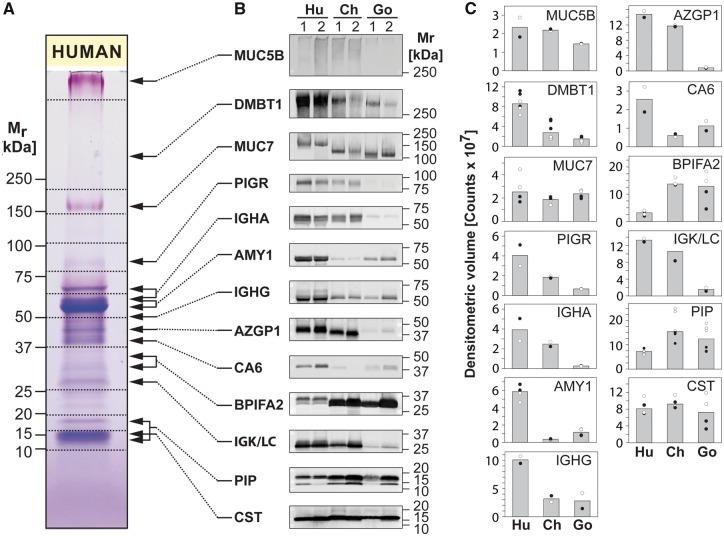
To solidify these observations quantitatively, we performed immuno-detection of 13 abundant salivary protein components common to all three species. Immunoblots confirmed the identities of all 13 proteins in the expected molecular mass range based on 1D GeLC-MS and 2D PAGE spot identifications (fig. 4B). The differences based on immunoblot band intensities (fig. 4C) showed similar trends as the estimated quantitative differences of corresponding spot groups in 2D gels (see fig. 3B andsupplementary fig. S5, Supplementary Material online). Densitometric analysis of immunoblots showed that AMY1, immunoglobulin G heavy chain (IGHG), and CA6 were expressed at higher levels in humans than in both great ape species. DMBT1, PIGR, immunoglobulin A heavy chain (IGHA), AZGP1, and IGK/LC were expressed highest in humans, less in chimpanzees, and least in gorillas. An opposite trend was found for BPIFA2 where humans showed lower protein levels than both great ape species. The shift in electrophoretic mobility for MUC7 was confirmed by immunoblot. Also the band for BPIFA2 in humans was found to be shifted compared with its homologs in great ape saliva. As the lengths and amino acid sequences for BPIFA2 are nearly identical among the three species (supplementary fig. S5, Supplementary Material online), it might likely be posttranslational modifications accounting for that shift in mobility. A possible explanation could be a difference in glycosylation as it has been reported that BPIFA2 in humans is glycosylated (Abdolhosseini et al. 2012).
Fig. 4.
Immunoblot analysis of human, chimpanzee, and gorilla saliva. (A) Equal amounts of protein in human saliva separated by 1D PAGE were stained with Coomassie blue and PAS stains to reveal general and glycosylated protein bands, respectively. (B) Nitrocellulose transfers containing saliva from two human (Hu), two chimpanzee (Ch), and two gorilla (Go) individuals (individual 1 and 2 of each species) were probed with antibodies against MUC5B, DMBT1, MUC7, PIGR, IGHA, AMY1, IGHG, AZGP1, CA6, BPIFA2, IGKC, IGLC, PIP, and CST, followed by detection of bound antibodies with Alexa Fluor 488-tagged IgG secondary antisera. (C) Intensities of protein bands were quantified by ImageQuant software and plotted as band volumes. Each data point for individual 1 (black circle, ●) and individual 2 (empty circle, ◯) represents a measurement from an independent immunoblot experiment. The column heights correspond to the respective median band volumes.
Immunoglobulin-related components (PIGR, IGHA, IGHG, and IGK/LC), all of which are components of the secretory immunoglobulin A (IgA) molecule that occurs in abundant levels in saliva, showed highest levels in human saliva and very low levels in gorillas. Since the antibodies used for immunoblotting were raised against human versions of their target proteins, it could be argued that they might have reacted less with the nonhuman primate protein analogs, which would then theoretically explain reduced immunoblot band intensities in gorillas. However, in the case of PIGR/secretory component, sequence alignments show a high degree of homology of the human version with its analogs in great apes (see Supplementary Material online, supplementary fig. S5, Supplementary Material online), yet there still appears a much weaker band in gorilla saliva. Also, the antibody used for detection of AZGP1 was raised against a defined peptide sequence that appears to be highly conserved in humans and great apes (supplementary fig. S5, panel AZGP1, Supplementary Material online). Thus, it is unlikely that the observed differences in expression of these components are mostly due to differences in antibody recognition. In addition, for PIGR and IGK/LC, low levels of expression in gorilla saliva are also supported by the 2D gel results (fig. 3A and B).
