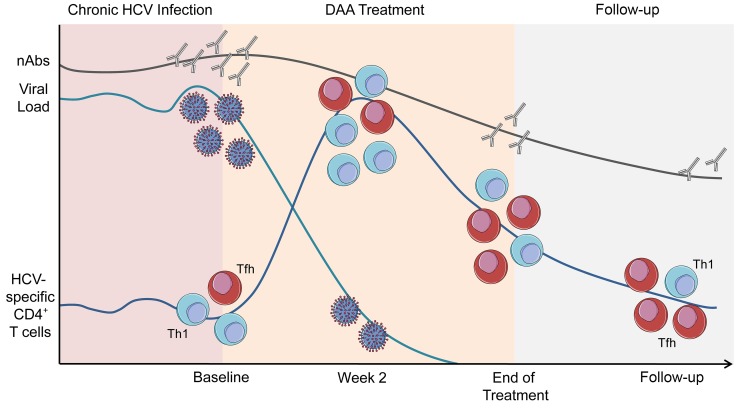
Abstract
BACKGROUND
Chronic hepatitis C virus (HCV) infection is characterized by a severe impairment of HCV-specific CD4+ T cell help that is driven by chronic antigen stimulation. We aimed to study the fate of HCV-specific CD4+ T cells after virus elimination.
METHODS
HCV-specific CD4+ T cell responses were longitudinally analyzed using MHC class II tetramer technology, multicolor flow cytometry, and RNA sequencing in a cohort of patients chronically infected with HCV undergoing therapy with direct-acting antivirals. In addition, HCV-specific neutralizing antibodies and CXCL13 levels were analyzed.
RESULTS
We observed that the frequency of HCV-specific CD4+ T cells increased within 2 weeks after initiating direct-acting antiviral therapy. Multicolor flow cytometry revealed a downregulation of exhaustion and activation markers and an upregulation of memory-associated markers. Although cells with a Th1 phenotype were the predominant subset at baseline, cells with phenotypic and transcriptional characteristics of follicular T helper cells increasingly shaped the circulating HCV-specific CD4+ T cell repertoire, suggesting antigen-independent survival of this subset. These changes were accompanied by a decline of HCV-specific neutralizing antibodies and the germinal center activity.
CONCLUSION
We identified a population of HCV-specific CD4+ T cells with a follicular T helper cell signature that is maintained after therapy-induced elimination of persistent infection and may constitute an important target population for vaccination efforts to prevent reinfection and immunotherapeutic approaches for persistent viral infections.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), the National Institute of Allergy and Infectious Diseases (NIAID), the European Union, the Berta-Ottenstein-Programme for Advanced Clinician Scientists, and the ANRS.
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, Cellular immune response, MHC class 2
Introduction
The development and approval of direct-acting antivirals (DAAs) targeting different viral proteins of the hepatitis C virus (HCV) has revolutionized the treatment of millions of chronically infected individuals worldwide (1). Although the impact of DAAs on health care cannot be overestimated, the development of these agents has also provided researchers with a fascinating novel tool to dissect immune responses to a pathogen that establishes persistent infection for decades and is eradicated from the host within weeks after initiation of antiviral therapy (2). Several landmark discoveries have been made on HCV immunity in this context, mostly demonstrating that HCV-induced alterations of the immune system are not rapidly restored after DAA-mediated virus clearance (3–5), as reviewed by Rehermann and Thimme (6). However, it has also been shown that HCV-specific CD8+ T cells acquire some characteristics of T cell memory and regain their ability to proliferate (7, 8). NK cell function appears to be partially restored (9), and the expression of peripheral and intrahepatic IFN-stimulated genes rapidly changes after DAA therapy (10). Moreover, analyses of bulk CD4+ and CD8+ T cells revealed that T cells expressing the CXC motif CXCR3, which is expressed on the vast majority of liver-infiltrating CD4+ T cells in chronic infection (11), are increased in the peripheral blood 1 to 2 weeks after treatment initiation, hinting toward an early emigration of liver-infiltrating lymphocytes to the blood (12). In line with these observations, levels of IP-10 (CXCL10, the ligand for CXCR3) are rapidly downregulated after DAA initiation (10). However, although HCV-specific CD4+ T cells are central regulators of HCV-specific immunity (13, 14), the fate of HCV-specific CD4+ T cells during and after DAA therapy has not been analyzed in previous studies. Specifically, it is unknown whether eliminating the virus results in reappearance of HCV-specific CD4+ T cells in the peripheral blood, as they are hardly detectable during chronic infection, possibly due to compartmentalization to the liver (14). Moreover, it has been shown that HCV-specific CD4+ T cells display an exhausted phenotype during chronic infection (15), and it remains to be demonstrated whether expression of inhibitory receptors is maintained after virus elimination and whether memory formation can be observed. In addition, mouse models of persistent viral infection revealed that virus-specific CD4+ T cells preferentially acquire a Th1 or a T follicular helper (Tfh) cell phenotype after viral infection (16, 17). It is unclear whether this differentiation fate can also be observed in a chronic viral infection in humans, however, and whether loss of the persistent antigen influences the differentiation pattern of the antigen-specific CD4+ T cell repertoire. Thus, in order to address these important issues, we comprehensively characterized the HCV-specific CD4+ T cell compartment in patients who are chronically infected with HCV undergoing DAA therapy. Importantly, our observations reveal dynamic changes of the frequency and the subset distribution of HCV-specific CD4+ T cells during antiviral therapy. We demonstrate that activation and inhibition markers are downregulated, while memory-associated markers are upregulated. In addition, transcriptional and phenotypic profiling reveals that CD4+ T cells with a Tfh signature are maintained months after eliminating the persistent antigen. Importantly, this coincides with a decline of germinal center activity and HCV-specific neutralizing antibodies (nAbs).
Results
Increase in HCV-specific CD4+ T cell frequencies within 2 weeks after initiation of antiviral therapy.
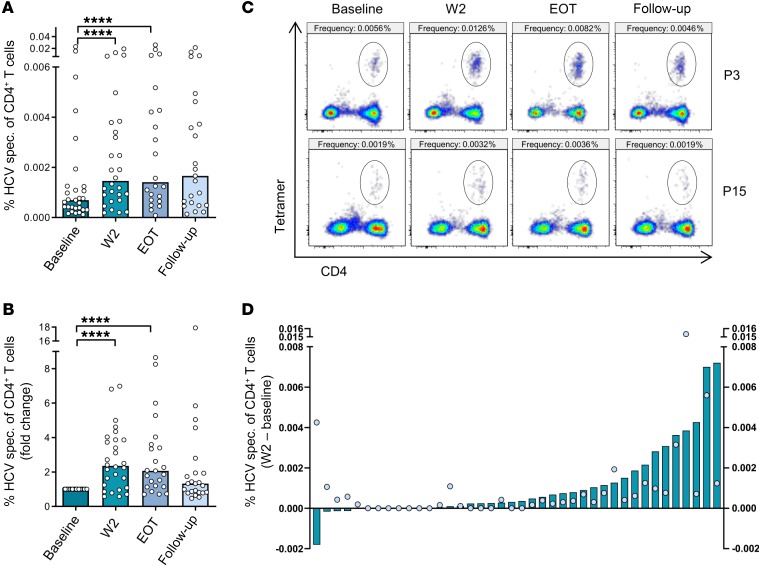
In order to monitor changes of HCV-specific CD4+ T cell frequencies throughout the course of antiviral therapy, we performed next-generation sequencing–based (NGS-based) HLA typing in a cohort of 248 patients with chronic HCV infection undergoing DAA therapy. Whole blood samples from patients who were HLA-DRB1*01:01–positive or HLA-DRB1*15:01–positive were taken before treatment initiation (baseline), 2 weeks after treatment initiation (W2), at the end of therapy (EOT), and 24 weeks after the end of treatment (follow-up) after written informed consent. MHC class II tetramer staining with bead-based enrichment was performed in 44 patients. In general, frequencies of HCV-specific CD4+ T cells were found to be very low and only detectable after bead-based tetramer enrichment in 29 of 44 patients at baseline (Figure 1, Figure 2A, Supplemental Figures 1 and 2, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI129642DS1). Interestingly, however, we observed a significant increase in HCV-specific CD4+ T cell frequencies as early as 2 weeks after initiating antiviral therapy (median at baseline and W2: 6.9 × 10–4 and 1.5 × 10–3, respectively; Figure 2, A–C). Following W2, CD4+ T cell frequencies tended to decrease and were found to be similar at follow-up compared with those at baseline (Figure 2, A–C). The increased frequency at W2 compared with baseline was observed in the majority of patients. While frequencies declined in 5 out of 40 patients (12.5%) who could be analyzed at both time points or remained undetectable in 7 out of 40 patients (17.5%), 28 out of 40 patients (70%) showed an increase in HCV-specific CD4+ T cell frequencies at W2 compared with baseline (Figure 2D). Importantly, detection rates of HCV-specific CD4+ T cells or changes in frequency from baseline to W2 were largely independent from the viral genotype; however, patients who were HLA-DRB1*15:01–positive were more likely to mount a detectable CD4+ T cell response (Supplemental Figure 3). As expected, viral titers and alanine transaminase levels rapidly declined after treatment initiation (Supplemental Figure 4). Collectively, these data suggest that DAA therapy can reinvigorate the circulating pool of HCV-specific CD4+ T cells.
