Abstract
Asthma is a common chronic respiratory disease that has a heritable component. Polymorphisms in the endoplasmic reticular protein orosomucoid-like protein 3 (ORMDL3), which regulates sphingolipid homeostasis, have been strongly linked with childhood-onset asthma. Despite extensive investigation, a link between ORMDL3 asthma–risk genotypes and altered sphingolipid synthesis has been lacking. In this issue of the JCI, Ono et al. establish a clear association between nonallergic childhood asthma, lower whole-blood sphingolipids, and asthma-risk 17q21 genotypes. These results demonstrate that genetic variants in ORMDL3 may confer a risk of developing childhood asthma through dysregulation of sphingolipid synthesis. As such, modulation of sphingolipids may represent a promising avenue of therapeutic development for childhood asthma.
Introduction
Asthma is a chronic disease of the airways characterized by reversible airway obstruction, airway inflammation, and airway hyperresponsiveness (AHR). It is a clinically heterogenous disease that varies by disease expression, age of onset, and inflammatory profiles. The asthma phenotype is thought to arise as a result of the interplay between genetic and environmental factors. Although asthma phenotypes have been linked to numerous chromosomal regions, some time ago, Moffatt and colleagues (1) demonstrated that variants on chromosome 17q21 were strongly and reproducibly associated with childhood-onset asthma. Moreover, they made the seminal observation that the identified SNPs were consistently associated with elevated orosomucoid-like protein 3 (ORMDL3) transcript levels, indicating that genetic variants regulating ORMDL3 expression contribute to the development of childhood asthma. Despite more than a decade of intensive investigation, elucidation of the mechanisms by which elevations in ORMDL3 may drive childhood asthma has eluded investigators.
ORMDL3 and de novo sphingolipid regulation
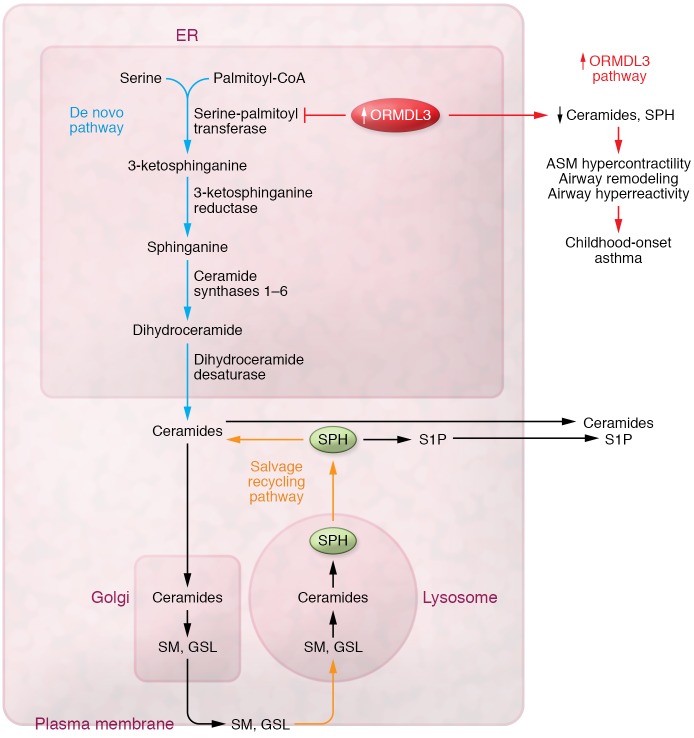
Given the evolutionarily conserved role of ORMDL family members in the regulation of sphingolipid homeostasis, it has been proposed that the mechanism(s) linking ORMDL3 polymorphisms to asthma pathogenesis are mediated through dysregulation of sphingolipid synthesis. Specifically, studies in yeast suggest that the ORMDL (1–3) family of proteins negatively regulate sphingolipid synthesis through inhibition of serine-palmitoyltransferase (SPT), the rate-limiting enzyme complex of de novo sphingolipid synthesis (2). The first step in de novo sphingolipid (SPH) synthesis begins with the condensation of serine and palmitoyl-CoA by SPT to produce 3-ketosphinganine. 3-ketosphinganine, is rapidly converted to sphinganine, which is further metabolized by distinct ceramide synthases to dihydroceramides (Figure 1). Dihydroceramide can then generate ceramides via dihydroceramide desaturases or get phosphorylated to sphinganine-1-phosphate. These analytes characterize the de novo sphingolipid synthesis pathway and are not generated through other pathways. As sphingolipids are essential in both the formation and integrity of cellular membranes and in the regulation of key cellular processes including proliferation and apoptosis, their levels are tightly regulated through a complex regulatory loop. In the face of external perturbations, the lysosome can salvage and recycle complex SPHs (sphingomyelins and glycosphingolipids) from plasma membranes to maintain physiologic SPH concentrations in the ER.
