Abstract
Skeletal muscles contain heterogeneous myofibers that are different in size and contractile speed, with type IIb myofiber being the largest and fastest. Here, we identify methyltransferase-like 21e (Mettl21e), a member of newly classified nonhistone methyltransferases, as a gene enriched in type IIb myofibers. The expression of Mettl21e was strikingly up-regulated in hypertrophic muscles and during myogenic differentiation in vitro and in vivo. Knockdown (KD) of Mettl21e led to atrophy of cultured myotubes, and targeted mutation of Mettl21e in mice reduced the size of IIb myofibers without affecting the composition of myofiber types. Mass spectrometry and methyltransferase assay revealed that Mettl21e methylated valosin-containing protein (Vcp/p97), a key component of the ubiquitin-proteasome system. KD or knockout of Mettl21e resulted in elevated 26S proteasome activity, and inhibition of proteasome activity prevented atrophy of Mettl21e KD myotubes. These results demonstrate that Mettl21e functions to maintain myofiber size through inhibiting proteasome-mediated protein degradation.—Wang, C., Zhang, B., Ratliff, A. C., Arrington, J., Chen, J., Xiong, Y., Yue, F., Nie, Y., Hu, K., Jin, W., Tao, W. A., Hrycyna, C. A., Sun, X., Kuang, S. Methyltransferase-like 21e inhibits 26S proteasome activity to facilitate hypertrophy of type IIb myofibers.
Keywords: Mettl21e, Myostatin, non-histone methylation, muscle atrophy, VCP/p97
Mammalian skeletal muscles are mainly composed of mature muscle cells (myofibers) that are heterogeneous in the expressions of myosin heavy chain (Myh) isoforms (1). Based on the expression of Myh7, Myh2, Myh1, or Myh4, rodent limb muscle myofibers are classified as type I, IIa, IIx/d, or IIb, respectively. These 4 types of myofibers also exhibit distinct physiologic and metabolic characteristics (2). The contractile speed of myofibers ranks in the order of IIb > IIx > IIa > I, whereas the fatigue resistance of myofibers is negatively correlated with the contractile speed (2). Biochemically and metabolically, I and IIa myofibers have high oxidative capacity with a large number of mitochondria. In contrast, IIx and IIb myofibers mainly utilize glycolysis to generate ATP. Besides the physiologic and metabolic differences, the size of myofibers are generally in the order of IIb > IIx > IIa > I (3). However, it is not clear why IIb myofibers have the largest size compared with the other types of myofibers.
The identity of a myofiber is controlled by several factors, including motor neuron innervation, hormonal regulation, progenitor cell origin, and myofiber type–specific gene expression (2). Each mature myofiber is monoinnervated by a single motor neuron, which transmits electrical impulses to a subset of myofibers (4). Firing of slow motor neurons (motor neurons innervating slow myofibers) generates sustained and low-amplitude elevations in intracellular calcium concentrations, leading to the activation of calcineurin (5). In contrast, fast motor neuron firing leads to brief calcium transients of high amplitude, which are insufficient to activate calcineurin (5). Activated calcineurin activates Nuclear factor of activated T-cells (NFAT) and myocyte enhancer factor-2 (MEF2) transcription factors to promote transcription of slow myofiber-related genes (6–9). Besides motor neurons, Twist Family BHLH Transcription Factor 2 (Twist2)-dependent progenitor cells are recently shown to maintain IIb myofibers (10). In addition, myofiber-type specific genes or microRNAs are shown to contribute to myofiber specifications, such as type I–specific ephrin-A3 and microRNA-208/microRNA-499 (11, 12), oxidative myofiber–enriched PGC1-α (13), glycolytic myofiber–specific T-Box 15 (14), and fast myofiber–enriched SRY-Box 6 (15). However, gene programs that regulate subtypes of fast myofibers are poorly understood.
Methyltransferases are enzymes that transfer a methyl group (CH3) from a donor, generally S-adenosyl-l-methionine (SAM), to lysine and arginine residuals of substrate proteins (16). Methylation of histone proteins are often related to changes in chromatin state and gene expression, whereas methylation of nonhistone proteins may modify their activity, stability, and molecular interactions (17). In humans, a group of methyltransferase-like 21 (METTL21) proteins (A–D) has recently been identified to mediate lysine methylation of molecular chaperones and eukaryotic translation elongation factor 1A (eEF1A) (18–22). Specifically, METTL21A (HSPA lysine methyltransferase, or HSPA-KMT) methylates several heat shock protein 70 (HSPA/Hsp70) proteins (20), METTL21B methylates eEF1A at Lys165 (K165) (21, 22), and METTL21C and METTL21D (VCP-KMT) methylate the ATP-dependent chaperone VCP at Lys315 (K315) (18, 19, 23). In addition, a yeast Mettl21-like protein is shown to methylate eEF1A (24), suggesting a potential role of Mettl21 family methyltransferases in protein synthesis, a pivotal process for muscle hypertrophy.
In searching for myofiber type–specific regulators, we analyzed microarray and RNA-sequencing data comparing gene expression in the skeletal muscles of control and myostatin (Mstn) knockout (KO) mice, which exhibit robust myofiber hypertrophy and increased abundance of IIb myofibers (25, 26). This analysis led to the identification of Mettl21e gene that was among the top up-regulated genes in MstnKO muscles. Intriguingly, the expression of Mettl21e is also increased in the Callipyge sheep model of muscle hypertrophy and in response to β-adrenergic agonist–induced muscle protein accretion in lambs (27, 28), suggesting a conserved link between Mettl21e and muscle hypertrophy in multiple species. In this study, we used loss-of-function assays in cell culture and in a novel mouse model to investigate the function of Mettl21e in vivo. Our results demonstrate that Mettl21e is a type IIb myofiber–enriched protein that functions to maintain myofiber size through suppression of 26S proteasome activity.
MATERIALS AND METHODS
Mice
Constitutive Mstn KO mice were generated by Dr. S. E. Jin-Lee (Johns Hopkins University, Baltimore, MD, USA) (29). Heterozygous Mstn+/− mice were bred to generate Mstn KO and wild-type (WT) littermates. Unless otherwise indicated, we used 2-mo-old mice for experiments. All procedures involving the use of animals were performed in accordance with the guidelines presented by Purdue University’s Animal Care and Use Committee.
Generation of Transcription activator-like effector nuclease-mediated Mettl21e KO mice
The Transcription activator-like effector nuclease (TALEN)-mediated Mettl21e KO mice were produced by Beijing ViewSolid Biotechnology (Beijing, China). The TALEN plasmid pCS5-eTALEN-T was designed to induce Mettl21e frameshift mutation. TALEN-left targets the sequence (5′-GGTCGCAGAGATCATGG-3′) of the sense strand, and TALEN-right targets the sequence (5′-AGTCGTTATCAGAGTTG-3′) of the antisense strand. Mutated mice were generated by pronuclear injection using standard methods. Founder mice were screened for the presence of mutation by sequencing the PCR products amplified by the primers for Mettl21e-sens (5′-ATGGACCTCACAGTAACTCACAT-3′) and Mettl21e-anti (5′-GCTTGCCACAATGGAGACAAG-3′). Mutant mice were mated with WT C57BL/6 mice to obtain heterozygous Mettl21eHe mice. Mettl21eHe mice were then mated at least 3 generations to obtain Mettl21eKO and WT mice.