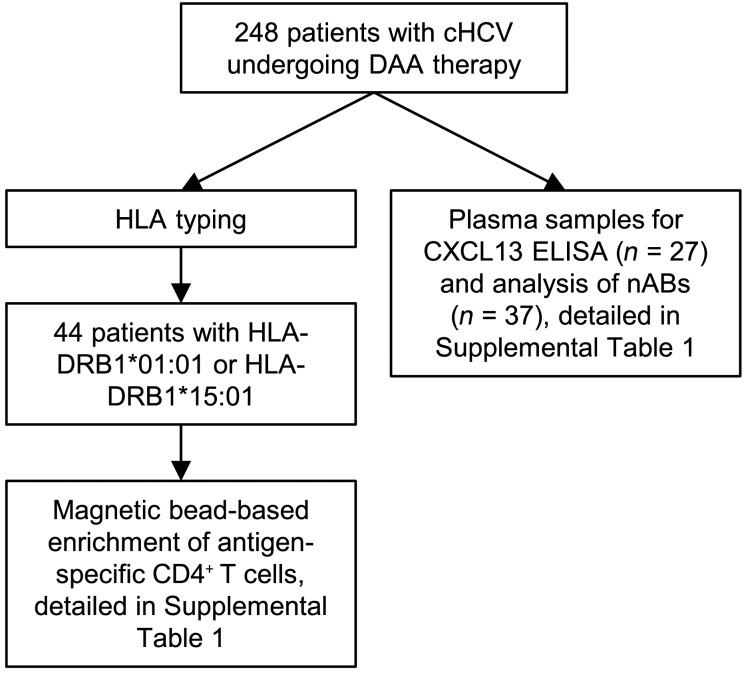
Figure 1. Flow chart outlining the study design and the composition of the cohort.
Patients with chronic HCV undergoing DAA therapy were HLA typed. Patients with HLA-DRB1-01:01 or HLA-DRB1-15:01 were used for magnetic bead–based enrichment of antigen-specific CD4+ T cells and used for further analysis. Plasma samples of 27 patients with chronic HCV undergoing DAA therapy were used for CXCL13 ELISA and 37 plasma samples for nAb analysis.
Figure 2. Frequency of HCV-specific CD4+ T cells increases shortly after initiation of DAA therapy.
(A and B) PBMCs from patients positive for HLA-DRB1*01:01 or HLA-DRB1*15:01, chronically infected with HCV undergoing DAA therapy were acquired before antiviral therapy (baseline), at W2 after initiation of therapy, at EOT, and at follow-up (24 weeks after EOT). Bead-based tetramer enrichment and surface staining were performed as described in the Methods section prior to analysis by flow cytometry. (A) Frequencies of HCV-specific CD4+ T cells within CD4+ T cells are shown as percentage and (B) as fold change compared with baseline frequencies (n = 29). (C) Representative pseudocolor flow cytometry plots with the corresponding frequency are shown for 2 patients (P3 and P15). (D) Frequencies of HCV-specific CD4+ T cells at baseline were subtracted from the frequencies at W2 to visualize the decrease or increase in the frequency. All patients analyzed at both time points are included in the analysis (n = 40). Dots represent the frequency at baseline and bars represent the calculated decrease or increase in the frequency (W2 – baseline). Each symbol represents 1 patient, bars represent medians (A and B). ****P < 0.0001, nonparametric distribution with Wilcoxon’s matched-pairs signed-rank test was applied between indicated groups. Due to multiple comparisons (n = 3), significance level was adjusted using Bonferroni’s correction and P values of < 0.01 were considered statistically significant. Thus, P values > 0.01 are not indicated.
Downregulation of inhibitory receptors and activation markers on HCV-specific CD4+ T cells during DAA therapy.
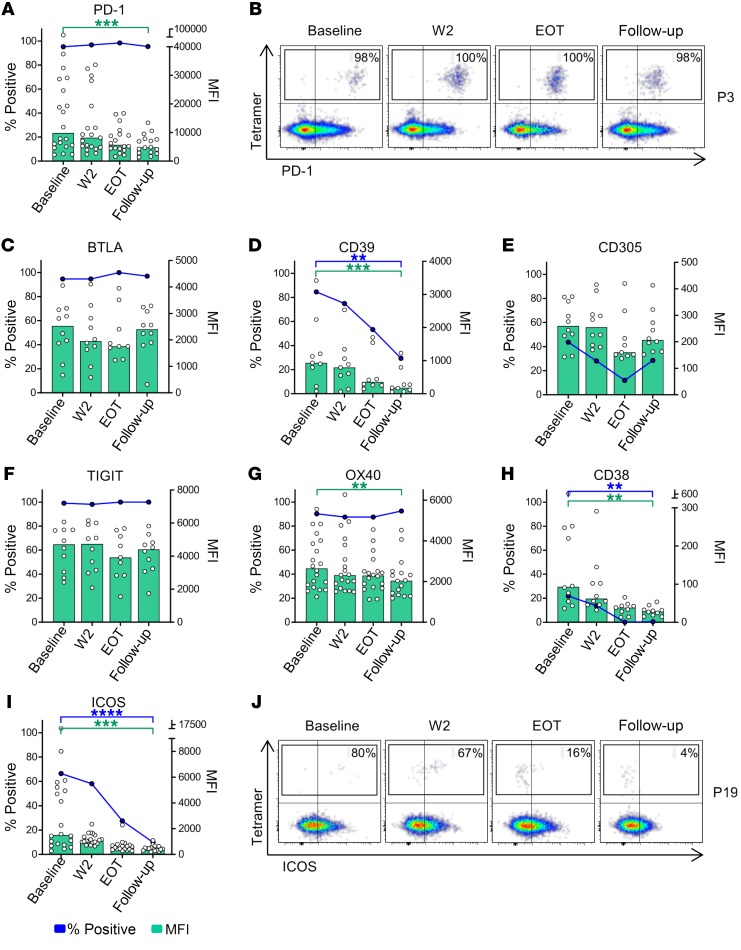
Due to the low frequencies of HCV-specific CD4+ T cells in the chronic phase of HCV infection, information on their ex vivo phenotype is limited. Although some data exist on the hierarchy of inhibitory receptors (15), data on activation markers are mostly lacking. Moreover, it is entirely unclear whether virus clearance after years of persistent infection alters the state of HCV-specific CD4+ T cells. In order to overcome this shortcoming, we analyzed the expression of several inhibitory receptors and activation markers on HCV-specific CD4+ T cells in chronic HCV infection and throughout antiviral therapy. The analyses of inhibitory receptors at baseline revealed high percentages of HCV-specific CD4+ T cells (median > 80%) expressing programmed cell death protein 1 (PD-1), B and T cell lymphocyte attenuator (BTLA), CD39, and T cell immunoreceptor with Ig and ITIM domains (TIGIT) in the chronic phase of the infection (baseline) while fewer cells expressed CD305 (Figure 3, A–F, blue dots). Interestingly, the expression of these receptors showed different dynamics during antiviral therapy. While CD39 was rapidly downregulated (percentage positive and median fluorescence intensity [MFI]), HCV-specific CD4+ T cells maintained expression of PD-1, BTLA, and TIGIT during the course of therapy (Figure 3, A–F, blue dots and lines). However, analyses of the PD-1 MFI revealed a significant reduction in the expression levels of PD-1 (Figure 3, A and B, green bars and scattered white dots). Thus, expression of the inhibitory receptors CD39 and PD-1 decreased during the course of antiviral therapy, while low-level PD-1 expression is maintained on HCV-specific CD4+ T cells after therapy. Owing to the loss of ongoing antigen stimulation during and after DAA therapy, we hypothesized that HCV-specific CD4+ T cells would also display changes in their expression patterns of activation markers. Among the analyzed activation markers, OX40 (CD134) was most strongly expressed in the chronic phase and was maintained throughout the course of therapy; however, similar to the expression pattern of PD-1, MFI decreased from baseline toward follow-up (Figure 3G). The activation markers ICOS and CD38 were less strongly expressed at baseline compared with OX40, but expression also significantly decreased during the course of therapy and was almost undetectable in the follow-up period (Figure 3, H–J). Collectively, these data reveal substantial changes in the ex vivo phenotype of HCV-specific CD4+ T cells after eliminating the persistent antigen.
