Abstract
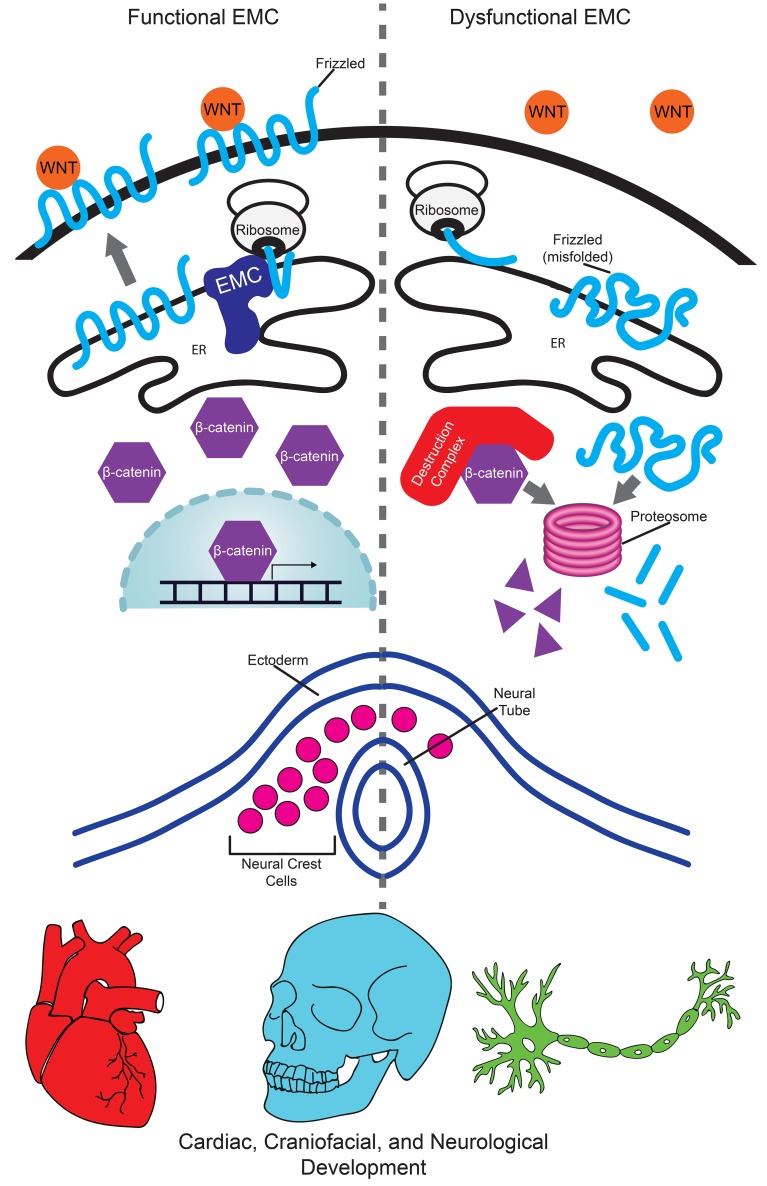
Multipass membrane proteins have a myriad of functions, including transduction of cell-cell signals, ion transport, and photoreception. Insertion of these proteins into the membrane depends on the endoplasmic reticulum (ER) membrane protein complex (EMC). Recently, birth defects have been observed in patients with variants in the gene encoding a member of this complex, EMC1. Patient phenotypes include congenital heart disease, craniofacial malformations, and neurodevelopmental disease. However, a molecular connection between EMC1 and these birth defects is lacking. Using Xenopus, we identified defects in neural crest cells (NCCs) upon emc1 depletion. We then used unbiased proteomics and discovered a critical role for emc1 in WNT signaling. Consistent with this, readouts of WNT signaling and Frizzled (Fzd) levels were reduced in emc1-depleted embryos, while NCC defects could be rescued with β-catenin. Interestingly, other transmembrane proteins were mislocalized upon emc1 depletion, providing insight into additional patient phenotypes. To translate our findings back to humans, we found that EMC1 was necessary for human NCC development in vitro. Finally, we tested patient variants in our Xenopus model and found the majority to be loss-of-function alleles. Our findings define molecular mechanisms whereby EMC1 dysfunction causes disease phenotypes through dysfunctional multipass membrane protein topogenesis.
Keywords: Development, Genetics
Keywords: Cardiovascular disease, Embryonic development, Monogenic diseases
Introduction
Multipass membrane proteins cross lipid bilayers multiple times with specific topologies essential for their function. The multipass membrane proteins that contain a lumenal N-terminus require the endoplasmic reticulum (ER) membrane protein complex (EMC) to overcome the energetic barrier necessary for initial topogenesis and membranous insertion. Multiple α-helical polypeptide chains stabilize the transmembrane regions within the membrane. These α-helical chains and the protein domains residing intra- or extracellularly allow these proteins to perform specialized signal transduction between cells and their environment.
The EMC is conserved from yeast to vertebrates, acting as a molecular chaperone for protein folding and membrane insertion (1–3). In invertebrate systems, this complex is essential for the proper synthesis, folding, and localization of several transmembrane proteins, including acetylcholine receptors and rhodopsin (4, 5). In humans, the 10 subunits of the EMC (EMC 1–10) are together sufficient to directly insert multipass transmembrane proteins into their membranous context, a finding determined in both cellular and cell-free systems (3, 6).
Interestingly, in studies of patients with various congenital malformations, geneticists have identified multiple alleles of EMC1 (ER membrane protein complex subunit 1). Initially, a variant was identified in a patient with retinitis pigmentosa (7). Subsequently, additional mutations have been identified in cohorts of young patients exhibiting combined neurodevelopmental, visual, and craniofacial abnormalities (8, 9). In trio exome sequencing studies of congenital heart disease (CHD), mutations in EMC1 were associated with cardiac pathologies, primarily aortic outflow abnormalities (10, 11). Of the many variants identified (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI129308DS1), half are missense mutations in which the impact on protein function is predicted but lacks experimental proof of pathogenicity. To this end, a vertebrate model for EMC1 dysfunction is crucial for translating our knowledge of EMC function into an understanding of a disease mechanism that can lead to such varied phenotypes.
In our high-throughput Xenopus tropicalis model, emc1 depletion led to multiple phenotypes that relate to the neural crest cell (NCC) lineage and other phenotypes that appear independent of NCCs. NCCs are a multipotent population of cells unique to vertebrates that originate early in development. These cells delaminate from the neural plate border and migrate in stereotyped patterns to diverse destinations within the developing embryo. NCCs then differentiate into a multitude of cell types, including chondrocytes, adipocytes, neurons, glia, and melanocytes, depending on their microenvironment (12, 13). Decades of research have revealed an intricate gene regulatory network that defines the induction, maintenance, migration, and subsequent differentiation of NCCs (13–18). Bone morphogenetic proteins, fibroblast growth factors, and WNTs have a demonstrated role in multiple steps of NCC function by regulating the expression of neural plate border specifiers (Msx1/2, Pax3/7, Dlx3/5) and neural crest specifiers (Snail1/2, Sox8/9/10, FoxD3, AP-2, Twist) that result in the proper migration and differentiation of this unique cell population (19–24).
Here, we demonstrate that depletion of emc1 leads to NCC dysfunction via the WNT pathway. Consistent with patient phenotypes and defects in the neural crest, Xenopus embryos depleted of emc1 have craniofacial abnormalities and alterations in the cardiac outflow tract. Our data are consistent with a model in which emc1 depletion diminishes WNT signaling, possibly via the Fzd receptor that is essential for NCC development. We also detected a neurological weakness that may be due to contributions of NCCs to the peripheral nervous system and/or misfolding of the acetylcholine receptor that is essential for neurotransmission. Finally, we exploited our Xenopus model to demonstrate that the identified patient variants are indeed pathogenic to function in the majority of cases. Our results illuminate the cellular and molecular basis for human disease phenotypes due to EMC dysfunction.
Results
Craniocardiac phenotypes with emc1 depletion.
Of the structural anomalies in patients with emc1 alleles, cardiac and craniofacial abnormalities are among the most prominent (8, 10, 11). We sought to test for phenocopy in Xenopus via emc1 depletion. Xenopus is an ideal model for this inquiry because gene dosages can be titrated to test the consequences of gain and loss of function. In the case of cardiac development, Xenopus has a septated atrium and cardiac trabeculations, allowing for closer human modeling compared with other aquatic models, and it is much higher throughput and less costly than mammalian models.
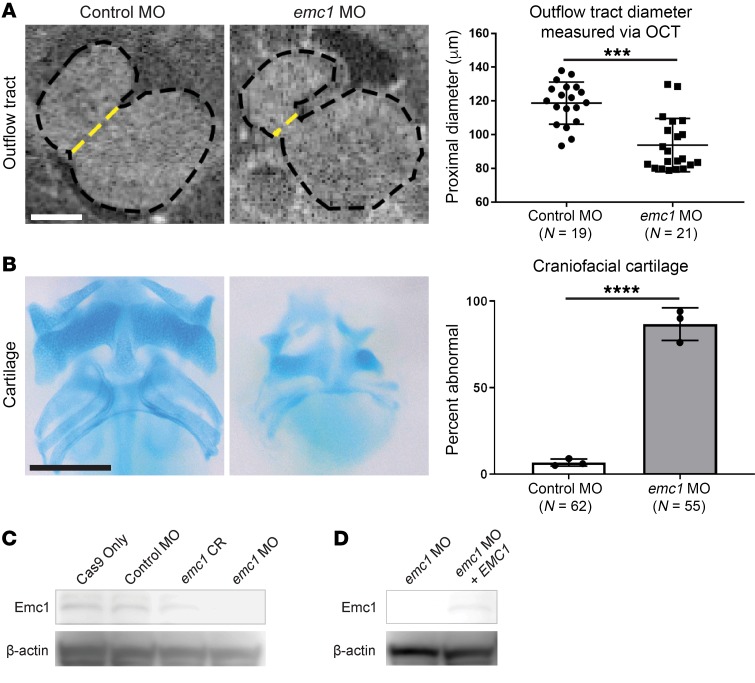
To begin, we tested whether depletion of emc1 using morpholino oligonucleotides (MOs) caused cardiac dysmorphology that could be related to the phenotypes seen in CHD patients with emc1 mutations. Remarkably, morphant embryos had narrower cardiac outflow tracts compared with controls (Figure 1A). Using optical coherence tomography, an imaging modality similar to ultrasound but using light instead of sound (25, 26), we could readily detect a change in the diameter of the outflow tract from 120 μm (n = 19, SD = 12 μm) in control embryos to 90 μm (n = 21, SD = 16 μm) in emc1 morphants.
