Abstract
Against the background of a growing demand for the implementation of environmentally friendly production processes, microorganisms are engineered for the large-scale biosynthesis of chemicals, fuels, or food and feed additives from sustainable resources. Since strain development is expensive and time-consuming, continuous improvement of molecular tools for the genetic modification of the microbial production hosts is absolutely vital. Recently, the CRISPR/Cas12a technology for the engineering of Corynebacterium glutamicum as an important platform organism for industrial amino acid production has been introduced. Here, this system was advanced by designing an easy-to-construct crRNA delivery vector using simple oligonucleotides. In combination with a C. glutamicum strain engineered for the chromosomal expression of the β-galactosidase-encoding lacZ gene, this new plasmid was used to investigate CRISPR/Cas12a targeting and editing at various positions relative to the PAM site. Finally, we used this system to perform codon saturation mutagenesis at critical positions in the mechanosensitive channel MscCG to identify new gain-of-function mutations for increased l-glutamate export. The mutations obtained can be explained by particular demands of the channel on its immediate lipid environment to allow l-glutamate efflux.
Keywords: CRISPR/Cas, recombineering, l-glutamate, Corynebacterium glutamicum, metabolic engineering, genome editing
Clustered regularly interspaced short palindromic repeats (CRISPR) together with CRISPR-associated (Cas) proteins constitute an immune system in bacteria and archaea that cut foreign DNA entering the cell.1 DNA degradation is achieved by DNA cleavage at specific positions via the Cas-encoded nuclease activity, which is guided to the target site by a short CRISPR-RNA (crRNA) complementary to one of the strands of the target DNA. The simplicity and programmability of the CRISPR/Cas system, established for a broad range of different organisms, has enabled its straightforward application to specifically target any genomic location. Its applicability for engineering bacteria was first demonstrated in the context of recombineering in Escherichia coli where 65% of recombinant cells could be recovered by specifically killing of nonmutated cells.2 Since then, this approach has been successfully used for manipulating numerous bacterial species, including Streptococcus pneumoniae,2Lactobacillus reuteri,3Clostridium beijerinckii,4 and Streptomyces coelicolor,5 and more recently also Corynebacterium glutamicum.6,7
While Cas9 is currently the best-characterized and most widely used endonuclease among available CRISPR editing methods, Cas12a (previously named Cpf1) has recently emerged as an alternative for Cas9. This can be reasoned by the cytotoxicity of Cas9 for a number of bacteria;6,8−10 possible reasons could be the generation of blunt ends by Cas9 and the lack of or reduced activity of nonhomologous end-joining (NHEJ) in bacteria.11 In contrast, Cas12a generates staggered cut DNA with 5′-overhangs, which can be easier repaired by prokaryotes. Recently, Cas12a found additional applications in CRISPR genome editing methods for bacteria, such as E. coli,12Mycobacterium smegmatis,12Cyanobacteria,13Pseudomonas putida,14 and Clostridium difficile.15
The first developed CRISPR/Cas method for bacteria taking advantage of Cas12a was developed for C. glutamicum, directly combined with single-stranded DNA (ssDNA) recombineering to relieve allosteric feedback inhibition of the γ-glutamyl kinase by l-proline.16C. glutamicum, generally recognized as safe (GRAS), has been used for decades in industry for the million ton-scale production of monosodium l-glutamate.17,18 Despite intensive efforts to understand l-glutamate synthesis in this organism in detail, it was only recently possible to solve the mystery of l-glutamate export by identifying the responsible mechanosensitive channel MscCG.19 In addition, C. glutamicum has also been engineered for the production of organic acids,20 alcohols,21,22 polyphenols,23,24 and specialty chemicals.25
Methodical milestones for the engineering of C. glutamicum include the availability of DNA transfer methods,26 a two-step homologous recombination method (the “sacB-system”),27 and, more recently, the introduction of RecT-mediated recombineering.28 The sacB-system, based on the conditionally lethal levansucrase-activity (SacB), is currently the method of choice for the site-specific introduction of genomic mutations. In principle, RecT-mediated recombineering with oligonucleotides (ssDNA) represents a true and less laborious alternative to the sacB-system, but applicability of this method requires a selection step since the frequency of obtaining recombinants in C. glutamicum is about 10–3.28 Here, biosensors, enabling direct screening for mutated cells with increased metabolite formation via fluorescence-activated cell sorting (FACS), were successfully used to isolate recombineered variants.29−32 However, depending on the application in question, development of a biosensor-based screen is not an option. In such cases, CRISPR/Cas12a mediated killing of nonrecombineered C. glutamicum cells is another option for quickly identifying variants with the desired genotype by selection, which holds the potential to decrease the experimental effort even further.
In this manuscript, we characterized the CRISPR/Cas12a system with regard to the introduction of point mutations into the genome of C. glutamicum and developed a simplified CRISPR/Cas12a protocol. Furthermore, we used this system to identify new gain-of-function mutations in the mechanosensitive channel MscCG leading to l-glutamate efflux.
Materials and Methods
Plasmid Constructions
In the used CRISPR/Cas12a system,7 vector pJYS2_crtYf is applied for crRNA delivery. Using this vector as a template, plasmid pJYScr was constructed comprising a BamHI/BstBI cloning site which facilitates the insertion of protospacer sequences into the delivery vector. Removal of the original crtYf protospacer sequence was performed by DNA restriction using BamHI and DraI. The respective restriction sites for these enzymes are located at position 3196 and 3244. Since the available vector pJYS2_crtYf contains three additional DraI sites,16 the selected DraI restriction site was not suitable for cloning. However, to make this site available for cloning, the restriction site was replaced by a BstBI site using a synthesized 313 bp XhoI-AflII fragment (Thermo Fisher Scientific, Waltham, MA, USA), comprising the parental sequence but replacing two nucleotides in the DraI site to create a BstBI restriction site. For plasmid construction, the delivery vector pJYS2_crtYf was cut with XhoI/AflII. Subsequently, the resulting plasmid backbone fragment was purified and combined with the synthesized fragment. Since the vector backbone contains an additional BstBI restriction site, site-directed mutagenesis was performed to introduce a C1165T mutation thereby eliminating this restriction site. The QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) was used for this purpose. However, we experienced that even after gel purification of vector DNA treated with BamHI/BstBI still intact vector was obtained after ligation with oligonucleotides. Therefore, the 49 bp BamHI/BstBI fragment was replaced by a 931 bp dummy fragment (nt 379847–383111 of BA000036) yielding vector pJYScr. This vector makes it easy to separate individual DNA fragments after BamHI/BstBI restriction for further processing. The DNA sequence of pJYScr and the vector itself are deposited at Addgene (#132719). As control experiment, pJYScr:crtYf bearing the same crRNA as pJYS2_crtYf was constructed to evaluate, whether the replacement of restriction sites has an effect on the editing efficiency in comparison to pJYS2_crtYf. In these experiments, 21 ± 7 cfu were obtained with the pJYScr:crtYf delivery vector and with the original vector pJYS2_crtYf 34 ± 10 cfu as escaper cells could be counted. This indicated that the vector backbone modifications did not affect the formation of an active Cas12a/crRNA complex in C. glutamicum cells.