Figure 1. Potential role of genetic variants in ORMDL3 and childhood-onset asthma.
The first and rate-limiting step in de novo SPH synthesis begins with the condensation of serine and palmitoyl-CoA by SPT to produce 3-ketosphingosine, which is rapidly converted to sphinganine. Sphinganine is then further metabolized by distinct ceramide synthases to dihydroceramides, which can then generate ceramides via dihydroceramide desaturases (DES1). Dihydroceramides can get phosphorylated to sphinganine-1-phosphate or get transported out of the ER to form complex sphingomyelins or glycosphingolipids. To maintain physiologic concentrations of SPHs in the ER, the lysosome can salvage and recycle complex SPHs (sphingomyelins, glycosphingolipids) from plasma membranes. As a consequence of increased ORMDL3 expression, de novo synthesis of SPHs is reduced. Reductions in whole-blood SPH levels have been associated with several features of childhood asthma including increased ASM contractility, airway remodeling, and airway hyperreactivity. GSL, glycosphingolipids; SM, sphingomyelins.
Given the role of ORMDL3 in the inhibition of de novo sphingolipid synthesis, it has been assumed that polymorphisms in ORMDL3 that elevate its expression should result in reductions in the levels of these lipids. However, given the complexity of this regulatory pathway, establishing a straightforward relationship between ORMDL3 expression levels and sphingolipid levels has proven more difficult than originally anticipated. The discrepancies in the ORMDL3 field may be due to differences in SPH levels in various biological samples, differences in analytical approaches to measure SPH profiles, and/or differences in approaches to modulate ORMDL3 expression.
A report by Ono et al. (3) in this issue of the JCI has shed some light on the relationship between ORMDL3, SPH levels, and asthma susceptibility. In the first set of analyses, the authors quantified plasma and whole-blood sphingolipid levels in control subjects and children with asthma, stratified by the level of blood eosinophils as a measure of allergic disease. Interestingly, the levels of select dihydroceramides (C18 and 18:1), ceramides (C18, C20, C24, and C24:1), and sphingomyelins (C18, C18:1, and C24:1) were significantly lower in the whole blood of children with asthma who had low eosinophil counts (nonallergic) as compared with levels in the age-matched control subjects and in children with asthma who had high eosinophil counts. These results provide the first demonstration that nonallergic forms of childhood asthma are indeed associated with lower sphingolipid levels, whereas allergic asthma is not associated with significant alterations in SPHs. This study supports the previous findings by Moffat (1) and others (4–6) that the association between ORMDL3 and asthma is primarily with nonatopic childhood-onset asthma. Dysregulated ORMDL3 appears to predominantly affect processes or responses involved in environmental exposures that occur in a critical window early in life. Viral infections (7) and other early life triggers of asthma (such as environmental tobacco smoke) can enhance genotype-specific differences in ORMDL3 transcript abundance between children with asthma and those without asthma. The lack of an association of ORMDL3 polymorphisms with atopic disease is curious, given that allergen exposure of mice upregulates ORMDL3 (8), and in some instances overexpression is associated with an allergen-induced asthma phenotype (9), but not in others (10).
ORMDL3 SNPs and SPH synthesis
To query the relationship between whole-blood SPH levels and 17q21 genotypes, Ono and authors determined the genotypes at 5 different 17q21 SNPs. Interestingly, two of these risk genotypes (rs7216389 and rs8076131), which have been associated with increased ORMDL3 transcripts in lung and whole blood, correlated with decreased whole-blood levels of four products of SPT enzymatic activity (dihydroceramides [C16, C18, C24, and C24:1] and three ceramides [C16, C18, and C20]), whereas the others (rs8067378, rs4065275, and rs12603332) did not correlate with whole-blood SPH levels (3). These results provide strong evidence that asthma-risk ORMDL3 genotypes are associated with dysregulated SPH homeostasis. Interestingly, other SPHs including sphingosine, S1P, and sphingomyelins, which have previously been thought to play a role in asthma (11), were not associated with any of the genotypes examined. When the subjects were stratified for asthma, the lead SNP (rs7216389) in the original GWAS (3) remained associated with two dihydroceramides (C16 and C18). The rs8076131 was linked with four dihydroceramides (C16, C18, C24, and C24:1) and three ceramides (C16, C18, and C20). One drawback of the Ono et al. study is that because the authors did not assess ORMDL3 expression in the blood, they were unable to infer a direct association between asthma-risk alleles, altered ORMDL3 expression, and SPH levels. Nonetheless, these findings provide evidence that the 17q21 asthma–risk alleles are associated with a specific pattern of decreased whole-blood sphingolipids (3).
