Abstract
Technological advances in rapid data acquisition have transformed medical biology into a data mining field, where new data sets are routinely dissected and analyzed by statistical models of ever-increasing complexity. Many hypotheses can be generated and tested within a single large data set, and even small effects can be statistically discriminated from a sea of noise. On the other hand, the development of therapeutic interventions moves at a much slower pace. They are determined from carefully randomized and well-controlled experiments with explicitly stated outcomes as the principal mechanism by which a single hypothesis is tested. In this paradigm, only a small fraction of interventions can be tested, and an even smaller fraction are ultimately deemed therapeutically successful. In this Review, we propose strategies to leverage large-cohort data to inform the selection of targets and the design of randomized trials of novel therapeutics. Ultimately, the incorporation of big data and experimental medicine approaches should aim to reduce the failure rate of clinical trials as well as expedite and lower the cost of drug development.
By simply noting facts, we can never succeed in establishing a science. Pile up facts or observations as we may, we shall be none the wiser.
— Claude Bernard, An Introduction to the Study of Experimental Medicine, 1865
Introduction
We are experiencing unprecedented growth in the amount of biological and medical information collected from human populations. Large prospective cohorts, such as the UK Biobank (1), the All of Us Research Program (2), and the China Kadoorie Biobank (3), are generating increasingly broad and detailed phenotypic descriptions of health trajectories for millions of individuals. Overall, initiatives in more than 30 countries have established more than 60 cohorts, each enrolling at least 100,000 individuals, collectively projected to include as many as 36 million participants (4). For a typical participant, a comprehensive picture of their physical state is provided by fine-grained data collected across various biological domains, including genetics, biomarker profiling, and biomedical imaging. Data from personal electronic devices are harvested continuously to capture physical activity, dietary habits, and social interactions. Streams of biological data are ultimately integrated with medical histories, made available by the rising adoption of electronic health records, to create complex models predictive of medical outcomes (5).
Perhaps not surprisingly, the rapid growth in health and genetic data has led to an explosion in the number of observations connecting physiological traits and diseases to genetic variants that may demark candidate targets for therapeutic intervention (Table 1). This increase in targets has been driven, at least partially, by widespread genome-wide association studies (GWAS) that measure statistical relationships between genetic and phenotypic variation among individuals in a population (6). GWAS and other analytical methods, applied to larger and larger data sets, have uncovered more and more genetic variants with smaller and smaller phenotypic contributions, and have informed our appreciation of the genetic complexity of human disease (6).
Table 1. Summary statistics of known gene-disease associations as reported in various database collections.
However, our ability to uncover genetic disease associations has far outpaced our ability to understand them and, even more so, to act on them. It is abundantly clear that only a small fraction of these associations can be functionally tested, and if we are to use these genetically inspired hypotheses for drug development and clinical testing, the list will need to be prioritized so as to avoid increasing the rates of failure in clinical trials.
Indeed, failures in clinical trials are far more frequent than successes. Among drugs entering clinical development, only about 10% will ultimately pass the stringent regulatory requirements necessary for a new-drug approval (7). The few successful trials bear the expense of all the failed ones, leading to the ever-increasing financial cost of drug development (8).
As the accumulation of human population data and the resulting gene-disease associations continues to increase, a question becomes central to the future of drug development: how can we mitigate the costs and improve on the success rate of clinical trials? Among the many strategies to tackle this question, a fundamental one is to reduce the number of candidate targets before they reach the clinical stage and to enrich them for the most promising hypotheses. Here we highlight four complementary avenues to achieve this goal. First, large and diverse cohorts provide greater power for discovery and fine mapping of likely causal variants, refining potential hypotheses of the effects of genetic variants. Second, the acquisition of intermediate phenotypes, bridging genetic variants and clinical manifestations, enriches our understanding of disease etiology and informs the design of more rational therapeutic strategies. Third, the development of more accurate and interpretable statistical approaches, especially those integrating orthogonal data types, helps prioritize targets and eliminates the least promising ones in silico. Finally, testing candidate interventions using deep and perturbed phenotyping in relatively small studies (i.e., experimental medicine) helps validate hypotheses, refine selection of patients and their appropriate dosages, and reduce the probability of failure at later clinical stages. The ability to move from hypotheses generated in large data sets to validation, integration, and hypothesis testing in small numbers holds the promise of a more efficient approach to drug development.
