Abstract
Reliable and cost-effective assays with adequate sensitivity are required to detect the DNA methylation profile in plants for scientific and industrial purposes. The proposed novel assay, named EpiHRMAssay, allows to quantify the overall methylation status at target loci and to enable high-throughput analyses. It combines in tube High Resolution Melting Analysis on bisulphite-treated templates with the in silico prediction of the melting profile of virtual epialleles using uMELTSM software. The predicted melting temperatures (Tm-s) of a set of epialleles characterized by different numbers of methylated cytosines (#mC) or different mC configurations were obtained and used to build calibration models, enabling the quantification of methylation in unknown samples using only the in tube observed melting temperature (Tm-o). EpiHRMAssay was validated by analysing the promoter region of CMT3, DDM1, and ROS1 genes involved in the regulation of methylation/demethylation processes and chromatin remodelling within a population of peach plants. Results demonstrate that EpiHRMAssay is a sensitive and reliable tool for locus-specific large-scale research and diagnostic contexts of the regulative regions of genes, in a broad range of organisms, including mammals. EpiHRMAssay also provides complementary information for the assessment of heterogeneous methylation and can address an array of biological questions on epigenetic regulation for diversity studies and for large-scale functional genomics.
Keywords: DNA methylation, High Resolution Melting Analysis, HRMA, in silico melting prediction, somaclonal variation, epigenetic regulation
Introduction
DNA methylation is one of the best described epigenetic mechanisms, with a role in genome protection against transposable elements and retroviruses, genomic imprinting, and heterochromatin-induced silencing [1, 2]. DNA methylation also provides instruction to gene expression machinery as to where and when the gene should be expressed [3]. In mammalian systems, the genomic DNA methylation pattern is global, with the conspicuous exception of short unmethylated regions called CpG islands [4]. In plants, DNA methylation can also occur in asymmetric sequence contexts, such as CpHpG and CpHpH (where H = A, C, or T) and irregular spread in a mosaic pattern among genic and intergenic regions [4–6]. In plants, at least three types of DNA methyltransferase regulate methylation (METHYLTRANSFERASE1, MET1), responsible for symmetric CpG methylation maintenance [7, 8], CHROMOMETHYLASE1, 2, and 3 (CMT1, 2, and 3), plant-specific DNA methyltransferases, responsible for the maintenance of DNA methylation at CpHpG sites [9, 10] and DOMAIN REARRANGED METHYLTRANSFERASES (DRMs), responsible for de novo DNA methylation at all sequence contexts [11]. The DECREASED DNA METHYLATION1 (DDM1) gene, a SWItch/Sucrose Non-Fermentable (SWI/SNF) family chromatin-remodelling factor, is required for normal genomic DNA methylation and ectopic gene and transposon silencing [12, 13], allowing MET1 access to its target region [8]. REPRESSOR OF SILENCING1 (ROS1) and DEMETER-like proteins DML2 and 3, are glycosylases regulating DNA demethylation, for protecting genes from potentially deleterious methylation [14].
Molecular tools for the detection of DNA methylation status are revolutionizing the understanding of molecular physiology and provide an opportunity to unravel the relationship between gradients of environmental factors and gene expression and/or somaclonal variations, determining genetic modification and trait diversity in species. Many PCR-based techniques using bisulphite-treated DNA templates have been developed for single-locus DNA methylation analysis [15]. Bisulphite sequencing is the most recurrently applied technique, particularly in plant [6, 9, 12], and provides information on the methylation pattern at single nucleotide resolution. However, its costs and labour intensiveness make this approach not always the most advantageous to DNA methylation analysis, particularly when the purpose is to obtain information about overall methylation status at specific template regions. Methods based on DNA melting properties after the bisulphite treatment of the template, such as methylation-sensitive melting curve analysis (MS-MCA) or methylation-sensitive high-resolution melting (MS-HRM), represent cost-effective alternative or complementary tools for methylation analysis in specific regions by PCR amplicon through melting behaviour of PCR products [16, 17]. Bisulphite treatment, which converts unmethylated cytosine to uracil, leaves unchanged methylated cytosines (mC); the uracils are amplified in subsequent PCR reaction as thymines, whereas mC residues are amplified as cytosines, thereby determining the methylation-dependent modification of DNA template. Amplicons from methylated template have a higher cytosine content and, therefore, a higher melting temperature than those of the amplicons from unmethylated templates. MS-MCA technique could efficiently detect qualitative differences among samples. However, for a quantitative analysis, this technique should be coupled with sequencing procedures and/or standard calibrators, i.e. amplicons with known methylation status. In plants, the variability of methylation patterns could increase the complexity of using of calibrators, because methylation occurs in different sequence contexts and at different frequencies [6].
DNA denaturation (melting) is the separation of dsDNA into two ssDNA. The temperature at which occurs the melting of the DNA (Tm) is defined as the temperature at which half of the DNA is in double helical state and the other half is in the random coil or single-stranded (ssDNA) state. The melting temperature (Tm) is dependent on several physicochemical properties, although the base-pairing and base-stacking interaction between complementary strands and adjacent bases are among of the main factors [18–20]. Among the several approaches used for DNA melting modelling, those based on both statistical mechanics (Poland–Scheraga type model) and nearest-neighbour thermodynamics have been widely employed and implanted in many bioinformatics tools such as POLAND thermodynamics algorithms, such as POLAND [21], MELTSIM [22], DINAMelt [23], Stitchprofiles [24], and more recently, uMELTSM [25]. This last is a web-based application tool, which implements a fastest algorithm for base-pairing probability profile [26] and several other features, including Mg2+ and DMSO concentrations [27] along with updated thermodynamics set. The in silico prediction of melting profiles is a convenient tool to design and optimize high-resolution melting experiments, particularly for mutation scanning and genotyping [28].
In this work, uMELTSM software, available at https://www.dna.utah.edu/umelt/umelt.html, was used to predict the melting profile of a range of virtual epialleles at target loci, characterized by a different number and/or position of mC. The in silico predicted melting temperature (Tm-s) of each epiallele and the respective number of mC were used to build a calibration model able to quantify the overall methylation status of unknown epialleles in each sample based only on the in tube observed melting temperature (Tm-o). This approach has been validated by analysing the methylation status of the CMT3, DDM1, and ROS1 promoter regions within a population of in vitro propagated Prunus persica L. Batsch plants.
