Abstract
Since the discovery of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system (Cas) as a tool for gene editing a plethora of locus-specific as well as genome-wide approaches have been developed that allow efficient and reproducible manipulation of genomic sequences. However, the seemingly unbound potential of CRISPR/Cas does not stop with its utilization as a site-directed nuclease. Mutations in its catalytic centers render Cas9 (dCas9) a universal recruitment platform that can be utilized to control transcription, visualize DNA sequences, investigate in situ proteome compositions and manipulate epigenetic modifications at user-defined genomic loci. In this review, we give a comprehensive introduction and overview of the development, improvement and application of recent dCas9-based approaches.
Keywords: CRISPR/Cas, dCas9, CRISPRi, CRISPRa, CasID, CASPEX, CRISPR imaging
Introduction
Prokaryotes not only employ a variety of innate defense mechanisms against foreign viral or plasmid DNA, such as restriction/modification systems or blocking of phage adsorption [1], but also possess a sophisticated adaptive defense mechanism [2] called clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas).
First observed in Escherichia coli, approximately half of bacteria (∼40%) and nearly all archaea (∼90%) are equipped with [3–5] these systems, which rely on small CRISPR-RNAs (crRNAs) to guide nucleases to invading nucleic acids. All CRISPR/Cas systems comprise a set of Cas genes, organized in operons, and a CRISPR-locus, harboring an array of foreign DNA-derived genome-targeting sequences (termed spacers) flanked by identical direct repeats [6].
In general, CRISPR/Cas-mediated adaptive immunity occurs in the following steps. First, upon phage infection or plasmid uptake, short stretches (∼30 bp) of exogenous DNA (termed protospacer) are recognized and integrated into the CRISPR-array. Second, transcription of this array, results in a pre-CRISPR-RNA (pre-crRNA), which is subsequently processed into small mature crRNAs containing a portion of the direct repeat sequence and the spacer. Third, the crRNA forms a ribonucleoprotein-complex with Cas protein(s) and, in case of type II systems, a trans-activating crRNA (tracrRNA). Finally, the complementarity between the spacer of the crRNA and the protospacer of the invading DNA and a short protospacer-adjacent motif (PAM) lead to binding of this complex and target degradation [6].
Based on the presence of unique Cas proteins, the modes of crRNA maturation and RNA-guided interference, CRISPR/Cas systems are subdivided into two classes and five main types (type I, II, III, V and VI). Class I systems include type I as well as type III systems and rely on multi-subunit effector proteins for target degradation. In type I systems, the endonucleases Cas6 or Cas5d facilitate the maturation of crRNA [7–9], which in turn interacts with a complex of five Cas proteins (CasA–E) called Cascade (CRISPR-associated complex for antiviral defense) for target recognition via complementary base pairing [10]. Upon target binding, conformational changes (R-loop formation) lead to recruitment of the nuclease Cas3, which facilitates DNA degradation [11]. In type III systems, Cascade is replaced by a complex consisting of repeat units of Csm or Cmr proteins and target degradation requires Cas10 [11–15].
Contrary to Class I, Class II CRISPR/Cas systems (types II, V, and VI) only require a single effector protein for target degradation. Cas9 is the major hallmark for type II CRISPR/Cas systems and forms a bi-lobed structure with a larger recognition lobe (REC lobe), a smaller nuclease lobe (NUC lobe) and a C-terminal PAM-interacting domain. Cas9 interacts with the repeat: anti-repeat sequences of the tracrRNA: crRNA duplex via a positively charged arginine-rich motif at the inner surface of the REC lobe. RNA-binding induces a conformational change and reorientation of the NUC lobe toward the REC lobe. The two lobes form a clam-like shape with a positively charged central channel, which accommodates both the tracrRNA: crRNA duplex and the target DNA. Furthermore, the two nuclease domains RuvC and HNH in the NUC lobe are positioned in a favorable way for subsequent target cleavage [16, 17].
For genome engineering purposes, the site-directed nuclease Cas9 has been used in combination with a fusion of the crRNA and tracrRNA into a single guide RNA (sgRNA), to induce a double-stranded break (DSB) at a desired locus in a great variety of cell types and organisms, including human, mouse, fly, worm and zebrafish [18–22]. DSBs are resolved by the hosts repair machinery either by non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ) or homology-directed repair (HDR). While the NHEJ-pathway results in a high frequency of insertions or deletions (indels) near the break-site, MMEJ leads to deletions only. Both pathways, however, cause premature stop codons or other frameshift mutations and eventually a knock-out of gene function [23]. Yet, in the presence of a homologous donor DNA, DSBs can be repaired while introducing defined sequences [24].
To increase the likelihood of HDR and thus the insertion of defined homologous DNA sequences, Cas9 nuclease domains have been mutated/engineered to convert Cas9 into a nicking enzyme with only one functional nuclease domain. The resulting single strand breaks are preferentially repaired by HDR and the frequency of NHEJ is decreased. The most commonly used point mutations to inactivate either of the two nuclease domains are D10A and H840A for RuvC and HNH, respectively [16, 17, 21, 25, 26]. The Cas9 RuvC domain shares structural similarities with an RNase H fold found in RuvC Holliday junction resolvases of prokaryotes, such as E. coli and Thermus thermophilus [27, 28]. These nucleases are characterized by four catalytic amino acids and cleave their target via a two-metal mechanism. In addition to the D10A mutation, it was shown that substituting either Glu762, His983 or Asp986 with alanine also resulted in a Cas9 nickase [17]. The catalytic center of the HNH domain comprises a ββα-metal fold and shares structural similarities to the phage T4 endonuclease VII and the Vibrio vulnificus nuclease. Contrary to the RuvC domain, HNH nuclease activity depends on three catalytic residues and is facilitated by a single-metal mechanism [29, 30]. Besides the H840A mutation, it was demonstrated that in a N863A mutant Cas9, HNH-mediated substrate cleavage is abolished [17].
