Abstract
Poly(R)-3-hydroxybutyric acid (PHB) is a biodegradable natural polymer produced by microorganisms and plants under nitrogen deprivation and physiological stress. Metabolic engineering and synthetic biology approaches are underway to develop strains that can produce PHB and its co-polymers. One of the major limitations to the scaling and success of strain development for biosynthesis of PHB is the absence of fast, accurate, quantitative and scalable methods to estimate PHB in polymer producing cells. In this study, a Nile red-based spectrofluorometric method is developed for absolute quantitation of PHB in recombinant Escherichia coli. The method is a modification of an existing Nile red-based method currently only used for relative quantitation. The two added steps of sonication and ethanol extraction increase the dynamic range of the assay and limit of detection/quantitation. Sonication of PHB standards provides uniform distribution of surface area to volume ratios. This ensures reproducibility and accuracy (lower %relative error) of quantitative staining of granules by Nile red even in a higher dynamic concentration range of 125–1000 µg/ml. Ethanolic extraction of the PHB bound Nile red allows higher recovery and accurate absolute quantitation. To reproduce high recovery and ensure accuracy and precision of the analytical method directly using cells, a protein digestion step was added. This accounted for fluorescence from over-expressed protein and resulted in screening of nonproducers of PHB amongst samples. Thus, the method developed is rapid, accurate, and reproducible, requires low sample volumes and processing compared to other conventional methods. This method is scalable to other PHA’s and diverse plastics.
Keywords: absolute quantitation, spectrofluorometry, nile red, sonication, ethanol extract, recombinant Escherichia coli
Introduction
Metabolic engineering and synthetic biology approaches for polyhydroxyalkanoates (PHA) are critical to the success of biopolymer and bioplastics technology. Fast and accurate absolute quantitation of these polymers is a critical step in strain development and real-time monitoring of bioprocess. Poly(R)-3-hydroxybutyric acid (PHB), has been quantitated previously using methods such as gravimetry [1, 2] and turbidimetry [2, 3]. These methods have major limitations for high throughput quantitation of PHB, including the need for large amounts of sample, organic solvent and laborious processing. Although gas chromatography [4] is rapid, methanolysis is generally water sensitive and results in incomplete recovery of PHB [2, 5]. Despite the ability to use small sample volumes (100 µl) and low processing time, Ultra violet-Visible (UV-Vis) Spectrophotometry spectrophotometry [6, 7] is limited to PHB estimation. Further the method is hazardous due to concentrated H2SO4 treatment and heating to form crotonic acid. A high-throughput High-performance liquid chromatography (HPLC) [8] method developed using alkaline hydrolysis of dried cells has been reported mainly for the compositional analysis but not for absolute quantitation. Major limitation of all reported techniques is sensitivity of measurements in the physiological micromolar range for cellular quantitation of PHA and noise from cellular components.
Nile red, a lipophilic dye that binds to intracellular lipids and PHA granules [9, 10] has been used to quantitate PHB (relative) in bacterial cells with reasonable accuracy using two-dimensional fluorescence spectroscopy and flow cytometry [11, 12]. A quantitative method has been previously reported [13–15] using PHA producing cells as a standard to establish a calibration curve. The main hurdles in accurate quantitation are physical properties of standard PHA granules i.e. surface area and volume. Another complication during quantitation in cells is the formation of inclusion bodies and background noise associated with the solvatochromic effect of Nile red fluorescence [11, 16].
Our study attempts to develop an improved method (Fig. 1) overcome all the limitations discussed above by including two additional steps in the Nile red-based fluorescence method. The steps of sonication and ethanol extraction improve the recovery and thus accuracy and sensitivity of PHB quantitation. An additional protein digestion step improved the accuracy of quantitation significantly in PHB producing recombinant Escherichia coli cells.
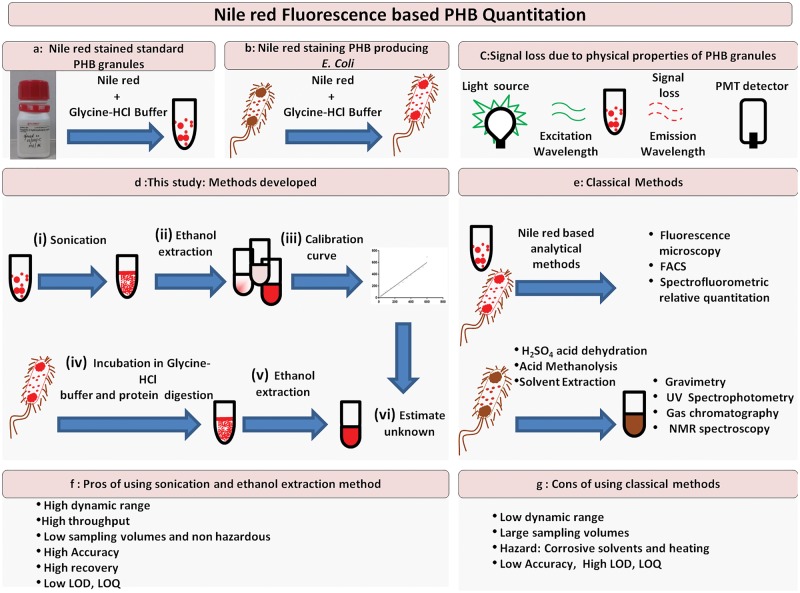
Figure 1:
Schematic workflow. Pictorial representation of workflows/methods for improved accuracy of quantitation of PHB. The principle of the assay is depicted in Panels a–c. Standard PHB suspensions in aqueous glycine-HCl buffer bind quantitatively to Nile red and have specific fluorescence spectral characteristics. Panel b shows how cells producing PHA can be, grown in the presence of Nile red that can bind to intracellular carbonosome, an intracellular PHA producing inclusion body. Panel c shows loss in emission signal of fluorescence dependent on physical properties of granules. Panel d represents the methodology developed in this study while panel e represents the classical method for quantitation using UV spectrophotometry. Panels f–g discuss the Pros and Cons of the methods.
