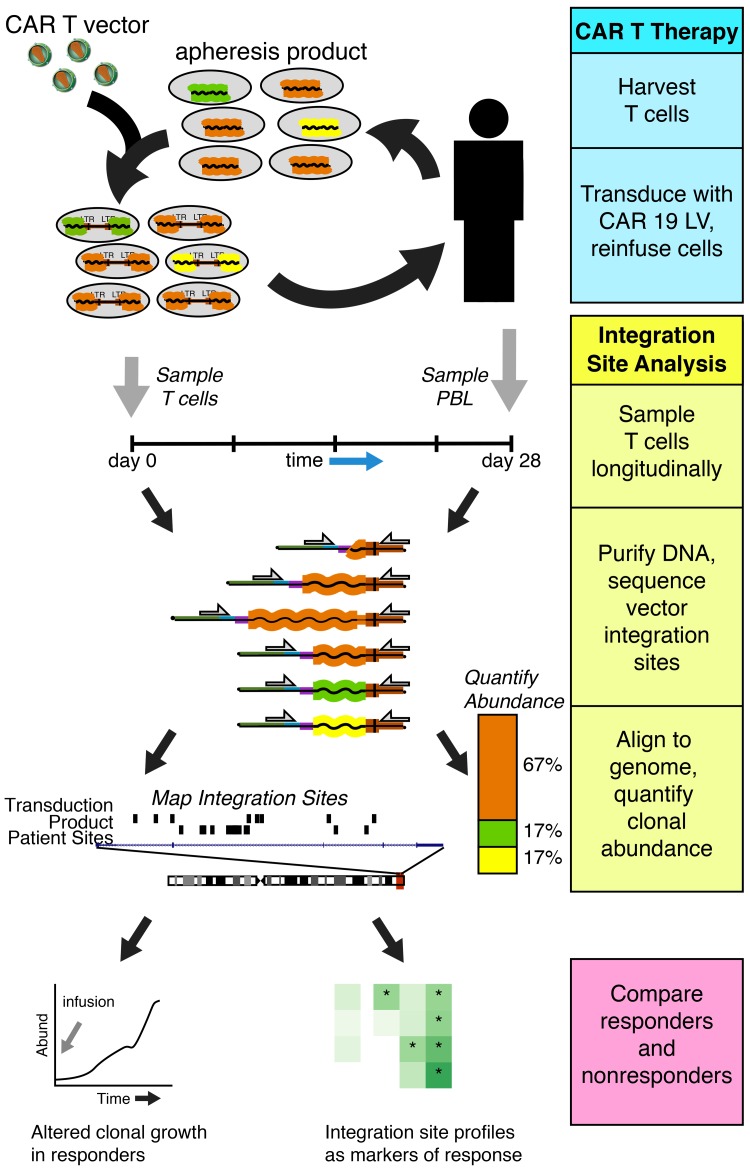
Abstract
Chimeric antigen receptor–engineered T cells targeting CD19 (CART19) provide an effective treatment for pediatric acute lymphoblastic leukemia but are less effective for chronic lymphocytic leukemia (CLL), focusing attention on improving efficacy. CART19 harbor an engineered receptor, which is delivered through lentiviral vector integration, thereby marking cell lineages and modifying the cellular genome by insertional mutagenesis. We recently reported that vector integration within the host TET2 gene was associated with CLL remission. Here, we investigated clonal population structure and therapeutic outcomes in another 39 patients by high-throughput sequencing of vector-integration sites. Genes at integration sites enriched in responders were commonly found in cell-signaling and chromatin modification pathways, suggesting that insertional mutagenesis in these genes promoted therapeutic T cell proliferation. We also developed a multivariate model based on integration-site distributions and found that data from preinfusion products forecasted response in CLL successfully in discovery and validation cohorts and, in day 28 samples, reported responders to CLL therapy with high accuracy. These data clarify how insertional mutagenesis can modulate cell proliferation in CART19 therapy and how data on integration-site distributions can be linked to treatment outcomes.
Keywords: Microbiology
Keywords: Immunotherapy
Introduction
Patient T cells engineered to express a CD19-specific chimeric antigen receptor (CART19) have proven effective in inducing long-term remissions in pediatric acute lymphoblastic leukemia (ALL), with complete response rates exceeding 80% (1–3), yet only 26% of patients with chronic lymphocytic leukemia (CLL) achieved stable complete remission (4, 5). For other treatment-refractory cancers, CAR T cells have shown dramatic successes in some but not all cases (6–8). Studies of responders and nonresponders (NRs) in CLL CART19 therapy (4, 5, 9–11) revealed that durable remission was associated with a higher peak expansion of CART19 after infusion and longer persistence. Cell products that showed greater proliferative capacity prior to infusion and enrichment in specific T cell subsets were particularly effective (5). RNA-Seq revealed that the gene expression in preinfusion T cells differed between complete responders (CRs) and NRs (5). Greater representation was seen in CRs for genes involved in early-memory T cell differentiation and IL-6/STAT3 responsiveness, whereas those from NRs exhibited gene sets enriched in effector T cell differentiation, exhaustion, aerobic glycolysis, and apoptosis. Here, we interrogate orthogonal data — the locations of vector-integration acceptor sites — in an effort to link genomic modifications, growth of gene-modified cells, and clinical outcomes.
Recently, we reported a case (patient 10) of insertional mutagenesis and clonal expansion in CART19 therapy associated with clinical success (12). Patient 10 had a complex clinical course with relapsed and refractory CLL; after 2 infusions of CART19, he was found to have a sharp clonal expansion associated with tumor elimination. We found that the CAR19 vector was integrated into the cellular TET2 locus, which encodes a methylcytosine dioxygenase involved in converting 5-methylcytosine to 5-hydroxymethylcytosine, a reaction that ultimately results in repair and replacement of the methylated base with an unmodified cytosine base. TET2 has previously been implicated in clonal expansion in cells of the hematopoietic lineage — it is the most commonly mutated gene in healthy individuals with clonal hematopoiesis (13–15). Analysis of TET2 mRNA in CAR-expressing T cells showed the presence of new mRNAs that spliced into the vector and terminated, truncating the TET2 protein to remove the encoded catalytic domain. Extensive follow-up studies found that the patient also harbored a polymorphism in his other TET2 allele that diminished protein function (12), so the 2 genetic lesions led to sharply reduced TET2 activity. When the CART19 compartment was dominated by TET2-disrupted clones, the majority of these cells exhibited a less-differentiated central memory phenotype; cells of this lineage are believed to show superior proliferation and antitumor activity compared with other subsets (16, 17). We and others have replicated these results by demonstrating that modulation of the TET2 pathway promotes the emergence of central memory T cells (12, 18, 19). Optimal proliferation, persistence, and antitumor potency of CAR- or T cell receptor–modified (TCR-modified) T cells depend on a young, central memory phenotype, and epigenetic programming through TET2 downregulation can enforce this state (12, 18, 19). We hypothesize that TET2 insertion improved therapeutic activity via preservation of a central memory phenotype in CART19.
Inactivation of TGFβRII using a dominant-negative allele (dnTGFβRII) has also been associated with enhanced T cell proliferation and activation (20, 21). Following up on these observations, we recently tested whether the antitumor efficacy of prostate-specific membrane antigen–directed (PSMA-directed) CAR T cells could be enhanced by coexpression of a dnTGFβRII. Abrogation of TGF-β signaling in anti-PSMA CAR T cells increased proliferation, effector cytokine production, long-term persistence, and the ability of these engineered lymphocytes to mediate tumor eradication in aggressive human prostate cancer mouse models (22). The clinical efficacy of PSMA-directed CAR T cells bearing a dnTGFβRII is currently being evaluated at University of Pennsylvania in a clinical trial (ClinicalTrials.gov NCT03089203).
We sought to investigate the hypothesis that insertional mutagenesis by CAR lentiviral vector integration in patient T cells provided information on pathways affecting cell proliferation and response to therapy. Many types of studies support the idea that genetic alterations can affect proliferation of nontransformed primary human cells. Direct studies based on genome-wide mutagenesis have revealed that changes in gene dosage over many human genes can alter cellular rates of proliferation, though responses were highly cell type–specific (23, 24). Evidence from human (25–34) and murine (35) stem cell gene therapy trials has provided examples of clonal expansion associated with insertional mutagenesis by gene-transfer vectors. In addition, integration of HIV DNA in latently infected cells is believed, in some cases, to alter T cell regulatory pathways and thus promote clonal expansion and, consequently, persistence of the latent HIV reservoir (36–38). In data from patients undergoing CART19 therapy, we noted clonal outgrowth in cells with integration sites in both TET2 and TGFBR2 (see below). These findings led us to conduct a detailed study of vector integration in CART19 from 40 treated patients to identify genes and pathways potentially influencing therapeutic cell proliferation.