In vitro tissue culture is an important industrial and scientific activity and has applications in plant propagation, breeding, biotechnology, and germplasm preservation. The in vitro conditions are stressful environments [29, 30] and plants grown in vitro might exhibit genotypic and phenotypic variability, defined somaclonal variation [31], even within the same plant clone. Studies on stability of plants grown in vitro have also revealed the involvement of epigenetic variations [32]. In vitro plant production can be achieved either through the proliferation of pre-existing axillary buds or by adventitious regeneration from somatic tissues [33], and both systems can generate homogeneous-like plants, although their impact on both heritable and epigenetic characters is different ([32] and references therein). The phenotypic and genotypic changes in plant can impact on plant identity [34] and thus the economic efficiency of the micropropagation industry.
Materials and methods
Plant material and DNA extraction
In vitro shoot cultures of P. persica L. Batsch, cultivar Rich Lady, were obtained from a single axillary bud excised in spring from an in vivo growing adult tree as previously reported [35]. The obtained shoot cluster was transferred into a vessel (Magenta, Sigma, Italy) containing 50 mL of a multiplication medium (MM), consisting in QL [36] macro salts, MS [37] micro salts and organics, 87.7 mM sucrose (Eridania, Italy), and 5.5 g L−1 agar (B & V, Italy). MM medium was supplemented by 1.11 µM BA, 16.3 µM adenine sulphate, 0.29 µM IBA, and 0.19 µM GA3. The pH was adjusted to 5.7 prior to sterilization in autoclave and cultures were maintained at 24 ± 2°C under a 16-h photoperiod and at the photon flux density of 40 µM m−2 s−1, provided by cool white fluorescent tubes (Fluora L58 vv/77, Osram, Italy). Sub-culturing on fresh medium was performed every 21 days. Leaf tissues were sampled from the donor plant grown in field, and the leaves and stems of cloned shoots grown in vitro were sampled at the 4th, 18th, and 23th sub-culture.
Total genomic DNA was extracted using modified CTAB protocol [38] and dissolved in TE buffer. RNase digestion was performed using the DNeasy Tissue Extraction Kit (Qiagen, Hilden, Germany), following the manufacturer protocol. DNA was quantified using Qubit® 2.0 Fluorometer (Life Technologies, Italy) and visualized on agarose electrophoretic gel.
Bisulphite treatment
Bisulphite treatment and recovery of samples were carried out with the EpiTect Bisulphite kit (Qiagen, Hilden, Germany) following the manufacturer’s instruction. Briefly, 1 µg of DNA in a 20 µL was used for each reaction and mixed with 85-µL bisulphite mix and 25-µL DNA protected buffer. Bisulphite conversion was performed on a thermocycler as follows: 95°C for 5 min, 60°C for 25 min, 95°C for 5 min, 60°C for 85 min, 95°C for 5 min, 60°C for 175 min, and 20°C overnight. The bisulphite-treated DNA was recovered using an EpiTect spin column according to the manufacturer’s instructions and eluted a final volume of 50 µL, to give a theoretical concentration of 20 ng µL−1 (assuming no loss of DNA during bisulphite conversion).
Unmethylated standard and synthesized epialleles for improving model calibration
Unmethylated standard (UMstd) epialleles for DDM1, CMT3, and ROS1 were obtained by the amplification of target regions from untreated genomic DNA template. PCR products were treated with bisulphite using EpiTect Bisulphite kit (Qiagen, Hilden, Germany), and used as unmethylated control template (UMstd) for HRMA assay. Standard epialleles of CMT3 were synthesized by GeneArt (Life Technologies, Carlsbad, CA, USA) as double-strand DNA fragments.
HRMA and digital MS-HRM
PCR amplifications were performed in capillaries on Light Cycler® 1.5 (Roche, Germany), using SensiMix kit (Quantace, USA) and LC-Green II plus dye (Idaho Technology, USA), in a 20 µL total volume containing: 4 µL Mix Buffer, the appropriate concentration of MgCl2 for each studied gene (0.75 mM for CMT3 and ROS1, and 1 mM for DDM1), and 100 nM of each primer specifically designed for DNA template and for DNA bisulphite treated (Table 1), 1 µL of LC-Green dye, 1.5 µL of Taq polymerase and 1 µL (20 ng µL−1) of DNA template and bisulphite-modified template. Only for the amplification of CMT3 fragments was 3% DMSO added to the reaction mixture. The amplification profile was set to 10 min at 95°C and 50 cycles of 10 s at 95°C, 10 s at the primer annealing temperature and 10 s at 72°C. HRMA was performed separately on an HR1 instrument (Idaho Technology, ID, USA) with a temperature ramp from 70° to 90°C, rising by 0.10°C s−1. Two technical replications and two independent experiments were run for each reaction. The melting curves were normalized using the software provided with HR1 instrument and visualized using Derivative and Difference Plot tools. Digital methylation-sensitive high-resolution melting (dMS-HRM, [39]) was performed using a dilution series from 1:50 to 1:100, starting from 1 ng µL−1 of template. Poisson distribution was used as a guideline to determine the expected distribution of templates. HRMA and PCR amplification profile was the same above described, except for cycles number, which was set at 60.
Table 1.
Genome localization of CpG-enriched region, primer sequences, amplicon size, and potential number of methylation site of each promoter gene
| Gene | Position (Peach Genome v2.0) | Primer sequence for bisulphite-treated DNA (5′-3′) | Length (bp) | No. of methylated sites (#mC) |
|||
|---|---|---|---|---|---|---|---|
| CpG | CpHpG | CpHpH | Total | ||||
| DDM1 | sd_7:16329264.16329474 | F- GCAGAGAGGTATATAAAGTAATTT | 211 | 8 | 4 | 9 | 21 |
| R- CAACCCACCAATA TCTCCATTCAC | |||||||
| CMT3 | sd_6:847278.847527 | F- GTTTGGTGGAGAGAGAAAGAGGG | 250 | 14 | 1 | 7 | 22 |
| R- TAAAAAAGAAAAT AAAGTTAAAGT | |||||||
| ROS1 | sd_7:1472992.14673292 | F-TAGAAGAAATTGAAGAGAATAGA | 301 | 9 | 11 | 19 | 39 |
| R- GTAAATATGGTAC AAATAGCAACAC | |||||||
Identification of CpG islands-enriched template region and design of primers
The DDM1, CMT3, and ROS1 promoter regions were identified in Peach Genome v1.0 [40] assembly (http://www.rosaceae.org/species/prunus_persica/genome_v1.0) available at GDR database and analysed with Methyl Primer Express® v1.0 software (Applied Biosystems Inc., Foster City, CA, USA) to find CpG-enriched template regions (Table 1). The CpHpG and CpHpH sites were manually identified and annotated. Primers were designed to not discriminate between methylated and unmethylated templates. The amplicons size range in the 200–300 bp window, a hypothetical compromise between sensitivity and screening resolution. The analysis of DDM1 gene revealed a single CG island (CGI), located from −729 to −125 upstream ATG site. The primers amplify a 211-bp amplicon positioned in the bottom strand and composed by 21 potential mC (Table 1). A single CGI was identified for CMT3 gene, located from −943 to −329 upstream ATG site. The primers amplify a 250-bp amplicon positioned in the bottom strand and composed by 22 potential mC (Table 1). For ROS1 gene, a single CGI, located from −1199 to −899 upstream ATG site was also identified. The primers amplify a 301-bp amplicon positioned on the top strand and composed by 39 potential mC (Table 1). The primers for bisulphite-treated template, amplicons length and number of mC are shown in Table 1. The primers for untreated DNA were the follows: DDM1, F-CAACCCACCAATATCTCCATTCAC, R-GCAGAG AGG CATATA-AAGTAACCC; CMT3, F-TAGACATATC CACACGCT TATTG, R-GTTTGGTGGAGA-GAGAAAGAGGG. The primers for ROS1 were the same used for bisulphite-treated template.
