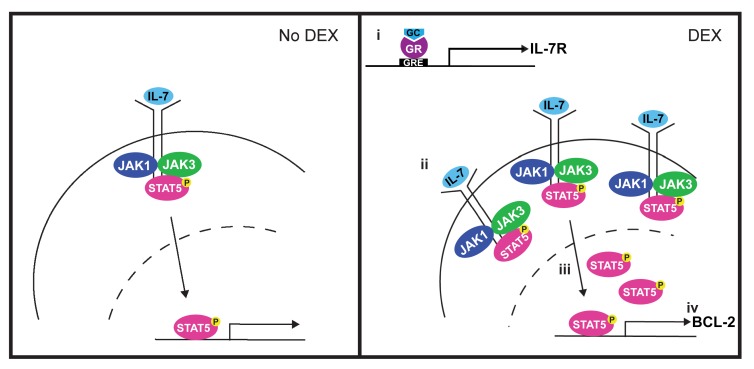
Abstract
Glucocorticoids (GCs) are a central component of therapy for patients with T cell acute lymphoblastic leukemia (T-ALL), and although resistance to GCs is a strong negative prognostic indicator in T-ALL, the mechanisms of GC resistance remain poorly understood. Using diagnostic samples from patients enrolled in the frontline Children’s Oncology Group (COG) T-ALL clinical trial AALL1231, we demonstrated that one-third of primary T-ALLs were resistant to GCs when cells were cultured in the presence of IL-7, a cytokine that is critical for normal T cell function and that plays a well-established role in leukemogenesis. We demonstrated that in these T-ALLs and in distinct populations of normal developing thymocytes, GCs paradoxically induced their own resistance by promoting upregulation of IL-7 receptor (IL-7R) expression. In the presence of IL-7, this augmented downstream signal transduction, resulting in increased STAT5 transcriptional output and upregulation of the prosurvival protein BCL-2. Taken together, we showed that IL-7 mediates an intrinsic and physiologic mechanism of GC resistance in normal thymocyte development that is retained during leukemogenesis in a subset of T-ALLs and is reversible with targeted inhibition of the IL-7R/JAK/STAT5/BCL-2 axis.
Keywords: Oncology
Keywords: Leukemias, Signal transduction, T cells
Introduction
T cell acute lymphoblastic leukemia (T-ALL) is a genetically heterogeneous disease characterized by a range of alterations involving transcription factors, cell-cycle regulators, and signal transduction effectors (1). Unlike B cell ALL (B-ALL), in which genetic factors are widely used to inform risk stratification and subsequent intensification of therapy (2), few genetic lesions in T-ALL have independent prognostic significance (1). As a result, efforts to implement risk-adapted therapeutic strategies have been limited by a lack of genetic biomarkers, highlighting the need for functional studies aimed instead at elucidating recurrent patterns of drug response and resistance across the spectrum of T-ALL.
Although outcomes for children with T-ALL have improved dramatically over the past several decades, children with relapsed T-ALL continue to face poor survival rates (3), suggesting that novel strategies are needed to improve the upfront efficacy of therapy in order to induce deeper remissions and decrease the likelihood of disease relapse. Glucocorticoids (GCs) are a central component of T-ALL therapy, and the initial response to GC therapy is an important predictor of long-term outcomes (4). For example, on the ALL-Berlin-Frankfurt-Münster (ALL-BFM) 95 trial, patients were stratified into groups of those who had a prednisone good response (PGR) and those who had a prednisone poor response (PPR) following 7 days of prednisone monotherapy. Patients with a PGR had an 8-year event-free survival rate of 81.3%, as opposed to only 55.1% for patients with a PPR (5). These data demonstrate that intrinsic differences in GC sensitivity exist at the time of disease diagnosis and that these differences can have long-term prognostic significance. Despite decades of clinical use, a comprehensive understanding of the mechanistic basis for differential intrinsic GC sensitivity is lacking. GCs act by binding to a cytoplasmic GC receptor (GR), which promotes translocation of the GR to the nucleus, where it binds to target gene loci and induces a transcriptional program that results in apoptosis in lymphoid cells (6). Unlike other agents used in the treatment of T-ALL, GCs are unique in that they also exist as endogenous hormones that play critical roles in normal T cell physiology. For example, endogenous GC activity has been shown to interact with T cell receptor (TCR) signaling to shape the developing T cell repertoire (7, 8) and to promote T cell homeostasis in the periphery following an immune response (9). Given these frequent encounters with GCs in normal physiology and the fact that GCs are potent inducers of apoptosis in both normal and transformed lymphoid cells, we reasoned that T cells must possess intrinsic mechanisms that allow them to resist GC-induced apoptosis under certain developmental and environmental conditions. Furthermore, we hypothesized that these mechanisms may be retained during leukemogenesis and exploited to confer resistance to GC therapy in T-ALL.