Importantly, by mutating both nuclease domains simultaneously, Cas9 can be engineered into a RNA-guided DNA-binding platform, which still binds DNA in a sequence-specific manner without cleaving the underlying target [26, 31]. To date, this catalytically dead Cas9 (dCas9) has been employed outside the gene editing context, for example, for genome visualization, transcriptional regulation, epigenetic manipulation and investigation of chromatin composition (Fig. 1). In this review, we present a current overview of those dCas9-based CRISPR/Cas methodologies.
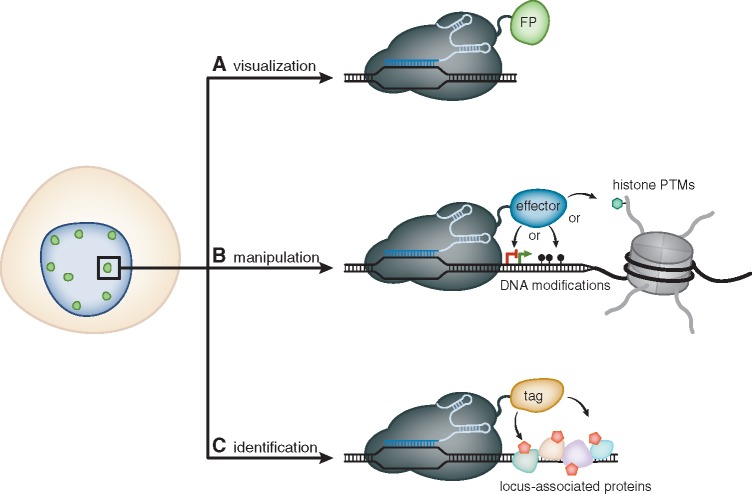
Figure 1:
Overview of dCas9-based applications to study and manipulate chromatin. (A) Catalytically inactive Cas9 (dCas9) represents a RNA-guided DNA-binding platform that can be harnessed to target FP to a pre-defined genomic sequence and allows to visualize spatiotemporal dynamics of chromatin in living cells. (B) Fused to a variety of effector proteins, dCas9 can be employed to directly alter the transcriptional state of specific genes or to precisely manipulate epigenetic marks, such as CpG-methylation or post-translational modifications of histones. (C) Additionally, dCas9 can be fused to biotin ligases (tag = BirA* or APEX2). This approach allows to biotinylate (red pentagons) locus-associated proteins and to subsequently identify them by mass spectrometry.
Earlier approaches
For decades, sequence-specific visualization of chromatin mainly relied on fluorescence in situ hybridization (FISH)-based approaches. Here, the target sequence is detected via complementary base pairing with an epitope- or fluorophore-labeled nucleic acid probe after the genomic DNA has been denatured. Using this technique on fixed cells, entire chromosomes, chromosome arms or single loci have been visualized. By combining different fluorophores, simultaneous detection of several loci or even entire sets of chromosomes can be achieved [32]. By omitting the DNA denaturation step, the FISH approach can be adapted for the visualization of RNA. Moreover, sophisticated methods, such as the fusion of MS2 aptamers (discussed in Section Site-specific visualization of genomic elements) to the target RNA or the microinjection of molecular beacons, facilitate imaging of individual RNA molecules in living cells [33]. In living cells, bulk chromatin can be visualized by cell-permeable DNA-dyes (e.g. DRAQ5), fluorescently labeled nucleotides (e.g. Cy5-dUTP) or histones fused to fluorescent proteins (FP) (e.g. H2B-GFP) [34–36]. Furthermore, specific genomic sequences that are characterized by a defined protein composition, such as centromeres or telomeres can be traced by fluorescently tagging their associated binding proteins [37, 38]. Since loci that meet this requirement are limited, lac or tet operator repeats were introduced at specific genomic regions via genome engineering. These sequences can then be recognized by Lac- or Tet-repressor proteins, respectively, fused to FP [39, 40]. However, since these large artificial sequences might interfere with chromatin dynamics, this method only provides indirect information about the native locus. Hence, live-cell approaches to examine the spatiotemporal dynamics of endogenous loci rely on DNA-interacting proteins, which bind their target in a sequence-specific manner.
Recognition and labeling of user-defined sequences was first realized utilizing zinc finger (ZnF) proteins [41]. This highly diverse group of proteins serves a large variety of biological functions, including transcriptional activation, protein folding, regulation of apoptosis and nucleic acid binding [42]. Notably, the classic Cys2His2 ZnF structural motif, which was first described in the transcription factor IIIA from Xenopus laevis, is conserved among higher eukaryotes and represents the predominant DNA-binding domain in humans [43–45].
Individual Cys2His2 ZnF domains comprise ∼30 amino acids, forming a ββα-motif, in which two cysteines and histidines coordinate a single Zn2+ ion. Target recognition is predominantly facilitated by the α-helix, which establishes contact to three bases within the major groove of the target DNA [46, 47]. Since each ZnF motif recognizes a distinct base triplet, tandem arrangement of up to six modules into a polydactyl ZnF protein (PZF) enables the recognition of unique genomic loci [48]. Fused to green fluorescent protein (GFP), PZFs were the first modular DNA-binding proteins, which were used to visualize repetitive genomic sequences in vivo [49]. It was shown that PZFs can be harnessed to trace major satellite sequences in mouse cells, as well as centromeric repeats in Arabidopsis, demonstrating that this method can be applied in different organisms. Although PZFs have additionally been successfully employed for genome editing and transcriptional regulation, it has been demonstrated that individual zinc fingers display a preference for GC-rich substrates and that neighboring modules affect each other’s target specificity. Hence, target sequence prediction is limited and newly designed PZFs have to be subjected to a rigorous selection process, rendering this approach laborious and expensive [50–52].
To overcome these limitations, fluorescently tagged designer transcription activator-like effectors (dTALEs) have been employed to substitute PZFs [53–57]. Contrary to PZFs, target recognition via dTALEs is facilitated via a simple repeat variable diresidue (RVD)-based code. The fact that one RVD specifically recognizes one nucleotide greatly simplifies the process of designing new dTALEs. Importantly, dTALEs are characterized by a high target specificity and, in fact, were shown to distinguish 1–2 nucleotide differences on the target sequence [58]. Hence, fluorescent dTALEs allow to detect individual chromosomes by single nucleotide polymorphisms (SNPs) [54]. Despite these evident advantages over PZFs, the highly repetitive central domain of dTALEs has been suggested to cause self-assembly and formation of protein aggregates, preventing an effective binding of the cognate target DNA. To this end, a recent study demonstrated that fusion of dTALEs to the chaperone-like protein thioredoxin (TRX) enhances their solubility in human cells [55]. However, the repetitive nature of the DNA-recognition domain renders it difficult to reassemble dTALEs for different sequences and necessitates the use of laborious cloning techniques [59–61].
