Abstract
Proteolytic processing of Bacillus thuringiensis (Bt) Cry protoxins by insect midgut proteases is critical to their insecticidal activities against target insects. Although transgenic Bt cotton expressing Cry1Ac and Cry2Ab proteins have been widely used for control of the cotton bollworm (Helicoverpa armigera) in the field, the proteolytic cleavage sites in the two protoxins targeted by H. armigera midgut proteases are still not clear. In this study, the proteolysis of Cry1Ac and Cry2Ab protoxins by midgut juice prepared from midgut tissue of H. armigera larvae was investigated. Cleavage of Cry1Ac protoxin by midgut proteases formed a major protein fragment of ~65 kDa, and N-terminal sequencing revealed that cleavage occurred at Arg28 in the fore-end of helix α-1 in domain I of Cry1Ac. Cleavage of Cry2Ab protoxin by midgut juice proteases produced a major protein fragment of ~50 kDa, and the cleavage occurred at Arg139 between helices α-3 and α-4 in domain I of Cry2Ab. The amino acids Arg28 of Cry1Ac and Arg139 of Cry2Ab were predicted as putative trypsin cleavage sites. Bioassay data showed that the toxicities (LC50s) of Cry1Ac and Cry2Ab protoxins were equivalent to those of their respective midgut juice-activated toxins in the susceptible SCD strain of H. armigera. Identification of the exact sites of N-terminal activation of Cry1Ac and Cry2Ab protoxins will provide a basis for a better understanding of the mode of action and resistance mechanisms based on aberrant activation of these protoxins in H. armigera.
Introduction
Bacillus thuringiensis (Bt) is a ubiquitous gram-positive bacterium, and during sporulation, Bt strains produce crystal proteins (Cry toxins) that are toxic to a variety of insects, such as lepidopterans, coleopterans, dipterans and hemipterans [1]. Bt Cry toxins have been extensively used in sprays and transgenic plants, which has contributed to the efficient control of different agricultural pests. They also have reduced the use of chemical insecticides and increased farmer profits [2–5].
The majority of Cry toxins are produced in insoluble and inactive forms as crystal inclusions composed of protoxins. After ingestion by target insect larvae, the crystals are solubilized in the alkaline environment of the larval midgut and are activated by midgut proteases [6]. Then, the activated toxins pass through the peritrophic matrix and sequentially bind to specific receptors located on the brush border membrane (BBM) surface of the cells, leading to membrane insertion and pore formation [7,8]. It is generally accepted that the activation of protoxins is one of the most important and essential steps to exert toxicity [9–11].
Considering the molecular weight of Cry proteins, two types of protoxins have been identified: short protoxins of approximately 70 kDa (such as Cry2Ab) and long protoxins of 130 kDa (such as Cry1Ac) [9]. In the case of the short protoxins, approximately 40 amino acids of the N-terminal end are removed during activation with midgut proteases, while for the long protoxins, in addition to N-terminal processing, this activation entails removal of 500–600 amino acids from the C-terminal end. Both cases result in activated Cry toxins of ~60 kDa [1,9,12]. The midgut proteases of lepidopteran larvae mainly belong to the serine protease class, such as trypsin-like and chymotrypsin-like proteases [13–15]. Such midgut proteases are likely to be responsible for protoxin activation. It was reported that improper activation, such as insufficient processing or over digestion, in some insect populations has resulted in insect resistance to Cry protoxin action [16].
The cotton bollworm, Helicoverpa armigera (Hübner), is one of the most invasive pests infesting cotton, maize and other crops. This insect originated from Africa, Asia, Europe and Australia; however, long-range migration and international trade helped this pest spread throughout South and Central America [17,18]. In China, the planting of transgenic cotton expressing only Cry1Ac since 1997 has been very successful in controlling H. armigera [19,20]. Although Bt cotton has remained useful against H. armigera, regular and significant increases in the frequency of resistant individuals to Cry1Ac have provided an early warning for the possibility of resistance evolution in northern China [21,22]. The amino acid sequence identity of Cry2A with Cry1Ac and other Bt toxins is less than 20% [23], so Cry2A exhibits a lower level of cross-resistance with other Bt toxins, such as Cry1Ac [24,25]. Therefore, to delay the evolution of resistance, Bt cotton pyramids that produce the Cry1Ac and Cry2Ab toxins have been successfully planted in the United States, Australia and India [26].