For construction of the individual crRNA delivery vectors, annealed oligonucleotides encoding loop region and spacer were used. For annealing, 10 μL of both oligonucleotides (200 nmol/mL H2O) were mixed, incubated for 10 min at 98 °C, at room temperature for 1 h, and subsequently stored on ice. The annealed oligonucleotides were then ligated with the 4332 bp BamHI/BstBI vector fragment of pJYScr prepared via gel purification. Then E. coli DH5α was transformed with the ligation mix. Spectinomycin resistant clones indicating a successful transformation event were identified on LB plates containing the antibiotic at a concentration of 100 mg/L. Oligonucleotides pJYS2_fw2 and pJYS2_rv2 to amplify a 995 bp PCR fragment and DNA sequencing were used to verify the presence the crRNA coding region in the vector. If desired, recombinant strains were cured from the pJYScr plasmids by cultivation at 34 °C without supplementation of antibiotics as described earlier.7
Strain Constructions
C. glutamicum ATCC13032 was used throughout this study. Integration of the lacZ gene under control of the synthetic PH36 promoter was achieved by isolating a 3190 bp SmaI-fragment from the plasmid pMK-RQ_PH36_LacZ synthesized by Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA). This fragment was inserted into the pK18mobsacB vector carrying the two flanking regions of the chromosomal noncoding region at nucleotide 1.404.651 (acc. no. NC_006958) as previously described.33 Using two homologous recombination events strain C. glutamicum WT::lacZ was obtained. DNA sequencing confirmed a correct chromosomal integration. C. glutamicum WT::lacZ colonies plated onto BHIS-X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) turned blue after 3 days, whereas the C. glutamicum wild type cells remained yellowish. The two constructed mscCG mutants devoid of the carboxyterminal end were constructed by homologous recombination using pK19mobsacB-420stopΔ and pK19mobsacB-424stopΔ (Supporting Table S1), each carrying two flanking fragments of ∼500 bp.34
Transformation and Recombination
Preparation of electrocompetent C. glutamicum pJYScr cells providing inherent expression of RecT and Cas12a was performed as described previously with only minor modifications.16 In short, a single colony from a BHI agar plate was used to inoculate a preculture of 4 mL BHISG-Kan and incubated overnight at 30 °C and 220 rpm. Subsequently, 500 μL of the preculture was transferred into 50 mL of BHISG-Kan supplemented with 1 mL/L Tween 80 and 4 g/L glycine. The culture was harvested when the optical density at 600 nm (OD600) reached approximately 1. Cells were chilled on ice for 20 min and centrifuged at 2,600 g and 4 °C for 10 min. Subsequently, two washing steps in 50 mL of 10% glycerol were performed. Competent cells were resuspended in 500 μL 10% glycerol, and aliquots of 90 μL were stored at −80 °C. For transformation, an aliquot was thawed on ice, mixed with 0.5 μg of the pJYScr derivative and 10 μg of PAGE purified oligonucleotide (Eurofins MWG Operon (Ebersberg, Germany), and transferred into 4 °C precooled electroporation cuvettes (width 2 mm). The oligonucleotides used were 59 bp in size and encoded a mutated PAM site, spacer region, and, if applicable, the mutation to be introduced. After electroporation, cells were immediately transferred to 900 μL of prewarmed BHISG medium and heat-shocked for 6 min at 46 °C. The cells were incubated to recover for 2 h at 30 °C with shaking at 170 rpm. Cells were plated on BHISG containing kanamycin and spectinomycin (BHISG-Kan-Spc-Xgal), and incubated for 3–4 days to determine the number of colony forming units and their respective color.
Media and l-Glutamate Formation
E. coli was grown on LB and C. glutamicum on BHI (Difco Laboratories, Detroit, USA). BHIS contained in addition 90 g/L sorbitol, BHISG moreover 10 g/L glucose, BHISG-Kan 15 mg/L kanamycin, BHISG-Kan-Spec 100 mg/L spectinomycin, and BHISG-Kan-Spc-Xgal 50 mg/L. For glutamate formation, precultures were grown overnight in 20 mL BHI. Cells were harvested by centrifugation, washed with 20 mL 0.9% NaCl and inoculated to an OD600 of 1 in defined CGXII medium (Eggeling and Bott 2005) containing 50 μg/L biotin. l-Glutamate in supernatants was determined as its o-phthaldialdehyde derivative via HPLC and fluorescence detection as previously described.35
Results
Simple Construction of crRNA Delivery Vectors Using Oligonucleotides
According to the protocol currently used, amplification of the entire pJYS2_crtYf vector is necessary for inserting the desired crRNA-encoding spacers.7 Despite different PCR conditions and the use of megaprimers, this procedure resulted in a low yield and often a doubling of the oligonucleotides. With the original protocol we were able to construct only 1 of 5 desired vectors, which motivated us to develop an improved crRNA delivery vector expediting necessary constructions. The starting vector pJYS2_crtYf encodes the loop region of the crRNA in combination with the specific crtYf-targeting sequence on a 49 bp BamHI-DraI-fragment. The single BamHI site is located between the strong promoter pJ23119 and the specific spacer, whereas one of the four DraI sites of the vector is located between the spacer and the T1rrnB terminator. Using a synthesized fragment and applying site-directed mutagenesis, we replaced the DraI site by a BstBI site and removed an existing BstBI site in the vector backbone. The resulting vector in principle allows replacing the crtYf-specific crRNA-encoding sequence on the 49 bp BamHI-BstBI fragment by any other sequence.