Genetically inspired target space
Many diseases arise from a complex interplay between genetics, environment, and time-dependent interactions between the two. Although heritability estimates are highly trait specific, most studies report some heritable component (9), suggesting that genetic studies may be useful for understanding pathophysiology and possibly identifying candidate targets for clinical development.
Genetic studies using pedigree- and linkage-based approaches (10, 11) have proved very effective for identifying genetic associations with Mendelian disorders (12). More recently, study designs (13, 14) enabled by inexpensive genotyping have mapped associations between genetic variants and thousands of diseases and quantitative traits (15). Driven by the growing size of sampled human populations and the diversity of measured phenotypic traits, as many as 32,000 gene-disease associations have been mapped so far (Table 1), and many more are expected in the near future.
An overview of all gene-trait connections discovered to date reveals a complex picture (Figure 1A). On one hand, many genes exhibit a high degree of pleiotropy and appear to be associated with many seemingly unrelated traits and diseases (Figure 1B). On the other hand, many traits and diseases are highly polygenic (Figure 1C). The complex gene and protein interactions that likely underlie pleiotropy and polygenicity are such that therapeutically intervening on a single gene-trait link without perturbing other neighboring connections is unlikely. Moreover, intervention on any significant fraction of these connections, aiming to alleviate a reasonable portion of human diseases, also stands as an intractable problem because of the sheer number of clinical trials that would be required.
Figure 1. The polygenic and pleiotropic space of GWAS associations.
The complexity of the gene-trait association network hinders the development of targeted interventions. (A) A representative network derived from the National Human Genome Research Institute (NHGRI)/European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) GWAS Catalog shows 6348 associations between 2939 genes and 650 traits (15). (B) Pleiotropic genes show associations with multiple phenotypic traits. (C) Polygenic traits are affected by multiple genes.
Despite recent progress in identifying variant-trait associations, there is rarely a direct path from a statistically significant variant-trait association to a testable therapeutic hypothesis, posing a major challenge for translational applications. Common variants, which drive a substantial portion of the heritability of complex traits (16), can occur in haplotypes, i.e., large chromosomal regions that tend to be inherited together. The non-independent segregation of variants within haplotypes, known as linkage disequilibrium (LD), makes it difficult to identify the causal variant(s) and thereby the mechanisms driving the association. Moreover, disease-associated loci are frequently found in noncoding genomic regions (17). As our current understanding of the regulatory landscape of the genome is incomplete, we often cannot infer directly the underlying gene(s) or other intermediate trait(s) that mediate the observed genetic association. Such knowledge is critical to generate clear hypotheses that are testable in a clinical setting. By prioritizing the collection of intermediate phenotypes — comprehensive molecular and physiological readouts — along with the development of advanced analytical methods that incorporate known regulatory features of the genome, it may nevertheless be possible to identify candidate targets with clear hypotheses that can be validated through carefully designed functional studies and clinical experiments.
Refining and replicating statistical associations
One of the biggest operational challenges in the analysis of large cohorts is replication of findings (18). This is a particularly difficult problem for associations with small effect sizes, as the ability to replicate a study depends on the existence of similarly sized or larger cohorts with equivalent phenotypic measurements. Computational strategies such as cross-validation (e.g., splitting a cohort into training and validation sets) can be used, but at the cost of decreasing power to detect novel associations. Within-cohort replication cannot address biases present in that cohort (e.g., access to health care, prevalence of smoking), as every random subset of the cohort used in cross-validations suffers from the same bias.