Figure 3. Longitudinal analysis of inhibitory receptors and activation markers on HCV-specific CD4+ T cells during antiviral therapy.
(A and C–I) Expression of different inhibitory receptors and activation markers on HCV-specific CD4+ T cells was assessed at the indicated time points before and during antiviral therapy. Median expression of the individual surface marker on HCV-specific CD4+ T cells is characterized by the blue dots and lines. The MFI of the individual samples and the median MFI are displayed as white scattered dots and green bars, respectively (n = 20 for PD-1, OX40, and ICOS; n = 10 for BTLA, CD38, CD305, TIGIT, and CD38). Each symbol represents 1 patient, bars represent medians. (B and J) Representative pseudocolor plots for expression of PD-1 and ICOS on HCV-specific CD4+ T cells from 2 patients (P3 and P19) are shown after gating on live, non-naive CD4+ T cells. **P < 0.01, ***P < 0.001, ****P < 0.0001; nonparametric distribution with Wilcoxon’s matched-pairs signed-rank test between indicated groups. Due to multiple comparisons (n = 3), significance level was adjusted using Bonferroni’s correction and P values of < 0.01 were considered statistically significant. Thus, P values > 0.01 are not indicated.
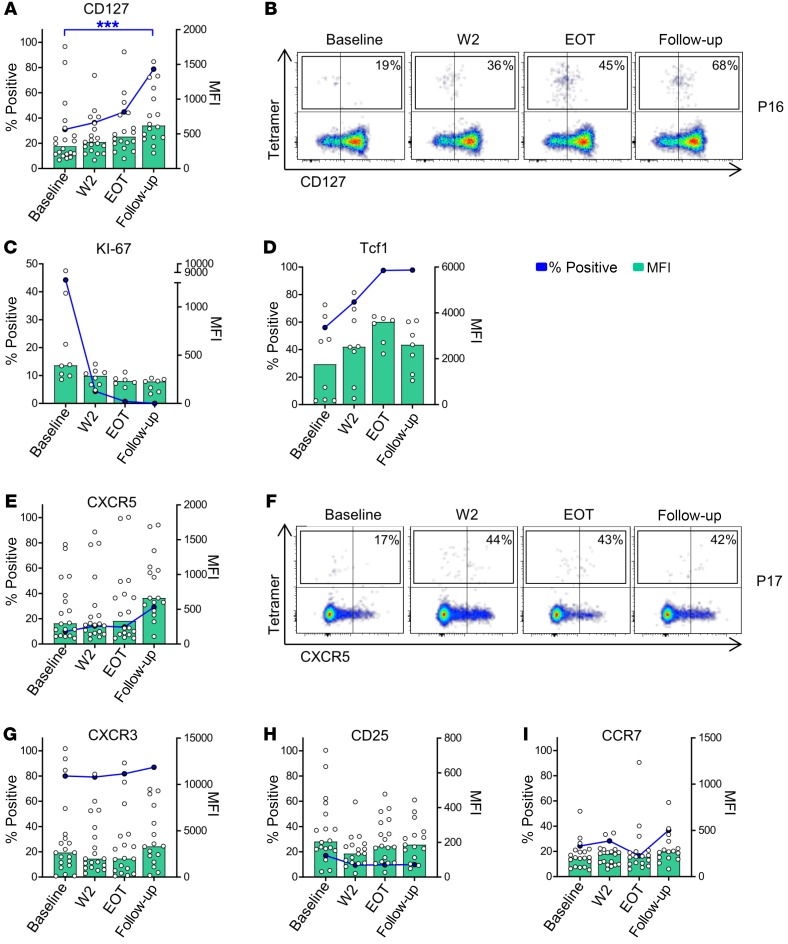
Antiviral therapy is associated with increased expression of memory-associated markers and changes of chemokine receptor expression on HCV-specific CD4+ T cells.
Next, we asked whether changes in expression of inhibition and activation markers are also accompanied by changes of markers that indicate T cell memory or helper-lineage affiliation of CD4+ T cells. These analyses revealed an upregulation of CD127 (Figure 4, A and B) on HCV-specific CD4+ T cells after antiviral therapy, indicating CD4+ T cell memory (18). This was associated with a loss of Ki-67 expression (Figure 4C) and an upregulation of the transcription factor T cell factor 1 (Tcf1) (Figure 4D), a transcription factor that facilitates memory development (19) and longevity of T cell immunity during persistent infection (20). With regard to chemokine receptors, we observed that few HCV-specific CD4+ T cells expressed CXCR5 at baseline, in line with our previous observation (11). However, CXCR5 expression increased at follow-up compared with baseline (Figure 4, E and F). In contrast, CXCR3 expression levels were high at baseline and were maintained during the course of antiviral therapy, while CCR7 and CD25 were expressed at low levels throughout the observation period, with a slight increase in CCR7 expression (Figure 4, G–I). Taken together, these data demonstrate that HCV-specific CD4+ T cells increasingly display characteristics of memory development as characterized by upregulation of CD127 and Tcf1.
Figure 4. Cytokine receptor, chemokine receptor, and transcription factor expression on HCV-specific CD4+ T cells.
(A, C–E, and G–I) Median expression of the individual surface marker on HCV-specific CD4+ T cells is characterized by the blue dots and lines. The MFI of the individual samples and the median MFI are displayed as white scattered dots and green bars, respectively (n = 20 for CD127, CXCR3, CCR7, CXCR5, and CD25; n = 8 for Ki-67 and Tcf1). Each symbol represents 1 patient, bars represent medians. (B and F) Representative pseudocolor plots for expression of CD127 and CXCR5 are shown after gating on live, non-naive CD4+ T cells from 2 patients (P16 and P17). *P < 0.05, ***P < 0.001; nonparametric distribution with Wilcoxon’s matched-pairs signed-rank test between indicated groups. Due to multiple comparisons (n = 3), significance level was adjusted using Bonferroni’s correction and P values of < 0.01 were considered statistically significant. Thus, P values > 0.01 are not indicated.
