Abstract
Objective
To implement an open-source tool that performs deterministic privacy-preserving record linkage (RL) in a real-world setting within a large research network.
Materials and Methods
We learned 2 efficient deterministic linkage rules using publicly available voter registration data. We then validated the 2 rules’ performance with 2 manually curated gold-standard datasets linking electronic health records and claims data from 2 sources. We developed an open-source Python-based tool—OneFL Deduper—that (1) creates seeded hash codes of combinations of patients’ quasi-identifiers using a cryptographic one-way hash function to achieve privacy protection and (2) links and deduplicates patient records using a central broker through matching of hash codes with a high precision and reasonable recall.
Results
We deployed the OneFl Deduper (https://github.com/ufbmi/onefl-deduper) in the OneFlorida, a state-based clinical research network as part of the national Patient-Centered Clinical Research Network (PCORnet). Using the gold-standard datasets, we achieved a precision of 97.25∼99.7% and a recall of 75.5%. With the tool, we deduplicated ∼3.5 million (out of ∼15 million) records down to 1.7 million unique patients across 6 health care partners and the Florida Medicaid program. We demonstrated the benefits of RL through examining different disease profiles of the linked cohorts.
Conclusions
Many factors including privacy risk considerations, policies and regulations, data availability and quality, and computing resources, can impact how a RL solution is constructed in a real-world setting. Nevertheless, RL is a significant task in improving the data quality in a network so that we can draw reliable scientific discoveries from these massive data resources.
Keywords: privacy-preserving record linkage, clinical research network, PCORnet
INTRODUCTION
The last few years have witnessed an increasing number of clinical research networks (CRNs), curating and using immense collections of electronic health records (EHRs) and administrative claims data. One prominent example is the national Patient-Centered Clinical Research Network (PCORnet) funded by Patient-Centered Outcomes Research Institute (PCORI).1,2 There are 9 PCORnet Clinical Research Data Networks (CDRNs) and 20 Patient-Powered Research Networks aiming to facilitate nationwide pragmatic clinical trials and comparative effectiveness studies. Each CDRN has a partnership with contributing health care systems, clinical practices, government agencies, third party payers, and academic institutions. For example, the OneFlorida Clinical Research Consortium (CRC), one of the 9 PCORnet CDRNs, includes 10 unique health care organizations (HCOs) that provide care for approximately 48% of Floridians through 4100 physicians, 914 clinical practices, and 22 hospitals area covering all 67 Florida counties.3 The centerpiece of OneFlorida is its Data Trust, a centralized data repository that contains longitudinal and robust patient-level records of ∼15 million Floridians from various sources, including Medicaid and Medicare programs, cancer registries, vital statistics, and EHR systems from its clinical partners. The amount of individual-level patient data collected by each CDRN is staggering. A recent estimate shows that PCORnet has data on more than ∼100 million patients nationwide.4
However, different data records of the same patient can come from different sources. For example, EHRs from providers and claims data from payers both have records on the same patient. Further, the same patient can seek care in different HCOs in the network. Thus, linking and resolving duplicates in a CRN is a significant task in improving the quality of its data resources. A recent study shows that the rate of duplicate records is high in EHR systems ranging from 0.16% to 15.47%.5 Considering a CRN like OneFlorida that involves 10 unique HCOs with ∼15 million patients, as well inclusive of public payer data such as Florida Medicaid, we potentially have more than 2.25 million duplicated patient records across the network.
Entity resolution/record linkage (ER/RL)—the process of finding non-identical duplicates and merging the duplicates into a single tuple (record)—is an information integration problem given that the same “real-world entities” (eg, patients) are often referred to in different ways in multiple data sources. Finding related patient records and creating links among them is an important task in a CRN. Without easy access to ER/RL methods that create linked datasets, the innovative use of sharing large datasets in a CRN for tasks such as cohort discovery will be limited. For example, duplicated patient records may over-represent key clinical features in cohort discovery results such as the number of affected patients, the severity of a disease, or the extent of a treatment. Nevertheless, linking patient records in a CRN is not a trivial task balancing among privacy protection needs, linking efficiency, and many other practical considerations such as partners’ business. The information used for efficient ER/RL always contains sensitive personal identifiers including names, social security number (SSN), addresses, and health care beneficiary numbers. A privacy-preserving solution is highly desired.
Privacy-preserving record linkage (PPRL) solutions or RL in general have a variety of options in terms of system architectures, matching algorithms, optimization strategies, and data management and transfer procedures. Although commercial RL systems do exist (eg, Health Data Link6) privacy risk considerations and regulations are different in the context of health research data use and across the different HCOs in a CRN. Discrepancies in HCOs’ policies, computing resources, data availability, and data quality can further impact how a RL solution is constructed in a CRN.
In this article, we describe our experience in a real-world design and implementation of an open-source software tool—OneFL Deduper—that performs deterministic PPRL across the OneFlorida network.
