Abstract
The nuclear factor erythroid 2–related factor 2 (Nrf2) signaling axis is a target of covalent drugs and bioactive native electrophiles. However, much of our understanding of Nrf2 regulation has been focused at the protein level. Here we report a post-transcriptional modality to directly regulate Nrf2-mRNA. Our initial studies focused on the effects of the key mRNA-binding protein (mRBP) HuR on global transcriptomic changes incurred upon oxidant or electrophile stimulation. These RNA-sequencing data and subsequent mechanistic analyses led us to discover a novel role of HuR in regulating Nrf2 activity, and in the process, we further identified the related mRBP AUF1 as an additional novel Nrf2 regulator. Both mRBPs regulate Nrf2 activity by direct interaction with the Nrf2 transcript. Our data showed that HuR enhances Nrf2-mRNA maturation and promotes its nuclear export, whereas AUF1 stabilizes Nrf2-mRNA. Both mRBPs target the 3′–UTR of Nrf2-mRNA. Using a Nrf2 activity–reporter zebrafish strain, we document that this post-transcriptional control of Nrf2 activity is conserved at the whole-vertebrate level.—Poganik, J. R., Long, M. J. C., Disare, M. T., Liu, X., Chang, S.-H., Hla, T., Aye, Y. Post-transcriptional regulation of Nrf2-mRNA by the mRNA-binding proteins HuR and AUF1.
Keywords: antioxidant response, zebrafish, mRNA-stabilization, mRNA-maturation, mRNA-trafficking
The nuclear factor erythroid 2–related factor 2 (Nrf2) signaling axis is the cell’s linchpin stress defense (1), whereby the central player, the Nrf2 transcription factor, controls the expression of numerous genes for cytoprotection and detoxification (2). Nrf2 up-regulation accompanies cell stimulation with either endogenous or exogenously introduced reactive electrophilic species (RES) and reactive oxidative species (ROS), which are increasingly appreciated as bona fide cell signaling cues (3). Beyond RES/ROS-stimulated conditions, Nrf2 activity at basal (i.e., nonstimulated) conditions is tightly regulated. However, the most well-understood modes of Nrf2 regulation—both at basal and stimulated cell states—are centered around Nrf2 protein-level control (2). Among the most notable players are Kelch-like ECH associated protein 1 (Keap1) and β-transducin repeat-containing protein (β-TrCP), which independently maintain low steady-state levels of Nrf2 protein in nonstimulated cells through Nrf2 proteasomal targeting (4, 5). Deregulation of protein-level control of Nrf2 transcriptional activity is common in various cancers (6). Recent efforts to modulate Nrf2 signaling by targeting protein-based Nrf2 regulators like Keap1 have underscored the pharmaceutical relevance of this pathway (7). In addition to protein-level regulation of Nrf2, some microRNAs (miRNAs) target this pathway either by direct Nrf2 targeting or targeting of other protein regulators, such as Keap1 (8). However, expression of these miRNAs tends to be highly tissue-/context-specific. A general regulatory mechanism (e.g., through interaction with a ubiquitously expressed protein) of Nrf2 activity at the mRNA level has not been reported.
Regulation of mRNA is a complex process that can be mediated through structural (9), sequence (10) or epitranscriptomic (11) elements within a transcript. The ubiquitously expressed mRNA-binding protein (mRBP) HuR (ELAVL1) is a postulated stress-relevant protein that binds to adenylate uridylate (AU)–rich sites within regulatory mRNA targets and regulates their expression via post-transcriptional mechanisms (12). These binding sites typically reside within 3′–UTRs of target transcripts. However, binding within introns, coding regions, and 5′–UTRs has also been observed. Regulation of mRNA targets by HuR can occur via direct binding or indirectly by miRNA-dependent mechanisms (13, 14).
State-of-the-art sequencing techniques such as photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) (15, 16) have revealed thousands of functionally diverse targets of HuR (17, 18). Although Nrf2-mRNA has been detected by PAR-CLIP analysis of HuR, no functional validations or interaction studies have been made, likely because the Nrf2 transcript appears as a low-frequency hit [with only 46 T-to-C/A-to-G conversions for HuR (marking cross-links to HuR), compared to hundreds to thousands of conversions for the most highly ranked transcripts such as AKT serine/threonine kinase 3 (AKT3) and DNA polymerase alpha 1 (POLA1)]. However, PAR-CLIP rankings generally correlate poorly with HuR target affinity (Supplemental Table S1); PAR-CLIP conversion number may be affected by varying expression levels of different targets or artifacts of the PAR-CLIP procedure. Accordingly, there remains a need to mechanistically investigate how HuR regulates important disease-relevant targets such as Nrf2-mRNA.
In addition to the difficulty in ranking importance of hits from high-throughput data sets, the multimodal regulatory activities of HuR render predicting functional consequences of a HuR-mRNA binding event difficult. HuR—distributed between the nucleus and cytosol in 10:1 ratio in HeLa cells (19)—largely modulates its target transcripts through alterations in mRNA stability, typically by stabilizing bound transcripts (20). Additional regulatory mechanisms employed by HuR on its target transcripts have also been reported [e.g., nuclear export of the cyclooxygenase-2 (COX-2) mRNA (21, 22) and splicing regulation of the death receptor FAS and the translational regulator eIF4Enif1 (23, 24)]. However, the generality of these nuanced regulatory roles remains largely unclear, and their importance in Nrf2 regulation is completely unknown.
HuR is strongly linked to disease as a key player in inflammation and cancers among other disorders (25). The growing appreciation of HuR as a major player in disease is underscored by recent efforts to screen for inhibitors of the HuR–RNA interaction (26–28). The noncovalent inhibitors identified from such screens disrupt target-transcript binding or HuR oligomerization (key to HuR function). Some HuR inhibitors have shown promise in arresting cell cycle and inducing apoptosis in cultured lung cancer cells (29). The mRNA-stabilizing ability of HuR is also affected by small-molecule stress inducers, such as the electrophilic prostaglandin A2 (PGA2) and lipopolysaccharide (LPS), which triggers oxidative stress (30–33). However, no study has directly compared the role of HuR regulation of the Nrf2 signaling axis under conditions of oxidative or electrophilic stress.
Understanding HuR-regulatory circuits is further complicated by the presence of multiple mRBPs that bind to target mRNAs at the same/similar sites as HuR but generally elicit antagonistic outputs to HuR binding. One such mRBP is AUF1 (HNRNPD), which shares significant target overlap with HuR based on PAR-CLIP analysis (34). Mechanistically, in contrast with the canonical role of HuR in promoting mRNA stability, AUF1 typically promotes degradation of target mRNAs (35). However, other regulatory mechanisms employed by AUF1 have recently come into focus: AUF1 can promote translation of myocyte enhancer factor 2C but suppresses translation of profilin 1 (34, 36). Crosstalk between HuR and AUF1 in regulating common mRNA targets has also been investigated for the cyclin-dependent kinase inhibitor p21 and cyclin D1 (19). Stability of both of these targets was regulated positively by HuR and negatively by AUF1. However, general understanding of these coregulatory events by these 2 mRBPs remains limited. Whether HuR and AUF1 bind to different sites or compete for the same sites within a specific co-regulated transcript also remains unclear (19). Nevertheless, AUF1 generally functions as the antipode of HuR in terms of target transcript regulation.
Herein, we describe novel post-transcriptional regulatory modes of the Nrf2 pathway, controlled by HuR and AUF1. Our initial RNA-sequencing (RNA-seq) analysis of HuR-dependent global transcriptomic changes in response to cell stimulation by H2O2 and 4-hydroxynonenal (HNE)—the prototypical oxidant (ROS) and electrophile (RES)—revealed that only the electrophile HNE causes significant induction of Nrf2-driven genes. Surprisingly, we found that HuR depletion generally diminished Nrf2 transcriptional activity in nonstimulated cells. Yet, upon electrophile stimulation, Nrf2 activity was more strongly up-regulated in HuR-depleted cells. Our RNA-seq data implicate the involvement of different subsets of Nrf2-driven genes underlying these 2 effects. Our further investigations into Nrf2 regulation by HuR in nonstimulated cells led us to discover a previously unrecognized regulatory loop controlling Nrf2 activity, which our data show is conserved from cultured cells to zebrafish. This newly identified post-transcriptional regulatory mode of Nrf2 activity potentially offers a novel alternative intervention to modulate Nrf2/antioxidant response (AR) in disease.
MATERIALS AND METHODS
Statistics and data presentation
For experiments involving cultured cells, samples generated from individual wells or plates were considered biologic replicates. For zebrafish experiments, each larval fish was considered a biologic replicate. In the figure legend for each experiment, the number of independent biologic replicates and how the data are presented in the figure (typically mean ± sem) are clearly indicated. Significance values calculated with unpaired Student’s t test are clearly indicated within data figures. Data were plotted/fit and statistics generated using Prism 7 or 8 (GraphPad, La Jolla, CA, USA).
Reagents
All HNE used in this study was HNE(alkyne) (referred to as HNE in the manuscript/figures for clarity) and was synthesized as previously reported (37). Unless otherwise indicated, all other chemical reagents were bought from MilliporeSigma (Burlington, MA, USA) at the highest availability purity. Tris(2-carboxyethyl)phosphine (TCEP) was from Chem-Impex International (Wood Dale, IL, USA). Puromycin was from Santa Cruz Biotechnology (Dallas, TX, USA). Actinomycin D was from MilliporeSigma. AlamarBlue was from Thermo Fisher Scientific (Waltham, MA, USA) and was used according to the manufacturer’s instructions. Minimal Essential Medium (MEM), Opti-MEM, Dulbecco’s PBS, 100× pyruvate (100 mM), 100× nonessential amino acids (11140-050), and 100× penicillin streptomycin (15140-122) were from Thermo Fisher Scientific. Protease inhibitor cocktail Complete EDTA-free was from Roche (Basel, Switzerland). 3XFlag peptide was from ApexBio Technology (Houston, TX, USA). Anti-Flag(M2) resin (A2220) was from MilliporeSigma. Talon (635503) resin was from Clontech Laboratories (Mountain View, CA, USA). Ni-NTA agarose (30210) was from Qiagen (Hilden, Germany). 2020 and LT1 transfection reagents were from Mirus Bio (Madison, WI, USA). DharmaFECT I and Duo were from Dharmacon (Lafeyette, CO, USA). Polyethylenimine was from Polysciences (Warrington, PA, USA). Venor GeM PCR-based mycoplasma detection kit was from MilliporeSigma. ECL substrate and ECL-Plus substrate were from Thermo Fisher Scientific and were used as directed. Acrylamide, ammonium persulfate, tetramethylethylenediamine, Precision Plus protein standard were from Bio-Rad (Hercules, CA, USA). All lysates were quantified using the Bio-Rad Protein Assay (Bio-Rad) relative to bovine serum albumin (BSA) as a standard (Bio-Rad). PCR was carried out using Phusion Hot start II (Thermo Fisher Scientific) as per the manufacturer’s protocol. All plasmid inserts were validated by sequencing at Cornell Biotechnology sequencing core facility (Ithaca, NY, USA). All sterile cell culture plasticware was from CellTreat Scientific Products (Pepperell, MA, USA).