Although Cry1Ac and Cry2Ab toxins have been widely used to control H. armigera, their proteolytic cleavage sites targeted by H. armigera midgut proteases are still not clearly defined. In the present work, we investigated the proteolysis of Cry1Ac and Cry2Ab protoxins by H. armigera midgut juice in vitro and detected the proteolysis by SDS-PAGE. Verification of N-terminal sequences of the activated toxins, ~65 kDa (for Cry1Ac) and ~50 kDa (for Cry2Ab), by Edman degradation sequencing analysis showed that the proteolysis of Cry1Ac and Cry2Ab protoxins occurred at Arg28 and Arg139, respectively. Determination of the cleavage sites provided a basis for further study of the mechanism of action and resistance caused by abnormal activation.
Materials and methods
Insect strain
The susceptible strain (SCD) of H. armigera was collected from the Ivory Coast, Africa, in the 1970s and has been maintained in laboratory conditions without exposure to Bt toxins or other insecticides for more than 40 years [27]. Larvae were reared on an artificial diet based on wheat germ and soybean power at 26 ± 1°C with a photoperiod of 16:8 h (Light:Dark).
Preparation of Bt toxins
Cry1Ac protoxin was produced from the HD-73 strain of B. thuringiensis subsp. kurstaki, and Cry2Ab protoxin was produced from the engineered HD-73- strain containing the cry2Ab gene. The Bt strains were grown for 36 h at 30°C in lysogeny broth (LB) medium until 50% of the crystal was released. The crystals were collected by centrifugation at 5,000 rpm for 20 min, and then the pellet was washed with sterile water, followed by 1 M NaCl. The crystals were dissolved in 100 mM Na2CO3 buffer (pH 10.5, containing 10 mM DTT). Purified Cry1Ac and Cry2Ab protoxins were checked by SDS-PAGE. The protein concentrations were determined by the method of Bradford [28], using bovine serum albumin (BSA) (Solarbio, Beijing, China) as a standard.
Midgut juice preparation
Fifth instar 2nd day larvae from the susceptible SCD strain were chilled on ice for 10 min before they were dissected. Ten midguts were dissected, and midgut tissues were separated from peritrophic membranes containing the food bolus. The content was homogenized with 2 ml of cold NaCl solution (0.15 M) and centrifuged at 12,000 rpm for 15 min at 4°C; the clear supernatants were collected. The protein concentration of midgut juice was quantified by the method of Bradford [28] and adjusted to 1 mg/ml; the stocks were then frozen at -80°C until use.
Digestion of Cry1Ac and Cry2Ab protoxins by midgut juice
Before use, the two protoxins were diluted to 0.5 mg/ml, and the 1 mg/ml midgut juice was diluted to six different concentrations. Ten microliters of each protoxin (5 μg) were mixed with 2 μl of six different concentrations of midgut juice (500, 250, 50, 25, 12.5, 2.5 μg protein/ml) in a final volume of 20 μl of 100 mM Na2CO3 buffer (pH 10.5). The mixtures were incubated at 37°C for 1 h. The reactions were stopped by adding 1 μl of 10 mM PMSF. The samples were separated by 10% SDS-PAGE and stained with Coomassie blue R-250 for 1 h. The proteins were visualized by destaining with 10% acetic acid.
N-terminal sequencing
Activated Cry1Ac and Cry2Ab toxins were prepared by incubating 1:50 (w/w) midgut juice/protoxin for 1 h at 37°C. A total of 30 μg of each protoxin was incubated with midgut juice; the reactions were stopped by adding 5×SDS-PAGE sample buffer and heated for 5 min at 95°C. Then, the samples were divided into six replicates and separated by 10% SDS-PAGE. After electrophoresis, the proteins were transferred to PVDF membrane (Bio-Rad, Hercules, CA, USA) using CAPS buffer (10 mM CAPS, 10% methanol, pH 11.0) at 350 mA (constant current) for 1 h at 4°C. Before staining, the PVDF membranes were soaked in methanol solution for 15 s, stained with 0.1% (w/v) biological dye ponceau S (containing 5% acetic acid) for 10 min, and washed with distilled water until the red background faded completely. The activated toxin bands were clearly marked and submitted for N-terminal sequencing based on Edman degradation using a Shimadzu automated protein/peptide sequencer (PPSQ-333A, Kyoto, Japan).
