Abstract
Genomic selection is routinely used worldwide in agricultural breeding. However, in Russia, it is still not used to its full potential partially due to high genotyping costs. The use of genotypes imputed from the low-density chips (LD-chip) provides a valuable opportunity for reducing the genotyping costs. Pork production in Russia is based on the conventional 3-tier pyramid involving 3 breeds; therefore, the best option would be the development of a single LD-chip that could be used for all of them. Here, we for the first time have analyzed genomic variability in 3 breeds of Russian pigs, namely, Landrace, Duroc, and Large White and generated the LD-chip that can be used in pig breeding with the negligible loss in genotyping quality. We have demonstrated that out of the 3 methods commonly used for LD-chip construction, the block method shows the best results. The imputation quality depends strongly on the presence of close ancestors in the reference population. We have demonstrated that for the animals with both parents genotyped using high-density panels high-quality genotypes (allelic discordance rate < 0.05) could be obtained using a 300 single nucleotide polymorphism (SNP) chip, while in the absence of genotyped ancestors at least 2,000 SNP markers are required. We have shown that imputation quality varies between chromosomes, and it is lower near the chromosome ends and drops with the increase in minor allele frequency. Imputation quality of the individual SNPs correlated well across breeds. Using the same LD-chip, we were able to obtain comparable imputation quality in all 3 breeds, so it may be suggested that a single chip could be used for all of them. Our findings also suggest that the presence of markers with extremely low imputation quality is likely to be explained by wrong mapping of the markers to the chromosomal positions.
Keywords: genomic selection, imputation, low-density chip, pigs, single nucleotide polymorphism
INTRODUCTION
The key step in animal breeding is the assessment of the animal’s breeding value that in other words is the value of its genes to progeny, its genetic potential, which allows to rank animals in the population in order to select the best-performing ones for desired traits and to include them in the breeding programs. The estimated breeding value (EBV) is an estimate of the genetic merit of the animal which would be passed on to its progeny calculated using mathematical methods. The estimation could be done based on the animal’s own phenotype, parent phenotypes, or the phenotypes of its progeny. Evaluation by parents is characterized by lower accuracy, while evaluation by progeny or by animal’s own phenotype is more accurate, but takes time and for terminal traits could be done only on slaughter (Liesbeth van der Waaij, 2014); thus, it appears to be challenging to obtain an accurate EBV for animals while they are still young. To enable early and accurate evaluation, genomic methods are adopted worldwide. Genomic selection (GS), first proposed by Meuwissen et al. in 2001 (Meuwissen et al., 2001), is an approach under which associations are found between phenotypes and a large set of single nucleotide polymorphism (SNP) markers (often tens of thousand) closely spaced across the genome so as for most or all quantitative trait loci (QTLs) to be in linkage disequilibrium (LD) with 1 marker in a sufficiently large reference population. The inferred associations are used further to calculate genomic EBVs in the animals genotyped for the large set of SNPs, but for which phenotypic data are absent (Goddard and Hayes, 2007; Ibáñez-Escriche et al., 2014; Samorè and Fontanesi, 2016). The implementation of GS makes it possible to assess the breeding values of young animals before they have progeny and reach maturity age, as well as breeding values for the traits associated with resistance/susceptibility to diseases, maternal traits, and the traits which are usually difficult or expensive to measure, or those which take a long time to be evaluated, or can be evaluated only on slaughter, thus promoting their efficient use in breeding programs (Lillehammer et al., 2011; Tribout et al., 2013; Abell et al., 2014; Ibáñez-Escriche et al., 2014). Genomic selection may also be advantageous when crossbred performance needs to be predicted from the performance of purebred animals, which is important, in particular, for pig breeding (Knol et al., 2016).
In commercial pig breeding, a 3-layer breeding pyramid based on the use of a 3-way crossing scheme aimed to take advantage of heterosis and separate selection in dam and sire lines has been used since the 1960s to 1970s. The 3 layers are represented by the nucleus layer which include high-quality purebred animals, the multiplier level at which purebred animals from different lines are multiplied and crossed to obtain the F1 crossbred animals, and the production (commercial) level, at which F1 sows are crossed with purebred sires, the progeny of these crosses being used for pork production. Selection takes place at the nucleus and partially at the multiplier levels (Visscher et al., 2000; Lopes, 2016; Lopez et al., 2016). The use of breeding programs of this kind leads to a certain genetic improvement lag, which is the time taken for genetic improvements achieved in the higher layer to reach the next layers (Bichard, 1971; See, 1995), between the nucleus level at which selection and testing mostly occurs and the commercial level at which terminal (slaughter) hybrids are produced which should have the desired qualities. However, according to Bichard (1971), notwithstanding the improvement lag, genetic improvement achieved at the lower, multiplier and commercial, levels depends directly on the rate of the genetic improvement achieved in the nucleus. Therefore, a high rate of genetic progress at the nucleus layer will accordingly lead to a higher rate of progress on subsequent levels. At the same time, it is known that genetic progress is affected both by the accuracy of breeding value prediction and the length of the generation interval (Falconer, 1989). Genomic selection allows to considerably improve the former and decrease the latter (Lillehammer et al., 2011; Baby et al., 2014), thus reducing the genetic lag size. Another point in which pig breeding may benefit from GS is the enhanced ability to predict crossbred performance based on purebred performance and to control inbreeding in purebred populations by including nonadditive effects in the estimations (Ibáñez-Escriche et al., 2014; Esfandyari et al., 2016; Hidalgo et al., 2016; Christensen et al., 2019), which is of much relevance considering the specificity of pig breeding schemes. Although, the practical implementation of GS in pig breeding has started relatively recently prompted by the development of the first commercial high-density (HD) SNP panel for genotyping (Ramos et al., 2009), the so far obtained results seem to be promising (Simianer, 2009; Lillehammer et al., 2011; Tribout et al., 2012; Ibáñez-Escriche et al., 2014).