Figure 1. Phenotypic assessment of Emc1 loss of function in Xenopus reveals craniofacial and cardiac dysmorphology.
1-cell–stage embryos were injected with either standard control MO or emc1 MO and phenotypically assessed at stage 45. (A) Representative images and measurements of 3 replicates of stage 45 control MO (n = 19) and emc1 MO (n = 21) embryo outflow tract morphology imaged with OCT imaging (dotted yellow line indicates measured diameter). Scale bar: 100 μm. (B) Representative images and percentages of 3 replicates of stage 45 control MO (n = 62) and emc1 MO (n = 55) embryo craniofacial cartilage stained with Alcian blue. Scale bar: 250 μm. (C) Immunoblot of pooled (n = 20) Emc1 protein in control and emc1 knockout/knockdown embryos. (D) Immunoblot of pooled (n = 20) Emc1 protein in emc1 knockdown and EMC1 rescued emc1 knockdown embryos. ****P < 0.0001, ***P < 0.0005 by (A) Student’s t test or (B) Fisher’s exact test. Bars indicate mean and SD.
Next, we examined craniofacial morphology. Using Alcian blue, which stains the cartilage elements of the tadpole head, we detected a striking deficit in the craniofacial skeleton. Each element of the craniofacial cartilage — mandibular, hyoid, and branchial cartilage elements — appeared equally lost in 87% (n = 55, SD = 9%) of morphant embryos compared with only 7% (n = 62, SD = 2%) of controls (Figure 1B). In addition, we noticed that morphants had an abnormal pigment cell morphology (Supplemental Figure 1). We confirmed these striking phenotypic abnormalities via phenocopying with a CRISPR-based knockout of emc1 (Supplemental Figure 2A). We assessed the efficacy of MO- and CRISPR-based approaches through immunoblotting for protein levels in morphant and crispant embryos compared with controls. Both methods reduced Emc1 protein levels compared with controls (Figure 1C). We also tested the specificity of the anti–Emc1 antibody by coinjecting wild-type human EMC1 mRNA along with the MO and demonstrating an increase of signal via immunoblot (Figure 2D). CRISPR efficacy was also confirmed through assessing the indels introduced in individual F0 mutants through Sanger sequencing and tracking of indels by decomposition (TIDE) analysis (ref. 27 and Supplemental Figure 2B). Given phenotypes in the cardiac outflow tract, craniofacial cartilage, and pigment cells, we considered NCCs as a potential unifying explanation for the abnormalities in each of these tissues based on their known contributions (28–30).
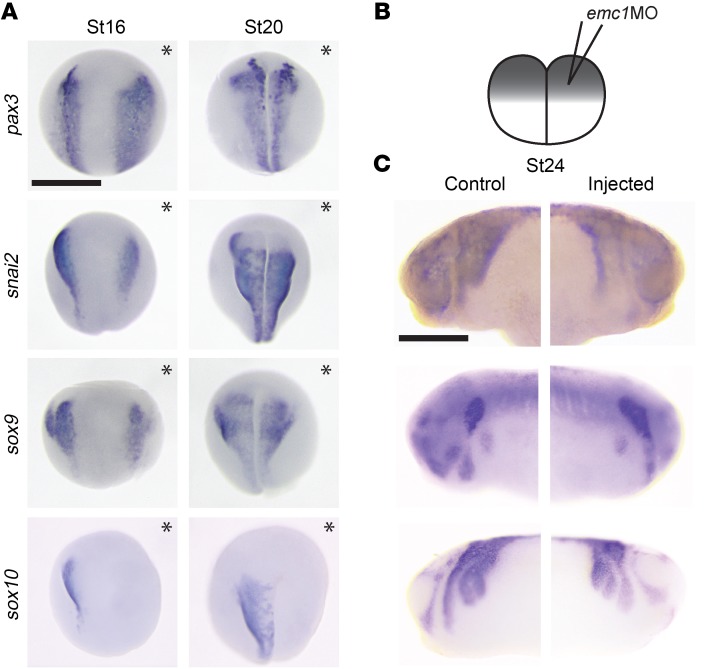
Figure 2. emc1 depletion results in neural crest gene expression abnormalities early in development.
Embryos were injected into 1 cell at the 2-cell stage with emc1 MO followed by interrogation of neural crest markers via WISH. (A) WISH for markers of NCC cell lineage revealed that expression of earlier markers (pax3, snai2, and sox9) was present at expected developmental stages (stages 16 and 20 shown) but displayed abnormal distribution; the later marker sox10 was almost entirely lost (n = 45 per marker per stage done in 3 replicates; injected halves of embryos indicated by asterisks). Scale bar: 500 μm. (B) Schematic of the experimental setup, in which injection of MO into 1 cell of a 2-cell embryo allowed for 1 side of the embryo to develop under the effects of the MO injection; the other half served as an internal control for developmental phenotypes. (C) Markers showed abnormal distribution at later stages (stage 24 shown), suggesting mispatterning of the embryonic rostrum (n = 45 per marker per stage done in 3 replicates). Scale bar: 500 μm.
emc1 is essential for NCCs.
After neurulation, NCCs delaminate from the neural plate border and undergo an extensive pattern of migration in response to environmental cues. After they arrive in their correct locations, they can differentiate into an extraordinary set of cell types, including those affected in our Xenopus emc1 depletion model. To assess NCCs at various time points in development, we used whole mount in situ hybridization (WISH) to evaluate a battery of NCC markers that define neural plate border formation, NCC specification, and NCC migration. For these experiments, we exploited an advantage of Xenopus where injection of 1 cell at the 2-cell stage can lead to targeting of either the left or right side of the embryo, providing the unaffected side as an internal control that can be easily established by coinjecting fluorescent tracers. In this manner, we depleted emc1 and examined early markers of the neural plate border and NCCs that appeared to be present at the expected stages of development (Figure 2). Although NCC markers were present, there were clear abnormalities including reduced signal. Impressively, at stages 16 and 20, sox10 was dramatically reduced (Figure 2A). At later stages, sox10 seemed to recover, although clear differences remained between emc1-depleted and control sides. To evaluate whether the early loss of NCC markers could be due to an increase in cell death, we used TUNEL (Supplemental Figure 4). Comparing control MO and emc1 MO embryos injected in 1 cell at the 2-cell stage, we did not detect a significant difference in positive TUNEL outside of the general variability seen in cell death in embryos. We detected TUNEL in the dorsal midline as previously described (31), and a positive control, nup85, led to a marked increase in TUNEL signal. Our previous work suggested that the nucleoporin encoded by nup85 is important for the survival of a subset of cell types (32). From these studies, we conclude that NCC specification, maintenance, migration, and differentiation are altered in emc1-depleted embryos.
emc1 depletion affects WNT signaling.