Bioassays
Cry1Ac and Cry2Ab midgut juice-activated toxins were prepared by incubating 20:1 (w/w) of protoxin:midgut juice at 37°C for 1 h. The toxicological responses of Cry1Ac and Cry2Ab protoxins and midgut juice-activated toxins were determined against neonates of the susceptible SCD strain of H. armigera with diet surface overlay bioassays as described previously [29]. Gradient concentrations of Bt protein solution were prepared by diluting the stock suspensions with 0.01 M phosphate buffer solution (PBS), pH 7.4. A liquid artificial diet (900 μl) was dispensed into each well of a 24-well plate. After the diet cooled, 100 μl of Bt protein solution was applied evenly to the diet surface in each well. A single unfed neonate (24 h old) was placed in each well, and mortality was recorded after 7 days. Forty-eight larvae were tested for each concentration. Larvae were considered dead if they died or weighed less than 5 mg at the end of bioassays. The LC50 values (the concentration of Bt protein that killed 50% of larvae) and the 95% fiducial limits of the LC50 for each strain were calculated through probit analysis of the mortality data using the PoloPlus program [30].
Results
Proteolysis of Cry1Ac and Cry2Ab protoxins and N-terminal sequencing
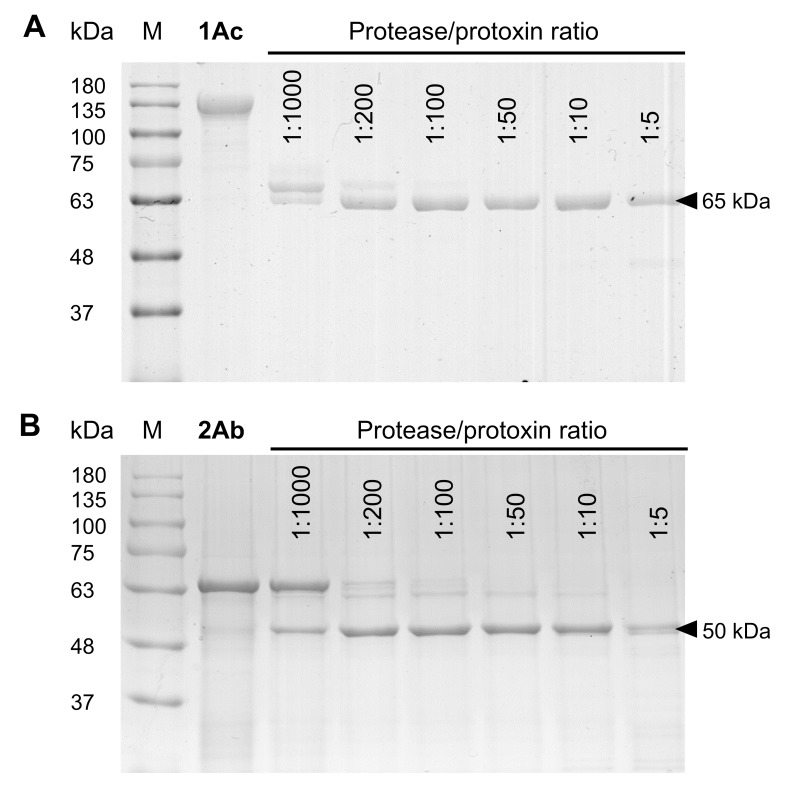
Protoxins were digested with H. armigera midgut juice at 37°C using different midgut juice/protoxin ratios. Analysis of SDS-PAGE showed that digesting Cry1Ac protoxin with low concentrations of midgut juice resulted in a doublet band of ~70 and ~65 kDa. As the ratio of midgut juice/protoxin increased, the ~70 kDa protein was completely converted to ~65 kDa, which remained stable until the concentration of midgut juice increased to 1:5. Slight degradation was observed (Fig 1A). For Cry2Ab protoxin, at low midgut juice concentrations, only a small amount of protoxin was converted to an ~50 kDa protein. As the ratio of midgut juice/protoxin increased, the amount of Cry2Ab protoxin significantly decreased until all protoxin was converted to activated toxin (Fig 1B).
Fig 1. SDS-PAGE analysis proteolytic digestion of protoxins by H. armigera larval midgut juice.
(A) The digestion of Cry1Ac protoxin. (B) The digestion of Cry2Ab protoxin. M, the BlueRay prestained protein marker (MDBio, Qingdao, China). Both protoxins were incubated with midgut juice at different midgut juice protein/protoxin ratios (total protein content of the gut juice added/5 μg protoxin protein) for 1 h at 37°C.