A key finding of the Ono et al. study was that plasma and whole-blood SPH levels and composition were fundamentally different. Whereas whole-blood levels of distinct SPH metabolites were reduced in children with nonallergic asthma, plasma sphingolipid levels in afflicted individuals tended to be higher than those in control subjects — however, none reached statistical significance, and there were no differences in plasma levels of any SPHs between allergic and nonallergic children (3). This finding may explain some discrepancies in the literature, as previous measurements of SPHs in patients with asthma have primarily been assessed in plasma or serum. Since whole blood contains red cells and leukocytes, which are absent in plasma, it is likely that the lower sphingolipid levels observed in the whole blood of nonallergic children with asthma reflect decreased blood cell membrane–associated sphingolipid levels. Whether these findings reflect a true association between structural alterations in leukocyte membrane composition and asthma susceptibility or are simply markers of a perturbed system remains unknown.
ORMDL3 polymorphisms and de novo SPH synthesis
Since the recycling/salvage pathway may generate SPHs independently of SPT activity (Figure 1), Ono et al. (3) explored the effect of the ORMDL3-risk allele on de novo synthesis in PBMCs from children with asthma. As circulating T and B cells express high levels of ORMDL3, PBMCs were incubated with stable isotype–labeled serine and sphinganine, and sphinganine-1-phosphate levels were examined. Both SPHs were lower in cells from children with asthma compared with levels in controls, with the lowest levels being observed in cells from children carrying the asthma-risk TT genotype at rs7216389. These findings provide the strongest link to date between genetic variants in ORMDL3 and de novo SPH synthesis in asthma.
Support for a link between altered de novo SPH synthesis and functional changes in airway responsiveness comes from several studies in mice, in which decreasing de novo sphingolipid synthesis, either by direct pharmacologic inhibition with myriocin or genetic haploinsufficiency of SPT, increases airway reactivity in the absence of allergic inflammation (12). Several studies in ORMDL3-transgenic mice support the connection between AHR and ORMDL3 (9). The functional airway changes observed in mice overexpressing human ORMDL3 might occur through the enhancement of airway smooth muscle (ASM) contraction, as ASM cells isolated from these mice exhibit enhanced proliferation and contractility (13). Alternatively, increased contractility may occur through ORMDL3-mediated upregulation of the levels of several remodeling genes, such as TGFB1 and ADAM8, which might contribute to stiffer airways (9). ORMDL3 may also regulate airway function through its ability to stimulate calcium release from the sarcoplasmic reticulum through Ca-ATPase 2b and to activate the ATF6α pathway of the ER unfolded protein response (UPR), which regulates SERC2b levels, thereby enhancing AHR (8, 14). Whether dysregulated ORMDL3 induces AHR through SPH-dependent processes or through a combination of cellular effects remains unknown. However, recent observations that lipid perturbation can directly activate the UPR independently of misfolded proteins suggests that these pathways may not be mutually exclusive (15).
As ORMDL3 is widely expressed in immune and structural cells that are relevant to asthma, the question arises as to whether expression of the risk allele has an effect on SPH synthesis in lung structural cells that is similar to that on circulating blood cells. To address this issue, Ono and colleagues (3) examined the association between the asthma-risk genotype and SPH synthesis in airway epithelial cells (AECs) from adults carrying either the rs7216389-TT or rs7216389-CC genotype grown at the air-liquid interface. They found that de novo–synthesized sphinganine and dihydroceramide C24 levels were reduced in epithelial cells from subjects with the risk-associated TT genotype compared with cells carrying the CC genotype. In contrast to the results observed in whole blood, no differences were detected in other sphingolipids: S1P, ceramide-C18, or sphingomyelin-C18, suggesting that the impact of ORMDL3 SNPs on the pattern of SPHs synthesized is cell-type dependent.