Materials and methods
Calibration curves for PHB standard using spectrofluorometry and UV spectrophotometry
A spectrofluorometric method for PHB quantitation was developed (Fig. 1; Supplementary Method S1) using Nile red fluorophore on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific; Cat no. Q32866) with LED light source. The blue LED light source in Qubit 2.0 was used in a raw mode for excitation; it has the maximum intensity at 470 nm with excitation filter ranging from 430 to 495 nm and emission filter ranging from 665 to 720 nm. Suspensions of 1000 µg/ml standard PHB in triplicate were prepared by suspending 10 mg of PHB (Sigma: cat no. 363502) in 10 ml of 0.1 M glycine-HCl buffer pH 3.0 in 30 ml Borosil flat bottom culture tube (Cat no. 9910010). Suspension was kept in ice bath for 20 min and moved to a sonication chamber with an ice bath. The suspension was probe sonicated at 80% amplitude for 10 min at a 10/20 cycle on Sonic Vibracell VCX130 (130 watt, 20 kHz) equipped with 6-mm stepped tip sonication probe. Suspensions with the same volume and concentration were also prepared and used without sonication. All experiments were performed in triplicate. Nonsonicated and sonicated standard suspensions were added to 1 µg/m; Nile red (Sigma: cat no. N3013) [in dimethylsulfoxide (DMSO) 0.5 mg/ml] and incubated for 2 h (Supplementary Fig. S1e) at room temperature (25°C). To establish that Qubit 2.0 can be used for Nile red-based quantitation, we measured the excitation and emission fluorescence spectrum of 1 ml (1000 µg/ml) of Nile red stained standard PHB suspension (Supplementary Fig. S1a) and its ethanolic extracts (Supplementary Fig. S1b) on Photon Technology Fluorescence QM40 Spectrophotometer in Quartz high precision cell (Hellma analytics Art. No.101-10-40). Since blue LED in Qubit 2.0 has maximum excitaton at 470 nm, emission fluorescence spectrum was measured with excitation at 470 nm. For calibration curves using standards, Nile red stained sonicated and nonsonicated PHB suspensions were diluted with 0.1 M glycine-HCl buffer to 1 ml using glass pipettes. 200 µl suspensions were aliquoted at various concentrations (in the range of 20 to 1000 µg/ml) and washed twice by vortexing in 200 µl of 0.1 M glycine-HCl buffer followed by centrifugation at 4000 rpm for 5 min in Eppendorf® 5430 R centrifuge. The pellet was resuspended in 200 µl glycine-HCl buffer and fluorescence measured. For ethanol extraction, 200 µl Nile red stained sonicated and nonsonicated granules were washed twice by vortexing in 200 µl 0.1 M glycine-HCl buffer followed by centrifugation at 12 000 rpm for 5 min. The Nile red bound to the PHB standard was released by resuspending stained granules in 200 µl ethanol. The fluorescence was measured after excitation at the determined wavelengths.
For UV spectrophotometric estimation, the suspension of PHB granules (20–100 µg/ml) in triplicate were centrifuged and resuspended in 100 µl ethanol and transferred into glass tubes. This procedure was repeated four times to ensure complete transfer of all granules. Ethanol was evaporated by heating at 60°C and drying under vacuum. Concentrated H2SO4 (1 ml) was added to the dried pellet, and capped tubes were heated for 10 min in a boiling water bath. The solution was cooled to room temperature (25°C) and transferred to a high precision quartz cuvette to measure absorbance at 230 nm on a Spectrophotometer (BioPhotometer Plus, Eppendorf).
Cloning of PHB producing operon
Gibson assembly [17, 18], a recombination based cloning method was used for constructing the plasmid ppct_phaC_Wt (Fig. 3a) used to make PHB in this study. The genes Propionyl-CoA Transferase pctap and PHA polymerase phaCcv from two different organisms were co-expressed in E. coli.
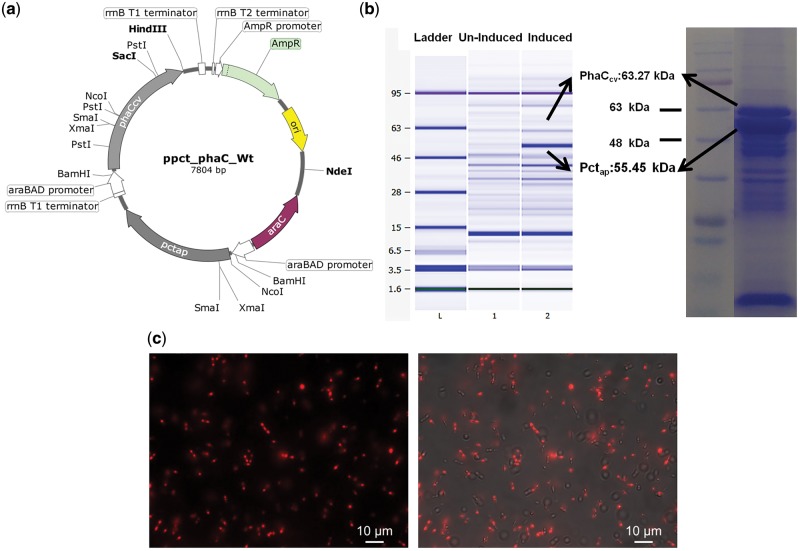
Figure 3:
Plasmid constructs and biosynthesis of PHB using recombinant E. coli (a) ppct_phaC_Wt Plasmid for Co-expression of PHB producing pctap and phaCcv genes with arabinose inducible araBAD promoter, (b) Over expression of proteins PhaCcv (mol. Wt.63.27 kDa) and Pctap (mol Wt. 55.45 kDa) in the PHB producing recombinant E.coli DH10β transformed with ppct_phaC_Wt recorded on an Agilent 2100 bioanalyzer and 10% SDS-PAGE (c) Fluorescent microscopy of Nile red stained intracellular carbonosomes in recombinant E.coli grown on glycerol supplemented media in the Rhodamine 20 channel (Right) and Rhodamine 20 channel superimposed with TL DIC (Left).