Genetic association studies have historically focused on White populations; however, recognition that diverse backgrounds can improve discovery and fine mapping (19) has led to efforts to study more diverse populations and recruit participants of diverse genetic backgrounds in biobank cohorts (20, 21). In addition, rare variants are more likely to be population specific (19), meaning that diverse cohorts will also improve power for discovery of new targets. While most of the observed variation in effect size across populations can be explained by low power, allele frequency differences, or differences in LD structure (22), we cannot rule out the possibility that some variants may have effect sizes that vary between populations (23), pointing to a genetic effect that may be specific to a particular environment.
Even with biobank-scale primary analysis in diverse populations, replication in independent cohorts will remain an important tool to strengthen true gene-disease associations and weaken false ones (18). Representatives of the 60 human cohorts with the largest number of worldwide participants have formed the International 100K Cohorts Consortium (4), whose primary mission is to facilitate exchange of knowledge and best practices and to devise a strategy for sharing data.
However, even when replicated, genetic evidence alone is insufficient to provide a clear path to intervention. Such a path typically requires a more detailed understanding of the molecular pathways that lead to disease development and relies in large part on deeper phenotypic profiling of the relevant populations.
Deep phenotyping
Our ability to intervene on a putative target is greatly assisted by a molecular understanding of disease pathogenesis and progression. Gaining such an understanding is a difficult task, as it requires collection of longitudinal biochemical and physiological data in patients, cell cultures, and laboratory animals, along with intense analytical labor needed for interpretation. Recent technological advancements have facilitated data collection across various levels of human biology and have provided us with rich data sets describing the molecular, biomarker, and physiological states of numerous cells, tissues, and organs. The comprehensive collection of such multilayer phenotypic data is often referred to as “deep phenotyping.”
At the molecular level, global initiatives, such as ENCODE (24), NIH Roadmap Epigenomics (25), BLUEPRINT (26), and GTEx (27), have quantified DNA methylation, chromatin accessibility, and gene regulation across a myriad of tissues and cell types in the human body. Additionally, efforts have begun to profile tissues at the single-cell level in normal conditions (28), as well as various developmental (29) and environmental contexts (30).
At the biomarker level, it is now possible to apply metabolomic, lipidomic, and proteomic analyses to large cohorts and acquire a comprehensive catalog of molecular species found in biofluids (31–35) and the microbiome (36). These multi-omics techniques gather data in a nontargeted manner to capture a large fraction of the biochemical space, including known as well as unknown molecular entities. Unbiased longitudinal measurements of biomarkers, collected before and after disease manifestation, can be instrumental for identifying potential causal mechanisms of disease pathology and have a considerable impact on the results of clinical testing: a recent analysis suggests that the success rate of clinical trials can be doubled by inclusion of at least one biomarker in patient selection (37).
At the physiological level, radiological imaging techniques are starting to provide noninvasive, high-resolution anatomical and functional information across all aspects of human physiology. Efforts to link brain imaging data to genetic and outcome data have already yielded new biological insights (38, 39). In addition, passive data collection from wearable devices allows for physiological monitoring at high temporal resolution (40).
While these early phenotyping efforts have already produced an unprecedented wealth of information, it is safe to say that this is just the beginning. In the future, the phenotypic data collected for human populations have the potential to scale up along every possible dimension: depth (number of traits captured), width (number of time points sampled), and height (number of individuals profiled). Integrating these highly dimensional data sets with genetics and clinical outcomes will be key to refining our mechanistic understanding of disease and prioritizing actionable therapeutic hypotheses (Figure 2). However, such integration will be challenging given the current lack of coherent conceptual frameworks and appropriate modeling techniques.