Identification of G + C content domains in the promoter region of analysed genes
The GC-rich domains in the DDM1, CMT3, and ROS1 sequences were searched and estimated by the web-based tool GC-Profile (http://tubic.tju.edu.cn/GC-Profile/), released by Gao and Zhang [41].
In silico HRMA
Melting curve simulations of virtual epialleles were performed in uMELTSM, testing different thermodynamic sets: MELTSIM [19], and those described by Huguet et al. [42] and Weber [43]. The in silico Tm-s values of melting peaks were calculated as the negative derivative of helicity with respect to temperature through the derivative plot function. Salt concentrations were set considering the experimental PCR conditions. However, to account for the effect of the intercalating dye on Tm-s, the monovalent cation concentration was adjusted to reproduce in tube observed Tm (Tm-o) of UMstd fragments. For DDM1 and ROS1, default values of α and σ parameters were used (as described in ‘Results’ section), whereas for CMT3, the σ parameter was set from 0 to 0.25. The melting temperature range was set between 70°C and 90°C, with maximum resolution of 0.01°C. Simulated melting curve data were downloaded and used for derivative plot graph reconstruction using SigmaPlot v10.0 software (SYSTAT, San Jose, CA, USA).
Cloning and sequencing of amplicons
Amplicon samples generated from DNA template and bisulphite-treated DNA were purified using PCR Purification kit (QIAGEN), cloned into pGEM-T vector system (Promega, Madison, WI, USA) and sequenced using universal primers. The sequences were aligned and displayed using CyMATE tool [41], freely available at www.cymate.org website.
Building of predictive models
For each analysed amplicon, the Tm-s value was obtained from simulated melting curve, and the whole set of Tm-s values was used to build a linear model for predicting the number of mC. For the promoter region of each gene, a linear dependence of the number of methylated cytosines (#mC) on the increase in melting temperature of the corresponding fragment (Tm) with respect to the melting temperature of the unmethylated standard fragment (UMstd Tm) was hypothesized. According to this hypothesis, the equation #mC = a + b (Tm-s − UMstd Tm-s) was used to describe this dependence and a linear regression procedure was used to obtain the model parameters (a and b) for each gene. To take into account the variability of Tm-s values due to the positions patterns of the mC, a proper set of virtual fragments was realized and employed for simulations for each gene (vide infra); all the simulated Tm values were then used to build each model. The goodness of the fitting procedure was evaluated by analysing the correlation coefficient value. These models were applied to predict the #mC for the investigated fragments using the corresponding experimental value of the Tm-o.
Results
HRM qualitative scanning of methylation status of samples
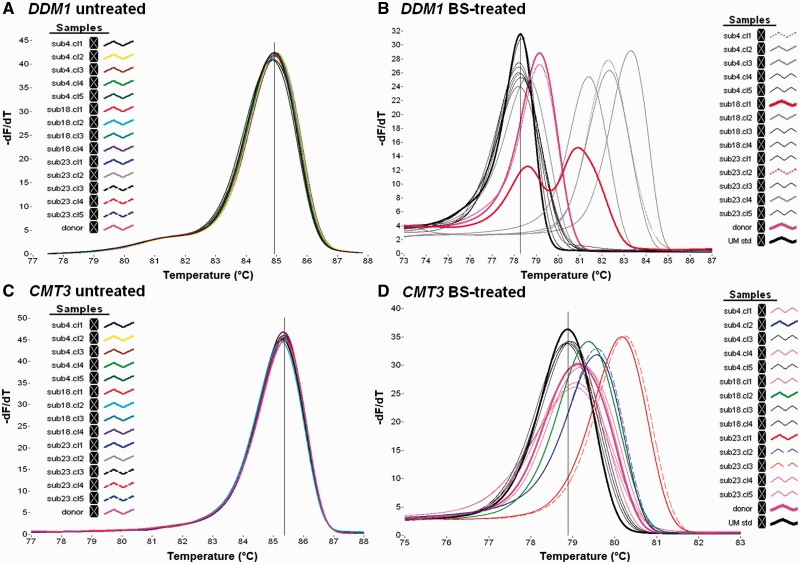
Sequence mutations, such as single nucleotide polymorphisms, might affect the prediction of methylation status, due to their effect on thermodynamic properties of DNA with the consequent effect on the melting profile and Tm value. Therefore, HRMA was first carried out on untreated DNA templates of each plant samples to check for the presence of polymorphisms within the promoter regions of the DDM1, CMT3, and ROS1 genes. The amplicons showed no significant Tm-o or melting shape differences among samples, excluding the occurrence of genetic variations (Fig. 1A and C, and Supplementary Fig. 1A). The HRM analysis that was carried out on bisulphite-treated DNA of the same plant samples showed wide differences in the melting curve profiles (Fig. 1B and D, and Supplementary Fig. 1B), revealing the presence of methylated epialleles. The Tm-o values of some amplicons resulted identical to those of DDM1, CMT3, and ROS1 UMstd and, therefore, their methylation status can be assigned based exclusively on the melting profile. Instead, overall methylation status of amplicons with different Tm-o values cannot be inferred, although, in theory, sample Tm-o values are expected to increase with sample methylation degree, as schematically shown in Supplementary Fig. 2A. The presence of a single melting peak for each analysed sample suggests a homogeneous pattern of methylation within in vitro plant clones, except for DDM1 amplicon of sub18.cl1 sample (Fig. 1B), showing two distinct peaks. This suggests a heterogeneous methylation pattern with a mixture of at least two different methylated epialleles (Fig. 1B and Supplementary Fig. 2B). In most of the samples, a reduction of the level of methylation occurred with respect to donor plant, as highlighted by the lower Tm-o value. In contrast, the level of methylation increased in a few samples in the promoter region of DDM1 and CMT3 genes (Fig. 1B and D) and only in one sample of the ROS1 gene promoter (Supplementary Fig. 1B).