Generation of plasmids
Sequences of all primers used for cloning and site-directed mutagenesis are listed in Supplemental Table S9. All plasmids generated were fully validated by Sanger sequencing at the Cornell Genomics Core Facility.
pCS2+8 Flag3HuR was generated by PCR amplification of human HuR (plasmid provided from the Hla lab), extension of the resulting product, and cloning into linearized pCS2+8 vector (plasmid 34931; Addgene, Watertown, MA, USA). For recombinant expression, the same procedure was used to clone HuR into a pET28a vector.
pLJM60 AUF1p42 was obtained from Addgene (plasmid 38242) and cloned with a Flag2 tag into pCS2+8 as described above. Using the resulting plasmid, AUF1p37 and AUF1p45 were generated by deletion of AUF1-exon 7 and addition of AUF1-exon 2, respectively. AUF1p40 was generated by addition of AUF1-exon 2 to AUF1p37. For recombinant expression, these constructs were cloned into a pET28a vector.
The pSGG luciferase reporter plasmid (38) containing the Nrf2 3′–UTR was obtained from Prof. Qun Zhou (University of Maryland School of Medicine, Baltimore, MD, USA). This plasmid was modified for intron-reporter assays by cloning fragments of the Nrf2 transcript with or without introns (amplified from genomic DNA or cDNA prepared from HEK293T cells, respectively) upstream of the luciferase coding sequence.
Short hairpin (sh)-RNA–resistant expression plasmids (for rescue experiments) were produced by PCR amplification of the starting plasmid with forward and reverse mutagenesis primers containing the desired mutations (Supplemental Table S9) followed by DpnI (NEB) treatment. These plasmids code for the same protein but contain mismatches (highlighted in red in the primer sequences; Supplemental Table S9) in the shRNA-targeting sequence, allowing expression of the ectopic protein in knockdown cells.
Cell culture
HEK293T [obtained from American Type Culture Collection (ATCC), Manassas, VA, USA] and mouse embryonic endothelial cells (MEECs; generated in the Hla lab) were cultured in MEM (51090036; Thermo Fisher Scientific) supplemented with 10% v/v fetal bovine serum (FBS; MilliporeSigma), penicillin/streptomycin (Thermo Fisher Scientific), sodium pyruvate (Thermo Fisher Scientific), and non-essential amino acids (Thermo Fisher Scientific) at 37°C in a humidified atmosphere of 5% CO2. Medium was changed every 2–3 d.
Generation of shRNA-based knockdown cell lines
HEK293T packaging cells (5.5 × 105 cells) were seeded in 6-well plates in antibiotic-free medium and incubated for 24 h. The cells were transfected with a mixture of 500 ng packaging plasmid (pCMV-R8.74psPAX2), 50 ng of envelope plasmid (pCMV-VSV-G), and 500 ng of pLKO vector containing the hairpin sequence (MilliporeSigma; Supplemental Table S2) using TransIT-LT1 (Mirus Bio) following the manufacturer’s protocol. shControl plasmid was obtained from Prof. Andrew Grimson (Cornell University). Eighteen hours post-transfection, the medium was replaced with medium containing 20% FBS and incubated for a further 24 h. Medium containing virus particles was harvested, centrifuged (800 g, 10 min), filtered through a 0.45-μm filter, and used for infection or frozen at −80°C for later use.
HEK293T or MEEC cells (5.5 × 106 cells) in 6-well plates were treated with 1 ml of virus-containing medium in a total volume of 6 ml of medium containing 8 μg/ml polybrene (MilliporeSigma). After 24 h, the medium was changed and the cells were incubated for 24 h. Following this period, the medium was changed to medium containing 2 μg/ml puromycin (Santa Cruz Biotechnology). Following selection, cells were assayed by Western blotting.
RNA sequencing
Following treatment of shHuR/shControl cells with HNE (50 μM) or H2O2 (225 μM) for 18 h, cells were lysed by the addition of Trizol (Thermo Fisher Scientific), and RNA was isolated following the manufacturer’s protocol. The quality of the RNA was assessed by Nanodrop spectrophotometry (A260/A280 ratio ∼2) and fragment analysis using a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA quality numbers for all samples were 10.0. The rest of the procedure, including data analysis, was performed by the RNA sequencing core service at Cornell University. Briefly, rRNA was subtracted by hybridization from total RNA samples (100 ng total RNA input) using the RiboZero Magnetic Gold H/M/R Kit (Illumina, San Diego, CA, USA). Following cleanup by precipitation, rRNA-subtracted samples were quantified with a Qubit 2.0 (RNA HS kit; Thermo Fisher Scientific). TruSeq-barcoded RNA-seq libraries were generated from half of the rRNA-subtracted samples with the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA). Each library was quantified with a Qubit 2.0 (dsDNA HS kit; Thermo Fisher Scientific), and the size distribution was determined with a Fragment Analyzer (Agilent Technologies) prior to pooling. Libraries were sequenced on a NextSeq500 instrument (Illumina). At least 60 M single-end 75 bp reads were generated per library. Reads were trimmed for low quality and adaptor sequences with cutadapt v.1.8 [parameters: –m 50 –q 20 –a 5′-AGATCGGAAGAGCACACGTCTGAACTCCAGTC-3′ –match-read-wildcards (39)]. Trimmed reads were mapped to rRNA sequences with bowtie2 v.2.2 to remove them [parameters: default mapping options, –al and –un to split matching and non-matching reads (40)]. Reads were mapped to the reference genome/transcriptome (GRCh37/hg19) using tophat v.2.1 [parameters: –library-type = fr-firststrand –no-novel-juncs –G < ref_genes.gtf > (41)]. Cufflinks v.2.2 (cuffnorm/cuffdiff) was used to generate fragments per kilobase of transcript per million mapped reads (FPKM) values and statistical analysis of differential gene expression (42). RNA-Sequencing data have been submitted to the Gene Expression Omnibus (GEO; accession number GSE127444).
Growth inhibition assays
HEK293T cells (4000 cells/well) were seeded in 96-well plates. After 24 h, cells were treated with the indicated molecules at the indicated concentrations and incubated for 48 h. AlamarBlue (Thermo Fisher Scientific) was added to each well, and the cells were incubated for a further 3 h, after which fluorescence (excitation 560 nm; emission 590 nm) was measured using a BioTek Cytation 3 microplate reader (BioTek, Winooski, VT, USA). To test whether reduced alamarBlue is oxidized by H2O2 over the timescales used, HEK293T cells in a 96-well plate were incubated with the manufacturer’s recommended concentration of alamarBlue for 3 h to reduce the dye. The growth medium containing reduced alamarBlue was collected, pooled, centrifuged to remove cells, and then incubated with the concentrations of H2O2 used in the viability experiment (Supplemental Fig. S2B), and fluorescence was measured over time as previously described. We observed ∼3% loss of fluorescence after 4 h in reduced alamarBlue treated with 1250 μM H2O2; the fluorescence of samples at all other concentrations was not significantly different from equivalent medium containing reduced alamarBlue but not treated with H2O2. Thus, this control experiment confirmed that our experimental conditions involving oxidants are compatible with redox-sensitive alamarBlue reagents employed in our growth inhibition assays.
Knockdown of HuR and Nrf2 with small interfering RNA
Small interfering (si)-RNAs targeting the open reading frame of human HuR or Nrf2 were obtained from Dharmacon (Supplemental Table S6), and nontargeting control siRNAs (Control siRNA-A or -E) were obtained from Santa Cruz Biotechnology. HEK293T (3.6 × 105 cells) in 6-well plates were transfected with siRNA using Dharmafect I (Dharmacon) for 48 h following the manufacturer’s protocol, then assayed. For cotransfection of siRNA and plasmid(s) for reporter assays, cells were transfected with Dharmafect Duo (Dharmacon) for 48 h following the manufacturer’s protocol, then assayed.
Western blotting
Cells were resuspended in RIPA buffer (Santa Cruz Biotechnology) supplemented with protease and phosphatase inhibitors and lysed by 3 cycles of rapid freeze-thaw. Lysates were cleared by centrifugation (20,000 g, 10 min, 4°C) and total protein concentration was determined by the Bradford assay. Typically, 20–40 μg of total protein was loaded per lane, separated by SDS-PAGE, transferred to PVDF, blocked, and incubated with the appropriate antibodies (Supplemental Table S10). Detection was carried out on a ChemiDoc-MP imaging system (Bio-Rad) using ECL Western blotting Substrate (Thermo Fisher Scientific) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Western blot data were quantitated using the Gel Analysis tool in ImageJ (National Institutes of Health, Bethesda, MD, USA). Bands of interest were integrated and normalized to the loading control.
Luciferase reporter assays
Luciferase reporter assays were carried out as previously described (43). Briefly, cells were cotransfected with firefly luciferase plasmid [pGL4.37 E364A for antioxidant response element:luciferase assays (Promega, Madison, WI, USA); pSGG containing the human Nrf2 3′–UTR sequence (from Prof. Qun Zhou, University of Maryland School of Medicine)] and Renilla luciferase plasmid [pGL4.75, E693A (Promega)] in a 40:1 ratio for 48 h using TransIT 2020 (Mirus Bio) or Dharmafect Duo (Dharmacon). For experiments involving ectopic protein expression, the 40:1 reporter plasmid mixture was cotransfected with ectopic protein expression plasmid in a 1:1 ratio. Cells were lysed for 15 min in passive lysis buffer (Promega) with gentle shaking, and homogenized lysate was transferred to the wells of an opaque white 96-well plate (Corning, Corning, NY, USA). Firefly and Renilla luciferase activity was measured on a BioTek Cytation3 microplate reader. For assays involving HNE treatment, medium was replaced with medium containing the indicated concentration of HNE at 30 h post-transfection, and cells were incubated for a further 18 h.
Real-time quantitative PCR
Real-time quantitative PCR (qPCR) was carried out as previously described (44). Total RNA was isolated from cells using Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. One microgram of total RNA (purity/integrity assessed by agarose gel electrophoresis and concentration determined by A260 nm using a BioTek Cytation3 microplate reader with a Take3 accessory) was treated with amplification-grade DNase I (Thermo Fisherr Scientific) and reverse transcribed using Oligo(dT)20 as a primer and Superscript III Reverse Transcriptase (Thermo Fisherr Scientific) following the manufacturer’s protocol. PCR was performed for 2 technical replicates per sample using iQ SYBR Green Supermix (Bio-Rad) and primers specific to the gene of interest (Supplemental Table S11) following the manufacturer’s protocol. Amplicons were chosen that were 150–200 bp in length and had no predicted off-target binding predicted by the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information, Bethesda, MD, USA). For genes with multiple splice variants, primers that amplified conserved sequences across all splice variants were chosen. Primers were validated using standard curves generated by amplification of serially diluted cDNA; primers with a standard curve slope between −0.8 and 1.2 and R2 ≥ 0.97 were considered efficient. Single PCR products were confirmed by melting analysis following the PCR protocol. Data were collected using a LightCycler 480 (Roche). Threshold cycles were determined using the LightCycler 480 software. Samples with a threshold cycle >35 or without a single, correct melting point were not included in data analysis. Normalization was carried out using a single housekeeping gene as indicated in each dataset and the ΔΔCt method.
Analysis of reporter mRNA levels
HEK293T cells (1.6 × 105) in 12-well plates were transfected with luciferase reporters as previously described for 48 h. Cells were then lysed in 250 μl of passive lysis buffer for 15 min with shaking, and 50 μl of lysate was taken to measure luciferase activity as described above. Trizol LS was added to the remaining lysate, and RNA was isolated as described above. Prior to RT, total RNA (1 μg) was treated with amplification-grade DNase I (Thermo Fisher Scientific) to remove any residual plasmid. RT and real-time qPCR was then carried out as above.