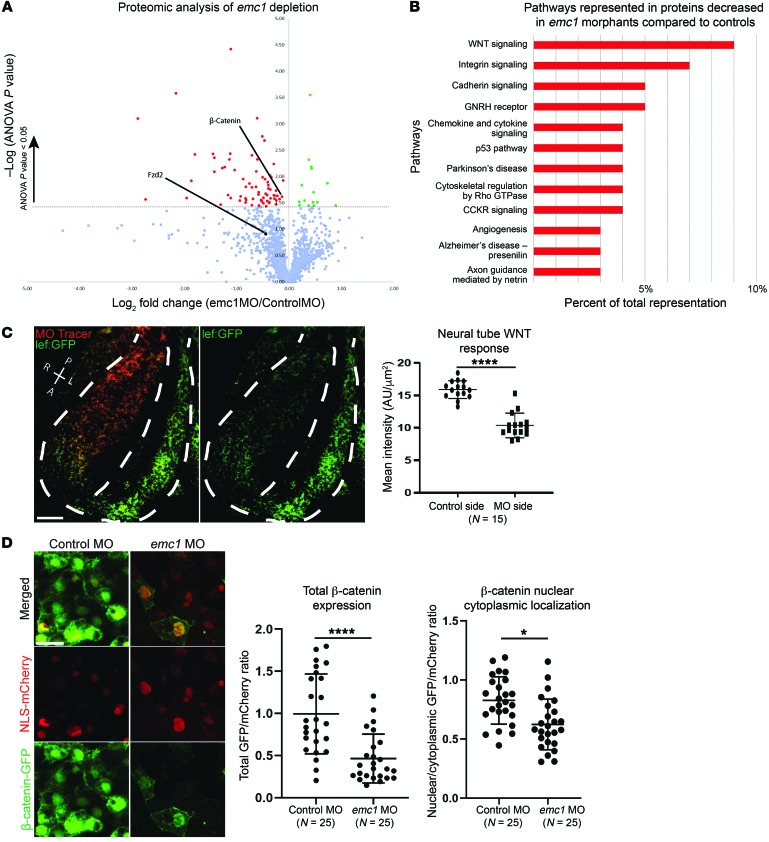
The EMC and EMC1 in particular are essential for numerous multipass transmembrane proteins; therefore, it is uncertain which target proteins affect neural crest development. To identity a potential target, we took an unbiased approach to analyze the global landscape of proteins in emc1 morphant embryos compared with controls. We performed label-free quantitative mass spectrometry (LFQMS) of protein lysates from stage 24 emc1 morphants and controls, and detected many changes in protein levels that corresponded to developmentally important processes. Using pathway analysis, WNT, integrin, and cadherin signaling were decreased in emc1 morphant embryos compared with controls (Figure 3, A and B, and Supplemental Table 2). In particular, we were struck by the decrease in β-catenin in this data set because β-catenin serves as the intracellular messenger for canonical WNT signaling (33, 34). Given that the WNT pathway is instrumental in the activation and maintenance of factors necessary for NCC development (35–37), we sought to investigate this pathway in embryos depleted of emc1. To accomplish this, we first assessed WNT response in the Xtr.Tg(WntREs:dEGFP)Vlemx transgenic line, which has 7 copies of a TCF/LEF1 DNA binding sequence driving GFP expression (38, 39). The neural tube has strong WNT activity at early stages and is easily scored for GFP expression in our 1 of 2 cell injected embryos (40). In the half of the neural tube depleted of emc1, there was a clear decrease in the WNT-responsive GFP signal (Figure 3C). Next, we assayed β-catenin levels and nuclear/cytoplasmic localization of β-catenin. In the WNT pathway, both β-catenin levels and the nuclear translocation of β-catenin are essential for signal transduction. To visualize β-catenin in cells, we exogenously delivered a GFP-tagged β-catenin ratiometrically with an NLS-mCherry and compared the relative amount of GFP versus mCherry signal and the relative nuclear localization of β-catenin and NLS-mCherry. We saw a mild reduction in the nuclear/cytoplasmic ratio of β-catenin, but we detected a dramatic reduction in the total GFP signal compared with mCherry, suggesting that β-catenin levels were lower overall in emc1-depleted embryos, a result consistent with our proteomics data (Figure 3D). From these data, we conclude that WNT signaling was reduced in emc1-depleted embryos based on a reporter gene assay and a reduction in β-catenin levels observed through our unbiased proteomics. However, Emc1 regulates multipass membrane proteins, so we suspected that the reduction in β-catenin was indirect. Although a number of multipass membrane proteins are important for neural crest migration (41–43), a role for both NCC migration and WNT signaling makes the Frizzled (Fzd) receptor a strong candidate (44–46). Although Fzd proteins did not appear in our proteomics data set, we could identify Fzd2 if we reduced the stringency, allowing for proteins to be identified based on only a single unique peptide rather than 2 or more unique peptides (Figure 3A, black dot). This may be due to the relative low abundance of the Fzd proteins and similarities in peptide sequences across the Fzds. We saw a consistent reduction in Fzd2 in our proteomics data set, although this did not reach statistical significance. However, based on these data, we sought to investigate Fzd proteins more carefully.
Figure 3. Proteomic analysis of emc1 knockdown uncovers affected developmental pathways including WNT signaling.
One-cell–stage embryos were injected with either standard control MO or emc1 MO, and LFQMS was carried out on stage 24 embryos. (A) Plot of mean protein levels from 3 biological replicates of 20 pooled emc1 morphants compared with control morphants as determined via LFQMS (statistically significantly increased proteins are shown in green, statistically significantly decreased proteins are shown in red, Fzd2 is shown as a black dot since it was only identified by one unique peptide sequence as opposed to the other proteins in the graph). (B) Plot of gene ontology terms for statistically significantly decreased proteins with human homologs displays enrichment for a subset of signaling pathways. (C) Representative image and quantitation of WNT signaling visualized in a transgenic Xenopus WNT reporter line as a comparison between mean fluorescence of emc1 MO-injected versus uninjected sides of the neural tube (white dotted line shows outline of neural tube and division between injected and uninjected sides). P, posterior; A, anterior; R, right; L, left. Scale bar: 100 μm. (D) Representative image and quantitation of β-catenin subcellular localization visualized in Xenopus neural tubes as a comparison between emc1 MO-injected versus control MO-injected embryos normalized to NLS-mCherry localization. Statistically significant protein changes assessed via ANOVA (A) with red and green points indicating P < 0.05. Scale bar: 10 μm. ****P < 0.0001, *P < 0.05 by Student’s paired t test (C) and Student’s t test (D). Bars indicate mean and SD.
EMC1 depletion affects multipass transmembrane proteins including Fzd.
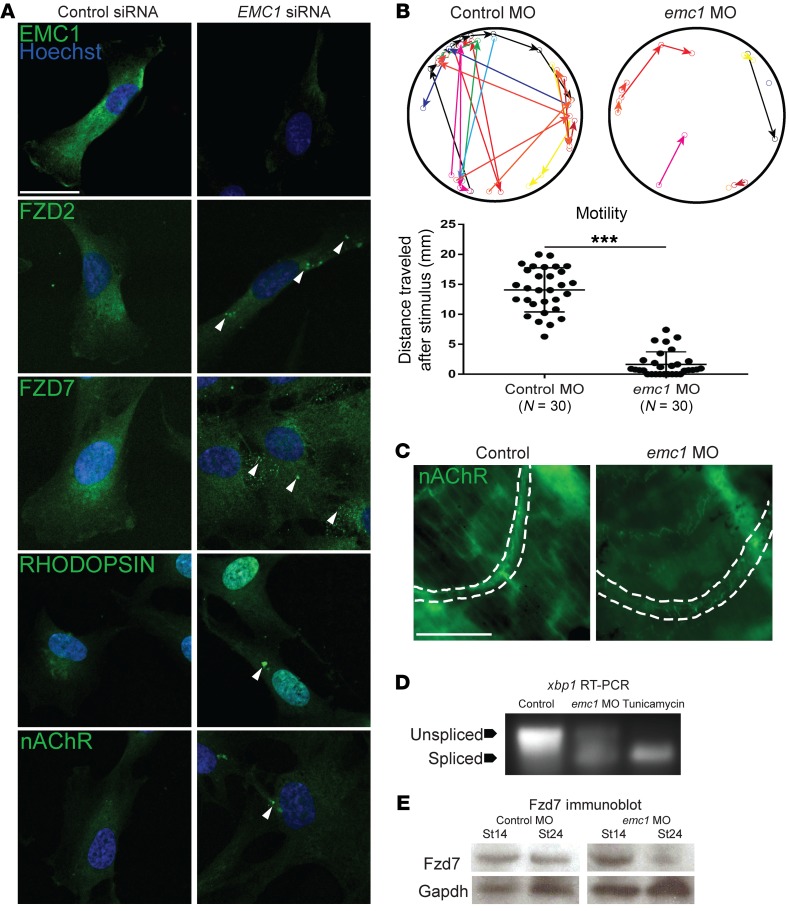
In WNT signaling, the Fzd receptor is a multipass transmembrane protein with a lumenal N-terminus that is essential for transducing signal from the WNT ligand to the β-catenin destruction complex that leads to the stabilization of β-catenin. β-Catenin then enters the nucleus to activate WNT responsive genes (33, 34). Since our unbiased proteomics suggested an effect on WNT signaling in the context of emc1 depletion, we sought to determine the relation between EMC1 and the Fzd receptor. We depleted EMC1 in hTERT retinal pigmented epithelial (RPE) cells using siRNA and assayed the localization of FZD2 and FZD7 by immunofluorescence. Compared with control siRNA-transfected RPE cells, EMC1 siRNA-transfected cells had an altered punctate localization of FZD2 and FZD7 reminiscent of previously observed misfolded proteins in the context of EMC disruption (ref. 6 and Figure 4A). We also expected that emc1 depletion should lead to misfolding and mislocalization of additional multipass transmembrane proteins. For example, we noted that multiple patients with emc1 alleles had visual defects including retinitis pigmentosa. One cause of retinitis pigmentosa is variants in the rhodopsin gene, a multipass transmembrane G protein–coupled receptor that is exceptionally sensitive to light. In EMC1-depleted cells, rhodopsin expression appeared as puncta rather than diffusely localized (Figure 4A). In addition, we also tested localization of the nicotinic acetylcholine receptor (nAChR), which has previously been shown to be affected in invertebrate models of EMC depletion (5). The dysfunction of this receptor could also be a compelling reason for the neurological weakness observed in patients with EMC1 variants. In RPE cells depleted of EMC1, nAChR also appeared to be mislocalized (Figure 4A).
Figure 4. Knockdown of EMC1 in human RPE cells and Xenopus affects multiple transmembrane proteins and embryo motility and activates ER stress.
(A) Immunofluorescence antibody labeling of EMC1 revealed a decrease in its expression after EMC1 siRNA treatment as compared with control siRNA treatments. Multipass membrane proteins (RHODOPSIN, nAChR, FZD2, FZD7) were abnormally localized (n = 10 high power fields per marker per condition done in 3 replicates). Scale bar: 20 μm. (B) Sample traces and measurement of control morphant (n = 30) and emc1 morphant (n = 30) tadpole movement over 10 seconds after stimulation (different colors differentiate distinct tadpoles) over 3 replicates. (C) Labeling of nAChR in the proximal tail of emc1-depleted stage 45 tadpoles showed sparse and less intense expression as compared with control counterparts. Scale bar: 50 μm. (D) Splicing assay for xbp1 in pooled (n = 30 per condition) stage 24 Xenopus embryos displayed increased splicing with emc1 MO depletion compared with control embryos repeated in 4 biological replicates. Tunicamycin treatment acted as positive control. (E) Immunoblotting for Fzd7 showed similar levels of Fzd7 in pooled (n = 30 per stage per condition) emc1 morphants as compared with control morphants at stage 14, but a marked decrease in levels at stage 24. ***P < 0.0005 by Student’s t test. Bars indicate mean and SD.