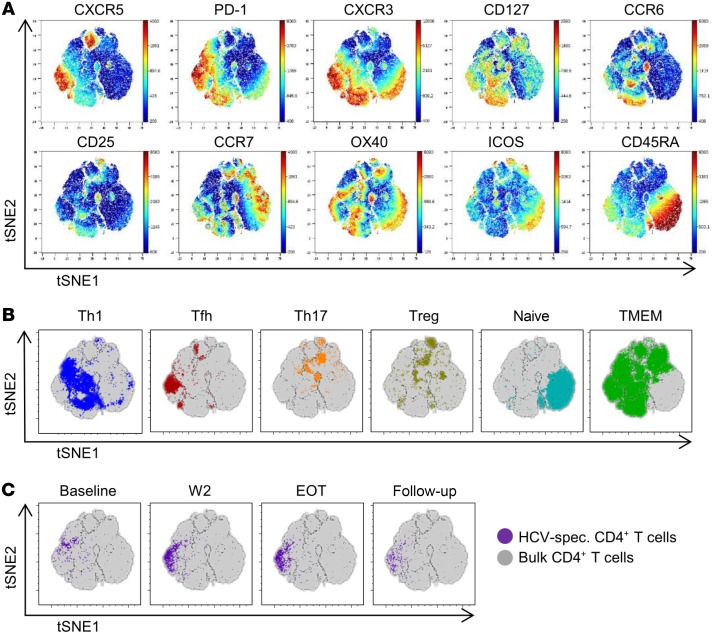
Shift of HCV-specific CD4+ T cells toward Tfh cells after initiation of antiviral therapy.
Commitment of virus-specific CD4+ T cells to various differentiation stages in chronic infection has been reported in murine chronic infection, including differentiation of Th1 or Tfh cells (17). To investigate the CD4+ T cell differentiation landscape during chronic HCV infection and DAA therapy, we costained CD4+ T cells with HCV-specific MHC class II tetramers and key markers (CCR6, CCR7, CXCR3, CXCR5, CD25, CD45RA, CD127, ICOS, OX40, and PD-1), allowing for a comprehensive analysis of the polarization of human T helper cells. The phenotypic complexity of CD4+ T cells was visualized using t-distributed stochastic neighbor embedding (tSNE) analyses on longitudinal stainings of a patient chronically infected with HCV with a strong HCV-specific CD4+ T cell response before, during, and after DAA therapy. The expression of phenotypic markers on the resulting tSNE map is shown in Figure 5A. As expected, markers associated with naive or central memory T cells (such as CD45RA, CCR7) and polarized effector responses (including CCR6, CXCR3, CXCR5) localized to different areas of the map. We thus speculated that this approach would allow distinguishing naive, memory, and individual T helper cell lineages. Therefore, we next gated T helper populations using canonical marker combinations and visualized their expression on the tSNE map (Figure 5B). Distinct areas of the map were occupied by naive, memory, Tfh, Th1, Th17 or Treg cells. These results indicated that localization on the tSNE map could be used to provide information on the underlying polarization state. To understand the polarization of HCV-specific CD4+ T cells, we identified their localization on the tSNE map before, during, and after therapy (Figure 5C). Interestingly, we observed a strong shift of HCV-specific CD4+ T cells after initiating therapy. Although the majority of HCV-specific CD4+ T cells colocalized with Th1-polarized areas of the map at baseline, a large population of HCV-specific CD4+ T cells shifted toward a Tfh-polarized area at W2 (Figure 5, B and C). Following W2, we observed a decline in the numbers of HCV-specific CD4+ T cells toward follow-up (as also seen in Figure 2), but the localization to the Tfh area was maintained. Very similar observations were made in other patients (Supplemental Figure 7).
Figure 5. Phenotypic shift of HCV-specific CD4+ T cells after initiation of antiviral therapy.
tSNE analysis of CD4+ T cells based on polychromatic flow cytometry data including CCR6, CCR7, CXCR3, CXCR5, CD25, CD45RA, CD127, ICOS, OX40, and PD-1 was performed in longitudinal samples from a representative patient analyzed in a single batch. HCV-specific CD4+ T cell responses were analyzed by tetramer staining. (A) The expression levels of individual surface markers are visualized on total CD4+ T cells using dot plots colored according to channel fluorescence intensity. Color scales are denoted adjacent to tSNE dot plots. (B) Localization of predefined CD4+ T helper lineages were assessed on the tSNE map. Specifically, Tfh cells (CXCR5+PD-1+), Th1 cells (CXCR5–CXCR3+CCR6–), Th17 cells (CXCR5–CXCR3–CCR6+), Treg cells (CD127–CD25+), naive T cells (CCR7+CD45RA+), and memory CD4+ T cells (CD127+CD45RA–) are displayed as overlays on the bulk CD4+ T cell population. (C) HCV-specific CD4+ T cells indicated by tetramer binding visualized on the tSNE map per time point analyzed, indicating phenotypic changes in high-dimensional space of the virus-specific CD4+ T cell response after treatment initiation.
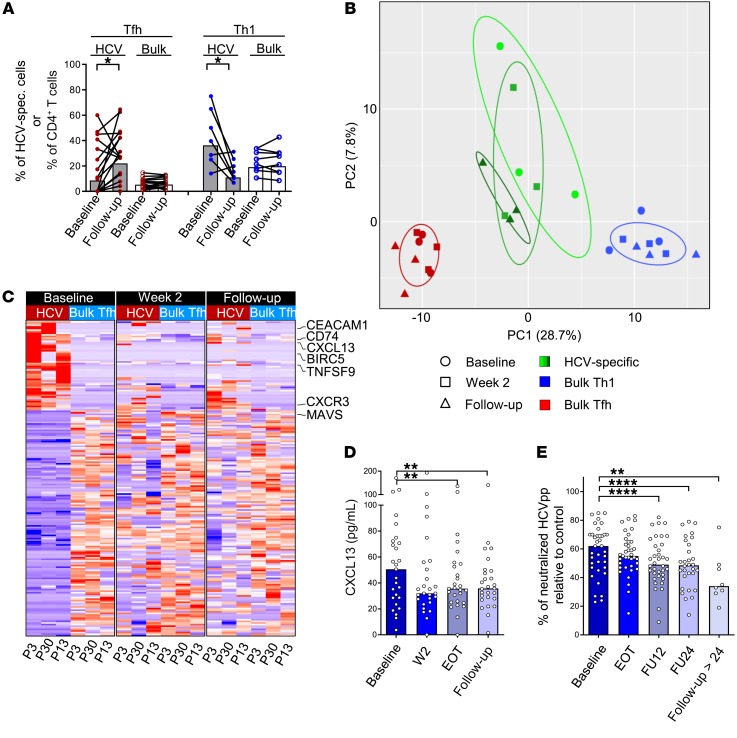
In order to follow up on this observation and investigate whether this shift toward a Tfh phenotype is a common feature of HCV-specific CD4+ T cells during antiviral therapy, we analyzed Tfh and Th1 signatures on HCV-specific and bulk CD4+ T cells at baseline and at follow-up in 16 and 8 patients, respectively. Interestingly, the percentage of HCV-specific CD4+ T cells with a Tfh phenotype significantly increased from baseline to follow-up (10.4% to 27.2% [median]; P = 0.035; Figure 6A). Specifically, 12 out of 16 patients displayed higher HCV-specific Tfh frequencies at follow-up compared with baseline, while 3 out of 16 had decreasing frequencies with 1 patient showing no changes (Figure 6A). Importantly, this effect was restricted to the HCV-specific CD4+ T cell population, as these changes were not observed on the bulk CD4+ T cell population, suggesting an antigen-specific effect (Figure 6A). In contrast, cells with a Th1 phenotype decreased throughout the course of therapy in 6 out of 8 patients, while 2 patients showed slight increases in Th1 frequencies (44.8% to 13.5% [median]; P = 0.039); again, this effect was only observed on HCV-specific CD4+ T cells (Figure 6A). Notably, we did not observe significant changes in the production of the lineage-defining cytokines IFN-γ (Th1) and IL-21 (Tfh) in HCV-specific CD4+ T cells during the course of therapy (Supplemental Figure 5). In sum, these analyses indicate that HCV-specific CD4+ T cells undergo major changes in lineage commitment after initiation of DAA therapy.
