Abstract
deoxynucleoside triphosphate (dNTPs) are the reduced nucleotides used as the building blocks and energy source for deoxyribonucleic acid (DNA) replication and maintenance in all living systems. They are present in highly regulated amounts and ratios in the cell, and their balance has been implicated in the most important cell processes, from determining the fidelity of DNA replication to affecting cell fate. Furthermore, many cancer drugs target biosynthetic enzymes in dNTP metabolism, and mutations in genes directly or indirectly affecting these pathways that are the cause of devastating diseases. The accurate and systematic measurement of these pools is key to understanding the mechanisms behind these diseases and their treatment. We present a new method for measuring dNTP pools from biological samples, utilizing the current state-of-the-art polymerase method, modified to a solid-phase setting and optimized for larger scale measurements.
Keywords: dNTP, analysis, solid-phase, method, nucleotide
Introduction
Deoxyribonucleic acid (DNA) carries the genetic information of all known living organisms, and its synthesis utilizes deoxynucleoside triphosphates (dNTPs), both as building blocks and as energy source for the polymerization reaction. dNTP pools depend on cell cycle and cell and tissue type, with proliferating cells typically having higher pools than post-mitotic cells [1]. dNTPs are typically present at very low concentrations, which are tightly regulated via the expression, regulation and localization of their biosynthetic and catabolic enzymes [2–4]. These enzymes form a complex, highly regulated network in animal cells, involving the nuclear, mitochondrial and cytoplasmic compartments. These interlinked pathways include the biosynthetic de novo pathway and the parallel cytoplasmic and mitochondrial salvage pathways [5, 6].
The absolute amounts and ratios of the different dNTPs in time and space have been shown to be critical for a wide range of cellular processes, including DNA replication fidelity and mutagenesis [7, 8], DNA repair, cell cycle progression and regulation, mitochondrial DNA (mtDNA) maintenance function and oncogenic and apoptotic processes [9]. Because of the mutagenic effects of imbalanced dNTP pools, they have been studied in detail in the context of cancer biology. However, accumulating data points to tight co-regulation of the mitochondrial and cytoplasmic pools, evidenced by defects in the core synthetic enzymes causing depletion or mutagenesis of mtDNA [10, 11]. These disorders present varying tissue specificity and different ages of onset, typically with multi-systemic devastating consequences [12–14]. Furthermore, disorders of mitochondrial DNA maintenance can secondarily affect the cytoplasmic dNTP pool balance [15, 16], underscoring the intimate crosstalk of the different compartmentalized dNTP pools. Preclinical evidence suggest that some mtDNA instability disorders could be treated with nucleosides [17–19]. However, knowledge of the regulation of the dNTP pools in pathological metabolic states or upon supplementation therapies is still scarce, and sensitive methodology is needed.
The most widely used approach to measure dNTP concentrations from biological samples is based on a polymerase assay using radioactively labelled substrate, first proposed in the late 1960s [20] and further applied and developed two decades later [21]. This traditional method is laborious and multistep, challenging its sensitivity. After different modifications and the replacement of the Klenow fragment for a thermostable polymerase to improve ribonucleotide discrimination [22], the most recent update for this method was developed for the measurement of mitochondrial dNTP pools [23]. The assay relies on the specificity of a polymerase to utilize the dNTPs present in the sample, and bind them on a designed template, together with a different radiolabelled dNTP which will be incorporated proportionally. The amount of the radioactive labelled products (newly synthesized oligonucleotides) together with a standard series of known dNTP concentrations, provides a quantitative result. However, the field has been hampered by the recently restricted commercial availability of specific materials needed for this protocol, motivating method development.
We present here the adaptation of the dNTP measurement into a solid-phase format, utilizing combined knowledge from the conventional polymerase-based dNTP measurement and the ‘solid-phase mini-sequencing’ principle of single-nucleotide detection [24, 25]. This allows a microtitre-plate-based, automatable approach, with an accurate measurement of a large number of samples, improving efficiency, safety and accuracy of the methodology.