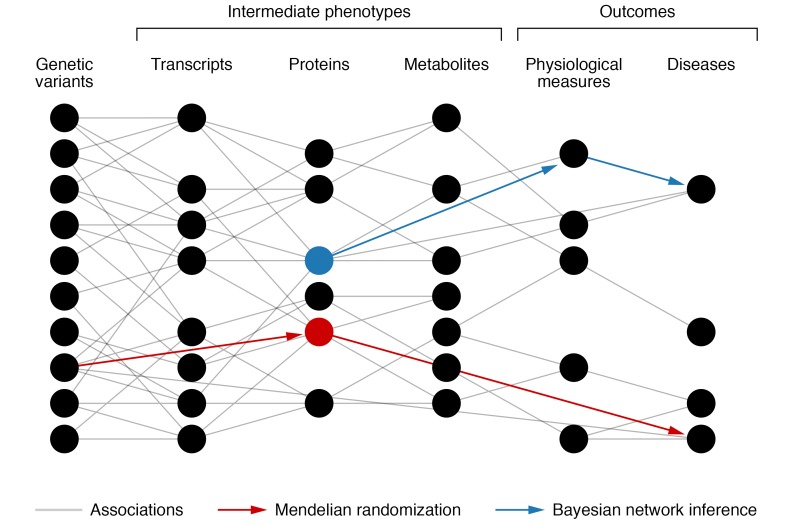
Figure 2. Use of deep phenotyping to limit the number of intervention hypotheses.
Association between genetic variation, intermediate traits, and outcomes. The large number of correlation connections (gray) can be reduced by introduction of sparsity into a network structure via Bayesian network inference (blue). Spurious correlations can be removed if outcomes are explained better by a different path through the network. Mendelian randomization (red) can also identify causal connections by using genetic variation within populations. Interventions on a red node or a blue node are more likely to succeed, as they mediate a path to a disease.
We are still in the early phase of our efforts to collect deep-omic phenotypes at scale, but the issue of replication should be carefully considered here as well. Apart from a few clinical biomarkers that have been routinely measured across large cohorts with carefully validated standardized procedures, no such standardization exists for most nonclinical biomarker measurements. Robust deep-omics measurement techniques that could be deployed on large cohorts are an active area of development (41, 42). Furthermore, application of these techniques across multiple cohorts would require a substantial multi-organizational effort to standardize sample preparation procedures, instrumentation, and quality control measures. These are important and necessary steps that will determine the usability of deep-omics data in the long run.
Integrative modeling and causal inference
The ultimate goal of computational modeling is to enable accurate predictions of a system’s behavior under perturbation. The complexity of biological systems, driven by a dense network of dynamic biochemical and regulatory interactions, has long hindered our ability to model them comprehensively. A variety of methods have been developed to make predictions based on correlations between molecular, physiological, and clinical measurements (43); however, transitioning from correlation to causation (a key ingredient for a successful clinical trial) remains a great challenge.
To address this challenge, a number of statistical approaches have been developed (44–46). These approaches, cumulatively referred to as mediation analyses, focus on the identification of intermediate phenotypes (e.g., biomarker levels in plasma) that might explain the association between an exposure (e.g., drug treatment) and an outcome (e.g., disease). The identification of such phenotypes is often critical for uncovering molecular mechanisms and provides considerable assistance in drug development. Among the various methods for mediation analysis, Mendelian randomization (MR) is of particular interest, as it takes advantage of natural genetic variation in human populations (46), allowing for stratification of individuals in a way that is analogous to a random assignment in clinical trials (47). In the most basic MR design, a robust genetic association of a variant (e.g., in the PCSK9 locus) with an intermediate phenotype (levels of LDL cholesterol) can be used as a proxy to estimate the effect of a drug exposure (statins) on an outcome (cardiovascular disease) (48–50).
Provided that the underlying assumptions are met, MR methods offer a powerful tool for identifying potential causal relationships. For instance, the directionality of the relationship between levels of LDL cholesterol and risk of coronary artery disease (CAD), as predicted by MR (51), is consistent with the results of clinical trials (52). Similar results have been obtained for HDL cholesterol and CAD (53–55), as well as vitamin D and type 2 diabetes (56–58). Causal predictions are particularly useful when clinical testing would be impractical or unethical — for instance, the effect of alcohol consumption on cardiovascular traits (59, 60). Encouraged by these early successes, the development of MR methods is an active area of research. One particular challenge is that many genetic associations have small effects on intermediate phenotypes, which can lead to inaccuracies in the causal effect estimates (61).