Figure 6. Emergence of a Tfh signature on HCV-specific CD4+ T cells after antigen removal.
(A) HCV-specific (gray bars and filled dots) and bulk CD4+ T cells (white bars and empty dots) were analyzed for coexpression of CXCR5 and PD-1 (indicating Tfh differentiation; red dots) or for CXCR5–CXCR3+CCR6– expression (indicating Th1 differentiation, blue dots) at baseline and follow-up (Tfh, n = 16; Th1, n = 8). (B and C) RNA sequencing was performed on HCV-specific CD4+ T cells, cTfh cells (CXCR5+PD-1+CXCR3–), and Th1 cells (CXCR3+CCR6–) of 3 patients (P3, P30, and P13) at 3 time points (baseline, W2, follow-up). (B) Principal component analysis was generated using the differentially expressed genes (FDR < 0.05; 297 genes) between bulk Th1 and bulk cTfh cells. (C) Differentially expressed genes between HCV-specific CD4+ T cells at baseline and bulk cTfh cells were used to generate a gene set of 198 genes. The heatmaps were generated using this gene set in longitudinal samples (baseline, W2, follow-up) to analyze changes of HCV-specific CD4+ T cells. Cutoff for generation of the heatmaps was FDR < 0.01. (D) CXCL13 levels, indicating germinal center activity, were measured by ELISA in the plasma of patients undergoing DAA therapy (n = 27). (E) Neutralizing antibodies were assessed in the plasma of patients using infection of Huh7.5.1 cells with lentiviral HCVpp bearing HCV envelope glycoproteins. Neutralization of genotype-matched HCVpp compared with control (100%) by individual sera is shown (n = 37). Each symbol represents 1 patient, bars represent medians. FU12, 12 weeks after the end of treatment; FU24, 24 weeks after the end of treatment. *P < 0.05, **P < 0.01, ****P < 0.0001; nonparametric distribution with Wilcoxon’s matched-pairs signed-rank test between indicated groups (A). Due to multiple comparisons (n = 3–4), significance level was adjusted using Bonferroni’s correction and P values of < 0.01 were considered statistically significant. Thus, P values > 0.01 are not indicated (D and E).
Emergence of a transcriptional Tfh cell signature after virus clearance.
In order to compare the HCV-specific CD4+ T cell population with Tfh cells in more detail, we performed RNA sequencing of HCV-specific CD4+ T cells, bulk Th1 cells, and bulk circulating Tfh (cTfh) cells that have previously been demonstrated to most closely resemble lymphoid tissue–derived Tfh cells. Circulating Tfh cells are defined as CD4+ T cells expressing CXCR5 and PD-1 in the absence of CXCR3 (21). In order to determine whether the phenotypic shift from Th1 toward Tfh can also be reproduced on the level of gene transcription, we performed RNA sequencing at different time points in 3 patients. For technical reasons, only patients with a sufficiently strong HCV-specific CD4+ T cell response at baseline (i.e., >50 HCV-specific CD4+ T cells within 10 × 106 to 20 × 106 PBMCs) could be included for sequencing. In order to identify genes that are differentially expressed between bulk Th1 and Tfh cells, we pooled the respective populations from all time points and identified 297 differentially expressed genes between cTfh and Th1 cells (false discovery rate [FDR] cutoff was set to 0.05, Supplemental Table 4). Using principal component analysis, we analyzed the proximity of the HCV-specific CD4+ T cells from baseline, W2, and follow-up to the bulk Th1 and cTfh populations. Importantly, in line with our phenotypic observations, we observed a convergence of the HCV-specific CD4+ T cells toward the cTfh population away from the Th1 population from baseline to follow-up (Figure 6B and Supplemental Figure 8). In addition, focusing on genes that were differentially expressed between HCV-specific CD4+ T cells at baseline and cTfh cells, we observed that the transcriptional profile of HCV-specific CD4+ T cells converged toward that of cTfh cells during antiviral therapy (Figure 6C and Supplemental Table 5). Importantly, we observed that IFN-stimulated genes such as CAECAM1 (22) but also CXCL13 (23) were among those genes that were rapidly downregulated on HCV-specific CD4+ T cells after treatment initiation. CXCL13, the ligand for CXCR5, has recently been established as a biomarker for germinal center activity (24). The appearance of HCV-specific cells with a Tfh phenotype in the circulation during and after antiviral therapy could be associated with a reduction of germinal center activity in the lymphoid tissues and the liver. Thus, we analyzed CXCL13 levels in the plasma of patients. Interestingly, we observed a decrease in CXCL13 in patients with chronic HCV infection after initiating antiviral therapy, suggesting a decline of overall germinal center activity (Figure 6D). In order to analyze HCV-specific germinal center activity, we analyzed genotype-specific nAb titers at baseline and after initiating antiviral therapy. Importantly, we observed that resolution of persistent infection also resulted in a decline of HCV-specific nAb titers (Figure 6E). Collectively, these data demonstrate that HCV-specific CD4+ T cells with transcriptional and phenotypic features of Tfh cells appear in the circulation after elimination of the persistent antigen, which is associated with decreasing levels of CXCL13 and HCV-specific nAbs, revealing that global and HCV-specific germinal center activity declines after virus clearance.
Discussion
Due to the difficulties in the detection of pathogen-specific CD4+ T cells in humans, little is known about their differentiation fate, their localization during infection, and their functional characteristics, particularly in the context of a persistent infection. However, given their central role in regulating cellular and humoral immunity and their relevance for vaccine development, a detailed knowledge of their characteristics and their biological properties is essential. Although some studies have analyzed different aspects of HCV-specific CD4+ T cells in the context of acute infection (11, 14, 25–27), information on their functional, phenotypic, and transcriptional properties in persistent infection is scarce. Most importantly, it remains entirely unclear how IFN-free therapy and elimination of the persistent antigen affects the HCV-specific CD4+ T cell population. This is a crucial question, as it has been demonstrated that DAA-mediated virus clearance does not protect from reinfection and rates of reinfection are high in populations at risk (28). Thus, the optimal therapeutic strategy in chronically infected patients would be antiviral therapy followed by prophylactic vaccination. And whereas the development of a prophylactic vaccine is challenging by itself (29, 30), it might be even more difficult in patients previously exposed to persistent infection and a subsequent “chronic imprint” on the pathogen-specific T cells (31). CD4+ T cells are of particular importance in this context, as they constitute prime targets for vaccines due to their potential to facilitate both CD8-mediated cytotoxicity and B cell–mediated humoral immunity. Interestingly, we observed changes in the HCV-specific CD4+ T cell compartment that indicate a trend toward memory development, most prominently characterized by upregulation of CD127 and Tcf1. Importantly, however, CD127 expression levels on HCV-specific CD4+ T cells after DAA therapy did not reach the levels that were observed on bulk CD4+ T cells (Supplemental Figure 6). Similarly, longitudinal analyses of activation markers and inhibitory receptors revealed a downregulation of several markers (CD38, CD39, ICOS, OX40, PD-1), while some of them were maintained, albeit at lower expression levels (i.e., PD-1 and OX40). In agreement with observations in HCV-specific CD8+ T cells, these changes indicate that some features of T cell exhaustion and activation are reversible even after decades of persistent infection, while others appear to be persistently imprinted (8, 32), preventing the development of classical T cell memory. Of note in this context, the phenotypic changes toward a memory-like phenotype were not accompanied by changes in the cytokine expression pattern of HCV-specific CD4+ T cells.