Primary myoblast isolation, culture, and differentiation
Primary myoblasts were isolated from 5-wk-old WT female mice. The hind limb skeletal muscles were minced and digested in type I collagenase and dispase B mixture (Roche Applied Science, Penzberg, Germany). The digestion was stopped with F-10 Ham’s medium containing 17% fetal bovine serum, and the cells were filtered from debris, centrifuged, and cultured in growth medium (F-10 Ham’s medium supplemented with 17% fetal bovine serum, 4 ng/ml basic fibroblast growth factor, and 1% penicillin-streptomycin) on uncoated dishes for 3 d when 5 ml growth medium was added each day. Then, the supernatant were collected, centrifuged, and trypsinized with 0.25% trypsin. After washing off the trypsin, primary myoblasts were seeded on collagen-coated dishes, and the growth medium was changed every 2 d. Myoblasts were induced to differentiate on matrigel-coated dishes and cultured in differentiation medium (DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin). Differentiation medium was replaced every day. Primary myoblasts were cultured in normal humidified tissue culture incubators with 5% CO2. To inhibit Mstn signaling, cells were treated with 200 ng/ml follistatin (SRP3045; MilliporeSigma, Burlington, MA, USA). To inhibit proteasome activity, differentiated myoblasts were treated with 25 nM bortezomib (B-1408; LC Laboratories, Woburn, MA, USA).
Immunostaining and image acquisition
Muscle samples were processed following the protocol described by Wang et al. (30). Myoblasts and myotubes were fixed with 4% paraformaldehyde and then blocked with blocking buffer (5% goat serum, 2% bovine serum albumin, 0.2% Triton X-100, and 0.1% sodium azide in PBS) for at least 1 h. Then, the samples were incubated with primary antibodies [1:200 in blocking buffer; sarcomeric myosin heavy chain antibody (clone MF20) was from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, Iowa, USA), and anti-DYKDDDDK epitope (FLAG) antibody (F1804) was from MilliporeSigma] overnight. After washing with PBS, the samples were incubated with respective secondary antibodies and DAPI for 45 min at room temperature. Fluorescent images were captured using a Leica DM 6000B fluorescent microscope (Leica Microsystems, Wetzlar, Germany).
RNA extraction and real-time quantitative PCR
Total RNA of muscles or myoblasts were extracted using Trizol Reagent (15596-018; Thermo Fisher Scientific, Waltham, MA, USA). RNA was treated with RNase-free DNase I (AM2224; Thermo Fisher Scientific) to remove genomic DNA. The purity and concentration of total RNA were measured by Nanodrop 3000 (Thermo Fisher Scientific). Random primers and Moloney murine leukemia virus reverse transcriptase were used to convert RNA into cDNA. Real-time PCR was performed using Roche Lightcycler 480 PCR System with SYBR Green Master Mix (04707516001; Roche Applied Science). Primers used were listed in Supplemental Table S1 and ref. 31. Ct value of 18S rRNA was used as internal control, and 2−ΔΔCt method was used to analyze the relative mRNA expression of various genes.
Single myofiber isolation
Extensor digitorum longus (EDL) and soleus (SOL) muscles were removed carefully and digested with 2 mg/ml collagenase type 1 (CLS-1; Worthington Biochemical, Lakewood, NJ, USA) in DMEM (MilliporeSigma) for 45 min at 37°C. Digestion was stopped by carefully transferring EDL or SOL muscles to a horse serum–coated Petri dish (60-mm) with DMEM. Myofibers were released by gently flushing muscles with a large bore glass pipette. The released single myofiber was washed in PBS and then transferred to a 0.2-ml PCR tube. The residual PBS was removed from the PCR tube. RNA of a single myofiber was extracted using a PicoPure RNA Isolation Kit (KIT0204; Thermo Fisher Scientific) according to the manufacturer’s protocol. Generally, we pipetted 50 µl extraction buffer into the PCR tube containing a single myofiber and incubated for 30 min at 42°C to extract cellular contents. Then, we gently mixed 50 µl 70% ethanol into the cell extracts, and the mixture was added into preconditioned purification column. After 2 rounds of wash, RNA was eluted with 11 µl elution buffer. The eluted RNA was used directly in reverse transcription to generate cDNA for real-time PCR analyses.
Protein extraction and Western blot analysis
Muscle samples and cultured myoblasts were washed with PBS and homogenized with radioimmune precipitation assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein concentrations were determined using Pierce BCA Protein Assay Reagent (Pierce Biotechnology, Rockford, IL, USA). Proteins (100 μg) were separated by 10% SDS-PAGE, electrotransferred onto PVDF membrane (MilliporeSigma), and incubated with specific primary antibodies. Glyceraldehyde 3-phosphate dehydrogenase (6C5) antibodies (1:1000 in 5% w/v nonfat dry milk) were from Santa Cruz Biotechnology (Dallas, TX, USA), and MF20 (1:100 in 5% w/v nonfat dry milk) was from Developmental Studies Hybridoma Bank. Immunodetection was performed using ECL Western blotting substrate (Pierce Biotechnology) and detected with FluoChem R imaging system (ProteinSimple, San Jose, CA, USA).
Adenovirus generation
The adenoviruses with short hairpin (sh)-RNAs, Mstn, or Mettl21e-FLAG were generated using the AdEasy system (240009; Agilent, Santa Clara, CA, USA). We subcloned the U6-shRNA cassettes from pLKO.1-U6-shRNA plasmids (Sh1: TRCN0000176763; Sh2: TRCN0000177496; Sh3: TRCN0000177877; Sh4: TRCN0000176682; MilliporeSigma) with primers (pLKO.1-f/r, Supplemental Table S1). Mstn ORF and Mettl21e-FLAG ORF were cloned with primers (Mstn-f/r for Mstn; Mettl21e-f/Mettl21e-FLAG-r for Mettl21e-FLAG; Supplemental Table S1). These cloned DNA sequences were inserted into pAdTrack-CMV plasmid (16405; Addgene, Watertown, MA, USA) (The cloned Mettl21e-FLAG ORF was also inserted into pcDNA3.1 for intracellular location detection) and then were digested by PmeI and transfected with the DH5a competent cell with pAdEasy-1. The following steps were exactly according to the methods generating the MyoD-overexpression adenovirus (31).
RNA sequencing
RNA was extracted from tibialis anterior (TA) muscles of three 8-wk-old WT and Mettl21eKO mice. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s recommendations. RNA was sequenced and analyzed by Novogene (Beijing, China) using the Illumina Hiseq4000 platform (Illumina, San Diego, CA, USA). The original data of RNA sequencing are deposited to the Gene Expression Omnibus (GEO) data set (GSE122024, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122024; Token: qrqjeqcqnfmxvct).