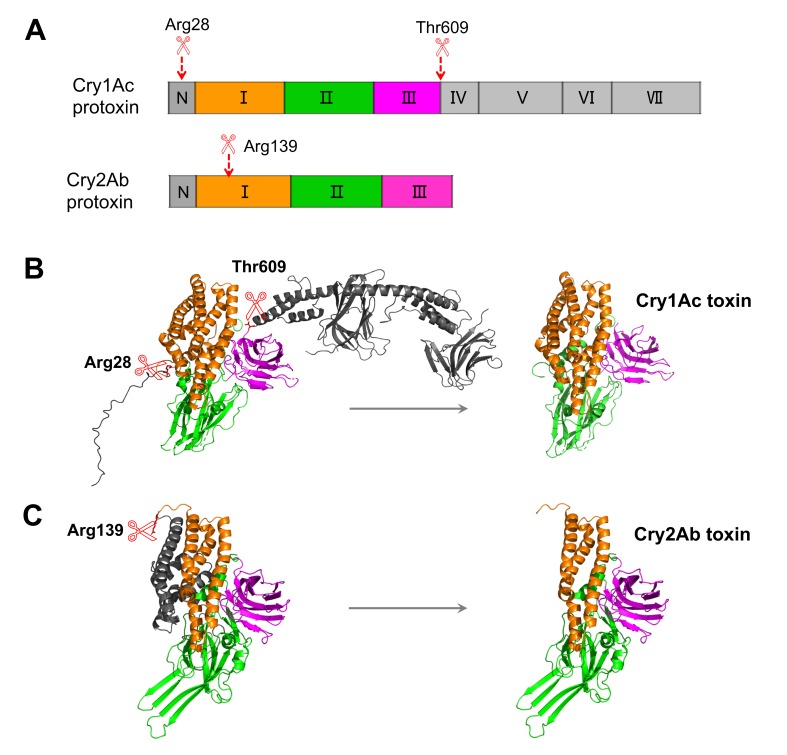
Ten cycles of Edman degradation reactions were executed to determine the N-terminal sequences of the activated Cry1Ac and Cry2Ab toxins. The amino acid sequences of the N-terminal end indicated that proteolysis by the midgut juice proteases of H. armigera cleaved Cry1Ac protoxin at Arg28 and Cry2Ab protoxin at Arg139 (Fig 2A); both residues are predicted as putative trypsin protease cleavage sites.
Fig 2. The structure of Cry1Ac and Cry2Ab.
(A) The schematic diagram of Cry1Ac and Cry2Ab protoxins. The red dotted arrows represent cleavage sites. (B) Protein 3D structures of Cry1Ac protoxin and activated toxin. (C) Protein 3D structures of Cry2Ab protoxin and activated toxin. The three-dimensional structures of Cry1Ac and Cry2Ab protoxins were based on homologous modeling, built with SYBYL-Orchestrar software and viewed by the PyMOL program. The removed parts are shown in gray. The orange, green and purple represent domains I, II and III, respectively. Arg28 and Thr609 represent the cleavage sites of protease hydrolysis of Cry1Ac protoxin from the N- and C-termini, and Arg139 represents the cleavage sites of Cry2Ab from the N-terminus.
Structural analysis of Cry1Ac and Cry2Ab protoxins and toxins
The 3-D Cry toxin family is the largest group of Cry toxins, and they are globular molecules containing three structural domains. As mentioned before, two types of 3d-Cry protoxins have been identified: Cry2Ab is a short protoxin, while Cry1Ac is a long protoxin. Although Cry1Ac and Cry2Ab share low amino acid sequence identity, they share a similar fold consisting of three domains. In addition to the three-domain toxic core, the Cry2Ab protoxin has an additional N-terminal end (Fig 2A). Domain I is a bundle composed of seven α-helices; our data indicate that digestion with the midgut proteases of H. armigera introduced a cleavage between helices α-3 and α-4 in domain I. This suggests that the region comprising the N-terminus and helices α-1 to α-3 of Cry2Ab was removed in the activated toxin (Fig 2C). However, the denaturant conditions of the SDS-PAGE analysis do not allow us to conclude if this region is completely detached from the protein or remains attached by noncovalent interactions. Compared with the structure of Cry2Ab protoxin, Cry1Ac protoxin has an additional long C-terminal end composed of four discrete domains (domains IV-VII) (Fig 2A). The activation of Cry1Ac cleaved the entire C-terminal region and 28 amino acids from the N-terminus in the fore-end of helix α-1 in domain I (Fig 2B), yielded an ~65 kDa activated toxin. Again, we want to state that these data would not allow us to conclude if some peptides from protoxin still remain attached to the activated toxin. More studies will be needed in the future to clearly demonstrate if activation is linked to protein degradation of the resulting peptides.