While the development of GS was initially fueled by the introduction of high-throughput SNP genotyping using microarray beadchips, which harbor tens of thousands of markers, like those designed by Illumina or Affimetrix (reviewed in Samorè, 2016), high-throughput genotyping also appears to be a factor that in certain way obstructs the broader use of GS in animal breeding, in particular in pig breeding. This is due to the fact that genotyping cost is still rather high, and, considering the large numbers of animals to be genotyped, relatively short generation intervals in pig herds, and lower, compared to dairy cows, economic value of selection candidates, the process may be somewhat cost-intensive and in certain cases even come to be economically unprofitable (Tribout et al., 2013; Ibáñez-Escriche et al., 2014).
One of the possible ways of reducing genotyping costs is the use of low-density chips (LD-chip) and imputation to infer HD genotypes from the LD panel data (Abell et al., 2014) using such methods as FImpute (Sargolzaei et al., 2014) or Beagle (Browning et al., 2018). Ninety percent imputation accuracy is sufficient to obtain genomic evaluations almost identical to evaluations obtained using HD genomes (Wellmann et al., 2013). Recently, several attempts to create LD-chips for use in cattle (Zhang and Druet, 2010; Judge et al., 2016; Aliloo et al., 2018; Korkuć et al., 2019), pig (Gualdrón Duarte et al., 2013; Wellmann et al., 2013; Xiang et al., 2015; Carillier-Jacquin et al., 2018; Grossi et al., 2018), as well as sheep (Ventura et al., 2016; Raoul et al., 2017; O’ Brien et al., 2019), and chicken (Wang et al., 2013; Herry et al., 2018) breeding have been published. The previous studies analyzing the prospects of using imputation from LD panels suggested different approaches for marker selection for LD-chips: random, uniform, or based on LD (Gualdrón Duarte et al., 2013; Judge et al., 2016; Grossi et al., 2018). It may also be useful to include the markers specific for a given population or associated with the traits or diseases important for local breeding programs among the markers in LD panel. It has been also pointed out that the most important factors that should be taken into account when designing an LD panel in order to increase imputation accuracy are the reference population size and the degree of kinship between the test and reference population, the rate of genotyped close relatives in the reference population, as well as the effects of minor allele frequencies (MAFs) for the imputed SNPs and LD threshold (Cleveland and Hickey, 2013; Grossi et al., 2018; Herry et al., 2018). Another important factor to consider when developing LD-chips is SNP density on the LD panel. The results obtained so far by other authors have demonstrated that panels containing about 3K SNP markers might be enough to impute HD genotypes with high confidence in multiple pig breeds (Wellmann et al., 2013; Grossi et al., 2018). Low-density panels may be also designed to include specific SNPs associated with desirable production, or reproduction traits, or disease susceptibility.
In Russia, pork constitutes an important component of the population’s diet, being the second most popular source of animal protein (about a third of the total meat consumption) (Rosstat, 2018). In recent years, a substantial increase in pork production has been observed. On the other hand, the efficiency of pork production is relatively low mainly due to the use of traditional pig breeding on farms, dependence on imported genetic material, imperfect recording systems, etc., thus reducing the pace of pork industry growth and imposing limitations on further development. The implementation of modern genetic selection approaches which allow prediction of breeding values with higher accuracy may significantly advance pig breeding, and as a consequence will aid in increasing the efficiency of the pork production industry, and the agricultural industry on the whole. However, the poorly developed genotyping environment in Russia, high costs of genotyping procedures, and small sample sizes result in unreasonably high genotyping costs thus counterbalancing the positive effects of GS. Hence, the question of reducing genotyping costs is of special relevance in our country.
Three pig breeds are mainly used in Russian pork production based on conventional 3-cross breeding schemes, namely, Duroc, Landrace, and Large White, the latter being the most frequently used one. In the present study, we aimed to develop an LD-chip that can be further used to advance pig breeding in Russia by aiding in the development of low-cost strategies for GS implementation. To our knowledge, it is the first attempt made for the combination of these 3 breeds based on a Russian pig population. We assessed imputation quality depending on the marker selection method, LD-chip size, and the presence of HD-genotyped relatives in the reference population. We have shown that with the block method, 300 markers are enough to obtain good imputation quality when both parents of the animal have HD genotypes, while in other cases at least 2,000 markers are required. Additionally, as part of the present work we investigated the role of SNP chromosomal position and MAF on the imputation quality.
MATERIALS AND METHODS
Animals and Data
Animal Care and Use Committee approval was not obtained for this study because no experimentation involving animals was part of the present work. Pig ear tissue samples for the study were obtained from the Unique Scientific Set (USS), the Gene Pool of Animal and Avian Genetic Material, ID-498808 (Dubrovitsy, Moscow Oblast, Russia). A total of 807 Landrace, 1,227 Large White, and 684 Duroc pig samples were taken into the study. The genealogy data for the animals used in the study as well as the data on sex, breed, and birth date were obtained from the breeders (OOO Breeding and Hybrid Center, Verkhnyaya Khava, Voronez Oblast, Russia). The complete animal data are provided in the e-Supplement (Supplementary Table S1).