Another important finding of our study was the observation that frequencies of HCV-specific CD4+ T cells significantly increased early after initiation of antiviral therapy. Due to the short lifespan of hepatitis C virions (approximately 3 hours) (33), inhibition of their replication by DAAs results in a rapid decline of viral loads in treated patients. Consequently, HCV-specific T cells are no longer required in the liver parenchyma to suppress viral replication. Based on the presence of liver-infiltrating HCV-specific T cells targeting the virus in chronic HCV infection (34), a rapid efflux of liver-infiltrating T cells after virus elimination is well conceivable. Hepatocyte-specific expression of IP-10 results in migration of CXCR3-expressing T cells to the liver in the chronic phase of the infection and has been shown to correlate with hepatic inflammation (35). After DAA initiation, IP-10 levels rapidly decrease (4), coinciding with the emergence of CXCR3-expressing T cells in the peripheral blood (12). Similarly, we and others have previously shown that CD4+ T cells with functional and phenotypic characteristics of Tfh cells accumulate in the chronically HCV-infected liver (11, 36) and have now identified an increase in HCV-specific CD4+ T cells with phenotypic and transcriptional characteristics of Tfh cells in the circulation after DAA therapy. Collectively, these observations suggest an active Tfh program in virus-specific CD4+ T cells during chronic infection at the site of infection (and possibly in lymphoid tissues) that becomes detectable in the circulation after termination of viral replication in the liver. Unfortunately, due to restrictions in accessing different tissues at various time points in humans, we cannot track the fate of the liver-infiltrating Tfh cells after DAA-induced termination of persistent infection to experimentally address whether the increase in Tfh cells in the circulation is associated with a reduction in intrahepatic Tfh responses.
The decline of germinal center activity after antiviral therapy, as characterized by decreases in CXCL13 and the HCV-specific nAbs, further suggests that HCV-specific Tfh cells may no longer actively promote germinal center function in lymphoid tissues or the liver and may be released into the circulation.
Coexpression of CXCR5 and PD-1 serves as a reliable marker to identify Tfh cells in lymphoid tissues and in the peripheral blood. However, the phenotype that most precisely characterizes functional Tfh cells in the periphery remains to be identified. In this regard, CXCR3 has emerged as a central marker to distinguish highly functional Tfh cells from those with little ability to provide B cell help. It has been speculated that expression of CXCR3 on Tfh cells may directly translate into the quality of the resulting antibody response. Analyses from bulk memory Tfh cells have demonstrated that Tfh cells lacking CXCR3 expression possess the strongest ability to provide B cell help in vitro (21, 37). Moreover, the frequency of these cells is positively associated with the presence of highly potent broadly neutralizing HIV-specific antibodies in HIV-infected individuals (21). In contrast, Tfh cells elicited after influenza vaccination as well as HCV-specific CD4+ T cells express CXCR3 (11, 38–40) that is maintained even after the loss of the persistent antigen. In case of the influenza vaccination, antibodies provide little protection and are rather short-lived (41, 42). Similarly, HCV-specific antibodies with neutralizing ability have been shown to emerge late after infection in patients developing chronic infection and are not sufficiently maintained after spontaneous virus clearance (43, 44). Here, we demonstrate that nAbs decrease after DAA-mediated virus clearance, suggesting that persistent HCV infection induces Tfh cells that can maintain HCV-specific germinal center responses but that these rapidly collapse after virus clearance. These observations support the hypothesis that Tfh cells with a Th1 differentiation bias, characterized by the expression of CXCR3, are suboptimal in enabling the generation of rapidly emerging, broadly neutralizing, and long-lasting antibody responses.
Regarding limitations of this study, it certainly lacks longitudinal analyses of liver samples and ideally also liver-draining lymph node samples throughout antiviral therapy to corroborate the observations in the peripheral blood. However, these samples are almost impossible to acquire even in individual patients, especially taking HLA requirements into account in order to perform in-depth analyses of HCV-specific CD4+ T cells.
Collectively, our data demonstrate that clearance of persistent HCV infection by direct-acting antivirals results in an appearance of HCV-specific Tfh cells in the circulation. These cells may be liver-derived and released into the circulation after eliminating the persistent antigen. In contrast with their Th1 counterparts, they are maintained in the periphery, months after virus elimination. Their appearance in the circulation coincides with the reduction of germinal center activity and precedes the reduction of HCV-specific nAb titers, indicating that these cells may be involved in maintaining HCV-specific humoral immunity during chronic infection. However, efforts to develop a vaccine to prevent reinfection will have to show whether these HCV-specific memory-like Tfh cells are able to induce long-lasting and protective nAb responses.
Methods
Study subjects.
A total of 248 patients chronically infected with HCV undergoing DAA therapy have been screened for inclusion in this study. Samples were obtained at baseline (before therapy), at W2 of therapy, at EOT, and 24 weeks after the end of therapy (follow-up). In selected patients, additional samples were taken at later time points (follow-up > 24 weeks). Patients were HLA typed by NGS using commercially available primers (GenDx) and run on a MiSeq system. Data were analyzed using the NGSengine software (GenDx). A total of 76 patients were included in this study; 44 were positive for either HLA DRB1*01:01 or DRB1*15:01. Plasma samples and PBMCs were analyzed at the time points indicated in each figure. All samples were frozen until usage. Patient characteristics are summarized in Supplemental Table 1.
Magnetic bead–based enrichment of antigen-specific CD4+ T cells.
The magnetic bead–based enrichment of antigen-specific CD4+ T cells was performed as described previously (11, 45) using anti-PE magnetic beads according to the manufacturer’s protocol (MACS Technology, Miltenyi Biotec). In brief, PBMCs from HLA-DRB1*01:01– or HLA-DRB1*15:01–positive donors were thawed and incubated with the respective PE-labeled MHC class II tetramer (see Supplemental Table 2). Magnetic beads were added, and the HCV-specific cells were enriched by magnetic cell separation. The pre-enriched sample, the depleted sample, and the enriched sample were stained with fluorochrome-conjugated antibodies and analyzed by flow cytometry. Five antigen-specific cells were considered the lower detection limit. The frequency of HCV-specific CD4+ T cells was calculated as follows: absolute number of tetramer-binding CD4+ T cells (enriched sample) divided by the absolute number of CD4+ T cells (pre-enriched sample) × 100, as previously described (11).
Extracellular staining and flow cytometry.
Pre-enriched, depleted, and enriched cell samples were stained 20 minutes at 4°C with antibodies (see Supplemental Table 3). All samples were acquired on an LSR Fortessa flow cytometer (BD Biosciences) and were analyzed with FlowJo v10 software (FlowJo, LLC). For the analysis of HCV-specific CD4+ T cells and bulk CD4+ T cells, dump gate–positive (CD14+, CD19+, viability dye+) cells and subsequently naive CD4+ T cells (CD45RA+CCR7+) were excluded.
Transcription factor staining.
Enriched samples were stained for 10 minutes at room temperature against surface markers (see Supplemental Table 3), then fixed and permeabilized (eBioscience Intracellular Fixation & Permeabilization Buffer Set, Thermo Fisher Scientific) and stained with antibodies against transcription factors for 30 minutes at 4°C. Antibodies are listed in Supplemental Table 3. All samples were acquired on an LSR Fortessa flow cytometer and were analyzed with FlowJo v10 software.
Intracellular cytokine staining.
Enriched samples were stimulated with PMA (20 ng/mL, Sigma-Aldrich Chemie GmbH) and ionomycin (0.4 μg/mL, Sigma-Aldrich Chemie GmbH). A total of 0.5 μL/mL GolgiPlug and 0.325 μL/mL GolgiStop (BD Biosciences) was added to each well. After 3.5 hours of incubation, the cells were stained for 15 minutes at room temperature with antibodies against surface markers, and then fixed and permeabilized (Cytofix/Cytoperm, BD Biosciences) and stained with antibodies against cytokines and CD4 for 20 minutes at room temperature. Antibodies are listed in Supplemental Table 3. All samples were acquired on an LSR Fortessa flow cytometer and were analyzed with FlowJo v10 software.
tSNE analysis.