Immunoprecipitation to pull down Mettl21e binding complex
Vectors containing Mettl21e-FLAG or GFP-FLAG were transduced into primary myoblasts (70–80% confluent) by adenovirus. Two days posttransduction, cells were induced to differentiation for 3 d and then were scraped with ice-cold PBS (from 10 × 100–mm plates for each plasmid) and were centrifuged. The cell pellet was completely resuspended with 1 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% NP40 with 1 time Protease inhibitor cocktail) on ice and sonicated with a 1-s pulse at 5-s intervals 10 times. After 5 min on ice, cell lysate was centrifuged with 14,000 rpm at 4°C, and then the supernatant (containing 5 mg proteins determined by the Pierce bicinchoninic acid assay) was transferred to 50 μl precleaned anti-FLAG magnetic beads slurry (M8823; MilliporeSigma). After incubation on rotator for 3 h in a cold room (4°C), the magnetic beads were washed 3 times with lysis buffer. Then, we added 500 μl ddH2O, and pipeted up and down several times to remove salt or solvent remainder (twice). Bead-captured proteins were eluted with 100 μl of 50 mM triethyl amine and 5 mM DTT on a thermal shaker (99°C, 5 min). The protein elution was centrifuged with CentriVap Concentrator (Labconco, Kansas City, MO, USA) to partially remove triethyl amine, adjusted to 100 μl and pH 8.0 with 1% acetic acid and 15 mM Iodoacetamide, and placed in the dark for 1 h. Before desalting the protein elution with C18 Zip tips (NT3C18.96; Glygen, Columbia, MD, USA), 1 μg trypsin was reacted with the 100 μl adjusted protein elution for 16 h at 37°C. The following steps for mass spectrometric data acquisition and analysis were according to the descriptions by Wang et al. (31).
Recombinant protein production and purification
pETDuet-1-derived plasmids incorporating His6-VCP, His6-Mettl21e, or His6-Hsp90ab1 were transformed into the Escherichia coli expression strain BL21 (DE3; Thermo Fisher Scientific). Cells were cultured in LB medium with 0.1 mg/ml ampicillin at 37°C in a shaking incubator at 220 rpm until the absorbance at 600 nm reached 0.6 optical density (OD). The culture was induced with 100 µM isopropyl β-d-thiogalactoside (Gold Biotechnology, St. Louis, MO, USA), and the temperature was lowered to 18°C for 18 h. Cells were harvested by centrifugation at 7000 g. Cell pellets were resuspended in lysis buffer [50 mM Tris (pH 7.5), 500 mM NaCl, 10% (w/v) glycerol, 30 mM imidazole, 3 mM 2-ME, 0.5% Triton X-100, 1 tablet cOmplete EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland), and 2 mM 4-benzenesulfonyl fluoride hydrochloride (Gold Biotechnology)]. Cells were lysed via probe sonication and underwent centrifugation at 100,000 g for 1 h. The soluble protein in the supernatant was rocked with Ni-NTA resin (Thermo Fisher Scientific) at 4°C. After 1 h, resin was washed with buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, and 30 mM imidazole) followed by an additional wash with the addition of 0.5 M KCl. Recombinant proteins were removed from resin with elution buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 300 mM imidazole). The eluted protein was concentrated in Amicon Ultra MWCO concentrators columns (MilliporeSigma) with appropriate molecular mass cutoffs. Proteins were divided into aliquots and stored at −80°C and were thawed on ice prior to use in assays. Concentrations of each protein were determined via Bradford protein assay (Thermo Fisher Scientific). Purity was confirmed by loading purified protein (1 µg) on 10% SDS-PAGE and either stained with Coomassie [0.25% (w/v) Coomassie Brilliant Blue R-250 (20278; Thermo Fisher Scientific), 80% methanol, and 20% acetic acid] or transferred to a nitrocellulose membrane (0.22 µm; GE Healthcare, Waukesha, WI, USA). The nitrocellulose membrane was blocked at 4°C overnight in 20% (w/v) nonfat dry milk in PBS with Tween 20 [137 mM NaCl, 2.7 mM KCl, 4 mM Na2HPO4, 1.8 mM KH2PO4 and 0.05% (v/v) Tween-20, pH 7.4]. The blocked membrane was incubated for 1 h at room temperature with α-His-hrp (1:10,000; MilliporeSigma) antibody in 5% (w/v) nonfat dry milk in PBS with Tween 20. The membranes were washed 3 times with PBS with Tween 20, and the protein bands were visualized using ECL (Pierce Biotechnology).
In vitro methyltransferase reaction
Methyltransferase reactions were performed in 50-µl volumes for 1 h at 37°C in methyltransferase reaction buffer [50 mM Tris (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM ATP], 13 µM [14C] SAM (2 µCi), 1 µM methyltransferase enzyme or 4 µM substrates, and varying concentrations of substrates or methyltransferase enzyme. The reactions were stopped by precipitating proteins with 50-µl 10% (v/v) Trichloroacetic acid (TCA) at 4°C for 1 h. The reactions were spotted onto glass fiber filters (45 µm; Whatman, Maidstone, United Kingdom), and the acid-insoluble material was retained during vacuum filtration. The reaction tubes were rinsed with an additional 50 µl 10% (v/v) TCA and applied to the filters. The filters were then washed with 1 ml of 10% (v/v) TCA followed by 1 ml 100% ethanol and left to dry for 10 min at room temperature. The dried filters were placed in scintillation vials with 10 ml Bio-Safe II biodegradable scintillation cocktail (Research Products International, Mt Prospect, IL, USA), and radioactivity was measured by scintillation counting.
Proteasome activity assay
TA muscles or differentiated myoblasts were homogenized with 0.5% NP-40. Then, the tissue or cell lysates were centrifuged at 4°C at 13,000 rpm for 10 min. The supernatants were collected and measured using a Proteasome Activity Assay Kit (ab107921; Abcam, Cambridge, MA, USA) according to the manufacturer’s protocol.
Statistical analysis
The data were presented with mean and sd. P values were calculated using unpaired 2-tailed Student’s t test for 2 group comparison and 1-way ANOVA for multiple group comparison. Values of P < 0.05 were considered to be statistically significant.
RESULTS
Mettl21e expression is elevated in hypertrophic Mstn KO muscles
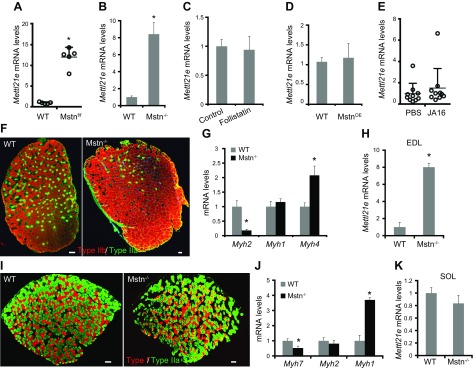
We analyzed microarray data comparing gene expression of gastrocnemius (GA) muscles from WT and Mstn conditional KO mice (25). Three months after Cre-mediated deletion of Mstn in adult mice, the expression of Mettl21e in GA muscles was 12-fold higher in Mstn KO mice than in WT mice (Fig. 1A). Consistently, a higher level of Mettl21e expression was detected in the TA muscles of constitutive Mstn KO mice relative to the same muscles of WT mice (Fig. 1B). These results suggest that the expression of Mettl21e is negatively correlated with Mstn but positively associated with muscle hypertrophy.
Figure 1.