MATERIALS AND METHODS
Setting and data sources
The OneFlorida Data Trust integrated various data sources from contributing organizations in the OneFlorida CRC including 10 unique HCOs: (1) 2 academic health centers (ie, University of Florida Health, UFHealth, and University of Miami Health System, UHealth), (2) 7 healthcare systems including Tallahassee Memorial Healthcare (TMH affiliated with Florida State University), Orlando Health (ORH), Adventist Health (AH, formerly known as Florida Hospital), Nicklaus Children’s Hospital (NCH, formerly known as Miami Children’s Hospital), Bond Community Health (BCH), Capital Health Plan (CHP), and Health Choice Network (HCN), and (3) CommunityHealth IT—a rural health network in Florida. In addition, we also obtained claims data from the Florida Medicaid (FLM) program. The Data Trust contains only a limited data set under the Health Insurance Portability and Accountability Act (HIPAA) and follows the PCORnet Common Data Model (CDM) v4.17 including patient demographics, enrollment status, vital signs, conditions, encounters, diagnoses, procedures, prescribing (ie, provider orders for medications), dispensing (ie, outpatient pharmacy dispensing), and lab results. Seven HCOs (UFHealth, UHealth, TMH, ORH, AH, NCH, and BCH) contributed EHRs, while CHP and FLM contributed claims data. The scale of the data is ever growing with over 450 million encounters, 900 million diagnoses, 1 billion prescribing records, and 1.1 billion procedures as of November 2018.8 As of the writing of this article, we have linked patients across 6 EHR sources (ie, UFHealth, UHealth, TMH, ORH, AH, and NCH), the entire FLM, and the tumor registry data (ie, linked using a different process) from UFHealth and ORH.
A pilot study for developing a rule-based privacy-preserving record linkage method
Previously, a PPRL solution9 was implemented at the Chicago Area Patient-Centered Outcomes Research Network (CAPriCORN),5 one of the 9 PCORI-funded CDRNs similar to our OneFlorida consortium. In their solution, Kho et al used 4 linkage rules to determine whether 2 records from different data sites are considered to “match”: (1) first name + last name + date of birth, (2) date of birth + SSN, (3) last name + SSN, or (4) 3 letter first name + 3 letter last name + soundex first name + soundex last name + date of birth + SSN. They then created hashes of these 4 combinations of patient identifiers using a cryptographic hash function.9
We adopted the PPRL approach proposed by Kho et al but developed new linkage rules because of the policies and data availability across OneFlorida partners, which constrained us to use only quasi-identifiers such as names, date of birth, sex, and race among others, rather than direct identifiers such as SSN. Previously, Kuzu et al used the North Carolina voter registration dataset to validate their RL method.10 Thus, we used the Florida Voter Registration System (FVRS) data11 to determine our linkage rules. The FVRS data is a public resource that contains voter identification information including names, date of birth, sex, race, mailing address, phone number, and email address among others, which are very similar to the set of patient demographic attributes available in EHR systems. Each voter in FVRS is assigned with a unique voter ID that is consistent across years and can be used to establish a gold-standard linked dataset. Even though individual quasi-identifiers are not unique, combinations of these quasi-identifiers can uniquely identify a patient. Thus, we examined different combinations of the quasi-identifiers available in the FVRS dataset to discover linkage rules that will be accurate in identifying unique patients.
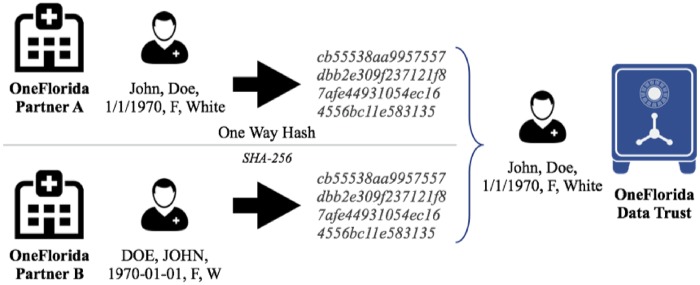
Based on the linkage rules discovered using FVRS, we conducted a pilot study to link and deduplicate patient records between UFHealth and FLM pediatric data. In the pilot study, we validated the linkage rules using random subsets of UFHealth and FLM data, where we manually reviewed the extracted records to identify false positives and false negatives based on additional identifiable information of the patients including direct identifiers such as SSN and Medicaid beneficiary number. We obtained approval from the UF Institutional Review Board (IRB) to access SSNs for a small number of patient records. We consider 2 records (A and B) in 2 different data sources to be about the same patient if one or more combinations of the quasi-identifiers (ie, linkage rules) have the same corresponding values (eg, A.name + A.dob + A.race = B.name + B.dob + B.race) and these combinations can uniquely identify the patient in the datasets. Thus, the problem of linking patient data is transformed into equivalence tests based on these linkage rules, as shown in Figure 1. We evaluated the performance of the linkage rules using standard information retrieval metrics in terms of precision and recall. In the context of RL, a true positive (TP) is where 2 patient records do truly belong to the same patient; a false positive (FP) is where 2 patient records do not belong to the same patient, however, the linkage rules indicated that they are; and a false negative (FN) is where the quasi-identifiers do not match, however, the 2 patient records do belong to the same patient. Further, a true negative (TN) is where 2 patient records do not belong to the same patient and it is consistent with the linkage rule results. Nevertheless, TNs are not necessary in calculating the precision (ie, TP/(TP+FP)) and recall (ie, TP/(TP + FN)) metrics.