In fact, during the course of our experiments, we noted that Xenopus embryos depleted of emc1 did not move as vigorously as control tadpoles. To quantitate this, we measured the distance a tadpole moved after stimulation with a thin plastic pipette tip. emc1 morphants moved considerably shorter distances than their stage-matched siblings in the control group (Figure 4B). To test whether this might be due to abnormal localization of the nAChR in Xenopus, we performed immunofluorescence for nAChR in tadpole tails, examining the locations where muscle contraction impulses were generated (Figure 4C). In control embryos, the nAChR signal was a sharp arc across the somitic muscle, whereas in emc1-depleted embryos, the nAChR signal appeared weaker and more discontiguous. From these results, we conclude that nAChR expression was reduced, contributing to the reduction in neurological muscle activity in tadpoles depleted of emc1.
Our results in human and Xenopus cells indicate that multipass membrane proteins were mislocalized and reduced in amount because of emc1 depletion. Under conditions of ER stress due to excessive misfolded proteins, the unfolded protein response (UPR) may lead to their degradation. In this context, Ire1 (inositol-requiring enzyme1) is activated, and its ribonuclease function leads to the atypical RNA splicing of xbp1. This splice product of xbp1 activates ER-associated degradation (ERAD) genes. Assaying the alternative isoform of xbp1 is an effective and straightforward way to test activation of the UPR that could lead to protein degradation. We performed RT-PCR flanking a region excised in the atypical isoform and compared emc1 morphants to control embryos (Figure 4D). In morphant embryos, we detected an additional shorter PCR band, similar to control embryos treated with tunicamycin that induced the UPR. By running these PCR products on a capillary gel, which has single base pair resolution, we could confirm that the spliced product was indeed increased in emc1 morphants and tunicamycin-treated embryos compared with controls and the predicted 26 base pair change in PCR product length (Supplemental Figure 3). From these data, we conclude that emc1-depleted embryos had an increase in the UPR that could lead to the degradation of multipass transmembrane proteins through ERAD.
To verify this possibility, we hypothesized that if Fzd proteins were being degraded downstream of increased UPR activity and subsequent ERAD, then blocking proteasomal degradation that mediates this protein clearance would result in a retention of otherwise lost Fzd protein. We tested this by using MG132 to block proteasomal degradation while blocking protein synthesis to exclude any changes occurring via changes in synthesis rates. Although Fzd2 levels in control cells remained relatively unchanged over this time course experiment, Fzd2 levels increased during proteasomal blockade when emc1 was depleted, further suggesting that the UPR and downstream ERAD could be functioning to clear misfolded Fzd (Supplemental Figure 5). Additionally, returning to our neural crest phenotype and WNT signaling in Xenopus embryos, the UPR should lead to reduced levels of the Fzd receptor. Indeed, by Western blot, Fzd7 protein was reduced in stage 24 morphants compared with controls (Figure 4E). In this case, we looked at Fzd7 levels because the role of Fzd7 has been well characterized in the development of NCCs (35).
Downstream activation of WNT can rescue NCC abnormalities.
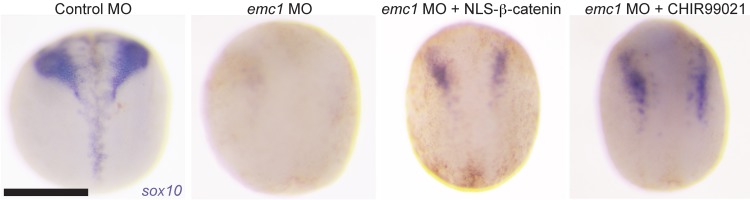
If the dysfunction seen in the NCCs stemmed from a reduction in WNT signaling, we hypothesized that activating WNT signaling downstream of transmembrane effectors should rescue the loss of the NCC marker sox10 in emc1 morphant embryos. We tested this through injection of mRNA encoding β-catenin to which a nuclear localization signal had been added (47). Excessive activation of WNT signaling causes a second axis to form (creating 2-headed embryos); thus, we titrated the dose of mRNA to minimize this effect while attempting to rescue the emc1 depletion phenotype. In 40% (n = 45, SD = 9%) of embryos coinjected with emc1 MO and NLS–β-catenin mRNA, we detected sox10 expression compared with 7% (n = 45, SD = 7%) of those injected with emc1 MO alone (Figure 5). As an alternative strategy, we also used the CHIR 99021 GSK3β inhibitor, which prevents β-catenin degradation and activates the WNT pathway downstream of the Fzd receptor. In emc1 morphant embryos incubated from stage 12 to 14 in media containing CHIR 99021, we detected a robust expression of sox10 in 33% (n = 30, SD = 6%) of embryos in contrast to 11% (n = 45, SD = 3%) of emc1 morphant embryos incubated from stage 12 to 14 in DMSO-containing media as a control (Figure 5).
Figure 5. Neural crest marker expression rescued in emc1 morphants by modulating β-catenin.
Expression of sox10 in control MO-injected embryos raised in media containing DMSO showed stereotyped expression of this marker at stage 20; loss of sox10 expression in stage 20 embryos was observed after injection with emc1 MO. sox10 expression was partially rescued by injection of NLS-fused β-catenin mRNA or the small molecular GSK3β inhibitor CHIR 99021. Three replicates of 15 embryos were analyzed for each condition. Scale bar: 500 μm.
Human-derived NCCs require EMC1 for proper development.
Our results indicate that depletion of emc1 leads to dysfunction of the EMC and misfolding of multipass transmembrane proteins, activating the UPR and protein degradation. In the case of the neural crest, decreased WNT signaling leads to the dysregulation of NCCs in Xenopus. To further relate our findings in Xenopus to observed human pathologies, we sought to translate our findings into a human cell model.
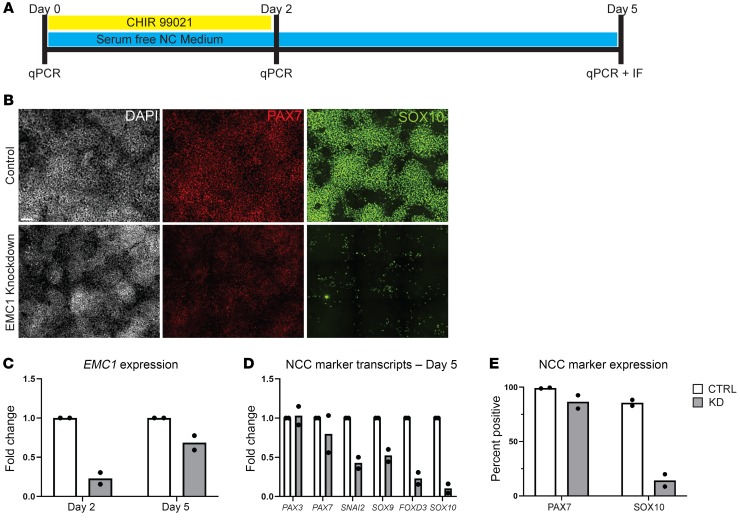
Human embryonic stem cell–derived (hESC-derived) NCCs have recently emerged as a powerful tool for modeling NCC specification and function (48). This human NCC model provides synchronized NCC development, enabling us to assess human NCC biology and directly test our insights into disease mechanism in a human system. Additionally, this model faithfully recapitulates NCC development as observed in multiple organisms, including the hierarchical expression of genes during neural crest formation, as well as the requirement of WNT/β-catenin signaling in NCC induction (49–53). We used a modified version of the standard protocol (48), which provides a transient WNT activation (2 days of CHIR 99021 treatment) in the initial phase of 5 days of culture (Figure 6A and ref. 54). As a first step, we characterized the expression of EMC1 in this model via qPCR. We found that EMC1 expression remained present during WNT activation and was maintained in hNCCs (Supplemental Figure 6). To assess the molecular events regulated by EMC1 during human neural crest formation, we used siRNA-mediated knockdown of EMC1 and monitored multiple markers associated with the progression from hESC to hNCC.
Figure 6. Effects of EMC1 depletion in human embryonic stem cell –derived NCCs.
(A) Schematic of the induction protocol used to generate hNCCs in which hESCs were exposed to CHIR 99021 for 2 days in serum-free medium prior to withdrawal of this agent and continued culture for 3 additional days. (B) Immunofluorescent antibody staining for NCC markers PAX7 and SOX10 at day 5 in EMC1 siRNA-treated cells as compared with control siRNA-treated cells performed in 2 separate biological replicates. Scale bar: 100 μm. (C) qPCR analysis at day 2 and day 5 showed a decrease in transcripts of EMC1 performed in 2 separate biological replicates. (D) qPCR analysis at day 5 performed in 2 separate biological replicates showed decreased levels of several NCC marker transcripts (PAX7, SNAI2, SOX9, FOXD3, SOX10), although other markers appeared to be unaffected (PAX3). (E) Quantified percentage of cells expressing each PAX7 and SOX10 protein performed in 2 separate biological replicates.
First, we assessed the knockdown efficiency of EMC1 siRNA via immunofluorescence and qPCR after the first 2 days and then at the end of the 5-day culture (Figure 6C and Supplemental Figure 7, A and B). We further confirmed the loss of FZD7 in these cells by immunofluorescence (Supplemental Figure 7, A and B). Next, we assessed the expression of PAX7 and SOX10 using immunofluorescence at the end of the hNCC differentiation protocol (Figure 6, B and E). Both PAX7 and SOX10 seemed noticeably reduced in the EMC1 siRNA samples compared with control siRNA. A large proportion of cells still expressed PAX7, although at an apparently much reduced level. Instead, SOX10 appeared to be expressed in very few cells (Figure 6, B and E). To extend our analysis to a wider collection of NCC markers, we performed RT-qPCR for PAX3, PAX7, SNAI2, SOX9, FOXD3, and SOX10 on day 5 samples treated with control or EMC1 siRNAs (Figure 6D). The mRNA expression of PAX3 seemed unaffected, and that of PAX7 appeared to be only slightly reduced by EMC1 siRNA treatment. A stronger effect was seen for the mRNA expression of the NCC markers SNAI2, SOX9, FOXD3, and SOX10 in EMC1 siRNA samples. These results indicate that EMC1 plays a critical role in neural crest development, from the expression of early markers, acquisition of mature markers, maintenance, and subsequent end tissue differentiation.