Materials and methods
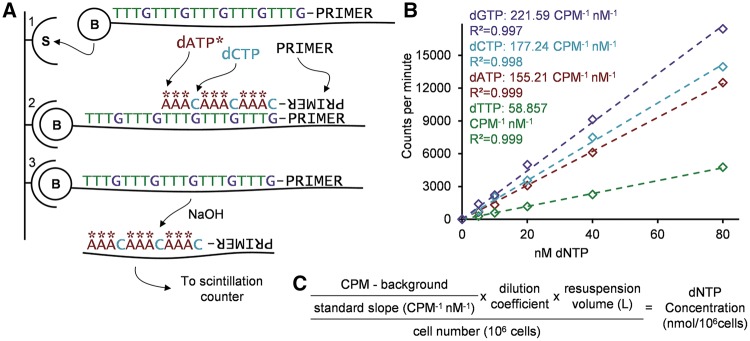
Figure 1A shows an example of the reaction in the setting of the present method, being used to measure deoxycytidine triphosphate (dCTP).
Figure 1:
(A) Schematic description of the reaction principle using dCTP measurement as an example. (A1) Affinity capture of biotinylated oligonucleotide OligoC to streptavidin-coated plate. (A2) Polymerization: priming of the template, followed by the proportional incorporation of the measured nucleotide (dCTP) and the radiolabelled nucleotide (dATP*). (A3) Alkaline release of the labelled chain and radioactive count. (B) Example of standard curves and slopes to be used in the data analysis, with the regression’s calculated R2 for each nucleotide measurement. (C) Equation for the analysis of the CPM obtained from the procedure, utilizing the resulting slope from the standards and the blank, as well as the recorded dilution factors and cell number to obtain the concentration of the nucleotide per million cells.
Biotinylated oligonucleotides and primer
Table 1 below shows the sequences of the primer and the oligonucleotides used in the reaction. The templates are labelled ‘Oligo’+N, with N indicating the base of the specific dNTP to be measured with that template and the primer being common to all reactions.
Table 1:
Detail of the primers and biotinylated oligonucleotides used for the reactions. The initial after 'Oligo' indicates the base of the dNTP to be measured
| Sequence (5ʹ-3ʹ) | Length | |
|---|---|---|
| Primer | CCTGTCTCATACACAGACAC | 20 bp |
| OligoA | [B]AAATAAATAAATAAATAAATGGACAGAGTATGTGTCTGTG | 40 bp |
| OligoT | [B]TTATTATTATTATTATTAGGACAGAGTATGTGTCTGTG | 38 bp |
| OligoC | [B]TTTGTTTGTTTGTTTGTTTGGGACAGAGTATGTGTCTGTG | 40 bp |
| OligoG | [B]TTTCTTTCTTTCTTTCTTTCGGACAGAGTATGTGTCTGTG | 40 bp |
The biotin-labelled oligonucleotides should be High-performance liquid chromatography (HPLC)-purified, and the primer diluted to 5 μM concentration for the reaction, aliquoted and stored at −20°C.
Reagents and equipment
The content in parenthesis indicates the name of the product/company used in the set-up
60% methanol in water, kept at −20°C.
50 mM NaOH (made fresh every 4 weeks).
Thermostable DNA polymerase (DyNAzyme II DNA Polymerase by ThermoFisher Scientific), with its 10x optimized Enzyme Buffer, stored at −20°C.
0.5 M dithiothreitol (DTT) in small aliquots stored at −20°C (single use because of instability).
dNTP mix stock, 40 mM dNTPs (10 mM each) (Bioline), and series dilutions until 1 µM stored at −20°C.
[8-3H(N)]-Deoxyadenosine 5ʹ-triphosphate Tetrasodium Salt in 1:1 ethanol:water mixture (PerkinElmer), stored at −20°C.
[Methyl-3H]-Deoxythymidine 5ʹ-Triphosphate Tetrasodium Salt in 1:1 ethanol:water mixture (PerkinElmer), stored at −20°C.
Phosphate buffered Saline (PBS).
TWEEN® 20 (Amresco).
TENT solution (40 mM Tris-HCl, 1 mM EDTA, 50 mM NaCl, 0.1% TWEEN® 20, pH 8.0–8.8).
Biotinylated oligonucleotides and primer (Sigma Aldrich), stock and dilutions in −20°C.
Streptavidin-coated 96-well plate (BioBind Streptavidin Strip Assembled Solid by Thermo Scientific).
Ultima Gold™ liquid scintillation cocktail for aqueous and non-aqueous samples (PerkinElmer).