Results
Patients analyzed.
Forty patients treated for ALL (n = 11, both pediatric and adult) or CLL (n = 29) were analyzed. Supplemental Table 1 summarizes patient data (supplemental material available online with this article; https://doi.org/10.1172/JCI130144DS1). On average, patients with ALL were younger (24 years versus 64 years for those with CLL (Supplemental Table 2). Outcomes were scored as CR, partial response (PR), partial response with transformed disease (PRtd), or NR; detailed criteria appear in the Methods section. In the following analysis, patients with CR or PRtd (CR/PRtd) were judged to represent clinically efficacious responses, while patients with PR or NR (PR/NR) were considered to have experienced clinical failure, as in previous work (5). A validation cohort of preinfusion samples from another 18 patients from a CLL CART19 trial was also analyzed (ref. 9 and our unpublished observations).
Samples for integration-site analysis were derived from the transduced final CAR T cell product prior to infusion and peripheral blood leukocytes (PBLs) collected from patients on day 28 after infusion. For some patients, additional time points were assessed as available (Supplemental Table 3).
Analyzing cell populations by sequencing host-vector junctions.
Locations of vector-integration sites in patient samples were determined by ligation-mediated PCR as described previously (26, 39–45). Samples of patient cell DNA were sheared by sonication, and DNA adapters were ligated to the broken ends. PCR was then used to amplify from the viral DNA end to the adapter. A total of 78.9 × 106 sequence reads were acquired in 184 preinfusion and postinfusion samples from the 40 patients, yielding approximately 145,600 unique integration sites mapping to the human genome. Analysis by the SonicAbundance method, which uses unique DNA fragment sizes to infer the numbers of cells sampled (41), indicated that approximately 198,700 gene-modified T cells were queried.
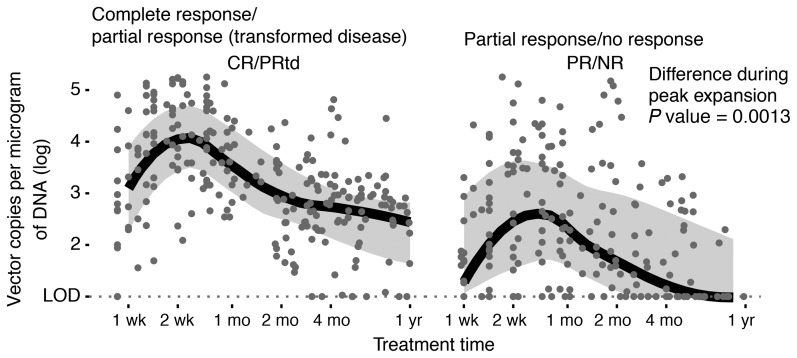
The average vector copy number (VCN) per microgram of DNA was quantified for all samples in the discovery cohort (Figure 1). Confirming previous analyses (1, 4, 5), peak expansion was greater for CR/PRtd versus PR/NR (P = 0.00047).
Figure 1. VCN analyzed by qPCR longitudinally, comparing CR/PRtd with PR/NR.
Peak expansion was assessed as maximal VCN 10–21 days after infusion. The difference in medians was tested using Wilcoxon’s rank-sum test.
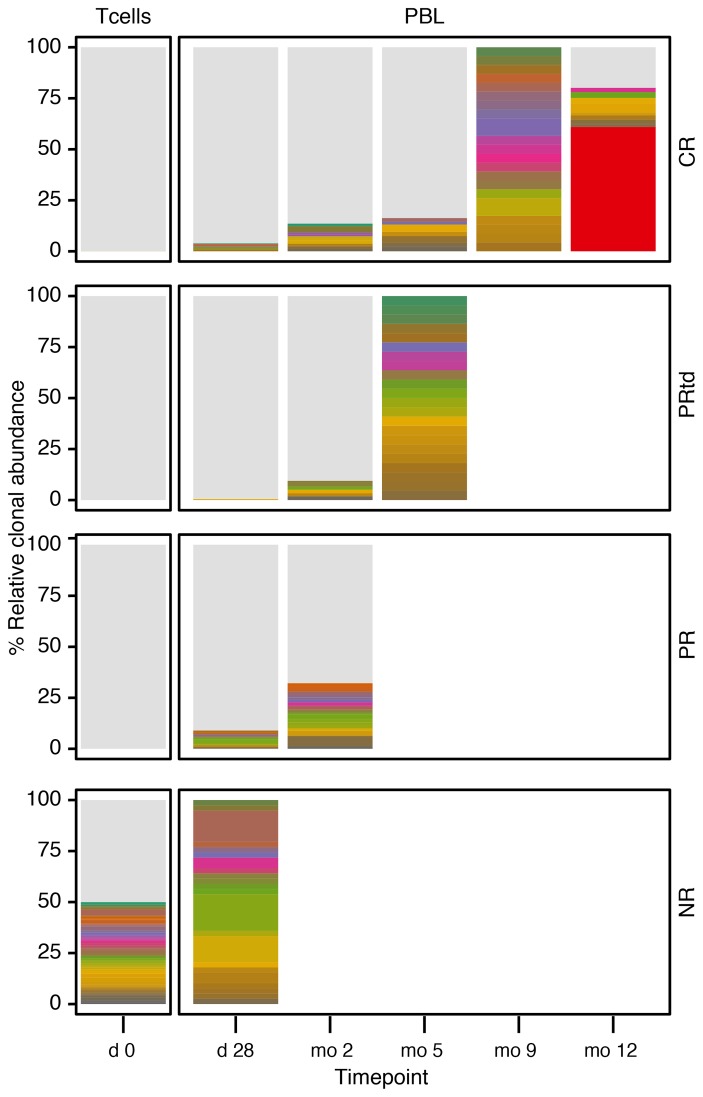
Integration sites were mapped to the human genome, and longitudinal evolution was assessed. Examples of longitudinal analysis of integration-site distributions are shown in Figure 2. Overall, integration was favored in active transcription units for both CR/PRtd and PR/NR, paralleling many studies of lentiviral integration (refs. 25, 39, 40, 42, and 46 and Supplemental Figure 1).
Figure 2. Examples of longitudinal analysis of integration-site distributions for CR, PRtd, PR, and NR patients.
Day 0 indicates the preinfusion T cell product. Later samples from patients were from PBLs. Each color indicates a different clone; the height of the bar indicates the relative abundance. No clones were shared among patients. Light gray indicates low-abundance clones; white indicates no samples available. The abundant clone in the CR patient (red) is in the gene ZNF573.
Criteria for assessing the association between integration-site placement and cell proliferation.
We used 4 criteria to evaluate whether integration of the CAR-encoding lentiviral vector may have influenced activity of the targeted host gene and, potentially, therapeutic cell proliferation. In the TET2 case, a notable feature of the expanded clone was its long-term persistence. Thus, for the 39 additional cases reported here, we tabulated genes at integration sites in cells that persisted the longest. Length of follow-up varied, ranging from at least 28 days to 5 years. We also compared the frequency of appearance of integration sites in each human transcription unit, reasoning that increasing abundance after infusion marked genes where loss of function increased expansion in vivo, and genes with reduced abundance after infusion marked genes that were important for proliferation in patients. We further tracked the most expanded clones, again with the goal of identifying clones showing behavior similar to the TET2 clone in patient 10.
A full list of genes called by each of the 4 criteria is presented in Supplemental Reports (1-4). Separate reports allow interrogation of specific subsets, including genes called in ALL patients, genes called in CLL patients, genes called in clinical responders only, and genes called over a pool of all patients studied. For each of the 4 analyses described below and presented in Figure 3, samples from both ALL and CLL patients were pooled to maximize statistical power, as we hypothesized that potential effects of insertional mutagenesis would be largely cell autonomous.