DNA Isolation and SNP Genotyping
Genomic DNA was isolated from the pieces of pig ear tissue using the DNA Extran-2 kit (Syntol, Russia) based on the Proteinase K lysis and isopropanol precipitation technique according to the manufacturer’s instructions. About 10 mg of tissue was used for DNA isolation for each sample.
The integrity of the obtained DNA samples was assessed by electrophoresis in 1% agarose gel; the quality of the samples was estimated based on the OD 260/280 and OD 260/230 ratios obtained using NanoDrop100 (Thermo Fisher Scientific, United States), and DNA quantity in the sample was estimated using the Qubit fluorometer (Invitrogen, United States).
Genotypes for all animals included in the analysis were obtained using the GeneSeek-Neogen GGP Porcine HD (INF Porcine 80K) BeadChip (Illumina, United States) consisting of approximately 70,000 SNPs evenly distributed throughout the pig genome at the average distance of 25 kbp. Raw fluorescence intensity data were processes with the aid of the GenomeStudio Genotyping module (Illumina, United States). The threshold genome calling significance level (GC score) was set at 20%, the other settings were as default. Genotypes quality control was performed using the PLINK 2.0 software (Purcell et al., 2007).
Breed Verification
Principal component analysis (PCA) was performed based on the genotype data to identify clusters (Fig. 1) using PLINK 2.0. Samples were clustered using the k-means method (k = 3). Clusters were annotated based on the most frequent breed. In total, 2,609 samples belonged to the correct cluster, but 43 Large White samples were located in the “Landrace” cluster, while 58 Landrace were in the “Large White” cluster, and also 1 Large White belonged to the “Duroc” cluster. All 102 samples that were not in the correct cluster were excluded from the subsequent analysis.
Figure 1.
Principal coordinate analysis (PCA) of high-density genotypes of pigs from 3 breeds. One point corresponds to a single animal. Landrace, Large White, and Duroc pigs are shown in green, orange, and violet, respectively.
Parentage Verification and Discovery
Parentage verification was performed using custom Python3 scripts. To identify the potential relatives based on the genomic data, all possible pairs of genotypes were considered. For each pair, the proportion of positions where genotypes were different among the positions that were homozygous in both compared samples was calculated. It was assumed that samples are in the parent–offspring relationship, if the proportion was less than 1% (Supplementary Fig. S2A). To exclude duplicate samples, the proportion of positions where genotypes were different among the positions that were heterozygous in at least 1 species was also calculated (Supplementary Fig. S2B). All pairs with the later proportion below 10% were considered as duplicates. In total, 2,270 “parent–offspring” pairs were identified. The 19 detected pairs of duplicates were removed from analysis. To identify who is parent within each pair, the birth dates were used. Assuming that the parent has to be at least 1 yr older than the offspring, 1,451 sire–offspring pairs and 795 dam–offspring pairs were assigned. In total, 1,643 samples had at least 1 parent (Table 1). The obtained pairs were compared with the pedigrees and for 1,427 animals, the pedigree was confirmed by genomic information, while for 203 animals contradictory results were obtained (Supplementary Table S1). Additionally, 38 new parentages were discovered. The pedigrees identified based on the genotype data were used in the subsequent analyzes.
Table 1.
Number of animals in the groups defined by the presence of genotyped parents for each breed
| Breed | Landraсe | Large White | Duroc | With at least 1 parent (total) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parents genotyped | Sire | Dam | Dam and Sire | No parents | Sire | Dam | Dam and Sire | No parents | Sire | No parents | |
| By pedigree | 266 | 72 | 271 | 128 | 589 | 98 | 330 | 158 | 29 | 647 | 1,655 |
| By genotypes | 285 | 91 | 252 | 138 | 568 | 77 | 351 | 163 | 32 | 644 | 1,643 |
| Confirmed1 | 344 | 150 | 193 | 118 | 638 | 147 | 281 | 144 | 25 | 644 | 1,427 |
| Total animals | 737 | 1,175 | 676 | ||||||||
1Confirmed stands for the pedigree relationships confirmed by genomic data.
Test and Reference Populations
To assess the accuracy of imputation, which characterizes the quality of the LD-chip, the samples were subdivided into the reference and test populations. In total, 2,587 samples including 675 Duroc, 737 Landrace, and 1,175 Large White were used. To perform the subsequent analyses, the test samples were assigned to 1 of the 5 groups according to whether or not the animals have genotyped ancestors in the reference population (relatives groups): 1—“No relatives,” 2—“Both parents,” 3—“Dam or Sire,” 4—“One parent & grandsire from a second parent,” and 5—“Only grandsire.”
In the full data set there were present no samples falling into the groups 4 and 5, but there were samples with dam or sire with grandsire (4a) and with both parents (5a), where at least one had grandsire (ST 2). Thus, the groups 4 and 5 were obtained from the groups 4a and 5a by removing the corresponding relatives from the reference set. Duroc pigs were represented only by groups 1 and 3.
Samples were distributed into the reference and test populations in accordance with the following considerations: samples in both populations should be equally represented by breed, and all relatives groups should be equally present in the test population, if possible. As a result, ~600 samples were selected in the test (~200 per breed) and ~1,500 samples (~500 per breed), in the reference populations (Supplementary Tables S2 and S3). The size of the reference set was limited by Duroc as the least represented breed. For Landrace and Large White, about 40 pigs were picked out into the test population from each of the 5 groups, and for the Duroc breed, all animals from the group 3 (32 samples) were taken and other 168 samples were randomly selected. The corresponding parents and grandparents of the test animals were included in or excluded from the reference population with other animals being randomly selected to obtain 500 samples from each analyzed breed.