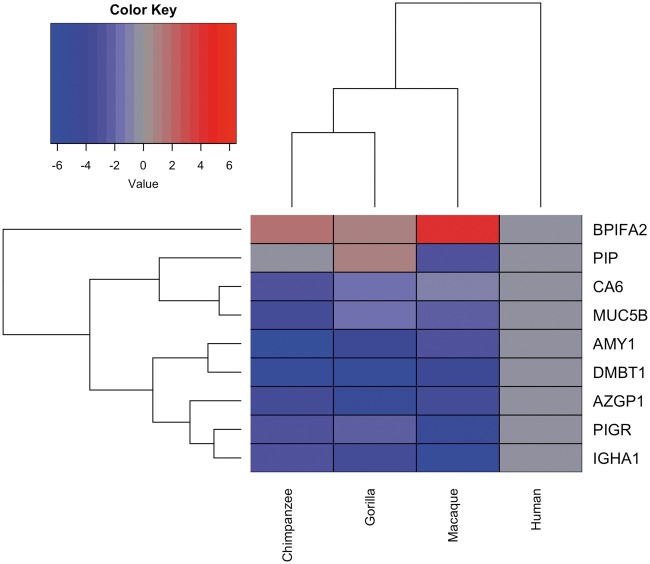
To obtain additional quantitative evaluation, we performed comparative nano-LC-ESI-MS/MS analysis specifically targeting the proteins found to be differently expressed by immunoblot analysis. We selected peptides that 1) are unique for the targeted proteins and 2) occurred both in the human and nonhuman primate versions of the proteins (supplementary fig. S6B, Supplementary Material online). As a phylogenetic outgroup, we included saliva from Rhesus macaques in the analysis. To ensure robustness of data, we analyzed saliva from at least five individuals of each species, with the exception of chimpanzees where only two individuals were available for collection. The cumulative peptide abundance data obtained for the different proteins are summarized in a heatmap (fig. 5). Significance values as well as quantitative protein and single peptide data are provided in supplementary table S3, and supplementary fig. S6A, Supplementary Material online. There was good agreement between immunoblot results and corresponding peptide abundances. Rhesus macaques did not appear as an outgroup in the dendrogram, an unexpected result considering their more distant phylogenetic relationship to great apes and humans. Instead, humans emerged as the outgroup. This trend is driven mainly by AMY1 and BPIFA2 that both showed human lineage-specific, albeit opposite, quantitative trends. Immunoglobulin-related components [(PIGR, IGHA, and immunoglobulin G light chains (IGLC)] as well as DMBT1, followed a similar trend. Overall, the results further corroborate our observations and raise the possibility that salivary protein profiles of humans may be more unique than expected solely based on phylogenetic distances of species.
Fig. 5.
Differences in protein abundances in saliva samples of humans, chimpanzees, gorillas, and macaques. (A) Protein abundances shown in a heatmap-like fashion represent the cumulative peptide abundances for each given protein as shown in Supplementary figure S6, Supplementary Material online. The heatmap shows the average fold-change of abundances of salivary proteins in nonhuman primates as compared with their respective abundances in human saliva. The normalized abundance data from multiple individuals of each species (number of individuals [n]: humans, n = 6; chimpanzees, n = 2; gorillas, n = 5; macaques, n = 5) and multiple peptides corresponding to the same protein were averaged to represent the cumulative abundance of each protein in a given species. Specifically, for constructing the heatmap, log2 of the ratio of the abundance of a given protein in a nonhuman species as compared its abundance in humans was calculated. Human protein abundances were set to log2(1) = 0, shown in gray. Proteins that are less abundant in nonhuman primates appear in blue, and those that are more abundant in red. Significance values are provided in Supplementary table S3, Supplementary Material online.
Discussion
Evolutionary adaptations to a new human-specific diet have resulted in obvious changes to human viscerocranium and dentition (Lucas et al. 2006; von Cramon-Taubadel 2011; Xia et al. 2015; Ledogar et al. 2016) as well as to alterations in the oral microbiome (Adler et al. 2013; Cornejo et al. 2013; Warinner et al. 2014). However, less is known about the degree to which the human salivary proteome may have undergone a similar evolutionary adaptation to dietary, technological (pounding, cutting, and cooking), environmental, and microbial pressures during hominin development. In this study, we discovered that salivary proteins differ between humans and nonhuman hominid species, despite their close genetic homology. Some of the changes observed here, for example, the increased levels of salivary amylase in humans, confirmed at the protein level what others had reported at the gene level (Perry et al. 2007, 2008) or by enzyme activity measurements (McGeachin and Akin 1982; Behringer et al. 2013; Pajic et al. 2019). We discovered here additional quantitative and qualitative differences of major salivary proteins among humans, great apes, and Old World monkeys that had not been previously recognized.