Given the heterogeneity of information and types of regulation within biological networks, multiscale models will be required to integrate information from different levels of biology (62, 63). A complete molecular description of network structure underlying human physiology does not exist, and we are left with all-by-all correlation structure between genes, proteins, metabolites, and physiological measures constructed from big data. A variety of methods (e.g., Bayesian networks, partial correlation networks) have been developed to reduce the complexity of these networks by removing spurious connections that are explained best by other connections in the network (43). These techniques produce sparse representations of a network where edges are the most likely causal relationships (Figure 2). We have not seen wide adoption of these methods to target discovery, but, combined with genetics, they could be of high value to clinical research.
Collectively, causal inference and integrative modeling can be instrumental in reducing the number of possible gene-disease associations to a more actionable subset. However, despite their early successes, current prediction methodologies are still in their infancy, and the predictions made by such methods can only be firmly established using experimentation and clinical testing.
Experimental medicine
Considering the inability of current computational models to predict accurately the effects of therapeutic interventions, our primary path to knowledge is through experimental testing. Animal models of disease are often used to perform rescue experiments that test the ability of a candidate intervention to revert or at least ameliorate the disease phenotype (64, 65). While proven to be extremely useful, animal models have known limitations due to their inability sometimes to recapitulate the physiological changes and response to therapy observed in humans (66, 67).
An alternative strategy is to learn about human physiology from individuals that carry loss-of-function mutations in promising target genes and can therefore be thought of as models of inhibition of those targets (68). Such “natural experiments” are found in populations that underwent strong founder events or elevated rates of consanguineous marriages that resulted in high rates of homozygosity for rare mutations, including those predicted to have severe loss-of-function effects (69–71). Deep phenotypic profiling of these individuals, who are effectively knockout models for one or more genes, can be used to investigate the physiological effects and safety implications of gene product inhibition, gain greater insights into biological pathways, explore gene modifiers, and establish gene dosage effects on disease outcomes. Early analyses of naturally occurring human knockouts in European and Pakistani populations have validated known drug targets and suggested new routes for intervention (e.g., NAV1.7 and pain, CCR5 and HIV, APOC3 and HDL cholesterol) (69, 72, 73).
Although affording important insights into human biology (74), the genomics of large-scale data, including those derived from human knockouts, is only one hand clapping. Many pathologies and indeed drug responses arise from genetics, environment, and time-dependent interactions. The full extent of these nongenetic contributions is hard to approximate, but most estimates suggest that somewhere between 60% and 80% of phenotypic variation is environmental (75). In one example, a maximal estimate of the contribution of genomics to variability in drug response in young healthy volunteers was approximately 30% (76). Data recorded on drug administration in the electronic health record are rarely confirmed by measurements of drug exposure or other objective assessments of adherence.
Understanding both the interindividual differences in network perturbations consequent to target engagement and how variable environmental conditions (77), including time of dosing (78), alter drug response within an individual is intrinsic to the development of a more precise approach to medicine. Such insights are dependent on experiments that test interventions in small groups of human subjects under basal and perturbed conditions in controlled environments. These experiments can afford deep and unbiased phenotypic characterization of their molecular, biomarker, and physiological responses. The data can provide unique insights into drug efficacy (79), identify biomarkers of drug susceptibility and response (80), and help refine patient selection for inclusion in clinical trials (81).
Importantly, experimental medicine addresses the greatest vulnerability in drug development — an accelerated passage through phase II, leading to poor estimates of drug efficacy due to shallow response measurements and low power, and thus to poor decisions about proceeding to phase III, the longest, most expensive, and most labor-intensive stage of clinical trials (82, 83). This bidirectional integration of such deep phenotypic data from experimental medicine with large observational data sets — i.e., human phenomic science (84) — promises to improve our understanding of drug action and variability in drug response. This knowledge will refine patient selection for large and expensive phase III trials, potentially limiting the size, duration, and cost of drug development.