Toxicity of the protoxins and digested toxins to H. armigera
The LC50 values for Cry1Ac protoxin and midgut juice-activated toxin against the SCD strain were 0.0056 and 0.0063 μg/cm2, respectively, and their 95% fiducial limits overlapped (Table 1), indicating that the toxicities of the two forms of Cry1Ac were not significantly different. The LC50 value for Cry2Ab protoxin (0.10 μg/cm2) was roughly equivalent to that of midgut juice-activated toxin (0.19 μg/cm2), although their 95% fiducial limits did not overlap (Table 1). As expected, the toxicities of Cry1Ac were approximately 20-fold higher than that of Cry2Ab to the SCD strain of H. armigera.
Table 1. Toxicity of Cry1Ac and Cry2Ab protoxins and their respective midgut juice-digested toxins against neonates of the susceptible SCD strain of H. armigera.
| Bt toxin | Na | Slopes ± SE | LC50 (μg/cm2) | 95% FLb |
|---|---|---|---|---|
| Cry1Ac protoxin | 336 | 1.46 ± 0.16 | 0.0056 | 0.0036–0.008 |
| Cry1Ac toxin | 336 | 1.58 ± 0.16 | 0.0063 | 0.0048–0.008 |
| Cry2Ab protoxin | 336 | 2.39 ± 0.21 | 0.10 | 0.08–0.12 |
| Cry2Ab toxin | 336 | 1.69 ± 0.17 | 0.19 | 0.15–0.25 |
aTotal number of individuals tested.
b95% fiducial limits of LC50.
Discussion
All 3d-Cry proteins are produced as protoxins, and the proteolysis of protoxins by midgut proteases to produce the activated toxin core is crucial to exert toxicity [8,9]. Here, we demonstrated that proteolytic processing of Cry1Ac and Cry2Ab protoxins with H. armigera midgut juice resulted in activated toxins of ~65 and ~50 kDa, respectively (Fig 1). For Cry1Ac-activated toxin, the N-terminal sequences indicated that the first 28 amino acids of the N-terminal end before helix α-1 in domain I were cleaved, while for Cry2Ab-activated toxin, cleavage occurred at Arg139, which is located in the loop between helices α-3 and α-4 in domain I (Fig 2). Determination of the cleavage sites would provide a basis for further study of the mechanism of action, and these data are helpful for investigating insect resistance mechanisms caused by abnormal proteolysis of these Cry protoxins.
In our work, we found that Cry1Ac protoxin was digested by H. armigera midgut proteases, finally yielding an ~65 kDa activated toxin, which was resistant to further proteolysis within a wide range of protease concentrations (Fig 1A). The N-terminal sequence of this product indicated that proteolysis occurred at Arg28 in the fore-end of α-1 in domain I (Fig 2B), sharing the same cleavage site that was described with commercial trypsin. Digestion of Cry1Ac protoxin by midgut proteases isolated from Pieris brassicae and Mamestra brassicae both produced a soluble form of the toxin with a molecular mass of ~60 kDa. N-terminal sequencing of these products indicated that proteolysis occurred at Arg28 in both cases [31]. Similarly, the same cleavage site was found after treatment with midgut juice from Adoxophyes spp. and Bombyx mori [32]. In contrast, midgut juice proteases that are present in Plutella xylostella larval midgut digested Cry1Ac protoxin at Leu46, located after helix α-1, while proteolytic activation of Cry1Ac by Spodoptera litura midgut proteases cleaved the protein at Gly66, located before helix α-2b [32].
Proteolytic activation of Cry2Ab protoxin by midgut juice of H. armigera resulted in an ~50 kDa protein (Fig 1B). Proteolysis occurred at residue Arg139, which is located on the loop structure between helices α-3 and α-4 in domain I (Fig 2C), and this site was predicted as a putative trypsin cleavage site. When Cry2Ab protoxin was cleaved by P. xylostella midgut proteases, a single band of 50 kDa was formed. Interestingly, further analysis showed that this single band was a mixture of two proteins, and the N-terminal sequences demonstrated that cleavages occurred at Arg139 and Leu144, predicted as trypsin and chymotrypsin cleavage sites, respectively [33]. The Cry2Ab and Cry2Aa amino acid sequences showed 87% identity [34], and the diversity in amino acid sequences indicated that the proteolysis of Cry2Ab might differ from that of Cry2Aa. Previously, cleavage of the Cry2Aa1 and Cry2Aa3 protoxins by Lymantria dispar and B. mori larvae midgut juice produced two major bands, 58 and 49 kDa toxins. N-terminal sequencing confirmed that the first cleavage site was at Tyr49, which is on the loop between α-0 and α-1, while the second cleavage site was at Leu144, which is on the loop between α-3 and α-4 [35,36].