Scintillation vials and beta counter (MicroBeta2 by PerkinElmer).
Microplate washer (Wellwash™ by Thermo Labsystems).
Speed vacuum concentrator.
Countess automated cell counter (Invitrogen).
dNTP isolation
Culture cells in Petri dishes to an ideal minimum of 1 x 106 cells. As dNTPs vary greatly between dividing and non-dividing cells and throughout the cell cycle, plan to harvest the cells in similar confluency and cell cycle stage. Wash the cells carefully with cold PBS and detach by trypsinization.
For normalization, count the cell concentration in the suspension. Then pellet the cells in a microcentrifuge at 250·g, and store the pellet at −80°C.
For the isolation, add 1.5 ml of cold (−20°C) 60% methanol in water and vortex thoroughly. Then incubate for at least 1 h at −80°C.
Pellet the insoluble cell contents for 15 min at maximum speed (20 000·g at 4°C); incubate the samples at 95°C for 3 min, cool down on ice and pellet again similarly.
Collect the supernatant and transfer it to a new tube. Desiccate the supernatant with a speed vacuum concentrator until fully dry.
Store the solid extract at −80°C until analysis.
Solid-phase radioactive polymerase reaction
Affinity capture of oligonucleotides: Each dNTP will be measured in a separate well. Mark the plates carefully for the four dNTP reaction, for the standard series and sample replicates. Each measurement replicate requires four wells (one per dNTP), and it is recommended to have at least duplicates at different dilutions to confirm linearity. The standard series of six concentrations will use 24 wells. Transfer 2.5 μl of the specific template oligonucleotide into each streptavidin-coated well, and add 47.5 μl of a 0.1% TWEEN® in PBS solution. Incubate the plate at 37°C for 1.5 h with gentle shaking (Table 2).
Discard the liquid from the wells, and wash the wells thoroughly four times at room temperature with 200 µl of TENT solution. An automated plate washer can be used. Tap the wells dry against tissue papers after the washes.
Standards: Prepare a standard dilution series of a commercial dNTP mixture, with the following concentrations: 80 nM, 40 nM, 20 nM, 10 nM, 5 nM, and a water blank.
Sample preparation: Dissolve the frozen solid nucleotide extract in cold sterile water (normally 100 µl, modify according to the cell number), vortex thoroughly and keep on ice for 10 min.
Prepare replicates of the different dilution coefficients (normally 1:5 and 1:10) for each sample to be measured, in a volume sufficient for four reactions per dilution (minimum 50 µl).
Reaction: Prepare master-mixes for each nucleotide to be measured according to Table 3. [3H]-dATP is used for measuring dTTP, dCTP and dGTP (the wells with OligoT, OligoC and OligoG) in the sample, while [3H]-dTTP is used for the dATP (OligoA) reaction mixture. Since the radiolabelled dNTPs are stored in ethanol:water mixture, the molar concentration of each batch should be calculated from the activity per volume and specific radioactivity (usually 1mCi/ml and 10–25 Ci/mmol), and the solvent of the required amount evaporated with a speed-vacuum concentrator and dissolved in water, as recommended by [23]. A 1:3 dilution of the radiolabel with cold dATP or dTTP is also described by [23] to lower the specific radioactivity and thus the costs without appreciable loss of sensitivity.
Incubate the plate at 55°C for 1 h, with a flotation device in a water bath.
Discard the contents of the wells and perform the washing process as in step 2.
Release the newly synthesized labelled strand by adding 60 µl of 50 mM NaOH to each well and incubate for 3 min at room temperature.
Transfer the eluted DNA to scintillation vials into 3 ml of scintillation cocktail and measure the radioactivity in a beta counter, with 1 min counting time per sample.
Data analysis: After exporting the data, analyse the counts-per-minute (CPM) values for each nucleotide independently. Subtract the blank value from all samples measuring that nucleotide. Next, generate a linear standard curve from the origin (Fig. 1B), and use the regression slope to calculate the concentration of the measured replicates based on their CPM values. Values outside the range of standards should be omitted and re-measured at a different dilution. Then use the cell number and dilution factors utilized to calculate the amount of dNTP per number of cells (Fig. 1C).