Site-specific visualization of genomic elements
CRISPR/Cas-based imaging (CRISPR imaging) can be regarded as the next generation of tools to study chromatin dynamics in living cells. Contrary to PZF- or dTALE-based approaches, target specificity is solely mediated by the sgRNA. Therefore, CRISPR imaging can easily be customized to visualize new sequences by simply exchanging the sgRNA without the need to replace the protein itself. By employing CRISPR imaging, repetitive sequences have been successfully visualized [62, 63]. Although such tandem repeats are present in virtually all eukaryotes, it might prove difficult to find one near a locus of interest. Tracing of single-copy loci, therefore, requires targeting of many different consecutive sequences in order to enrich the signal over background levels [63]. In this context, CRISPR imaging is likely to be superior, as it only requires the introduction of small RNAs, whereas for dTALE- or PZF-based approaches, different proteins for each target sequence have to be expressed. This is illustrated by the fact that labeling of a non-repetitive sequence via dTALEs has not been realized, so far. Moreover, recent studies presented a method that employs a cocktail of restriction enzymes and controlled micrococcal nuclease (MNase) digests to generate a genome-wide library of sgRNAs [64, 65]. Further refinement of these methods could allow the production of sgRNA-libraries, which are specific for whole chromosomes or non-repetitive subsets of chromosomes. Together with suitable means to deliver these libraries into living cells (e.g. via lentiviral transduction), CRISPR imaging could facilitate to monitor the spatiotemporal dynamics of chromosomes in real time.
Since the first demonstration of CRISPR imaging, great efforts have been made to further optimize this system (Fig. 2). One issue of visualizing single-copy loci via CRISPR imaging is a rather low signal-to-noise ratio, caused by freely diffusing dCas9-eGFP molecules (Fig. 2A and B). To this end, it has been recently demonstrated that the SunTag (SUperNova tag) is suitable to significantly amplify the fluorescence signal at targeted loci [66, 67]. This system comprises a peptide, which contains up to 24 tandemly arranged copies of a short peptide epitope (GCN4), and a GFP-tagged cognate single-chain variable fragment antibody (scFv). By combining the SunTag with CRISPR imaging of telomeres, it was shown that a ∼20-fold signal enhancement is possible. Due to this bright signal, it would be feasible to visualize genomic loci with lower light illumination, thus reducing phototoxic effects during long-term imaging. Moreover, high signal-to-noise ratios enabled the visualization of two low-repeat loci in human embryonic kidney (HEK293T) cells [67].
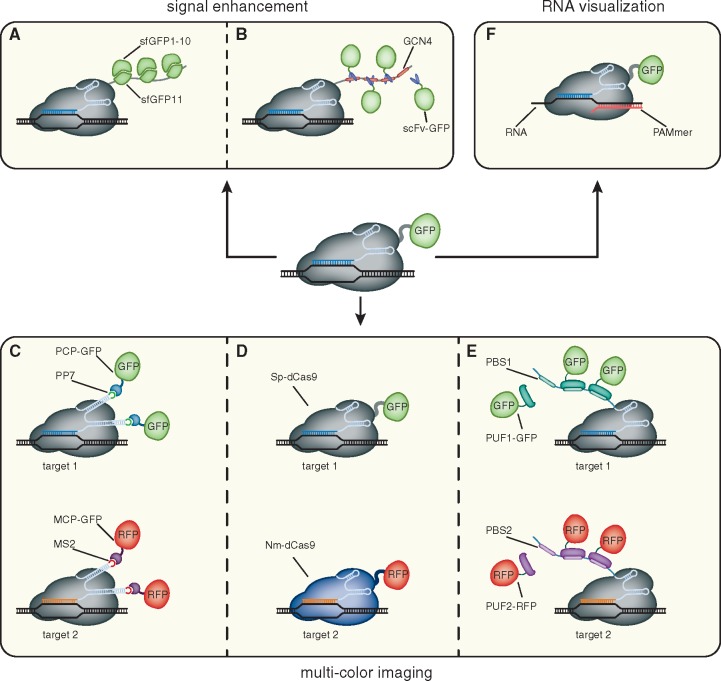
Figure 2:
Expanding the CRISPR imaging toolkit. (A, B) To enhance the signal-to-noise ratio of CRISPR imaging, dCas9 is fused to arrays of small peptide epitopes (GCN4 and sfGFP11). These epitopes then either recruit fluorescent molecules (scFv-GFP) or are complemented (sfGFP1–10), reconstituting fluorescence. (C–E) Multi-color CRISPR imaging can be achieved by either co-expressing differentially labeled orthogonal dCas9 proteins (Sp-dCas9-GFP and Nm-dCas9-RFP) or by fusion of RNA aptamers (PP7 or MS2) to the sgRNA and co-expressing the cognate, fluorescently tagged binding proteins (PCP-GFP and MCP-GFP, respectively). Moreover, sgRNAs can be appended by PUF-binding sites (PBS1 or PBS2, respectively). These sites are then recognized by differentially tagged PUF proteins (PUF1-GFP and PUF2-RFP). (F) By substituting the PAM sequence in the form of an oligonucleotide (PAMmer), dCas9 can be targeted to single stranded RNA molecules.