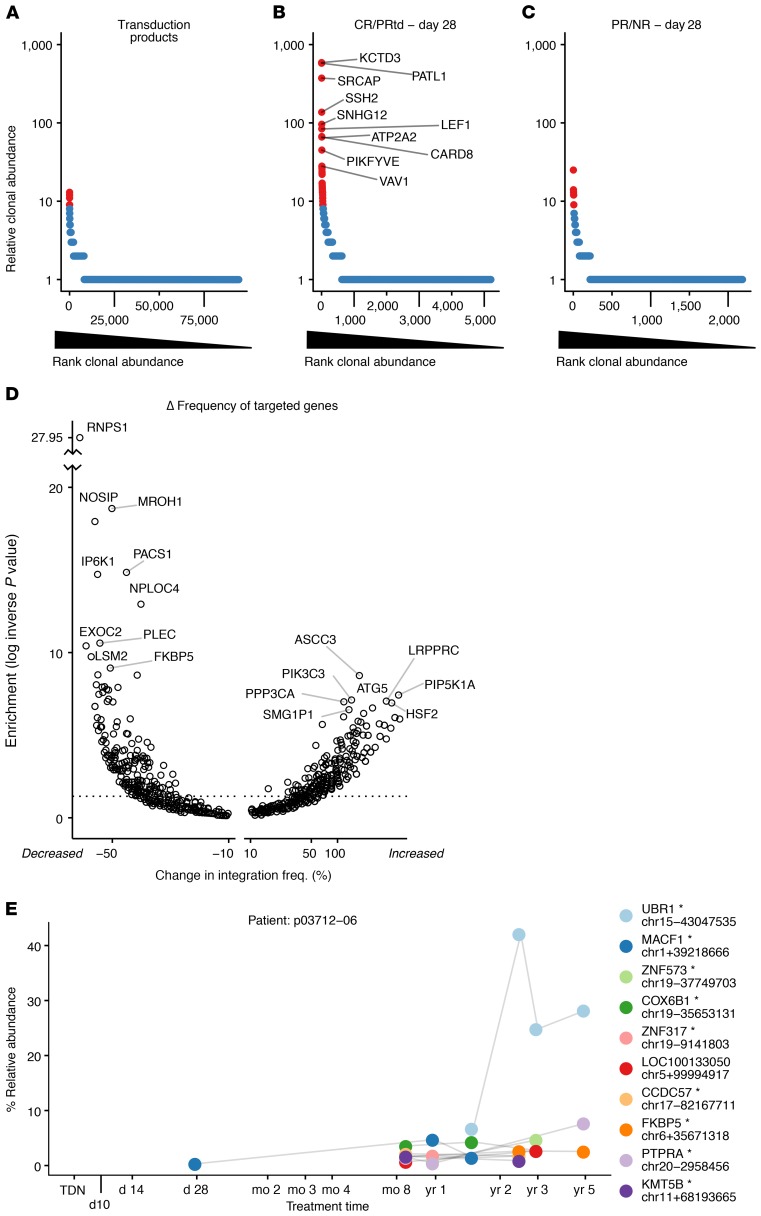
Figure 3. Clonal behavior of CART19 assessed by tracking sites of integrated vectors.
(A–C) Rank-abundance plots summarizing clonal abundance for the transduction products (A); CR/PRtd assayed on day 28 (B); and PR/NR assayed on day 28 (C). Cell clones (identified by identical integration sites) were ordered by most abundant (left) to least abundant (right) as assessed by SonicAbundance (41). Highly abundant clones (red) were scored as the top 1% of all expanded clones, corresponding to at least 9 cells representing each clone. The top 10 most-abundant clones for CR/PRtd on day 28 are labeled with gene symbols. A Venn diagram showing overlap among the top 30 most-expanded clones in A–C appears in Supplemental Figure 2. (D) Volcano plot showing genes where integration frequency was enriched or depleted during growth in patients. The total number of integration sites in each transcription unit was quantified for the preinfusion and posttransplant samples and normalized within each group. The 2 values were subtracted to obtain a normalized percent change (x axis). Fisher’s exact test (corrected for multiple comparisons using the Benjamini-Hochberg method) was used to assess enrichment or depletion (y axis). The x axis shows the percent change in frequency; the y axis shows the inverse log of the P value. (E) Example of a longitudinal expanded clone, from patient p03712-06. The x axis shows the time points sampled; the y axis shows the percent relative abundance of each clone determined by SonicAbundance. The nearest gene and the chromosomal location of each integration site (mapped on hg38) are indicated for the 10 most-abundant clones. The asterisk indicates integrated within the indicated transcription unit. TDN, transduction.
Genes at integration sites associated with clonal expansion.
We assessed genes marked by integration in expanded clones using the SonicAbundance method to count the numbers of cell genomes recovered (41). Figure 3, A–C, shows rank-abundance curves, where the most abundant 1% of clones from CART19-treated patients are shown in red. The figure compares clonal expansion in the transduction product (Figure 3A), day 28 samples from CR/PRtd patients (Figure 3B), and day 28 samples from PR/NR (Figure 3C). There are notable expanded clones in the postinfusion patient samples compared with the transduction product samples, with more pronounced expansions in the clinically successful cases (CR/PRtd; Figure 3B). Genes marked by integration in the expanded clones mostly differed among the 3 groups (Supplemental Figure 2).
If insertional mutagenesis indeed promotes clonal expansion, then over the 40 patients studied, one might expect genes targeted in expanded clones to host integration events in multiple patients. An analysis was conducted examining recurrence of genes at integration sites in the top 5% of expanded clones over all patients, and the frequency was found to be much higher than expected by chance (Supplemental Figure 3). Examples of genes at expanded clones in multiple patients included NPLOC4, KDM2A, PACS1, PCNX1, and RNF157. The finding of recurrent expansion of clones with vector integration in specific genes strengthens the idea that insertional mutagenesis can promote clonal expansion.
Genes with increasing or decreasing frequency of vector integration following cell infusion.
We also reasoned that if integration in or near a specific gene was promoting or inhibiting persistence, then integration sites in those genes should be detected with altered frequency after growth of cells over time in treated patients. Figure 3D shows a volcano plot comparing the genes that expanded or diminished most in frequency following growth in patients.
We checked these criteria by asking whether TET2 was identified in an analysis excluding patient 10, because patient 10 was found to have a hypomorphic mutation in his other TET2 allele, raising the possibility that therapeutic expansion was a highly unusual event (12). We reran the analysis after removing data on the expanded clone in patient 10 and found that TET2 was still called due to increased frequency of unique integration sites after cell infusion compared with the preinfusion product (with the patient 10 site: OR 2.97, P = 0.0067; without the patient 10 site: OR 2.78, P = 0.0115). Therefore, identification of TET2 is a common feature of our CART19 trials and not a function of the unusual genetic background of patient 10.
Genes at integration sites associated with longitudinal clonal persistence.
Another notable feature of the TET2 clone in patient 10 was the long-term persistence of the clone. Among the other 39 patients, most of the long-term persisting cells were from CR/PRtd, indicating an association with therapeutic success. An example of this is shown in Figure 3E, where patient 6 from the UPCC03712 trial (ClinicalTrials.gov NCT01747486) showed long-term clonal expansion over 5 years, with the expanded clone reaching 40% of all vector-modified clones. The long-term persisting clone contained a vector integrated within the UBR1 transcription unit, suggesting formation of a reduced-function allele. The gene encodes a member of the ubiquitin ligase family that is expressed in lymphoid cells and is important in protein degradation.
Genes targeted by integration suggest pathways modulating CART19 proliferation.
To begin to identify the cellular functions affected by potential insertional mutagenesis and clonal expansion, genes marked by integration and called as enriched by each of the above 4 criteria in a pool of all patients were queried for their membership in gene ontology categories. Results are summarized in Supplemental Figure 4, A and B. Notably, affected pathways included those involved in phosphotidyl inositol regulation, cAMP, TCR, and covalent chromatin modification. Examples of well-known genes in these pathways identified here include those encoding the methylase DNMT1 and the demethylase TET2; the methyl 5′-C-phosphate-G-3′–binding (CpG-binding) proteins MECP2 and MBD3; the histone lysine methyltransferases ASH1L, DOT1L, EHMT1, KMT2C, KMT2D, KMT5B, and SETD2; the lysine demethylases KDM4A and KDM6A; the cAMP-responsive chromatin regulators CREBBP and SRCAP; the transcriptional regulator ZNF573; and the rapamycin-targeted pathway proteins MTOR and FKBP5.