Conclusion
The highly regulated world of clinical trials relies on blinded randomized experiments to test whether a single intervention is a safe and effective means to improve human health. Recent advances in genomic technologies, biochemistry, imaging, and automation are generating an unprecedented amount of data that, in turn, produce an overwhelming number of therapeutic hypotheses that could be taken into clinical trials. Importantly, while mining big data does create a deluge of hypotheses, it also offers a path to navigate through them. Large data sets, along with rigorous computational methods, enable validation, integration, and causal analysis of multiple lines of evidence to support or refute a hypothesis, improve our understanding of disease mechanisms, and identify a development path most likely to succeed. Furthermore, small-scale validation experiments afforded by experimental medicine provide a better understanding of candidate interventions and help to design better strategies for large-scale clinical testing. Outstanding challenges include the development of capacity for replication of experimental medicine data sets and the recognition of and adjustment for sources of bias in cohort data (e.g., ethnic and social diversity). Ultimately, the incorporation of big data and experimental medicine approaches into a standard practice should help reduce the failure rate of clinical trials and lower the cost of drug development.
Acknowledgments
GAF is the McNeil Professor of Translational Medicine and Therapeutics, and is supported by grants from the National Institutes of Health (1U54TR001623 and HL141912) and a Merit Award from the American Heart Association. This work was also supported by Calico Life Sciences LLC.
Version 1. 01/13/2020
Electronic publication
Version 2. 02/03/2020
Print issue publication
Footnotes
DLT’s present address is: Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, United Kingdom; and Medical Genomics and Metabolic Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland.
Conflict of interest: EM, DLT, AS, MC, AB, and NVB are full-time employees of Calico Life Sciences LLC, and GAF is an advisor to the company.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):575–581.https://doi.org/10.1172/JCI129196.
Contributor Information
Eugene Melamud, Email: eugene@calicolabs.com.
D. Leland Taylor, Email: lelandtaylor@gmail.com.
Anurag Sethi, Email: anurag@calicolabs.com.
Madeleine Cule, Email: cule@calicolabs.com.
Anastasia Baryshnikova, Email: abaryshnikova@calicolabs.com.
Danish Saleheen, Email: saleheen@pennmedicine.upenn.edu.
Nick van Bruggen, Email: nvb@calicolabs.com.
References
- 1.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. All of Us Research Program 2019. National Institutes of Health. https://allofus.nih.gov/ Accessed November 26, 2019.
- 3. China Kadoorie Biobank 2019. University of Oxford. https://www.ckbiobank.org Accessed November 26, 2019.
- 4. International 100K Cohorts Consortium 2019. International 100K Cohorts Consortium. https://ihcc.g2mc.org Accessed November 26, 2019.
- 5.Rajkomar A, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visscher PM, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowden H, Munro J. Trends in clinical success rates and therapeutic focus. Nat Rev Drug Discov. 2019;18(7):495–496. doi: 10.1038/d41573-019-00074-z. [DOI] [PubMed] [Google Scholar]
- 8.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Ge T, Chen C-Y, Neale BM, Sabuncu MR, Smoller JW. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 2017;13(4):e1006711. doi: 10.1371/journal.pgen.1006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- 12.Chong JX, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97(2):199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu DJ, Leal SM. Estimating genetic effects and quantifying missing heritability explained by identified rare-variant associations. Am J Hum Genet. 2012;91(4):585–596. doi: 10.1016/j.ajhg.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huffman JE. Examining the current standards for genetic discovery and replication in the era of mega-biobanks. Nat Commun. 2018;9(1):5054. doi: 10.1038/s41467-018-07348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojcik GL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sankar PL, Parker LS. The Precision Medicine Initiative’s All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med. 2017;19(7):743–750. doi: 10.1038/gim.2016.183. [DOI] [PubMed] [Google Scholar]
- 21.Gaziano JM, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Zanetti D, Weale ME. Transethnic differences in GWAS signals: a simulation study. Ann Hum Genet. 2018;82(5):280–286. doi: 10.1111/ahg.12251. [DOI] [PubMed] [Google Scholar]