It is believed that the mode of action of Cry toxins in lepidopteran starts with proteolytic activation by midgut proteases, the correct activation of the protoxin is an essential step for proper insecticidal activity [10], and low susceptibility may be caused by improper processing of Cry toxins [16]. It has been reported that a mutant Cry1Ac protoxin that affects its proteolytical processing at the N-terminal end by Manduca sexta midgut juice resulted in 25-fold lower toxicity than the properly activated toxin lacking the first 28 amino acids [10]. A major midgut protease (T1) that activated Cry1A protoxins was found to be absent in the midgut extracts from the 133-r and 198-r strains of Plodia interpunctella that are resistant to Cry1A action, and it was confirmed that the absence of T1 was genetically linked to Cry1A resistance [37]. Another protease-mediated Cry1Ac resistance mechanism was found in a Heliothis virescens strain with reduced protoxin processing [38]. Additionally, a laboratory-selected strain of H. armigera showed 72-fold resistance to Cry1Ac protoxin; the larval midgut juice from this resistant strain displayed improper processing of the protoxin, which was likely caused by the down regulation of HaSP2 protease [39]. In addition, 99% reduced transcription of the serine protease HaTryR was reported to be related to Cry1Ac resistance in H. armigera [40]. Clearly, almost all of the results described above indicated that protease-mediated resistance to Cry toxins was associated with at least one major serine protease. However, the composition of insect midgut proteases is highly variable, and the proteolytic activation of Cry protoxins is a complex process. Therefore, the role of each protease in this process is difficult to predict. Moreover, there has been no conclusive evidence that in vivo, a single midgut protease can activate Cry protoxins alone.
Previous studies have shown that Cry1Ac and Cry2Ab do not share receptors in H. armigera [41–43]; thus, it is believed that there is a low risk of cross-resistance between the two toxins, which is mainly based on receptor alterations. However, if a single or few common proteases account for the activation of both Cry1Ac and Cry2Ab protoxins, there may be a risk of cross-resistance between these two toxins. In the future, it is necessary to identify the exact midgut proteases of H. armigera that are responsible for activation of Cry protoxins and determine whether few specific proteases or a pool of proteases participate in this process.
Acknowledgments
We thank Alejandra Bravo for polishing on an earlier version of this paper.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Work presented in the report was supported by the grant from Ministry of Agriculture and Rural Affairs of China to YY(grant number 2016ZX08011002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schnepf E, Crickmore N, Rie JV, Lereclus D, Baum J, Feitelson J, et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998; 62: 775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendelsohn M, Kough J, Vaituzis Z, Matthews K. Are Bt crops safe? Nat Biotechnol. 2003; 21:1003–1009. 10.1038/nbt0903-1003 [DOI] [PubMed] [Google Scholar]
- 3.Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321: 1676–1678. 10.1126/science.1160550 [DOI] [PubMed] [Google Scholar]
- 4.Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P. Bacillus thuringiensis: A century of research, development and commercial applications. Plant Biotechnol J. 2011;9: 283–300. 10.1111/j.1467-7652.2011.00595.x [DOI] [PubMed] [Google Scholar]
- 5.Comas C, Lumbierres B, Pons X, Albajes R. No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: A meta-analysis of 26 arthropod taxa. Transgenic Res. 2014;23: 135–143. 10.1007/s11248-013-9737-0 [DOI] [PubMed] [Google Scholar]
- 6.Tojo A, Aizawa K. Dissolution and degradation of Bacillus thuringiensis δ-endotoxin by gut juice protease of the silkworm Bombyx mori. Appl Environ Microbiol. 1983;45: 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo A, Gómez I, Conde J, Muñozgaray C, Sánchez J, Miranda R, et al. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochimi Biophys Acta. 2004;1667: 38–46. [DOI] [PubMed] [Google Scholar]