LD-Chip Development
Single nucleotide polymorphism markers which passed the following filters: SNP genotype having been called in at least 90% of samples, Hardy–Weinberg P-value > 10e−06 (test), and MAF > 2% in each breed separately were kept for the analysis. The 3 filters removed about 590, 61, and 3,907 markers, respectively, for Landrace, 245, 78, and 5,274 markers, respectively, for Large White, and 380, 285, and 9,351 markers, respectively, for Duroc. Filtration was made using the PLINK 2.0 software. Finally, the remaining 39,801 markers located on autosomes with the known position which were common in all 3 breeds were used to design the LD panel.
The 3 commonly used algorithms to develop an LD-chip, namely, “Random,” “Uniform,” and “Block” were applied. The number of SNP per chromosome was proportional to the chromosome length and equal for each method. Eight virtual panels containing 100, 300, 600, 1,000, 2,000, 3,000, 6,000, and 12,000 SNPs were constructed using each method. In total, 24 LD-chips were tested as a part of this work.
“Random” method.
Single nucleotide polymorphisms were selected per chromosome using random uniform distribution function by Python3.
“Uniform” method.
Chromosomes were divided into blocks, where each block contained an equal number of SNPs. Total number of blocks corresponded to the panel density. Then, the central SNP for each block was selected.
“Block” method.
This method is based on the one suggested in Judge et al. (2016) and implies the selection of representative SNPs based on LD (r2) structure and MAF. In more detail, each chromosome was divided into n − 2 blocks, where n is equal to the predefined number of SNPs. This was made in order to include extra SNPs at the peripheries of the chromosomes. Minor allele frequency and the average r2 between target SNP and other SNPs in the same block were estimated and then were standardized within the block. To select the best SNP, the SNPs were ranked based on the sum of the standardized average r2 and MAF. Finally, SNPs with high indexes in the block were chosen. To select extra SNPs into the peripheral blocks, the partial correlation between the already selected SNP and other SNPs in the block was calculated. The highest-ranking SNP for the MAF index plus the average partial correlations (standardized to have equal variances) was chosen as the second most informative SNP.
All LD panels were constructed using custom scripts written in Python3 using the pandas, numpy, sklearn packages.
Imputation
The quality of LD panels was estimated by the imputation quality. Imputation was carried out with the aid of FImpute version 2.2 using both scenarios: “Population” and “Population + Family,” with default settings. Imputation for each panel and breed was undertaken separately. The imputation quality was characterized by correlation (r) between the imputed and real genotypes (Browning and Browning, 2009), and by allele discordance rate (ADR). Both values were estimated for SNPs not included in the LD panel. The r and ADR parameters were calculated using custom python scripts.
Detection of Misplaced SNPs
To detect possible SNP misplacement, R2 for all pairs of SNPs were estimated using the PLINK 2.0 software. Marker with R2 < 0.3 with all SNPs within the 2,000 kbp window around the marker and with R2 > 0.5 with the SNPs in any other region were marked as SNPs with presumably wrong position.
RESULTS AND DISCUSSION
We genotyped 807 Landrace, 1,227 Large White, and 684 Duroc pigs using the GeneSeek-Neogen GGP Porcine HD (INF Porcine 80K) BeadChip (Illumina, United States). Genotype call rates ranged from 0.69 to 0.95. Three animals for which the genotype call rates were below 0.9 were excluded from the analysis. After the quality control, 737, 1,175, and 675 Landrace, Large White, and Duroc pig samples, respectively, remained and were used in the subsequent analysis. Principal component analysis revealed clear separation of the obtained genotypes into 3 breeds (Fig. 1). The parental data were available for 83%, 87%, and 4% of Landrace, Large White, and Duroc pigs, respectively; for 73%, 78%, and 4%, respectively, the sire was genotyped, and for 47%, 36%, and 0%, respectively, the dam was genotyped (Table 1). We validated 86% parentages for the genotyped animals for which parent genotypes were available and discovered 216 new parentage relationships (see Materials and Methods section and Supplementary Table S1).
Based on the previous works (Judge et al., 2016), we considered 3 different algorithms for marker selection for the LD-chip: the random, uniform, and block (see Materials and Methods section). In the case of each method, we designed 8 virtual panels consisting of 100, 300, 600, 1,000, 2,000, 3,000, 6,000, and 12,000 SNP markers (Supplementary Table S4).
The animals from each breed were divided into the reference and test sets. Test set was designed so as to include animals that have no relatives, 1 parent, both parents, only 1 parent and grandfather, and only grandfather in the reference set, with about 40 animals per each indicated group (Supplementary Tables S2 and S3). Then, for each given LD-chip design, we masked all markers except those that belonged to the LD-chip in the test set, imputed them using FImpute, and assessed the imputation quality using ADR and allelic correlation. FImpute gives better results when run for each breed individually than when combined data set is used, with the relative difference in ADR values being 1% to 2%. Therefore, we used the former approach. For all breeds and all panel sizes larger than 100 SNP, the block method performed significantly better than the random method (t-test, P-value < 0.05), and significantly better than the uniform method (t-test, P-value < 0.05), so in our work, we focused on the block method (Fig. 2).
Figure 2.
Dependence of allele discordance rate (ADR) on the panel size. Mean ADRs (in log scale) for animals from the test set are shown. Only animals without ancestors in the reference set were used. Different breeds are shown on different panels; random, uniform, and block methods are shown by dotted, dashed, and solid lines, respectively. FImpute was run in the “Population + Family” mode. Horizontal dashed line corresponds to 0.05 ADR.