Phenotypic analysis of CD4+ T cell responses was performed longitudinally in patients. Patient samples analyzed in 1 experimental batch were analyzed to visualize high-dimensional phenotypes on a 2-dimensional map using tSNE, as previously described (46, 47). Briefly, equal numbers (n = 12,500) of live singlet CD4+ T cells were sampled per individual time point. Two-dimensional tSNE representation was calculated (1000 iterations, eta 200; perplexity = 25) using the single-cell expression information from 10 antibody costainings (CCR6, CCR7, CXCR3, CXCR5, CD25, CD45RA, CD127, ICOS, OX40, PD-1) and analyzed using Cytobank and FlowJo v10.5.3 software.
RNA sequencing.
HCV-specific CD4+ T cells were enriched by magnetic bead–based enrichment and surface stained with antibodies (see Supplemental Table 3). Live cells were sorted on a FACSMelody cell sorter (BD Biosciences). HCV-specific CD4+ T cells were sorted as CD4+tetramer+ non-naive cells. cTfh cells were sorted as CD4+CXCR5+PD-1+CXCR3– non-naive cells and Th1 cells were sorted as CD4+CXCR3+CCR6– non-naive cells. Fifty cells each were sorted into 1× reaction buffer per time point per patient and processed with SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech Laboratories, Inc., A Takara Bio Company). The libraries were sequenced at the EMBL Genomics Core Facility using the Illumina NextSeq 500 platform with 75-bp paired-end reads. Raw read counts per gene (using the gencode v19 gene models) are available as Supplemental Data.
RNA sequencing data alignment and differential analysis.
RNA sequencing reads were mapped in the human reference genome (build 37, version hs37d5) using STAR (version 2.5.2b) (48) using a 2-pass alignment. The alignment call parameters used were as follows: --sjdbOverhang 200 --runThreadN 8 --outSAMtype BAM Unsorted SortedByCoordinate --limitBAMsortRAM 100000000000 --outBAMsortingThreadN=1 --outSAMstrandField intronMotif --outSAMunmapped Within KeepPairs --outFilterMultimapNmax 1 --outFilterMismatchNmax 5 --outFilterMismatchNoverLmax 0.3 --twopassMode Basic --twopass1readsN -1 --genomeLoad NoSharedMemory --chimSegmentMin 15 --chimScoreMin 1 --chimScoreJunctionNonGTAG 0 --chimJunctionOverhangMin 15 --chimSegmentReadGapMax 3 --alignSJstitchMismatchNmax 5 -1 5 5 --alignIntronMax 1100000 --alignMatesGapMax 1100000 --alignSJDBoverhangMin 3 --alignIntronMin 20 --clip3pAdapterSeq CTGTCTCTTATACACATCT --readFilesCommand gunzip -c.
Other parameters were as default, or only pertinent for particular samples. Duplicate reads were marked using sambamba (version 0.6.5) (49) using 8 threads and were sorted by position using SAMtools (version 1.6) (50). BAM file indexes were generated using sambamba. Quality control analysis was performed using the SAMtools flagstat command, and the rnaseqc tool (version v1.1.8.1) (51) with the 1000 Genomes assembly and gencode 19 gene models. Depth of coverage analysis for rnaseqc was turned off. The quality statistics for each sample are reported in Supplemental Tables 6 and 7. FeatureCounts (version 1.5.1) (52) was used to perform gene specific read counting over exon features based on the gencode V19 gene model. The quality threshold was set to 255 (which indicates that STAR found a unique alignment). Strand-unspecific counting was used. For total library abundance calculations, during FPKM/TPM expression values estimations, all genes on chromosomes X, Y, MT, and rRNA and tRNA were omitted, as they possibly introduce library size estimation biases. Differential expression analyses were performed using DESeq2 (version 1.14.1) (53) and heatmaps were visualized using the ComplexHeatmap package (version 1.99.5) (54). Genes with an FDR less than 0.05 were considered for further analysis.
RNA sequencing data are deposited in the EGA under the accession number EGAS00001003950.
CXCL13 ELISA.
CXCL13 Quantikine ELISA (R&D Systems Europe, Ltd) was performed according to the manufacturer’s protocol. In brief, standards and plasma samples were pipetted into preassigned wells. After 2 hours of incubation and washing, anti-CXCL13 conjugate was added to the wells. After another 2 hours of incubation, the plate was washed again and substrate solution was applied for 30 minutes. The enzyme reaction was stopped by stop solution. The ELISA plate was measured at 450 nm and 570 nm with a TECAN Spark (Tecan GmbH). Concentrations were calculated using Magellan software (Tecan GmbH).
nAbs.
Lentiviral HCV pseudoparticles (HCVpp) bearing patient-derived HCV envelope glycoproteins from viral isolates (strains H77, genotype 1a; HCV-J, genotype 1b, genotype 2b; UKN3A1.28, genotype 3a) were produced, and HCVpp entry as well as neutralization were performed as described previously (55). Briefly, genotype-matched HCVpp were incubated with control serum or decomplemented patient sera at a dilution of 1 to 200 for 1 hour at 37°C before incubation with Huh7.5.1 cells. After 72 hours, HCVpp entry was quantified by measuring the luciferase activity as previously described (55). HCVpp incubated with serum from anti-HCV seronegative individuals served as a negative control.
Statistics.
Statistical analyses and graphical visualization were performed using GraphPad Prism 8 software. For analysis of multifunctional expression of cytokines, SPICE software was applied (56). To compare changes between cHCV and the treatment and posttreatment time points, nonparametric tests (Wilcoxon’s matched-pairs signed-rank test) were applied, as there was no Gaussian distribution of the data as confirmed by the Kolmogorov-Smirnov test. If multiple statistical tests were used (in general, 3 statistical tests: baseline versus W2, baseline versus EOT, baseline versus follow-up; in Figure 6E, 4 statistical tests were used), the statistical significance level of 0.05 was adjusted using Bonferroni’s correction. The adjusted significance level is detailed in the corresponding figure legend. P values less than 0.05 in single testing were considered significant, whereas in case of multiple testing with 3 tests, an adjusted P value of 0.01 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Study approval.
This study was approved by the ethics committee of the University Hospital Freiburg (344/13 and 227/15). Written informed consent was obtained from all individuals prior to inclusion in the study.
Author contributions
TB was responsible for study concept and design. MS, DW, KZ, CF, and KJ acquired data. TB, MS, NI, ZG, BB, and CC analyzed and interpreted data. TB and MS drafted the manuscript. BB, RT, TFB, MH, and CNH engaged in critical revision of the manuscript for important intellectual content. MS and ZG were responsible for statistical analysis. TB, CNH, MH, RT, and TFB obtained funding. FE, TFB, and RE provided technical or material support. RT and TB supervised the study.
Supplementary Material
Acknowledgments
The authors thank Sebastian Merker and Katharina Doernbrack for excellent administrative assistance and Sebastian Zehe, Özlem Sogukpinar-Beheshti, Elisa Wüstrich, and Patricia Otto-Mora for excellent technical assistance; Francis V. Chisari (Scripps Research Institute, La Jolla, California, USA) and Charles M. Rice (Rockefeller University, New York, New York, USA) for providing the Huh7.5.1 cell line; Robert H. Purcell and Jens Bukh (NIAID, Bethesda, Maryland, USA) for providing the cDNA for the H77 strain; T. Jake Liang (Liver Disease Branch, NIDDK, Bethesda, Maryland, USA) for expression constructs of the HCV-J strain; and Jonathan Ball (University of Nottingham) for providing the UKN3A strain. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, project 272983813) TRR179-TP04 (to TB), TRR179-TP01 (to MH and RT), TRR179-TP02 (to CNH), and TRR179-flexible funds (to BB) and BO 3361/4-1 (to TB). TB was supported by the Berta-Ottenstein-Programme for Advanced Clinician Scientists, Faculty of Medicine, University of Freiburg. NIAID grant U19-AI123862 (to TFB), ARC-IHU (to TFB), European Union grants ERC-AdG-2014-671231-HEPCIR and H2020-2015-667273-HEP-CAR (to TFB and RT), and ANRS grant ECTZ87384 (to TFB) also supported this work.