Table 2:
Composition of the affinity capture reaction mixture
| Volume (μl) | Final concentration | |
|---|---|---|
| PBS/TWEEN® solution | 47.5 | |
| 5 μM oligonucleotide | 2.5 | 0.25 μM |
| Total volume | 50.0 |
Table 3:
Composition of the polymerase reaction mixture
| Volume (μl) | Final concentration | |
|---|---|---|
| 10x polymerase buffer | 5 | 1x |
| 0.5 M DTT | 0.5 | 5 mM |
| 15 μM radiolabelled dNTP | 2.5 | 0.75 μM |
| 5 μM primer | 2.5 | 0.25 μM |
| Thermostable polymerase | a | 0.025 U/μl |
| Sample | 12.5 | |
| Water | 28.5 | |
| Total volume | ∼50 |
calculated from the concentration of polymerase.
Results
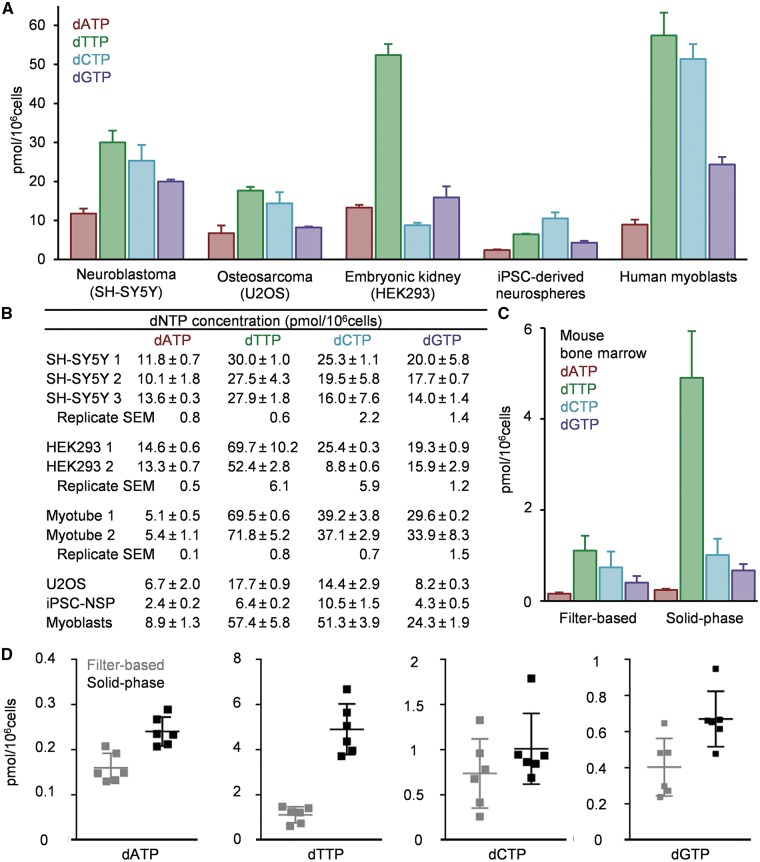
The assay was performed on an array of different cell lines. The standard curves show consistently strong linearity within the measurement range, with R2 values for the regression ∼0.99 (Fig. 1B). Each line or cell type presented a particular dNTP pool profile (Fig. 2A). The measurements show good reproducibility with same samples measured on different days (SH-SY5Y 1 vs. 2 and HEK293 1 vs. 2), as well as with independently cultured (and differentiated in case of myotubes) samples from a same cell line (SH-SY5Y 1–2 vs. 3 and myotubes 1–2) (Fig. 2B).
Figure 2:
(A) Illustrative dNTP concentration profiles from an array of cell lines. Error bars represent the standard deviation of two technical replicates. (B) Concentration values obtained from the measurement of the same sample on different days (SH-SY5Y 1 vs. 2 and HEK293 1 vs. 2) and from independently cultured samples of the same cell line (SH-SY5Y 1–2 vs. 3 and Myotubes 1 vs. 2), as well as the values for the cell lines plotted above, ± standard deviation between two technical replicates. The SEM is also calculated to assess the variability between the replicates. (C–D) Comparison between the dNTP concentration values of identical isolates of mouse bone marrow cells (n = 6) measured with the traditional filter-based protocol or with the present solid-phase methodology in technical duplicates. Data in (C) is represented as means of the values of each bone marrow isolate from a separate mouse. Each data point in (D) represents the value for a single mouse individual obtained from technical duplicates. Error bars represent standard deviation. iPSC, induced pluripotent stem cells; NSP, neurospheres.