Testing pathogenicity of patient variants.
With a plausible pathogenesis mechanism that appeared consistent between Xenopus and human NCC models, we hoped to address the pathogenicity of patient variants. Many of the identified alleles for EMC1 are missense with in silico predicted loss of function, and our vertebrate animal model offers a platform to test these predictions. Previous work has identified a series of alleles for EMC1 (Supplemental Table 1). In patients with CHD, consisting primarily of cardiac outflow tract abnormalities, variants were all heterozygous and missense. However, in the cases of skeletal abnormalities with neurodevelopmental disorders and/or vision deficits, patients all bore homozygous variants save for a case in which the allele for EMC1 was again a missense mutation. Therefore, testing for loss of function of EMC1 alleles offered the potential to reveal genotype-phenotype correlations that may prove invaluable for clinical interpretation of variant burden in patients. We thus sought to determine whether pathogenicity could be tested in our Xenopus model.
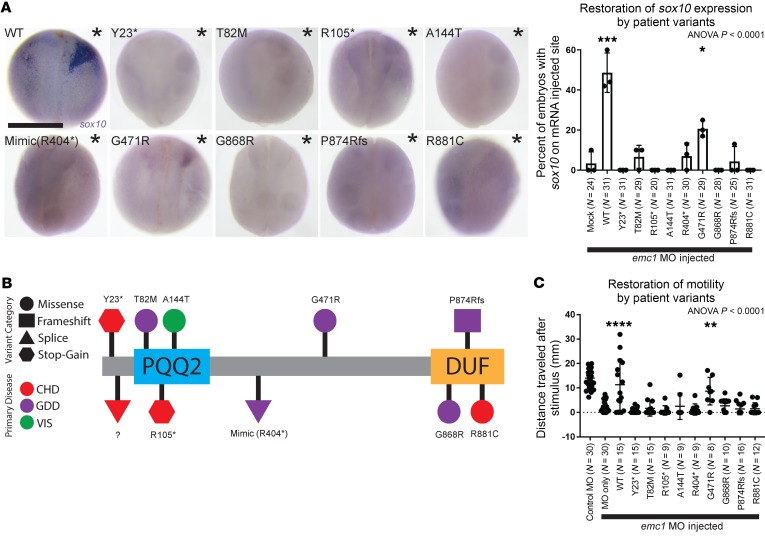
To accomplish this, we assayed the expression of sox10 as a marker of effects on NCCs and tadpole motility as a marker of broader neurodevelopmental function. For NCC assessment, we depleted emc1 throughout the embryos via MO-mediated knockdown at the 1-cell stage, and then introduced either wild-type mRNA or EMC1-variant mRNA into 1 of the 2 cells at the 2-cell stage. We anticipated that the wild-type mRNA and variants without functional detriment would rescue the phenotype on 1 half of the embryo. To mimic a splice site mutation c1212+1G>A, we introduced a stop codon immediately after exon 11 because experimental evidence has suggested that the variant c.1212+1G>A results in premature protein truncation (9). The remaining splice site mutation (c.96-2A>T) was not tested because we were unsuccessful in determining the resultant change in transcript via conventional in vitro methods. Alternatively, for motility assessment, we coinjected the emc1 MO with either wild-type or EMC1-variant mRNA in 1-cell–stage embryos. For both assays, only mRNA from the wild-type and c.1411G>C variant appeared to consistently rescue sox10 expression, tadpole movement (Figure 7), and the nAChR signal within the neuromuscular junctions of tadpole tails (Supplemental Figure 8). Based on this result, we would predict that the c.1411G>C allele retains function or represents a relatively mild allele as compared with the others. Interestingly, the patient phenotype with this allele was not notably different from other patients in this EMC1 cohort, but our results would suggest that other alleles may be worth exploring to explain the disease phenotype. Finally, it appears that a genotype-phenotype correlation may exist where compound heterozygosity leads to skeletal and neurodevelopmental abnormalities and heterozygosity leads to CHD, a hypothesis worth testing as additional alleles are identified.
Figure 7. Xenopus model of emc1 loss of function provides a platform for testing patient variant pathogenicity.
Injection of wild-type EMC1 mRNA and to a lesser extent p.Gly471Arg (c.1411G>C) variant mRNA restored sox10 expression and tadpole movement in emc1-depleted embryos, whereas the mRNA of all other variants did not. (A) Representative images and percentages of embryos with sox10 expression after injection with emc1 MO at the 1-cell stage followed by injection with wild-type (n = 31) or variant EMC1 mRNA (n = 31 for Y23*, 29 for T82M, 20 for R105*, 31 for A144T, 30 for R404*, 29 for G471R, 28 for G868R, 25 for P874Rfs, and 31 for R881C) in 1 cell of the 2-cell stage over 3 biological replicates (injected halves of embryos are indicated by asterisks). Scale bar: 500 μm. (B) Schematic diagram of EMC1 protein with domain annotations and locations and effects of patient variants. CHD, congenital heart disease; GDD, global developmental delay; VIS, vision impairment; PQQ2, pyrrolo-quinoline quinone redox coenzyme domain; DUF, domain of unknown function. (C) Measurement of tadpole motility after coinjection with emc1 MO and wild-type (n = 15) or variant EMC1 mRNA (n = 15 for Y23*, 15 for T82M, 9 for R105*, 9 for A144T, 9 for R404*, 8 for G471R, 10 for G868R, 16 for P874Rfs, and 12 for R881C) at the 1 cell stage over 3 biological replicates. ANOVA P values were calculated as a comparison of mock injection to all other groups (A) and MO only to all other groups (C). ****P < 0.0001, ***P < 0.0005, **P < 0.005, *P < 0.05 by post hoc Tukey’s test of multiple comparisons of mock injection to all other groups (A) and MO only to all other emc1 MO-injected groups (C). Bars indicate mean and SD.
Discussion
The importance of EMC1 on human health has been established by the multiple alleles identified in disease subjects. Also, the EMC has an established role in multipass membrane protein synthesis, folding, and insertion into cell membranes. However, a connection between these findings was lacking. In fact, given the multitude of multipass transmembrane proteins that rely on the EMC, one might expect that EMC dysfunction would lead to very early embryonic lethality rather than specific congenital malformations. However, multiple human genetic studies have associated tissue-specific human diseases to proteins that are otherwise critical for basic cell biological processes (7–11). For example, ribosomopathies lead to craniofacial phenotypes via dysfunction of the neural crest (55, 56). Nucleoporin deficiencies can lead to CHD or steroid-resistant nephrotic syndrome (32, 57, 58). EMC1 is an addition to this (far from comprehensive) list of proteins that are fundamental to the function of every cell, yet when dysfunctional can lead to tissue-specific diseases such as retinitis pigmentosa, craniofacial malformations, CHD, and neurological weakness.
In our search for a mechanism to explain these phenotypes, we recapitulated the craniofacial and cardiac phenotypes of patients with EMC1 variants in our Xenopus model. We also noted a melanocyte phenotype, which all could be explained by the diverse cellular contributions of NCCs. Gratifyingly, we detected remarkable parallels between our Xenopus neural crest results and our human NCC model derived from embryonic stem cells. In both cases, early markers of NCC development seemed altered, and the maintenance of these cells was clearly abnormal when EMC1 was depleted. In both cases, we detected a striking reduction in sox10 expression, which is critical for neural crest development (59, 60). From these results, we propose that the craniofacial and CHD defects seen in patients with EMC1 variants is due to fundamental defects in neural crest development. Importantly, beyond its importance in NCC development, SOX10 is critical for later proliferation and differentiation of nervous system Schwann cells and oligodendrocytes, which serve to myelinate neuronal axons (59, 60). Furthermore, human genetic studies have associated SOX10 loss of function with myelinopathies that have varying manifestations, possibly due to the specific location of mutations in SOX10 (61–63). Thus, an interesting possibility is a similar contribution of SOX10 loss in patients with damaging EMC1 variants, although whether this is via WNT signaling or some other mechanism is uncertain. Although one such patient was reported to have nerve conduction studies within normal limits (9), this may still provide an intriguing avenue for additional studies.
Since multiple multipass membrane proteins are necessary for NCCs, defining which upstream protein(s) affect NCCs with EMC1 depletion was far from obvious. The stringent approach of LFQMS is powerful for identifying protein changes, but the requirement for proteins to be sufficiently abundant to capture multiple unique peptide sequences and assign protein identity in this method makes it difficult to rule out changes in proteins that are not represented in the data. Our unbiased approach did identify the WNT pathway, and directed our studies to the Fzd receptor as a strong candidate, yet we cannot rule out contributions to observed phenotypes from other multipass membrane proteins (41, 42, 64–68). Interestingly, WNT signaling plays a role in multiple embryological functions, including axis formation (69) and head induction (70), yet we do not see these phenotypes in emc1 morphants. This may be due to timing. Fzd protein levels are not markedly reduced until stage 24, a time point that is critical for neural crest development, but too late for axis or head formation. Whether the perdurance of Fzd protein to this time point is due to functional maternal proteins or timing of the UPR will be an interesting avenue for future study. Although we cannot rule out the contribution of other Fzd proteins, we focused our studies on Fzd2 and Fzd7 because of the detection of decreased Fzd2 across all emc1 morphant LFQMS replicates and the previously identified role in NCC development (35). Yet, in considering which FZD proteins might be most likely to contribute to disease phenotypes downstream of EMC1 loss of function, FZD2 and FZD7 are likely candidates. Across the stages of development analyzed in our emc1 loss-of-function Xenopus model, fzd2 and fzd7 are the most highly expressed fzds, which may indicate a particular importance for Fzd2 and Fzd7 in the critical developmental signaling occurring at these developmental stages (71). Interestingly, mutations in FZD2 have been implicated as causes of omodysplasia and Robinow syndrome (72–74). The overlap in craniofacial malformation phenotypes between patients with EMC1 and FZD2 variants may further suggest the importance of FZD2 loss downstream of EMC dysfunction, especially with respect to craniofacial development.