Version 1. 11/07/2019
In-Press Preview
Version 2. 01/21/2020
Electronic publication
Version 3. 02/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):998–1009.https://doi.org/10.1172/JCI129642.
Contributor Information
Maike Smits, Email: maike.smits@uniklinik-freiburg.de.
Katharina Zoldan, Email: katharina.martina.zoldan@uniklinik-freiburg.de.
Naveed Ishaque, Email: n.ishaque@dkfz-heidelberg.de.
Zuguang Gu, Email: z.gu@dkfz-heidelberg.de.
Katharina Jechow, Email: katharina.jechow@bihealth.de.
Dominik Wieland, Email: dominik.wieland@web.de.
Christian Conrad, Email: christian.conrad@bihealth.de.
Roland Eils, Email: roland.eils@charite.de.
Catherine Fauvelle, Email: catherinefauvelle@hotmail.fr.
Thomas F. Baumert, Email: Thomas.Baumert@unistra.fr.
Florian Emmerich, Email: florian.emmerich@uniklinik-freiburg.de.
Bertram Bengsch, Email: bertram.bengsch@uniklinik-freiburg.de.
Christoph Neumann-Haefelin, Email: christoph.neumann-haefelin@uniklinik-freiburg.de.
Maike Hofmann, Email: maike.hofmann@uniklinik-freiburg.de.
Robert Thimme, Email: robert.thimme@uniklinik-freiburg.de.
Tobias Boettler, Email: tobias.boettler@uniklinik-freiburg.de.
References
- 1.Pawlotsky JM, Feld JJ, Zeuzem S, Hoofnagle JH. From non-A, non-B hepatitis to hepatitis C virus cure. J Hepatol. 2015;62(suppl 1):S87–S99. doi: 10.1016/j.jhep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R, et al. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res. 2018;248:53–62. doi: 10.1016/j.virusres.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Strunz B, et al. Chronic hepatitis C virus infection irreversibly impacts human natural killer cell repertoire diversity. Nat Commun. 2018;9(1):2275. doi: 10.1038/s41467-018-04685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengst J, et al. Direct-acting antiviral-induced hepatitis C virus clearance does not completely restore the altered cytokine and chemokine milieu in patients with chronic hepatitis C. J Infect Dis. 2016;214(12):1965–1974. doi: 10.1093/infdis/jiw457. [DOI] [PubMed] [Google Scholar]
- 5.Bolte FJ, et al. Intra-hepatic depletion of mucosal-associated invariant T cells in hepatitis C virus-induced liver inflammation. Gastroenterology. 2017;153(5):1392–1403.e2. doi: 10.1053/j.gastro.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehermann B, Thimme R. Insights from antiviral therapy into immune responses to hepatitis B and C virus infection. Gastroenterology. 2019;156(2):369–383. doi: 10.1053/j.gastro.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin B, et al. Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol. 2014;61(3):538–543. doi: 10.1016/j.jhep.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 8.Wieland D, et al. TCF1+ hepatitis C virus-specific CD8+ T cells are maintained after cessation of chronic antigen stimulation. Nat Commun. 2017;8:15050. doi: 10.1038/ncomms15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serti E, et al. Successful interferon-free therapy of chronic hepatitis C virus infection normalizes natural killer cell function. Gastroenterology. 2015;149(1):190–200.e2. doi: 10.1053/j.gastro.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meissner EG, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest. 2014;124(8):3352–3363. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raziorrouh B, et al. Virus-specific CD4+ T cells have functional and phenotypic characteristics of follicular T-helper cells in patients with acute and chronic HCV infections. Gastroenterology. 2016;150(3):696–706.e3. doi: 10.1053/j.gastro.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Meissner EG, et al. Rapid changes in peripheral lymphocyte concentrations during interferon-free treatment of chronic hepatitis C virus infection. Hepatol Commun. 2017;1(7):586–594. doi: 10.1002/hep4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Missale G, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98(3):706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze Zur Wiesch J, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209(1):61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raziorrouh B, et al. Inhibitory molecules that regulate expansion and restoration of HCV-specific CD4+ T cells in patients with chronic infection. Gastroenterology. 2011;141(4):1422–1431, 1431.e1. doi: 10.1053/j.gastro.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Hale JS, et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38(4):805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boettler T, et al. Exogenous OX40 stimulation during lymphocytic choriomeningitis virus infection impairs follicular Th cell differentiation and diverts CD4 T cells into the effector lineage by upregulating Blimp-1. J Immunol. 2013;191(10):5026–5035. doi: 10.4049/jimmunol.1300013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinstry KK, et al. Effector CD4 T-cell transition to memory requires late cognate interactions that induce autocrine IL-2. Nat Commun. 2014;5:5377. doi: 10.1038/ncomms6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeannet G, Boudousquié C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A. 2010;107(21):9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Utzschneider DT, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45(2):415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Locci M, et al. Human circulating PD-1+CXCR3–CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitenshtein A, et al. CEACAM1-mediated inhibition of virus production. Cell Rep. 2016;15(11):2331–2339. doi: 10.1016/j.celrep.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denton AE, et al. Type I interferon induces CXCL13 to support ectopic germinal center formation. J Exp Med. 2019;216(3):621–637. doi: 10.1084/jem.20181216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Havenar-Daughton C, et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci U S A. 2016;113(10):2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlach JT, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117(4):933–941. doi: 10.1016/S0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 26.Ulsenheimer A, et al. Detection of functionally altered hepatitis C virus-specific CD4 T cells in acute and chronic hepatitis C. Hepatology. 2003;37(5):1189–1198. doi: 10.1053/jhep.2003.50194. [DOI] [PubMed] [Google Scholar]
- 27.Lucas M, et al. Tracking virus-specific CD4+ T cells during and after acute hepatitis C virus infection. PLoS One. 2007;2(7):e649. doi: 10.1371/journal.pone.0000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossi C, et al. Hepatitis C virus reinfection after successful treatment with direct-acting antiviral therapy in a large population-based cohort. J Hepatol. 2018;69(5):1007–1014. doi: 10.1016/j.jhep.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Bailey JR, Barnes E, Cox AL. Approaches, progress, and challenges to hepatitis C vaccine development. Gastroenterology. 2019;156(2):418–430. doi: 10.1053/j.gastro.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoukry NH. Hepatitis C vaccines, antibodies, and T cells. Front Immunol. 2018;9:1480. doi: 10.3389/fimmu.2018.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 32.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35(3):400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann AU, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-α therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 34.Neumann-Haefelin C, et al. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology. 2008;47(6):1824–1836. doi: 10.1002/hep.22242. [DOI] [PubMed] [Google Scholar]
- 35.Harvey CE, et al. Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J Leukoc Biol. 2003;74(3):360–369. doi: 10.1189/jlb.0303093. [DOI] [PubMed] [Google Scholar]
- 36.Spaan M, et al. CD4+ CXCR5+ T cells in chronic HCV infection produce less IL-21, yet are efficient at supporting B cell responses. J Hepatol. 2015;62(2):303–310. doi: 10.1016/j.jhep.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 37.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bentebibel SE, et al. ICOS(+)PD-1(+)CXCR3(+) T follicular helper cells contribute to the generation of high-avidity antibodies following influenza vaccination. Sci Rep. 2016;6:26494. doi: 10.1038/srep26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herati RS, et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci Immunol. 2017;2(8):eaag2152. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray GT, et al. Intraseason waning of influenza vaccine effectiveness. Clin Infect Dis. 2019;68(10):1623–1630. doi: 10.1093/cid/ciy770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 43.Underwood AP, et al. Understanding the determinants of BnAb induction in acute HCV infection. Viruses. 2018;10(11):E659. doi: 10.3390/v10110659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestka JM, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104(14):6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amir el-AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bengsch B, et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48(5):1029–1045.e5. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 55.Fofana I, et al. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology. 2012;143(1):223–233.e9. doi: 10.1053/j.gastro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roederer M, Nozzi JL, Nason MC. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79(2):167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.