Propionyl-CoA Transferase pctap from Acetobacter pasteurianus 386B (Sequence ID: HF677570.1) was chemically synthesized and cloned into pBADHis_C_A238 plasmid to give pBAD_pct (Supplementary Fig. S1a). The Ter-Pro sequence containing rrnB terminator-araBAD promoter sequence was chemically synthesized separately as a linear fragment. Gene synthesis and cloning services for pBAD_pct was provided by GeneArt® Gene Synthesis services. DNA was received in lyophilized form and handled as per manufacturer’s instructions. Genomic DNA was isolated from Chromobacterium violaceum ATCC 12472 (Sequence ID: AE016825.1) from 5 ml overnight culture using Qiagen DNeasy Blood & Tissue Kits (Cat. no. 69506). phaCcv (Supplementary Fig. S1c) and Ter-Pro (Supplementary Fig. S1c) with overhang for Gibson assembly were PCR amplified from C. violaceum genomic DNA and Ter-Pro sequence respectively using Q5 High-Fidelity DNA polymerase (Cat. no. NEB M0491). All PCR amplifications were performed on Applied Biosystems Veriti® Thermo Cycler using primers listed in Supplementary Table S4.
Gibson assembly was carried in 10 µl reaction mixture containing 40 ng pBAD_Pct double digested with KpnI/kHindIII, 2-fold excess of phaCcv and Ter-Pro PCR products. The assembly mix was incubated at 50°C for 60 min and stored at −20°C for subsequent transformation. Chemically competent E. coli DH10β were transformed with assembly mix and cells were plated on Luria Bertani Agar, Miller (HiMedia Cat no. M1151) with ampicillin (100 µg/ml). Plate was incubated overnight for growth at 37°C. Isolated ampicillin resistant colonies were picked and the transformed plasmid ppct_phaC_Wt was confirmed by restriction digestions with restriction enzyme ScaI followed by sequencing. Cloning procedure was designed and simulated on SnapGene software (from GSL Biotech; available at snapgene.com, RRID: SCR_015053).
Expression of PHB producing operon
Single colony of a E. coli DH10β transformed with ppct_phaC_Wt was inoculated in 10 ml Luria Bertani Broth (LB) media and incubated overnight at 37°C with shaking at 200 rpm. Overnight culture was sub-cultured in 10 ml LB media supplemented with 2 mg/ml glycerol. Culture was induced with 0.025% L-arabinose to express PHB producing genes after 3 h of growth. A 1 ml culture was harvested by centrifugation at 10000 rpm for 5 min, 9 h after induction and resuspended in 1 ml of PBS while adjusting cell density (OD600) to 1.0. Cells were washed by centrifugation at 10000 rpm for 5 min with 20 mM Tris-HCl (pH: 8.0) and resuspended in 100 µl lysis buffer (20 mM Tris-HCl and 2 mM Ethylene Diamine Tetra Acetic Acid (EDTA) containing 1% Triton X-100). Lysozyme (1 mg/ml) was added to the cells in lysis buffer and incubated with shaking (500 rpm) at 37°C for 1 h. The soluble fraction of cell lysate was analyzed on an Agilent Bioanalyzer protein 80 chip. Soluble fraction did not show the over expression of phaCcv, since it is an insoluble protein in its active form. Therefore complete cell lysate was prepared by heating cell pellet in 100 µl 1X SDS-PAGE sample buffer (80 mM Tris-Cl pH: 6.8, 2% SDS, 10% glycerol, 5% β-Mercaptoethanol and 0.002% Bromophenol blue) for 5 min at 95°C. Cell lysate was centrifuged at 8000 rpm for 5 min and 10 µl loaded on 10% Sodium Dodecyl Sulfate -PolyAcrylamide Gel Electrophoresis (SDS-PAGE) gel in Tris-glycine running buffer pH: 8.3 (25 mM Tris-Cl, 250 mM Glycine, 0.1% SDS).
Quantitation of PHB using spectrofluorometry from recombinant E. coli
Single colony of a E. coli DH10β transformed with ppct_phaC_Wt was inoculated in 10 ml LB media and incubated overnight at 37°C with shaking at 200 rpm. Overnight culture was sub-cultured in 10 ml LB media supplemented with no extra carbon source, glycerol, and glucose. 1 µg/ml Nile red was added to LB media while sub-culturing for staining PHB producing cells. Culture was induced with 0.025% L-arabinose to express PHB producing genes after 3 h of growth. Un-induced culture was used as a control, grown in similar conditions with Nile red. 200 µl culture was harvested at regular intervals for 24 h by centrifugation at 10000 rpm for 5 min. Cells were washed with PBS pH 7.4 by vortexing and followed by centrifugation at 10 000 rpm for 5 min Cells can be stored in PBS at 4°C if assay is not carried out immediately after harvesting. Cells were washed twice with 200 µl of 0.1 M glycine-HCl buffer by vortexing followed by centrifugation at 4000 rpm. Pellet was resuspended in 200 µl of 0.1 M Glycine-HCl buffer at pH 3.0 and incubated for 2 h. Fluorescence was measured in a Qubit 2.0 fluorometer [10, 11, 14]. For ethanol extract, the cell suspension in glycine-HCl buffer was centrifuged at 10000 rpm for 5 min and supernatant was discarded. 200 µl ethanol was added to the pellet to extract PHB bound Nile red. The fluorescence of the ethanolic extract was measured in a Qubit 2.0 fluorometer (Supplementary Method S2).