- 8.Pardo-López L, Soberón M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev. 2013;37: 3–22. 10.1111/j.1574-6976.2012.00341.x [DOI] [PubMed] [Google Scholar]
- 9.de Maagd RA, Bravo A, Crickmore N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001;17: 193–199. 10.1016/s0168-9525(01)02237-5 [DOI] [PubMed] [Google Scholar]
- 10.Bravo A, Sanchez J, Kouskoura T, Crickmore N. N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac insecticidal toxin. J Biol Chem. 2002;277: 23985–23987. 10.1074/jbc.C200263200 [DOI] [PubMed] [Google Scholar]
- 11.Gómez I, Sánchez J, Muñoz-Garay C, Matus V, Gill SS, Soberón M, et al. Bacillus thuringiensis Cry1A toxins are versatile proteins with multiple modes of action: Two distinct pre-pores are involved in toxicity. Biochem J. 2014;459: 383–396. 10.1042/BJ20131408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choma CT, Surewicz WK, Carey PR, Pozsgay M, Raynor T, Kaplan H. Unusual proteolysis of the protoxin and toxin from Bacillus thuringiensis Structural implications. Eur J Biochem. 1990;189: 523–527. 10.1111/j.1432-1033.1990.tb15518.x [DOI] [PubMed] [Google Scholar]
- 13.Christeller JT, Laing WA, Markwick NP, Burgess EPJ. Midgut protease activities in 12 phytophagous lepidopteran larvae: Dietary and protease inhibitor interactions. Insect Biochem Mol Biol. 1992;22: 735–746. [Google Scholar]
- 14.Milne R, Kaplan H. Purification and characterization of a trypsin-like digestive enzyme from spruce budworm (Choristoneura fumiferana) responsible for the activation of δ-endotoxin from Bacillus thuringiensis. Insect Biochem Mol Biol. 1993;23: 663–673. 10.1016/0965-1748(93)90040-y [DOI] [PubMed] [Google Scholar]
- 15.Keller M, Sneh B, Strizhov N, Prudovsky E, Regev A, Koncz C, et al. Digestion of δ-endotoxin by gut proteases may explain reduced sensitivity of advanced instar larvae of Spodoptera littoralis to CryIC. Insect Biochem Mol Biol. 1996;26: 365–373. 10.1016/0965-1748(95)00102-6 [DOI] [PubMed] [Google Scholar]
- 16.Oppert B. Protease interactions with Bacillus thuringiensis insecticidal toxins. Arch Insect Biochem Pathol. 1999;42: 1–12. [DOI] [PubMed] [Google Scholar]
- 17.Tay WT, Soria MF, Walsh T, Thomazoni D, Silvie P, Behere GT, et al. A brave new world for an old world pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. PLoS One. 2013;8: e80134 10.1371/journal.pone.0080134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones CM, Parry H, Tay WT, Reynolds DR, Chapman JW. Movement ecology of pest Helicoverpa: Implications for ongoing spread. Annu Rev Entomol. 2019;64: 274–295. [DOI] [PubMed] [Google Scholar]
- 19.Wu KM, Guo YY, Lv N, Greenplate JT, Deaton R. Efficacy of transgenic cotton containing a cry1Ac gene from Bacillus thuringiensis against Helicoverpa armigera (Lepidoptera: Noctuidae) in northern China. J Econ Entomol. 2003;96: 1322–1328. 10.1093/jee/96.4.1322 [DOI] [PubMed] [Google Scholar]
- 20.Lu YH, Wu KM, Jiang YY, Guo YY, Desneux N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature. 2012;487: 362–365. 10.1038/nature11153 [DOI] [PubMed] [Google Scholar]
- 21.Zhang HN, Yin W, Zhao J, Jin L, Yang YH, Wu SW, et al. Early warning of cotton bollworm resistance associated with intensive planting of Bt cotton in China. PLoS One. 2011;6: e22874 10.1371/journal.pone.0022874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L, Zhang HN, Lu YH, Yang YH, Wu KM, Tabashnik BE, et al. Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat Biotechnol. 2015;33: 169–174. 10.1038/nbt.3100 [DOI] [PubMed] [Google Scholar]
- 23.Morse RJ, Yamamoto T, Stroud RM. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure. 2001;9: 409–417. 10.1016/s0969-2126(01)00601-3 [DOI] [PubMed] [Google Scholar]
- 24.Carrière Y, Crickmore N, Tabashnik BE. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015;33: 161–168. 10.1038/nbt.3099 [DOI] [PubMed] [Google Scholar]