One critical endogenous factor in the T-ALL microenvironment is the cytokine IL-7. In addition to promoting the survival and differentiation of developing thymocytes (10), the IL-7 receptor/JAK/STAT5 (IL-7R/JAK/STAT5) signaling pathway contributes to T-ALL pathogenesis and disease maintenance (11–13). We previously demonstrated that over half of primary treatment–naive T-ALL patient samples were intrinsically resistant to the glucocorticoid dexamethasone (DEX) when cultured in the presence of IL-7. Of these DEX-resistant samples, half could be sensitized to DEX with inhibition of JAK signaling (14). Interestingly, the majority of samples with JAK/STAT5-mediated DEX resistance lacked activating mutations in components of the IL-7R/JAK/STAT5 pathway. In the present study, we analyzed a larger cohort of fresh diagnostic samples obtained from pediatric patients enrolled in the frontline Children’s Oncology Group (COG) T-ALL clinical trial AALL1231, with the goal of establishing the mechanistic basis for this DEX resistance phenotype. In this cohort, we demonstrated that one-third of primary diagnostic samples were resistant to DEX specifically when cultured in the presence of IL-7. Furthermore, we found that subsets of normal developing thymocytes also had this IL-7–induced DEX resistance phenotype. Through functional analyses, we elucidated a mechanism by which GCs paradoxically induced their own resistance by augmenting the prosurvival activity of the IL-7R/JAK/STAT5 pathway in distinct subsets of developing thymocytes and T-ALLs cells. Taken together, these data suggest that IL-7 facilitates GC resistance in developing thymocyte populations and that subsets of T-ALL cells retain this capacity to utilize IL-7 as a means of resisting GC-induced apoptosis. These findings have significant therapeutic implications, as they suggest that inhibition of the IL-7R/JAK/STAT5 pathway or its transcriptional targets may enhance GC efficacy in patients who have a poor initial response to GC therapy.
Results
JAK signaling mediates DEX resistance in a subset of T-ALLs.
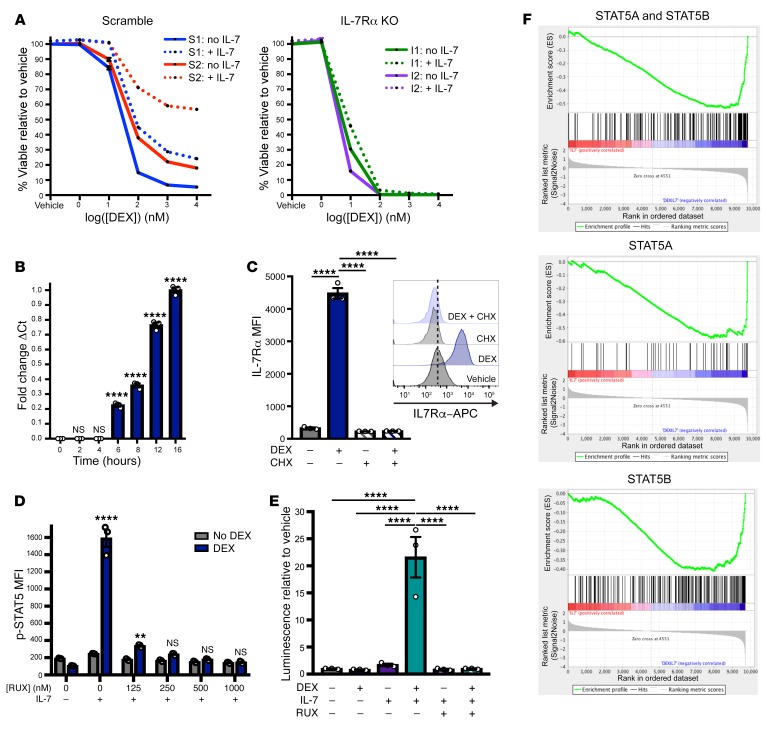
In a large independent cohort consisting of 73 samples from patients enrolled in COG AALL1231, we validated our previous finding that in vitro DEX sensitivity was highly variable across T-ALLs (Figure 1A) (14). Importantly, these differences in DEX sensitivity were not dose dependent but persisted at saturating concentrations of DEX (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI130189DS1). Using the definition “DEX-resistant” as samples that retained greater than 50% cell viability following DEX exposure, we found that 63% of primary diagnostic samples were intrinsically DEX resistant when cultured in the presence of IL-7 (Figure 1A). Binding of IL-7 to the IL-7R results in the recruitment of JAK1 and JAK3, which subsequently become phosphorylated to create docking sites for STAT5 (15). Consistent with our previous findings (14), inhibition of JAK signaling with the JAK1/2 inhibitor ruxolitinib (RUX) was sufficient to overcome DEX resistance in 54% of these DEX-resistant samples (P < 0.0001 for DEX versus DEX plus RUX and for RUX versus DEX plus RUX) (Figure 1B).