The expression of Mettl21e is associated with Mstn KO in fast muscles. A) Mettl21e expression in GA muscles from WT and postdevelopmental Mstn KO mice. B) Mettl21e expression in TA muscles from WT and constitutive Mstn KO mice. C) Mettl21e expression in differentiated myoblasts treated with follistatin. D) Mettl21e expression in myoblasts with Mstn overexpression. E) Mettl21e expression in GA muscles with anti-Mstn antibody JA16 or PBS. F) Immunostaining of type IIb (Myh4, Red) and type IIa (Myh2, Green) myofibers in EDL muscles of WT and Mstn KO mice. G, H) qPCR analysis of Myh genes (G) and Mettl21e (H) in EDL muscles of WT and Mstn KO mice. Scale bars, 100 µm. I) Immunostaining of type I (Myh7, Red) and type IIa (Myh2, Green) myofibers in SOL muscles of WT and Mstn KO mice. Scale bars, 100 µm. J, K) qPCR analysis of Myh genes (J) and Mettl21e (K) in SOL muscles of WT and Mstn KO mice. The values in A and E were originated from the published microarray data. Error bars represent mean + sd of 6 mice (B, G, H, J, K) or 5 independent biologic experiments (C, D). *P < 0.05 (Student’s t test).
To determine whether Mettl21e expression is directly regulated by Mstn downstream signaling, we examined acute responses to perturbations of Mstn signaling. We first treated primary myoblasts with an Mstn inhibitor (follistatin) in differentiation medium for 5 d. The treatment did not alter the expression of Mettl21e (Fig. 1C). We also used adenoviral vectors to overexpress Mstn or GFP (control) in primary myoblasts cultured in growth medium. Overexpression of Mstn did not affect the expression of Mettl21e (Fig. 1D). We further analyzed microarray data comparing gene expression of GA muscles of mice injected with PBS or an Mstn antibody (clone JA-16 or PF-354) for 4 d, at which time point muscle hypertrophy did not occur (32). The short-term injection of JA16 did not change the expression of Mettl21e (Fig. 1E), though it blocked Mstn signaling (32). These results suggest that Mettl21e expression is associated with muscle hypertrophy rather than directly regulated by Mstn downstream signaling.
Muscle hypertrophy is often associated with changes in myofiber type distribution. We found that there was an apparent increase in the abundance of IIb myofibers in the Mstn KO EDL muscles relative to WT muscles (Fig. 1F), accompanied by an increased mRNA level of Myh4 (Fig. 1G), which encodes the IIb isoform Myh. Interestingly, the mRNA levels of Mettl21e were also evaluated by 8-fold in Mstn KO compared with WT EDL muscles (Fig. 1H). In contrast, Mettl21e levels in the SOL muscle that lacks IIb myofibers were comparable in Mstn KO and WT mice (Fig. 1I–K). These results indicate that the up-regulation of Mettl21e in the Mstn KO muscles is correlated to an increase in IIb myofibers.
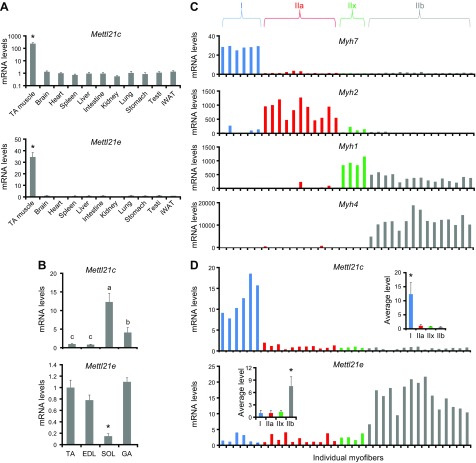
Mettl21e is enriched in type IIb myofibers
We next directly determined the expression of Mettl21e in different types of myofibers. We used Mettl21c as a control, which is specifically expressed in type I myofibers (23). We found that both Mettl21c and Mettl21e were primarily expressed in the skeletal muscle of mice (Fig. 2A). Within various muscle groups, the levels of Mettl21c were found to be SOL > GA > TA and EDL (Fig. 2B), a pattern consistent with the relative abundance of type I myofibers in these muscles. In contrast, the expression of Mettl21e was much higher in TA, EDL, and GA muscles than in SOL muscles, which lack type IIb myofibers (Fig. 2B). To directly examine the expression of Mettl21e in various types of myofibers, we isolated individual myofibers from SOL and EDL muscles and performed quantitative (q) PCR analysis on single myofibers. All individual myofibers predominantly express one of Myh isoforms, and based on the relative levels of Myh7, Myh2, Myh1, and Myh4, individual myofibers can be readily grouped into type I (blue column), IIa (red column), IIx (green column), or IIb (gray column) myofibers (Fig. 2C). Interestingly, all IIb myofibers also expressed a medium level of Myh1 (Fig. 2C). Strikingly, whereas Mettl21c expression was restricted to type I myofibers, Mettl21e expression was mainly enriched in IIb myofibers (Fig. 2D). On average, the mRNA level of Mettl21c was 12-fold in type I myofibers as in the other types (IIa, IIx, IIb) of myofibers, and Mettl21e expression was about 7-fold in type IIb myofibers as in the other types (I, IIa, IIx) of myofibers (Fig. 2D, inset). The single-cell–analysis results demonstrate that Mettl21e is enriched in type IIb myofibers.
Figure 2.
The expression Mettl21e in different types of myofibers. A) qPCR showing relative mRNA levels of Mettl21c and Mettl21e in different tissues. B) qPCR analysis of Mettl21c and Mettl21e in fast-twitch (TA, EDL, and GA) and slow-twitch (SOL) muscles; n = 6 independent biologic experiments with 3 technical repeats. C) qPCR analysis of Myh genes in isolated myofibers from SOL and EDL muscles of 4 mice. Individual myofibers were grouped into type I (blue), IIa (red), IIx (green), and IIb (gray). D) qPCR analysis of Mettl21c and Mettl21e in individual myofibers. Inset: the average expression of Mettl21c and Mettl21e in different types of myofibers. Error bars represent mean + sd *P < 0.05 (1-way ANOVA).
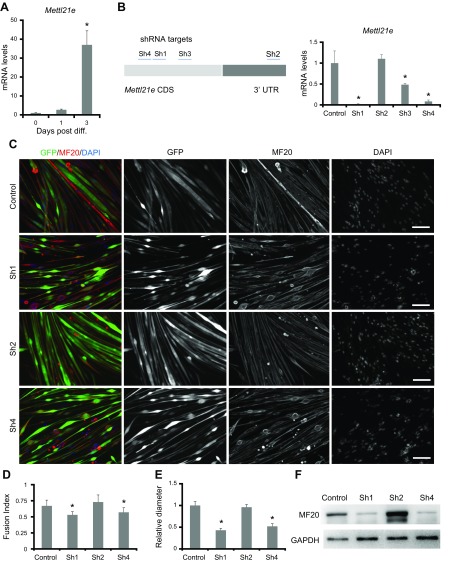
Mettl21e regulates myotube size in myoblast culture
Mettl21e expression was up-regulated by 37-fold at 3 d postinduction of differentiation in primary myoblasts (Fig. 3A), suggesting it mainly functions in differentiated myotubes. We thus performed shRNA to knock down (KD) Mettl21e in myotubes. Four adenoviral shRNA-GFP vectors (Sh1–Sh4) were constructed; 3 of them target coding DNA sequence, and 1 targets 3′UTR (Fig. 3B). Sh1–Sh4 reduced the expression of Mettl21e by 99%, 0%, 52%, and 92%, respectively (Fig. 3B). Sh1 and Sh4 were thus chosen to KD Mettl21e, and Sh2 was used as a negative control. Myoblasts were transduced with the Sh1, Sh2, Sh4, or control (GFP only) adenoviral vectors for 2 d and then induced to differentiate for 3 d. As expected, myotubes treated with Sh2 were morphologically similar to control myotubes (Fig. 3C). In contrast, myotubes treated with Sh1 or Sh4 were thinner than control myotubes (Fig. 3C). Quantitative analysis showed that Mettl21eKD decreased the fusion index and the diameter of myotubes (Fig. 3D, E). Mettl21eKD myotubes also expressed lower levels of Myh than did the control myotubes (Fig. 3F). Our cell culture data suggest a function of Mettl21e in regulating myofiber size.