- 23.Veturi Y, et al. Modeling heterogeneity in the genetic architecture of ethnically diverse groups using random effect interaction models. Genetics. 2019;211(4):1395–1407. doi: 10.1534/genetics.119.301909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30(3):224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 27.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. doi: 10.1101/121202. Regev A, et al. The Human Cell Atlas. bioRxiv. Published May 8, 2017. Accessed November 26, 2019. [DOI]
- 29. doi: 10.1101/630996. Cuomo ASE, et al. Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. bioRxiv. Published May 8, 2019. Accessed November 26, 2019. [DOI] [PMC free article] [PubMed]
- 30.Lareau CA, et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol. 2019;37(8):916–924. doi: 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49(4):568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 32.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Vries PS, et al. Whole-genome sequencing study of serum peptide levels: the Atherosclerosis Risk in Communities study. Hum Mol Genet. 2017;26(17):3442–3450. doi: 10.1093/hmg/ddx266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20(2):273–286. doi: 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen L, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doherty A, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. doi: 10.1038/s41467-018-07743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bittremieux W, et al. Quality control in mass spectrometry-based proteomics. Mass Spectrom Rev. 2018;37(5):697–711. doi: 10.1002/mas.21544. [DOI] [PubMed] [Google Scholar]
- 42.Beger RD, et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective.”. Metabolomics. 2016;12(10):149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9(8):796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glymour C, Zhang K, Spirtes P. Review of causal discovery methods based on graphical models. Front Genet. 2019;10:524. doi: 10.3389/fgene.2019.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thoemmes F, Ong AD. A primer on inverse probability of treatment weighting and marginal structural models. Emerging Adulthood. 2016;4(1):40–59. doi: 10.1177/2167696815621645. [DOI] [Google Scholar]
- 46.Pingault J-B, et al. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19(9):566–580. doi: 10.1038/s41576-018-0020-3. [DOI] [PubMed] [Google Scholar]
- 47.Walker VM, Davey Smith G, Davies NM, Martin RM. Mendelian randomization: a novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol. 2017;46(6):2078–2089. doi: 10.1093/ije/dyx207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 49.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ference BA, et al. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375(22):2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 51.Ference BA, et al. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol. 2012;60(25):2631–2639. doi: 10.1016/j.jacc.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 52.Cholesterol Treatment Trialists’ (CTT) Collaboration, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376(9753):1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmes MV, et al. Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J. 2015;36(9):539–550. doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keene D, Price C, Shun-Shin MJ, Francis DP. Effect on cardiovascular risk of high density lipoprotein targeted drug treatments niacin, fibrates, and CETP inhibitors: meta-analysis of randomised controlled trials including 117 411 patients. BMJ. 2014;349:g4379. doi: 10.1136/bmj.g4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye Z, et al. Association between circulating 25-hydroxyvitamin D and incident type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2015;3(1):35–42. doi: 10.1016/S2213-8587(14)70184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krul-Poel YHM, et al. Effect of vitamin D supplementation on glycemic control in patients with type 2 diabetes (SUNNY Trial): a randomized placebo-controlled trial. Diabetes Care. 2015;38(8):1420–1426. doi: 10.2337/dc15-0323. [DOI] [PubMed] [Google Scholar]