Microscopy standard
Two aliquots of 1000 µg/ml standard PHB suspension were prepared with 10 mg of PHB in 10 ml of 0.1 M glycine-HCl buffer pH 3.0 in 30 ml Borosil flat bottom culture tube. Suspensions were kept in an ice bath for 20 min and moved to a sonicator chamber also with an ice bath. Suspension was probe sonicated at 80% amplitude for 10 min at a 10/20 cycle on Sonic Vibracell VCX130 (130 watt, 20 kHz) equipped with 6-mm stepped tip sonication probe. 1 µg/ml Nile red in DMSO (0.5 mg/ml) was added to both nonsonicated and sonicated suspension and incubated for 2 h. 20 µl was spread over microscopy glass slide, covered with a cover slip and examined on an Axio Observer.Z1 inverted fluorescence microscope under 63x/1.4 oil immersion with DIC and 20 Rhodamine channel. Microscopy images were analyzed for granule surface area and volume using ZEN lite (Blue version) software from ZEISS.
Cell microscopy
Overnight culture was sub-cultured in 10 ml LB media supplemented with 2 mg/ml glycerol, with and without Nile red. Culture was induced with 0.025% L-arabinose to express PHB producing genes after 3 h of growth. To visualize intracellular carbonosome, 100 µl culture without Nile red was harvested after 12 h (9 h after induction), and cells were washed by centrifugation at 10 000 rpm with 100 µl PBS buffer and resuspended in 1 ml distilled water. On a microscopy glass slide, 20 µl cells suspended in water were smeared and heat fixed. Smear was flooded with 1% aqueous solution of Nile red and incubated for 10 min at 55°C. Excess staining solution was removed and smear was washed gently with water. Smear was flooded with 8% (v/v) aqueous acetic acid and incubated for 1 min at room temperature. The excess was removed and smear was washed gently with water. Cover slip was placed and the stained cells were examined with Axio Observer Z1 inverted fluorescence microscope under 63x/1.4 oil immersion with DIC and 20 Rhodamine channel.
A 200 µl of growing culture sub-cultured with Nile red was harvested, washed twice with 200 µl of 0.1 M glycine-HCl buffer by vortexing followed by centrifugation at 4000 rpm, resuspended in 200 µl 0.1 M glycine-HCl buffer pH 3.0, and incubated for 2 h at room temperature. Over a microscopy glass slide, 20 µl suspensions was spread, covered with cover slip, and examined with Axio Observer Z1 inverted fluorescence microscope under 63x/1.4 oil immersion with DIC and 20 Rhodamine channel. Microscopy images were analyzed for granule surface area and volume using ZEN lite (Blue version) software from ZEISS.
Quantitation of PHB using UV spectrophotometry from recombinant E. coli
For UV spectrophotometric estimation, 100 µl of culture was harvested at regular time intervals. Cells were washed with PBS pH 7.4 followed by centrifugation at 10 000 rpm for 5 min. This was followed by washing with glycine-HCl buffer. Pellet was resuspended in glycine-HCl buffer and incubated for 2 h at room temperature (25°C). Cell suspension was centrifuged at 12 000 rpm at room temperature for 10 min and resuspended in 1 ml chloroform. Separated chloroform extract was transferred to clean glass tubes and chloroform was allowed to evaporate at 50°C and complete drying was achieved under vacuum. To the dried pellet, 1 ml concentrated H2SO4 was added and the capped tubes were heated for 10 min in a boiling water bath. The solution was cooled and transferred to Quartz high precision cell and absorbance was measured at 230 nm on a Spectrophotometer (BioPhotometer Plus, Eppendorf).
Estimation of fluorescence contributions from expressed proteins in growing cells
Nile red (1 µg/ml) in DMSO (0.5 mg/ml) was added to the LB media while sub culturing cells. Culture was induced with 0.025% L-arabinose to express PHB producing genes at 3 h. Cells were harvested by centrifuging at 10 000 rpm at 12 and 24 h after inoculation. Cell pellets were resuspended in 1 ml of PBS and the cell density was adjusted (OD600 = 1.0). Cells were washed with 20 mM Tris-HCl (pH: 8.0) and resuspended in 100 µl lysis buffer (20 mM Tris-HCl and 2 mM EDTA containing 1% Triton X-100). Lysozyme (1 mg/ml) and trypsin (1 mg/ml) was added followed by incubation with shaking (500 rpm) at 37°C for 1 h. The soluble fraction of cell lysate was analyzed on the Agilent Bioanalyzer protein 80 chip. Insoluble fraction was mixed with 200 µl 0.1 M glycine-HCl buffer and incubated for 2 h. Treated insoluble fraction was centrifuged and the supernatant was discarded. To the treated insoluble fraction, 1 ml of ethanol was added and fluorescence was measured from an ethanolic extract. Replicates of the same culture without protein digestion were quantitated for PHB using the established spectro-fluorometric method.
Curve fitting, visualization and analysis of the cell density over time were done using GraphPad Prism Version 6.01 (GraphPad Software, San Diego, CA, USA, www.graphpad.com). Statistical analysis was done and performance characteristics calculated to validate and compare all calibration methods for precision, accuracy, and sensitivity [19].