Genes enriched in the 4 categories were also queried for enrichment in cancer-associated genes (Supplemental Tables 4 and 5). We compared the sets of vector-marked genes to the allOnco list, a broad collection of cancer-associated genes designed for preliminary surveys (47), the Catalogue of Somatic Mutations in Cancer–Cancer Gene Census and The Cancer Genome Atlas cancer gene lists, a list of genes commonly disrupted in lymphoid cancers, and a list of genes implicated in clonal hematopoiesis. All 4 categories of genes at integration acceptor sites showed significant enrichment in at least some of the categories (Supplemental Table 5). For example, the cancer-associated gene VAV1 is the most strongly affected over the above 4 criteria when comparing pooled CR/PRtd with PR/NR (Supplemental Report 4 and Supplemental Table 2). Taken together, these findings suggest that insertional mutagenesis of genes known to be involved in growth control can indeed influence CART19 growth. There was no outgrowth of T cells harboring integration sites near genes previously identified as involved in adverse events in stem cell gene therapy (e.g., LMO2, CCND2, MDS/EVI1) (27–29, 48).
Distributions of vector-integration sites relative to mapped features in the human genome.
We next focused on whether global features of the integration-site distributions could be associated with outcome. Of particular interest are features of the posttransduction/preinfusion product that forecast later clinical responses, because these could be biomarkers useful for optimization of cell-manufacturing methods.
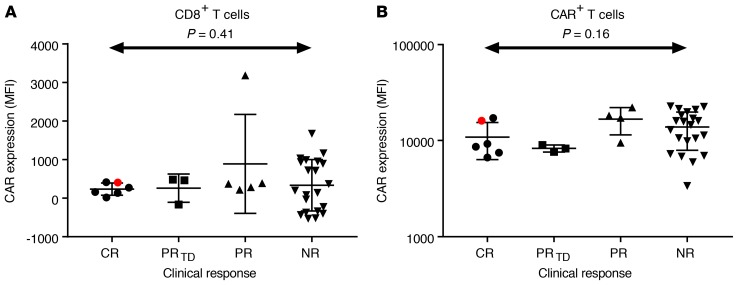
One possible model for an association between vector integration–site locations and clinical response could be via expression of the CAR19 transgene. That is, if integration in different chromosomal locations resulted in different expression levels and if optimal expression was important for clinical response, then integration targeting could be linked to outcome. We thus compared the levels of surface CAR19 expression measured by flow cytometry as mean fluorescence intensity for CR/PRtd and PR/NR in CD8+ cells (Figure 4A) and bulk CAR+ cells (Figure 4B) in preinfusion products. No significant differences were observed. The preinfusion cells from patient 10, who harbored the TET2 clonal expansion, did not show notably higher levels of surface CART19 protein expression. We thus disfavor the model that integration in different chromosomal sites in preinfusion products leads to differential CART19 expression, which in turn dictates outcome. Another possibility is that the percentage of CAR+ cells differed between CR/PRtd and PR/NR after transduction, but this was tested and found not to be the case (5).
Figure 4. Levels of CAR expression do not distinguish the patient response groups.
(A) CAR expression on CD8+ T cells, measured as MFI (y axis) compared by response group. No significant difference was detected among groups (1-way ANOVA). Red shows patient 10, who harbored the TET2 expansion. (B) As in A, but measured on bulk CAR+ T cells. Again no significant difference was detected (1-way ANOVA).
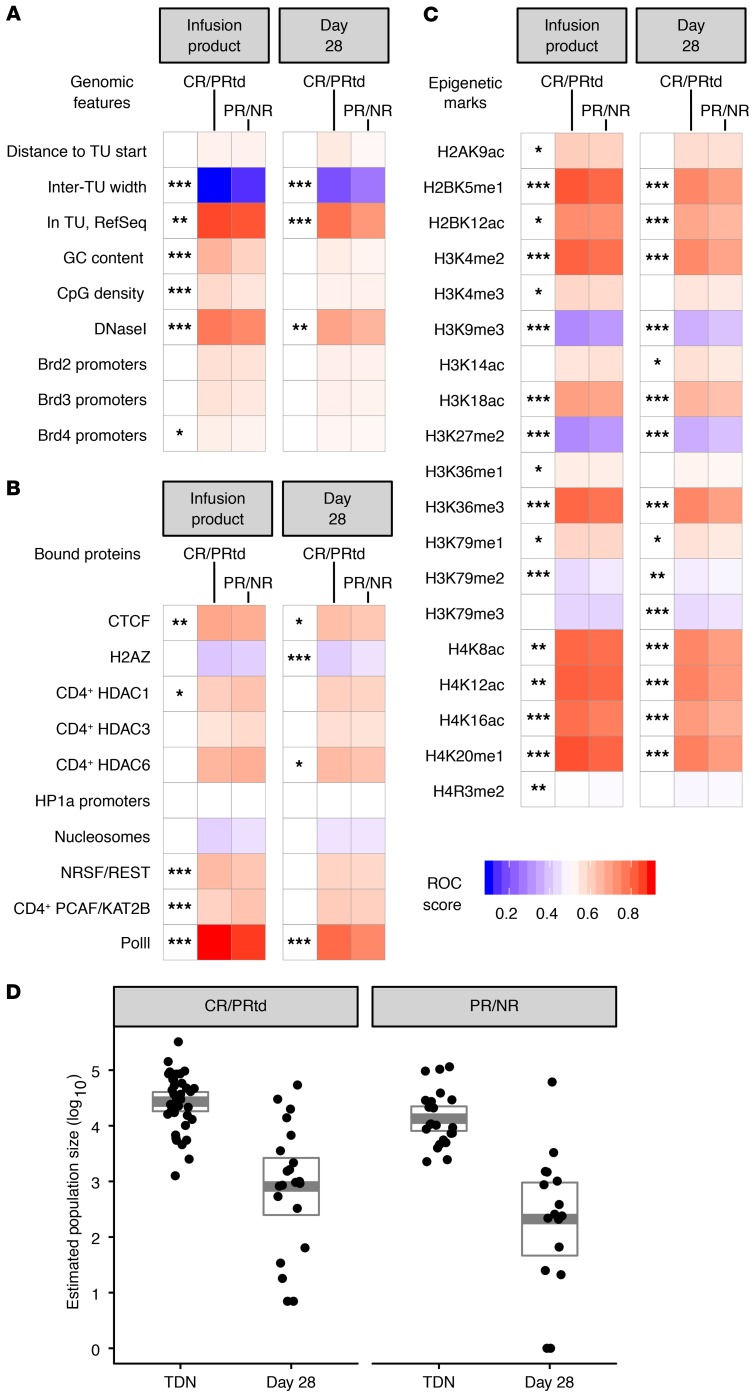
We next assessed integration-site placement relative to a large number of features to allow global modeling of distributions. Samples were categorized as “preinfusion” and “day 28 after infusion” and as “CR/PRtd” and “PR/NR.” Lentiviral integration is favored in active transcription units (Supplemental Figure 1 and refs. 39, 40, 42, 43, and 49), so different states of transcriptional activity in patient T cells before transduction may potentially result in differing patterns of integration targeting. Global integration-site distributions in CART19 generally paralleled those seen with HIV and lentiviral vectors in previous studies (50–52). Overall, 81.5% of integration sites were in annotated transcription units. The relationship of integrated vectors to genomic features (Figure 5A), bound proteins (Figure 5B), or sites of epigenetic modification (Figure 5C) was compared with random distributions to assess biases in integration-site distributions (40, 53). As expected, integration was favored near DNAse I hypersensitive sites, CpG islands, and regions of high gene density. Several bound proteins correlated with favored integration, including CTCF and Pol II, while the histone H2AZ correlated negatively. Comparison with epigenetic marks mapped previously in T cells (Figure 5C) showed that integration was positively associated with marks of gene activity, such as H4K20 monomethylation, H3K4 monomethylation and dimethylation, and multiple sites of acetylation, while integration was negatively associated with heterochromatic marks, such as H3K9 dimethylation and trimethylation and H3K27 dimethylation and trimethylation.
Figure 5. Frequency of integration near chromosomal features is associated with outcome.
(A) Genomic features, (B) chromosome-bound proteins, and (C) epigenetic marks associated with vector-integration frequency are shown for transduction products and day 28 peripheral blood samples. CR/PRtd and PR/NR are compared (columns) to mapped chromosomal features (rows). Associations were calculated by an ROC area method (41, 45). Values of the ROC area can vary between 0 (negatively associated) and 1 (positively associated), with 0.5 indicating no association. All epigenetic features were assessed within a 10 kb window. Asterisks beside the heatmap indicate comparisons between clinical response groups; separate analyses were conducted for transduction product on left and day 28 samples on right. P values were calculated using Wald’s test with a χ2 distribution; no correction for multiple comparisons was applied. (D) Right: box plot representation of Chao1 estimated population sizes for responders (CR and PRtd), comparing the transduction product and day 28 samples (PR and NR). Left: box plot representations of Chao1 estimated population sizes for nonresponders, comparing the transduction products and day 28 samples (CR/PRtd-TDN ~ PR/NR-TDN: P = 0.033, CR/PRtd-TDN ~ CR/PRtd-day 28: P < 0.001, PR/NR-TDN ~ PR/NR-day 28: P < 0.001, calculated using Wilcoxon’s test with a Benjamini-Hochberg correction for multiple comparisons) (20, 67–72). *P < 0.05, **P < 0.01, ***P < 0.001. TU, transcription unit; TDN, transduction.