Similar to the SunTag, a split version of super-folder GFP (sfGFP) has been adopted as an epitope tag for signal enhancement in targeted gene activation and fluorescence imaging [68]. For this approach, sfGFP is split between the 10th and 11th β-strand, resulting in the non-fluorescent sfGFP1–10 and sfGFP11, which serves as a short epitope. A tandem array, comprising up to seven repeats of the sfGFP11 epitope (dCas9-sfGFP11x7), can be fused to a protein of interest (POI) and subsequently recognized by the co-expressed sfGFP1–10 fragment. Upon self-complementation, the chromophore matures and sfGFP regains its fluorescence [69]. By expressing dCas9-sfGFP11x7 together with VP64-sfGFP1–10, it was demonstrated that this system is capable to drastically increase the expression level of the targeted CXCR4 gene. Hence, it would be interesting to evaluate, whether the sfGFP11-tag can be adopted for CRISPR imaging. Whereas the SunTag system still might cause background fluorescence due to unbound scFv-GFP molecules, this GFP-derived epitope tag would be particularly promising, since sfGFP1–10 can be overexpressed without causing background signals. Moreover, a similar tag has also been derived from super-folder Cherry (sfCherry11), which could enhance the versatility of this approach [68].
In comparison to dTALE-based visualization of genomic elements, one inherent drawback of the original CRISPR imaging technique is the fact that simultaneous multi-color labeling of several loci is not feasible. To overcome this limitation, orthogonal dCas9 proteins from the three different bacterial species Streptococcus pyogenes (Sp-dCas9), Neisseria meningitidis (Nm-dCas9) and Streptococcus thermophilus (St1-dCas9) were harnessed (Fig. 2D) [70]. Importantly, target recognition of these dCas9 variants is constrained by different PAM sequences, enabling multiplexed sgRNA-guided recruitment to multiple genomic sites [71]. Hence, by simultaneously co-expressing differently tagged dCas9 orthologs with their cognate sgRNAs, each specific for a distinct locus, it was possible to resolve the inter- and intrachromosomal distances between two repetitive sequences in living human cells. In addition, another study repurposed the small Cas9 ortholog from Staphylococcus aureus (Sa-dCas9) for multiplexed CRISPR imaging and demonstrated that a combination of Sp-dCas9-eGFP and Sa-dCas9-mCherry is capable to resolve two different loci, which are separated by less than 300 kb [72].
Although representing a significant improvement in CRISPR imaging, orthogonal CRISPR/Cas systems are characterized by more complex PAM requirements. For instance, whereas Sp-dCas9 recognizes a 5´-NGG-3´-sequence, Nm-dCas9 is only targeted to sequences followed by a 5´-NNNNGATT-3´ motif, thus restricting the flexibility toward target sequences. To circumvent this issue, a second strategy for multi-color CRISPR imaging has been developed (Fig. 2C). Here, the S. pyogenes sgRNA is fused to the RNA aptamers MS2 or PP7 that are specifically bound by the bacteriophage coat proteins MCP (MS2 coat protein) and PCP (PP7 coat protein), respectively [73–75]. The resulting scaffold RNA (scRNA) is still capable to guide dCas9 to the desired target and can be detected by fluorescently tagged coat proteins. A similar approach combined dCas9-mediated DNA-recognition with Pumilio-assisted RNA-binding (Fig. 2E) [76]. This technique, termed Casilio, utilizes the fact that the RNA-binding domain of the Drosophila protein Pumilio (PUF) can be reprogrammed to bind specific 8-mer RNA sequences (PUF-binding site, PBS) [77]. Therefore, appending different sgRNAs with PUF-binding sites, specific for differently tagged PUF domains, facilitates simultaneous tracing of multiple genomic loci.
Recently, the scope of CRISPR imaging has been further expanded by demonstrating that dCas9 can be harnessed as a tool to visualize endogenous, unmodified RNA (Fig. 2F) [78]. Although the type II CRISPR/Cas system has evolved to target double-stranded DNA, Cas9 can be guided to RNA, when the PAM is provided in trans by an oligomer (PAMmer) that is partially complementary to the targeted RNA [79]. However, due to high background signals, so far RNA could only be visualized, when large quantities accumulated within stress granules. In this case, fusing dCas9 to the aforementioned sfGFP11-tag is likely to enhance the signal-to-noise ratio, thus facilitating the tracking of less abundant RNAs. It is worth to mention that an additional Cas protein, Cas13, has recently been harnessed for targeted RNA visualization, as well as RNA knockdown and editing [80, 81]. This CRISPR effector is a member of the Class II type VI system, which specifically targets single-stranded RNA in an crRNA-guided manner and requires two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains for RNA degradation [82]. Similar to Cas9, Cas13 can be converted into a catalytically inactive variant (dCas13) by substituting catalytic residues within the HEPN domains. By fusing dCas13 to a monomeric superfolder GFP (msfGFP), accumulation of actin beta (ACTB) transcripts to stress granules could be tracked in living cells [80]. Interestingly, when fused to a hyperactive mutant of adenosine deaminase acting on RNA 2 (ADAR2), dCas13 has been demonstrated to correct 33 out of 34 tested disease-relevant G-to-A mutations [81]. This ability to manipulate proteins on the RNA-level could be a putative treatment for a variety of diseases.
Collectively, CRISPR imaging offers high flexibility and represents a time- and cost-effective alternative to visualization techniques, based on modular DNA-binding proteins. Especially the potential to trace non-repetitive sequences renders CRISPR imaging a versatile tool to study chromatin dynamics.
Transcriptional regulation and epigenetic manipulation
The eukaryotic transcriptional landscape is regulated by a complex network of epigenetic modifications, transcription factors and chromatin organization. To dissect cause and consequence of any of these elements, it is crucial to have tools at hand that allow their precise manipulation. Since the first description of Cas9-mediated genome engineering, the CRISPR/Cas system has in parallel been refined for site-specific transcriptional regulation and manipulation of epigenetic marks and represents a powerful tool to interrogate the mechanistic relationship between chromatin state and regulation of gene expression (Fig. 1B).
The applicability of the CRISPR/Cas system to specifically repress gene transcription has first been described in E. coli. Termed CRISPR interference (CRISPRi), DNA-bound dCas9 can either interfere with transcription elongation by physically blocking RNA polymerase progression or hinder the binding of essential transcription factors [31, 83]. In prokaryotes, this method can lead to up to a ∼1000-fold reduction of mRNA levels. In mammalian cells, however, only a modest ∼2-fold reduction of reporter transcription was observed [31]. Since these initial studies, the CRISPRi system has been further refined. For a more efficient repression of transcription in mammalian cells, dCas9 can be either fused to the repressor domain Krüppel-asociated box (KRAB) or four-linked mSin3 interaction domains (SID4X). Using these fusion proteins, endogenous genes, such as octamer-binding protein 4A (OCT4A), sex determining region Y (SRY)- box 2 (SOX2), C-X-C chemokine receptor type 4 (CXCR4) and cluster of differentiation 71 (CD71), have been specifically and efficiently repressed [84–86].