NMR spectroscopy
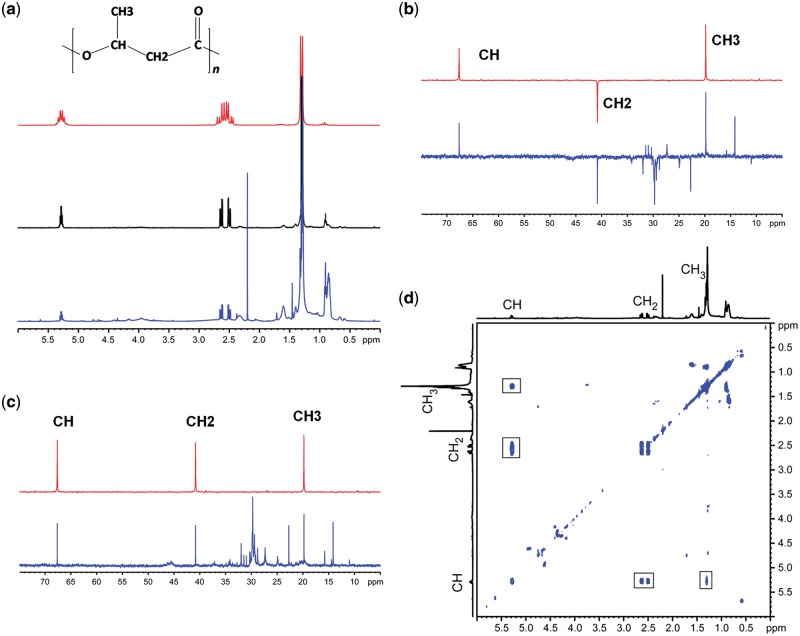
Transformed E. coli DH10β cells were cultured in 800 ml LB media supplemented with glycerol and glucose as substrate. The cells were harvested after 24 h for extraction of PHB using combination of Trypsin-sodium hypochlorite method and solvent extraction. The solvent mixture was evaporated in a Rotavapor® R-100 and the extracted polymer was dissolved in CDCl3. Nuclear Magnetic Resonance (NMR) Spectroscopy experiments were carried out at 25°C on Bruker Avance 200 and Bruker Avance 500 spectrometers, using BBO probes with z-gradients. 1 H, 13 C, and 13 C DEPT spectra were recorded with 32, 10 000 and 3200 scans, respectively. Flip angle of 30° and interscan delay of 3 s was employed for 1D 1 H and 13 C spectra. The diffusion filtered NMR spectrum was recorded using pulsed field gradient stimulated echo experiments, setting diffusion time to 80 ms, encode/decode gradient pulses to 2300 μs and a gradient strength of 48 G/cm. The high gradient strength is sufficient to filter out signals from all low molecular weight species in the solution, however the signals from PHB is observed in the spectrum due to the slow diffusion of the polymer. The diffusion coefficient determined for PHB is ∼ 2.35 × 10−11 m2/s which is typical for polymeric species. The 2D correlation spectroscopy (COSY) experiment was recorded with 2 K × 128 data points and 16 scans. The connectivity pattern between the different types of protons clearly shows that the polymeric species is PHB.
Results
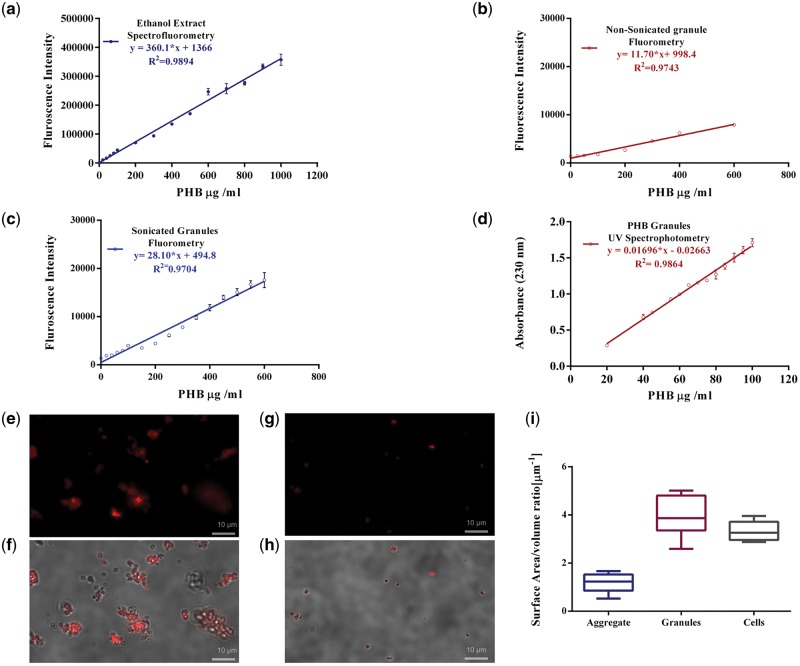
The analytical calibration curves (n = 14) of fluorescence intensity measurements of PHB granules stained with Nile red, sonicated and extracted into ethanol were linear in the broad range of 125–1000 µg/ml (Fig. 2a). Fluorescence intensity measurements were made on a Qubit 2.0 with appropriate excitation (470 nm, slits fixed at 20 nm) and emission (600–640 nm) wavelengths (Supplementary Fig. S1a and b). The standard error of mean (SEM) values are indicated in Table 1. The linearity was assessed by calculating the regression equation (y = ax ± b) and the correlation coefficient (r2) by the least squares method. The linear regression equation was described by equation (1) and r2= 0.9894
| (1) |
Figure 2:
Development of analytical method for PHB. Calibration curves for PHB standard and physical parameters of standard PHB granules. (a) Spectrofluorometric calibration curve of ethanol extract from sonicated Nile red stained PHB standard granules in glycine-HCl buffer (excitation wavelength: 470 nm; emission wavelength: 640 nm) in the concentration range of 20 to 1000 μg/ml, (b) Spectrofluorometric calibration curve of ethanol extract from Nonsonicated Nile red stained PHB standard granules in glycine-HCl buffer (excitation wavelength: 470 nm; emission wavelength: 640 nm) in the concentration range of 25 to 600 μg/ml, (c) Spectrofluorometric Calibration curve for sonicated PHB granules stained with Nile red in Glycine HCl buffer using (excitation wavelength: 470 nm; emission wavelength: 600 nm) in the concentration range of 20 to 600 μg/ml, (d) calibration curve for PHB standards using crotonic acid based UV spectrophotometry (absorbance wavelenght: 230 nm) in concentration range of 20 to 100 μg/ml. Image panels depict Fluorescence microscopy and DIC images of Nile red stained granules incubated in glycine-HCl buffer for (e–f) nonsonicated PHB granules stained (g–h) sonicated PHB granules, (i) Differential surface area to volume ratios for nonsonicated aggregates, sonicated PHB granules and cells.