Figure 1.
A deterministic record linkage process.
An open-source rule-based privacy-preserving record linkage tool—OneFL deduper
Based on the pilot study, we created OneFL Deduper, an open source (available at https://github.com/ufbmi/onefl-deduper) PPRL tool developed at the University of Florida (UF). The tool was implemented in Python 3. The RL process is split into 2 steps: (1) a hasher, and (2) a linker. The hasher is run by each individual partner at their sites locally and uses the 2 linkage rules (ie, R1: first name + last name + date of birth + sex; and R2: first name + last name + date of birth + race) we developed through the pilot studies above to generate a pair of unique hashes (one for each rule). The individual quasi-identifiers (ie, first name, last name, date of birth, sex, and race) are normalized into a consistent format before generating the hashes. Cryptographic hash functions such as SHA256 are one-way functions, where minor differences in the input string (eg, “Joe” vs. “joe” in the name) will result in different hash values. Normalizing input data is thus necessary to ensure that the hashes are consistently generated across the data sources. We made a few efforts to standardize the input data.
All textural input fields are converted to lower case.
Special characters (eg, periods, hyphens, and spaces) in the name fields are removed.
The date of birth field is standardized and formatted as “year-month-date” (eg, “1982-12-17”).
The sex and race fields are transformed into the standardized PCORnet CDM representation of the values (eg, “male” is set to “m”).
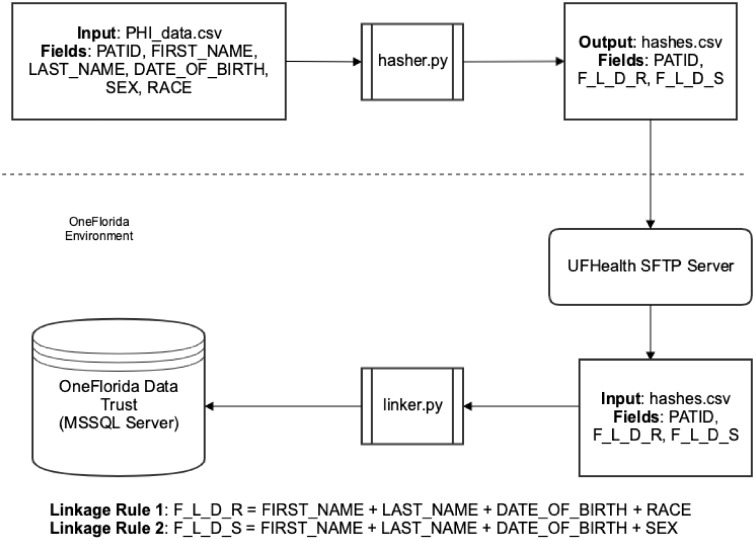
As shown in Figure 2, the hasher takes a csv file with clear-text PHI, where each patient record is uniquely identified with a local patient identifier (ie, a local PATID specific to the data partner); and then generates a unique hash for each linkage rule using a salted SHA256 algorithm. The hasher outputs a new csv file that contains the local PATIDs and the hash values of R1 and R2. Each data partner in OneFlorida then securely transmits the output hash csv file to the OneFlorida data coordinating center (ie, UF) using the Secure File Transfer Protocol (SFTP).
Figure 2.
The record linkage workflow of the OneFL Deduper tool.
The OneFlorida data coordinating center processes the received hash csv files using the linker tool in the OneFL Deduper. The linker process begins by comparing the hash values of each rule in the incoming csv hash file to those that were already received, processed, and stored in the database. The linker also generates a universally unique identified (UUID) using the uuid1() function in Python (ie, a RFC 4122 compliant UUID generator) for each new patient record (ie, any record that cannot be linked to an existing patient record in the database).
The reconciliation process for patient demographics
Once the patient records from the data partners are linked, we are left with multiple demographic information of the same patient, which are often inconsistent across data sources. Thus, we implemented a reconciliation process to obtain the most accurate and complete demographic information from the multiple data partners. The reconciliation process begins by collecting the most recent encounter for that patient from each partner. For each demographic variable (ie, sex, sexual orientation, gender identify, race, ethnicity, and patient’s preferred spoken language), we look for the most complete and recent data across the different sources: (1) use known values over incomplete (ie, unknown, no information, and NULL) and (2) use the most recent (ie, based on the most recent encounter date) value if both sources have known values. Taking “race” as an example, if the race is unknown in UFHealth but known in FLM (eg, “Asian”), the value from FLM would be selected; if the race is populated with known values in both UFHealth and FLM, we would choose the race from the partner who has the most recent encounter of that patient.