Of note, our LFQMS pathway analysis was also enriched for cadherin and integrin signaling. These pathways may be dependent on WNT signaling but also have established roles in neural crest delamination and migration (75). Therefore, each of these 3 pathways may have played a role in the neural crest defects seen in our Xenopus and human NCC models. Our rescue of sox10 expression with β-catenin further supports the importance of WNT signaling in this process, but cadherins and integrins are likely to play an important role in NCC function in the context of EMC1 dysfunction.
Although our unbiased approach was highly effective in identifying a target for NCCs in emc1-depleted embryos, other phenotypes such as the visual and weakness phenotypes may be explained by clear candidates. In the case of retinitis pigmentosa, the multipass transmembrane protein rhodopsin was a clear candidate, and our results indicated that rhodopsin localization was affected in EMC1-depleted RPE cells. We made a similar finding for nAChR, which coupled with the neural crest phenotype could explain the neurological weakness seen in patients and our Xenopus model.
Together, our results in Xenopus and human embryonic stem cells provide plausible mechanisms for many of the phenotypes seen in our patients. This has immediate implications for patients because chemical WNT activators can rescue some but are unlikely to rescue all of their phenotypes. Importantly, we also established a robust assay in Xenopus for testing EMC1 variants for pathogenicity that could assist in the future diagnostics of patient alleles that are not clearly detrimental to function. In this assay, all but one of the previously described variants appeared to be loss-of-function alleles when tested in our Xenopus system. The exception, the c.1411G>C allele, appeared to be functional in our rescue of both the neural crest and motility. The clinical phenotype of the patient with the c.1411G>C allele was consistent with that of other patients with EMC1 alleles, but our functional testing would suggest retention of function for this variant. Although it is possible that the effects of this variant are subtler at the early developmental stages assayed in this study, other rare hemizygous and compound heterozygous variants were identified in this patient. Some of these variants may yet be relevant to patient disease and merit further study. For example, the compound heterozygous variants identified in the URB1 gene described in this patient could be the pathogenic basis of disease. These results highlight the need for functional testing of putative disease-causing variants, and Xenopus offers a robust and high-throughput platform for this endeavor. Interestingly, if we exclude the c1411G>C allele from consideration, then the mode of inheritance more closely relates to disease phenotype. Patients heterozygous for EMC1 variants have CHD, whereas patients with homozygous variants have neuromuscular and visual deficits.
In summary, our results indicate that disruption of the EMC complex can cause human disease due to WNT signaling dysfunction in the neural crest. This result is supported both by our high-throughput Xenopus model and our human NCC model derived from embryonic stem cells. Additional patient phenotypes can be explained by the role of the EMC on other multipass transmembrane proteins. We note that our proteomics approach also enriched for other pathways that might also lead to human disease, and as additional patients with EMC variants are identified, these pathways may provide a rich avenue for further inquiry.
Methods
Xenopus husbandry.
Xenopustropicalis were raised, housed, and cared for in-house. We induced ovulation and collected embryos by in vitro fertilization as previously described (76). Embryos were raised in 1/9MR + gentamicin. Staging of Xenopus tadpoles was performed according to Nieuwkoop and Faber (77).
Xenopus MOs, mRNA, and CRISPR.
Using standard protocols, we injected MO (emc1: 5′-ATGCCTTTGCTCAATTTGTTTCTGG-3′, control: 5′-CCTCTTACCTCAGTTACAATTTATA-3′, Gene Tools), fluorescent tracers, and/or mRNA into 1- or 2-cell Xenopus embryos, and assayed gene expression through WISH. We injected 10 ng MO into 1-cell–stage embryos and 5 ng MO into 1 cell of a 2-cell–stage embryo. We injected 100 pg mRNA for β-catenin–GFP and NLS-mCherry localization and rescue experiments. β-Catenin–GFP mRNA was generated from Addgene plasmid 16839 (47). NLS-ctnnb1-GFP was generated from Addgene plasmid 16838(47). NLS-mCherry mRNA was generated from Addgene plasmid 49313 (78). EMC1 mRNA was generated from NM_015047.3 in a pCSDest backbone. mRNAs were generated using the SP6 mMessage machine kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Variants were generated using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs) following the manufacturer’s instructions. For CRISPR-based experiments, CRISPR/Cas9-mediated genome editing in Xenopus tropicalis embryos was used as previously described (79). A CRISPR sgRNA targeting exon 2 of emc1 based on gene model XM_018090190.1 was designed to generate F0 knockout embryos (5′-GGAGCGCAGCAGGCGGCCCC-3′). For targeted loss-of-function experiments, 400 pg sgRNA along with 1.6 ng Cas9 (CP03, PNA Bio) in a 2nl volume were injected into 1-cell–stage embryos. We acquired Xtr.Tg(WntREs:dEGFP)Vlemx Xenopus from the National Xenopus Resource (38).
WISH.
WISH was carried out as previously described (80). Briefly, Xenopus embryos were fixed in 4% paraformaldehyde and dehydrated through washes in methanol. Embryos were then rehydrated in PBS with 0.1% tween-20. Embryos were then hybridized with digoxigenin-labeled RNA probes complementary to target genes. Embryos were then washed and blocked prior to incubation with anti–DIG-Fab fragments (Roche) overnight at 4 degrees. BM purple (MilliporeSigma) was used to visualize expression prior to post-fixation in 4% paraformaldehyde with 0.1% glutaraldehyde.
Optical coherence tomography imaging/measurements of cardiac outflow tract.
Stage 45 Xenopus embryos were immobilized and imaged using a Thorlabs Telesto 1325 nm spectral domain–OCT system. The cardiac 2D cross-sectional images were obtained as part of a volumetric scan, of which the maximal diameter of the proximal outflow tract was identified and measured using the integrated Thorlabs software.
Whole mount Alcian blue cartilage staining.
Stage 45 embryos were fixed in 100% ethanol for 48 hours at room temperature and then washed briefly in acid alcohol (1.2% HCl in 70% ETOH). A 0.25% Alcian blue solution in acid alcohol was used to stain the embryos over 48 hours at room temperature. Specimens were then washed in acid alcohol several times, rehydrated into H2O, and bleached for 2 hours in 1.2% hydrogen peroxide under a bright (2500 lux) light. They were then washed several times in 2% KOH and left rocking overnight in 10% glycerol in 2% KOH. Samples were processed through 20%, 40%, 60%, and 80% glycerol in 2% KOH.
Motility assay.
Stage 45 tadpoles were individually placed into wells of a standard 48-well culture dish. Prior to imaging, individual wells were aligned for video capture and tadpoles were allowed to reach a resting state for at least 5 minutes. Tadpoles were then gently prodded using a capillary pipette tip. Video of tadpole movement was captured over 30 seconds after this stimulation. Videos were acquired using an Accu-Scope Excelis camera mounted on a Nikon SMZ 745T stereomicroscope. Videos were analyzed using Kinovea 0.8.15 (https://www.kinovea.org/). Analysis consisted of marking a center point of the tadpole head in each frame of a 30-second video and plotting this on a circular map. Average motion was determined via converting marked tracking points to vectors and measuring the sum of these vectors over the 30-second recorded time.
Proteomics.
Xenopus embryos were injected with 10 ng of either control MO or emc1 MO and allowed to develop until stage 24. Embryos were homogenized in RIPA buffer (MilliporeSigma) using a pestle. Lysates were centrifuged to remove embryonic debris. Chloroform-methanol/water protein precipitation was performed to recover a dried protein pellet. Resuspension and LFQMS were carried out as previously described (81). LC-MS/MS data were processed with Progenesis QI Proteomics software (Nonlinear Dynamics, version 2.0); protein identification was carried out using the Mascot search algorithm. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (82) with the data set identifier PXD012770 and 10.6019/PXD012770.
Gene ontology.
Ontology analysis was carried out using PANTHER (http://www.pantherdb.org). The input was all genes with human orthologues whose protein product was statistically significantly decreased in emc1 morphants as compared with control MO-injected embryos. Ontology analysis was assessed on the level of pathway ontology.
siRNA-mediated knockdown in RPE cells.
hTERT RPE-1 cells were obtained from ATCC. siRNA-mediated knockdown was performed using Silencer Select siRNA directed against EMC1 (Thermo Fisher Scientific, 4392420) or a control siRNA (Thermo Fisher Scientific, 4390843) transfected with RNAiMAX (Thermo Fisher Scientific) per manufacturer instructions. Cells were fixed for imaging, lysed for immunoblotting, or exposed to small molecule–based inhibition 24 hours after transfection.
Splicing assays for evaluation of UPR.
xbp1 splicing was assayed in stage 24 embryos as previously described (83) with primers modified for use in Xenopus (5′-GACTGCTCGGGACAGGAAAA-3′ and 5′-GCCCAACAAGAGATCAGACTCA-3′). Additionally, the methods were also adapted for use with fluorescently labeled 5′ primers for fragment analysis as previously described (79).
TUNEL staining.
TUNEL staining was carried out as previously described (84) on embryos that were injected in 1 of 2 cells at the 2-cell stage.