Whether disruption of ORMDL3 solely in AECs is sufficient to induce the asthma phenotype is a matter of debate. In support of a prominent role for the AEC, studies in which mice with a global Ormdl3 deletion were reconstituted with ORMDL3 transcripts only in the bronchial epithelium exhibited enhanced Alternaria-induced AHR (16). However, the contribution of ORMDL3 expression in the airway epithelium (AE) was called into question when mice lacking ORMDL3 expression only in the AE unexpectedly showed increased AHR when compared with WT mice (17). Taken together, these results suggest that ORMDL3 expression in the AE is necessary, but not necessarily sufficient, to regulate airway function. Alternatively, other ORMDL family members may compensate for the loss of SPT regulation and AHR in AE lacking ORMDL3. The specific nature of the allergens (Alternaria versus OVA) utilized in the respective studies may also play a role in the discrepancies observed between studies, as these allergens have very different characteristics (16, 17). Further studies will be required to identify the specific cell types (structural versus immune cells) that are critically involved in ORMDL3-mediated regulation of AHR.
Concluding remarks
Collectively, the Ono et al. study in this issue of the JCI provides direct evidence that genetic variants in ORMDL3 are linked to SPH synthesis disruption and asthma susceptibility (3). Whether the risk is conferred directly or indirectly through sphingolipid-triggered mechanisms or through other ORMDL3-associated effects (UPR, ER stress, remodeling gene induction, or other as-yet unidentified pathways) remains to be elucidated. However, these pathways are not necessarily mutually exclusive, as bidirectional interactions between SPHs and the UPR have been reported. More studies are needed to tease out the role of altered ORMDL3 and SPHs in individual cell types and how these individual cellular changes alone or in combination elicit asthma in the presence and absence of allergic tendencies and at different windows of susceptibility. Nonetheless, these studies have moved us one step closer to understanding the relevance of this strong genetic signal for asthma and should inform the development of asthma prevention and treatment strategies for children with the 17q21 high-risk genotype.
Version 1. 01/13/2020
Electronic publication
Version 2. 02/03/2020
Print issue publication
Footnotes
Conflict of interest: The author declares that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):604–607. https://doi.org/10.1172/JCI134333.
See the related article at Decreased sphingolipid synthesis in children with 17q21 asthma–risk genotypes.
References
- 1.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Breslow DK, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463(7284):1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono JG, et al. Decreased sphingolipid synthesis in children with 17q21 asthma–risk genotypes. J Clin Invest. 2020;130(2):921–926. doi: 10.1172/JCI130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acevedo N, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24(3):875–890. doi: 10.1093/hmg/ddu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like three (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64(4):629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perzanowski MS, et al. Distinct serum sphingolipid profiles among school-aged children with exercise-induced wheeze and asthma persistence. Am J Respir Crit Care Med. 2017;195(8):1068–1070. doi: 10.1164/rccm.201609-1884LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calışkan M, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368(15):1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller M, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA. 2012;109(41):16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller M, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debeuf N, et al. The ORMDL3 asthma susceptibility gene regulates systemic ceramide levels without altering key asthma features in mice. J Allergy Clin Immunol. 2019;144(6):1648–1659.e9. doi: 10.1016/j.jaci.2019.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolly PS, Rosenfeldt HM, Milstien S, Spiegel S. The roles of sphingosine-1-phosphate in asthma. Mol Immunol. 2002;38(16-18):1239–1245. doi: 10.1016/S0161-5890(02)00070-6. [DOI] [PubMed] [Google Scholar]
- 12.Worgall TS, et al. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5(186):186ra67. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Miller M, Unno H, Rosenthal P, Sanderson MJ, Broide DH. Orosomucoid-like three (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca2+ oscillations in asthma. J Allergy Clin Immunol. 2018;142(1):207–218.e6. doi: 10.1016/j.jaci.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19(1):111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 15.Ho N, Xu C, Thibault G. From the unfolded protein response to metabolic diseases - lipids under the spotlight. J Cell Sci. 2018;131(3):jcs199307. doi: 10.1242/jcs.199307. [DOI] [PubMed] [Google Scholar]
- 16.Löser S, et al. Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease. J Allergy Clin Immunol. 2017;139(5):1496–1507.e3. doi: 10.1016/j.jaci.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller M, et al. Cutting edge: targeting epithelial ORMDL3 increases, rather than reduces, airway responsiveness and is associated with increased sphingosine-1-phosphate. J Immunol. 2017;198(8):3017–3022. doi: 10.4049/jimmunol.1601848. [DOI] [PMC free article] [PubMed] [Google Scholar]