Figure 3.
Knockdown (KD) of Mettl21e induces myotube atrophy in vitro. A) Relative levels of Mettl21e in myoblasts during differentiation; n = 5 independent biologic experiments with 3 technical repeats. B) Relative levels of Mettl21e in myotubes treated with the adenoviral shRNAs; n = 5 independent biologic experiments with 3 technical repeats. C) Myotubes treated with shRNAs were stained by MF20 (red), and nuclei were counterstained by DAPI (blue). Scale bar, 100 µm. Myoblasts were incubated with adenoviruses for 1 d and cultured in virus-free growth medium for 1 more day before differentiating for 2 d. D, E) Fusion index (D) and relative diameter (E) quantified based on staining in C. Fusion index measures the percentage of myonuclei present in multinucleated myofibers. Only GFP+ cells were used for quantification; n = 5 independent biologic experiments, with 5 different areas analyzed in each experiment. F) Western blot analysis showing the effect of Mettl21e KD on Myh protein expression. Error bars represent mean + sd *P < 0.05 (Student’s t test).
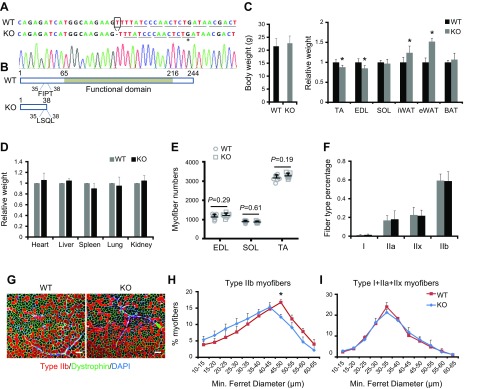
Mutation of Mettl21e decreases the size of type IIb myofibers in mice
We then generated Mettl21e KO mice using TALEN technique, creating a single nucleotide deletion in the 5′ region of the Mettl21e gene (Fig. 4A). This frameshift mutation generated 4 mutated amino acids (35FIPT38 to 35LSQL38) and a stop codon (TGA), leading to a truncated Mettl21e protein with only 38 amino acids (Fig. 4B). The truncated Mettl21e protein does not contain the functional domain of Mettl21e, indicative of a loss-of-function mutation (Fig. 4B). Importantly, the weights of 2 representative fast muscles (TA and EDL) were significantly lower in the Mettl21eKO mice compared with WT littermates (Fig. 4C). Interestingly, the decreases in muscle weight is accompanied by an increase in the weights of subcutaneous and epididymal white adipose tissues (Fig. 4C). As a result, there was no distinguishable difference in the overall body weight and tissue weights of other tissues between WT and Mettl21eKO mice (Fig. 4C, D).
Figure 4.
Loss of Mettl2e decreases type IIb myofiber size. A) Sequencing chromatograms of targeted region of Mettl21e gene. Reading frame is underlined. The stop codon is highlighted by line and dot in mutated Mettl21e gene. B) Predicted Mettl21e protein in WT and Mettl21eKO mice. Numbers indicate the sites of amino acids in annotated protein. Conserved functional domain of Mettl21e is in gray box. C, D) Whole body weights (C) and relative weights (D) of different tissues; n = 7 pairs of mice. E) Myofiber numbers in skeletal muscles of WT and Mettl21eKO mice; n = 7 pairs of mice. F) Quantification of myofiber types in EDL muscles. G) Immunostaining of type IIb (Red) myofibers in WT and Mettl21eKO mice. Scale bars, 100 µm. H, I) Size distribution of type IIb myofibers (H) or the other types (I + IIa + IIx) of myofibers (I) in TA muscles of WT and Mettl21eKO mice; n = 5 pairs of mice. Error bars represent mean + sd *P < 0.05 (Student’s t test).
We further investigated the histology of fast-twitch muscles. Numbers of myofiber were comparable between WT and Mettl21eKO mice in several different muscles (Fig. 4E). Staining for Myh isoforms showed that Mettl21eKO did not change myofiber type composition in EDL muscles (Fig. 4F). However, type IIb myofibers had smaller size in Mettl21eKO mice than in WT mice (Fig. 4G). Analysis of size distribution of type IIb myofibers showed that Mettl21eKO myofibers had a peak at 40–45 µm, whereas the WT myofiber diameter peaked at 45–50 µm (Fig. 4H). In contrast, diameters of the other myofiber types (type I + IIa + IIx) were comparable between WT and Mettl21eKO mice (Fig. 4I). These loss-of-function studies provide compelling evidence that Mettl21e is essential for maintaining type IIb myofiber size but dispensable for myofiber patterning.
Mettl21e methylates Vcp and modulates the activity of 26S proteasome
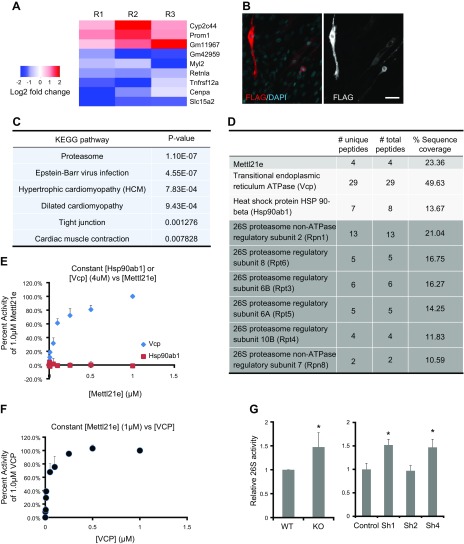
We performed RNA-sequencing analysis to better understand how Mettl21e KO alters TA muscle gene expression. Compared with TA muscles of WT mice, there were only 3 genes that were up-regulated and 6 genes that were down-regulated by more than 2-fold (fold change >2 and P < 0.01) in Mettl21eKO samples in triplicate (Fig. 5A). The small number of genes whose expression is affected by Mettl21e KO suggests a post-transcriptional regulatory function of Mettl21e. Consistently, we found that Mettl21e was localized exclusively in the cytoplasm by using FLAG antibody to stain C2C12 myoblasts transfected with a pcDNA3.1-Mettl21e-FLAG plasmid that encodes a Mettl21e-FLAG fusion protein (Fig. 5B).
Figure 5.