In addition to CRISPRi, the CRISPR/Cas system has also been harnessed to specifically activate gene expression. In E. coli, CRISPR-mediated gene activation (CRISPRa) was realized by fusing the ω-subunit of RNA polymerase to dCas9. Targeted to the promoter of a reporter gene, this fusion protein led to the recruitment of the polymerase holoenzyme, resulting in up to 3-fold increase in gene expression [83]. In eukaryotic cells, CRISPRa is commonly achieved by fusing dCas9 to heterologous transcription activation domains, such as VP64 or p65 [84, 87–89]. To achieve significant activation of endogenous genes, however, it is often necessary to recruit dCas9-effector to neighboring sites via multiple different sgRNAs [88, 89]. By screening more than 20 effector proteins with known transcriptional roles, one study led to the development of a powerful hybrid activation domain [90]. This tripartite domain, termed VPR, consists of tandemly linked VP64, p65 and the Epstein-Barr virus R transactivator (Rta), which synergistically enhance gene expression. Targeting dCas9-VPR fusion proteins to endogenous genes revealed that, compared to dCas9-VP64 alone, transcription can be amplified up to ∼300-fold. Moreover, this dCas9-VPR approach was successfully tested in diverse eukaryotic cells, such as human, mouse, fly and yeast.
Analogous to signal amplification in CRISPR imaging approaches, both dCas9 and the sgRNA have been engineered to indirectly target effector proteins to specific loci. As mentioned before, co-expressing dCas9-sfGFP11x7 with VP64-sfGFP1–10 and a sgRNA targeting the CXCR4 locus in K562 cells, resulted in a ∼45-fold increase of CXCR4 expression, when compared to dCas9-VP64 [68]. Additionally, a SunTag array, consisting of 10 copies of the GCN4 epitope, was fused to dCas9. This construct was successfully used to recruit scFv-VP64 fusions to CXCR4 and CDKN1B loci in K562 cells, resulting in significant activation of gene expression [66]. The efficiency of both CRISPRi and CRISPRa approaches highly depends on the genomic position to where dCas9 is recruited. To elucidate targeting rules for CRISPR-based modulation of transcription, one study performed an extensive screen, targeting a set of 49 genes with a total of 54.810 sgRNAs. It was demonstrated that for CRISPRi (via dCas9-KRAB), active sgRNAs cluster in a window from -50 bp to +300 bp relative to the transcription start site (TSS). For CRISPRa (via dCas9-SunTag/VP64), a peak in transcription activation was observed, when dCas9 was targeted between -400 bp to -50 bp upstream of the TSS [91].
Besides engineering the dCas9 protein, several studies demonstrated that the sgRNA can be modified to recruit effector proteins to a desired genomic locus. In a first attempt, two MS2 RNA stem-loops were incorporated at the 3´ end of the sgRNA, which subsequently can be recognized by MCP-VP64 fusion proteins. Although this system was capable to activate ZFP42 expression, it was outperformed by dCas9-VP64 [92]. To further improve sgRNA-mediated manipulation of transcription, the synergistic activation mediator (SAM) technology was developed. For this, MS2 aptamers were inserted at the tetraloop and the stem-loop 2 of the sgRNA. Notably, at these positions, the sgRNA does not interact with the dCas9 protein. MS2 then recruits MCP fused to p65 and HSF1 (heat shock factor 1). Combining this modified sgRNA with a dCas9-VP64 fusion resulted in drastically increased expression levels of targeted genes and was shown to simultaneously upregulate 10 genes [114]. In contrast to RNA-hairpin structures, the Casilio technique introduces specific 8-mer sequences (PBS; see above) that are recognized by PUF domains. Using this approach, it was demonstrated that recruitment of PUF-p65-HSF1 results in robust activation of SOX2 and OCT4 [76].
Direct fusions of dCas9 with epigenetic effectors have been successfully employed to activate transcription of previously silenced genes. For instance, recruiting the catalytic core of the human acetyltransferase p300 to promoters or enhancers leads to robust transcriptional activation and directly implicates H3K27ac in this process [93]. Additionally, targeted DNA demethylation, mediated by dCas9-ten-eleven translocation 1 (TET1) fusions results in gene reactivation [94–96]. Analogous to targeted activation, the CRISPR/Cas system has been utilized to recruit DNA de novo methyltransferases or histone demethylases to specific gene regulatory regions, resulting in a local repressive epigenetic state and thus gene silencing [97–99].
As for CRISPRi and CRISPRa, dCas9-based epigenetic editing approaches have been further improved to amplify the enzymatic activity at the target site and to enable the use of more modular strategies. The dCas9-SunTag system was employed to recruit epigenetic effectors to influence the transcriptional state of specific genes. Whereas targeting the catalytic domain of TET1 (scFv-TET1CD) to the imprinted H19 locus led to DNA demethylation and gene expression, dCas9-SunTag-DNMT3A was able to increase CpG methylation and thereby repress HOXA5 transcription [100, 101]. Furthermore, a recent study used MS2 stem loops incorporated into the sgRNA to recruit the epigenetic effector proteins fused to MCP. This setup allowed the efficient and specific demethylation and of the RANKL and MAGEB2 gene using the TET1 catalytic domain. Interestingly, demethylation was not observed directly at the target site but in a distance of 100–300 bp pointing toward so far uncharacterized mechanisms that restrict access of dCas9 to the target site or cellular pathways that protect certain CpGs from demethylation [96]. Another approach utilized dCas9 fused to a GFP-binding nanobody (GBP), enabling specific and effective recruitment of GFP-tagged epigenetic effectors to manipulate local DNA methylation [102].