Figure 1. JAK/STAT inhibition overcomes DEX resistance in a subset of primary T-ALL samples and in the T-ALL cell line CCRF-CEM.
(A) Viability relative to vehicle control of cells from 73 primary diagnostic T-ALL samples exposed to 2.5 μM DEX for 48 hours in the presence of 25 ng/mL IL-7. The red line indicates the 50% viability cutoff used to define DEX resistance. (B) Viability of cells from the 46 DEX-resistant primary diagnostic T-ALL samples in A exposed to 2.5 μM DEX and/or 0.5 μM RUX for 48 hours in the presence of 25 ng/mL IL-7, relative to vehicle control cells. ****P < 0.0001, by 1-way ANOVA with Tukey’s method for multiple comparisons adjustment. (C) Viability of CCRF-CEM cells exposed for 72 hours to DEX in the absence or presence of increasing concentrations of IL-7. The experiment was performed in technical triplicate. (D) Viability of CCRF-CEM cells exposed to DEX in the presence of 25 ng/mL IL-7 in the absence or presence of increasing concentrations of RUX. The experiment was performed in technical triplicate. The “No IL-7” (black line) and the 25 ng/mL IL-7 (red line) conditions were replotted from Figure 1C. (E) Heatmap of Bliss independence scores calculated as the average of technical triplicates for the combination of DEX and RUX in CCRF-CEM cells cultured in the presence of 25 ng/mL IL-7 for 72 hours, in which positive values, indicated in red, are indicative of a synergistic interaction. All CCRF-CEM cell data are representative of 3 independent experiments.
To facilitate further studies aimed at investigating the mechanistic basis for DEX resistance mediated by JAK signaling, we next evaluated the human T-ALL cell line CCRF-CEM for its utility as a model system in which to study the DEX resistance phenotype observed in these patients’ primary T-ALL samples. This analysis revealed a dose-dependent reduction in DEX sensitivity with increasing concentrations of IL-7 (Figure 1C and Supplemental Figure 1B). Consistent with the primary samples, RUX was sufficient to completely restore DEX sensitivity in CCRF-CEM cells in the presence of IL-7 (Figure 1D). Furthermore, Bliss independence analysis indicated a synergistic interaction between DEX and RUX (Figure 1E). To ensure that this sensitization effect was due specifically to JAK1 inhibition by RUX and not to off-target effects, we also used the JAK3 inhibitor tofacitinib, which should similarly inhibit IL-7R signaling, and the JAK2 inhibitor CHZ868, which should not inhibit IL-7R signaling. In this analysis, tofacitinib phenocopied the effects of RUX to overcome IL-7–induced DEX resistance, whereas CHZ868 had no effect on cell viability (Supplemental Figure 1C), suggesting that on-target inhibition of JAK1 or JAK3 is sufficient to abrogate IL-7–induced DEX resistance.
DEX exposure augments IL-7R/JAK/STAT5 pathway activity.