Figure 1.
Derivate plot curves from the HRMA assay of the DDM1 and CMT3 gene promoter regions, as resulted by the amplification of untreated bisulphite DNA template (A and C, respectively) and from bisulphite-treated template (B and D, respectively). (B, D) The cursor indicates the Tm-o value of unmethylated standard (UMstd).
Development of calibration models by using in silico Tm-s prediction
For the quantification of the overall methylation status of the investigated samples, calibration models were developed for each analysed template region through the following procedures:
generation of a set of virtual epialleles for the target template regions;
evaluation of the variability of Tm-s values within and between the generated classes of epialleles, using the in silico melting prediction software uMELTSM;
optimization of the in silico melting parameters using standard epialleles (optional); and
building of calibration model through a fitting procedure based on the hypothesis of a linear dependence between the number of methylated cytosines (#mC) and the increase in the melting temperature of the corresponding fragment (Tm) with respect to the melting temperature of the standard fragment (UMstd Tm).
Generating a set of virtual classes of epialleles for the target regions
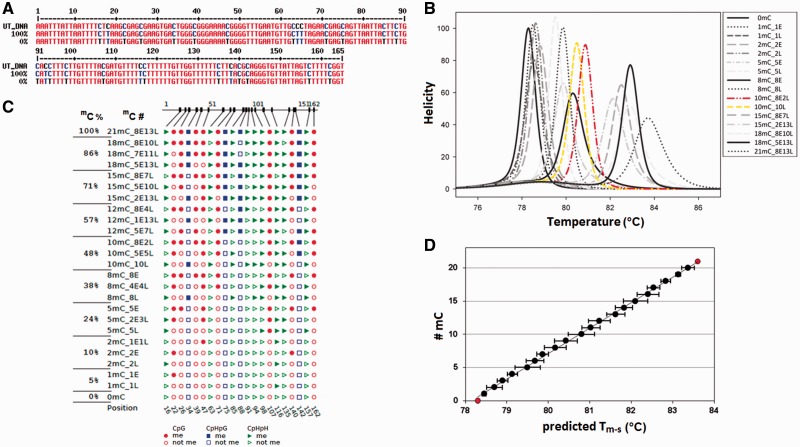
After the identification of potential methylation sites, CpG (named E in this article), CpHpG and CpHpH (named L) by Methyl Express tool and fine-tuned by hand (Supplementary Fig. 3), an in silico bisulphite treatment was simulated on the target genomic sequences of DDM1, CMT3, and ROS1, by converting every cytosine into uracil, obtaining the respective unmethylated epialleles. Then, a set of virtual classes of epialleles covering the complete range of methylation sites number (from unmethylated to fully methylated) was generated by computationally changing the methylation status (mC) of every putative methylated cytosine (Fig. 2A and Supplementary Figs 4A and 5A). According to the potential number of methylated sites, a total of 22, 23, and 40 classes of epialleles was generated for each target region of DDM1, CMT3, and ROS1, respectively. Excluding the unmethylated and fully methylated classes of epialleles, which can only assume one possible configuration, all the other classes can assume a set of multiple configurations, with a fixed number of mC, but a different mC distribution pattern among the available sites. Therefore, each configuration is characterized by its own melting profile and a specific Tm-s value (Fig. 2B and C, and Supplementary Figs 4B, C and 5B, C). uMELTSM software predicts the trend of the average Tm value of each class of epiallel (Fig. 2D and Supplementary Figs 4D and 5D), concomitant with the increase in the number of methylated cytosines, as theoretically expected from the increase of C:T replacing (Fig. 2C and Supplementary Figs 4C and 5C). Indeed, the bisulphite treatment of DNA does not convert the methylated cytosine in uracil and the base is amplified as cytosine, thus increasing the GC content and the melting temperature of the amplicon. However, the increase of Tm-s values is not only dependent on the absolute number of mC, but also on their distribution among the CpG, CpHpG, and CpHpH sites, as visible from the variability of mean Tm-s associated with each class of epialleles (Fig. 2C and D, and Supplementary Figs 4C, D and 5C, D).
Figure 2.
(A) The UT_DNA genomic sequence of the DDM1 amplicon aligned with the unmethylated (0%) and the full-methylated (100%) sequence after in silico bisulphite treatment. The primer sequences were excluded from the alignment. (B) The derivative melting plots predicted with uMELTSM software and the MELTSIM algorithm. In yellow and red line are shown the derivative melting plot of 10L and 8E2L, respectively. (C) The CyMATE visualization of representative configurations used to build of calibration model for each class of epialleles. The classes of epialleles are indicated by percentage of methylated cytosine (mC%) and the methylated site number (#mC). The configurations in each class differ for the distribution of methylation at E (CpG) and/or L (CpHpG/CpHpH) sites. (D) #mC values as a function of the predicted mean Tm-s (±SD) values of each epiallele class.
Evaluating the variability of Tm-s values within and between class of epialleles
For building a robust calibration model that properly correlates the Tm-s value and mC number, the Tm-s variability generated by the different configurations in each class of epialleles should be evaluated, also determining the maximum and minimum Tm-s values. Generally, the smaller is the value of the Tm-s difference between the extreme configurations within each class compared with the difference between classes, the more accurate will be the calibration model. A reasonable range of Tm-s values for each class of epialleles was estimated, considering a subset of configurations. According to nearest-neighbour thermodynamic criteria, the variation of Tm-s values among different configurations in each class mainly depends on the thermal stability conferred by the mC neighbour nucleotides, more specifically by the G nucleotide present at CpG site compared to the A or T nucleotides presents at CpHpG or CpHpH sites. Therefore, a subset of configurations for each class was chosen based on the frequency of methylation at E sites (CpG) or L sites (CpHpG/CpHpH), allowing to reasonably estimate the range of variability. For example, in the epialleles class 10mC for DDM1 amplicon, the highest Tm-s value of 81.20°C was predicted for the configuration 8E2L (8E is the maximum possible number of mC at CpG site), whereas the lowest value, 80.50°C, for the 10L (all the 10 mC positioned on CpH sites), with a difference of 0.70°C between the two configurations (Fig. 2B). Based on this criterion, a number of configurations ranging from 3 to 7 was considered for each class of epialleles and for each analysed region, and the respective Tm-s values were used for building the calibration model.