Next, we tested whether the presence of the dense genotypes of close relatives could assist imputation. We compared the results obtained using FImpute in the “Population” and “Population + Family” modes (Fig. 3). For animals which ancestors were not genotyped, the differences between the modes were negligible, but the presence of a single parent and grandsire resulted in the significant improvement of imputation in the family mode (from 0.08 to 0.06 ADR for 600 SNP panel, t-test, P-value < 10−10). We achieved more than 95% allelic concordance rate and 0.93 correlation with the 600 SNP chip for Landrace and Large White breeds, when both parents had dense genotypes, while in the absence of dense ancestral genotypes the comparable imputation quality required the use of at least 2,000 SNP chip (Fig. 3D–F). In the case of Duroc, we had almost no animals with genotyped ancestors, so we were not able to assess the imputation quality for this breed in case when dense parental genotypes are present. But, taking into account that in the case of the animals without genotyped parents, imputation quality was similar in the 3 breeds, we expected that the presence of dense parental genotypes should have the similar effect in Duroc pigs as it had in the 2 other analyzed breeds.
Figure 3.
Imputation quality depending on the presence of genotyped ancestors in the reference set. Different breeds are shown on different panels: Landrace—A and D; Large White—B and E; Duroc—C and F. Test subsets are shown by different lines: dotted line—No relatives; dashed line—Only grandsire; densely dotted line with circle—Dam or Sire; densely dotted line with triangle—Parent & grandsire; solid line—Both parents. Horizontal dashed line corresponds to 0.05 ADR. (A–C) FImpute method: “Population,” (D–F) FImpute method: “Population + Family.”
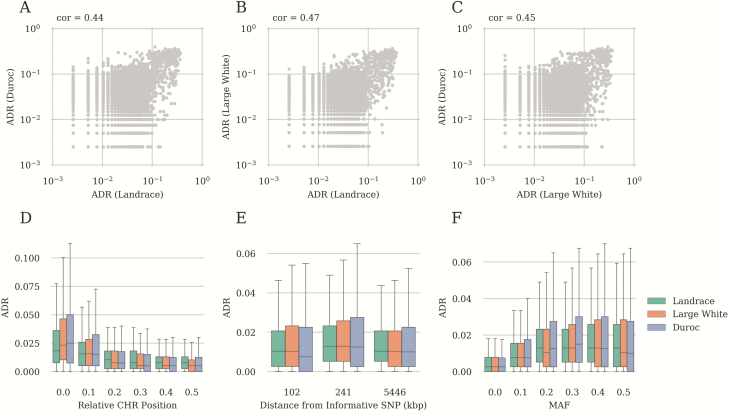
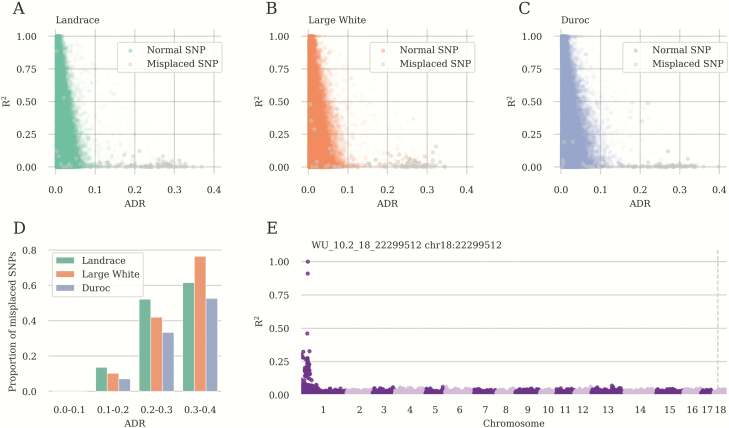
In the course of the work, we observed that imputation quality varied considerably across SNPs and exhibited strong correlation between breeds (Spearman correlation coefficient > 0.44; Fig. 4A–C). Hence, we assumed that there should exist some common factors that influence imputation quality in all breeds. These factors might be the SNP chromosomal position, distance to the LD-chip markers, or correlation with them. While MAF also affected ADR (Fig. 4F), it exhibited very low between-breed correlation (Spearman correlation coefficient varied from 0.01 to 0.05). To assess the potential contribution of these factors, we compared ADR between different chromosomes, depending on the relative position of the SNP on the chromosome, depending on the distance to the LD-chip markers for the 3,000 SNP panel, and depending on MAF (Fig. 4A–C; Supplementary Fig. S1). Allele discordance rate varied significantly between chromosomes (Supplementary Fig. S1, Kruskal–Wallis test, P-value < 3 × 10−38). We also observed that imputation quality decreased to the periphery of the chromosomes, notwithstanding that we selected extra SNPs at the periphery according to the block algorithm (Fig. 4D, t-test, P-value < 2 × 10−13) similar observation having been made by Badke et al. (2013). At the same time, the distance to the informative SNP (the closest SNP from LD-chip) did not affect the imputation quality (Fig. 4E, t-test, P-value = 1). Increased MAF resulted in the reduced imputation quality (Fig. 4F, t-test between MAF 0.0 and MAF 0.1 groups, P-value < 1 × 10−5). We also compared the correlation of the markers with informative SNPs and ADR for the same markers (Fig. 5A–C). Most of SNPs with extremely high ADR exhibited almost no correlation with the informative SNP. Such low correlation could be explained either by recombination hotspots or by wrong mapping of SNPs to chromosomal positions. To check the latter, we compared allelic correlation across all pairs of markers. We have found that 18% to 30% of SNPs with ADR > 0.1 exhibited higher correlation with the chromosomal region other than its own location (Fig. 5E, see Materials and Methods section) and this proportion increased with increasing ADR threshold (Fig. 5D). Therefore, the conclusion was made that the majority of markers with the extremely high ADR (above 0.3) are likely to have wrong chromosomal positions.