One general difference was the overall lower protein content of human saliva compared with great apes and Old World monkeys. Higher viscosity of saliva from nonhuman primates was noted earlier (Levine et al. 1978; Herzberg et al. 1979). We argued that the observed differences in protein concentration are unlikely due to the sampling technique (see Results section). Further insight could be gained if salivary gland output could be measured in great apes. Unfortunately, very little is known about anatomy and histology of salivary glands in nonhuman hominid species. It has been stated, however, that the submandibular gland in the chimpanzee differs from its human counterpart in being composed of almost exclusively mucous acini (Bourne and De Bourne 1972). Did human salivary glands evolve to produce a more watery saliva to accommodate a human diet which drastically differs from that of great apes (Wrangham et al. 1999; Ungar and Sponheimer 2011)? Great apes and Old World monkeys chew on their fiber-rich food for longer periods of time, whereas humans swallow food faster, an ability arguably supported by the cooking of food (Smith et al. 2015). Thus, watery consistency of saliva could aid in faster processing of dry food in the oral cavity, and easier swallowing. It might also be advantageous to keep the mouth environment moist in arid savannah-like environments where hominins evolved. A more watery saliva may also facilitate human vocalization and language as it is well known that speaking and singing abilities are impaired in individuals who suffer from a dry mouth (Sreebny 2000). Of course, those possibilities remain speculative, warranting further work to reveal the functional implications of the observed differences in water content and viscosity between human and nonhuman primate saliva.
For most of the salivary proteins that differed between humans and great apes, sequence alignments showed a high degree of homology (supplementary figs. S5 and S6, Supplementary Material online). The few differences in amino acid sequence are unlikely to account for the observed differences at the protein level. Differences in gene copy numbers between humans and great apes, such as those that account for the higher levels of salivary amylase in humans (Perry et al. 2007, 2008), have not been reported for any of the other salivary proteins (Gokcumen et al. 2013). Across humans, however, copy number variants for DMBT1, and CST genes have been documented (de Sousa-Pereira et al. 2013; Polley et al. 2015, 2016). Hence, it is plausible that copy numbers for these genes might also differ between humans, great apes, and other nonhuman primates, which is a testable hypothesis that can be investigated in further studies focusing on those genes. A special case are the salivary mucins MUC5B and MUC7, which contain densely O-glycosylated PTS-rich repeat segments, a characteristic hallmark of the mucin family of proteins (Dekker et al. 2002). For both of these salivary mucin genes, subexonic PTS repeat number polymorphisms have been documented in humans (Biesbrock et al. 1997; Lang et al. 2007). In the case of MUC7, our group recently showed that subexonic copy number repeats differ among humans and between humans and non-human primates, and are highly polymorphic within the primate lineage (Xu et al. 2016, 2017). Another interesting case is the gene encoding for the surfactant protein latherin in the great apes, which in humans has been declared a “dying gene,” called BASE (breast cancer and salivary gland expression; Bingle et al. 2004), due to a human-specific frameshift mutation in this gene (Hahn and Lee 2005). Our failure to detect any BASE-related peptides by MS analysis suggests that a latherin/BASE protein product might not be expressed in human saliva. Most other quantitative differences observed here cannot readily be explained by gene exonic differences and, thus, might be caused by transcriptional, posttranscriptional, translational, or posttranslational mechanisms. This is consistent with other proteome-level studies in different tissues and different organisms where similar observations were made (Laurent et al. 2010; Khan et al. 2013; Wang et al. 2018) including the original classical study by King and Wilson (1975). We argue that most of the findings of the present study would likely have been missed if only looking from a genetic perspective and that the influence of posttranscriptional and posttranslational diversification on evolutionary processes should not be undervalued (Diz et al. 2012; Baer and Millar 2016).