Optimization of in silico melting parameters by using standard epialleles
A melting profile with a single peak was predicted by uMELTSM for almost all selected configurations of DDM1 and ROS1 epialleles (Fig. 2B and Supplementary Fig. 5B). In contrast, many configurations of CMT3 amplicon displayed multiple melting domains and/or complex shapes, independent of the thermodynamic set used in uMELTSM (data not shown). No Tm-s value can be assigned to these configurations, causing their exclusion from the calibration procedure and thus limiting the reliability of the modelling. In the sequence of CMT3, a GC-rich domain with four nested CpG sites seems to affect the predicted melting profile of many configurations, resulting in a complex curve shape (Supplementary Fig. 6A). The frequency of occurrence of complex profiles is higher for those configurations retaining methylation at all the four CpG sites irrespective of the methylation status of the other cytosines surrounding the domain and particularly for the epialleles classes ranging from 5 to 15 mC. For example, the melting profile of the configurations 5E (5mC class) and 14E1L (15mC class) showed two melting peaks, whereas the configuration 10E (10mC class) showed an asymmetric profile with a skew toward high temperatures (Supplementary Fig. 6B). In the classes with more than 15 mC, the frequency drops down (data not shown). The complexity of melting domain might be resolved or attenuated in uMELTSM by modifying the values of σ parameter, accounting for the cooperativity of melting domains. Increasing the σ value from the default value up to 0.25 modify the melting curves, resulting in a single melting peak (Supplementary Fig. 6C). The effect of CpG methylation within the GC-rich domain was experimentally verified using synthetic amplicons for representative configurations of the 10mC class (10E and 2E8L) and 15mC class (14E1L and 7E8L), including also the fully methylated class 22mC (14E8L). The melting profile resulting from in tube HRMA of the 14E1L and 10E configurations clearly showed a single and slightly broadened peak (Supplementary Fig. 6D), supporting the hypothesis that a suitable setting of σ parameter may improve the goodness of the simulation and resolve melting complexity, at least in this situation. The value of σ parameter that best approximates the experimental curves was 0.25; thus, it has been set for melting prediction in all other epiallele classes.
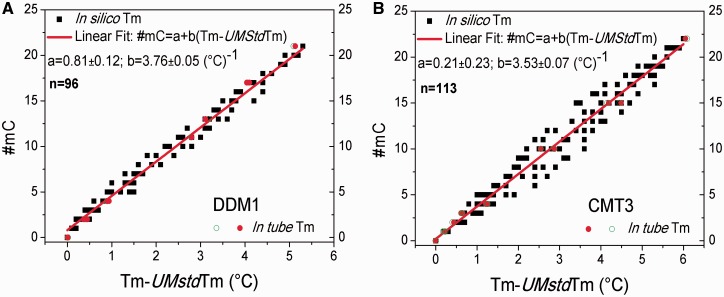
Building of calibration model through a fitting procedure
For each analysed amplicon, all the Tm-s values obtained from the set of selected epialleles were used to build a model for predicting the mC number, under the hypothesis of linear correlation between #mC and Tm increase #mC = a + b (Tm-s − UMstd Tm-s). The data used to build the models for DDM1, CMT3 amplicons are shown in Fig 3A and B, respectively, along with the corresponding fitted curves; the fitting parameters and linear correlation coefficient values are reported in the captions. The same is shown in Supplementary Fig. 7 for ROS1; the fitting parameters and correlation coefficient value are reported in the figure caption. The observed values of the correlation coefficient (R = 0.993, 0.978, and 0.991, for DDM1, ROS1, and CMT3, respectively) confirmed the hypothesis of linear dependence between the considered variables (Fig. 3 and Supplementary Fig. 7). MELTSIM thermodynamic set were used for the melting prediction of the three promoter fragments. Recently, other two thermodynamic sets were released as described by Huguet et al. [42] and Weber [43], both available in uMELTSM. Obtained Tm-s values were then used to build predicting models. As shown in Supplementary Fig. 7, where all the three model calibrations for DDM1 amplicon are shown, the three thermodynamic sets performances are similar without any significant differences. The same was observed also for CMT3 and ROS1 (data not shown) and, therefore, the choice of MELTSIM set has been arbitrary. However, we cannot exclude that in other sequence context, the accuracy of the three thermodynamic sets might be significantly different.
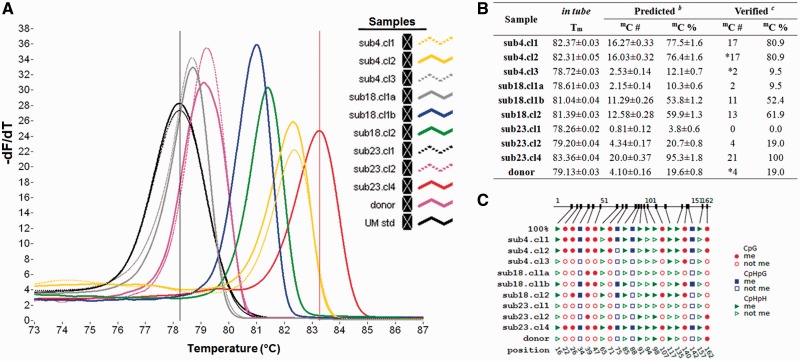
Figure 4.
(A) The derivative melting curves generated from the set of samples with different #mC analysed for the DDM1 locus. The Tm-o values correspond to the curve peaks for each DDM1 amplicons (coloured curves). Dark and pink curves represent the UMstd and sub23.cl4 sample, respectively. (B) The Tm-o values detected for each sample and generated from the comparison between numbers and percentages of the predicted mC. (C) The position of methylated sites, as observed through the sequencing of amplicons using CyMATE representation.
Figure 3.
The #mC values obtained from simulated melting profiles (black squares) and from experiments (red and green empty circles) as a function of (Tm − UMstd Tm) for DDM1(A) and CMT3(B). The red line is the result of a linear fitting procedure of the simulated data under the hypothesis that the linear dependence #mC = a + b (Tm − UMstd Tm) holds (see text). Fitting parameters for DDM1(n = number of values used to build the model = 96): a = 0.81 ± 0.12 and b = 3.76 ± 0.05 (°C)−1 with R = 0.993. Fitting parameters for CMT3(n = 113): a = 0.21 ± 0.23 and b = 3.53 ± 0.07 (°C)−1 with R = 0.978. The P value corresponding to the observed R value of both built models is lower than 0.0001, which confirms the high significance of the hypothesized linear relationship.