Figure 4.
Factors influencing the quality of imputation from low density chips. (A–C) Allele discordance rate relationship calculated for Landrace and Duroc (A), Landrace and Large White (B), and Landrace and Duroc (C). Correlation values are shown at the top of corresponding plots. (D) Allele discordance rate across relative chromosome positions, 0.5—is the center of the chromosome and 0.0—is the beginning of the end of the chromosome. (E) Allele discordance rate depending on the distance from the informative SNP, kbp. Single nucleotide polymorphisms were divided by bins with the same sample size. Labels of bins correspond to maximum distance (kbp) in the group. (F) Dependence of imputation quality on MAF. Minor allele frequency values divided into 5 equal sized bins. Different breeds are shown in different colors. Results were obtained using the LD panel with 3,000 SNP markers.
Figure 5.
Misplaced SNP markers lead to poor imputation quality. (A) The dependence of ADR on R2 between target and informative SNPs for Landrace imputed SNPs. (B) The dependence of ADR on R2 between target and informative SNPs for Large White imputed SNPs. (C) The dependence of ADR on R2 between target and informative SNPs for Duroc imputed SNPs. In (A–C), SNPs with wrong mapping are indicated with gray color. (D) Proportion of misplaced SNPs in ADR range per breed. (E) Manhattan plot for R2 between the target SNP WU_10.2_18_22299512 located at the 22299512 bp position on the chromosome 18 and other SNPs across the whole genome. Position of the target SNP is shown by the vertical gray dashed line. Results were obtained using the LD panel containing 3,000 SNP markers.
In such a way, we have made here the first attempt to develop an LD-chip which could be utilized when making steps toward the implementation of GS approaches in pig breeding in Russia. The 3 breeds commonly used in pork production in Russia, namely, Landrace, Duroc, and Large White were considered. The previous studies on genotype imputation from LD panels were mainly focused on the Yorkshire, Pietrain, and Landrace pig breeds (e.g., Wellmann et al., 2013; Badke et al., 2014; Xiang et al., 2015), while Duroc and Large White pigs have been directly considered only in a single study each (Grossi et al., 2018 and Carillier-Jacquin, 2018, respectively). We concentrated on these breeds in our work since they play the central role in Russian pork production, and we believe that the designed LD-chips may be of value for selection within these breeds on pig farms. Using same LD panels in our study we could obtain almost similar imputation quality in all 3 studied breeds. Therefore, it appears possible to use a single LD-chip for all 3 breeds, which may be rather advantageous.
We have observed in our study that imputation accuracy for individual SNPs differed for different chromosomes, the observation which was also made previously in several studies (Badke et al., 2013; Xiang et al., 2015; Carillier-Jacquin, 2018) where imputation was performed both by the Beagle and FImpute software and which was explained by the difference in the average LD level for each individual chromosome (Xiang et al., 2015).
Taken as a whole, the results obtained in the present work correspond well with those reported previously. Similar to some earlier studies performed both in pigs and in other agricultural animal and poultry species (Badke et al., 2013; Judge et al., 2016; Herry et al., 2018; O’Brien, 2019), we have demonstrated here that the block method for SNP maker selection performed much better that the random and uniform approaches. In agreement with Huang et al. (2012), Cleveland and Hickey (2013), and Wellmann et al. (2013), the presence of dense parental genotypes improved imputation accuracy, which was particularly so in the case of virtual LD panels containing very low SNP number (e.g., 300 SNP, 600 SNP). Similar to the results obtained recently by Grossi et al. (2018) the minimum panel size which allowed somewhat reasonable imputation quality was the 300 SNP panel; however, sufficiently high imputation accuracy in this case could be achieved only when both parents were genotyped using HD panels. It should be noted that in the previous studies, 450 SNP (Cleveland and Hickey, 2013) and 384 SNP (Huang et al., 2012; Wellmann et al., 2013) LD panels were regarded as the most low density ones. Grossi et al. (2018) suggested 3,000 SNP markers to be sufficient to accurately impute HD genotypes; however, in the present work we observed that 2,000 SNP LD panel might be sufficient to obtain high imputation accuracies even when no HD-genotyped relatives were present. In general, the imputation quality for the corresponding panel sizes achieved in this work is similar to the results obtained using pig populations from other countries and/or for other species (Zhang and Druet, 2010; Huang et al., 2012; Wellmann et al., 2013; Judge et al., 2016; Carillier-Jacquin, 2018; Herry et al., 2018; Grossi et al., 2018; O’Brien et al., 2019).
CONCLUSIONS
We have demonstrated that in our data set, the block method for SNP marker selection outperforms the random and uniform methods. We have designed 2 virtual LD panels including 300 and 2,000 markers. Based on our findings, we recommend to use 300 SNP markers when both parents of the animal have HD genotypes and to use 2,000 SNP markers in all other cases. We have shown that in both cases allelic concordance rate reaches about 0.95. We have shown that while imputation quality is influenced by chromosomal position and MAF, the main factor that explains most SNPs with extremely bad imputation quality is mismapping of markers to the chromosomal positions in investigated animals. Hence, sufficient improvement in imputation quality could be achieved through the correction of chromosomal positions of these markers.
Supplementary Material
The authors declare no conflict of interest.
Footnotes
The work was supported by the Ministry of Science and Higher Education of the Russian Federation (grant no. 14.604.21.0182; identification no. RFMEFI60417X0182).