However, we substituted the crtYf-fragment by a larger fragment of 931 bp resulting in the vector pJYScr of 5.252 kb, which enables easy preparation of the 4.321 kb BamHI-BstBI vector fragment for the insertion of oligonucleotides encoding the desired crRNA (Figure S1). This new vector for genome editing has been made available through Addgene (#132719).
To validate the functionality of pJYScr in a CRISPR/Cas12a selection experiment, a crRNA targeting murE of C. glutamicum was cloned into the vector. The murE gene encodes the essential UDP-N-acetylmuramoylalanyl-d-glutamate 2,6-diaminopimelate ligase.36 The corresponding oligonucleotides comprising the PAM site 5′-GTTG-3′ (nt 255–258) within murE, were annealed and ligated with the 4.321 kb BamHI-BstBI fragment of pJYScr, resulting in pJYScr:murE. After transformation of C. glutamicum pJYS1-petFU (providing constitutive Cas12a-activity) with 1 μg of the pJYScr:murE plasmid, only 81 ± 26 cfu as escaper cells were obtained demonstrating the formation of an active Cas12a/crRNA complex in the cells. A control experiment, in which C. glutamicum wild type cells without pJYS1-petFU were transformed with the same vector yielded 3.2 × 105 cfu.
Efficiency of CRISPR/Cas12a-Assisted ssDNA-Recombineering in C. glutamicum
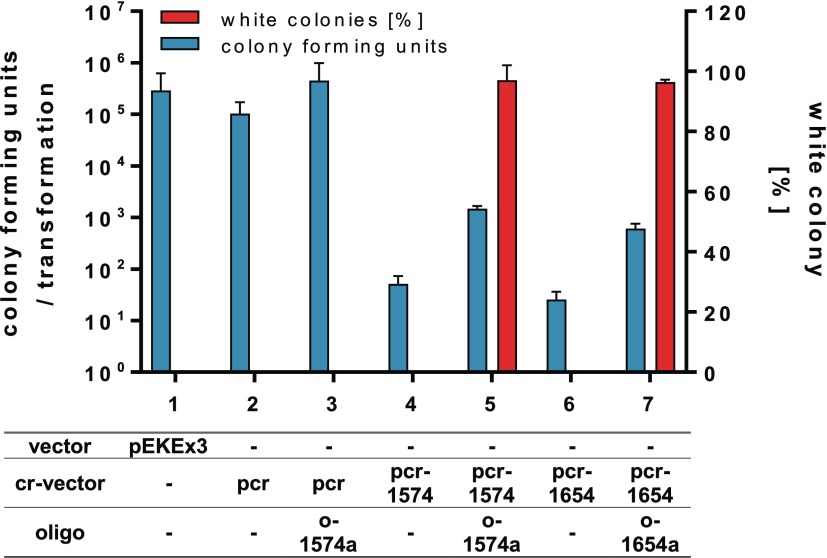
With the aim to explore the capabilities of combining ssDNA-recombineering with CRISPR/Cas12a-mediated elimination of nonrecombinant cells, we first constructed a C. glutamicum test strain carrying a chromosomal integration of the lacZ gene encoding for the β-galactosidase from E. coli (C. glutamicum WT::lacZ). In this particular strain, lacZ-expression is under control of the synthetic promoter pH36,37 enabling easy identification of generated lacZ mutants on agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) by blue-white screening. Subsequently, this strain was transformed with pJYS1-petFU providing constitutive Cas12a- as well as RecT-activity to this strain for the following recombineering-CRISPR/Cas12a experiments.16 When transformed with the empty pJYScr-vector with or without the recombineering oligonucleotide o-1574a, high transformation rates comparable to that with the standard pEKEx3 vector as a control were obtained (Figure 1). A high transformation rate is essential to obtain recombinants in the combined recombineering-CRISPR/Cas12a-killing approach. Noteworthy, the transformation rates obtained also confirmed the nontoxicity of Cas12a opposed to Cas9 as it has been already described for various organisms13 including C. glutamicum.16 As crRNA delivery vector targeting the chromosomal lacZ gene, we have built pJYScr-lacZ-1574 using the intragenic PAM site 5′-(T)TT(C)-3′ with a guide sequence of 21 nts. When C. glutamicum WT::lacZ pJYS1-petFU was transformed with this vector, only 51 ± 21 cells survived underlining the efficiency of Cas12a targeting in C. glutamicum (Figure 1). To assay for DNA-editing plus targeting of unmutated cells, C. glutamicum WT::lacZ pJYS1-petFU was transformed with pJYScr-lacZ-1574 in combination with the editing oligonucleotide o-1574a (all recombinogenic oligos are listed in Supporting Table S2). This oligonucleotide was designed to replace the PAM site and the adjacent codon TCG (5′-TTTCG-3′) by 5′-TATAG-3′, which results in an inactivated PAM site and the introduction of a stop codon within the open reading frame of lacZ. This experiment yielded 1500 ± 172 colonies of which more than 97% were white (Figure 1). Comparable numbers could be obtained when targeting a second PAM-site (5′-(T)TT(A)-3′) within lacZ using the oligonucleotide o-1654a and the simultaneous introduction of a stop codon by substituting a single nucleotide (Figure 1).
Figure 1.