Quantification of methylation status of experimentally investigated samples using in silico-developed calibration models
The calibration models that were built using in silico generated Tm values were used to estimate the overall methylation status of plant samples based on the in tube detected Tm-o. The experimental melting profiles of DDM1 show Tm-o values ranging from the highest one obtained for sub23.cl4 amplicon (83.36 ± 0.04°C) to the lowest one obtained for UMstd and sub23.cl1 (78.26 ± 0.02°C) (Fig. 3). Applying the proper calibration model to the in tube detected Tm-o, several methylated cytosines varying from mC = 20.0 for sub23.cl4 to mC = 0.81 for sub23.cl1 were predicted. Regarding the sub18.cl1, showing two melting peaks, to determine whether the two peaks arise from PCR artefacts or represent a mixture of different methylated epialleles, a dMS-HRM approach was applied on PCR products. Two differently methylated alleles were isolated (named α and β), and the proportional amount of each allele in the sample was estimated using the approach described in Hetzl et al. [44] (Supplementary Fig. 9). The respective calibration models were also applied to CMT3 and ROS1 amplicons (Supplementary Figs 10 and 11). As expected, the fully methylated amplicon of configuration 14E8L of class 22mC of CMT3 showed the highest Tm-o (84.96 ± 0.04°C), whereas the lowest was detected for sub4.cl1 and the donor plant, with a predicted number of only 1 out of 22 methylation sites. In the case of ROS1, the clones showed low levels of methylation, except for sub23.cl4 (Tm-o of 83.76 ± 0.05°C), and a respective predicted number of 26.65 mC.
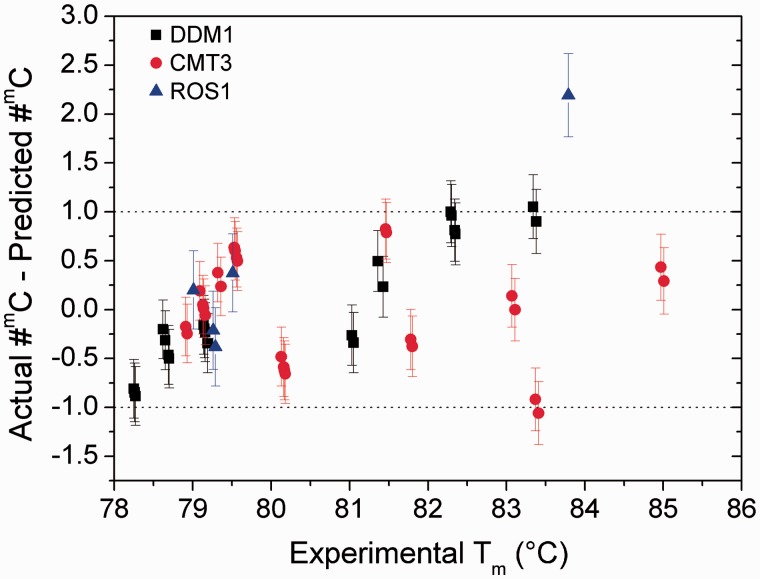
The reliability of the predictions was evaluated by comparing observed #mC, verified by bisulphite sequencing, and predicted #mC of experimentally investigated samples. The results of bisulphite sequencing of all the tested amplicons confirmed the trustworthiness of the prediction models, particularly for the DDM1 and CMT3 amplicons. In fact, the predicted #mC number differs, within the errors, from the observed ones of less than ± 1, which is the smallest possibly uncertainty of a discrete quantity, thus confirming the affordability of the predictions obtained by the models (Fig. 5). This was also true for ROS1 amplicons, although the error in #mC prediction increases up to about ±2 for sub23.cl4 (Fig. 5).
Figure 5.
Predicted #mC values were obtained from experimental Tm-o values for the investigated fragments (with known mC, named actual #mC) using the built models. The difference between the actual and predicted #mC values is reported for all of the genes as a function of observed Tm-o.
EpiHRMAssay pipeline
All components of the EpiHRMAssay pipeline workflow, inputs, and outputs have been summarized in Supplementary Fig. 12. The EpiHRMAssay pipeline consists of three main sets of sequential steps. Based on a user-defined of target regions, fine-tuned by hand for asymmetric mC number (CpHpG and CpHpH), and designed parameters, the pipeline retrieves annotated primer pairs and a list of in silico Tm-s predicted values for epialleles libraries that are used to generate predicting models. Based on the primers pairs, EpiHRMAssay generates a HRM qualitative scanning of the methylation states of target region. Based on in silico-developed calibration models, EpiHRMAssay quantifies the methylation status of experimentally investigated samples.
Discussion
The analysis of cytosine methylation at specific DNA regions, such as promoters and/or gene regulatory elements, is critical for understanding the role of epigenetic events in plant development and environmental adaption. Rapid, affordable, and cost-effective methods allowing a high-throughput screening of a large number of individuals are required to unravel the complexity of DNA methylation pattern, dynamically regulated at organ, tissue, or cellular level and affected by developmental and/or environmental cues. Despite the several methods and approaches that have been developed for DNA methylation analysis, plant methylation studies rely almost exclusively on bisulphite-sequencing approaches, which allow the high-resolution mapping of methylated cytosine both at target loci and at whole-genome level. However, bisulphite sequencing is an expensive and time-consuming procedure and might be surrogated by more rational approaches, particularly when the purpose is to obtain information on methylation status at certain DNA regions or to scan one target gene in a large number of samples. Among the several PCR-based methods, those based on HRMA have proven to be valid alternatives or complementary tools for bisulphite sequencing, e.g. in the quality assessment of methylation differences between samples or in the evaluation of epiallele proportion in heterogeneous methylated samples [16, 17, 45–48]. The basis of this method is that PCR products generated from bisulphite-treated DNA templates with different contents of mC show differences in melting temperature, which can be visualized by DNA intercalating dyes and melting analysis in a thermal cycler coupled with a fluorometer [16, 49–51].
The EpiHRMAssay described in this work is a further branch of the application of HRMA-based methods. The procedure involves the generation of a representative set of epialleles covering the entire range of methylation degree; the prediction of the Tm values by melting simulation software, such as uMELTSM, and their use for the development of a calibration model to quantify the overall methylation status of unknown samples based only on the in tube detected melting temperature (Tm-o). Although the procedure does not identify the specific position of mC sites, it provides the opportunity to estimate the overall methylation status of target region using a rapid and relative simple assay to detect and resolve samples with heterogeneous methylation, which is often difficult to manage by bisulphite-sequencing approaches.