Finally, in order to compare the performance of the methodology with the traditional one from in vivo samples, instead of cell lines, we measured identical aliquots of freshly isolated mouse bone marrow extracts with a modified version of the protocol in [23] for whole cell lysates, and with the present method. The resulting values (Fig. 2C–D) show very similar dNTP concentration landscapes, with a tendency of increased absolute value, particularly strong, concerning the measurement of dTTP.
Discussion
We report here a development to the current state-of-the-art methodology for quantitative measurement of the four canonical dNTPs from biological samples. The method allows the performance of the measurement in a solid-phase multiwell setting, and the microtitre-plate format allows automation of the pipetting and washing steps, as well as circumventing time-consuming primer preparation and filter-based steps from the previous methods [23]. Furthermore, it allows utilization of optimized reaction buffers and template sequences. All these features dramatically reduce the required analysis steps, time and manual work required, diminishing the potential sources of error. Importantly, it also increases method safety, reducing direct contact with hazardous compounds.
The main observable difference in the results obtained with the new methodology compared to the conventional filter-based method is the tendency to higher absolute concentrations, particularly for dTTP. The consistent standard curve linearity, the nature of dTTP as the most concentrated canonical dNTP in the cell and the reduction of steps that could potentially lead to loss of sample in the method, as well as the unlikely possibility of consistent contamination or signal acquisition specific for dTTP, prompts us to propose that the higher values may be a true improvement in the sensitivity of the method.
The current shortcoming of the methodology is the increase of the reaction volume due to the nature of the well, leading to a larger requirement of sample and reagents. Moreover, the normalization strategy is a challenge in the field [1], due to the inaccuracy of cell counting and difficulties comparing results from different publications. The most widely used normalization method is cell number, although other strategies are possible, such as utilizing the absorbance of an alkaline lysate, proportional to cell mass [26]. The consistency of the results obtained by cell number normalization lead us to utilize it due to its simplicity, but a more sensitive approach should be considered for more challenging biological samples, such as protein or DNA content measurement, or evaluation of cell volume for molar concentration presentation.
The methanol-based nucleotide isolation is commonly used for dNTP measurements and hence was chosen for this methodology. However, methanol extracts have been described to present residual levels of enzymes that could interfere with the polymerase reaction [27]. The boiling step between precipitations should aid the inactivation of the remaining enzymatic activities [28], but in further development of assay sensitivity, other alternatives excluding this possibility could be considered, such as perchlorate/trioctylamine extraction/neutralization [21, 29].
Nucleotide metabolism is one of the main targets of chemotherapeutic drugs in cancer, despite the fact that current understanding on nucleotide metabolism and tissue-specific features of dNTP regulation is superficial. Furthermore, mitochondrial dysfunction has turned out to be a common cause of inherited degenerative conditions, also causing rare and devastating mitochondrial disorders in children, where nucleoside therapies are emerging as a potential therapeutic option [12]. Our sensitive, quantitative and efficient method should be a highly useful tool to provide evidence of dNTP metabolism in different biological samples, in pathological states and in treatment of cancer and metabolic diseases.
Funding
This work was supported by grants from the Academy of Finland, Sigrid Jusélius Foundation, European Research Council and University of Helsinki.
Conflict of interest statement. None declared.