We also compared these distributions for samples from clinical successes (CR/PRtd) and failures (PR/NR). Biases toward annotations related to gene activity were strong in all samples, but the strength of the associations varied. The most random patterns were in the PR/NR day 28 samples, where the associations were weaker over many of the forms of annotation assessed; differences could also be detected in the posttransduction/preinfusion samples (asterisks to the left of the “day 28” and “infusion product” panels [Figure 5, A–C]).
Several further summaries of population structure were developed from integration-site data for use in multivariate models. These included inferred population sizes of CAR19-modified T cells (Chao 1), diversity (Shannon Index), evenness (Gini Index), and the count of clones contributing to the most abundant 50% of clones sampled (UC50).
The population sizes of marked clones dropped sharply from those seen in the preinfusion product to the day 28 time point for both CR/PRtd and PR/NR (considering pooled ALL and CLL patient data), indicating that most marked T cell clones do not persist long term (Figure 5D; P = 0.013 for CR/PRtd and P = 2 × 10–6 for PR/NR). Population sizes were larger on day 28 for CR/PRtd compared with PR/NR (P = 0.046). On day 0, populations trended toward larger in CR/PRtd but did not achieve significance (P = 0.149). For CLL data analyzed in isolation, the population size (inferred from Chao1) was significantly larger for CR/PRtd versus PR/NR on day 28 (P = 0.008).
We also compared the diversity of T cell marking as reported by integration-site analysis to that reported by TCR sequencing of CAR19-sorted T cells (Supplemental Figure 5). Samples ranged widely in TCR diversity in the preinfusion product, and this was not correlated with the diversity of the integration-site population, likely reflecting differing quality in the starting T cells that did not strongly affect efficient, high-level lentiviral transduction. After transplantation and growth for 28 days, TCR on CAR19+ cells and integration-site diversity varied together, reflecting the extent of outgrowth of the vector-marked cells.
Biomarkers in the transduction product associated with outcome could be useful in optimizing therapeutic strategies; therefore, we sought to aggregate all of these metrics into a global multivariate model associating outcome with integration-site distributions.
Multivariate models predicting outcome based on integration-site data.
To develop predictive tools, we constructed a least absolute shrinkage and selection operator (LASSO) logistic regression model linking integration-site distributions and outcomes. For this, we studied the CLL patients in order to focus on a consistent clinical condition and because the ALL patients included only 2 NRs and 7 responders. For transduction products, 11 CLL CR/PRtd were compared with 18 PR/NR. For day 28 samples, 11 CR/PRtd were compared with 10 PR/NR (for some of the PR/NR, no cells were available to analyze by day 28). Patient groups were compared over 91 features of the integration-site distributions (Supplemental Table 6). Variables included population metrics (n = 7, including Richness, Chao1, Gini, etc.), genomic features (n = 24, including GC content, CpG islands, percentage within transcription units, etc.), and epigenetic features measured in T cells (n = 60, including different histone methylation and acetylation profiles, etc.).
Because many of these variables are highly correlated, a dimension-reduction step was used in which principal components were constructed to summarize the variance in the data (Supplemental Figure 6, A and B). Twenty-eight principal components were used to classify the posttransduction/preinfusion products, and twenty were used to classify the day 28 postinfusion samples. Model performance was assessed by leave-one-out cross-validation. Models were selected that provided the lowest misclassification rate after penalization for increasing numbers of model components.
The misclassification rate for the optimal model using integration-site sequence data from transduced preinfusion products was 21%. For the day 28 samples, the misclassification rate was only 4%. Removal of all clones associated with the TET2 gene, followed by a rerunning of the model, did not result in altered misclassification rates or weighting of model components, showing that the results were not driven by integration at TET2. Hence, a robust signal exists in each integration-site data set associated with outcome.
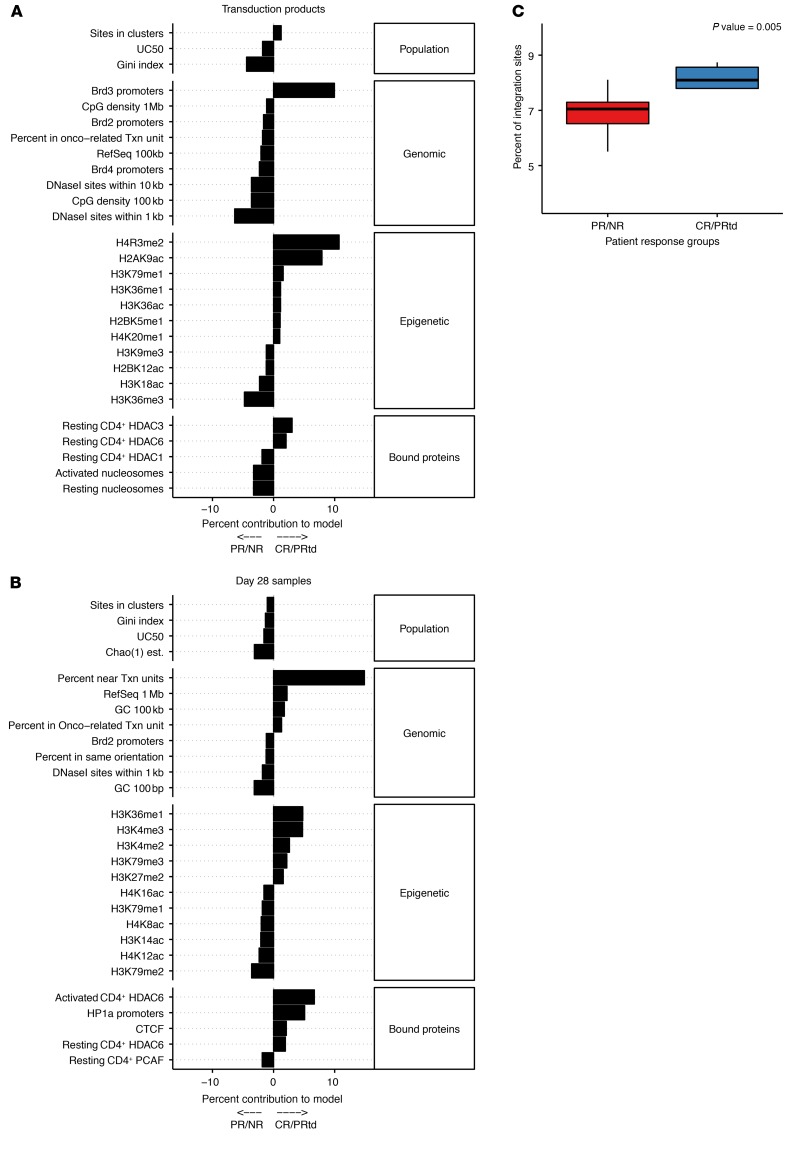
For the posttransduction/preinfusion model (Figure 6A), the most influential positive variables predictive of outcome included proximity to the epigenetic modifications H4R3me2 and H2AK9ac and proximity to BRD3 promoters. BRD3 is associated with hyperacetylated chromatin and is reported to allow transcription through nucleosome-bound DNA, indicating a robustly active transcriptional state (54); H4R3 dimethylation is a repressive mark (55) that is proposed nevertheless to activate key genes involved in promoting cancer cell proliferation (56, 57). For the day 28 model (Figure 6B), the most influential variable contributed positively and was the percentage of integration sites near but not in transcription units. This may reflect the dual effects of favored initial integration in active chromatin in robustly transcribing cells balanced against negative selection for integration in transcription units that disrupts function during cell growth (58). Thus, the models disclose unanticipated associations of integration-site profiles and genomic annotation linked to response.
Figure 6. Predicting clinical outcome from integration-site data.