Efficiency of genome targeting and editing in C. glutamicum. The transformation assays to transform C. glutamicum WT::lacZ pJYS1-petFU contained 500 ng plasmid and 10 μg oligonucleotide where indicated. The transformants were plated on X-Gal plates for blue-white screening. The results are averages from at least two independent experiments, and error bars depict standard deviations. In control experiments, C. glutamicum was either transformed with the cloning vector pEKEx3 (column 1), the empty vector pJYScr (column 2) inscribed as pcr, or empty vector plus oligo o-1574a targeting a PAM site in the chromosomal lacZ gene (column 3). When the crRNA was provided by vector pJYScr-lacZ-1574 (pcr-1574, column 4) or vector pJYScr_lacZ-1654 (pcr-1654, column 6), only few cells survived, whereas addition of the corresponding oligonucleotides (o-1574a, column 5 or o-1654a, column 7) for PAM site-inactivation and introduction of a stop codon resulted in a drastic increase of surviving colonies with >96% of having the desired mutation.
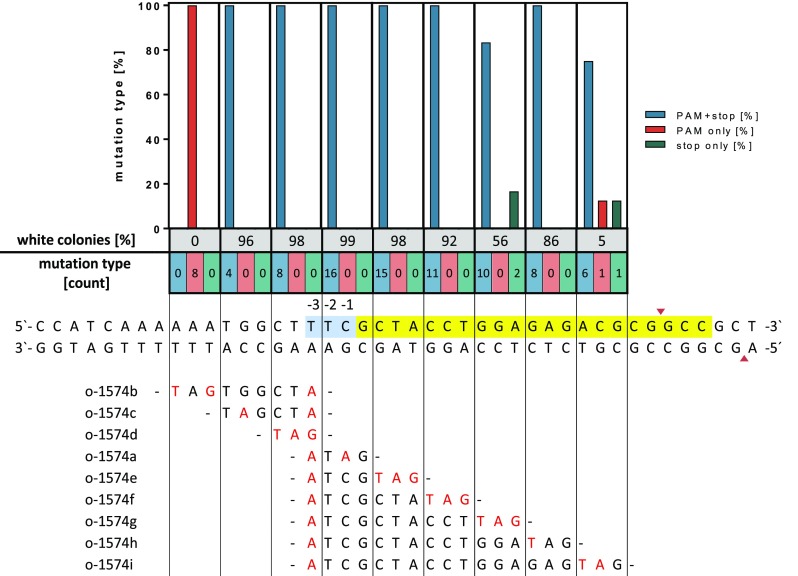
These results raised the question at which frequency a reliable introduction of mutations at different positions relative to the PAM site is possible with this system. To answer this, nine different oligonucleotides with a length of 59 nt were designed for introducing a stop codon over a stretch of 18 nt along with a mutation inactivating the PAM site. Oligonucleotides used for recombineering and introduction of the desired mutations in the lacZ gene and the PAM site are given in Table S2. Experiments conducted with these oligonucleotides yielded no white colonies when oligonucleotide o-1574b was used, which would introduce two nucleotides substitutions upstream of the PAM-site (Figure 2). In this case, sequencing of eight randomly picked blue clones revealed indeed that only the PAM site was mutated. When performing recombineering-CRISPR/Cas12a with the editing oligonucleotides o-1574c, -d, -a, e, and -f, more than 96% of the surviving C. glutamicum variants were white in all cases indicating a successful introduction of the stop codon into the lacZ gene. This observation was supported by DNA sequencing of randomly selected clones, which confirmed correct incorporation of both mutations in all clones analyzed. However, when using the recombinogenic oligonucleotides o-1574g, o-1574h, or o-1574i designed for introducing a stop codon further downstream of the PAM site, introduction of the desired mutations was still possible, but only at a reduced frequency ranging from 75% to 90% (blue bar), depending on the number of substitutions necessary to generate a stop codon. Interestingly, DNA-sequencing revealed that in a few cases only the stop codon was successfully introduced whereas the PAM site was still intact (given in Figure 2 as “stop only”). We obtained a similar result of such partially successful editing events in experiments targeting a second PAM site in lacZ (Table S2). In these cases, possibly only a short sequence of the genomic DNA was exchanged for the respective oligonucleotide during recombineering. These clones escape Cas12a cleavage when only mismatches in the PAM-distal region of the DNA target are present.38,39 Collectively, these data suggest that site-directed mutagenesis using the combined recombineering/Cas12a selection strategy is possible over a stretch of four nucleotides upstream and ten nucleotides downstream of the PAM sequence.
Figure 2.
Genome editing efficiency relative to the PAM site in C. glutamicum using the combined recombineering-CRISPR/Cas12a selection method. Shown is a portion of the chromosome-integrated lacZ gene with the PAM sequence highlighted in blue and the protospacer highlighted in yellow. The Cas12a-cleavage site yielding staggered ends is indicated by red triangles. The relevant DNA sequence of the oligonucleotides for site-directed mutagenesis is shown at the bottom with all mutations highlighted in red. The fraction of white colonies obtained is given as percentage share. From each assay, the lacZ gene of randomly selected white clones was sequenced (n = 4–16). Either the PAM site was mutated and the stop codon was introduced (blue bars), only the PAM site was mutated (red bars), or only the stop codon was introduced (green bars). The respective numbers for each type of mutation (PAM+stop/stop only/PAM only) assessed via DNA sequencing are also given as numbers below the colored bars. Detailed DNA sequencing results are given in Supporting Table S1.
Inspired by these results, the mutagenic oligonucleotide o-1574fe-random was designed, which carries a continuous stretch of 18 random nucleotides spanning the previously targeted PAM sequence plus five nucleotides upstream and ten nucleotides downstream of that PAM site (Supporting Table S3). After performing the recombineering-CRISPR/Cas12a selection, only eight white colonies were obtained. DNA sequencing of the lacZ gene of all clones revealed a mutated and thus nonfunctional PAM sequence in all cases with four clones carrying additional mutation(s) downstream of the targeted position (nucleotides +11 to +16, Supporting Table S3). Apparently, when considering the low number of obtained colonies one can assume, that with the given oligonucleotide bearing such a long random sequence, recombineering does not work. Hence, this method cannot be used to exchange a longer stretch of DNA and is limited to the introduction of three consecutive nucleotide substitutions into the genome of C. glutamicum.