To confirm that IL-7 induces DEX resistance in CCRF-CEM cells via signaling through IL-7R, we first used CRISPR/Cas9 genome editing to generate clonal populations of scrambled control and IL-7Rα–KO CCRF-CEM cells (Supplemental Figure 2A). In the KO clones, loss of IL-7R expression was sufficient to restore DEX sensitivity in the presence of IL-7 (Figure 2A). We next asked whether IL-7R signaling interferes with GR activation and/or function. First, to determine whether exposure to IL-7 alters the availability of the GR for DEX binding, we assessed GR protein expression in CCRF-CEM cells treated with DEX with or without IL-7. Under these conditions, we found that DEX exposure effectively induced comparable levels of GR expression regardless of the presence of IL-7 (Supplemental Figure 2B). Furthermore, upon exposure to DEX in the absence or presence of IL-7, GR effectively translocated to the nucleus and became phosphorylated on Ser211 (Supplemental Figure 2C), a modification that has been shown to correlate with the capacity to activate or repress transcription (16). Finally, to determine whether IL-7 interferes with induction of the GR transcriptional program, we performed RNA-Seq on 4 scrambled control CCRF-CEM cell clones exposed to vehicle control or DEX with or without IL-7 for 4 hours to identify primary GR target genes. Under these conditions, IL-7 did not interfere with GR-mediated transcript induction or repression (Supplemental Figure 2D and Supplemental Table 1). Taken together, these data suggest that GR activity remains intact in the presence of IL-7.
Figure 2. DEX exposure augments IL-7R expression and downstream JAK/STAT signaling.
(A) Viability of scrambled control (S1 and S2) and IL-7Rα KO (I1 and I2) CCRF-CEM cell clones exposed for 72 hours to DEX in the absence (solid lines) or presence (dotted lines) of 25 ng/mL IL-7. The experiment was performed in technical triplicate. (B) Fold change relative to the 16-hour time point of the ΔCt of IL7RA transcript levels relative to GAPDH as determined by qPCR, performed in technical triplicate in CCRF-CEM cells exposed to 1 μM DEX and 100 ng/mL IL-7 for the indicated duration. (C) MFI of IL-7Rα in CCRF-CEM cells treated with or without 1 μM DEX and/or 10 μg/mL CHX in technical triplicate for 24 hours. Inset shows representative histograms of IL-7Rα in CCRF-CEM cells treated with or without 1 μM DEX and/or 10 μg/mL CHX for 24 hours. (D) MFI of p-STAT5 in CCRF-CEM cells treated with or without 1 μM DEX for 24 hours in the absence of IL-7 followed by a 1-hour exposure to vehicle control or RUX prior to a 15-minute stimulation with 100 ng/mL IL-7 in technical triplicate. Significance is relative to the DEX-treated condition in the absence of IL-7 stimulation. (E) Relative luminescence of CCRF-CEM cells transfected with the STAT5 reporter construct and treated with or without 1 μM DEX, 100 ng/mL IL-7, and 500 nM RUX in technical triplicate for 36 hours prior to lysis and measurement of luciferase activity. (F) GSEA plots of STAT5 gene expression signatures comparing scramble control clones (n = 4) treated with 100 ng/mL IL-7 versus the combination of 1 μM DEX and 100 ng/mL IL-7 for 16 hours. **P < 0.01 and ****P < 0.0001, by 1-way ANOVA with Tukey’s method for multiple comparisons adjustment (B–E). With the exception of the RNA-Seq experiment, all data are representative of 3 independent experiments.
In some mature T cell populations, GCs have been shown to induce expression of IL-7Rα (17–20). Specifically, ChIP studies in both murine (21) and human (22) lymphocytes have demonstrated that the GR is recruited to a GR-binding motif in a noncoding sequence upstream of the IL-7Rα promoter, and deletion of this region is sufficient to abrogate GR-induced upregulation of IL-7Rα (17), suggesting that IL-7Rα upregulation occurs as a direct transcriptional effect of activated GR. To determine whether DEX modulates IL-7Rα expression in CCRF-CEM cells, we measured IL7RA transcript levels before and after DEX exposure and found a time-dependent increase in IL7RA transcript expression (Figure 2B) that occurred in the absence and presence of IL-7 (Supplemental Figure 2E). DEX exposure also increased IL-7Rα protein expression at the cell surface relative to expression levels in untreated cells (P < 0.0001). This increase was inhibited in the presence of the translation inhibitor cycloheximide (CHX), suggesting that the increase in cell-surface IL-7Rα reflects de novo protein synthesis rather than relocalization of existing protein (Figure 2C). Given our finding that inhibition of JAK signaling is sufficient to sensitize cells to DEX in the presence of IL-7, we next asked whether the increase in cell-surface IL-7R expression is associated with an increased capacity for JAK signaling downstream of the IL-7R. We