- 58.Pittas A, Dawson-Hughes B, Staten M. Vitamin D supplementation and prevention of type 2 diabetes. Reply. N Engl J Med. 2019;381(18):1785–1786. doi: 10.1056/NEJMc1912185. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Smith GD, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5(3):e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho Y, et al. Alcohol intake and cardiovascular risk factors: a Mendelian randomisation study. Sci Rep. 2015;5:18422. doi: 10.1038/srep18422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgess S, Thompson SG, CRP CHD Genetics Collaboration Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 62.Walpole J, Papin JA, Peirce SM. Multiscale computational models of complex biological systems. Annu Rev Biomed Eng. 2013;15:137–154. doi: 10.1146/annurev-bioeng-071811-150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qu Z, Garfinkel A, Weiss JN, Nivala M. Multi-scale modeling in biology: how to bridge the gaps between scales? Prog Biophys Mol Biol. 2011;107(1):21–31. doi: 10.1016/j.pbiomolbio.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fluri F, Schuhmann MK, Kleinschnitz C. Animal models of ischemic stroke and their application in clinical research. Drug Des Devel Ther. 2015;9:3445–3454. doi: 10.2147/DDDT.S56071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King A, Bowe J. Animal models for diabetes: understanding the pathogenesis and finding new treatments. Biochem Pharmacol. 2016;99:1–10. doi: 10.1016/j.bcp.2015.08.108. [DOI] [PubMed] [Google Scholar]
- 66.Lynch VJ. Use with caution: developmental systems divergence and potential pitfalls of animal models. Yale J Biol Med. 2009;82(2):53–66. [PMC free article] [PubMed] [Google Scholar]
- 67.Martić-Kehl MI, Schibli R, Schubiger PA. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur J Nucl Med Mol Imaging. 2012;39(9):1492–1496. doi: 10.1007/s00259-012-2175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. doi: 10.1101/530881. Minikel EV, et al. Evaluating potential drug targets through human loss-of-function genetic variation. bioRxiv. Published January 29, 2019. Accessed November 26, 2019. [DOI] [PMC free article] [PubMed]
- 69.Saleheen D, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544(7649):235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47(5):448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 71.Narasimhan VM, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352(6284):474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox JJ, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444(7121):894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y, et al. The role of a mutant CCR5 allele in HIV–1 transmission and disease progression. Nat Med. 1996;2(11):1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 74.Zanoni P, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351(6278):1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lakhani CM, Tierney BT, Manrai AK, Yang J, Visscher PM, Patel CJ. Repurposing large health insurance claims data to estimate genetic and environmental contributions in 560 phenotypes. Nat Genet. 2019;51(2):327–334. doi: 10.1038/s41588-018-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fries S, et al. Marked interindividual variability in the response to selective inhibitors of cyclooxygenase-2. Gastroenterology. 2006;130(1):55–64. doi: 10.1053/j.gastro.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 77.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 78.Ruben MD, Smith DF, FitzGerald GA, Hogenesch JB. Dosing time matters. Science. 2019;365(6453):547–549. doi: 10.1126/science.aax7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.et al. Opportunities and challenges in cardiovascular pharmacogenomics: from discovery to implementation. Circ Res. 2018;122(9):1176–1190. doi: 10.1161/CIRCRESAHA.117.310965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller MC, 3rd, Mohrenweiser HW, Bell DA. Genetic variability in susceptibility and response to toxicants. Toxicol Lett. 2001;120(1–3):269–280. doi: 10.1016/s0378-4274(01)00279-x. [DOI] [PubMed] [Google Scholar]
- 81.Dienstmann R, Rodon J, Tabernero J. Biomarker-driven patient selection for early clinical trials. Curr Opin Oncol. 2013;25(3):305–312. doi: 10.1097/CCO.0b013e32835ff3cb. [DOI] [PubMed] [Google Scholar]
- 82.FitzGerald GA. Testing cardiovascular drug safety and efficacy in randomized trials. Circ Res. 2014;114(7):1156–1161. doi: 10.1161/CIRCRESAHA.114.301809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Catella-Lawson F, et al. Effects of specific inhibition of cyclooxygenase-2 on sodium balance, hemodynamics, and vasoactive eicosanoids. J Pharmacol Exp Ther. 1999;289(2):735–741. [PubMed] [Google Scholar]
- 84.FitzGerald G, et al. The future of humans as model organisms. Science. 2018;361(6402):552–553. doi: 10.1126/science.aau7779. [DOI] [PubMed] [Google Scholar]
- 85.Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stenson PD, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Piñero J, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]