This conclusion is supported by the results of two additionally performed experiments using oligonucleotides o-1574e-mix and o-1574g-mix, which were both designed for the saturation mutagenesis of a selected codon (Supporting Table S3). Twenty-four white colonies obtained by recombineering-CRISPR/Cas12a selection using o-1574e-mix were randomly picked and sequenced. All variants were characterized by a mutated PAM site and 16 of them carried additional mutation(s) in the desired codon leading to 12 amino acid substitutions on the protein level without a discernible preference for specific substitutions (Supporting Table S3). Similarly, targeted DNA sequencing of 24 white clones resulting from the genome editing experiment using oligonucleotide o-1574g-mix yielded 13 variants bearing mutations in the desired codon, leading to eight different amino acids at this particular position (Supporting Table S3).
Site-Directed Mutagenesis Targeting mscCG in C. glutamicum
An efficient l-glutamate export is an important prerequisite for achieving high l-glutamate titers with C. glutamicum. In case of this bacterium, the mechanosensitive channel MscCG (533 aa) is mainly responsible for l-glutamate efflux.19 However, l-glutamate efflux only occurs in response to changes in the structure of this channel, induced either by growth under biotin limitation or addition of penicillin, ethambutol, or fatty acid ester surfactants to the medium.40 In addition, an altered lipid composition may cause efflux.41,42 According to the current model, these cultivation/production conditions mentioned increase membrane tension and thus activate the MscCG channel.43 Interestingly, several gain-of-function (GOF) variants of MscCG are known causing constitutive l-glutamate efflux.19,44 These mutational hot spots are located in the pore-lining helix of the N-terminal domain (1–286 aa), which has been also shown to be responsible for l-glutamate excretion in response to the above-mentioned treatments.44 Furthermore, two additional GOF mutants, characterized by mutations in the C-terminal extracytoplasmic domain are known. In one case, insertion of an IS-element at position 419 was described, resulting in the introduction of a stop codon at position 424.19 The other GOF mutant bears an P424L-substitution.19 Finally, a third mutant devoid of the entire cytoplasmic domain (amino acids 420–533) has been described.44 We were interested in studying consequences of mutations in this particular region at positions 422 and 423 of MscCG and used the recombineering-CRISPR/Cas12a method to assay additional amino acid substitutions at these particular positions. Both targeted amino acid positions of MscCG are predicted to be located at the transition of the fourth transmembrane helix to the periplasmic space.19 Presumably, this site determines the overall structure of MscCG and is crucial for the interaction of the channel protein with other components of the cell wall.
C. glutamicum pJYS1petFU was transformed with pJYScr_mscCG1269c providing the crRNA and with 10 μg of the mutagenic oligo o-mscCG1269-V422nnn for site saturation mutagenesis (Table S4). The assay yielded 43 clones of which 16 were randomly chosen and sequenced. In these clones, altogether seven different amino acid substitutions at amino acid position V422 of MscCG could be identified. Similarly, position E423 was also targeted, eventually yielding 27 clones surviving CRISPR/Cas12a selection after recombineering. In 19 clones, DNA sequencing revealed introduction of eight different amino acids and one stop codon. In addition, one variant with an undesired deletion of two nucleotides in the PAM sequence was found. This frameshift mutation inevitably resulted in a stop codon (E423AQstop). Recombinant strains were cured from the respective plasmids and subsequently assayed for l-glutamate accumulation in the culture supernatant.
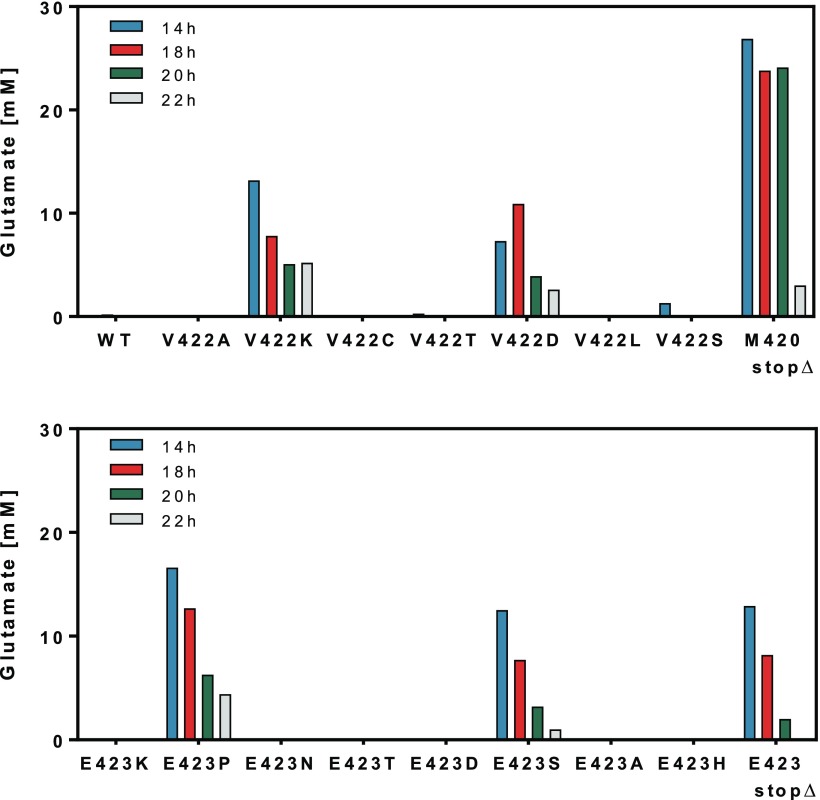
As previous studies have shown, l-glutamate formation, even in described MscCG mutants, is very much dependent on biotin concentration, medium composition and sugar concentration.19,44,45 For instance, the known MscCG mutant devoid of the extracytoplasmic domain44 excretes l-glutamate only at low biotin concentrations, whereas the mutant carrying the IS-element insertion at position 419 accumulates l-glutamate even in the presence of excess biotin.19 For assaying the l-glutamate accumulation capabilities of the newly constructed MscCG-variants obtained by recombineering-CRISPR/Cas12a selection, we chose stringent conditions in defined medium with biotin excess (50 μg/L) in combination with a low glucose concentration (40 g/L). Under these conditions, the C. glutamicum wild type does not excrete l-glutamate (Figure 3a). The MscCG variants V422A, V422C, V422T, V422L, and V422S did not accumulate l-glutamate. In contrast, in the supernatant of the MscCG-variants V422 K and V422D, in which the valine residue was substituted by charged amino acids, a substantial l-glutamate accumulation of up to 13 mM could be detected (Figure 3a). However, variants E423 K and E423D did not show l-glutamate accumulation, as did mutants E423N, E423T, E423A, and E423H (Figure 3b). In contrast, mutant E423P, E423S as well as the strain with the MscCG variant E423stop promoted l-glutamate export. Noteworthy, l-glutamate concentrations decreased with time due to reuptake and utilization of this amino acid as known from previous studies.45
Figure 3.