References
- ?>1. Gandhi VV, Samuels DC.. A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells. Nucleosides, Nucleotides and Nucleic Acids 2011;30:317–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ?>2. Mathews CK. DNA precursor metabolism and genomic stability. FASEB J 2006;20:1300–14. [DOI] [PubMed] [Google Scholar]
- ?>3. Hofer A, Crona M, Logan DT. et al. DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol 2012;47:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ?>4. An S, Kumar R, Sheets ED. et al. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008;320:103–06. [DOI] [PubMed] [Google Scholar]
- ?>5. Pontarin G, Fijolek A, Pizzo P. et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci USA 2008;105:17801–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ?>6. Wang L. Mitochondrial purine and pyrimidine metabolism and beyond. Nucleosides, Nucleotides and Nucleic Acids 2016;35:578–94. [DOI] [PubMed] [Google Scholar]
- ?>7. Davidson MB, Katou Y, Keszthelyi A. et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J 2012;31:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ?>8. Buckland RJ, Watt DL, Chittoor B. et al. Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity. PLoS Genet 2014;10:e1004846.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ?>9. Mathews CK. Deoxyribonucleotides as genetic and metabolic regulators. FASEB J 2014;28:3832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bourdon A, Minai L, Serre V. et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet 2007;39:776–80. [DOI] [PubMed] [Google Scholar]
- 11. Tyynismaa H, Ylikallio E, Patel M. et al. A heterozygous truncating mutation in RRM2B causes autosomal-dominant progressive external ophthalmoplegia with multiple mtDNA deletions. Am J Hum Genet 2009;85:290–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gorman GS, Chinnery PF, DiMauro S. et al. Mitochondrial diseases. Nat Rev Dis Primers 2016;2:16080.. [DOI] [PubMed] [Google Scholar]
- 13. Ylikallio E, Suomalainen A.. Mechanisms of mitochondrial diseases. Ann Med 2012;44:41–59. [DOI] [PubMed] [Google Scholar]
- 14. Nunnari J, Suomalainen A.. Mitochondria: in sickness and in health. Cell 2012;148:1145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikkanen J, Forsström S, Euro L. et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab 2016;23:635–48. [DOI] [PubMed] [Google Scholar]
- 16. Dalla Rosa I, Cámara Y, Durigon R. et al. MPV17 loss causes deoxynucleotide insufficiency and slow DNA replication in mitochondria. PLoS Genet 2016;12:e1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cámara Y, González-Vioque E, Scarpelli M. et al. Administration of deoxyribonucleosides or inhibition of their catabolism as a pharmacological approach for mitochondrial DNA depletion syndrome. Hum Mol Genet 2014, 10.1093/hmg/ddt641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garone C, Garcia-Diaz B, Emmanuele V. et al. Deoxypyrimidine monophosphate bypass therapy for thymidine kinase 2 deficiency. EMBO Mol Med 2014;6:1016–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez-Gomez C, Levy RJ, Sanchez-Quintero MJ. et al. Deoxycytidine and deoxythymidine treatment for thymidine kinase 2 deficiency. Ann Neurol 2017;81:641–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Solter AW, Handschumacher RE.. A rapid quantitative determination of deoxyribonucleoside triphosphates based on the enzymatic synthesis of DNA. BBA Sect Nucleic Acids Protein Synth 1969;174:585–90. [DOI] [PubMed] [Google Scholar]
- 21. Sherman PA, Fyfe JA.. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal Biochem 1989;180:222–26. [DOI] [PubMed] [Google Scholar]
- 22. Ferraro P, Franzolin E, Pontarin G. et al. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res 2010;38:e85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martí R, Dorado B, Hirano M.. Measurement of mitochondrial dNTP pools. Methods Mol Biol 2012;837:135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Syvänen AC, Hultman T, Aalto-Setälä K. et al. Genetic analysis of the polymorphism of the human apolipoprotein E using automated solid-phase sequencing. Genet Anal Biomol Eng 1991;8:117–23. [DOI] [PubMed] [Google Scholar]
- 25. Suomalainen A, Syvänen A-C.. Quantitative analysis of DNA sequences by PCR and solid-phase minisequencing In: Walker JM, Rapley R. (eds), Molecular Biomethods Handbook. 2nd edn.Totowa, NJ: Humana Press Inc, 2007, 169–78. [Google Scholar]
- 26. Frangini M, Franzolin E, Chemello F. et al. Synthesis of mitochondrial DNA precursors during myogenesis, an analysis in purified C2C12 myotubes. J Biol Chem 2013;288:5624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. North TW, Bestwick RK, Mathews CK.. Detection of activities that interfere with the enzymatic assay of deoxyribonucleoside 5’-triphosphates. J Biol Chem 1980;465:6640–6645. [PubMed] [Google Scholar]
- 28. Pontarin G, Gallinaro L, Ferraro P. et al. Origins of mitochondrial thymidine triphosphate: dynamic relations to cytosolic pools. Proc Natl Acad Sci USA 2003;100:12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khym JX. An analytical system for rapid separation of tissue nucleotides at low pressures on conventional anion exchangers. Clin Chem 1975;21:1245–52. [PubMed] [Google Scholar]