A total of 91 features spanning population metrics, genomic features, and epigenetic features from 29 patients were used in LASSO logistic regression to build a classification model. Results are from leave-one-out cross-validation of models based on /preinfusion products (A) and day 28 peripheral blood samples (B). Bar plots indicate the contribution of different features to classification in each model. Positive values indicate correlation with a positive clinical outcome, while negative contributions indicate a correlation with negative clinical outcomes. (C) Vector integration in the CR/PRtd sample is favored in transcription units preferentially active in the T cells from CR/PRtd. RNA-Seq data were analyzed to identify the top 500 genes that were preferentially active in preinfusion products from CR/PRtd versus PR/NR. The frequency of integration in these genes (y axis) was then compared among integration sites from CR/PRtd versus PR/NR (x axis). Median values were compared using the Mann–Whitney U test.
We then sought to test the posttransduction/preinfusion model on an independent validation data set. For this, we analyzed 18 posttransduction/preinfusion samples from a trial in which CART19 therapy for CLL was augmented with the Bruton’s tyrosine kinase inhibitor ibrutinib (59). Therapeutic success was scored 3 months after treatment by bone marrow morphology/flow cytometry analysis. The model called outcome correctly in 13 of 18 cases (72% accuracy). A statistical test was conducted to assess whether the model was just guessing that the validation cohort showed the same proportions of responders and NRs as the discovery cohort; a binomial test showed the proportions to be distinct (P = 0.047; 1-sided binomial test), supporting robust function of the model. Therefore, the posttransduction/preinfusion LASSO regression model was generalizable to patients not initially used to construct the model, despite several differences in the patient cohort and outcome scoring.
What feature of the preinfusion product accounts for the difference in integration-site distributions between CR/PRtd and PR/NR? One possible explanation could be that gene activity is different in the initially harvested T cells from CR/PRtd versus PR/NR. To investigate this, we compared RNA-Seq data for preinfusion products from CR/PRtd and PR/NR patients and identified genes that were preferentially transcribed in CR/PRtd. Comparison with integration-site data showed that these genes were more often targeted for integration in CR/PRtd versus PR/NR (Figure 6C). Thus, differential gene activity provides one potential mechanism for differential integration targeting in the cell products from CR/PRtd versus PR/NR. Another question centers on whether the T cell subsets in preinfusion products differed between responders and NRs, and thus differential subset-specific transcription might have influenced integration target site selection. Previous data indicate that differential representation of T cell subsets in preinfusion products was correlated with outcome (5). A specific test of the patients studied here did not show a link between the percentage of central memory cells, which was implicated as important in the patient 10 TET2 case, and outcome in ALL and CLL (Supplemental Figure 7). Hence, we do not think that differing proportions of central memory T cell subsets in the infusion product fully explain the differing integration-site distributions and clinical outcomes. However, it remains possible that vector integration–targeting is reporting other types of compositional differences in cell products from CR/PRtd and PR/NR patients.
Discussion
We observed differences in distributions of lentiviral vector–integration sites in CAR T cells that distinguish patients showing positive clinical responses (CR/PRtd) from those showing limited or no responses (PR/NR). We attribute these differences to several mechanisms. For the first of these, transcriptional activity in the initially transduced cell pool differs, and this is associated with differences in integration targeting. This is consistent with reports that lentiviruses favor integration in active transcription units (25, 39, 40, 42, 46), and transcriptional differences have been reported to distinguish preinfusion cell products from CR/PRtd versus PR/NR (5). Other mechanisms appear to involve insertional mutagenesis of the T cell genome, so that cells with certain genetic modifications proliferate more rapidly than others. In extreme cases, this may influence therapeutic outcome, as in the TET2 example (12). More globally, differential proliferation may mark genes or pathways that influence proliferation, regardless of effects on outcome — thus, manipulation of these pathways may allow optimization of therapeutic efficacy in patients where proliferation is limiting — as is often the case in CLL. These mechanisms are discussed below.
Aggregating 91 different measures of the integration-site distributions in posttransduction/preinfusion products into a multivariate model allowed prediction of the outcome correctly in the training set with 79% accuracy using leave-one-out cross-validation. Comparison to outcome in another CLL trial not used to generate the model allowed correct prediction with 72% accuracy. This shows that there is a signal in the preinfusion product associated with success prior to cell infusion into patients. Comparison with RNA-Seq data suggested that differential transcriptional programs resulted in differential integration targeting in CR/PRtd versus PR/NR. Hypotheses explaining the role of some of the features selected by the model are readily proposed, whereas others are of unclear importance, suggesting topics for future research. H4R3me2, the most influential variable in the posttransduction/preinfusion model, is a repressive mark associated with favored methylation (55), but it is also associated with increasing proliferation of certain cancer cell types (56, 57). Proximity of integration sites to BRD3-responsive promoters was another factor positively associated with outcome. BRD3 is expressed in T cells and implicated in immune signaling (60); possibly, BRD3-responsive promoters are in a state conducive to integration in T cells that can be programmed for efficacious tumor targeting. For the day 28 model, the enrichment for integration sites near but not in transcription units was the most influential; possibly, integration in transcription units is most commonly disruptive of cell growth, so that integration sites outside of transcription units tend to accumulate with the robust growth characteristic of effective therapy. These examples show that the LASSO regression model provides multiple hypotheses for mechanisms linking T cell biology and therapeutic efficacy.
Two previous studies identified genes where alterations in activity promoted therapeutic proliferation of CAR T cells, and these genes were identified by insertional mutagenesis here. In the case of TET2, the gene was found in our integration-site survey to be called by 3 of our criteria. This was not driven solely by the single patient (patient 10) reported previously (12), because after removal of the expanded clone in patient 10 from the data set, TET2 was still called as an integration-marked gene. In the second case, the gene encoding the TGFβRII (TGFBR2) has been shown to modulate immunotherapy outcomes, and dominant-negative forms of the receptor, when introduced into CAR T cells, improved function (22). Here, a cell with an integrated vector in TGFBR2 was among our top 1% of expanded clones. These findings strengthen the idea that insertional mutagenesis in T cells can indeed modulate functions regulating proliferation in patients.
Multiple mechanisms may link CAR T cell proliferation and insertional mutagenesis. Analysis of the TET2 insertion in patient 10 suggested that altering CART19 to favor a central memory phenotype promoted long-term proliferation and function. Several of the identified pathways and genes mentioned above may also promote proliferation directly, as indicated by the enrichment in integration sites in cancer-associated genes (Supplemental Table 5). In addition, some of the integration target genes are proapoptotic (STK4, PIKFYVE), so inhibiting these functions by insertional mutagenesis may promote cell survival. These 3 mechanisms each provide targets for experimental optimization to promote CAR T cell function. Of course, care must be taken to ensure that any proproliferative modifications do not lead to excessive proliferation and frank transformation.
Several studies have investigated genes important for proliferation of T cells in different settings, yielding mostly different gene sets. Shifrut et al. performed a genome-wide CRISPR screen for genes involved in T cell proliferation ex vivo, including in a CAR-immunotherapy-based model (24). We compared their findings to ours and found no genes in common that promoted proliferation and a modest sharing of genes that, when mutant, interfered with proliferation (nominal P value = 0.012; Fisher’s exact test comparing our depletion list to genes called as depleted in replicate studies in ref. 24). Studies of HIV latency have also found persistent T cell clones marked with integration sites in specific genes (e.g., BACH2, MKL2, STAT5B) (36–38, 61, 62). Of these, only STAT5B scored in the CART19 data as a gene where integration was found more commonly in the posttransduction/preinfusion sample rather than in patient samples, suggestive of a role for proliferation in patients. TET2 was not called as notably affected in Shifrut et al. and has not emerged as a gene important in HIV latency. Evidently, the requirements for proliferation and persistence of T cells in each setting are sufficiently different to select mostly different gene sets, emphasizing the value of studying the samples recovered from CART19 patients, as described here.
Genes and pathways identified here may be modulated in CART19 to improve therapeutic outcomes. Small-molecule modulators are available for many of the pathways marked by insertional mutagenesis. For example, several of the genes called as targets for insertional mutagenesis encode kinases (e.g., STK4, CAMK2D, PIKFYVE, CDK8, MAPK14, and TGFBR2), which are the targets of known inhibitors. Any of the genes identified can also be downmodulated using shRNAs or CRISPR knockouts. Thus, the data presented here provide a rich source of starting points to improve CAR T cell function.
The integration-site analysis of preinfusion transduction products and application of the multivariate model may allow evaluation of products during the manufacturing process and before reinfusion. For example, cell collection and isolation methods, expansion conditions, and transduction protocols could potentially be optimized using this assay; cell batches could also be tested for suitability before expensive full-scale manufacturing and infusion into patients.