To validate this approach, three DNA amplicons belonging to the promoter regions of three genes were chosen, each characterized by different sequence length and methylation sites number. Amplicon length is a crucial parameter for the correct application of EpiHRMAssay approach and should be set to ensure a linear correlation between the increase in the predicted Tm-s with respect to the UMstd Tm and mC number. Clearly, this could be easily achieved by testing and designing a priori the candidate amplicon using melting prediction tools. In general, the setting of amplicon size in the range of 150–250 bp was an adequate compromise between sensitivity and resolution. The analysis of amplicons characterized by a balanced distribution of methylated sites and a reduced Tm-s differences within classes of epialleles (as observed in DDM1 amplicon) allows the building of accurate calibration model without a need for the adjustment of in silico melting parameters. However, in some cases, as for CMT3 amplicon, the melting prediction procedure can be improved by the adjustment of melting parameters using a few standard fragments. Moreover, EpiHRMAssay could be simplified by developing computer scripts to automatically generate the set of epiallele configurations, implementing a set of thermodynamic algorithms.
In this work, a representative example of the potential application of EpiHRMAssay procedure for the epigenetic screening of somaclonal variants produced by in vitro culture is also reported. For the first time, the methylation levels for DDM1, CMT3, and ROS1 are reported in shoots of in vitro cloned agamically propagated plants. No genetic polymorphism was detected in plants that were grown in vitro for long period and agamically generated from a single axillary bud of the ‘donor’ plant. However, according to our findings, we can definitely assert that methylation status can vary among the cloned shoots of the same genotype, and methylation changes are random; in fact, there is a different degree of the methylation events among the studied genes independent of the number of sub-cultures. When the methylation status of the ‘donor’ plant is not maintained in cloned shoots, in the major, the level of methylation (66%) was reduced, and only in a lower percentage did this level increase (25%). In addition, EpiHRMAssay resolves allelic heterogeneous methylation, detecting in the same reaction both low-methylated and high-methylated alleles, as relived by the two distinct melting peaks of DDM1 of the clone sub18.cl1, which yields two different amplicons with low and high Tm-o, respectively. DDM1, CMT3, and ROS1 methylation occurrences are relevant events in the epigenetic regulation of plant development because these genes are involved in chromatin remodelling [12, 13], in plant-specific maintenance of DNA methylation at CpHpG sites [9, 10], and in regulation of DNA methylation, respectively. The results obtained with the in vitro grown shoots suggest that some ‘normal’ plants can already present an epigenetic mutation that is not detectable by a visual analysis. Although many aspects of the mechanisms that result in somaclonal variation remain undefined, many phenotypic changes in in vitro propagated plants might be attribute to epigenetic variations [52], thus, the EpiHRMAssay represents a powerful tool for their assessment. Although mutations in DNA sequence are considered the cause of genetic variability, it is also becoming clear the prominent role played by epigenetic changes occurring throughout the in vitro culture process [53], even in mammalian cells and tissues, involving the interplay among DNA methylation, histone modification, RNA interference, and gene silencing [8–10, 54–56]. In our study, we have used whole leaf and stem tissue containing a representative sample population of tissue and cells; therefore, we consider that the observed unambiguous results reflect the methylation status of the whole plant.
Locus-specific methods, which are sensitive, specific, and cost-effective, are widely used to quantify the DNA methylation status for human diagnostic procedures ([57] and references therein). Whole-genome bisulphite sequencing is very accurate in organism where high-quality reference genome is available, allowing to recognize the substitutions of unmethylated C present in the regulatory regions of genes. However, this is only feasible for few individuals because of costs and data management, this last requiring dedicated algorithms and a long working time. If the diagnostic and research screening must be run on target regions on one or more individuals of unknown sequence the use of EpiHRMAssay in species without a reference genome sequence, EpiHRMAssay reduces strongly time and costs of investigation.
We speculate that by massive-parallel amplification procedure and reading of melting temperature, the overall cost and time required for methylation status identification of defined region of certain genes will be dramatically decreased and the procedure will be easy to handle. The Tm-s values obtained through uMELTSM software provide a rapid, high-throughput, inexpensive data set for the generation of epiallele libraries generating a calibration model, providing us the opportunity to identify the methylation status of a defined specific gene region when Tm-o values from post-PCR HRMA analysis are compared. The flexibility to generate fully customized sequences means that EpiHRMAssay procedure can address an array of biological questions on epigenetic regulation for diversity studies, as well as for large-scale functional genomics.
Conclusion
Epigenetics describes the heritable changes in gene function that occur independent of the DNA sequence. The molecular basis of gene regulation by epigenetic events is complex, but essentially involves modifications in the DNA itself or in DNA-associated proteins. Bisulphite sequencing methods usually rely on the availability of highly accurate genomic sequencing data and this could be a disadvantage for the use of these methods whether the sequences are unknown; the results reported in this work suggest that EpiHRMAssay can have a suitable application in the assessment of DNA methylation profiles of still non-sequenced organisms. The favourable ease and rapidity of use of EpiHRMAssay compared to other methods makes it a powerful tool for the detection of the methylation degree in unknown sequences of plant samples. Currently, for global methylation analysis, there are methods that measure the overall status of methylated cytosines in genome, while EpiHRMAssay is suitable for methylation analysis of the region of the desired template of certain gene to identify unknown methylation hotspots and/or methylated CpG islands, regardless of the methylation status. EpiHRMAssay will be useful in plant methylation studies but may also have potential utility in mammalian methylation diagnostics of established standards for specific target DNA, helping to understand the biological role of DNA methylation and its role in mammalian disease.
Supplementary data
Supplementary data is available at Biology Methods and Protocols online.
Supplementary Material
Acknowledgements
The study was partially funded by Ministry of Agricultural, Food and Forestry Policies (Italy), FRU.MED (D.M. 3690/7303/08) Project, and by ‘Fondazione Anna Maria Catalano – ONLUS’.