Our findings here provide a necessary basis to assess in future studies whether the differences in the human salivary proteome observed here might have been caused by natural selection. Diet is one obvious factor in which humans fundamentally diverged from nonhuman primates (Lucas et al. 2006; Smith et al. 2015; Carmody et al. 2016). To what degree those salivary proteins are involved in dietary or gustatory functions needs to be investigated. One candidate is CA6 because it assists in taste perception as was implied by its original name “gustin” (Thatcher et al. 1998). Whether the presence of latherin in primate saliva and its presumed loss in human saliva has to do with its surfactant properties potentially aiding in mastication of fiber-rich diet in primates (Vance et al. 2013) also remains to be determined. It could also be that latherin may not be required in humans anymore because of the human-typical loss of a fur coat where in other mammals latherin facilitates the evaporation of sweat at the surface of the pelt (Kennedy 2011; Vance et al. 2013). The importance of social grooming in all nonhuman primates, where individuals meticulously comb through the fur coat of social partners using their hands as well as their mouths, might also be worthy of consideration in context with the loss of latherin in humans. Cross-species comparisons of mammalian saliva, which are already pursued by others (Mau et al. 2009; Karn et al. 2013; Gutierrez et al. 2014; Blanchard et al. 2015; de Sousa-Pereira et al. 2015), will help to provide answers to these questions.
Besides diet, pathogenic pressure is another important driving force for evolutionary adaptation (Varki 2012). Whether any of the salivary proteins that show human-specific features evolved driven by pathogen challenges that came along with the evolution of the genus Homo into a top predator starting 2 million years ago and with the Neolithic shift towards animal husbandry or sedentarism in crowded dwellings (Wolfe et al. 2007) is an intriguing hypothesis that can be examined. Genome-wide association studies have already suggested the involvement of a number of salivary proteins in disease susceptibility, including MUC5B (Roy et al. 2014), DMBT1 (Polley et al. 2016), MUC7 (Watson et al. 2009), and AMY1 (Falchi et al. 2014; Usher et al. 2015). The remaining possibility that uncontrolled environmental or dietary factors might have influenced the results of this study cannot be fully refuted, lest by a larger study investigating the salivary proteomes of globally diverse populations of humans and of nonhuman primate populations living in their natural environments. Our data at present are limited in that we compared only the most abundant proteins in human and nonhuman primate saliva in a limited number of individuals of each species. Larger studies will be required, including a global proteomic comparison of saliva as well as a histologic and transcriptomic comparison of salivary glands from humans and nonhuman primates. Nevertheless, cumulative evidence provided in this study opens up the possibility that certain properties and components of human and nonhuman primate saliva might have evolved in a lineage-specific manner, and that salivary proteomes in humans and nonhuman primates could be hitherto unnoticed hotbeds of evolutionary activity.
Materials and Methods
Saliva Collection
Saliva collection was performed according to the protocols approved by the Health Science Institutional Review Board (No. ORB0511008E) and the Institutional Animal Care and Use Committee at the University at Buffalo (No. ORB25049N). From humans and chimpanzees, saliva was collected by expectoration. Chimpanzees were previously trained by the care taker to voluntarily expectorate into a plastic cup. Gorilla (Western lowland gorilla) and Rhesus macaque saliva was collected with a soft disposable plastic Pasteur pipette (VWR, Radnor, PA). Gorillas were trained to open their mouth upon the caretaker giving a signal. Only Rhesus macaques were under sedation while saliva samples were collected. Neither of the animals or human individuals were closely related. All samples were immediately transferred into a polypropylene tube that was kept on ice. Sodium azide was added to a final concentration of 0.1% w/v, and saliva samples were centrifuged at 12,000 × g for 15 min at 4°C to remove particulate matter. The thus clarified saliva supernatant was recovered and aliquots were stored frozen at −80°C until further analysis. Samples were prepared as described previously (Walz et al. 2009). Extremely viscous samples from macaques had to be sheared by use of a syringe fitted with a 22-gauge needle to break their viscosity and render them pipettable. Preliminary tests did not find evidence that the slightly different modalities of sample collection influenced the sample composition to any noticeable degree. Protein concentrations were determined by the bicinchoninic acid (BCA) protein assay (Pierce, Thermo Scientific, Rockland, IL) using bovine serum albumin as the standard.
1D PAGE
Saliva samples from human, chimpanzee, and gorilla individuals were denatured under reducing conditions. Equal amounts of total protein (15 µg per lane for Coomassie and periodic acid Schiff (PAS) stain, 1 µg per lane for silver stain) were subjected to separation by SDS-PAGE using 8–16% gradient Tris-glycine mini gels (Novex, Invitrogen, Carlsbad, CA), and stained for proteins and glycans as previously described (Heo et al. 2013). Stained gels were imaged using a flat-bed scanner in the transparent mode (ImageScanner III, GE Healthcare). For 1D DIGE (Unlü et al. 1997; Tonge et al. 2001), fluorochrome-labeled salivary proteins (5 µg of total protein per species) from human (Cy3, green), chimpanzee (Cy2, blue), and gorilla (Cy5, red) were combined, denatured under reducing conditions, and coseparated on 8–16% gradient Tris-glycine mini gels. Fluorescent signals were detected using a fluorescence laser scanner (Typhoon 9400, GE Healthcare). Further details are described in supplementary materials and methods, Supplementary Material online.