LITERATURE CITED
- Abell C. E., Dekkers J. C., Rothschild M. F., Mabry J. W., and Stalder K. J.. 2014. Total cost estimation for implementing genome-enabled selection in a multi-level swine production system. Genet. Sel. Evol. 46:32. doi: 10.1186/1297-9686-46-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliloo H., Mrode R., Okeyo A. M., Ni G., Goddard M. E., and Gibson J. P.. 2018. The feasibility of using low-density marker panels for genotype imputation and genomic prediction of crossbred dairy cattle of East Africa. J. Dairy Sci. 101:9108–9127. doi: 10.3168/jds.2018-14621 [DOI] [PubMed] [Google Scholar]
- Baby S., Hyeong K. E., Lee Y. M., Jung J. H., Oh D. Y., Nam K. C., Kim T. H., Lee H. K., and Kim J. J.. 2014. Evaluation of genome based estimated breeding values for meat quality in a Berkshire population using high density single nucleotide polymorphism chips. Asian Australas. J. Anim. Sci. 27:1540–1547. doi: 10.5713/ajas.2014.14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badke Y. M., Bates R. O., Ernst C. W., Fix J., and Steibel J. P.. 2014. Accuracy of estimation of genomic breeding values in pigs using low-density genotypes and imputation. G3 (Bethesda). 4:623–631. doi: 10.1534/g3.114.010504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badke Y. M., Bates R. O., Ernst C. W., Schwab C., Fix J., Van Tassell C. P., and Steibel J. P.. 2013. Methods of tagSNP selection and other variables affecting imputation accuracy in swine. BMC Genet. 14:8. doi: 10.1186/1471-2156-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichard M. 1971. Dissemination of genetic improvement through a livestock industry. Anim. Sci. 13:401–411. doi: 10.1017/S0003356100010606 [DOI] [Google Scholar]
- Browning B. L., and Browning S. R.. 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84:210–223. doi: 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Zhou Y., and Browning S. R.. 2018. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103:338–348. doi: 10.1016/j.ajhg.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillier-Jacquin C., Bouquet A., Labrune Y., Brenaut P., Riquet J., and Larzul C.. 2018. Using 1K SNP panel for genomic selection in 3 French pig breeds: accuracy of imputation and estimation of genomic breeding values using 1K SNP panel, designed for several breeds in French pig populations. In: World Congress on Genetics Applied to Livestock Production p. 294. [Google Scholar]
- Christensen O. F., Nielsen B., Su G., Xiang T., Madsen P., Ostersen T., Velander I., and Strathe A. B.. 2019. A bivariate genomic model with additive, dominance and inbreeding depression effects for sire line and three-way crossbred pigs. Genet. Sel. Evol. 51:45. doi: 10.1186/s12711-019-0486-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland M. A., and Hickey J. M.. 2013. Practical implementation of cost-effective genomic selection in commercial pig breeding using imputation. J. Anim. Sci. 91:3583–3592. doi: 10.2527/jas.2013-6270 [DOI] [PubMed] [Google Scholar]
- Esfandyari H., Bijma P., Henryon M., Christensen O. F., and Sørensen A. C.. 2016. Genomic prediction of crossbred performance based on purebred Landrace and Yorkshire data using a dominance model. Genet. Sel. Evol. 48:40. doi: 10.1186/s12711-016-0220-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S. 1989. Introduction to quantitative genetics. Longman, Scientific & Technical; Wiley, Burnt Mill, Harlow, Essex, England, New York. [Google Scholar]
- Goddard M. E., and Hayes B. J.. 2007. Genomic selection. J. Anim. Breed. Genet. 124:323–330. doi: 10.1111/j.1439-0388.2007.00702.x [DOI] [PubMed] [Google Scholar]
- Grossi D. A., Brito L. F., Jafarikia M., Schenkel F. S., and Feng Z.. 2018. Genotype imputation from various low-density SNP panels and its impact on accuracy of genomic breeding values in pigs. Animal 12:2235–2245. doi: 10.1017/S175173111800085X [DOI] [PubMed] [Google Scholar]
- Gualdrón Duarte J. L., Bates R. O., Ernst C. W., Raney N. E., Cantet R. J., and Steibel J. P.. 2013. Genotype imputation accuracy in a F2 pig population using high density and low density SNP panels. BMC Genet. 14:38. doi: 10.1186/1471-2156-14-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herry F., Hérault F., Picard Druet D., Varenne A., Burlot T., Le Roy P., and Allais S.. 2018. Design of low density SNP chips for genotype imputation in layer chicken. BMC Genet. 19:108. doi: 10.1186/s12863-018-0695-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A. M., Bastiaansen J. W., Lopes M. S., Calus M. P., and de Koning D. J.. 2016. Accuracy of genomic prediction of purebreds for cross bred performance in pigs. J. Anim. Breed. Genet. 133:443–451. doi: 10.1111/jbg.12214. [DOI] [PubMed] [Google Scholar]
- Huang Y., Hickey J. M., Cleveland M. A., and Maltecca C.. 2012. Assessment of alternative genotyping strategies to maximize imputation accuracy at minimal cost. Genet. Sel. Evol. 44:25. doi: 10.1186/1297-9686-44-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez-Escriche N., Forni S., Noguera J. L., and Varona L.. 2014. Genomic information in pig breeding: science meets industry needs. Livest. Sci. 166:94–100. doi: 10.1016/j.livsci.2014.05.020 [DOI] [Google Scholar]