Mettl21e methylates Vcp and regulates 26S proteasomes activity. A) Heat map showing genes changed in Mettl21eKO muscles (fold change >2 and P < 0.01) in 3 biologic replicates, including 3 up-regulated and 6 down-regulated. Blue indicates the change level of down-regulated genes, and red shows the change level of up-regulated genes. B) Immunostaining of FLAG in C2C12 myoblasts transfected with Mettl21e-FLAG plasmid. Scale bar, 50 µm. Scale bar, 100 µm. C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of Mettl21e-associated proteins identified by mass spectrometry. D) Mettl21e-associated proteins related to proteasome. E) In vitro methyltransferase kinetic analysis of Vcp or Hsp90ab1 as a substrate of Mettl21e. Vcp or Hsp90ab1 was held constant at 4 µM in the presence of 13 µM [14C] SAM and increasing concentrations of Mettl21e. The data are represented as a percentage of counts per minute (CPMs, representing methyl groups transferred) at 1 µM of Mettl21e. Error bars represent mean + sd of 3 experiments performed in duplicate. F) In vitro methyltransferase kinetic analysis of Vcp as a substrate of Mettl21e. Mettl21e was held constant at 1 µM in the presence of 13 µM [14C] SAM and increasing concentrations of Vcp as the substrate. The data are represented as a percentage of CPMs at 1 µM of Vcp. Error bars represent mean + sd of 3 experiments performed in duplicate. G) The activity of 26S proteasome in TA muscles of WT and Mettl21eKO mice and in control and Mettl21eKD myotubes. Error bars represent mean + sd of 5 independent biologic experiments with 3 technical repeats. *P < 0.05 (Student’s t test).
We next sought to identify Mettl21e-associated proteins that could be responsible for the regulation of myofiber size. We used FLAG antibody to immunoprecipitate protein complexes from myocytes transduced with Mettl21e-FLAG or GFP-FLAG adenoviral vectors. Using mass spectrometry, we identified a list of proteins specifically in Mettl21e-associated complexes but not in GFP-associated complexes (Supplemental Table S2). To narrow down the list of potential substrates, we did Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway analysis of the identified proteins, and the top enriched pathway is proteasome (Fig. 5C). We then prioritized the Mettl21e-associated proteins that are related to proteasome complexes, including 6 subunits of 26S proteasome [regulatory particle non-ATPase 1 (Rpn1), Rpn8, regulatory particle triple-A ATPase 3 (Rpt3), Rpt4, Rpt5, and Rpt6] and 2 chaperone proteins [Vcp and heat shock protein HSP 90-beta (Hsp90ab1)] together with Mettl21e (Fig. 5D).
Given the established role of Vcp and Hsp90 in 26S proteasome function (33, 34) and the recent report that Mettl21 proteins preferentially interact with chaperone proteins (18), we examined whether Vcp or Hsp90ab1 is subject to Mettl21e-mediated methylation using in vitro methylation assay. Recombinant Vcp or Hsp90ab1 protein was held constant at 4 µM in the presence of [14C] SAM and increasing concentrations of recombinant Mettl21e, and the incorporation of methyl groups into proteins was measured as TCA-insoluble radioactivity. The incorporated radioactivity indicates that Vcp but not Hsp90ab1 is dose-dependently methylated by Mettl21e (Fig. 5E). Conversely, we held recombinant Mettl21e at 1 µM with increasing concentrations of Vcp. The radioactivity curve confirms that Mettl21e methylates Vcp dose dependently (Fig. 5F).
Vcp is involved in the regulation of the ubiquitin-proteasome system (35). To determine if Mettl21e methylation of Vcp leads to alterations in the activity of proteasome, we performed 26S proteasome activity assay. KO of Mettl21e increased the activity of 26S proteasome by 47% in TA muscles (Fig. 5G). In addition, Mettl21eKD myotubes had about 50% higher proteasome activities than the control myotubes (Fig. 5G). These results indicate that Mettl21e methylates Vcp and modulates the activity of 26S proteasome.
Inhibition of proteasome activity prevents atrophy of Mettl21e KD myotubes
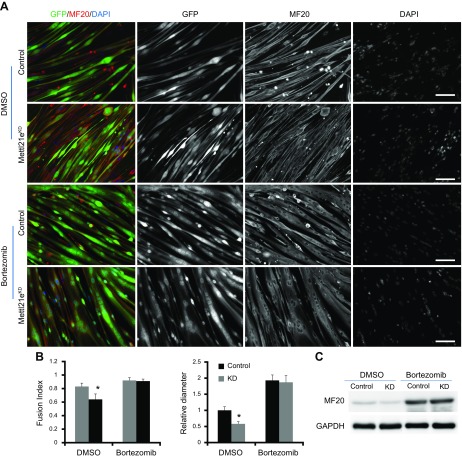
To verify that Mettl21e regulates myotube size through proteasome, we inhibited proteasome activity in control and Mettl21eKD myotubes using bortezomib, a Food and Drug Administration–approved 26S proteasome inhibitor. To this end, myoblasts were transduced with the adenoviral-GFP vector (control) and Sh1 (Mettl21eKD) at d 1 after serum withdrawal. After differentiation for an extra 2 d, myotubes were treated with DMSO or 25 nM bortezomib for 2 d. In the absence of the inhibitor, Mettl21eKD myotubes consistently exhibited thinner morphology than did the control KD myotubes (Fig. 6A). In contrast, there was no distinguishable difference between control and Mettl21eKD myotubes in the presence of bortezomib (Fig. 6A). Quantifications of fusion index and relative myotube diameter showed that bortezomib treatment blunted the differences between control and Mettl21eKD myotubes (Fig. 6B). Consistently, bortezomib treatment increased the protein levels of Myh in control myotubes and normalized the levels of Myh in Mettl21eKD myotubes (Fig. 6C). Together, we conclude that Mettl21e maintains myofiber size through a proteasome-dependent pathway.
Figure 6.
Bortezomib rescues myofiber atrophy induced by Mettl21e KD. A) Control and Mettl21eKD cell were treated with DMSO or Bortezomib for 2 d. The differentiated myoblasts were stained by MF20 (Red), and nuclei were counterstained by DAPI (Blue). Scale bars, 100 µm. B) Fusion index and relative diameters of myotubes counted according to the staining inA. Only GFP+ cells were used for quantification. C) Western blots analysis of Myh. Error bars represent mean + sd of 5 independent biologic experiments. *P < 0.05 (Student’s t test).
DISCUSSION
Mettl21e is one of the top up-regulated genes in Mstn KO fast-twitch muscles (25), but we show that it is not directly regulated by Mstn. Instead, increased expression of Mettl21e was due to increases in the abundance of IIb myofibers in Mstn KO fast-twitch muscles (36). This finding also explains the up-regulation of Mettl21e in hypertrophic muscles of Callipyge sheep (27), in which a substantial increase of type IIb myofibers in hypertrophic muscles was reported (37, 38). Interestingly, the counterpart of Mettl21e in humans is denoted as a unitary pseudogene (39). Human METTL21EP was possibly inactivated by a splice-junction mutation (AG to TA) located at the acceptor-splice site of its second intron. This splice-junction mutation is accompanied by the lack of detectable type IIb myofibers in human skeletal muscles (40).
Mettl21e is located in cytoplasm. This cytoplasmic localization indicates that Mettl21e is not a histone methyltransferase that functions to regulate gene transcription. Our mass spectrometry data show that Mettl21e physically interacts with subunits of 26S proteasome and its associated chaperone proteins. Mettl21eKO or Mettl21eKD increases the proteasome activity, therefore reducing myofiber size. Moreover, inhibition of proteasome activity rescues Mettl21eKD-induced myofiber atrophy, demonstrating that Mettl21e regulates myofiber size through proteasome.