Gel-Enhanced Liquid Chromatography Mass Spectrometry
GeLC-MS was performed as previously described (Lundby and Olsen 2011). After staining the entire track of the gel, each lane was cut into 13–16 longitudinal consecutive slices (supplementary fig. S2, Supplementary Material online). Following in-gel trypsin digestion, peptides were analyzed by LC-ESI-MS/MS and proteins were identified as described below.
Immunoblotting and Densitometric Analysis
Immunoblotting was performed as previously described (Heo et al. 2013). The primary antibodies against MUC5B, DMBT1, MUC7, PIGR, IGHA, AMY, IGHG, AZGP1, CA6, BPIFA2, IGKC/LC, PIP, and CST, and their dilutions used are described in supplementary materials and methods, Supplementary Material online.
Two-Dimensional Gel Electrophoresis
Sample preparation, first-dimension separation of equal amounts of proteins by isoelectric focusing and second dimension separation by gel electrophoresis were performed as previously described (Walz et al. 2009) with modifications described in supplementary materials and methods, Supplementary Material online. Gels were scanned using a fluorescence laser scanner (Typhon 9400) at the appropriate wavelengths for the respective cyanine fluorochrome. For 2D gels that were designated for spot identifications, the gels were counter-stained with Deep Purple (Amersham Biosciences, GE Healthcare) and scanned again to reveal all protein spots. Further details of image capture and densitometric analysis are provided in supplementary materials and methods, Supplementary Material online.
Mass Spectrometric and Chromatographic Methods and Bioinformatics
In-gel and in-solution trypsin digestion of peptides is described in supplementary materials and methods, Supplementary Material online. Three mass spectrometry platforms were used to acquire data: Peptide extracts from 1) 2D gel spots were analyzed by peptide mass fingerprinting using a micro MX-TOF mass spectrometer (Waters), 2) 1D gel slices were analyzed by reverse phase LC-MS/MS using a nanoACQUITY UPLC system coupled to a Q-ToF Premier mass spectrometer (Waters), and 3) in-solution digests were analyzed by reverse phase LC-MS/MS using an EASY-nanoLC 1000 LC system hyphenated to a QExactive Plus mass spectrometer (Thermo Scientific). For assigning protein IDs to 1D bands or 2D spots, the protein ID with the significant highest score was assigned. Additionally, exponentially modified protein abundance (emPAI) index scores in addition to Mascot scores were considered as indicators for protein abundance when more than one protein was assigned (Ishihama et al. 2005).
The details describing all chromatographic and mass spectrometric setup and parameter specifics as well as bioinformatic database search parameters are described in the supplementary materials and methods, Supplementary Material online. Clustal Omega tool (Sievers et al. 2011) was used to compare and align reference protein sequences retrieved from the UniProtKB database. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD015850.
Statistical Analysis
The GraphPad Prism software (Version 7 for Windows, GraphPad Software, San Diego, CA) was used to perform statistical analysis. Differences between total salivary protein concentration measurements were analyzed using the two-tailed Mann-Whitney U test, the difference between the total protein concentration of human saliva and those of nonhuman primates collectively was analyzed using Kruskal-Wallis statistic, and P < 0.05 was used as the cutoff for significance. For constructing the heatmap we applied a two-pronged unbiased hierarchical clustering approach using the heatmap.2 function of the openly available graphics software R (g.plot version 3.1).
Supplementary Material
Acknowledgments
The authors wish to thank Dr Molakala S. Reddy for his advice, and Mrs Lubov Neznanova for excellent technical help. We are grateful to Dr Kurt Volle, Alicia Dubrava, and fellow gorilla caretakers of the Buffalo Zoo for collecting saliva from gorillas, and to Carmen Presti for obtaining saliva from chimpanzees. Saliva from macaques was provided by the Southwest National Primate Research Center which is funded by the NIH Office of Research Infrastructure Programs (ORIP)/OD P51 OD011133, and by the Yerkes National Primate Research Center which is funded by ORIP/OD P51OD011132. This study was supported by grants NIDCR R01 DE019807, R21 DE025826, and NCI U01 CA221244 (S.R.) and, in part, by Roswell Park Cancer Institute and NCI grant P30 CA016056.