RNA immunoprecipitation-PCR
RNA immunoprecipitation (RIP)-PCR was carried out following previously described methods (45). HEK293T cells (4.5 × 106) in 10-cm-diameter plates were transfected with the indicated constructs (mixed 1:3 with empty plasmid) or empty plasmid alone (8 μg total DNA per plate) with polyethylenimine (21 g/plate). Medium was changed 24 h post-transfection, and the cells were incubated for 48 h total. Cells were washed once with PBS (Thermo Fisher Scientific), harvested by trypsinization, and washed thoroughly with PBS.
Cell pellets were resuspended in polysome lysis buffer [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Chem-Impex International) pH 7.0, 100 mM KCl (Thermo Fisher Scientific), 5 mM MgCl2 (Thermo Fisher Scientific), 0.5% Nonidet-P40, Complete EDTA-free Protease Inhibitor Cocktail (Roche), 0.2% vanadyl ribonuceloside complex (New England Biolabs)] and frozen at −80°C for at least 30 min. The lysate was thawed, centrifuged twice (20,000 g, 10 min each), and the protein concentration was determined using Bradford Assay. Three milligrams of total protein was diluted to 0.5 mg/ml in NT2 buffer [50 mM Tris pH 7.4, 150 mM NaCl (Thermo Fisher Scientific), 1 mM MgCl2 (Thermo Fisher Scientific), 0.05% Nonidet-P40, 0.2% vanadyl ribonuceloside complex] and incubated with 50 μl of Flag M2 agarose (MilliporeSigma) for 3 h at 4°C with end-over-end rotation. The resin was washed at least 4 times (5 min/wash) with NT2 buffer containing 300 mM NaCl, and a portion of the resin was retained for Western blot analysis. The resin was resuspended in 100 μl of NT2, supplemented with 0.1% SDS and 3 mg/ml proteinase K (Santa Cruz Biotechnology), and incubated at 55°C for 30 min. RNA was isolated using Trizol reagent following the manufacturer’s protocol and analyzed by real-time qPCR as previously described.
Analysis of mRNA stability
HEK293T shHuR/shAUF1 and shControl cells (9.6 × 105 cells) in 6-well plates were treated with 5 μg/ml actinomycin D (MilliporeSigma) and harvested in Trizol as described above at the indicated time points. The level of Nrf2-mRNA remaining was assessed by real-time qPCR as previously described.
Nuclear-cytosolic fractionation for RNA isolation
This procedure was adapted from a reported protocol (46). HEK293T cells (9.6 × 105 cells) in 6-well plates were harvested by tryspinization and washed twice with cold PBS. Cell pellets were then resuspended in RSB buffer [10 mM Tris (Chem-Impex International) pH 7.4, 10 mM NaCl (Thermo Fisher Scientific), 3 mM MgCl2 (Thermo Fisher Scientific)], incubated on ice for 3 min, and centrifuged (1500 g, 3 min, 4°C). Cells were resuspended in RSBG40 buffer [10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 10% glycerol (MilliporeSigma), 0.5% Nonidet P-40, 0.5 mM DTT (VWR International, Radnor, PA, USA), and 100 U/ml RNaseOUT (Thermo Fisher Scientific)] and lysed with gentle pipetting up and down. The suspension was centrifuged (4500 g, 3 min, 4°C), and the supernatant was saved as the cytosolic fraction. The nuclear fraction (pellet) was again resuspended in RSBG40 and 10× detergent solution [final concentrations: 3.3% sodium deoxycholate (Chem-Impex International), 6.6% Tween 20 (Thermo Fisher Scientific)] was added. The nuclei were pipetted up and down several times and the suspension was incubated on ice for 5 min and then centrifuged (4500 g, 3 min, 4°C). The supernatant was pooled with the previous supernatant, and the combined sample was used as the cytosolic fraction. The nuclei were washed once more with RSBG40, centrifuged (9500 g, 3 min, 4°C), and resuspended thoroughly in RSBG40. Trizol LS was then added to each sample and RNA was isolated, treated with amplification-grade DNase I, and subjected to qPCR as described above.
Zebrafish husbandry, microinjection, and imaging
All zebrafish procedures were performed in accordance with the guidelines of U.S. National Institutes of Health and were approved by Cornell University’s Institutional Animal Care and Use Committee (IACUC). Tg(-3.4gstp1:GFP)it416b fish were obtained from Riken Brain Science Institute, (Wako, Japan; National BioResource Project, Zebrafish). Transgenic fish were crossed with wild-type fish to generate a mixture of heterozygous and wild-type progeny for experiments. Fertilized eggs at the single-cell stage were injected with 8 ng of morpholino oligonucleotides (MOs) targeting zHuR (elavl1a) or zAUF1 (hnrnpd) or a random control MO (Gene Tools, Philomath, OR, USA; Supplemental Table S7). Fish were screened for green fluorescence protein (GFP) expression by imaging with a Leica M205-FA fluorescence stereoscope (Leica Microsystems, Wetzlar, Germany) ∼24 h postfertilization. Following imaging, fish were euthanized, washed twice in PBS with 0.1% Tween-20, and transferred to 4% formaldehyde for further immunofluoresence analysis (see below). GFP expression was detected using red fluorescent antibody staining because of high background signal in the GFP (ex: 488 nm; em: 520–550 nm) channel at this zebrafish developmental stage, which prevents accurate quantitation.
Whole-mount immunofluorescence
Whole-mount immunostaining of zebrafish larvae was carried out as previously described (47). All incubation steps were performed with gentle rocking. Euthanized fish were washed twice with PBST (PBS + 0.1% Tween-20) and fixed in 4% formaldehyde in PBS at 4°C overnight or up to 7 d. Fish were then washed twice for 30 min with PDT (PBST, 0.3% Triton X-100, 1% DMSO), blocked for 1 h at room temperature in blocking buffer (PBST, 10% FBS, 2% BSA), and incubated with primary antibody (Supplemental Table S10) in blocking buffer for 2 h at room temperature or overnight at 4°C. Fish were washed twice with PDT, reblocked for 1 h at room temperature, and incubated with secondary antibody (Supplemental Table S10) in blocking buffer for 1.5 h at room temperature. Following secondary antibody incubation, fish were washed twice with PDT and then imaged. Fluorescence was quantified using the “Measure” tool in ImageJ (National Institutes of Health, Bethesda, MD, USA).
Expression and purification of His6-HuR
BL21-CodonPlus cells (Agilent Technologies) were transformed with pET28a plasmid encoding His6-TEV-HuR. A single colony was divided into several starter cultures (5 ml) and grown in lysogeny broth medium containing chloramphenicol and kanamycin overnight at 37°C with shaking. Cultures were then each diluted into 1 L of lysogeny broth containing kanamycin and grown at 37°C with shaking to optical density (OD)600 nm = 0.5, at which point isopropyl-β-D-thiogalactoside (1 mM final concentration; GoldBio, St. Louis, MO, USA) was added to induce expression. The temperature was then reduced to 18°C, and cultures were grown overnight with shaking. Further steps were carried out at 4°C. Cells were harvested by centrifugation (4000 g, 20 min, 4°C), resuspended in lysis buffer [20 mM Tris pH 7.5, 500 mM KCl, 2 mM MgCl2, 10 mM imidazole (Thermo Fisher Scientific), 5% glycerol, 5 mM β-mercaptoethanol (βME), and 0.5 mM phenylmethylsulfonyl fluoride (Enzo Life Sciences, Farmingdale, NY, USA)] and lysed by 2 passages through an Emlusiflex cell disruptor (Avestin Europe, Mannheim, Germany). Debris was cleared by centrifugation (20,000 g, 30 min, 4°C). Streptomycin sulfate (1% wt/vol) was added dropwise over 20 min with gentle stirring, and precipitated material was cleared by centrifugation (20,000 g, 30 min, 4°C). The lysate then was incubated with nickel resin (Qiagen) for 1 h with gentle agitation. The resin was washed progressively with wash buffers (20 mM Tris pH 7.5; 150 mM KCl; 2 mM MgCl2; 50, 100, 200 mM imidazole; and 5 mM βME). Protein was eluted with elution buffer [20 mM Tris pH 7.5, 150 mM KCl, 2 mM MgCl2, 300 mM imidazole (Thermo Fisher Scientific), 5 mM βME], concentrated, and loaded onto a size-exclusion chromatography column (ÄKTA purification system, Hiload 26/600 Superdex 75 prep grade; GE Healthcare, Chicago, IL, USA) and run in storage buffer (20 mM Tris pH 7.5, 200 mM KCl, 2 mM MgCl2, 5% glycerol, 5 mM TCEP). The protein was collected, concentrated, divided into aliquots, flash frozen in liquid nitrogen, and stored at −80°C in single-use aliquots to avoid freeze/thaw.
Expression and purification of His6-AUF1
BL21-CodonPlus cells (Agilent Technologies) were transformed with pET28a His6-AUF1 (p37) and grown as described above. The purification procedure was the same as for His6HuR except for the buffers: lysis buffer [50 mM NaH2PO4 pH 7.6, 300 mM NaCl, 5 mM imidazole, 5 mM βME, and 1 mM phenylmethylsulfonyl fluoride, 0.5% Nonidet P40, 0.15 mg/ml lysozyme, 0.1 mg/ml RNase A (MilliporeSigma)]; wash buffers (50 mM NaH2PO4 pH 7.6; 150 mM NaCl; 20, 50, 100, mM imidazole; and 5 mM βME); elution buffer (50 mM NaH2PO4 pH 7.6, 150 mM NaCl, 250 mM imidazole, 5 mM βME). The eluate was supplemented with 0.1 mg/ml RNase A and rotated end over end for 1 h at room temperature. The resulting mixture was then dialyzed against storage buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.6, 150 mM KCl, 2 mM MgCl2, 2 mM TCEP] for 2 h at 4°C, then dialysis buffer was changed and dialysis was allowed to proceed overnight at 4°C. The protein was then concentrated and stored (with 5% glycerol added to storage buffer) at −80°C in single-use aliquots to avoid freeze/thaw.
[32P]-End-labeling of RNA oligos
RNA oligos (IDT, Coralville, IA, USA; Supplemental Table S8) were resuspended in RNase-free water to 200 μM. RNA was labeled with γ-[32P] ATP (PerkinElmer, Waltham, MA, USA) in a reaction containing (final concentrations) 20 pmol RNA, 20 pmol γ-[32P] ATP, 1× polynucleotide kinase buffer (New England Biolabs), and 0.4 U/μl polynucleotide kinase (New England Biolabs) in a total volume of 50 μl. The reaction was incubated at 37°C for 1 h, after which 50 μl of RNase-free water and 300 μl of Trizol LS were added, and the mixture was left at room temperature for 5 min. CHCl3 (200 μl) was added, the mixture was shaken for 15 s, left at room temperature for 2 min, and then spun down briefly to collect the contents. The aqueous layer was collected and mixed with an equal volume of CHCl3, shaken, and spun down briefly. The aqueous layer was again collected, supplemented with 15 μg of GlycoBlue (Ambion) and ammonium acetate to a final concentration of 500 mM, and 3 volumes of 100% EtOH were added. The mixture was vortexed and allowed to precipitate overnight at −20°C, then centrifuged (20,000 g, 15 min, 4°C). The labeled RNA pellet was washed twice with 75% EtOH in RNase-free water, dried briefly, and resuspended in RNase-free water. Labeled RNA was stored at −80°C in single-use aliquots to avoid freeze/thaw.