Table 1:
Performance characteristic of Nile red-based PHB estimation methods using fluorescence spectroscopy
| Accuracy |
Precision |
|||||||
|---|---|---|---|---|---|---|---|---|
| Method Detail | %REa | ANOVAb | % RSDc | SEMd (µg/µl) | Sensitivity (Slope) | LODe (µg/ml) | LOQf (µg/ml) | Dynamic |
| Range | ||||||||
| F(df) | ||||||||
| (µg/ml) | ||||||||
| Sonication | 9.59 | 1657.4, (1, 13) | 2.27 | 7.95 | 360.12 | 41.12 | 124.6 | 124.6–1000 |
| ethanol extraction | ||||||||
| spectrofluorometry | P = 3E-15 | |||||||
| Ethanol extraction | 14.53 | 541.6, (1, 13) | 4.99 | 8.58 | 28.54 | 46.80 | 141.83 | 141.83–600 |
| Spectrofluorometry | ||||||||
| P = 5.5E-12 | ||||||||
| Sonication spectroflurometry | 19.85 | 229.3, (1, 6) | 2.10 | 2.00 | 11.70 | 62.73 | 190.09 | 190.09–600 |
| P = 5.2E-06 | ||||||||
| UV Spectrophotometery | 24.96 | 1463.6, (1, 11) | 2.11 | 1.01 | 0.0169 | 6.15 | 18.64 | 18.64–100 |
| P = 4.7E-13 | ||||||||
Percentage relative error;
Analysis of Variance;
Relative standard deviation;
Standard error of mean;
Limit of detection;
Limit of quantitation.
Where ‘y’ is fluorescence emission at 640 nm and ‘x’ is the concentration of the standard in µg. Calibration curves were established for the method using sonication alone (Fig. 2c) and nonsonicated granule suspensions with ethanol extraction (Fig. 2b). The methods excluding either of the added steps, sonication or ethanol extraction show higher % relative error (Table 1) and estimate higher concentrations than actual PHB standard (Supplementary Fig. S3f). Sonication prevents aggregates (Fig. 2e–h) and controls size distribution and surface area to volume ratios (Supplementary Tables S1–S3). This results in uniform staining of granules with optimally sized granules with Nile red (Fig. 2i, Supplementary Fig. S2f–h).
The result of the method with both the added steps of sonication and extraction was comparable to the traditional quantitation method, based on degradation of PHB to crotonic acid using concentrated sulfuric acid followed by absorbance measurements at 230 nm. However, the maximum measurable value was an order of magnitude higher (dynamic range of UV-Vis spectroscopy: 20–100 µg/ml). The spectrophotometric calibration curve (Fig. 2d) was described by the regression equation
| (2) |
where ‘y’ is the absorbance at 230 nm and ‘x’ is the concentration of the standard in µg. The curve was highly linear with the regression coefficient r2 being 0.9666. The performance characteristics based on precision calculated as RSD (Table 1) was comparable with Nile red-based method; however, accuracy and sensitivity were compromised. The validation of the analytical methods with permutations of the additional steps was also performed. Performance characteristics of all the developed methods are tabulated (Table 1). The inclusion of the sonication and ethanol extraction steps had a statistically significant effect on precision, accuracy and sensitivity of measurement of true values. The standard measurement errors, RSD and standard deviations were lower. The method also had the lowest limits of detection and quantitation (Limit of detection (LOD) = 41.12 µg/ml; Limit of quantitation (LOQ) = 124.6 µg/ml). The dynamic range was high (124.6–1000 µg/ml). The accuracy of the finalized method was the best with lowest relative error (9.59%). This underscores the impact based on systemic errors vis a vis random errors. Further, based on variance analysis using Analysis of Variance (ANOVA), the effect of sonication had the highest statistical significance as reflected in the F-statistic (F1, 13 = 1657.83, P = 4.26252e−15). The ANOVA output of the linear regression model described (Table 1, Supplementary Table S5a–d) also confirmed linearity. Variance analysis resulted in correlation coefficient with a P-value <0.05 (Supplementary Table S4). r2 values >0.999 indicate a good correlation of linearity through all the concentrations used and a homoscedastic distribution of replicates in the calibration curve assembly.
The Gibson assembly method was successful in stitching the two genes pctap and PhaCcv that constitute the ppct_phaC_Wt (Fig. 3a). Propionyl-CoA Transferase (pctap) transfers a CoA group to the monomer and PHA polymerase (phaCcv) polymerizes it to PHB. Transcription and translation of these genes into functional proteins PhaCcv (MW 63.27 kDa) and Pctap (Mol Wt. 55.45 kDa) (Fig. 3b) were confirmed by overexpression studies in arabinose-induced cultures. The presence of intracellular PHB was confirmed with Nile red fluorescent microscopy (Fig. 3c).