References
- Abdolhosseini M, Sotsky JB, Shelar AP, Joyce PB, Gorr SU.. 2012. Human parotid secretory protein is a lipopolysaccharide-binding protein: identification of an anti-inflammatory peptide domain. Mol Cell Biochem. 359(1–2):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJA, Townsend G, Sołtysiak A, Alt KW, et al. 2013. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 45(4):450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredouani A, Stocchero M, Culeddu N, Moustafa JE, Group D, Tichet J, Balkau B, Brousseau T, Manca M, Falchi M.. 2016. Metabolomic profile of low-copy number carriers at the salivary alpha-amylase gene suggests a metabolic shift toward lipid-based energy production. Diabetes 65(11):3362–3368. [DOI] [PubMed] [Google Scholar]
- Azen EA, Oppenheim FG.. 1973. Genetic polymorphism of proline-rich human salivary proteins. Science 180(4090):1067–1069. [DOI] [PubMed] [Google Scholar]
- Baer B, Millar AH.. 2016. Proteomics in evolutionary ecology. J Proteomics 135:4–11. [DOI] [PubMed] [Google Scholar]
- Baum BJ, Yates JR 3rd, Srivastava S, Wong DT, Melvin JE.. 2011. Scientific frontiers: emerging technologies for salivary diagnostics. Adv Dent Res. 23(4):360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer V, Borchers C, Deschner T, Mostl E, Selzer D, Hohmann G.. 2013. Measurements of salivary alpha amylase and salivary cortisol in hominoid primates reveal within-species consistency and between-species differences. PLoS One 8(4):e60773.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbrock AR, Bobek LA, Levine MJ.. 1997. MUC7 gene expression and genetic polymorphism. Glycoconj J. 14(4):415–422. [DOI] [PubMed] [Google Scholar]
- Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ.. 2004. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 13(2):422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard AA, Ezzati P, Shamshurin D, Nistor AC, Leygue E, Wilkins JA, Myal Y.. 2015. Towards further defining the proteome of mouse saliva. Proteome Sci. 13(1):10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne GH, De Bourne MNG.. 1972. The histology and histochemistry of the chimpanzee tissues and organs In: Bourne GH, editor. The chimpanzee. Baltimore (MD: ): University Park Press; p. 1–76. [Google Scholar]
- Carmody RN, Dannemann M, Briggs AW, Nickel B, Groopman EE, Wrangham RW, Kelso J.. 2016. Genetic evidence of human adaptation to a cooked diet. Genome Biol Evol. 8(4):1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, et al. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol. 30(4):881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BW, Ruhl S.. 2018. Glycan recognition at the saliva - oral microbiome interface. Cell Immunol. 333:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Pedersen AM, Villa A, Ekstrom J, Proctor GB, Vissink A, Aframian D, McGowan R, Aliko A, Narayana N, et al. 2015. The functions of human saliva: a review sponsored by the World Workshop on Oral Medicine VI. Arch Oral Biol. 60(6):863–874. [DOI] [PubMed] [Google Scholar]
- de Sousa-Pereira P, Amado F, Abrantes J, Ferreira R, Esteves PJ, Vitorino R.. 2013. An evolutionary perspective of mammal salivary peptide families: cystatins, histatins, statherin and PRPs. Arch Oral Biol. 58(5):451–458. [DOI] [PubMed] [Google Scholar]
- de Sousa-Pereira P, Cova M, Abrantes J, Ferreira R, Trindade F, Barros A, Gomes P, Colaco B, Amado F, Esteves PJ, et al. 2015. Cross-species comparison of mammalian saliva using an LC-MALDI based proteomic approach. Proteomics 15(9):1598–1607. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rossen JW, Büller HA, Einerhand AW.. 2002. The MUC family: an obituary. Trends Biochem Sci. 27(3):126–131. [DOI] [PubMed] [Google Scholar]
- Diz AP, Martinez-Fernandez M, Rolan-Alvarez E.. 2012. Proteomics in evolutionary ecology: linking the genotype with the phenotype. Mol Ecol. 21(5):1060–1080. [DOI] [PubMed] [Google Scholar]
- Falchi M, El-Sayed Moustafa JS, Takousis P, Pesce F, Bonnefond A, Andersson-Assarsson JC, Sudmant PH, Dorajoo R, Al-Shafai MN, Bottolo L, et al. 2014. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet. 46(5):492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen O, Tischler V, Tica J, Zhu Q, Iskow RC, Lee E, Fritz MH, Langdon A, Stutz AM, Pavlidis P, et al. 2013. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A. 110(39):15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez AM, Ceron JJ, Fuentes-Rubio M, Tecles F, Beeley JA.. 2014. A proteomic approach to porcine saliva. Curr Protein Pept Sci. 15(1):56–63. [DOI] [PubMed] [Google Scholar]
- Hahn Y, Lee B.. 2005. Identification of nine human-specific frameshift mutations by comparative analysis of the human and the chimpanzee genome sequences. Bioinformatics. 21(Suppl 1):i186–i194. [DOI] [PubMed] [Google Scholar]