Immunofluorescence and microscopy for Xenopus and RPE cell culture.
For Xenopus and RPE cells, samples were first briefly permeabilized by 0.1% tween-20 followed by fixation in 4% PFA. Samples were mounted in Prolong Glass (Thermo Fisher Scientific). Immunostained cells or embryos were imaged on either a Zeiss Observer outfitted with optical interference (Apotome) microscopy (Figure 4C and Supplemental Figure 8) or a Zeiss LSM880 confocal microscope (Figure 3, C and D, and Figure 4A). Antibodies are listed in Supplemental Table 3. Fluorescent images were processed and analyzed with Fiji (85). Whole-mounted, craniofacial cartilage, WISH, and TUNEL stained embryos were imaged with a Canon EOS 5d digital camera mounted on a Zeiss discovery V8 stereomicroscope.
Image analysis.
All fluorescent micrographs were processed and analyzed using Fiji (85). Quantitation of fluorescence for the WNT response assay was carried out by comparing the mean fluorescence of single 10,000 μm2 regions within each half of the neural tube between injected and uninjected sides of the embryo. Quantitation of the nuclear to cytoplasmic ratio of β-catenin–GFP was assessed by manually delimiting the bounds of nuclei and measuring signal intensity per unit area for each nucleus and cytoplasmic compartment of 5 cells per neural tube. This measure was normalized to the same measurement made for localization of NLS-mCherry in the same cells. Fluorescence intensity of EMC1 and FZD7 in hNCCs was determined by measuring total signal intensity in 45-μm2 high-power fields and dividing by the number of cells in the field as determined by counting distinct nuclei.
Immunoblot analysis.
For Xenopus and RPE cells, pooled embryos or cells were lysed in RIPA buffer and immunoblots were performed with Bolt 4%–12% Bis-Tris plus gels and running buffer (Thermo Fisher Scientific) using standard methods (see complete unedited blots in the supplemental material).
Use of small-molecule inhibitors in Xenopus and RPE cells.
To inhibit GSK3β function during development in emc1 morphants, CHIR99021 (Tocris Bioscience) was added to culture media at a concentration of 10 μM between stages 10 and 20, at which point they were collected. After transfection of control or EMC1 siRNA, RPE cells were subjected to cycloheximide (Cell Signaling) treatment in the culture media at a concentration of 25 mg/mL and/or MG132 (Sigma-Aldrich) treatment in the culture media at a concentration of 10 μM prior to lysis. To induce UPR response in embryos, they were incubated in culture media containing tunicamycin (Sigma-Aldrich) at a concentration of 2.5 mg/mL.
Neural crest induction.
Human embryonic stem cells (WA01) were obtained from WiCell Research Institute, Inc. For neural crest induction, human ESC/iPSC colonies were treated with Accutase cell detachment solution (STEMCELL Technologies) and resuspended in induction medium plus 3 μM CHIR 99021 and 10μM ROCK inhibitor Y-27632 (Tocris Bioscience) plated at the 20,000 cells/cm2 density on Matrigel-coated surfaces. Induction medium contained DMEM/F12 medium supplemented with 2% B27 supplement (Gibco), 1X Glutamax (Gibco), and 0.5% bovine serum albumin (wt/vol) (Sigma-Aldrich). CHIR 99021 and ROCK inhibitor Y-27632 were added for the first 2 days; induction media were used without CHIR 99021 and ROCK inhibitor for the next 3 days.
siRNA-mediated knockdown in human ES/NC cells.
siRNA-mediated knockdown was performed using SMART Pool siRNA for EMC1 (Dharmacon, catalog no. L-014146-01) with RNAiMAX (Thermo Fisher Scientific, catalog no. 13778075). Reverse transfection was performed as per manufacturer’s instructions. Briefly, hES cells were plated in NC induction media as described above in a 96-well plate preplated with siRNA/RNAiMAX mixture. For each well, 1.2 picomoles of siRNA was used for EMC1 SMART Pool and non-targeting control siRNA (Thermo Fisher Scientific, catalog no. AM4611). Media were changed after every 24 hours, and cells were harvested at day 2 and day 5 for RNA extraction and immunofluorescence.
Gene expression analysis for human ES/NC cells.
Total RNAs were extracted using TRIzol reagent/DirectZol kit (Zymoresearch). Total RNA (0.5–1 μg) was reverse-transcribed using PrimeScript reverse transcriptase kit (Takara/Clontech Laboratories). Quantitative polymerase chain reaction (QPCR) was carried out using the SYBR Premix ExTaqII SYBR Green premix (Takara/Clontech Laboratories) on Applied Biosystems QuantStudio 6 Real-Time PCR System (Applied Biosystems). Primer concentration of 300 nM was used. For statistical analysis, 2 biological replicates were measured for each experiment using ΔΔCT method relative to control siRNA transfected cell state and normalized to RPL13 reference gene. Primers are listed in Supplemental Table 3.
Immunofluorescence for hES/hNC cells and image processing and cell count analysis for hES and hNC cells.
Immunofluorescence for PAX7 and SOX10 was performed on cells cultured in a 96-well plate as previously described (41). Images were taken using a Nikon Eclipse Ti inverted microscope under the same exposure settings and analyzed using Nikon Elements software (Nikon). Images were processed in Adobe Photoshop and were simultaneously adjusted. Nikon Elements software (with General Analysis) (Nikon) was used for cell counts and cell intensity analysis. For cell count analysis, under general analysis, bright spot detection method was used, with clustered object parameter, with variable object (cell) sizes depending on the sample, determined manually for each sample to detect every cell in the image under each channel. Each experiment was done in 2 biological replicates. Immunofluorescence for EMC1 and FZD7 was performed on cells cultured in 4- or 8-well chamber slides (Thermo Fisher Scientific) and mounted in Prolong Glass (Thermo Fisher Scientific). Images were acquired using a Zeiss LSM880 confocal microscope. Fluorescent images were processed and analyzed with Fiji (85). Each experiment was done in 3 biological replicates. Antibodies are listed in Supplemental Table 3.
Statistics.
Statistical significance of abnormalities was tested using Fisher’s exact tests for comparison of percentages of tadpoles displaying a particular phenotype, 2-tailed Student’s t tests (paired or unpaired) for comparison of measured quantities, or 1-way ANOVA with post hoc Tukey’s test for multiple comparisons in GraphPad Prism 8.2.1. Correction for multiple comparisons was accomplished by calculating multiplicity-adjusted P values as part of the post hoc Tukey’s test. P values less than 0.05 were deemed significant.
Study approval.
All of our experiments with Xenopus tropicalis were approved by the Institutional Animal Care and Use Committee at Yale University in New Haven, Connecticut.
Author contributions
JM, JC, and MKK conceived of and designed experiments presented and analyzed data from each of the experiments. JM, JC, and EKM performed all Xenopus experiments. RMC, MSP, and MIGC conceived, designed, performed, and analyzed experiments in human embryonic stem cells. JM and WYH performed cell culture experiments in RPE cells. JM and JC analyzed data with MKK. The manuscript was written by JM and MKK. All authors critically evaluated the manuscript.
Supplementary Material
Acknowledgments
We thank the patients and their families who are the inspiration for this study. We also wish to thank Michael Slocum and Sarah Kubek for animal husbandry. We are grateful to the National Xenopus Resource (RRID:SCR_013731) for provision of the Xtr.Tg(WntREs:dEGFP)Vlemx (NXR_1094) transgenic line. This work was possible due to the resources and analysis provided by the Discovery Proteomics Core at Yale. We are indebted to the Yale CCMI core for fluorescence microscopy resources. JM and WYH were supported by the Yale MSTP NIH T32GM07205 Training Grant, the Yale Predoctoral Program in Cellular and Molecular Biology T32GM007223 Training Grant, and the Paul and Daisy Soros Fellowship for New Americans. RMC is supported by NIH F32DE027862. MSP and MIGC were supported by NIH R01DE017914. EKM is supported by a grant from the Hartwell Foundation and is a Hartwell Fellow. This work was supported by the NIH R01HD081379 to MKK. MKK was a Mallinckrodt Scholar.
Version 1. 01/06/2020
Electronic publication
Version 2. 02/03/2020
Print issue publication
Funding Statement
JM and WYH
JM and WYH
RMC
MSP and MIGC
EKM
MKK
MKK
JM and WYH
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):813–826.https://doi.org/10.1172/JCI129308.
Contributor Information
Jonathan Marquez, Email: jonathan.marquez@yale.edu.
June Criscione, Email: june.criscione@yale.edu.