Our data shows that Mettl21e methylates Vcp, which is reported as a substrate of human METTL21C and METTL21D (18, 19, 23). Vcp is an important element of the ubiquitin-proteasome system (35, 41). It acts as a molecular chaperone to bring ubiquitinated substrates to the 26S proteasome for degradation (33, 41). It also maintains the activity of 26S proteasome through binding and antagonizing the inhibitory function of the proteasome inhibitor PI31 (42). However, the effect of methylation of Vcp on proteasome has yet to be determined (19). Current knowledge indicates that nonhistone methylation regulates protein functions through the crosstalk with other post-translational modification or through changing protein-protein interactions (17, 43). It is possible that Mettl21e methylates Vcp to affect its binding with the other proteins and thus affects proteasome activity. It is also possible that Mettl21e directly methylates one of the 26S proteasome subunits to regulate proteasome activity. Understanding the detailed mechanisms will extend our knowledge about molecular regulation of proteasome activity.
Mettl21e KO muscle showed a relatively milder phenotype in comparison with Mettl21e KD myotubes. We mutated Mettl21e by a frame-shifting indel, resulting in a premature termination codon and a truncated protein that does not contain the methyltransferase domain. It is possible that the truncated protein retains some methyltransferase-independent function in regulating myofiber size. In addition, the most recent studies report a nonsense-induced transcriptional compensation mechanism, in which mRNA containing premature termination codon is degraded through nonsense-mediated RNA decay to induce transcriptional up-regulation of genes similar to itself (44, 45). In this regard, other members of the Mettl21 family proteins may have partially compensated for the Mettl21e loss-of-function.
Myofiber size may be regulated at both cellular (myogenic differentiation and myoblast fusion) (46) and molecular (protein synthesis and degradation) level. Our data do not support a role of Mettl21e in myogenic differentiation. First, Mettl21e expression increases by 35-fold at 3 d after differentiation, suggesting that Mettl21e mainly functions after myogenic differentiation. Second, Mettl21e KD did not inhibit myoblast differentiation. However, we could not completely rule out a role of Mettl21e in myoblast fusion based on the observation that Mettl21e KD reduces myoblast fusion. As myoblast fusion was also increased with inhibition of proteasome, it is possible that that the reduced myoblast fusion after Mettl21e KD is a secondary effect of reduced proteasome activity.
Our finding helps to explain the myofiber-size paradox, in which oxidative myofibers are smaller than glycolytic myofibers despite the higher myonuclei content (per volume) and higher rates of protein synthesis in the oxidative myofibers (3, 47, 48). One explanation for this paradox is due to differential rate of protein degradation, which is mainly mediated by the ubiquitin-proteasome system. This system includes 2 discrete and successive processes: conjugating substrate proteins with multiple ubiquitin molecules and the subsequent degradation of the tagged proteins by the 26S proteasome (49). The conjugation of ubiquitin to lysine residues of substrate proteins requires 3 ATP-dependent enzymatic steps (49). E3 is the key enzyme in the conjugation process. Two muscle-specific E3, muscle atrophy F box and muscle RING finger 1, are found to have a higher expression in oxidative muscles than in glycolytic muscles (3, 50). Thus, the higher rates of protein degradation in the oxidative muscles may have underlined their smaller size. Here, we provide evidence that type IIb myofibers also employ an active mechanism to maintain their larger size through Mettl21e-mediated inhibition of 26S proteasome activity.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Fengfeng Zhang (ViewSolid Biotech, Beijing, China) for generating the methyltransferase-like 21e knockout mice, Chris Bidwell (Purdue University, West Lafayette, IN, USA) for discussion and sharing Callipyge sheep data, Jessie Ellis (Purdue University) for technical assistance, Jun Wu (Purdue University) and Mary Larimore (Purdue University) for mouse colony maintenance, and members of the Kuang laboratory for valuable comments. This work was supported by a grant from the U.S. National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (R01AR071649) and Innovation Fund for Medical Sciences (CIFMS) from Chinese Academy of Medical Sciences (2016-I2M-1-012). The authors declare no conflicts of interest.
Glossary
- EDL
extensor digitorum longus
- FLAG
DYKDDDDK epitope
- GA
gastrocnemius
- KO
knockout
- KD
knockdown
- Mettl21e
methyltransferase-like 21e
- Mstn
myostatin
- Myh
myosin heavy chain
- SAM
S-adenosyl-l-methionine
- sh
short hairpin
- SOL
soleus
- TA
tibialis anterior
- TALEN
transcription activator-like effector nuclease
- TCA
trichloroacetic acid
- Vcp
valosin-containing protein
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
C. Wang and S. Kuang conceived the project, designed the experiments, and prepared the manuscript; C. Wang, B. Zhang, A. C. Ratliff, J. Arrington, J. Chen, Y. Xiong, F. Yue., Y. Nie, and W. Jin performed the experiments and analyzed the data; and K. Hu, W. A. Tao, C. A. Hrycyna, and X. Sun provided key reagents and technical assistance.