- Heo SM, Choi KS, Kazim LA, Reddy MS, Haase EM, Scannapieco FA, Ruhl S.. 2013. Host defense proteins derived from human saliva bind to Staphylococcus aureus. Infect Immun. 81(4):1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg MC, Levine MJ, Ellison SA, Tabak LA.. 1979. Purification and characterization of monkey salivary mucin. J Biol Chem. 254(5):1487–1494. [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M.. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4(9):1265–1272. [DOI] [PubMed] [Google Scholar]
- Karn RC, Chung AG, Laukaitis CM.. 2013. Shared and unique proteins in human, mouse and rat saliva proteomes: footprints of functional adaptation. Proteomes 1(3):275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MW. 2011. Latherin and other biocompatible surfactant proteins. Biochem Soc Trans. 39(4):1017–1022. [DOI] [PubMed] [Google Scholar]
- Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y.. 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342(6162):1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC.. 1975. Evolution at two levels in humans and chimpanzees. Science 188(4184):107–116. [DOI] [PubMed] [Google Scholar]
- Kirkbride HJ, Bolscher JG, Nazmi K, Vinall LE, Nash MW, Moss FM, Mitchell DM, Swallow DM.. 2001. Genetic polymorphism of MUC7: allele frequencies and association with asthma. Eur J Hum Genet. 9(5):347–354. [DOI] [PubMed] [Google Scholar]
- Lang T, Hansson GC, Samuelsson T.. 2007. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A. 104(41):16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent JM, Vogel C, Kwon T, Craig SA, Boutz DR, Huse HK, Nozue K, Walia H, Whiteley M, Ronald PC, et al. 2010. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10(23):4209–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledogar JA, Dechow PC, Wang Q, Gharpure PH, Gordon AD, Baab KL, Smith AL, Weber GW, Grosse IR, Ross CF, et al. 2016. Human feeding biomechanics: performance, variation, and functional constraints. PeerJ. 4:e2242.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MJ, Herzberg MC, Ellison SA, Shomers JP, Sadowski GA.. 1978. Biochemical and immunological comparison of monkey (Macaca arctoides) and human salivary secretions. Comp Biochem Physiol B. 60(4):423–431. [DOI] [PubMed] [Google Scholar]
- Lucas PW, Ang KY, Sui Z, Agrawal KR, Prinz JF, Dominy NJ.. 2006. A brief review of the recent evolution of the human mouth in physiological and nutritional contexts. Physiol Behav. 89(1):36–38. [DOI] [PubMed] [Google Scholar]
- Lundby A, Olsen JV.. 2011. GeLCMS for in-depth protein characterization and advanced analysis of proteomes. Methods Mol Biol. 753:143–155. [DOI] [PubMed] [Google Scholar]
- Manconi B, Castagnola M, Cabras T, Olianas A, Vitali A, Desiderio C, Sanna MT, Messana I.. 2016. The intriguing heterogeneity of human salivary proline-rich proteins. J Proteomics 134:47–56. [DOI] [PubMed] [Google Scholar]
- Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA.. 2010. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One 5(10):e13352.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel ID. 1987. The functions of saliva. J Dent Res. 66(Spec Iss):623–627. [DOI] [PubMed] [Google Scholar]
- Marsh PD, Do T, Beighton D, Devine DA.. 2016. Influence of saliva on the oral microbiota. Periodontol 2000 70(1):80–92. [DOI] [PubMed] [Google Scholar]
- Mau M, de Almeida AM, Coelho AV, Südekum KH.. 2011. First identification of tannin-binding proteins in saliva of Papio hamadryas using MS/MS mass spectrometry. Am J Primatol. 73(9):896–902. [DOI] [PubMed] [Google Scholar]
- Mau M, Kaiser TM, Südekum KH.. 2009. Evidence for the presence of carbonic anhydrase 29-kDa isoenzyme in salivary secretions of three ruminating species and the gelada baboon. Arch Oral Biol. 54(4):354–360. [DOI] [PubMed] [Google Scholar]
- McGeachin RL, Akin JR.. 1982. Amylase levels in the tissues and body fluids of several primate species. Comp Biochem Physiol A Comp Physiol. 72(1):267–269. [DOI] [PubMed] [Google Scholar]
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ.. 2007. Salivary proteome and its genetic polymorphisms. Ann N Y Acad Sci. 1098(1):22–50. [DOI] [PubMed] [Google Scholar]