References
- 1. Vaillant I, Paszkowski I. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol 2007;10:528–33. [DOI] [PubMed] [Google Scholar]
- 2. Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 2007;39:61–69. [DOI] [PubMed] [Google Scholar]
- 3. Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 2011;12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genetics 2008;9:465–76. [DOI] [PubMed] [Google Scholar]
- 5. Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006;126:1189–201. [DOI] [PubMed] [Google Scholar]
- 6. Becker C, Hagmann J, Müller J, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011;480:245–49. [DOI] [PubMed] [Google Scholar]
- 7. Kankel MW, Ramsey DE, Stokes TL, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003;163:1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer P. DNA methylation systems and targets in plants. FEBS Lett 2011;585:2008–15. [DOI] [PubMed] [Google Scholar]
- 9. Stroud H, Do T, Du J, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 2014;21:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindroth AM, Cao X, Jackson JP, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001;292:2077–80. [DOI] [PubMed] [Google Scholar]
- 11. Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 2002;12:1138–44. [DOI] [PubMed] [Google Scholar]
- 12. Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev 1998;12:1714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem 2003;278:823–28. [DOI] [PubMed] [Google Scholar]
- 14. Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, et al. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 2008;67:671–81. [DOI] [PubMed] [Google Scholar]
- 15. Candiloro IL, Mikeska T, Hokland P, et al. Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenet Chrom 2008;1:7 doi:10.1186/1756-8935-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White HE, Hall VJ, Cross NCP. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem 2007;53:1960–62. [DOI] [PubMed] [Google Scholar]
- 17. Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 2007;35:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 2004;33:415–40. [DOI] [PubMed] [Google Scholar]
- 19. Blake RD, Delcourt SD. Thermal stability of DNA. Nucleic Acids Res 1998;26:3323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yakovchuk P, Protozonova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 2006;34:564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res 1994;22:2760–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blake RD, Bizzaro JW, Blake JD, et al. Statistical mechanical simulation of polymeric DNA melting with MELTSIM. Bioinformatics 1999;15:370–75. [DOI] [PubMed] [Google Scholar]
- 23. Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 2005;33:577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tøstesen E, Jerstad GI, Hovig E. Stitchprofiles.uio.no: analysis of partly melted DNA conformations using stitch profiles. Nucleic Acids Res 2005;33:573–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dwight Z, Palais R, Wittwer CT. uMELT: prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics 2011;27:1019–20. [DOI] [PubMed] [Google Scholar]
- 26. Tøstesen E, Liu F, Hovig E, et al. Speed-up of DNA melting algorithm with complete nearest neighbor properties. Nucleic Acids Res 2003;33:573–76. [DOI] [PubMed] [Google Scholar]
- 27. von Ahsen N, Wittwer CT, Schutz E. Oligonucleotide melting temperatures under PCR conditions: nearest-neighbor corrections for Mg(2+), deoxynucleotide triphosphate, and dimethylsulfoxide concentrations with comparison to alternative empirical formulas. Clin Chem 2001;47:1956–61. [PubMed] [Google Scholar]
- 28. Liew M, Pryor R, Palais R, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 2004;50:1156–64. [DOI] [PubMed] [Google Scholar]
- 29. Cassells AC, Curry RF. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: implications for micropropagators and genetic engineers. Plant Cell Tiss Org Cult 2001;64:145–57. [Google Scholar]
- 30. von Aderkas P, Bonga J. Influencing micropropagation and somatic embryogenesis in mature trees by manipulation of phase change, stress and culture environment. Tree Physiol 2000;20:921–28. [DOI] [PubMed] [Google Scholar]
- 31. Larkin PJ, Scowcroft WR. Somaclonal variation – a novel source of variability from cell cultures for plant improvement. Theor App Genet 1981;60:197–214. [DOI] [PubMed] [Google Scholar]
- 32. Miguel C, Marum L. A epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot 2011;62:3713–25. [DOI] [PubMed] [Google Scholar]
- 33. George EF. Plant Propagation by Tissue Culture; Part 1: The Technology. Edington: Exegetics Ltd, 1996. [Google Scholar]
- 34. Valledor L, Hasbún R, Meijón M, et al. Involvement of DNA methylation in tree development and micropropagation. Plant Cell Tiss Org Cult 2007;91:75–86. [Google Scholar]
- 35. Gentile A, Jaquez Gutierrez M, Martinez J, et al. Effect of metaTopolin on micropropagation and adventitious shoot regeneration in Prunus rootstock. Plant Cell Tiss Org Cult 2014;118:373–81. [Google Scholar]
- 36. Quoirin M, Lepoivre P. Improved media for ‘in vitro' culture of Prunus sp. Acta Hortic 1977;78:437–42. [Google Scholar]
- 37. Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 1962;15:473–97. [Google Scholar]
- 38. Muleo R, Colao MC, Miano D, et al. Mutation scanning and genotyping by high resolution DNA melting analysis in olive germplasm. Genome 2009;52:252–60. [DOI] [PubMed] [Google Scholar]
- 39. Snell C, Krypuy M, Wong E, et al. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res 2008;10:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verde I, Abbott AG, Scalabrin S. The high quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 2013;45:487–96. [DOI] [PubMed] [Google Scholar]
- 41. Gao F, Zhang C-T. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res 2006;34:W686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huguet JM, Bizarro CV, Forns N, et al. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc Natl Acad Sci USA 2010;107:15431–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weber G. Optimization method for obtaining nearest-neighbour DNA entropies and enthalpies directly from melting temperatures. Bioinformatics 2015;3:871–77. [DOI] [PubMed] [Google Scholar]
- 44. Hetzl J, Foerster AM, Raidl G, et al. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 2007;51:526–36. [DOI] [PubMed] [Google Scholar]
- 45. Candiloro ILM, Mikeska T, Dobrovic A. Assessing combined methylation-sensitive high resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics 2011;6:500–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lim AM, Candiloro ILM, Wong N, et al. Quantitative methodology is critical for assessing DNA methylation and impacts on correlation with patient outcome. Clin Epigenetics 2014;6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kristensen LS, Mileska T, Krypuy M., et al. Sensitive melting analysis after sensitive melting analysis after real time- methylation specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res 2008;36:e42 doi:10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mikeska T, Candiloro ILM, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics 2010;2:561–73. [DOI] [PubMed] [Google Scholar]
- 49. Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin Chem 2001;1:1183–89. [PubMed] [Google Scholar]
- 50. Guldberg P, Worm J, Grønbæk K. Profiling DNA methylation by melting analysis. Methods 2002;27:121–27. [DOI] [PubMed] [Google Scholar]
- 51. Dahl C, Grønskov K, Larsen LA, et al. A homogeneous assay for analysis of FMR1 promoter methylation in patients with fragile X syndrome. Clin Chem 2007;53:790–93. [DOI] [PubMed] [Google Scholar]
- 52. Miyao A, Nakagome M, Ohnuma T, et al. Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol 2012;53:256–64. [DOI] [PubMed] [Google Scholar]
- 53. Us-Camas R, Rivera-Solís G, Duarte-Aké F, et al. In vitro culture: an epigenetic challenge for plants. Plant Cell Tiss Org Cult 2014;118:187–201. [Google Scholar]
- 54. Li X, Wang X, He K, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 2008;20:259–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanurdzic M, Vaughn MW, Jiang H, et al. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 2008;6:2880–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiaofeng C, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 2002;12:1138–44. [DOI] [PubMed] [Google Scholar]
- 57. Kurdyukov S, Bullock M. DNA methylation analysis: choosing the right method. Biology 2016;5:3 doi:10.3390/niology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.