l-Glutamate accumulation in the supernatant of C. glutamicum variants with different mutations in the mechanosensitive channel MscCG gene at position V422 (top) and E423 (bottom). In addition, MscCG variants with stop codons at positions M420 (M420stopΔ) and E423 (E423stop), respectively, were also characterized with regard to the accumulation of l-glutamate. l-Glutamate concentrations for each variant were determined after 14, 18, 20, and 22 h.
Interestingly, the MscCG variant E423AQstop did not cause l-glutamate accumulation as expected (data not shown). We also used the conventional sacB-system to delete two different portions of the extracytoplasmic domain of MscCG. In mutant M420stopΔ methionine in position 420 was replaced by a stop codon and the carboxyterminal end of 113 amino acids deleted. Similarly, the C. glutamicum strain carrying the shortened MscCG variant P424stopΔ missing the carboxyterminal 109 amino acids was constructed. With variant M420stopΔ, l-glutamate concentrations of up to 27 mM could be detected, and with mutant P424stopΔ concentrations of up to 130 mM could be determined in the supernatant (Supporting Table S5). These observations are in line with previous data also implying that presence of the carboxyterminal domain has a negative impact on l-glutamate channeling activity.44
Discussion
CRISPR/Cas selection is a powerful technology to perform genome editing in living cells. On the basis of this system, methods for inhibiting transcription (CRISPRi), activation of gene expression, or modification of nucleobases have been developed.46 In bacteria, for the most part, methods introducing double-strand breaks using the crRNA/Cas-complex to eliminate unaltered strain variants find an application.47 The main challenge during combined recombineering-selection experiments using RecT-recombineering and CRISPR/Cas represents balancing activities of all individual components including the recombinase, the crRNA and the endonuclease involved. The need to ensure the availability of the respective components in appropriate quantities at the right time initially led to the development of CRISPR/Cas methods for E. coli, which require up to three vectors or three independent induction systems.48,49 However, these established Cas9-based systems were not suitable for applications in C. glutamicum, for the most part due to the cytotoxicity of the Cas9 endonuclease.
However, a CRISPR/Cas genome editing system based on the Cas12a endonuclease originating from Francisella novicida was introduced for C. glutamicum.16 This genome editing method is more suitable for this bacterium as (i) Cas12a is not toxic for C. glutamicum; (ii) induction of gene expression (to establish endonuclease or recombinase activity and crRNA formation) is not necessary due to a fortunate choice of constitutive promoters;16 and (iii) the crRNA, 43 nt in length, is about half the size of a typical Cas9 crDNA.39 However, the construction of the crRNA delivery vector for a distinct protospacer sequence remained laborious and time-consuming, lowering the overall applicability of this method. The reconstructed vector pJYScr presented here, overcomes this bottleneck as it allows for fast and simple vector assembly to enable crRNA delivery. With CRISPathBrick, a similar strategy was developed by the Koffas group, which allows for the modular assembly of CRISPR-bricks to be used for CRISPR-Cas9 applications in E. coli.50 In this study, CRISPR/Cas12a-assisted recombineering with ssDNA oligonucleotides using this new vector was assessed by performing experiments to introduce point mutations up- and downstream of PAM-sites in a chromosomally integrated lacZ gene. Interestingly, the results are consistent with previous experiments in Mycobacterium smegmatis, showing that any targeting distal from the PAM site results in a drastically decreased recombineering efficiency.12 Nonetheless, the results obtained in this study confirm the potential of CRISPR/Cas12a-assisted recombineering using ssDNA oligonucleotides for the efficient and precise introduction of nucleotide substitutions into the genome of C. glutamicum. This was demonstrated when performing site-directed mutagenesis of the gene encoding the MscCG channel of C. glutamicum. We focused on positions 422 and 423 of the MscCG protein located at the transition of the fourth transmembrane helix to the periplasmic space.19 It is striking that only substitution by charged amino acids residues resulted in an open-channel conformation at position 422. Given that the hydrophobic head groups of cardiolipin and phosphatidylglycerol are present in C. glutamicum in this region,41 the direct local membrane environment may be altered, which in turn creates the necessary conditions for the amino-terminal part of MscCG to mediate the l-glutamate efflux.44 In contrast, amino acid substitutions E423P and E423S, presumably causing drastic structural alterations, support l-glutamate efflux at position 422. Against the background that complete deletion of the periplasmic domain also fosters l-glutamate efflux,44 there are two obvious possible explanations. Again, the immediate channel environment could be affected, which could result in a deformed channel. Alternatively, the extracytoplasmic domain could be forced away from the channel opening, thus enabling the efflux of l-glutamate. Such models where a periplasmic domain functions as an efflux-controlling lid and where this lid further interacts with the extracellular matrix have been described for eukaryotes.51
Taken together, the crRNA delivery vector pJYScr introduced here simplifies plasmid constructions and expands the number of CRISPR/Cas12a applications in C. glutamicum. In particular in combination with recombineering, CRISPR/Cas12a is a powerful tool when performing site-directed mutagenesis in the genome of C. glutamicum by selectively eliminating unedited cells as presented in this study.
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No 638718)
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.9b00361.