- Judge M. M., Kearney J. F., McClure M. C., Sleator R. D., and Berry D. P.. 2016. Evaluation of developed low-density genotype panels for imputation to higher density in independent dairy and beef cattle populations. J. Anim. Sci. 94:949–962. doi: 10.2527/jas.2015-0044 [DOI] [PubMed] [Google Scholar]
- Knol E. F., Nielsen B., and Knap P. W.. 2016. Genomic selection in commercial pig breeding. Anim. Front. 6:15–22. doi: 10.2527/af.2016-0003 [DOI] [Google Scholar]
- Korkuć P., Arends D., and Brockmann G. A.. 2019. Finding the optimal imputation strategy for small cattle populations. Front. Genet. 10:52. doi: 10.3389/fgene.2019.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesbeth van der Waaij K. O. 2014. Textbook. Animal breeding and genetics for BSc students. Centre for Genetic Resources and Animal Breeding and Genomics Group, Wageningen University and Research Centre. [updated November 22, 2015; accessed September 1, 2019]. https://wiki.groenkennisnet.nl/display/TAB/. [Google Scholar]
- Lillehammer M., Meuwissen T. H., and Sonesson A. K.. 2011. Genomic selection for maternal traits in pigs. J. Anim. Sci. 89:3908–3916. doi: 10.2527/jas.2011-4044 [DOI] [PubMed] [Google Scholar]
- Lopes M. 2016. Genomic selection for improved crossbred performance. PhD thesis. Wageningen University, The Netherlands. [Google Scholar]
- Lopez B. M., Kang H. S., Kim T. H., Viterbo V. S., Kim H. S., Na C. S., and Seo K. S.. 2016. Optimization of swine breeding programs using genomic selection with ZPLAN. Asian Australas. J. Anim. Sci. 29:640–645. doi: 10.5713/ajas.15.0842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., and Goddard M. E.. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A. C., Judge M. M., Fair S., and Berry D. P.. 2019. High imputation accuracy from informative low-to-medium density single nucleotide polymorphism genotypes is achievable in sheep1. J. Anim. Sci. 97:1550–1567. doi: 10.1093/jas/skz043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., . et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P., Affara N. A., Amaral A. J., Archibald A. L., Beever J. E., Bendixen C., Churcher C., Clark R., Dehais P., . et al. 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4:e6524. doi: 10.1371/journal.pone.0006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul J., Swan A. A., and Elsen J. M.. 2017. Using a very low-density SNP panel for genomic selection in a breeding program for sheep. Genet. Sel. Evol. 49:76. doi: 10.1186/s12711-017-0351-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosstat. 2018. Russian statistical yearbook. Statistical handbook. Rosstat, Moscow. [Google Scholar]
- Samorè A. B., and Fontanesi L.. 2016. Genomic selection in pigs: state of the art and perspectives. Ital. J. Anim. Sci. 15:211–232. doi: 10.1080/1828051X.2016.1172034 [DOI] [Google Scholar]
- Sargolzaei M., Chesnais J. P., and Schenkel F. S.. 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 15:478. doi: 10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See M. T. 1995. What the commercial pork producer needs to know about genetic improvement. Animal Science Facts, North Carolina State University, ANS02-820S. p. 4. [Google Scholar]
- Simianer H. 2009. The potential of genomic selection to improve litter size in pig breeding programs. In: Proc 60th Annual Meeting of the European Association of Animal Production p. 210–216. [Google Scholar]
- Tribout T., Larzul C., and Phocas F.. 2012. Efficiency of genomic selection in a purebred pig male line. J. Anim. Sci. 90:4164–4176. doi: 10.2527/jas.2012-5107 [DOI] [PubMed] [Google Scholar]
- Tribout T., Larzul C., and Phocas F.. 2013. Economic aspects of implementing genomic evaluations in a pig sire line breeding scheme. Genet. Sel. Evol. 45:40. doi: 10.1186/1297-9686-45-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura R. V., Miller S. P., Dodds K. G., Auvray B., Lee M., Bixley M., Clarke S. M., and McEwan J. C.. 2016. Assessing accuracy of imputation using different SNP panel densities in a multi-breed sheep population. Genet. Sel. Evol. 48:71. doi: 10.1186/s12711-016-0244-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P., Pong-Wong R., Whittemore C., and Haley C.. 2000. Impact of biotechnology on (cross)breeding programes in pigs. Livest. Prod. Sci. 65:57–70. doi: 10.1016/S0301-6226(99)00180-3 [DOI] [Google Scholar]
- Wang C., Habier D., Peiris B. L., Wolc A., Kranis A., Watson K. A., Avendano S., Garrick D. J., Fernando R. L., Lamont S. J., . et al. 2013. Accuracy of genomic prediction using an evenly spaced, low-density single nucleotide polymorphism panel in broiler chickens. Poult. Sci. 92:1712–1723. doi: 10.3382/ps.2012-02941 [DOI] [PubMed] [Google Scholar]
- Wellmann R., Preuß S., Tholen E., Heinkel J., Wimmers K., and Bennewitz J.. 2013. Genomic selection using low density marker panels with application to a sire line in pigs. Genet. Sel. Evol. 45:28. doi: 10.1186/1297-9686-45-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Ma P., Ostersen T., Legarra A., and Christensen O. F.. 2015. Imputation of genotypes in Danish purebred and two-way crossbred pigs using low-density panels. Genet. Sel. Evol. 47:54. doi: 10.1186/s12711-015-0134-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., and Druet T.. 2010. Marker imputation with low-density marker panels in Dutch Holstein cattle. J. Dairy Sci. 93:5487–5494. doi: 10.3168/jds.2010-3501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.