References
- 1.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wideman JG. The ubiquitous and ancient ER membrane protein complex (EMC): tether or not? F1000Res. 2015;4:624. doi: 10.12688/f1000research.6944.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitwood PJ, Juszkiewicz S, Guna A, Shao S, Hegde RS. EMC is required to initiate accurate membrane protein topogenesis. Cell. 2018;175(6):1507–1519.e16. doi: 10.1016/j.cell.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satoh T, Ohba A, Liu Z, Inagaki T, Satoh AK. dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. Elife. 2015;4:e06306. doi: 10.7554/eLife.06306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard M, Boulin T, Robert VJ, Richmond JE, Bessereau JL. Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc Natl Acad Sci U S A. 2013;110(11):E1055–E1063. doi: 10.1073/pnas.1216154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guna A, Volkmar N, Christianson JC, Hegde RS. The ER membrane protein complex is a transmembrane domain insertase. Science. 2018;359(6374):470–473. doi: 10.1126/science.aao3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu-Safieh L, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23(2):236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harel T, et al. Monoallelic and biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am J Hum Genet. 2016;98(3):562–570. doi: 10.1016/j.ajhg.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geetha TS, et al. A novel splice variant in EMC1 is associated with cerebellar atrophy, visual impairment, psychomotor retardation with epilepsy. Mol Genet Genomic Med. 2018;6(2):282–287. doi: 10.1002/mgg3.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martik ML, Bronner ME. Regulatory logic underlying diversification of the neural crest. Trends Genet. 2017;33(10):715–727. doi: 10.1016/j.tig.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142(2):242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad MS, Charney RM, García-Castro MI. Specification and formation of the neural crest: Perspectives on lineage segregation. Genesis. 2019;57(1):e23276. doi: 10.1002/dvg.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szabó A, Mayor R. Mechanisms of neural crest migration. Annu Rev Genet. 2018;52:43–63. doi: 10.1146/annurev-genet-120417-031559. [DOI] [PubMed] [Google Scholar]
- 16.Rothstein M, Bhattacharya D, Simoes-Costa M. The molecular basis of neural crest axial identity. Dev Biol. 2018;444(suppl 1):S170–S180. doi: 10.1016/j.ydbio.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pla P, Monsoro-Burq AH. The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev Biol. 2018;444(suppl 1):S36–S46. doi: 10.1016/j.ydbio.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Muñoz WA, Trainor PA. Neural crest cell evolution: how and when did a neural crest cell become a neural crest cell. Curr Top Dev Biol. 2015;111:3–26. doi: 10.1016/bs.ctdb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A. 2010;107(8):3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayor R, Young R, Vargas A. Development of neural crest in Xenopus. Curr Top Dev Biol. 1999;43:85–113. doi: 10.1016/s0070-2153(08)60379-8. [DOI] [PubMed] [Google Scholar]
- 21.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7(3):291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46(11):673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- 23.Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin Cell Dev Biol. 2005;16(6):647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Stuhlmiller TJ, García-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69(22):3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deniz E, Jonas S, Hooper M, N Griffin J, Choma MA, Khokha MK. Analysis of craniocardiac malformations in xenopus using optical coherence tomography. Sci Rep. 2017;7:42506. doi: 10.1038/srep42506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deniz E, Mis EK, Lane M, Khokha MK. CRISPR/Cas9 F0 screening of congenital heart disease genes in Xenopus tropicalis. Methods Mol Biol. 2018;1865:163–174. doi: 10.1007/978-1-4939-8784-9_12. [DOI] [PubMed] [Google Scholar]
- 27.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation. 2012;84(1):25–40. doi: 10.1016/j.diff.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoki Y, et al. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259(1):19–33. doi: 10.1016/S0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 30.Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235(5):1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo W, Gautier J. A role for programmed cell death during early neurogenesis in xenopus. Dev Biol. 2003;260(1):31–45. doi: 10.1016/S0012-1606(03)00222-7. [DOI] [PubMed] [Google Scholar]
- 32.Braun DA, et al. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest. 2018;128(10):4313–4328. doi: 10.1172/JCI98688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13(12):767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 35.Abu-Elmagd M, Garcia-Morales C, Wheeler GN. Frizzled7 mediates canonical Wnt signaling in neural crest induction. Dev Biol. 2006;298(1):285–298. doi: 10.1016/j.ydbio.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 36.Maj E, et al. Controlled levels of canonical Wnt signaling are required for neural crest migration. Dev Biol. 2016;417(1):77–90. doi: 10.1016/j.ydbio.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396(6709):370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 38.Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. Genesis. 2012;50(3):155–163. doi: 10.1002/dvg.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran HT, Vleminckx K. Design and use of transgenic reporter strains for detecting activity of signaling pathways in Xenopus. Methods. 2014;66(3):422–432. doi: 10.1016/j.ymeth.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Borday C, Parain K, Thi Tran H, Vleminckx K, Perron M, Monsoro-Burq AH. An atlas of Wnt activity during embryogenesis in Xenopus tropicalis. PLoS One. 2018;13(4):e0193606. doi: 10.1371/journal.pone.0193606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairchild CL, Gammill LS. Tetraspanin18 is a FoxD3-responsive antagonist of cranial neural crest epithelial-to-mesenchymal transition that maintains cadherin-6B protein. J Cell Sci. 2013;126(pt 6):1464–1476. doi: 10.1242/jcs.120915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairchild CL, Conway JP, Schiffmacher AT, Taneyhill LA, Gammill LS. FoxD3 regulates cranial neural crest EMT via downregulation of tetraspanin18 independent of its functions during neural crest formation. Mech Dev. 2014;132:1–12. doi: 10.1016/j.mod.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J, et al. Transmembrane protein 198 promotes LRP6 phosphorylation and Wnt signaling activation. Mol Cell Biol. 2011;31(13):2577–2590. doi: 10.1128/MCB.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Gijn ME, Blankesteijn WM, Smits JF, Hierck B, Gittenberger-de Groot AC. Frizzled 2 is transiently expressed in neural crest-containing areas during development of the heart and great arteries in the mouse. Anat Embryol. 2001;203(3):185–192. doi: 10.1007/s004290000152. [DOI] [PubMed] [Google Scholar]
- 45.Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128(19):3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- 46.Abbruzzese G, et al. The Wnt receptor Frizzled-4 modulates ADAM13 metalloprotease activity. J Cell Sci. 2015;128(6):1139–1149. doi: 10.1242/jcs.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JR, Moon RT. Analysis of the signaling activities of localization mutants of β-catenin during axis specification in Xenopus. J Cell Biol. 1997;139(1):229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung AW, Murdoch B, Salem AF, Prasad MS, Gomez GA, García-Castro MI. WNT/β-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development. 2016;143(3):398–410. doi: 10.1242/dev.130849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.García-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297(5582):848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- 50.Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389(6654):966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 51.LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125(13):2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- 52.Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc Natl Acad Sci U S A. 1997;94(25):13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8(2):167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Gomez GA, Prasad MS, Sandhu N, Shelar PB, Leung AW, García-Castro MI. Human neural crest induction by temporal modulation of WNT activation. Dev Biol. 2019;449(2):99–106. doi: 10.1016/j.ydbio.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffin JN, Sondalle SB, Del Viso F, Baserga SJ, Khokha MK. The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS Genet. 2015;11(3):e1005018. doi: 10.1371/journal.pgen.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noack Watt KE, Achilleos A, Neben CL, Merrill AE, Trainor PA. The roles of RNA polymerase I and III subunits Polr1c and Polr1d in craniofacial development and in Zebrafish models of Treacher Collins Syndrome. PLoS Genet. 2016;12(7):e1006187. doi: 10.1371/journal.pgen.1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braun DA, et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet. 2016;48(4):457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Viso F, et al. Congenital heart disease genetics uncovers context-dependent organization and function of nucleoporins at cilia. Dev Cell. 2016;38(5):478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16(2):165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15(1):66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoue K, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36(4):361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 62.Inoue K, et al. Translation of SOX10 3′ untranslated region causes a complex severe neurocristopathy by generation of a deleterious functional domain. Hum Mol Genet. 2007;16(24):3037–3046. doi: 10.1093/hmg/ddm262. [DOI] [PubMed] [Google Scholar]
- 63.Lupski JR. Interruption of SOX10 function in myelinopathies. Ann Neurol. 2010;68(2):121–123. doi: 10.1002/ana.22123. [DOI] [PubMed] [Google Scholar]
- 64.Ruest LB, Clouthier DE. Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev Biol. 2009;328(1):94–108. doi: 10.1016/j.ydbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonano M, et al. A new role for the Endothelin-1/Endothelin-A receptor signaling during early neural crest specification. Dev Biol. 2008;323(1):114–129. doi: 10.1016/j.ydbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Shin MK, Levorse JM, Ingram RS, Tilghman SM. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402(6761):496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 67.Barriga EH, Maxwell PH, Reyes AE, Mayor R. The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J Cell Biol. 2013;201(5):759–776. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Theveneau E, et al. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19(1):39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heasman J, et al. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79(5):791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 70.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391(6665):357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 71.Owens NDL, et al. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 2016;14(3):632–647. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saal HM, et al. A mutation in FRIZZLED2 impairs Wnt signaling and causes autosomal dominant omodysplasia. Hum Mol Genet. 2015;24(12):3399–3409. doi: 10.1093/hmg/ddv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagasaki K, et al. Nonsense mutations in FZD2 cause autosomal-dominant omodysplasia: Robinow syndrome-like phenotypes. Am J Med Genet A. 2018;176(3):739–742. doi: 10.1002/ajmg.a.38623. [DOI] [PubMed] [Google Scholar]
- 74.White JJ, et al. WNT signaling perturbations underlie the genetic heterogeneity of Robinow syndrome. Am J Hum Genet. 2018;102(1):27–43. doi: 10.1016/j.ajhg.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012;366(1):34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 76.del Viso F, Khokha M. Generating diploid embryos from Xenopus tropicalis. Methods Mol Biol. 2012;917:33–41. doi: 10.1007/978-1-61779-992-1_3. [DOI] [PubMed] [Google Scholar]
- 77. Hubrecht-Laboratorium (Embryologisch Instituut). Normal table of Xenopus laevis (Daudin): A systematical chronological survey of the development from the fertilized egg till the end of metamorphosis, 2nd ed. Nieuwkoop PD, Faber J, eds. Amsterdam, Netherlands: North Holland Publishing Company; 1967. [Google Scholar]
- 78.Kirchmaier S, Lust K, Wittbrodt J. Golden GATEway cloning — a combinatorial approach to generate fusion and recombination constructs. PLoS One. 2013;8(10):e76117. doi: 10.1371/journal.pone.0076117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK. CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol. 2015;408(2):196–204. doi: 10.1016/j.ydbio.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375(6534):787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 81.Klein ZA, et al. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron. 2017;95(2):281–296.e6. doi: 10.1016/j.neuron.2017.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci. 2009;122(pt 16):2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 84.Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol. 1998;203(1):36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- 85.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.