In summary, these data suggest that insertional mutagenesis may specify multiple genes, pathways, and mechanisms potentially involved in therapeutic proliferation of CART19. These findings provide multiple potential approaches to optimizing T cell engineering for optimal function in immunotherapy protocols.
Methods
Human subjects.
Specimens were acquired from patients diagnosed with CLL or ALL who were enrolled in clinical trials for CART19 therapy (ClinicalTrials.gov NCT01626495, NCT01747486, and NCT01029366) (1, 4) or CART19 therapy in combination with ibrutinib (ClinicalTrials.gov NCT02640209). All ethical regulations were followed. Given that limited numbers of patients have been treated by CART19 therapy, sample sizes and duration of data collection were dictated by patient availability. The study design was necessarily open label, with the goal of asking whether features of vector-integration site distributions were associated with outcome.
Patients were assigned to outcome categories as follows. CR patients exhibited robust in vivo proliferation of CAR T cells, coincident with rapid clearance of leukemia from the blood and bone marrow and, in certain cases, significant reductions in nodal disease burden (4, 5, 10, 11). Sustained remission in some patients (e.g., CLL patients) was associated with durable persistence and function of CAR T cells and eradication of the leukemia clone, as determined by deep-sequencing analysis of the immunoglobulin heavy chain locus (4). Overall, CAR T cell persistence is shorter in many patients with ALL relative to CLL patients who respond, even though rates of complete remission are higher in ALL (63, 64). A small subset of CLL patients whom we have previously studied exhibited T cell expansion and tumor elimination kinetics similar to what was observed in CR patients, but they were designated as PRtd (5, 65). In contrast to CR and PRtd patients, CART19 expansion as well as persistence were less robust in individuals who exhibited a typical PR and minimal in NR patients (4, 5).
Sequencing sites of vector integration.
Our standard operating procedures have been described elsewhere (44, 45). Briefly, each genomic DNA sample (typically 300–500 ng of DNA) was sheared by sonication and ligated with unique linkers. Nested PCR was used to amplify DNA from the LTR of the integrated vectors to the linker. LTR-specific sequences included PCR1 primer CTTAAGCCTCAATAAAGCTTGCCTTGAG and PCR2 primer caagcagaagacggcatacgagatXXXXXXXXXXXXAGTCAGTCAGCCAGACCCTTTTAGTCAGTGTGGAAAATC, where lowercase nucleotides denote the Illumina P7 sequence for binding to the Illumina platform, Xs denote a 12-nucleotide barcode, and uppercase nucleotides match to the vector LTR sequence. Linker-specific primers annealed in a nested fashion to 1 of 96 sequences previously presented (44) and contained the Illumina P5 sequence appended to the beginning of the PCR2 primer. All samples were analyzed in quadruplicate independently to suppress founder effects in the PCR. Different adapters were used for each sample to suppress PCR crossover. DNA molecules were bar coded at both ends, and only molecules with 2 correct bar codes were accepted for analysis, minimizing the effects of PCR recombination. Cycling conditions were as follows: initial melt for 1 minute at 95°C; 25 cycles (for PCR1) or 20 cycles (for PCR2) of 30 seconds at 95°C, anneal at 80°C, and extend at 70°C for 5 cycles and then 67°C for the remaining cycles; a final extension for 4 minutes at 72°C; and a hold at 4°C. Amplified products were then purified using AMPure XP beads (Beckman Coulter) at a ratio of 0.7 (beads to sample, v/v) and quantified using KAPA library quantification qPCR. Amplicons were sequenced on an Illumina platform with 300-cycle kits (v2 chemistry).
Flow cytometric analysis of CAR expression.
Retrospective CART19 infusion products from CLL patients were thawed and rested in 24-well tissue culture plates (BD) at a concentration of 1 × 106 cells/mL. Cells were then preincubated with Aqua Blue dead cell exclusion dye (Invitrogen) in serum-free conditions and subsequently surface stained with commercially available flow cytometry antibodies against CD45, CD3, and CD8 (Biolegend). CAR expression was measured with an Alexa Fluor 647–conjugated anti-idiotypic antibody (a gift from thank B. Jena and L. Cooper [MD Anderson Cancer Center]), as previously described (66). All flow cytometry reagents were titrated prior to use. Fluorescence-minus-one controls were created for each antibody used in the above panel to set positive and negative gates. Samples were acquired on an LSRFortessa flow cytometer (BD). Data were analyzed using FlowJo version 10 software. CAR expression levels were reported as the geometric mean of the mean fluorescence intensity of cells labeled with the CAR anti-idiotype reagent. Sources of antibodies used are listed in Supplemental Table 7.
Bioinformatic analysis.
The integration-site sequence data were worked up using the INSPIIRED pipeline as described (26, 39–45). Briefly, DNA sequences were demultiplexed and trimmed of synthetic or LTR-specific sequences. Remaining sequences were filtered against the vector sequence to removed internal fragments sequenced not blocked by the INSPIIRED protocol. After filtering, unique sequences were independently aligned to the hg38 reference genome using BLAT. Alignment for R1 and R2 sequences were then joined together and filtered for quality alignments, yielding unique sites of integration or multihit locations. Data were stored within a SQL database for analysis to produce individual patient reports or to be used for further research. Integration sites with host-derived upstream sequences matching the positive-filtering “bit” sequence (TCTAGCA) were ignored so as not to confound the analysis with mispriming events.
Minimal population sizes were estimated with Chao1 and jackknifing. This showed that for each sample, we analyzed only a portion of the full population, averaging 17% of the Chao1 minimal population size estimate.
Comparison of RNA-Seq data to integration-site distributions.
RNA-Seq read-count data for CART19 patient transduction products were acquired from supplemental data in ref. 5. Gene expression was ranked by the average reads per kilobase million (RPKM) intensity across all patient-stimulated data, and genes were binned into 10 evenly sized groups based on their rank order. The percentage of integration sites within each bin from each patient was calculated and then averaged across response groups. Differential expression was determined from the difference in RPKM values of paired stimulated and mock-stimulated data for each patient, followed by averaging across response groups. Highly differentially expressed genes were then ranked by those favoring expression in PR/NR patients and those favoring expression in CR/PRtd patients. The percentage of integration sites observed was calculated for each patient from the 500 genes most favoring expression within CR/PRtd. Genes and patients were only considered if they were present in both studies. Significant differences between groups were assessed using nonparametric Wilcoxon’s tests. Data were deposited in NCBI’s Database of Genotypes and Phenotypes (phs001707.v1.p1).
Data availability.
Sequence data were deposited in the NCBI’s Sequence Read Archive (SRA BioProject PRJNA510570). Source code for manuscript analysis has been deposited in an archived format in Zenodo, https://doi.org/10.5281/zenodo.3366188
Statistics.
All statistical analyses are fully specified in the archived analytical code, available at https://doi.org/10.5281/zenodo.3366188 Sample sizes appear in Supplemental Table 1. Effects of multiple comparisons in the analysis of genomic clusters and orientation bias were controlled using the Benjamini-Hochberg FDR method. A P value of less than 0.05 was considered significant. Student’s t test was 2 tailed. For 1-way ANOVA, no post tests were applied because the overall P value was not significant. The LASSO regression model was generated using the glmnet package in R. Further information on statistical methods appears in Supplemental Report 1.
Study approval.
All clinical trials analyzed were approved by the institutional review board of the University of Pennsylvania. Written informed consent was received by all patients prior to enrollment in the study.
Author contributions
CLN, SSM, JKE, SR, JAF, SFL, JJM, and FDB performed experiments or analyzed data. All authors helped design experiments and assisted with interpretation of data. DLP, NF, SIG, SAG, SLM, DLS, BLL, and CHJ designed and/or carried out clinical trials. CLN, JAF, SFL, JJM, and FDB wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Minnal Gupta, Irina Kulikovskaya, Rachel Reynolds, and Angela Kim of the Translational and Correlative Studies Laboratory of the University of Pennsylvania for isolation of genomic DNA, and Anne Lamontagne and Alex Malykhin of the Clinical Cell and Vaccine Production Facility of the University of Pennsylvania. This work was supported by Novartis Institute for Biomedical Research (to CHJ, JAF, SFL, JJM, and FDB); NIH grants R01CA241762 (to JJM, JAF, SFL, and FDB) and T32CA009140 (to JAF); the Gabrielle’s Angel Foundation (to JAF); and NIH grants P01CA214278 (to JAF, DLP, SG, BLL, SFL, and CHJ), K08CA194256 (to SG), P30CA016520 (to BLL), R01CA226983 (to CHJ), U54CA244711 (to JAF and CHJ) R01AI082020, R01AI045008, and R01AI117950 (to FDB). We also acknowledge support from the Penn Center for AIDS Research (P30AI045008) and the PennCHOP Microbiome Program (to FDB). The authors thank B. Jena and L. Cooper (MD Anderson Cancer Center) for providing the CAR anti-idiotype detection reagent. C. Imai, D. Campana and others at St. Jude Children’s Research Hospital designed, developed and provided, under material-transfer agreements, the CD19-directed CAR used in this study.