Figure S1: Plasmid map of the pJYScr vector; Table S1: Plasmids and oligonucleotides used for plasmid constructions; Table S2: Recombinogenic oligos and obtained mutations at two lacZ locations; Table S3: Recombinogenic oligos for the insertion of a random stretch of nucleotides and codon saturation mutagenesis; Table S4: Recombinogenic oligonucleotides used for introducing mutations in mscCG and the mutations obtained; Table S5: l-Glutamate accumulation in the culture supernatant of constructed mscCG deletion mutants (PDF)
Author Contributions
L.E. designed the experiments, K.K. and C.K.S. conducted the experiments. L.E. and J.M. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Park H. M.; Liu H.; Wu J.; Chong A.; Mackley V.; Fellmann C.; Rao A.; Jiang F.; Chu H.; Murthy N.; Lee K. (2018) Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat. Commun. 9, 1–12. 10.1038/s41467-018-05641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.; Bikard D.; Cox D.; Zhang F.; Marraffini L. A. (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. H.; Van Pijkeren J. P. (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42, 1–11. 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M.; Hocq R.; Collas F.; Chartier G.; Wasels F.; Wijaya H. S.; Werten M. W. T.; Wolbert E. J. H.; Kengen S. W. M.; van der Oost J.; Ferreira N. L.; López-Contreras A. M. (2019) Adaptation and application of a two-plasmid inducible CRISPR-Cas9 system in Clostridium beijerinckii. Methods 10.1016/j.ymeth.2019.07.022. [DOI] [PubMed] [Google Scholar]
- Huang H.; Zheng G.; Jiang W.; Hu H.; Lu Y. (2015) One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim. Biophys. Sin. 47, 231–243. 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- Cho J. S.; Choi K. R.; Prabowo C. P. S.; Shin J. H.; Yang D.; Jang J.; Lee S. Y. (2017) CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab. Eng. 42, 157–167. 10.1016/j.ymben.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Qian F.; Yang J.; Liu Y.; Dong F.; Xu C.; Sun B.; Chen B.; Xu X.; Li Y.; Wang R.; Yang S. (2017) CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 8, 15179. 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K. E.; Ungerer J.; Cobb R. E.; Zhao H.; Pakrasi H. B. (2016) CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 15, 1–8. 10.1186/s12934-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.; Li Y.; Shi Z.; Hemme C. L.; Li Y.; Zhu Y.; Van Nostrand J. D.; He Z.; Zhou J. (2015) Efficient genome editing in clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 81, 4423–4431. 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H.; Doudna J. A. (2015) Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol. Cell 58, 568–574. 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- Richardson C. D.; Ray G. J.; DeWitt M. A.; Curie G. L.; Corn J. E. (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344. 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Yan M.-Y.; Yan H.; Ren G.; Zhao J.-P.; Guo X.; Sun Y.-C. (2017) CRISPR-Cas12a-Assisted Recombineering in Bacteria. Appl. Environ. Microbiol. 83, 1–13. 10.1128/AEM.00947-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J.; Pakrasi H. B. (2016) Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci. Rep. 6, 39681. 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Wang Q.; Jiang Y.; Wen Z.; Yang L.; Wu J.; Yang S. (2018) Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb. Cell Fact. 17, 41. 10.1186/s12934-018-0887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W.; Zhang J.; Cui G.; Wang L.; Wang Y. (2018) Multiplexed CRISPR-Cpf1-Mediated Genome Editing in Clostridium difficile toward the Understanding of Pathogenesis of C. difficile Infection. ACS Synth. Biol. 7, 1588–1600. 10.1021/acssynbio.8b00087. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Qian F.; Yang J.; Liu Y.; Dong F.; Xu C.; Sun B.; Chen B.; Xu X.; Li Y.; Wang R.; Yang S. (2017) CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S. (1972) Amino-acid production by the fermentation process. Nature 240, 211. 10.1038/240211a0. [DOI] [PubMed] [Google Scholar]
- Eggeling L.; Bott M. (2015) A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 3387–94. 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- Nakamura J.; Hirano S.; Ito H.; Wachi M. (2007) Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce L-glutamic acid production. Appl. Environ. Microbiol. 73, 4491–8. 10.1128/AEM.02446-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschalka S.; Blombach B.; Bott M.; Eikmanns B. J. (2013) Bio-based production of organic acids with Corynebacterium glutamicum. Microb. Biotechnol. 6, 87–102. 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B.; Riester T.; Wieschalka S.; Ziert C.; Youn J. W.; Wendisch V. F.; Eikmanns B. J. (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77, 3300–3310. 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojima T.; Noburyu R.; Sasaki M.; Tajima T.; Suda M.; Yukawa H.; Inui M. (2015) Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 1165–1172. 10.1007/s00253-014-6223-4. [DOI] [PubMed] [Google Scholar]
- Kallscheuer N.; Vogt M.; Stenzel A.; Gätgens J.; Bott M.; Marienhagen J. (2016) Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38, 47–55. 10.1016/j.ymben.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Milke L.; Ferreira P.; Kallscheuer N.; Braga A.; Vogt M.; Kappelmann J.; Oliveira J.; Silva A. R.; Rocha I.; Bott M.; Noack S.; Faria N.; Marienhagen J. (2019) Modulation of the central carbon metabolism of Corynebacterium glutamicum improves malonyl-CoA availability and increases plant polyphenol synthesis. Biotechnol. Bioeng. 116, 1380–1391. 10.1002/bit.26939. [DOI] [PubMed] [Google Scholar]
- Kogure T.; Inui M. (2018) Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 102, 8685–8705. 10.1007/s00253-018-9289-6. [DOI] [PubMed] [Google Scholar]
- Schwarzer A.; Pühler A. (1991) Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Nat. Biotechnol. 9, 84–7. 10.1038/nbt0191-84. [DOI] [PubMed] [Google Scholar]