Using the developed OneFL Deduper tool, we first linked the entire UFHealth and FLM pediatric data in July 2017. In subsequent data refresh cycles, we linked ORH and UHealth in November 2017 and added MCH, FLH, and TMH in July 2018.
RESULTS
Linkage rules learned from the Florida voter registration data and the validation study
We adopted a deterministic (or rule-based) RL approach; thus, we first used 2 snapshots of the FVRS data from 2015 and 2016 (ie, January 2015 and January 2016) to discover the best performing linkage rules in terms of precision and recall. Because of privacy risk considerations, we did not use any direct identifiers such as SSN. Instead, we aimed to find combinations of quasi-identifiers (ie, first name, last name, middle name, date of birth, gender, race, residence address city, and zip code) that can uniquely identify an individual based on the prevalence of these data elements in EHRs.12 Further, we favored precision over recall—minimizing false positive rate, while accepting a reasonable number of false negatives. More details about the learning process can be found in Supplementary Material. Based on the FVRS data, we found that the combinations of (1) R1: first name + last name + date of birth + sex had a duplication rate of 0.04%; and (2) R2: first name + last name + date of birth + race, had a duplication rate of 0.03%, the lowest among all the rules that we explored.
We validated these 2 linkage rules first using a random subset of UFHealth Medicaid patients (ie, 2511 patient records that were linked using one of the two rules between UFHealth and FLM data). We manually reviewed these records to identify false positives (ie, 2 records that do not refer to the same patient but linked inaccurately) based on additional identifiable information such as addresses, phone numbers, and direct identifiers such as SSN and Medicaid beneficiary number. Out of the 2511 records we found that there were 69 false positives (ie, a precision of 97.25%).
In a follow-up study, we extracted another 2 random subsets of the data from UFHealth Medicaid, where 1000 records were matched using one of the two rules, while another 1000 records were matched based on SSN and Medicaid IDs. Out of the 1000 individuals that were matched based on the 2 linkage rules, 7 were false positives (ie, a precision of 99.3%). Out of the 1000 records that were matched based on SSN and Medicaid IDs, 245 were false negatives (ie, a recall of 75.5%).
Analyses of the linked patient cohorts
We used the validated Deduper tool to link and deduplicate patient records across 7 data partners (ie, 6 health care systems including UFHealth, UHealth, ORH, TMH, AH, and NCH and the Florida Medicaid program, FLM). Further, TMH has 2 different EHR systems—TMA for inpatient and TMC for outpatient—where the same patient is not linked across the 2 systems. Thus, we considered TMH as 2 different data sources. Table 1 shows the demographics of the patient records in OneFlorida for the sites that we have linked.
Table 1.
Demographics of patients in OneFlorida (OneFlorida Overall vs. Florida Medicaid vs. OneFlorida EHR vs. Linked)a
| Characteristic | OneFlorida overallb, N = 13 550 611 (100%) | Florida Medicaid, N = 6 306 397 (100%) | OneFlorida EHR, N = 5 544 189 (100%) | Linked, N = 1 700 025 (100%) |
|---|---|---|---|---|
| Age | ||||
| <18 | 4 896 640 (36%) | 2 459 613 (39%) | 1 834 676 (33%) | 602 351 (35%) |
| 18–44 | 4 299 368 (32%) | 2 226 040 (35%) | 1 488 685 (27%) | 584 643 (34%) |
| 45–64 | 2 221 412 (16%) | 764 278 (12%) | 1 186 108 (21%) | 271 026 (16%) |
| 65–84 | 1 682 608 (12%) | 638 952 (10%) | 848 156 (15%) | 195 500 (11%) |
| >85 | 450 583 (3%) | 217 514 (3%) | 186 564 (3%) | 46 505 (3%) |
| Unknown | 345 837 (2%) | 66 (0%) | 345 771 (2%) | Not Applicable |
| Gender | ||||
| Male | 5 625 002 (42%) | 2 777 510 (44%) | 2 113 766 (38%) | 733 726 (43%) |
| Female | 6 919 184 (51%) | 3 527 658 (56%) | 2 425 229 (44%) | 966 297 (57%) |
| Unknown | 670 191 (5%) | 1295 (0%) | 668 892 (12%) | BTb (0%) |
| Race | ||||
| American Indian or Alaska Native | 26 154 (0%) | 15 216 (0%) | 6961 (0%) | 3977 (0%) |
| Asian | 194 358 (1%) | 85 665 (1%) | 84 194 (2%) | 24 499 (1%) |
| Black or African American | 2 441 710 (18%) | 1 382 871 (22%) | 556 772 (10%) | 502 067 (30%) |
| Native Hawaiian or Other Pacific Islander | 7037 (0%) | BTc (0%) | 5102 (0%) | 1934 (0%) |
| White | 5 685 296 (42%) | 2 073 237 (33%) | 2 524 512 (46%) | 1 087 547 (64%) |
| Multiple race | 6819 (0%) | NAd (0%) | 3959 (0%) | 2860 (0%) |
| Unknown | 1 767 273 (13%) | 365 185 (6%) | 1 391 807 (25%) | 10 281 (1%) |
| Other | 3 057 785 (23%) | 2 384 288 (38%) | 610 242 (11%) | 63 255 (4%) |
| Ethnicity | ||||
| Non-Hispanic | 7 352 643 (54%) | 3 456 108 (55%) | 2 714 255 (49%) | 1 182 280 (70%) |
| Hispanic | 3 360 818 (25%) | 1 905 626 (30%) | 986 277 (18%) | 468 915 (28%) |
| Unknown | 1 885 808 (14%) | 365 185 (6%) | 1 484 555 (27%) | 36 068 (2%) |
Data as of July 26, 2018 including data from January 2012 to March 2018.