Interestingly, several studies have shown that epigenetic editing is only transient and does not result in stable memory once the targeting system is removed [103–105]. However, a recent study demonstrated that combinatorial targeting of multiple effector domains involved in the silencing of endogenous retroviruses results in the stable silencing of gene expression even after the engineered repressors where removed [106]. This underlines the importance of the concerted action and recruitment of multiple epigenetic pathways components for the establishment of efficient long-term epigenetic memory.
Investigating chromatin composition
Besides visualizing the spatiotemporal dynamics of chromatin and manipulating the transcriptional state of endogenous genes, it is crucial to assess the protein composition of specific genomic loci to fully decipher the function of chromatin. To this end, several methods have been developed that facilitate the profiling of genome-wide or local binding sites of chromatin proteins, as well as detecting protein-protein associations.
Chromatin immunoprecipitation (ChIP) represents a powerful and well established method to study the distribution of a DNA-binding protein along the genome. For this, DNA-protein complexes are crosslinked in vivo. Subsequently, the chromatin is fragmented and DNA fragments that are bound by the POI are immunoprecipitated via specific antibodies. Finally, DNA-POI crosslinks are reversed and the released DNA is assayed by next-generation sequencing (ChIP-seq), PCR (ChIP-qPCR) or microarray (ChIP-chip) to determine the sequences bound by the POI [107]. However, these classical ChIP approaches are often limited by the availability of suitable antibodies and only provide information on the global distribution of POIs. To identify proteins that are associated with a specific genomic locus, engineered DNA-binding molecule-mediated ChIP (enChIP) has been developed (Fig. 3A) [108]. Here, FLAG-tagged dCas9 is recruited to a locus of interest, chromatin is crosslinked, fragmented and dCas9-bound fragments are enriched via FLAG-tag specific antibodies. After reversion of crosslinks, the isolated complexes can then be analyzed by mass spectrometry. Another approach to identify the chromatin composition of individual genomic loci, termed CasID, combines proximity-dependent biotin identification (BioID) with the target specificity of the CRISPR/Cas system (Fig. 3B) [109, 110]. For this, dCas9 is fused to the promiscuous biotin ligase BirA*. The culture medium of BirA*-dCas9 expressing cells is then supplemented with exogenous biotin, resulting in the biotinylation of proteins that are located in the near proximity (∼10 nm) of the targeted locus. After affinity purification via streptavidin-coated beads, biotinylated proteins can be analyzed by mass spectrometry. Using this technique, known and unknown proteins of telomeres, minor and major satellites could be identified. Recently, a similar method employed a fusion of dCas9 with the engineered ascorbate peroxidase APEX2 (CASPEX) [111]. Comparable to BirA*, APEX2 also ligates biotin to nearby lysine residues, the labeling radius, however, is smaller. Whereas CasID has only been tested on repetitive sequences, CASPEX successfully identified proteins at the single-copy loci hTERT and c-MYC. In addition to CRISPR-mediated recruitment of biotin ligases, dCas9, carrying a biotin acceptor tag, has been employed to study chromatin composition. Co-expressing BirA leads to in vivo biotinylation of this modified dCas9. Subsequent enrichment of targeted loci via streptavidin allowed to comprehensively study proteins and long-range DNA interactions associated with the β-globin cluster in K562 cells [112].
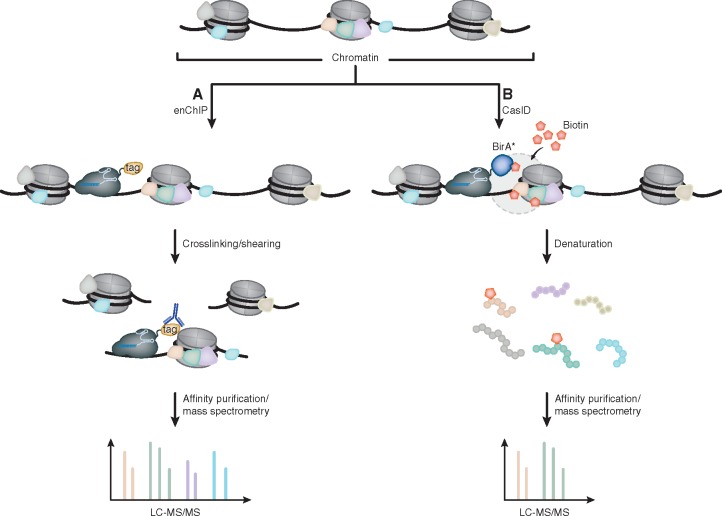
Figure 3:
Identification of locus associated proteins by dCas9. (A) For enChIP, dCas9 is fused to a FLAG-tag and targeted to a locus of interest. Chromatin is then crosslinked and fragmented. dCas9-bound chromatin fragments are subsequently isolated by FLAG-specific antibodies and analyzed via mass spectrometry. (B) Contrary to enChIP, CasID requires the expression of dCas9 fused to the promiscuous biotin ligase BirA*. After the culture medium has been supplemented with exogenous biotin, BirA* catalyzes the addition of biotin to lysine residues of proteins that are in close proximity to the dCas9-BirA* fusion protein. Lysis of the cells and denaturation of proteins is then followed by affinity purification of biotinylated peptides, which are identified via tandem MS.
Summary
The recent advances in dCas9-based approaches constitute powerful and versatile tools to investigate molecular pathways on a site-specific level. dCas9 as a recruitment platform of transcriptional activators and epigenetic modifiers offers the unique opportunity to dissect the cause and effect of epigenetic modifications on transcriptional regulation and vice versa. Epigenome-wide association studies (EWAS) are generating hundreds and thousands of epigenomic markers associated with human disease. dCas9-based epigenomic editing will be invaluable to single out the disease causing changes from indirect effects opening the doors to new treatment options and a better understanding of human pathogenesis.
While Chromatin precipitation and sequencing (ChIP-seq) approaches have been invaluable to investigate the localization of individual proteins throughout the genome, it is the complex interplay of multiple proteins and pathways that govern the regulation of gene expression. Technologies such as CasID and CASPEX will allow unprecedented insights into the complex regulatory mechanisms that control gene expression by unraveling the locus-specific changes in chromatin composition in response to signalling, differentiation and disease progression. Similarly, visualization of genomic sequences using dCas9 will greatly enhance our understanding of nuclear architecture dynamics during cellular differentiation and cell cycle in living cells.