EMSAs
These experiments were modeled on previously reported procedures (48). End-labeled RNA probes were mixed 1:10 with unlabeled probe. The binding reaction contained (final concentrations) 10 mM Tris pH 7.5, 100 mM KCl, 1 mM EDTA, 0.1 mM DTT, 5% glycerol, 0.01 mg/ml BSA, 0.5 nM RNA, and varying concentrations of HuR or AUF1p37. This mixture was incubated for 30 min at room temperature. The gel (10% 71:1 acrylamide:bisacrylamide Tris-borate-EDTA (TBE) gel) was prerun at 220 V for 20 min in chilled 0.5× Tris-borate-EDTA (TBE) buffer. The reactions were then loaded and run at 220 V for 12 min. The gels were exposed to an IP screen (GE Healthcare) at −80°C for 24–36 h, then imaged on a Typhoon FLA 7000 IP Imager (GE Healthcare). The bound RNA signal was quantitated using the Gel Analysis tool in ImageJ and fit to a one-site binding model (Eq. 1 below) using Prism 7.
 |
where y is the bound RNA signal, Bmax is the plateau, x is the concentration of the mRBP, and Kd(app) is the dissociation constant.
RESULTS
HuR depletion perturbs the global transcriptomic status in electrophile-stimulated and nonstimulated cells
HuR has been implicated as a stress-responsive protein: cytoplasmic translocation of HuR is promoted by several small-molecule stressors [e.g., H2O2, arsenite, and the cyclopentenone PGA2, a Michael acceptor RES (30, 31)]. Treatment with PGA2 also increases the affinity of HuR for p21 mRNA (30). Intrigued by reports of the stress-relevance of HuR, we launched gene-expression profiling studies in HuR-knockdown cells, in the presence or absence of small-molecule stress signals. HNE—a native RES with a reactive core chemically similar to that of PGA2—was selected as a representative bioactive RES (49, 50), and H2O2 was selected as a representative ROS. HuR-knockdown HEK293T cells were first generated (Supplemental Table S2). Relative to a nontargeted shControl, HuR was knocked down by 70% in these cells (Supplemental Fig. S1A). We then treated these shControl and shHuR cells with HNE (25 μM, 18 h) or H2O2 (225 μM, 18 h) and subsequently sequenced their RNA, post rRNA depletion. The chosen concentrations of HNE/H2O2 correspond to the approximate EC75 for growth inhibition, measured over 48 h (Supplemental Fig. S2). Significant differential expression was evaluated with CuffDiff (42), wherein gene-level pairwise comparisons having q value (P value corrected for multiple tests) < 0.05 are considered significantly differentially expressed (SDE).
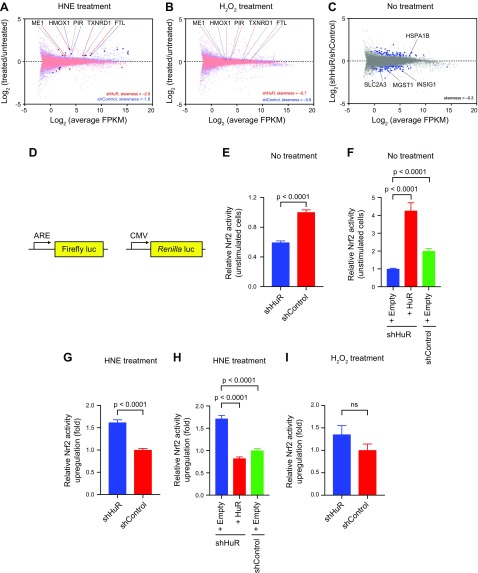
Compared with nontreated control, HNE-stimulated shControl cells showed considerable up-regulation of 11 genes out of 14 total genes SDE (79%). The data skewed in the positive direction overall, indicating a general stimulation of gene transcription [Fig. 1A (data in blue) and Supplemental Fig. S3 and Supplemental Tables S3 and S4]. In contrast, HNE-treated shHuR cells (compared with nonstimulated shHuR cells) showed up-regulation of only 58% of the 12 genes SDE, and the dataset skewed in the negative direction overall [Fig. 1A (data in red) and Supplemental Fig. S3 and Supplemental Tables S3 and S4]. Thus, depletion of HuR compromises the ability of cells to mount positive transcriptional responses following electrophile stimulation. By contrast, neither shHuR nor shControl cells mounted as robust a response to H2O2 as to HNE. Compared with the number of genes SDE in HNE-stimulated shControl and shHuR cells (14 and 12, respectively), only 2 genes were SDE in H2O2-treated shControl cells, and no genes were SDE in H2O2-treated shHuR cells (Fig. 1B and Supplemental Table S3). Both control and HuR-knockdown datasets with H2O2 treatment skewed slightly in the negative direction (Supplemental Fig. S3 and Supplemental Tables S3 and S4).
Figure 1.
RNA-seq expression profiling indicates context-specific HuR regulation in global and Nrf2-specific transcriptional activities. A, B) Differential expression from RNA-seq analyses in shHuR or shControl HEK293T cells following HNE (A, 25 μM, 18 h) or H2O2 (B, 225 μM, 18 h) stimulation relative to respective nonstimulated cells. Genes SDE are denoted with dark, opaque points; Nrf2-driven genes SDE in at least 1 comparison are labeled. Dashed lines to gene names indicate that the gene was not SDE in that comparison. See also Supplemental Figs. S1A, S2, and S3 and Supplemental Tables S2 and S3. C) Differential expression from RNA-seq analysis in nonstimulated shHuR cells relative to shControl cells. Genes SDE are denoted with blue points. Nrf2-driven genes found to be SDE are labeled. See also Supplemental Tables S3–S5. D) Nrf2 activity reporter system in which firefly luciferase is driven by Nrf2 and a constitutive Renilla luciferase is coexpressed as an internal normalization control. E) Relative Nrf2 activity in unstimulated shHuR and shControl cells (mean ± sem, n = 24); shControl set to 1.0. F) Relative Nrf2 activity was measured as in F, but ectopic HuR was expressed in non-HNE-stimulated shHuR cells (mean ± sem, n = 7 for shHuR+HuR and n = 8 for other conditions) and shHuR + Empty vector was set to 1.0. See also Supplemental Fig. S4. G) Relative Nrf2 activity up-regulation in HNE-stimulated shHuR (25 μM, 18 h) and shControl (mean ± sem, n = 16) lines was calculated by measuring (Nrf2 activity + HNE/Nrf2 activity +DMSO); shControl was set to 1.0. See also Supplemental Fig. S4. H) Relative Nrf2 activity up-regulation was measured as in G, but ectopic HuR was expressed in HNE-stimulated (25 μM, 18 h) shHuR cells (mean ± sem, n = 8); shHuR + Empty vector was set to 1.0. See also Supplemental Fig. S4. I) Nrf2 activity in H2O2-stimulated (225 μM, 18 h) shHuR and shControl (mean ± sem, n = 7 for shHuR and 8 for shControl) lines was calculated by measuring (Nrf2 activity with H2O2/Nrf2 activity with water); shControl was set to 1.0. See also Supplemental Fig. S4. All P values were calculated with Student’s t test. For (A, B), data were derived from 2 independent biologic replicates per treatment condition. Skewness was calculated with Prism. FPKM, fragments per kilobase of transcript per million mapped reads; ns, not significant.
HuR is a context-specific regulator of Nrf2 transcriptional activity
Beyond HuR-dependent effects on global transcriptional status, we compared the expression of Nrf2-driven genes in shHuR and shControl cells following HNE stimulation. We identified 1 gene, pirin (PIR) that was not significantly up-regulated in shHuR knockdown lines upon HNE treatment but was up-regulated in shControl lines upon HNE stimulation. However, we also detected a further 4 Nrf2-driven genes [malic enzyme 1 (ME-1), thioredoxin reductase 1 (TXNRD1), ferritin light chain (FTL), and heme oxygenase 1(HMOX1)] that were up-regulated in both shHuR and shControl cells treated with HNE (Fig. 1A and Table 1). Intriguingly, 3 of these genes were up-regulated to a greater extent in shHuR cells than in shControl cells: ME-1, TXNRD1, and FTL showed 1.05- to 1.2-fold higher fold-change (FC)(treated/untreated) in shHuR cells vs. shControl cells (Fig. 1A and Table 1). This trend contrasts with the overall down-regulation of global transcriptional activity following HNE-stimulation in shHuR-lines (0.98 average fold change, −2.9 skewness) compared with shControl-lines [1.0 average fold change, 1.8 skewness (Fig. 1A and Supplemental Fig. S3)]. We did not observe SDE of these genes between nonstimulated shHuR and shControl cells (Fig. 1C and Supplemental Table S3), but we were able to identify 3 other Nrf2-driven genes (solute carrier family 2 member 3 (SLC2A3), insulin induced gene 1 (INSIG1), and microsomal glutathione S-transferase 1 (MGST1)), each down-regulated by about 55% in shHuR cells relative to shControl cells under nontreated conditions (Fig. 1C and Supplemental Table S3). One gene was significantly up-regulated in shHuR relative to shControl lines, heat shock protein family A (Hsp70) member 1B (HSPA1B). For all genes SDE, there was no correlation between the fold change in expression we observed and the number of total HuR-specific PAR-CLIP conversion events (Supplemental Table S5) (17). Thus, these changes in regulation of Nrf2-controlled transcripts are not dependent on the proclivity of the SDE transcripts to bind HuR. From this analysis, we conclude that the consensus effects, at least, are likely attributable to changes in Nrf2 regulation, as we show further below. Taken together, these data give good initial evidence that HuR is a context-dependent regulator of Nrf2 activity, modulating different subsets of Nrf2-driven genes in electrophile-stimulated vs. nonstimulated conditions.
TABLE 1.
Fold changes for selected Nrf2-driven genes of interest upon HNE stimulation relative to nontreated
| shHuR |
shControl |
|||
|---|---|---|---|---|
| Gene | FC (treated/untreated) | q | FC (treated/untreated) | q |
| ME-1 | 1.6 | 0.04 | 1.4 | 0.41 (NS) |
| TXNRD1 | 1.6 | 0.02 | 1.4 | 0.18 (NS) |
| FTL | 1.9 | 0.02 | 1.6 | 0.02 |
| HMOX1 | 2.6 | 0.02 | 2.8 | 0.02 |
| PIR | 1.5 | 0.13 (NS) | 1.9 | 0.02 |
Q values calculated with CuffDiff. See also Supplemental Tables S3–S5. FC. fold change; NS, not significant.