Only patients from the 7 partners who were involved in the record linkage process are counted. The total number of patients including those from partners that were not involved in the initial record linkage process is 16 974 878.
BT: below threshold (n < 11).
There is no multiple race option in Medicaid data.
Before deduplication, OneFlorida contained 16 974 878 (including patients from partners that were not involved in this initial RL process) patient records. The linkage and deduplication process reduced the population by 10.79%, to 15 143 179 patients. A total of 1 700 025 patients have data in more than 2 sources; and 1 543 317 patient records were found in 2 different sources, 148 671 in 3 different sources, 7760 in 4 different sources, 260 in 5 sources, 18 in 6 sources, and 1 patient record existed across 7 sources. As shown in Table 2, excluding the Florida Medicaid data, ORH and AH have the biggest overlaps (ie, 202 502 patients) due to their proximity. Both of these clinical partners are in the Orlando, Florida area. Patient records from TMC and TMA have the next highest overlaps (ie, 117 119 patients) as they are the inpatient and outpatient data feeds, respectively, from the same health care system (ie, TMH).
Table 2.
Patient overlaps across two different data sources within OneFlorida
| UFHealth | ORH | UHealth | FLM | TMA | TMC | AH | NCH | |
|---|---|---|---|---|---|---|---|---|
| UFHealth | X | 18 331 | 13 194 | 481 151 | 12 522 | 26 970 | 47 841 | 2979 |
| ORH | X | 3092 | 130 720 | 730 | 2345 | 202 502 | 1826 | |
| UHealth | X | 197 360 | 745 | 1915 | 7583 | 48 414 | ||
| FLM | X | 41 856 | 96 342 | 347 134 | 225 801 | |||
| TMA | X | 117 119 | 1462 | 391 | ||||
| TMC | X | 4505 | 811 | |||||
| AH | X | 3142 | ||||||
| NCH | X |
Abbreviations: AH: Adventist Health; FLM: Florida Medicaid; NCH: Nicklaus Children’s Hospital; ORH: Orolando Health; TMA: Tallahassee Memorial Healthcare outpatient; TMC: Tallahassee Memorial Healthcare inpatient; UFHealth: University of Florida Health; UHelath: Univesritry of Miami Health.
Excluding data from FLM, a total of 14 387 unique patients were seen across 3 health care partners in OneFlorida. The largest overlaps (ie, 6891 patients) were across TMA, TMC, and UFHealth. UFHealth also has significant patient overlaps (ie, 3980) with ORH and AH. On the other hand, 4 groups of 3 partners (ie, TMA/AH/MCH, ORH/TMA/MCH, and UFHealth/TMA/MCH) have no overlap. The results are expected as these partners are geographically apart (ie, MCH is located in south Florida; TMA is located in northwest Florida; and UFHealth and ORH are located in center Florida).
We also examined the prevalence of 19 chronic conditions using the Chronic Conditions Data Warehouse (CCW) chronic condition (CC) algorithms.13,14Table 3 shows the numbers of patients with these 19 chronic condition categories from a linked Florida Medicaid cohort (n = 1 018 333), where the patient is a Florida Medicaid beneficiary and has visited a health care provider within the OneFlorida network. Note that, we only deduplicated the records at the patient level and then linked associated patient records (eg, diagnoses and procedures) from the different data sources under the same patient. Thus, we still have, for example, duplicated diagnoses from the same patient encounter but different data sources (eg, the same encounter data can come from both a EHR and FLM). To demonstrate the values of linked data, we used only FLM diagnosis data, only OneFlorida EHR diagnoses, and combined sources of diagnoses (ie, can come from multiple EHR sources and/or FLM if the patient sought care in multiple health care systems in OneFlorida) to identify the specific CCs for the same linked patient cohort, respectively. It is obvious as shown in Table 3 that even with the same cohort, the number of identified CCs varies significantly depending on the data source we used.