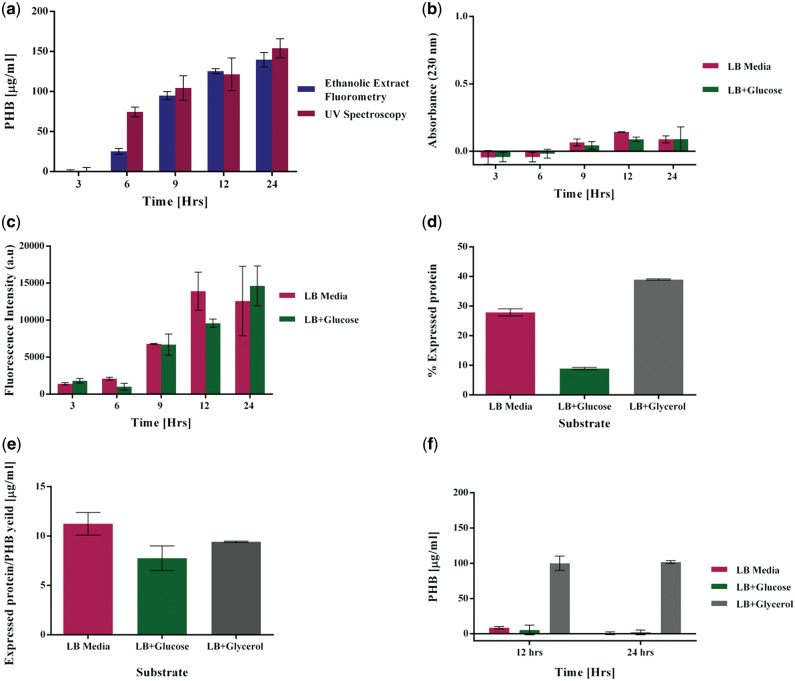
PHB quantitation was comparable (very low standard deviation) using both the methods over a time frame of 24 h from a growing culture of recombinant E. coli cells. Using glycerol as primary carbon source, the cells were seen to produce a maximum of 139.73 ± 9.02 µg/ml using our method as opposed to 154.02 ± 6.87 µg/ml with the crotonic acid method (Fig. 5a). All fluorescence measurements of induced cultures were normalized to that of uninduced control culture (Supplementary Fig. S3a and b). When LB media with and without glucose was used, no PHA signals were identified in the NMR data (Supplementary Fig. S4) indicating no PHB formation. An absorbance corresponding to 38 µg/ml of PHB over estimated PHB in the crotonic acid-based method (Fig. 5b). Fluorescence intensities of 14633 a.u and 12579 a.u were measured, respectively, estimating falsely an amount of PHB equivalent to 36.84 ± 4.32 and 31.14 ± 13.01 µg/ml after 24 h of growth (Fig. 5c). NMR experiments confirmed that recombinant E. coli growing on glycerol produced PHB (Fig. 4 a–d). Since the NMR results did not detect any PHA in the spectrum of E. coli grown on glucose, it was critical to identify the source of contribution of fluorescent intensity in these samples. Since PHB is known to form carbonosomes around themselves with PHA producing proteins, we hypothesized that the expressed proteins were contributing to the fluorescence. The relative over expressed protein contributed to 8.9, 38.9 and 27.8% of the total cell protein when cells were grown on glucose, glycerol and LB media, respectively (Fig. 5d). Quantitation of expressed protein suggests an eleven fold (Fig. 5e) ratio to PHB produced when grown on glycerol. Even in the absence of PHB biosynthesis, when cells are grown on LB with and without glucose, 7.75 and 9.41 µg of protein are produced per mL culture. It was evident on lysozyme digestion that in cells grown on glycerol, the over expressed protein contributed to 24% of the total fluorescence. All the fluorescence intensity (100%) emitted by the cells grown in LB with and without glucose supplementation was due to expressed protein (Fig. 5f) accounting for lack of PHA signals in the NMR spectrum. This indicates potential carbonosome formation and entrapment of some amounts of Nile red that contribute to incorrect or over estimation of PHB. The performance characteristic of the cell samples using the Nile red-based method with an additional step of protein digestion was higher in accuracy and precision as compared to the classical UV spectrophotometric method (Table 2).
Figure 5:
PHB quantitation from recombinant E. coli. (a) Comparison of PHB quantitation from recombinant E.coli grown on 2 mg/ml glycerol for 24 h using the classical UV spectrophotometric method and the method developed in our study. (b) Over estimation of PHB grown on LB media and LB Media supplemented with glucose using the UV spectrophotometric method. (c) Contributing fluorescence intensities of PHB producing cells grown on LB media without any supplement and LB media supplemented with glucose over estimating PHB. (d) Percentage of over expressed protein to the total protein after 24 h of growth. (e) Ratio of over expressed protein to the total PHB produced after 24 h of growth. (f) PHB quantitation in cell samples after protein digestion.
Figure 4:
NMR Spectroscopy for structural confirmation (a) 1H NMR spectrum of extract from recombinant E. coli grown on glycerol (blue, 500 MHz), diffusion filtered spectrum of the extract (Black, 500 MHz) and spectrum of PHB standard (Red, 200 MHz) in CDCl3 at 25C. The diffusion filtered spectrum selects signals from macromolecules and eliminates signals from low molecular weight species clearly shows the presence of PHB (b) DEPT spectra (c) 13C spectra of cell extract and PHB standard recorded on a 500 MHz spectrometer. Spectra of cell extract and PHB standard are show in blue and red, respectively. The different types of carbons in PHB are indicated (d) COSY spectrum peaks in 2D marked with square.
Table 2:
Performance characteristics of Nile red-based PHB quantitation in PHB producing E.coli cells compared to UV spectrophotometery
| Accuracy |
Precision |
|||
|---|---|---|---|---|
| Method Detail | %REa | ANOVAb | % RSDc | SEMd |
| F(df) | µg/µl | |||
| Ethanol extraction spectrofluorometry | 39.43 | 21.02, (1, 4) | 12.36 | 4.52 |
| P = 0.019 | ||||
| Protein digestion, | 17.22 | 0.12, (1, 4) | 5.531 | 3.47 |
| Ethanol extraction | ||||
| Spectrofluorometry | ||||
| P = 0.73 | ||||
Percentage relative error;
Analysis of Variance;
Relative standard deviation;
Standard error of mean.
Discussion
Nile red-based spectrofluorometry is a widely used technique for relative quantitation of PHB. Limitations mainly arise out of the suspension assay either due to surface to volume ratio in standard PHB granules or background fluorescence of hydrophobic moieties in cells. In this study (Fig. 1c), using Nile red-based fluorescence analytics, an analytical method for absolute quantitation of PHB using standards has been developed that includes two additional steps to improve accuracy. The method has a higher dynamic range, lower relative error, LOD, LOQ as compared to the classical method. The crotonic acid-based assay is also unsafe due to use of corrosive agents and heating at high temperatures. The inaccuracies in suspension assay, mainly attributable to the aggregation of PHB granules and microparticle size and distribution are corrected for through controlled sonication protocols. Fluorescence measurements after sonication cycles break up the clumped granules and increase the exposed area of binding Nile red (Fig. 2e–i). The recovery is improved through the extraction into ethanol.