Taken together, the CRISPR/Cas system represents a revolution that goes far beyond gene editing/genome engineering approaches. By inactivating the catalytic activity of Cas9, the enzyme becomes a universal and site-specific recruitment platform that opens a plethora of new applications for basic as well as medical research.
References
- 1. Samson JE, Magadan AH, Sabri M. et al. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 2013;11:675–87. [DOI] [PubMed] [Google Scholar]
- 2. Barrangou R, Fremaux C, Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709.. [DOI] [PubMed] [Google Scholar]
- 3. Grissa I, Vergnaud G, Pourcel C.. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007;35:W52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ishino Y, Shinagawa H, Makino K. et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987;169:5429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiedenheft B, Sternberg SH, Doudna JA.. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012;482:331–338. [DOI] [PubMed] [Google Scholar]
- 6. Ratner HK, Sampson TR, Weiss DS.. Overview of CRISPR–Cas9 biology. Cold Spring Harb Protoc 2016;2016:pdb.top088849.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carte J, Wang R, Li H. et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 2008;22:3489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garside EL, Schellenberg MJ, Gesner EM. et al. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA 2012;18:2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nam KH, Haitjema C, Liu X. et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 2012;20:1574–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jore MM, Lundgren M, van Duijn E. et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 2011;18:529–36. [DOI] [PubMed] [Google Scholar]
- 11. Westra ER, van Erp PB, Kunne T. et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 2012;46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hrle A, Su AA, Ebert J. et al. Structure and RNA-binding properties of the type III-A CRISPR-associated protein Csm3. RNA. Biol 2013;10:1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Samai P, Pyenson N, Jiang W. et al. Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell 2015;161:1164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Staals RH, Agari Y, Maki-Yonekura S. et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 2013;52:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Staals RH, Zhu Y, Taylor DW. et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 2014;56:518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jinek M, Jiang F, Taylor DW. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014;343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nishimasu H, Ran FA, Hsu PD. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014;156:935–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dickinson DJ, Goldstein B.. CRISPR-based methods for caenorhabditis elegans genome engineering. Genetics 2016;202:885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henao-Mejia J, Williams A, Rongvaux A. et al. Generation of genetically modified mice using the CRISPR-Cas9 genome-editing system. Cold Spring Harb Protoc 2016;2016:pdb.prot090704.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Housden BE, Perrimon N.. Cas9-mediated genome engineering in Drosophila melanogaster. Cold Spring Harb Protoc 2016;2016:pdb.top086843.. [DOI] [PubMed] [Google Scholar]
- 21. Mali P, Yang L, Esvelt KM. et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vejnar CE, Moreno-Mateos MA, Cifuentes D. et al. Optimized CRISPR-Cas9 system for genome editing in zebrafish. Cold Spring Harb Protoc 2016;2016:pdb.prot086850.. [DOI] [PubMed] [Google Scholar]
- 23. McVey M, Lee SE.. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 2008;24:529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu Rev Biochem 2010;79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cong L, Ran FA, Cox D. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jinek M, Chylinski K, Fonfara I. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ariyoshi M, Vassylyev DG, Iwasaki H. et al. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell 1994;78:1063–72. [DOI] [PubMed] [Google Scholar]
- 28. Gorecka KM, Komorowska W.. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biertümpfel C, Yang W, Suck D.. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 2007;449:616–20. [DOI] [PubMed] [Google Scholar]
- 30. Li C-L, Hor L-I, Chang Z-F. et al. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J 2003;22:4014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qi LS, Larson MH, Gilbert LA. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013;152:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolzer A, Kreth G, Solovei I. et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005;3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raj A, van Oudenaarden A.. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys 2009;38:255–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanda T, Sullivan KF, Wahl GM.. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 1998;8:377–85. [DOI] [PubMed] [Google Scholar]
- 35. Manders EM, Kimura H, Cook PR.. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J Cell Biol 1999;144:813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin RM, Leonhardt H, Cardoso MC.. DNA labeling in living cells. Cytometry A 2005;67:45–52. [DOI] [PubMed] [Google Scholar]
- 37. Krawczyk PM, Borovski T, Stap J. et al. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci 2012;125:2127.. [DOI] [PubMed] [Google Scholar]
- 38. Shelby RD, Hahn KM, Sullivan KF.. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J Cell Biol 1996;135:545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinett CC, Straight A, Li G. et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 1996;135:1685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roukos V, Voss TC, Schmidt CK. et al. Spatial dynamics of chromosome translocations in living cells. Science 2013;341:660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gersbach CA, Perez-Pinera P.. Activating human genes with zinc finger proteins, transcription activator-like effectors and CRISPR/Cas9 for gene therapy and regenerative medicine. Expert Opin Ther Targets 2014;18:835–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laity JH, Lee BM, Wright PE.. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 2001;11:39–46. [DOI] [PubMed] [Google Scholar]
- 43. Miller J, McLachlan AD, Klug A.. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 1985;4:1609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubin GM, Yandell MD, Wortman JR. et al. Comparative genomics of the eukaryotes. Science 2000;287:2204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tupler R, Perini G, Green MR.. Expressing the human genome. Nature 2001;409:832–3. [DOI] [PubMed] [Google Scholar]
- 46. Elrod-Erickson M, Rould MA, Nekludova L. et al. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure 1996;4:1171–80. [DOI] [PubMed] [Google Scholar]
- 47. Pavletich NP, Pabo CO.. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 1991;252:809–17. [DOI] [PubMed] [Google Scholar]
- 48. Liu Q, Segal DJ, Ghiara JB. et al. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A 1997;94:5525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindhout BI, Fransz P, Tessadori F. et al. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res 2007;35:e107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeFrancesco L. Move over ZFNs. Nat Biotechnol 2011;29:681–4. [DOI] [PubMed] [Google Scholar]
- 51. Ramirez CL, Foley JE, Wright DA. et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods 2008;5:374–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Segal DJ, Dreier B, Beerli RR. et al. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5’-GNN-3' DNA target sequences. Proc Natl Acad Sci U S A 1999;96:2758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma H, Reyes-Gutierrez P, Pederson T.. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci U S A 2013;110:21048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyanari Y, Ziegler-Birling C, Torres-Padilla M-E.. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 2013;20:1321–4. [DOI] [PubMed] [Google Scholar]
- 55. Ren R, Deng L, Xue Y. et al. Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res 2017;27:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thanisch K, Schneider K, Morbitzer R. et al. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res 2014;42:e38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yuan K, Shermoen AW, O’farrell PH.. Illuminating DNA replication during Drosophila development using TALE-lights. Curr Biol 2014;24:R144–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boch J, Scholze H, Schornack S. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009;326:1509–12. [DOI] [PubMed] [Google Scholar]