Table 3.
Counts of patients with chronic diseases on the linked population (n = 1 018 333; ie, linked between Florida Medicaid and OneFlorida EHR partners) using different sources of diagnosis data
| Chronic condition | Florida Medicaida, n = 1 018 333 | OneFlorida EHRsb, n = 1 018 333 | Combinedc, n = 1 018 333 |
|---|---|---|---|
| Acquired hypothyroidism | 81 673 (8%) | 70 379 (7%) | 125 566 (12%) |
| Acute myocardial infarction | 15 850 (2%) | 12 051 (1%) | 24 300 (2%) |
| Alzheimer’s disease | 11 202 (1%) | 7110 (1%) | 15 443 (2%) |
| Alzheimer’s disease and related disorders or senile dementia | 40 851 (4%) | 26 991 (3%) | 55 516 (5%) |
| Anemia | 250 461 (25%) | 169 120 (17%) | 336 642 (33%) |
| Asthma | 309 059 (30%) | 179 582 (18%) | 376 757 (37%) |
| Atrial fibrillation | 35 100 (3%) | 43 489 (4%) | 64 659 (6%) |
| Benign prostatic hyperplasia | 16 486 (2%) | 17 367 (2%) | 29 481 (3%) |
| Cataract | 63 148 (6%) | 33 893 (3%) | 79 839 (8%) |
| Chronic kidney disease | 138 860 (24%) | 121 889 (12%) | 202 784 (20%) |
| Colorectal cancer | 7215 (1%) | 8321 (1%) | 12 224 (1%) |
| Depression | 200 519 (20%) | 125 713 (12%) | 266 725 (26%) |
| Diabetes | 149 736 (15%) | 142 063 (14%) | 220 196 (22%) |
| Endometrial cancer | 2513 (0%) | 3073 (0%) | 4596 (0%) |
| Female/male breast cancer | 11 183 (1%) | 17 338 (2%) | 22 387 (2%) |
| Glaucoma | 40 572 (4%) | 23 923 (2%) | 52 715 (5%) |
| Heart failure | 67 358 (7%) | 52 318 (5%) | 95 951 (9%) |
| Hip/pelvic fracture | 10 270 (1%) | 9183 (1%) | 16 038 (2%) |
| Hyperlipidemia | 228 970 (22%) | 183 421 (18%) | 344 687 (34%) |
| Hypertension | 276 004 (27%) | 291 548 (29%) | 431 844 (42%) |
| Ischemic heart disease | 105 227 (10%) | 93 831 (9%) | 164 186 (16%) |
| Lung cancer | 8164 (1%) | 8436 (1%) | 12 725 (1%) |
| Obstructive pulmonary disease and bronchiectasis | 191 343 (19%) | 92 037 (9%) | 237 353 (23%) |
| Osteoporosis | 29 113 (3%) | 24 942 (2%) | 46 278 (5%) |
| Prostate cancer | 6212 (1%) | 11 805 (1%) | 15 016 (1%) |
| Rheumatoid arthritis/osteoarthritis | 14 4254 (14%) | 108 015 (11%) | 205 444 (20%) |
| Stroke/transient ischemic attack | 52 889 (5%) | 37 066 (4%) | 74 630 (7%) |
Using diagnosis data only from the Florida Medicaid program.
Using diagnosis data only from individual EHR sources.
Using diagnosis data from both Florida Medicaid and HER sources.
Further, we examined more closely the clinical encounters and associated services (eg, medications) for the UFHealth Medicaid population (n = 481 151). In OneFlorida, medication prescription data typically come from EHR sources; while dispensing data are from claims data sources (eg, Florida Medicaid). Within the UFHealth Medicaid population, Table 4 shows the number of prescription records from only UFHealth and from UFHealth plus other EHR sources (ie, a UF Health Medicaid patient can seek care in other OneFlorida healthcare systems) compared with the number of dispensing records from Florida Medicaid.
Table 4.