HuR-depleted cells manifest altered HNE-promoted antioxidant responsivity
These interesting findings, which overall are consistent with the poorly understood context dependence of Nrf2 signaling (51), led us to examine HuR-dependent regulation of Nrf2 activity in greater detail. Specifically, we examined the extent to which cellular Nrf2 responsivity to HNE is HuR-dependent. Nrf2 activity was measured using a luciferase reporter under the transcriptional control of a Nrf2-activatable promoter, normalized to a constitutively expressed Renilla luciferase control (Fig. 1D). Interestingly, shHuR cells under non-RES-stimulated conditions consistently featured a significant 2-fold suppression of Nrf2 activity compared with shControl cells (Fig. 1E), which is consistent with suppression of some Nrf2-driven genes revealed by our RNA-seq analysis. Upon ectopic expression of HuR [optimized to give only ∼2.5-fold above endogenous HuR levels (Supplemental Fig. S4A), as HuR overexpression showed a 5-fold elevation in AR levels in unstimulated cells, relative to empty vector–transfected cells (Supplemental Fig. S4B)] in these shHuR cells, close to normal basal Nrf2 activity levels were restored (Fig. 1F).
Whole-cell stimulation with HNE (25 μM; 18 h) resulted in 2-fold higher Nrf2 activity levels in HuR-depleted cells compared with shControl cells (Fig. 1G and Supplemental Fig. S4C). Under the same treatment conditions, ectopic expression of HuR in shHuR cells restored the extent of HNE-stimulated Nrf2 activity up-regulation to that observed in shControl cells (Fig. 1H). Thus, the suppression of Nrf2 activity in nonstimulated shHuR cells, as well as their greater HNE-induced Nrf2 activity up-regulation observed in both our reporter assay (Fig. 1E, G) and our RNA-seq data (Fig. 1A, C), are HuR-specific effects.
We validated these findings in MEECs. When the cells were depleted of HuR (Supplemental Fig. S5A), they also featured suppressed Nrf2 activity in the nonstimulated state (Supplemental Fig. S5B), and greater Nrf2 activity up-regulation following HNE-stimulation (25 μM; 18 h) (Supplemental Fig. S5C). Consistent with our RNA-seq data, no significant difference in Nrf2 activity was observed between shHuR and shControl HEK293T cells following H2O2 stimulation (Fig. 1I and Supplemental Fig. S4C).
HuR modulation of antioxidant responsivity is Nrf2-dependent
To confirm that the HuR-associated HNE-responsive effects observed above are Nrf2-dependent, we used siRNA (Supplemental Fig. S1B and Supplemental Table S6) to simultaneously knock down HuR and Nrf2. Multiple Nrf2 siRNAs led to ∼40% suppression of Nrf2 activity on average (Supplemental Fig. S6A), validating the ability of our reporter assay to measure changes in Nrf2 activity. This result is also consistent with the generally narrow dynamic range of Nrf2 activity assays, as we further explain below. We chose to use AR for the Nrf2 siRNA validation because of known issues previously encountered with Nrf2 antibodies that remain unresolved (52). Knockdown of either HuR or Nrf2 in nonstimulated cells suppressed Nrf2 activity by about 50% compared with control cells; simultaneous knockdown of HuR and Nrf2 did not lead to any further suppression (Supplemental Fig. S6B). Upon HNE treatment, Nrf2 depletion completely arrested the ability of cells to up-regulate Nrf2 activity in the presence or absence of simultaneous HuR knockdown (Supplemental Fig. S6C). Thus, the effects of HuR knockdown on Nrf2 signaling measured above are Nrf2-dependent.
The Nrf2-dependence of this HuR regulatory event is an important finding, especially given the emerging success and promise in therapeutic targeting of Nrf2-dependent pathways (53). However, the multifaceted regulatory modalities of Nrf2 remain poorly understood, and Nrf2-regulation at the post-transcriptional level remains an untapped arena. Thus, we chose to delve deeper—through the following series of experiments—to understand this novel post-transcriptional mechanism of Nrf2-mRNA regulation by HuR in nonstimulated cells.
HuR and AUF1 coregulate Nrf2 activity in nonstimulated cells
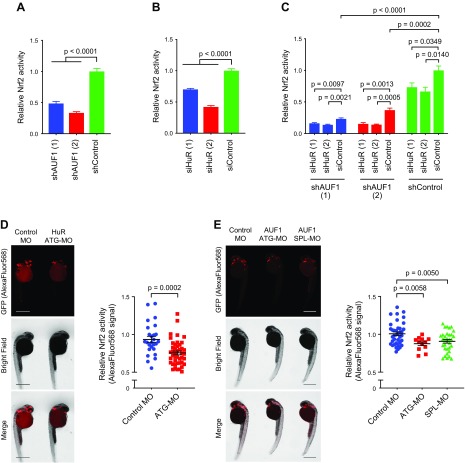
Considering that AUF1 is generally considered to be the antipode of HuR in regulating target transcripts and that AUF1 binds to similar AU-rich elements as HuR within target transcripts, we probed the effects of both HuR and AUF1 on Nrf2 activity. We generated multiple HEK293T shAUF1 lines [each expressing a single shRNA sequence targeting AUF1 mRNA (Supplemental Table S2)] to deplete AUF1 levels. Because there are 4 isoforms of human AUF1 (AUF1p37, AUF1p40, AUF1p42, and AUF1p45), shRNAs targeting sequences conserved across all the isoforms were used (Supplemental Table S2). Relative to a nontargeted shControl, total AUF1 was knocked down by 50–75% (Supplemental Fig. S1C). Because AUF1 typically destabilizes target transcripts, we expected that AUF1 knockdown would promote Nrf2 activity (20, 35). Contrary to this prediction, knockdown of AUF1 suppressed Nrf2 activity by 50–60% (Fig. 2A).
Figure 2.
Depletion of HuR or AUF1 in cells and larval zebrafish suppresses Nrf2 activity. A, B) Nrf2 activity in HEK293T cells depleted of AUF1 and HuR, respectively [mean ± sem of n = 12 (AUF1) and n = 8 (HuR) independent replicates per condition]. See also Supplemental Fig. S1B, C. C) Nrf2 activity upon simultaneous knockdown of HuR and AUF1 (mean ± sem of n = 4 independent replicates). See also Supplemental Fig. S7A. D, E) Nrf2 activity in Tg(gstp1:GFP) zebrafish upon knockdown of zHur and zAuf1. Larvae are stained with a red fluorescent antibody because green background fluorescence at this developmental stage prevents accurate quantitation of the GFP reporter signal. Inset: quantitation (mean ± sem) of mean fluorescence intensity measured using the Measure tool of ImageJ [sample sizes analyzed: D: n = 26 (Control MO), 44 (ATG-MO); E: n = 38 (Control MO), 12 (ATG-MO), 32 (SPL-MO)]. Each point represents a single fish. P values were calculated with Student’s t test. Scale bars, 500 μm. See also Supplemental Figs. S1D, E and S8. ATG-MO, translation initiation blocking morpholino oligonucleotide; SPL-MO, splice blocking morpholino oligonucleotide.
We investigated the effects of simultaneous knockdown of HuR, using the siRNAs from above, and AUF1, achieved by shRNA. Cells transfected with siHuR alone featured 40–50% suppression of Nrf2 activity (Fig. 2B) relative to cells transfected with siControl. Fold suppression of Nrf2 activity upon HuR knockdown in shAUF1 (1) cells (30% on average) was not significantly different from fold suppression of Nrf2 activity upon HuR knockdown in shControl cells (Fig. 2C and Supplemental Fig. S7A). However, a minor increase in suppression of Nrf2 activity [relative to that observed in shAUF1 (1) and shControl cells] occurred in shAUF1 (2) (Fig. 2C and Supplemental Fig. S7A). These observations likely indicate that HuR- and AUF1-regulation of Nrf2 activity function through independent mechanisms. However, because Nrf2/AR has multiple upstream effectors in addition to HuR/AUF1, and because these proteins are occasionally believed to work in tandem (54), we further address the issue of functional (in)dependence more directly below.
Modulation of Nrf2 activity by HuR and AUF1 is relevant at the organismal level
To extend the relevance of our findings, we turned to zebrafish (Danio rerio). HuR and AUF1 are both conserved in zebrafish (zebrafish gene names elavl1a and hnrnpd, respectively; we refer to these genes below as zHur and zAuf1, respectively, for clarity). The key regulators of Nrf2 pathway are also conserved in zebrafish (1). We used antisense MOs to deplete zHur and zAuf1 in developing embryos (Supplemental Table S7) of the established Nrf2 activity-reporter strain, Tg(gstp1:GFP) (55). An MO-blocking translation initiation of zHur (56) successfully depleted zHur levels at 24 h postfertilization by ∼25% (Supplemental Fig. S1D). This is likely an underestimate of knockdown efficiency given that the commercially available HuR antibody detects other Hu-family proteins not targeted by this MO. zAuf1 has only 2 isoforms. We designed 2 MOs to deplete zAuf1: one to block translation initiation (ATG-MO), and one to block a splice site (SPL-MO). Interestingly, each MO depleted only 1 isoform of zAuf1 (Supplemental Fig. S1E).
Larvae heterozygotic for the Nrf2-activity reporter were injected with MOs at the single-cell stage. Twenty-four hours following MO injection, we used immunofluorescent (IF) staining to assess GFP protein levels. Knockdown of either zHur or zAuf1 (by either MO for zAuf1) led to a significant 10–15% decrease in reporter protein fluorescence (Fig. 2D, E). Although the magnitude of this suppression is modest, the dynamic range of Nrf2-responsivity in these fish (like many other such systems) is narrow: knockdown of nfe2l2a [the zebrafish Nrf2 homolog which drives AR (57)] led to only a 20% suppression of GFP levels in these fish (Supplemental Fig. S8). Therefore, these data are consistent with our initial cell-based data (Fig. 2A, B) and confirm that Nrf2 activity suppression promoted by depletion of HuR or AUF1 is functionally relevant in a whole vertebrate animal.
HuR and AUF1 bind Nrf2-mRNA in cells
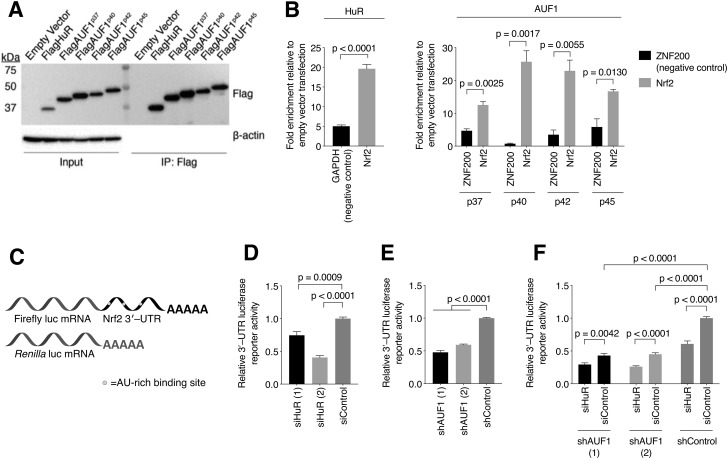
We next confirmed that HuR/AUF1 and Nrf2-mRNA interact. RIP experiments were performed using HEK293T cells ectopically expressing either Flag-HuR or Flag-AUF1 isoforms. Real-time qPCR analysis of eluted mRNA revealed that Nrf2 mRNA coeluted with both HuR and all AUF1 isoforms (Fig. 3A, B), confirming that both HuR and AUF1 bind to Nrf2-mRNA in cells. This interaction is further shown using purified recombinant proteins below.
Figure 3.