- Jäger W.; Schäfer A.; Pühler A.; Labes G.; Wohlleben W. (1992) Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 174, 5462–5. 10.1128/jb.174.16.5462-5465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S.; Siedler S.; Marienhagen J.; Bott M.; Eggeling L. (2013) Recombineering in Corynebacterium glutamicum combined with optical nanosensors: A general strategy for fast producer strain generation. Nucleic Acids Res. 41, 6360–6369. 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S.; Schendzielorz G.; Stäbler N.; Krumbach K.; Hoffmann K.; Bott M.; Eggeling L. (2012) A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 13, R40. 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling L.; Bott M.; Marienhagen J. (2015) Novel screening methods-biosensors. Curr. Opin. Biotechnol. 35, 30. 10.1016/j.copbio.2014.12.021. [DOI] [PubMed] [Google Scholar]
- Siedler S.; Khatri N. K.; Zsohár A.; Kjærbølling I.; Vogt M.; Hammar P.; Nielsen C. F.; Marienhagen J.; Sommer M. O. A.; Joensson H. N. (2017) Development of a Bacterial Biosensor for Rapid Screening of Yeast p-Coumaric Acid Production. ACS Synth. Biol. 6, 1860–1869. 10.1021/acssynbio.7b00009. [DOI] [PubMed] [Google Scholar]
- Jang S.; Jang S.; Xiu Y.; Kang T. J.; Lee S.-H.; Koffas M. A. G.; Jung G. Y. (2017) Development of Artificial Riboswitches for Monitoring of Naringenin In Vivo. ACS Synth. Biol. 6, 2077–2085. 10.1021/acssynbio.7b00128. [DOI] [PubMed] [Google Scholar]
- Blombach B.; Hans S.; Bathe B.; Eikmanns B. J. (2009) Acetohydroxyacid synthase, a novel target for improvement of L-lysine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 75, 419–27. 10.1128/AEM.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A.; Tauch A.; Jäger W.; Kalinowski J.; Thierbach G.; Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Marienhagen J.; Kennerknecht N.; Sahm H.; Eggeling L. (2005) Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J. Bacteriol. 187, 7639–7646. 10.1128/JB.187.22.7639-7646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheim J.; Kranz A.; Krumbach K.; Sokolowsky S.; Eggeling L.; Noack S.; Bocola M.; Bott M.; Marienhagen J. (2017) Mutations in MurE, the essential UDP-N-acetylmuramoylalanyl-d-glutamate 2,6-diaminopimelate ligase of Corynebacterium glutamicum: effect on l-lysine formation and analysis of systemic consequences. Biotechnol. Lett. 39, 283–288. 10.1007/s10529-016-2243-8. [DOI] [PubMed] [Google Scholar]
- Yim S. S.; An S. J.; Kang M.; Lee J.; Jeong K. J. (2013) Isolation of fully synthetic promoters for high-level gene expression in corynebacterium glutamicum. Biotechnol. Bioeng. 110, 2959–2969. 10.1002/bit.24954. [DOI] [PubMed] [Google Scholar]
- Swarts D. C.; van der Oost J.; Jinek M. (2017) Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233. 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I.; Richter H.; Bratovič M.; Le Rhun A.; Charpentier E. (2016) The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–21. 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Hirasawa T.; Wachi M. (2016) Glutamate Fermentation-2: Mechanism of L-Glutamate Overproduction in Corynebacterium glutamicum. Adv. Biochem. Eng./Biotechnol. 159, 57–72. 10.1007/10_2016_26. [DOI] [PubMed] [Google Scholar]
- Hoischen C.; Krämer R. (1990) Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J. Bacteriol. 172, 3409–16. 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nampoothiri K. M.; Hoischen C.; Bathe B.; Möckel B.; Pfefferle W.; Krumbach K.; Sahm H.; Eggeling L. (2002) Expression of genes of lipid synthesis and altered lipid composition modulates L-glutamate efflux of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 58, 89–96. 10.1007/s00253-001-0861-z. [DOI] [PubMed] [Google Scholar]
- Nakayama Y.; Hashimoto K.-I.; Sawada Y.; Sokabe M.; Kawasaki H.; Martinac B. (2018) Corynebacterium glutamicum mechanosensitive channels: towards unpuzzling “glutamate efflux” for amino acid production. Biophys. Rev. 10, 1359–1369. 10.1007/s12551-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita C.; Hashimoto K.-I.; Kumagai K.; Maeda T.; Takada A.; Yabe I.; Kawasaki H.; Wachi M. (2013) L-Glutamate secretion by the N-terminal domain of the Corynebacterium glutamicum NCgl1221 mechanosensitive channel. Biosci., Biotechnol., Biochem. 77, 1008–13. 10.1271/bbb.120988. [DOI] [PubMed] [Google Scholar]
- Becker M.; Börngen K.; Nomura T.; Battle A. R.; Marin K.; Martinac B.; Krämer R. (2013) Glutamate efflux mediated by Corynebacterium glutamicum MscCG, Escherichia coli MscS, and their derivatives. Biochim. Biophys. Acta, Biomembr. 1828, 1230–40. 10.1016/j.bbamem.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Arazoe T.; Kondo A.; Nishida K. (2018) Targeted Nucleotide Editing Technologies for Microbial Metabolic Engineering. Biotechnol. J. 13, e1700596. 10.1002/biot.201700596. [DOI] [PubMed] [Google Scholar]
- Jiang W.; Bikard D.; Cox D.; Zhang F.; Marraffini L. A. (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne M. E.; Moo-Young M.; Chung D. A.; Chou C. P. (2015) Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Appl. Environ. Microbiol. 81, 5103–14. 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C.; Pedersen L. E.; Sommer M. O. A.; Nielsen A. T. (2016) CRMAGE: CRISPR Optimized MAGE Recombineering. Sci. Rep. 6, 19452. 10.1038/srep19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress B. F.; Toparlak Ö. D.; Guleria S.; Lebovich M.; Stieglitz J. T.; Englaender J. A.; Jones J. A.; Linhardt R. J.; Koffas M. A. G. (2015) CRISPathBrick: Modular Combinatorial Assembly of Type II-A CRISPR Arrays for dCas9-Mediated Multiplex Transcriptional Repression in E. coli. ACS Synth. Biol. 4, 987–1000. 10.1021/acssynbio.5b00012. [DOI] [PubMed] [Google Scholar]
- Wilson M. E.; Maksaev G.; Haswell E. S. (2013) MscS-like mechanosensitive channels in plants and microbes. Biochemistry 52, 5708–22. 10.1021/bi400804z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.