Prescribing vs. dispensing records on the linked UFHealth Medicaid population
| Data from UFHealth only (prescribing) | Data from UFHealth and other EHRs (prescribing) | Linked UFHealth Medicaid patients (dispensing) | |
|---|---|---|---|
| Total number of records | 15 499 512 | 15 936 878 | 26 412 702 |
| Min | 1 | 1 | 1 |
| Max | 7810 | 9104 | 15 221 |
| Average | 47 | 48 | 74 |
| Median | 12 | 13 | 25 |
| Standard deviation | 134 | 136 | 151 |
DISCUSSION
The increasing adoption of EHR systems and proliferation of electronic clinical data offer unprecedented opportunities for both cohort identification to accelerate participant recruitment for clinical studies, especially pragmatic trials, and real-world evidence (RWE) data for observational studies and for data science projects. Further, as the national conversation on biomedical research continues to shift towards promoting data sharing and reuse, there is a surge of national efforts on building large scale CRNs with robust data infrastructures including PCORnet funded by PCORI, the National Center for Advancing Translational Sciences (NCATS)’s Clinical and Translational Service Award Accrual to Clinical Trails (CTSA ACT), and the Observational Health Data Sciences and Informatics (OHDSI) consortium. The innovative use of these massive data resources will be undermined without easy access to ER and RL tools; nevertheless, development of such a tool needs to be carefully calibrated according to the polices, regulations, and privacy risk considerations from the stakeholders of these networks. A privacy-preserving solution is thus significant. A hashing-based privacy-preserving approach is not bullet-proof. It is possible (although not computationally feasible or cost-effective) for attackers to carry out dictionary attack, although they will need to obtain the secret random seeds for each partner’s data. We are also making the assumption that all parties are honest-but-curious (ie, all parties follow the protocol honestly, but each party could be curious in exploring the data they have access to), especially the data coordinating center (ie, UF in our case). The data coordinating center in our scenario has all the secret random seeds; thus, it is easier for the coordinating center to carry out dictionary attack. However, since we combine 4 quasi-identifiers in each rule, permutating all possible combinations is a extremely large search space.
Implications on generating more complete patient profiles
Patients’ health records are in disparate sources and efficient tools linking and integrating these sources provide a more complete picture of individual patients’ health status and clinical characteristics such as comorbidities and disease histories. As shown in Table 3, on the same patient population, using different sources (eg, EHRs vs. claims) of diagnosis data, the prevalence of chronic conditions varies drastically. Patients’ disease profiles are severely underestimated using individual sources. For example, the estimated number of patients with diabetes bumped from 15% (ie, with Medicaid data alone) and 14% (ie, with only EHR data) to 22% using combined EHRs and claims sources. Estimates using individual sources are less reliable since patients would not only seek care through different health care systems but also are on different payer programs. As shown in Table 4, on the same UFHealth Medicaid population, other partners’ EHR systems contributed 437 366 addition prescription records; nevertheless, data from claims are still the most comprehensive source for patient medication information that can potentially add >65% more medication records comparting with data from the EHRs.
Furthermore, individuals’ health outcomes are multifaceted in nature and influenced by a complex interplay among different domains of influence (ie, biological, behavioral, environment, and health care system) as well as different levels of influence (ie, individual, interpersonal, community, and societal) within those domains.15 Nevertheless, barriers to linking, integrating, and efficiently exploiting health information across different sites and domains slow down health care research and the development of precision health programs. It is utterly important to create integrated data infrastructure, where an accurate and reliable RL method and tool is the necessary first step. Only when data about individual patients from different sources are linked, we then have the ability to explore factors beyond individual levels such as the social and environmental determinates of health.
Implications on improving the data quality
RL also enables us to discover and address data quality issues, especially identifying inconsistency between different data sources. For example, we examined the linked UFHealth Medicaid patients, and compared the patient information obtained from different sources. After eliminating likely outliers (ie, 8128 patients who had more than 702 encounters—3 standard deviations), 24 patients had 701 encounters. We examined their disease profiles and paid special attentions to the inconsistencies between different sources. For example, we observed a number of cases where there is often no documentation of an obesity diagnosis in the patients’ Medicaid data even through their body mass index (BMI) in their EHRs indicate either overweight or obesity. These findings are consistent with those of others who have previously reported poor documentation in claims data.16,17 However, RL provides a key opportunity to cross-referencing different sources—a key method for the evaluation of data quality.18
Limitations
There are certain limitations we have to recognize in this study. First, we tailored our linkage rules to favor precision over recall because of the use cases we considered for the OneFlorida CRC. The primary function of OneFlorida (and the PCORnet more broadly) is to provide cohort discovery services to accelerate recruitment in pragmatic trials. Thus, a small number of duplicates in the cohort identification queries is not a mission-critical barrier as additional screening and consenting processes are always needed to confirm and enroll eligible and willing participants. Nevertheless, no linkage and deduplication at all would lead to duplicated recruitment efforts across different sites and wasted resources. Further, we chose the deterministic approach because of the simplicity in implementing it comparing to alternatives such as a probabilistic linkage approach in a privacy-preserving setting.19,20
Second, we have to recognize that our work and most existing RL-related work in the literature are on the patient level. More fine-grained ER and RL solutions might be necessary in certain cases, especially for observational studies. For example, if we are to use linked patient medication data to measure medication adherence, we need to have more careful considerations in dealing with the duplicated prescription and dispensing data from different sources (eg, multiple EHRs and claims) as these duplicates might lead to over-estimated adherence measures. Similarly, neglecting duplicated encounter-level data in the linked population will lead to biased utilization measures of health care services, which are critical in health services research topics such as cost-effectiveness analyses.