- 25.Welch KL, Unnithan GC, Degain BA, Wei JZ, Zhang J, Li XC, et al. Cross-resistance to toxins used in pyramided Bt crops and resistance to Bt sprays in Helicoverpa zea. J Invertebr Pathol. 2015;132: 149–156. 10.1016/j.jip.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 26.Tabashnik BE, Thierry B, Yves C. Insect resistance to Bt crops: Lessons from the first billion acres. Nat Biotechnol. 2013;31: 510–521. 10.1038/nbt.2597 [DOI] [PubMed] [Google Scholar]
- 27.Yang YH, Yang YJ, Gao WY, Guo JJ, Wu YH, Wu YD. Introgression of a disrupted cadherin gene enables susceptible Helicoverpa armigera to obtain resistance to Bacillus thuringiensis toxin Cry1Ac. B Entomol Res. 2009;99: 175–181. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72: 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Wang HD, Liu SY, Liu LP, Tay WT, Walsh TK, et al. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem Mol Biol. 2017;87: 147–153. 10.1016/j.ibmb.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 30.Software LeOra. Polo Plus, a user’s guide to probit and logit analysis. LeOra Software; 2002. Berkeley, CA, USA. [Google Scholar]
- 31.Lightwood DJ, Ellar DJ, Jarrett P. Role of proteolysis in determining potency of Bacillus thuringiensis Cry1Ac δ-endotoxin. Appl Environ Microbiol. 2000;66: 5174–5181. 10.1128/aem.66.12.5174-5181.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogiwara K, Indrasith LS, Asano S, Hori H. Processing of δ-endotoxin from Bacillus thuringiensis subsp. kurstaki HD-1 and HD-73 by gut juices of various insect larvae. J Invertebr Pathol. 1992;60: 121–126. 10.1016/0022-2011(92)90084-h [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Pan ZZ, Zhang J, Liu B, Zhu YJ, Chen QX. Proteolytic activation of Bacillus thuringiensis Cry2Ab through a belt-and-braces approach. J Agric Food Chem. 2016;64: 7195–7200. 10.1021/acs.jafc.6b03111 [DOI] [PubMed] [Google Scholar]
- 34.Liang Y, Dean DH. Location of a lepidopteran specificity region in insecticidal crystal protein CryIIA from Bacillus thuringiensis. Mol Microbiol. 1994;13: 569–575. 10.1111/j.1365-2958.1994.tb00451.x [DOI] [PubMed] [Google Scholar]
- 35.Audtho M, Valaitis AP, Alzate O, Dean DH. Production of chymotrypsin-resistant Bacillus thuringiensis Cry2Aa1 δ-endotoxin by protein engineering. Appl. Environ. Microbiol. 1999;65: 4601–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohsawa M, Tanaka M, Moriyama K, Shimazu M, Asano S, Miyamoto K, et al. A 50-kilodalton Cry2A peptide is lethal to Bombyx mori and Lymantria dispar. Appl Environ Microbiol. 2012;78: 4755–4757. 10.1128/AEM.07123-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oppert B, Kramer KJ, Beeman RW, Johnson D, Mcgaughey WH. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J Biol Chem. 1997;272: 22346–23473. [DOI] [PubMed] [Google Scholar]
- 38.Karumbaiah L, Oppert B, Jurat-Fuentes JL, Adang MJ. Analysis of midgut proteinases from Bacillus thuringiensis-susceptible and -resistant Heliothis virescens (Lepidoptera: Noctuidae). Comp Biochem Physiol B Biochem Mol Biol. 2007;146: 139–146. 10.1016/j.cbpb.2006.10.104 [DOI] [PubMed] [Google Scholar]
- 39.Rajagopal R, Arora N, Sivakumar S, Rao NG, Nimbalkar SA, Bhatnagar RK. Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem J. 2009;419: 309–316. 10.1042/BJ20081152 [DOI] [PubMed] [Google Scholar]
- 40.Liu CX, Xiao YT, Li XC, Oppert B, Tabashnik BE, Wu KM. Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci Rep. 2014;4: 7219 10.1038/srep07219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu XJ, Yu LY, Wu YD. Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl Environ Microbiol. 2005;71: 948–954. 10.1128/AEM.71.2.948-954.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao YT, Zhang T, Liu CX, Heckel DG, Li XC, Tabashnik BE, et al. Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci Rep. 2014;4: 6184 10.1038/srep06184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tay WT, Mahon RJ, Heckel DG, Walsh TK, Downes S, James WJ,et al. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily a protein. PLoS Genet. 2015;11: e1005534 10.1371/journal.pgen.1005534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.