HuR and AUF1 bind directly to the 3′–UTR of Nrf2-mRNA in cells. A) RIP was carried out by expressing flag-tagged HuR or AUF1 in HEK293T cells and subjecting lysates to Flag IP. B) Nrf2-mRNA coeluting with enriched proteins was detected with real-time qPCR (mean ± sem of n = 4 independent replicates). C) 3′–UTR reporter system consisting of Nrf2-mRNA fused to a firefly luciferase transcript and normalized to Renilla luciferase. D, E) Knockdown of HuR and AUF1, respectively, reduces the 3′–UTR reporter activity in HEK293T cells (mean ± sem of n = 8 independent replicates for each bar). F) Knockdown of both proteins leads to a further suppression of the 3′–UTR reporter activity (mean ± sem of n = 4 independent replicates). See also Supplemental Fig. S7B. All P values were calculated with Student’s t test. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ZNF200, zinc finger protein 200.
HuR and AUF1 bind common sites in the Nrf2 3′–UTR with nanomolar affinities
The interaction of HuR and AUF1 with Nrf2-mRNA was further characterized in vitro by EMSA. Because HuR and AUF1 most commonly bind to sites within UTRs of target transcripts, 3 candidate binding sites within the 3′–UTR of Nrf2-mRNA were selected (Supplemental Fig. S9A and Supplemental Table S8) based on reported consensus binding motifs of HuR and AUF1 (34, 58). Both HuR and AUF1p37 bound most of these sites with nanomolar affinity (Supplemental Fig. S9B, C and Table 2), with the exception of Nrf2 site 2, which showed negligible binding to HuR. These affinities are on par with those previously reported for HuR (59) and AUF1 (60) to their target mRNA-binding sites, implying that the interactions with Nrf2-transcript we characterized are physiologically relevant (Supplemental Table S1).
TABLE 2.
Dissociation constants measured for HuR and AUF1p37 binding to Nrf2 3′–UTR sites
| Site | HuR Kd(app) (nM) | AUF1p37 Kd(app) (nM) |
|---|---|---|
| Nrf2 site 1 | 20 ± 5 | 6 ± 2 |
| Nrf2 site 2 | – | 25 ± 4 |
| Nrf2 site 3 | 75 ± 20 | 670 ± 82 |
Regulation depends on the 3′–UTR of Nrf2-mRNA
At this juncture, we hypothesized that suppression of Nrf2 activity in HuR-deficient cultured cells and larval fish stems is caused, at least in part, by loss of regulation at the 3′–UTR of Nrf2-mRNA. To test this hypothesis, we utilized a luciferase reporter in which the 3′–UTR of Nrf2 is fused to a firefly luciferase transcript (38). A constitutively expressed Renilla luciferase was used as an internal normalization control (Fig. 3C).
Cells deficient of HuR showed significantly reduced luciferase activity (30–60%) compared with controls (Fig. 3D). We found a similar degree (40–50%) of reporter suppression in shAUF1 cells (Fig. 3E). The magnitude of these effects on the 3′–UTR reporter (∼50%) is similar to the extent of Nrf2 activity suppression we observed in both HuR- (∼50%) and AUF1-knockdown cells (∼60%; Fig. 2A, B). Thus, regulation of the Nrf2 3′–UTR makes a significant contribution to the mechanism through which knockdown of AUF1 and HuR affects Nrf2 activity. Importantly, these findings further substantiate that the effects of HuR/AUF1 knockdown on Nrf2 activity occur specifically at the Nrf2-mRNA level, ruling out effects of HuR/AUF1 on protein-level regulation of Nrf2.
Simultaneous knockdown of AUF1 and HuR resulted in a further decrease in luciferase activity relative to AUF1 knockdown alone (Fig. 3F). Specifically, HuR knockdown suppressed 3′–UTR reporter levels by ∼45% regardless of the presence or absence of AUF1 (Supplemental Fig. S7B). Because this assay is a more direct readout of Nrf2-mRNA regulation by HuR than our Nrf2 activity reporter assay above (Fig. 2C and Supplemental Fig. S7A), we conclude that HuR and AUF1 act independently on Nrf2-mRNA.
Nrf2-mRNA stability is reduced by AUF1 knockdown but not HuR knockdown
Having established that HuR and AUF1 bind directly to Nrf2-mRNA and modulate Nrf2 activity, we sought to understand the specific mechanisms underlying this novel regulatory event. A common mode of HuR/AUF1 regulation involves stabilization or destabilization of the target upon binding (20).
We began by considering that HuR positively regulates Nrf2 transcript stability. Levels of endogenous, mature Nrf2-mRNA were reduced in shHuR cells (20–25% relative to shControl cells; Supplemental Fig. S10A). mRNA levels of the 3′–UTR reporter transcripts were also significantly suppressed by 55% in siHuR-treated cells (2) (Supplemental Fig. S10B), which featured a stronger suppression in the luciferase reporter assay than siHuR (1) (Fig. 3D). We expected that the half-life of Nrf2-mRNA in these cells would be similarly reduced. However, endogenous mature Nrf2 mRNA showed a nonsignificant (20% with 25% error) difference in half-life between shHuR and shControl cells [t1/2 = 98 min (k = 0.007 ± 0.002/min) and 112 min (k = 0.006 ± 0.0005/min), respectively] (Supplemental Fig. S10C), which, regardless of statistical significance, is not sufficient in magnitude to explain the 2-fold suppression in Nrf2 activity we consistently observed above.
Recent transcriptome-wide analysis of the stability of AUF1-targeted transcripts revealed the ability of AUF1 to positively regulate about 25% of target transcripts. Because our data showed that Nrf2 activity is suppressed in shAUF1 cells, we hypothesized that the stability of Nrf2-mRNA would be reduced in these cells. Consistent with this hypothesis, the half-life of Nrf2-mRNA was reduced by 20–60% upon knockdown of AUF1 (Supplemental Fig. S10D). These data also substantiate that our half-life measurements are sufficiently robust to reliably measure 2-fold changes in the stability of Nrf2-mRNA. The average suppression of half-life (50%) in shAUF1 cells is sufficient to explain the majority of the effect of AUF1 knockdown on Nrf2 activity. Thus, AUF1 positively regulates Nrf2 activity by stabilizing Nrf2-mRNA. Collectively, the mechanistic differences between the effect of AUF1 and HuR (which we elaborate on below) further point to independent effects of HuR and AUF1 on Nrf2 activity.
HuR enhances Nrf2 transcript splicing
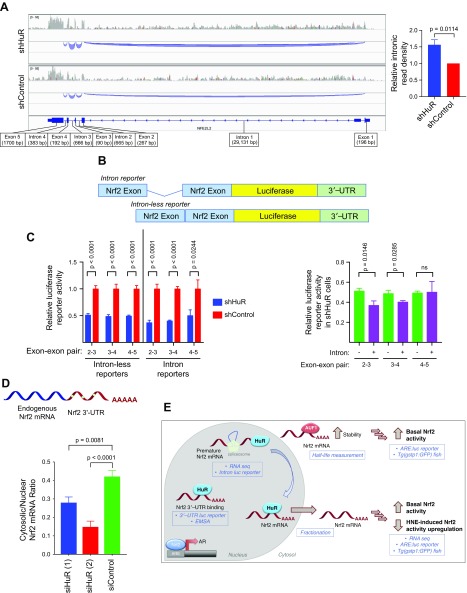
Having ruled out significant effects on Nrf2-mRNA stability upon HuR knockdown, we investigated other potential means by which HuR regulates Nrf2 activity. In addition to its classic role in stabilizing target mRNAs, HuR also regulates maturation of some of its targets (23). We reasoned that modulations in Nrf2-mRNA processing (e.g., splicing, translocation) could lead to decreased levels of mature Nrf2-mRNA and contribute to the suppression in Nrf2 activity we observed. Importantly, premature Nrf2-mRNA does not contribute to the pool of functional Nrf2-mRNA (competent for translation into Nrf2 protein) because it is not exported from the nucleus and cannot be translated to give a functional protein (Supplemental Fig. S11A).
We first turned to our RNA-seq data for evidence of misregulation of maturation. Because we prepared our RNA-seq library using rRNA depletion and deep-sequenced the resulting library, we were able to examine the levels of intronic RNA present in the Nrf2 transcript. Nrf2-mRNA is composed of 5 exons with 4 intervening introns. The first intron of Nrf2 is extremely long at ∼30,000 bp (Supplemental Fig. S11B), putting it in the longest 10% of human introns (61). When we examined the levels of intronic RNA in the Nrf2 transcript in shHuR and shControl cells, we found that Nrf2 transcripts in shHuR cells contained higher levels of intronic content (Fig. 4A). Thus, knockdown of HuR appears to suppress the maturation of Nrf2-mRNA, leading to an accumulation of unspliced, premature Nrf2-mRNA (Fig. 4A, inset).
Figure 4.
HuR knockdown suppresses Nrf2-mRNA splicing and nuclear export of Nrf2-mRNA. A) Integrative Genomics Viewer (71) view of splicing tracks for Nrf2 (NFE2L2) from RNA-seq analysis of shHuR and shControl HEK293T cells. The area of the blue tracks corresponds to the levels of intronic RNA detected. Shown is 1 representative replicate per cell line. Inset at right: areas of splicing tracks integrated with ImageJ (mean ± sem, n = 4 introns). See also Supplemental Fig. S11. B) Reporters to read out the effect of introns were constructed by fusing a portion of Nrf2-mRNA (with or without the intron) upstream of a firefly luciferase reporter. As with the 3′–UTR reporter (Fig. 3C), effects on this construct can be assayed by measuring luciferase activity in the lysates of cells expressing these reporters. C) Reporter levels upon HuR knockdown in nonstimulated HEK293T cells for both the intron-less reporter (left) and the intron reporter (right) (mean ± sem of n ≥ 7 per set). Inset at right: Comparison of reporter activity in shHuR cells (i.e., blue bars from the main plot in C upon introduction of introns. D) Real-time qPCR was used to measure the ratio of endogenous Nrf2-mRNA in nuclear and cytosolic extracts of HEK293T cells depleted of HuR [mean ± sem of n = 8 for siHuR (1, 2) and n = 7 for siControl]. E) Model of post-transcriptional regulation of Nrf2-mRNA by HuR and AUF1. HuR regulates Nrf2-mRNA maturation and nuclear export, and AUF1 stabilizes Nrf2-mRNA. Shown in blue text/boxes is the experimental evidence supporting each facet of this regulatory program. All P values were calculated with Student’s t test.
To probe this effect further and bolster our conclusions from the RNA-seq data, we utilized a firefly luciferase reporter with either an exon–intron–exon (intron reporter) sequence or an exon–exon (intronless reporter) sequence from Nrf2-mRNA fused upstream of the luciferase reporter. The Nrf2 3′–UTR was also retained downstream of the luciferase reporter. Reporters were constructed containing 3 different Nrf2 exon pairs, with or without the intervening introns (Fig. 4B). Regulation of these mRNA reporters can be assessed by measuring firefly luciferase enzyme activity in lysates of cells expressing these reporters (normalized to a constitutively expressed Renilla luciferase control). Because of the length of the first intron of Nrf2 (Supplemental Fig. S11B), this intron was not investigated in this assay.