CONCLUSIONS
The OneFL Deduper was developed as a standalone application that can be readily adopted in environments other than OneFlorida or PCORnet. Access to privacy-preserving RL methods and tools is mission-critical for these national CRNs that are developing massive collections of electronic data on their patients. Privacy risks, organizational policies and regulations, data availability and quality, and computing resources all have significant impact on how a PPRL solution is constructed in a real-world setting. Investigators who are using the data from these CRNs shall be aware of the caveats and inherent biases from these linked datasets. More fine-grained ER and RL solutions are also needed; thus; warrant further investigations.
FUNDING
This work was supported in part by National Institutes of Health (UL1TR001427), the OneFlorida Cancer Control Alliance (funded by James and Esther King Biomedical Research Program, Florida Department of Health 4KB16), the OneFlorida Clinical Research Consortium (CDRN-1501-26692) funded by the Patient Centered Outcomes Research Institute (PCORI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or PCORI.
AUTHOR CONTRIBUTIONS
JB, BH, and ES designed the initial concepts and framework for the proposed RL solution; AS, AL, and TM carried out the implementation and testing of the linkage software; GL, AS, JB, and AL carried out the validation studies. JB and AL wrote the initial draft of the manuscript. ES, YG, and BH edited the manuscript and provided critical feedback.
Supplementary Material
ACKNOWLEDGMENTS
We like to thank the partners in the OneFlorida Clinical Research Consortium who have participated and provided critical support in the implementation of the record linkage process.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. J Am Med Inform Assoc 2014; 21 (4): 576–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corley DA, Feigelson HS, Lieu TA, et al. Building data infrastructure to evaluate and improve quality: PCORnet. J Oncol Pract 2015; 11 (3): 204–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shenkman E, Hogan W.. OneFlorida Clinical Research Consortium. Patient-Centered Outcomes Research Institute; 2015. http://www.pcori.org/research-results/2015/oneflorida-clinical-research-consortium Accessed February 11, 2017. [Google Scholar]
- 4.PCORnet. PCORnet Data. The Natioanl Patient-Centered Clinical Research Network; 2018. https://pcornet.org/pcornet-data/ Accessed December 18, 2018. [Google Scholar]
- 5. Kho AN, Hynes DM, Goel S, et al. CAPriCORN: Chicago area patient-centered outcomes research network. J Am Med Inform Assoc 2014; 21 (4): 607–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.“Health Data Link.” Health Data Link. 2018. https://www.healthdata.link/ Accessed December 18, 2018.
- 7.PCORnet. PCORnet Common Data Model (CDM). 2018. https://pcornet.org/pcornet-common-data-model/ Accessed December 18, 2018.
- 8.OneFlorida. OneFlorida Clinical Research Consortium Data Summary. 2018. http://onefloridaconsortium.org/data/ Accessed December 18, 2018.
- 9. Kho AN, Cashy JP, Jackson KL, et al. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc 2015; 22 (5): 1072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuzu M, Kantarcioglu M, Durham EA, et al. A practical approach to achieve private medical record linkage in light of public resources. J Am Med Inform Assoc 2013; 20 (2): 285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.“Florida Department of State.” Voter Information Lookup. 2018. https://dos.myflorida.com/elections/for-voters/check-your-voter-status-and-polling-place/ Accessed December 18, 2018.
- 12. Culbertson A, Goel S, Madden MB, et al. The building blocks of interoperability. A multisite analysis of patient demographic attributes available for matching. Appl Clin Inform 2017; 8: 322–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CMS. Original CCW Chronic Condition Algorithms Reference List. 2006. file:///Users/bianjiang/Downloads/original-ccw-chronic-condition-algorithms-reference-list.pdf Accessed December 18, 2018.
- 14.CMS. Original CCW Chronic Condition Algorithms. 2010. https://www.ccwdata.org/documents/10280/19139421/original-ccw-chronic-condition-algorithms.pdf Accessed December 18, 2018.
- 15.NIMHD. National Institute on Minority Health and Health Disparities Research Framework. 2018. https://www.nimhd.nih.gov/about/overview/research-framework.html Accessed December 18, 2018.
- 16. Chan KS, Fowles JB, Weiner JP.. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev 2010; 67 (5): 503–27. [DOI] [PubMed] [Google Scholar]
- 17. Schlegel DR, Ficheur G.. Secondary use of patient data: review of the literature published in 2016. Yearb Med Inform 2017; 26: 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weiskopf NG, Weng C.. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 20 (1): 144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sayers A, Ben-Shlomo Y, Blom AW, et al. Probabilistic record linkage. Int J Epidemiol 2016; 45 (3): 954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doidge JC, Harron K.. Demystifying probabilistic linkage: Common myths and misconceptions. Int J Popul Data Sci 2018; 3 (1): 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.