REFERENCES
- 1.Schiaffino S., Gorza L., Sartore S., Saggin L., Ausoni S., Vianello M., Gundersen K., Lømo T. (1989) Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J. Muscle Res. Cell Motil. 10, 197–205 [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino S., Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 [DOI] [PubMed] [Google Scholar]
- 3.Van Wessel T., de Haan A., van der Laarse W. J., Jaspers R. T. (2010) The muscle fiber type-fiber size paradox: hypertrophy or oxidative metabolism? Eur. J. Appl. Physiol. 110, 665–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanes J. R., Lichtman J. W. (1999) Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22, 389–442 [DOI] [PubMed] [Google Scholar]
- 5.Dolmetsch R. E., Lewis R. S., Goodnow C. C., Healy J. I. (1997) Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386, 855–858 [DOI] [PubMed] [Google Scholar]
- 6.Naya F. J., Mercer B., Shelton J., Richardson J. A., Williams R. S., Olson E. N. (2000) Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 275, 4545–4548 [DOI] [PubMed] [Google Scholar]
- 7.Wu H., Rothermel B., Kanatous S., Rosenberg P., Naya F. J., Shelton J. M., Hutcheson K. A., DiMaio J. M., Olson E. N., Bassel-Duby R., Williams R. S. (2001) Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 20, 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan P. G., Chen L., Nardone J., Rao A. (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 10.Liu N., Garry G. A., Li S., Bezprozvannaya S., Sanchez-Ortiz E., Chen B., Shelton J. M., Jaichander P., Bassel-Duby R., Olson E. N. (2017) A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat. Cell Biol. 19, 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark D. A., Coffey N. J., Pancoast H. R., Arnold L. L., Walker J. P., Vallée J., Robitaille R., Garcia M. L., Cornelison D. D. (2015) Ephrin-A3 promotes and maintains slow muscle fiber identity during postnatal development and reinnervation. J. Cell Biol. 211, 1077–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rooij E., Quiat D., Johnson B. A., Sutherland L. B., Qi X., Richardson J. A., Kelm R. J., Jr., Olson E. N. (2009) A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 14.Lee K. Y., Singh M. K., Ussar S., Wetzel P., Hirshman M. F., Goodyear L. J., Kispert A., Kahn C. R. (2015) Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat. Commun. 6, 8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quiat D., Voelker K. A., Pei J., Grishin N. V., Grange R. W., Bassel-Duby R., Olson E. N. (2011) Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc. Natl. Acad. Sci. USA 108, 10196–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copeland R. A., Solomon M. E., Richon V. M. (2009) Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8, 724–732 [DOI] [PubMed] [Google Scholar]
- 17.Biggar K. K., Li S. S. (2015) Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5–17 [DOI] [PubMed] [Google Scholar]
- 18.Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9, e1003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G. M., Egge-Jacobsen W., Falnes P. O. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038 [DOI] [PubMed] [Google Scholar]
- 20.Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., Falnes P. O. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamey J. J., Wienert B., Quinlan K. G. R., Wilkins M. R. (2017) METTL21B is a novel human lysine methyltransferase of translation elongation factor 1A: discovery by CRISPR/Cas9 knockout. Mol. Cell. Proteomics 16, 2229–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malecki J., Aileni V. K., Ho A. Y. Y., Schwarz J., Moen A., Sørensen V., Nilges B. S., Jakobsson M. E., Leidel S. A., Falnes P. O. (2017) The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiederstein J. L., Nolte H., Günther S., Piller T., Baraldo M., Kostin S., Bloch W., Schindler N., Sandri M., Blaauw B., Braun T., Hölper S., Krüger M. (2018) Skeletal muscle-specific methyltransferase METTL21C trimethylates p97 and regulates autophagy-associated protein breakdown. Cell Rep. 23, 1342–1356 [DOI] [PubMed] [Google Scholar]
- 24.Jakobsson M. E., Davydova E., Małecki J., Moen A., Falnes P. O. (2015) Saccharomyces cerevisiae eukaryotic elongation factor 1A (eEF1A) is methylated at Lys-390 by a METTL21-like methyltransferase. PLoS One 10, e0131426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welle S., Cardillo A., Zanche M., Tawil R. (2009) Skeletal muscle gene expression after myostatin knockout in mature mice. Physiol. Genomics 38, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caetano-Anollés K., Mishra S., Rodriguez-Zas S. L. (2015) Synergistic and antagonistic interplay between myostatin gene expression and physical activity levels on gene expression patterns in triceps Brachii muscles of C57/BL6 mice. PLoS One 10, e0116828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming-Waddell J. N., Olbricht G. R., Taxis T. M., White J. D., Vuocolo T., Craig B. A., Tellam R. L., Neary M. K., Cockett N. E., Bidwell C. A. (2009) Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PLoS One 4, e7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubik R. M., Tietze S. M., Schmidt T. B., Yates D. T., Petersen J. L. (2018) Investigation of the skeletal muscle transcriptome in lambs fed β adrenergic agonists and subjected to heat stress for 21 d. Transl. Anim. Sci. 2, S53–S56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McPherron A. C., Lawler A. M., Lee S. J. (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Yue F., Kuang S. (2017) Muscle histology characterization using H&E staining and muscle fiber type classification using immunofluorescence staining. Bio Protoc. 7, e2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Wang M., Arrington J., Shan T., Yue F., Nie Y., Tao W. A., Kuang S. (2017) Ascl2 inhibits myogenesis by antagonizing the transcriptional activity of myogenic regulatory factors. Development 144, 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welle S., Burgess K., Mehta S. (2009) Stimulation of skeletal muscle myofibrillar protein synthesis, p70 S6 kinase phosphorylation, and ribosomal protein S6 phosphorylation by inhibition of myostatin in mature mice. Am. J. Physiol. Endocrinol. Metab. 296, E567–E572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai R. M., Li C. C. (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3, 740–744 [DOI] [PubMed] [Google Scholar]
- 34.Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. (2003) The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22, 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloppsteck P., Ewens C. A., Förster A., Zhang X., Freemont P. S. (2012) Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim. Biophys. Acta 1823, 125–129 [DOI] [PubMed] [Google Scholar]
- 36.Amthor H., Macharia R., Navarrete R., Schuelke M., Brown S. C., Otto A., Voit T., Muntoni F., Vrbóva G., Partridge T., Zammit P., Bunger L., Patel K. (2007) Lack of myostatin results in excessive muscle growth but impaired force generation. Proc. Natl. Acad. Sci. USA 104, 1835–1840; erratum: 104, 4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carpenter C. E., Rice O. D., Cockett N. E., Snowder G. D. (1996) Histology and composition of muscles from normal and callipyge lambs. J. Anim. Sci. 74, 388–393 [DOI] [PubMed] [Google Scholar]
- 38.Bidwell C. A., Waddell J. N., Taxis T. M., Yu H., Tellam R. L., Neary M. K., Cockett N. E. (2014) New insights into polar overdominance in callipyge sheep. Anim. Genet. 45(Suppl 1), 51–61 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z. D., Frankish A., Hunt T., Harrow J., Gerstein M. (2010) Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 11, R26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smerdu V., Karsch-Mizrachi I., Campione M., Leinwand L., Schiaffino S. (1994) Type IIx myosin heavy chain transcripts are expressed in type IIb fibers of human skeletal muscle. Am. J. Physiol. 267, C1723–C1728 [DOI] [PubMed] [Google Scholar]
- 41.Meyer H., Weihl C. C. (2014) The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 127, 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemen C. S., Marko M., Strucksberg K. H., Behrens J., Wittig I., Gärtner L., Winter L., Chevessier F., Matthias J., Türk M., Tangavelou K., Schütz J., Arhzaouy K., Klopffleisch K., Hanisch F. G., Rottbauer W., Blümcke I., Just S., Eichinger L., Hofmann A., Schröder R. (2015) VCP and PSMF1: antagonistic regulators of proteasome activity. Biochem. Biophys. Res. Commun. 463, 1210–1217 [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Wen H., Shi X. (2012) Lysine methylation: beyond histones. Acta Biochim. Biophys. Sin. (Shanghai) 44, 14–27 [DOI] [PubMed] [Google Scholar]
- 44.Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J. (2019) PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263 [DOI] [PubMed] [Google Scholar]
- 45.El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G. L. M., Takacs C. M., Lai S. L., Fukuda R., Gerri C., Giraldez A. J., Stainier D. Y. R. (2019) Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reza M. M., Subramaniyam N., Sim C. M., Ge X., Sathiakumar D., McFarlane C., Sharma M., Kambadur R. (2017) Irisin is a pro-myogenic factor that induces skeletal muscle hypertrophy and rescues denervation-induced atrophy. Nat. Commun. 8, 1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng B. S., Kasper C. E., Edgerton V. R. (1994) Cytoplasm-to-myonucleus ratios and succinate dehydrogenase activities in adult rat slow and fast muscle fibers. Cell Tissue Res. 275, 39–49 [DOI] [PubMed] [Google Scholar]
- 48.Goldberg A. L. (1967) Protein synthesis in tonic and phasic skeletal muscles. Nature 216, 1219–1220 [DOI] [PubMed] [Google Scholar]
- 49.Lecker S. H., Goldberg A. L., Mitch W. E. (2006) Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 17, 1807–1819 [DOI] [PubMed] [Google Scholar]
- 50.Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.