- Pajic P, Pavlidis P, Dean K, Neznanova L, Romano RA, Garneau D, Daugherity E, Globig A, Ruhl S, Gokcumen O.. 2019. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 8:e44628.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. 2008. Copy number variation and evolution in humans and chimpanzees. Genome Res. 18(11):1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S, Louzada S, Forni D, Sironi M, Balaskas T, Hains DS, Yang F, Hollox EJ.. 2015. Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc Natl Acad Sci U S A. 112(16):5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S, Prescott N, Nimmo E, Veal C, Vind I, Munkholm P, Fode P, Mansfield J, Skyt Andersen P, Satsangi J, et al. 2016. Copy number variation of scavenger-receptor cysteine-rich domains within DMBT1 and Crohn’s disease. Eur J Hum Genet. 24(9):1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, et al. 2014. Muc5b is required for airway defence. Nature 505(7483):412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S. 2012. The scientific exploration of saliva in the post-proteomic era: from database back to basic function. Expert Rev Proteomics 9(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannapieco FA. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 5(3):203–248. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Carmody RN, Dutton RJ, Wrangham RW.. 2015. The significance of cooking for early hominin scavenging. J Hum Evol. 84:62–70. [DOI] [PubMed] [Google Scholar]
- Sreebny LM. 2000. Saliva in health and disease: an appraisal and update. Int Dent J. 50(3):140–161. [DOI] [PubMed] [Google Scholar]
- Thatcher BJ, Doherty AE, Orvisky E, Martin BM, Henkin RI.. 1998. Gustin from human parotid saliva is carbonic anhydrase VI. Biochem Biophys Res Commun. 250(3):635–641. [DOI] [PubMed] [Google Scholar]
- Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M.. 2001. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1(3):377–396. [DOI] [PubMed] [Google Scholar]
- Ungar PS, Sponheimer M.. 2011. The diets of early hominins. Science 334(6053):190–193. [DOI] [PubMed] [Google Scholar]
- Unlü M, Morgan ME, Minden JS.. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071–2077. [DOI] [PubMed] [Google Scholar]
- Usher CL, Handsaker RE, Esko T, Tuke MA, Weedon MN, Hastie AR, Cao H, Moon JE, Kashin S, Fuchsberger C, et al. 2015. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet. 47(8):921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Hof W, Veerman EC, Nieuw Amerongen AV, Ligtenberg AJ.. 2014. Antimicrobial defense systems in saliva. Monogr Oral Sci. 24:40–51. [DOI] [PubMed] [Google Scholar]
- Vance SJ, McDonald RE, Cooper A, Smith BO, Kennedy MW.. 2013. The structure of latherin, a surfactant allergen protein from horse sweat and saliva. J R Soc Interface 10(85):20130453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 2012. Nothing in medicine makes sense, except in the light of evolution. J Mol Med. 90(5):481–494. [DOI] [PubMed] [Google Scholar]
- von Cramon-Taubadel N. 2011. Global human mandibular variation reflects differences in agricultural and hunter-gatherer subsistence strategies. Proc Natl Acad Sci U S A. 108(49):19546–19551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz A, Odenbreit S, Stühler K, Wattenberg A, Meyer HE, Mahdavi J, Borén T, Ruhl S.. 2009. Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics 9(6):1582–1592. [DOI] [PubMed] [Google Scholar]
- Wang SH, Hsiao CJ, Khan Z, Pritchard JK.. 2018. Post-translational buffering leads to convergent protein expression levels between primates. Genome Biol. 19(1):83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warinner C, Rodrigues JF, Vyas R, Trachsel C, Shved N, Grossmann J, Radini A, Hancock Y, Tito RY, Fiddyment S, et al. 2014. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 46(4):336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AM, Ngor WM, Gordish-Dressman H, Freishtat RJ, Rose MC.. 2009. MUC7 polymorphisms are associated with a decreased risk of a diagnosis of asthma in an African American population. J Investig Med. 57(8):882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J.. 2007. Origins of major human infectious diseases. Nature 447(7142):279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangham RW, Jones JH, Laden G, Pilbeam D, Conklin-Brittain N.. 1999. The raw and the stolen. Cooking and the ecology of human origins. Curr Anthropol. 40(5):567–594. [PubMed] [Google Scholar]
- Xia J, Zheng J, Huang D, Tian ZR, Chen L, Zhou Z, Ungar PS, Qian L.. 2015. New model to explain tooth wear with implications for microwear formation and diet reconstruction. Proc Natl Acad Sci U S A. 112(34):10669–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Taskent RO, Alachiotis N, Flanagan C, DeGiorgio M, Blekhman R, Ruhl S, Gokcumen O.. 2017. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol Biol Evol. 34(10):2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Thamadilok S, Redwood E, Fox S, Blekhman R, Ruhl S, Gokcumen O.. 2016. Recent evolution of the salivary mucin MUC7. Sci Rep. 6(1):31791.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.