- 59. Morbitzer R, Elsaesser J, Hausner J. et al. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res 2011;39:5790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reyon D, Tsai SQ, Khayter C. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012;30:460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schmid-Burgk JL, Schmidt T, Kaiser V. et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol 2013;31:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anton T, Bultmann S, Leonhardt H. et al. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus 2014;5:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen B, Gilbert LA, Cimini BA. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013;155:1479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Köferle A, Worf K, Breunig C. et al. CORALINA: a universal method for the generation of gRNA libraries for CRISPR-based screening. BMC Genomics 2016;17:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lane AB, Strzelecka M, Ettinger A. et al. Enzymatically generated CRISPR libraries for genome labeling and screening. Dev Cell 2015;34:373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tanenbaum ME, Gilbert LA, Qi LS. et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014;159:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ye H, Rong Z, Lin Y.. Live cell imaging of genomic loci using dCas9-SunTag system and a bright fluorescent protein. Protein Cell 2017;8:853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kamiyama D, Sekine S, Barsi-Rhyne B. et al. Versatile protein tagging in cells with split fluorescent protein. Nat Comms 2016;7:11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cabantous S, Terwilliger TC, Waldo GS.. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 2005;23:102–7. [DOI] [PubMed] [Google Scholar]
- 70. Ma H, Naseri A, Reyes-Gutierrez P. et al. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A 2015;112:3002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Esvelt KM, Mali P, Braff JL. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 2013;10:1116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen B, Hu J, Almeida R. et al. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res 2016;44:e75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fu Y, Rocha PP, Luo VM. et al. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat Comms 2016;7:11707.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qin P, Parlak M, Kuscu C. et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Comms 2017;8:14725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang S, Su JH, Zhang F. et al. An RNA-aptamer-based two-color CRISPR labeling system. Sci Rep 2016;6:26857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng AW, Jillette N, Lee P. et al. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res 2016;26:254–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen Y, Varani G.. Engineering RNA-binding proteins for biology. FEBS J 2013;280:3734–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nelles DA, Fang MY, O'Connell MR. et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 2016;165:488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. O’connell MR, Oakes BL, Sternberg SH. et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014;516:263–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Abudayyeh OO, Gootenberg JS, Essletzbichler P. et al. RNA targeting with CRISPR–Cas13. Nature 2017;550:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cox DBT, Gootenberg JS, Abudayyeh OO. et al. RNA editing with CRISPR-Cas13. Science 2017;358:1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Abudayyeh OO, Gootenberg JS, Konermann S. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016;353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bikard D, Jiang W, Samai P. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 2013;41:7429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gilbert LA, Larson MH, Morsut L. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kearns NA, Genga RMJ, Enuameh MS. et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 2013;141: 219–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Konermann S, Brigham MD, Trevino A. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 2013;500:472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cheng AW, Wang H, Yang H. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 2013;23:1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maeder ML, Linder SJ, Cascio VM. et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013;10:977–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Perez-Pinera P, Kocak DD, Vockley CM. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 2013;10:973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chavez A, Scheiman J, Vora S. et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gilbert LA, Horlbeck MA, Adamson B. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014;159:647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Konermann S, Brigham MD, Trevino AE. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2014;517:583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hilton IB, D'Ippolito AM, Vockley CM. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choudhury SR, Cui Y, Lubecka K. et al. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016;7:46545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu XS, Wu H, Ji X. et al. Editing DNA methylation in the mammalian genome. Cell 2016;167:233–47.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu X, Tao Y, Gao X. et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2016;2:16009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kearns NA, Pham H, Tabak B. et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 2015;12:401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. McDonald JI, Celik H, Rois LE. et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open 2016;5:866.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Vojta A, Dobrinić P, Tadić V. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016;44:5615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Huang YH, Su J, Lei Y. et al. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol 2017;18:176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Morita S, Noguchi H, Horii T. et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 2016;34:1060–5. [DOI] [PubMed] [Google Scholar]
- 102. Anton T, Bultmann S.. Site-specific recruitment of epigenetic factors with a modular CRISPR/Cas system. Nucleus 2017;8:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Braun SMG, Kirkland JG, Chory EJ. et al. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat Commun 2017;8:560.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Keung AJ, Joung JK, Khalil AS. et al. Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet 2015;16:159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Thakore PI, Black JB, Hilton IB. et al. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 2016;13:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Amabile A, Migliara A, Capasso P. et al. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 2016;167:219–32.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Collas P. The current state of chromatin immunoprecipitation. Mol Biotechnol 2010;45:87–100. [DOI] [PubMed] [Google Scholar]
- 108. Fujita T, Fujii H.. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun 2013;439:132–6. [DOI] [PubMed] [Google Scholar]
- 109. Roux KJ, Kim DI, Raida M. et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 2012;196:801.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Schmidtmann E, Anton T, Rombaut P. et al. Determination of local chromatin composition by CasID. Nucleus 2016;7:476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Myers SA, Wright J, Zhang F. et al. CRISPR/Cas9-APEX-mediated proximity labeling enables discovery of proteins associated with a predefined genomic locus in living cells. 2017; doi.org/10.1101/159517. [DOI] [PMC free article] [PubMed]
- 112. Liu X, Zhang Y, Chen Y. et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell 2017;170:1028–43.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Konermann S, Brigham MD, Trevino AE. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2014;517:583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mali P, Aach J, Stranges PB. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013b;31:833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]