Expression of the intronless reporters in shHuR cells led to a 50% decrease in reporter activity relative to shControl cells (Fig. 4C), which is consistent with our 3′–UTR luciferase reporter data above (Fig. 3D). Because this effect (∼50% reduction in shHuR cells) was similar across all 3 reporters containing different exons, it is likely attributable to the 3′–UTR (the common factor between the 3 reporters). However, 2 of the intron-containing reporters showed a further suppression upon HuR knockdown relative to their intronless counterparts (Fig. 4C, inset at right), which is consistent with our RNA-seq data (Fig. 4A). The greatest suppression effects were observed for introns 2 and 3, whereas intron 4 was not strongly suppressed in shHuR cells in either the RNA-seq or the reporter data.
Overall, these data agree with PAR-CLIP results indicating nucleotide conversions (i.e., HuR binding) within Nrf2 introns as well as the presence of multiple putative binding sites in the introns based on consensus HuR-binding sequences (17). These data collectively indicate that HuR enhances the splicing and maturation of Nrf2-mRNA. The magnitude of this effect is on par with the suppression of endogenous Nrf2-mRNA levels we observed above.
HuR regulates nuclear export of Nrf2-mRNA
Nuclear export of mature mRNA is intrinsically linked to the splicing/maturation process because maturation is a prerequisite for export. Coupling of these processes is an appreciated mechanism to enhance gene expression (62). mRNA that is not exported because of improper or incomplete splicing is rapidly degraded in the nucleus (63). Given the ability of HuR to shuttle between the nucleus and cytosol (21), we hypothesized that HuR governs Nrf2-mRNA export from the nucleus. Indeed, for some mRNA targets, such as COX-2, HuR plays a key role in regulating export from the nucleus (22). To test whether a similar regulatory program is at play for Nrf2-mRNA, we employed nuclear/cytosolic fractionation in shHuR/shControl cells coupled with qPCR detection of endogenous, mature Nrf2-mRNA. These experiments revealed that the cytosolic/nuclear ratio of endogenous Nrf2-mRNA decreased by 35% on average in HuR-deficient cells (Fig. 4D), indicating a decrease in Nrf2-mRNA nuclear export upon depletion of HuR. Faster turnover of nuclear mRNA may contribute to the reduction in overall levels of Nrf2-mRNA upon HuR knockdown (Supplemental Fig. S10A) and to the slight reduction in Nrf2-mRNA half-life we observed (Supplemental Fig. S10C). In sum, we conclude that the principal mechanisms by which HuR regulates Nrf2-mRNA are control of Nrf2-mRNA splicing/maturation and nuclear export of the mature transcript. Importantly, these results collectively point to a novel regulatory program that functions specifically at the Nrf2-mRNA level to modulate Nrf2 activity (Fig. 4E).
DISCUSSION
We began by exploring HuR-dependent global transcriptomic changes in cells stimulated with RES/ROS, using H2O2 and HNE as representative native ROS/RES. Contrary to previous reports of the sensitivity of HuR to oxidative stress (64), we found no transcriptional responses specifically attributable to HuR following H2O2 stimulation (Fig. 1B). Differences in cell type may explain this observed discrepancy, as we observed a muted transcriptional response to H2O2 stimulation overall in our HEK293T cell lines. Conversely, HNE-stimulation of HuR knockdown cells elicited a general down-regulation of global transcriptional activity compared with control cells (Fig. 1A and Supplemental Fig. S3). Given the important roles that pleiotropic covalent drugs and native RES play on Nrf2 signaling (2, 3), we homed in on differentially expressed Nrf2-driven genes. This analysis revealed an interesting bifurcation between HuR-dependent effects on Nrf2-driven genes in nonstimulated shHuR cells vs. HNE-stimulated cells. Nrf2-driven genes that were more strongly up-regulated in HNE-stimulated shHuR cells (ME-1, TXNRD1, and FTL) were not SDE in nonstimulated shHuR cells relative to shControl cells (Fig. 1A, C). However, a separate subset of Nrf2-driven genes (SLC2A3, INSIG1, and MGST1) was suppressed in nonstimulated shHuR cells (Table 1). Together with our Nrf2 activity reporter data (Fig. 1F, H), which was consistent with the majority of our RNA-seq analysis, these findings highlight context-specific complexities inherent in the regulation of the Nrf2 pathway by HuR that echo recent reports in the field (51). Because of these intricacies, we chose to focus our further investigation on nonstimulated cells.
For both HuR and AUF1, which we also identified as a novel regulator of Nrf2 activity, we found 2- to 3-fold suppression in Nrf2 activity in both HuR and AUF1 knockdown cells relative to control cells (Figs. 1F and 2A, B and Supplemental Fig. S5). These changes are indicative of significant regulatory events because we and others have previously documented the narrow dynamic range (2- to 4-fold) of Nrf2 activity modulation in various readouts, including luciferase reporter assays, flow cytometry–based analyses, as well as real-time qPCR and Western blot analyses that assay endogenous Nrf2-driven genes (37, 44, 65, 66). Consistent with this logic, knockdown of Nrf2 gave a similar fold change in Nrf2 activity to HuR knockdown. There was no further decrease in Nrf2 activity when HuR and Nrf2 were simultaneously knocked down, clearly showing that the fold change observed in Nrf2 activity is the maximum possible in the system, and that HuR functions by modulating Nrf2 activity. Two- to 3-fold changes are also typical magnitudes of response in other pathways regulated by electrophiles. For instance, knockdown of the electrophile sensor Pin1 elicits only about 30% decrease in viability upon treatment with HNE relative to knockdown control (67). Our findings that HuR knockdown induced Nrf2 activity suppression extends to whole zebrafish depleted of HuR or AUF1 (Fig. 2D, E) as well as to MEECs (Supplemental Fig. S5B) demonstrate a broader generality that is consistent with the fact that HuR and AUF1 are ubiquitously expressed. These novel general regulatory roles of HuR/AUF1 on Nrf2-mRNA stand in contrast with previously reported Nrf2-mRNA regulatory events by miRNAs that are highly cell/context specific (8). Thus, in terms of cell type generality and magnitude, our study goes a long way in demonstrating that Nrf2-mRNA regulation is an important modulator of Nrf2 activity.
HuR and AUF1 can function in concert to destabilize mRNAs, such as p16INK4 (54), and enhance the translation of mRNAs, such as TOP2A (34), among other examples. However, we found that HuR and AUF1 act independently on Nrf2-mRNA, despite similar binding affinities to common sites we identified within the 3′–UTR (Table 2). The distinct, orthogonal mechanisms by which HuR and AUF1 regulate Nrf2-mRNA—control of Nrf2-mRNA splicing and export as well as stabilization of Nrf2-mRNA, respectively—agree with our Nrf2 activity reporter data indicating independent effects of HuR and AUF1 on Nrf2/AR axis. These effects could not have been predicted based on the previously reported PAR-CLIP binding data alone. The complexities we uncovered in this system collectively speak to the need for careful mechanistic evaluation of on-target effects of HuR/AUF1 knockdown, particularly when common binding sites are involved.
As is appreciated in the mRBP field (17, 35), both HuR and AUF1 regulate their targets through multiple mechanisms. Although the magnitude of the UTR-reporter suppression (Fig. 3D) and nuclear-export suppression (Fig. 4D) we observed in HuR knockdown cells at first glance seem sufficient to explain the magnitude of Nrf2 activity suppression, our data testify that additional regulatory modes beyond UTR regulation are at play. For instance, using multiple approaches, we identified Nrf2-mRNA splicing as a subtle but significant component of HuR regulation of Nrf2-mRNA (Fig. 4A–C). These alternate/complementary mechanisms may explain why we unexpectedly observed a reduction in the global pool of Nrf2-mRNA without a similar fold change in the half-life of Nrf2-mRNA upon HuR-knockdown, for instance. Our mechanistic interrogations ultimately established that reduced splicing and reduced nuclear export of Nrf2-mRNA are the principal mechanisms by which Nrf2 activity is suppressed upon HuR-knockdown. These mechanisms are likely linked as splicing is generally a prerequisite for nuclear export (62), although the details of this mechanistic coupling for Nrf2-mRNA remain to be explored. Nevertheless, down-regulation of both splicing/maturation and nuclear export of Nrf2-mRNA upon HuR-knockdown likely explains why we observed a suppression in the pool of mature Nrf2-mRNA (Supplemental Fig. S10A). Overall, the mechanisms we identified strongly and clearly point to on-target, Nrf2-mRNA–specific events that support HuR/AUF1 regulation of Nrf2 signaling.
In sum, these data highlight an important intersection between proven disease-relevant players: HuR, AUF1, and Nrf2 are all up-regulated in cancers (68, 69); therapeutic targeting of Nrf2/AR axis through exploitation of protein regulators, such as Keap1, continues to be a promising small-molecule intervention (7, 70). The ubiquitous expression of HuR and AUF1, along with the conservation of their regulation of Nrf2 activity across multiple cell types and whole organisms underscore the importance of this newly identified regulatory program. Because this regulation of Nrf2/AR occurs specifically at the mRNA level, it potentially offers an orthogonal therapeutic strategy to modulate this conserved pathway of validated pharmacological significance.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Jen Grenier [Cornell RNA Sequencing Core (RSC) Cornell University] for assistance with RNA sequencing. The authors also thank Alexandra Van Hall-Beauvais and Gene Hu (Cornell University) for assistance with initial studies of HuR. The following organizations have supported this study: American Heart Association predoctoral fellowship (17PRE33670395 to J.R.P.); Swiss National Science Foundation (SNSF); Swiss Federal Institute of Technology Lausanne (EPFL); Novartis Medical-Biological Research Foundation; National Centre of Competence in Research Chemical Biology (NCCR) Chemical Biology; and U.S. National Institutes of Health (NIH) Director’s New Innovator (Office of the Director, 1DP2GM114850 to Y.A.). The authors declare no conflicts of interest.
Glossary
- βME
β-mercaptoethanol
- AR
antioxidant response
- BSA
bovine serum albumin
- COX-2
prostaglandin-endoperoxidase synthase 2
- EIF4ENIF1
eukaryotic translation initiation factor 4E nuclear import factor 1
- EMSA
electrophoretic mobility shift assay
- FAS
Fas cell surface death receptor
- FBS
fetal bovine serum
- FTL
ferritin light chain
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescence protein
- HNE
4-hydroxynonenal
- INSIG1
insulin induced gene 1
- Keap1
Kelch-like ECH associated protein 1
- LPS
lipopolysaccharide
- ME-1
malic enzyme 1
- MEEC
mouse embryonic endothelial cell
- MEM
Minimal Essential Medium
- MGST1
microsomal glutathione S-transferase 1
- miRNA
microRNA
- MO
morpholino oligonucleotide
- mRBP
mRNA-binding protein
- Nrf2
nuclear factor erythroid 2–related factor 2
- PAR-CLIP
photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation
- PGA2
prostaglandin A2
- POLA1
DNA polymerase alpha 1
- qPCR
quantitative PCR
- RES
reactive electrophilic species
- RIP
RNA immunoprecipitation
- RNA-seq
RNA-sequencing
- ROS
reactive oxidative species
- SDE
significantly differentially expressed
- sh
short hairpin
- si
small interfering
- SLC2A3
solute carrier family 2 member 3
- TCEP
Tris(2-carboxyethyl)phosphine
- TOP2A
DNA topoisomerase II alpha
- TXNRD1
thioredoxin reductase 1
- UTR
untranslated region
- ZNF200
zinc finger protein 200
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
J. R. Poganik was responsible for the investigation, formal analysis, writing (initial draft and later editing), and visualization; M. J. C. Long was responsible for the investigation (initial short hairpin RNA experiments, microinjection of zebrafish embryos), formal analysis, and writing (critical review and editing); M. T. Disare was responsible for the investigation (assisted with cloning plasmids and protein purification); X. Liu provided resources [synthesized HNE(alkyne)]; S.-H. Chang provided resources (MEEC cells); T. Hla provided resources and discussions of HuR biology; Y. Aye was responsible for the conceptualization, writing, supervision, project administration, and funding acquisition; and all authors assisted with final proofing of the manuscript.