Version 1. 12/17/2019
Electronic publication
Version 2. 02/03/2020
Print issue publication
Version 3. 02/18/2020
Corrected publish date in pdf file
Funding Statement
JAF
JAF
D.L.P, S.G., B.L.L., SFL, C.H.J.
B.L.L.
C.H.J.
S.G.
F.D.B.
F.D.B.
F.D.B.
F.D.B.
C.H.J., S.F.L., J.J.M., F.D.B.
Footnotes
Conflict of interest: CLN, JAF, DLP, SIG, BLL, CHJ, SFL, JJM, and FDB hold patent applications (US 20180258149, US 20180140602, US 20150283178) related to CAR T cell therapy. JAF, SIG, CHJ, SFL, JJM, and FDB receive research funding from Novartis. JAF, SIG, and SFL also receive research funding from Tmunity Therapeutics. JJM obtains additional research support from Incyte Corporation. SIG is a cofounder of Carisma Therapeutics and discloses consultancy. BLL and CHJ are cofounders and equity holders in Tmunity Therapeutics. BLL is a consultant for Novartis as well as CRC Oncology and is a member of the scientific advisory boards of Avectas, Brammer Bio, Cure Genetics, and Incysus. JJM is a consultant for Shanghai Unicar-Therapy Bio-medicine Technology Co. Ltd, Simcere of America Inc., IASO Biotherapeutics, and Poseida Therapeutics and is a member of the scientific advisory board of IASO Biotherapeutics.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):673–685.https://doi.org/10.1172/JCI130144.
Contributor Information
Scott Sherrill-Mix, Email: shescott@upenn.edu.
John K. Everett, Email: everj@pennmedicine.upenn.edu.
Shantan Reddy, Email: sgujja@mail.med.upenn.edu.
Noelle Frey, Email: noelle.frey@uphs.upenn.edu.
Carl H. June, Email: cjune@upenn.edu.
Simon F. Lacey, Email: simon.lacey@uphs.upenn.edu.
J. Joseph Melenhorst, Email: jos.melenhorst@uphs.upenn.edu.
References
- 1.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davila ML, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6(224):224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee DW, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter DL, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7(303):303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraietta JA, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24(5):563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali SA, et al. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood. 2016;128(13):1688–1700. doi: 10.1182/blood-2016-04-711903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brudno JN, et al. T cells genetically modified to express an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J Clin Oncol. 2018;36(22):2267–2280. doi: 10.1200/JCO.2018.77.8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster SJ, et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N Engl J Med. 2017;377(26):2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraietta JA, Schwab RD, Maus MV. Improving therapy of chronic lymphocytic leukemia with chimeric antigen receptor T cells. Semin Oncol. 2016;43(2):291–299. doi: 10.1053/j.seminoncol.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalos M, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95):95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraietta JA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558(7709):307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buscarlet M, et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130(6):753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 16.Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118(1):294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graef P, et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity. 2014;41(1):116–126. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Carty SA, et al. The loss of TET2 promotes CD8+ T cell memory differentiation. J Immunol. 2018;200(1):82–91. doi: 10.4049/jimmunol.1700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyrakis PA, et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature. 2016;540(7632):236–241. doi: 10.1038/nature20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N, Bevan MJ. TGF-β signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat Immunol. 2012;13(7):667–673. doi: 10.1038/ni.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26(5):579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Kloss CC, et al. Dominant-negative TGF-β receptor enhances PSMA-targeted human CAR T cell proliferation and augments prostate cancer eradication. Mol Ther. 2018;26(7):1855–1866. doi: 10.1016/j.ymthe.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sack LM, et al. Profound tissue specificity in proliferation control underlies cancer drivers and aneuploidy patterns. Cell. 2018;173(2):499–514.e23. doi: 10.1016/j.cell.2018.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shifrut E, et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175(7):1958–1971.e15. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavazzana M, Bushman FD, Miccio A, André-Schmutz I, Six E. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov. 2019;18(6):447–462. doi: 10.1038/s41573-019-0020-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang GP, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115(22):4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe SJ, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacein-Bey-Abina S, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363(4):355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deichmann A, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117(8):2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzwaelder K, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117(8):2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kustikova OS, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17(9):1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wünsche P, et al. Mapping active gene-regulatory regions in human repopulating long-term HSCs. Cell Stem Cell. 2018;23(1):132–146.e9. doi: 10.1016/j.stem.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Kustikova OS, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109(5):1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 40.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2(11):e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28(6):755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2(8):E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman E, et al. INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol Ther Methods Clin Dev. 2017;4:39–49. doi: 10.1016/j.omtm.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry CC, et al. INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol Ther Methods Clin Dev. 2017;4:17–26. doi: 10.1016/j.omtm.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craigie R, Bushman FD. Host factors in retroviral integration and the selection of integration target sites. Microbiol Spectr. 2014;2(6):MDNA3-0026-2014. doi: 10.1128/microbiolspec.MDNA3-0026-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12(1):51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 48.Braun CJ, et al. Gene therapy for Wiskott-Aldrich syndrome — long-term efficacy and genotoxicity. Sci Transl Med. 2014;6(227):227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 49.Marshall HM, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2(12):e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang GP, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17(5):844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine BL, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci U S A. 2006;103(46):17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tebas P, et al. Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV. Blood. 2013;121(9):1524–1533. doi: 10.1182/blood-2012-07-447250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ocwieja KE, et al. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011;7(3):e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30(1):51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2010;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu R, et al. PHD finger protein 1 (PHF1) is a novel reader for histone H4R3 symmetric dimethylation and coordinates with PRMT5-WDR77/CRL4B complex to promote tumorigenesis. Nucleic Acids Res. 2018;46(13):6608–6626. doi: 10.1093/nar/gky461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng X, et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene. 2017;36(9):1223–1231. doi: 10.1038/onc.2016.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poletti V, et al. Preclinical development of a lentiviral vector for gene therapy of X-linked severe combined immunodeficiency. Mol Ther Methods Clin Dev. 2018;9:257–269. doi: 10.1016/j.omtm.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fraietta JA, et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood. 2016;127(9):1117–1127. doi: 10.1182/blood-2015-11-679134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren W, et al. Bromodomain protein Brd3 promotes Ifnb1 transcription via enhancing IRF3/p300 complex formation and recruitment to Ifnb1 promoter in macrophages. Sci Rep. 2017;7:39986. doi: 10.1038/srep39986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis. 2007;195(5):716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 62.Cesana D, et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat Commun. 2017;8(1):498. doi: 10.1038/s41467-017-00609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maude SL, Teachey DT, Porter DL, Grupp SA. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood. 2015;125(26):4017–4023. doi: 10.1182/blood-2014-12-580068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown JR, Porter DL, O’Brien SM. Novel treatments for chronic lymphocytic leukemia and moving forward. Am Soc Clin Oncol Educ Book. 2014:e317-25. doi: 10.14694/EdBook_AM.2014.34.e317. [DOI] [PubMed] [Google Scholar]
- 65.Evans AG, et al. Evolution to plasmablastic lymphoma evades CD19-directed chimeric antigen receptor T cells. Br J Haematol. 2015;171(2):205–209. doi: 10.1111/bjh.13562. [DOI] [PubMed] [Google Scholar]
- 66.Jena B, et al. Chimeric antigen receptor (CAR)-specific monoclonal antibody to detect CD19-specific T cells in clinical trials. PLoS One. 2013;8(3):e57838. doi: 10.1371/journal.pone.0057838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Casper J, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46(D1):D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited in the NCBI’s Sequence Read Archive (SRA BioProject PRJNA510570). Source code for manuscript analysis has been deposited in an archived format in Zenodo, https://doi.org/10.5281/zenodo.3366188