The bigger challenge of transcending from standards to PHB measurements in cells was overcome by correcting for noise due to fluorescing protein moieties in the cell with appropriate controls before releasing the PHB-bound Nile red through ethanol extraction (Supplementary Fig. S3f). To correct for the lipid background fluorescence from PHB producing cells, induced cells (Supplementary Fig. S3a) was normalized to uninduced cells (Supplementary Fig. S3b). The noise related to protein overexpression (Table 2) that overestimated PHB was lowered by a simple lysozyme digestion step. Ethanolic extracts of hydrophobic granules and Nile red also allow the use of inexpensive polypropylene tubes instead of quartz cuvettes, in a compact LED-based fluorometer. This is further suitable for high throughput scale up if microtiter plate readers are used. The method is also scalable to other PHAs (Supplementary Fig. S1c and d).
Thus, the method developed for absolute PHB quantitation in standards and cells in this study is rapid, accurate, and sensitive and a significant improvement over the already existing methods, accounts for interference from lipophilic cell constituent, carbonosome formation, and protein. Further, it eliminates inconsistencies due to PHB granule aggregation and solvatochromism. This method can be scaled for higher throughput in systems biology and metabolic engineering approaches. The use of Nile red-based estimations are also scalable for PHAs, plastics [20] in the environment, and multiple cell types.
Supplementary data
Supplementary data is available at Biology Methods and Protocols Journal online.
Authors’ contributions
M.P.R. and A.R. conceived and designed the research. S.R. performed the NMR Experiments and S.R. and P.R.R. analyzed the NMR data. M.R. designed the primers and cloning strategy and performed Gibson assembly experiments to construct the plasmids, microbiological, and analytical experiments. M.P.R. and A.R. analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgement
We would like to thank Mrs. Nimisha Parekh at CSIR-NCL for fluorescence imaging.
Funding
This work was supported in part by grants from Department of Science and Technology (GAP292826) and Department of Biotechnology (GAP302126). M.P.R is supported through a UGC Senior Research Fellowship (NET Sr. no. 2061130639).
Conflict of interest statement. None declared.
References
- 1. Lemoigne M. Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull Soc Chem Biol 1926;8:770–82. [Google Scholar]
- 2. Serafim LS, Lemos PC, Levantesi C. et al. Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J Microbiol Methods 2002;51:1–18. [DOI] [PubMed] [Google Scholar]
- 3. Williamson DH, Wilkinson JF.. The isolation and estimation of the poly-hydroxy-butyrate inclusions of Bacillus species. J Gen Microbiol 1958;19:198–209. [DOI] [PubMed] [Google Scholar]
- 4. Braunegg G, Sonnleitner B, Lafferty RM.. A rapid gas chromatographic method for the determination of poly-beta-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol 1978;6:29–37. [Google Scholar]
- 5. Koller M, Rodríguez-Contreras A.. Techniques for tracing PHA-producing organisms and for qualitative and quantitative analysis of intra- and extracellular PHA. Eng Life Sci 2015;15:558–81. [Google Scholar]
- 6. Law JH, Slepecky RA.. Assay of poly-β-hydroxybutyric acid. J Bacteriol 1961;82:33–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slepecky RA, Law JH.. A rapid spectrophotometric assay of alpha, beta-unsaturated acids and beta-hydroxy acids. Anal Chem 1960;32:1697–9. [Google Scholar]
- 8. Watanabe Y, Ichinomiya Y, Shimada D. et al. Development and validation of an HPLC-based screening method to acquire polyhydroxyalkanoate synthase mutants with altered substrate specificity. J Biosci Bioeng 2012;113:286–92. [DOI] [PubMed] [Google Scholar]
- 9. Greenspan P, Fowler SD.. Spectrofluorometric studies of the lipid probe, Nile red. J Lipid Res 1985;26:781–9. [PubMed] [Google Scholar]
- 10. Spiekermann P, Rehm BHA, Kalscheuer R. et al. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 1999;171:73–80. [DOI] [PubMed] [Google Scholar]
- 11. Gorenflo V, Steinbüchel A, Marose S. et al. Quantification of bacterial polyhydroxyalkanoic acids by Nile red staining. Appl Microbiol Biotechnol 1999;51:765–72. [DOI] [PubMed] [Google Scholar]
- 12. Pcc S, Tyo KE, Zhou H, Stephanopoulos GN.. High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli. Appl Environ Microbiol 2006;72:3412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuriani R, Vigneswari S, Azizan MNM. et al. A high throughput Nile red fluorescence method for rapid quantification of intracellular bacterial polyhydroxyalkanoates. Biotechnol Bioproc Eng 2013;18:472–8. [Google Scholar]
- 14. Berlanga M, Montero MT, Fernández-Borrell J. et al. Rapid spectrofluorometric screening of poly-hydroxyalkanoate-producing bacteria from microbial mats. Int Microbiol 2006;9:95–102. [PubMed] [Google Scholar]
- 15. Degelau A, Scheper T, Bailey JE. et al. Fluorometric measurement of poly-β hydroxybutyrate in Alcaligenes eutrophus by flow cytometry and spectrofluorometry. Appl Microbiol Biotechnol 1995;42:653–7. [Google Scholar]
- 16. Greenspan P, Mayer EP, Fowler SD.. Nile Red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 1985;100:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibson DG, Young L, Chuang R-Y. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009;6:343–5. [DOI] [PubMed] [Google Scholar]
- 18. Gibson DG, Glass JI, Lartigue C. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010;329:52–6. [DOI] [PubMed] [Google Scholar]
- 19. Rodríguez LC, Campa[Nbreve]Ta AMG, Linares CJ. et al. Estimation of performance characteristics of an analytical method using the data set of the calibration experiment. Anal Lett 1993;26:1243–58. [Google Scholar]
- 20. Maes T, Jessop R, Wellner N. et al. A rapid-screening approach to detect and quantify microplastics based on fluorescent tagging with Nile Red. Sci Rep 2017;7:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.