REFERENCES
- 1.Ma Q. (2013) Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 53, 401–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes J. D., Dinkova-Kostova A. T. (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218 [DOI] [PubMed] [Google Scholar]
- 3.Parvez S., Long M. J. C., Poganik J. R., Aye Y. (2018) Redox signaling by reactive electrophiles and oxidants. Chem. Rev. 118, 8798–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taguchi K., Motohashi H., Yamamoto M. (2011) Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123–140 [DOI] [PubMed] [Google Scholar]
- 5.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J. D. (2013) Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32, 3765–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nioi P., Nguyen T. (2007) A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun. 362, 816–821 [DOI] [PubMed] [Google Scholar]
- 7.Copple I. M. (2012) The Keap1-Nrf2 cell defense pathway--a promising therapeutic target? Adv. Pharmacol. 63, 43–79 [DOI] [PubMed] [Google Scholar]
- 8.Cheng X., Ku C.-H., Siow R. C. M. (2013) Regulation of the Nrf2 antioxidant pathway by microRNAs: new players in micromanaging redox homeostasis. Free Radic. Biol. Med. 64, 4–11 [DOI] [PubMed] [Google Scholar]
- 9.Strobel E. J., Watters K. E., Loughrey D., Lucks J. B. (2016) RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr. Opin. Biotechnol. 39, 182–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cléry A., Blatter M., Allain F. H. (2008) RNA recognition motifs: boring? not quite. Curr. Opin. Struct. Biol. 18, 290–298 [DOI] [PubMed] [Google Scholar]
- 11.Roundtree I. A., Evans M. E., Pan T., He C. (2017) Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Von Roretz C., Di Marco S., Mazroui R., Gallouzi I. E. (2011) Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA 2, 336–347 [DOI] [PubMed] [Google Scholar]
- 13.Simone L. E., Keene J. D. (2013) Mechanisms coordinating ELAV/Hu mRNA regulons. Curr. Opin. Genet. Dev. 23, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S. H., Hla T. (2011) Gene regulation by RNA binding proteins and microRNAs in angiogenesis. Trends Mol. Med. 17, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitzer J., Hafner M., Landthaler M., Ascano M., Farazi T., Wardle G., Nusbaum J., Khorshid M., Burger L., Zavolan M., Tuschl T. (2014) PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 539, 113–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T., Xiao G., Chu Y., Zhang M. Q., Corey D. R., Xie Y. (2015) Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res. 43, 5263–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N. (2011) Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352 [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee N., Corcoran D. L., Nusbaum J. D., Reid D. W., Georgiev S., Hafner M., Ascano M., Jr., Tuschl T., Ohler U., Keene J. D. (2011) Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J. L., Gorospe M. (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23, 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan C. M., Steitz J. A. (2001) HuR and mRNA stability. Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X. C., Steitz J. A. (1998) HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doller A., Akool el-S., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2008) Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 28, 2608–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izquierdo J. M. (2008) Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 283, 19077–19084 [DOI] [PubMed] [Google Scholar]
- 24.Chang S. H., Elemento O., Zhang J., Zhuang Z. W., Simons M., Hla T. (2014) ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 111, 18309–18314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srikantan S., Gorospe M. (2012) HuR function in disease. Front. Biosci. 17, 189–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meisner N. C., Hintersteiner M., Mueller K., Bauer R., Seifert J. M., Naegeli H. U., Ottl J., Oberer L., Guenat C., Moss S., Harrer N., Woisetschlaeger M., Buehler C., Uhl V., Auer M. (2007) Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 3, 508–515 [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Bhattacharya A., Ivanov D. N. (2015) Identification of small-molecule inhibitors of the HuR/RNA interaction using a fluorescence polarization screening assay followed by NMR validation. PLoS One 10, e0138780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X., Lan L., Wilson D. M., Marquez R. T., Tsao W. C., Gao P., Roy A., Turner B. A., McDonald P., Tunge J. A., Rogers S. A., Dixon D. A., Aubé J., Xu L. (2015) Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol. 10, 1476–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muralidharan R., Mehta M., Ahmed R., Roy S., Xu L., Aubé J., Chen A., Zhao Y. D., Herman T., Ramesh R., Munshi A. (2017) HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci. Rep. 7, 9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X., Wang W., Fan J., Lal A., Yang D., Cheng H., Gorospe M. (2004) Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J. Biol. Chem. 279, 49298–49306 [DOI] [PubMed] [Google Scholar]
- 32.Lin S., Wang W., Wilson G. M., Yang X., Brewer G., Holbrook N. J., Gorospe M. (2000) Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 20, 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cok S. J., Acton S. J., Sexton A. E., Morrison A. R. (2004) Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 279, 8196–8205 [DOI] [PubMed] [Google Scholar]
- 34.Yoon J. H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K. M., Noh J. H., White E. J., Martindale J. L., Yang X., Kang M. J., Wood W. H., III, Noren Hooten N., Evans M. K., Becker K. G., Tripathi V., Prasanth K. V., Wilson G. M., Tuschl T., Ingolia N. T., Hafner M., Gorospe M. (2014) PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 5, 5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gratacós F. M., Brewer G. (2010) The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 1, 457–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panda A. C., Abdelmohsen K., Yoon J. H., Martindale J. L., Yang X., Curtis J., Mercken E. M., Chenette D. M., Zhang Y., Schneider R. J., Becker K. G., de Cabo R., Gorospe M. (2014) RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol. Cell. Biol. 34, 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long M. J., Lin H. Y., Parvez S., Zhao Y., Poganik J. R., Huang P., Aye Y. (2017) β-TrCP1 is a vacillatory regulator of Wnt signaling. Cell Chem. Biol. 24, 944–957.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M., Yao Y., Eades G., Zhang Y., Zhou Q. (2011) MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res. Treat. 129, 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17, 10–12 [Google Scholar]
- 40.Langmead B., Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parvez S., Long M. J., Lin H. Y., Zhao Y., Haegele J. A., Pham V. N., Lee D. K., Aye Y. (2016) T-REX on-demand redox targeting in live cells. Nat. Protoc. 11, 2328–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parvez S., Fu Y., Li J., Long M. J., Lin H. Y., Lee D. K., Hu G. S., Aye Y. (2015) Substoichiometric hydroxynonenylation of a single protein recapitulates whole-cell-stimulated antioxidant response. J. Am. Chem. Soc. 137, 10–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenenbaum S. A., Lager P. J., Carson C. C., Keene J. D. (2002) Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26, 191–198 [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Zhu W., Levy D. E. (2006) Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods 39, 356–362 [DOI] [PubMed] [Google Scholar]
- 47.Sorrells S., Toruno C., Stewart R. A., Jette C. (2013) Analysis of apoptosis in zebrafish embryos by whole-mount immunofluorescence to detect activated Caspase 3. J. Vis. Exp. 82, e51060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellman L. M., Fried M. G. (2007) Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schopfer F. J., Cipollina C., Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs A. T., Marnett L. J. (2010) Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 43, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chorley B. N., Campbell M. R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S. R., Bell D. A. (2012) Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 40, 7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau A., Tian W., Whitman S. A., Zhang D. D. (2013) The predicted molecular weight of Nrf2: it is what it is not. Antioxid. Redox Signal. 18, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuadrado A., Rojo A. I., Wells G., Hayes J. D., Cousin S. P., Rumsey W. L., Attucks O. C., Franklin S., Levonen A.-L., Kensler T. W., Dinkova-Kostova A. T. (2019) Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 18, 295–317 [DOI] [PubMed] [Google Scholar]
- 54.Chang N., Yi J., Guo G., Liu X., Shang Y., Tong T., Cui Q., Zhan M., Gorospe M., Wang W. (2010) HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell. Biol. 30, 3875–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsujita T., Li L., Nakajima H., Iwamoto N., Nakajima-Takagi Y., Ohashi K., Kawakami K., Kumagai Y., Freeman B. A., Yamamoto M., Kobayashi M. (2011) Nitro-fatty acids and cyclopentenone prostaglandins share strategies to activate the Keap1-Nrf2 system: a study using green fluorescent protein transgenic zebrafish. Genes Cells 16, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X., Lu Y.-C., Dai K., Torregroza I., Hla T., Evans T. (2014) Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood 123, 1384–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. (2009) The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 29, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker A., Epis M. R., Porter C. J., Hopkins B. R., Wilce M. C., Wilce J. A., Giles K. M., Leedman P. J. (2012) Sequence requirements for RNA binding by HuR and AUF1. J. Biochem. 151, 423–437 [DOI] [PubMed] [Google Scholar]
- 59.Sengupta S., Jang B. C., Wu M. T., Paik J. H., Furneaux H., Hla T. (2003) The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 278, 25227–25233 [DOI] [PubMed] [Google Scholar]
- 60.DeMaria C. T., Sun Y., Long L., Wagner B. J., Brewer G. (1997) Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J. Biol. Chem. 272, 27635–27643 [DOI] [PubMed] [Google Scholar]
- 61.Sakharkar M. K., Chow V. T., Kangueane P. (2004) Distributions of exons and introns in the human genome. In Silico Biol. (Gedrukt) 4, 387–393 [PubMed] [Google Scholar]
- 62.Valencia P., Dias A. P., Reed R. (2008) Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 105, 3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore M. (2002) Nuclear RNA turnover. Cell 108, 431–434 [DOI] [PubMed] [Google Scholar]
- 64.Mehta M., Basalingappa K., Griffith J. N., Andrade D., Babu A., Amreddy N., Muralidharan R., Gorospe M., Herman T., Ding W.-Q., Ramesh R., Munshi A. (2016) HuR silencing elicits oxidative stress and DNA damage and sensitizes human triple-negative breast cancer cells to radiotherapy. Oncotarget 7, 64820–64835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Y., Li W., Kong A.-N. T. (2012) Anti-oxidative stress regulator NF-E2-related factor 2 mediates the adaptive induction of antioxidant and detoxifying enzymes by lipid peroxidation metabolite 4-hydroxynonenal. Cell Biosci. 2, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levonen A.-L., Landar A., Ramachandran A., Ceaser E. K., Dickinson D. A., Zanoni G., Morrow J. D., Darley-Usmar V. M. (2004) Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 378, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aluise C. D., Rose K., Boiani M., Reyzer M. L., Manna J. D., Tallman K., Porter N. A., Marnett L. J. (2013) Peptidyl-prolyl cis/trans-isomerase A1 (Pin1) is a target for modification by lipid electrophiles. Chem. Res. Toxicol. 26, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdelmohsen K., Gorospe M. (2010) Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menegon S., Columbano A., Giordano S. (2016) The dual roles of NRF2 in cancer. Trends Mol. Med. 22, 578–593 [DOI] [PubMed] [Google Scholar]
- 70.Hur W., Gray N. S. (2011) Small molecule modulators of antioxidant response pathway. Curr. Opin. Chem. Biol. 15, 162–173 [DOI] [PubMed] [Google Scholar]
- 71.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.