first exposed CCRF-CEM cells to DEX for 24 hours and then stimulated them with IL-7 and measured the induction of phosphorylated STAT5 (p-STAT5) as a downstream effector of JAK signaling (Supplemental Figure 2F). Under these conditions, DEX exposure resulted in significantly more robust p-STAT5 induction in response to IL-7 stimulation relative to untreated cells (P = 0.0002). Furthermore, pretreating these cells with RUX for 1 hour prior to IL-7 stimulation was sufficient to abrogate the increased JAK/STAT5 signaling following DEX exposure (Figure 2D and Supplemental Figure 2G). Importantly, IL-7 stimulation, either with or without DEX pretreatment, did not induce phosphorylation of other STAT proteins that are activated downstream of other γ chain cytokine receptors (Supplemental Figure 2H). Given this specific increase in STAT5 activation upon IL-7 stimulation following DEX exposure, we then asked whether the increase in IL-7Rα expression was associated with a sustained increase in STAT5 transcriptional activity. To test this, we performed transient transfection of CCRF-CEM cells with a STAT5-luciferase reporter construct and assessed STAT5-induced luciferase activity in cells exposed to DEX in the presence or absence of IL-7 for 36 hours. This analysis revealed a significant induction of STAT5 transcriptional activity in cells exposed to the combination of DEX and IL-7 relative to either DEX or IL-7 alone (P < 0.0001 versus both DEX alone and IL-7 alone). Furthermore, the addition of RUX was sufficient to overcome this increase in transcriptional activity (Figure 2E). To further confirm this increase in STAT5 transcriptional output, we performed RNA-Seq on scrambled control CCRF-CEM cell clones treated with DEX, with or without IL-7, for 4 hours. Using gene set enrichment analysis (GSEA) with gene sets derived from published STAT5 ChIP-Seq data in human CD4+ T cells (23), we found enrichment for transcriptional targets of STAT5A and STAT5B (P < 0.0001; FDR <0.0001); STAT5A alone (P = 0.002; FDR = 0.001); and STAT5B alone (P < 0.0001; FDR = 0.001) in cells exposed to DEX plus IL-7 relative to IL-7 alone (Figure 2F and Supplemental Table 2), further supporting the finding that, in the presence of IL-7, DEX exposure augments activation of the STAT5 transcriptional program.
BCL-2 is a critical mediator of IL-7–induced DEX resistance.
On the basis of these findings, we hypothesized that STAT5 may be an important downstream mediator of DEX resistance in the presence of IL-7. To directly interrogate the role of STAT5 in this context, we used CRISPR/Cas9 genome editing to generate clonal populations of STAT5A-KO, STAT5B-KO, and STAT5A/B double-KO CCRF-CEM cells (Figure 3A). Importantly, deletion of 1 or both STAT5 isoforms did not affect GR expression relative to expression in scrambled control clones (Supplemental Figure 3A). Exposure of STAT5-KO cells to DEX in the presence or absence of IL-7 revealed that deletion of STAT5B, but not STAT5A, was sufficient to significantly attenuate DEX resistance in the presence of IL-7 (Figure 3B and Supplemental Figure 3B). These data suggest that transcriptional targets of STAT5B represent candidate DEX resistance genes. To identify these candidate genes, we performed RNA-Seq on scrambled control and STAT5A/B-KO CCRF-CEM cell clones treated with DEX with or without IL-7 for 16 hours. Using these data, we found that deletion of STAT5A/B did not affect the capacity for DEX-induced upregulation of IL7RA (Supplemental Figure 3C), further confirming that IL-7R upregulation is a GR-dependent but STAT5-independent transcriptional event. Using differential expression analysis, we identified the top differentially expressed genes between scrambled control cells treated with DEX alone or with a combination of DEX and IL-7 (Supplemental Table 3). We then compared this gene list with that of the core enrichment genes from the STAT5B gene set (Figure 2F and Supplemental Table 2) to identify STAT5B target genes that are differentially expressed in cells exposed to DEX relative to the combination of DEX and IL-7. This analysis identified the antiapoptotic family member BCL2 (log fold change = 1.48 for DEX plus IL-7 relative to DEX alone) and the Rho guanine nucleotide exchange factor ARHGEF3 (log fold change = 1.64) as 2 candidate mediators of DEX resistance in the presence of IL-7 (Figure 3C). Consistent with their presence on both of these gene lists, targeted analysis of the RNA-Seq data revealed that these genes were induced by the combination of DEX and IL-7 relative to treatment with DEX or IL-7 alone only in the scrambled control clones but not in the STAT5A/B-KO clones (Figure 3, D and E).
Figure 3. STAT5B, but not STAT5A, mediates the upregulation of BCL-2 expression in cells exposed to the combination of DEX and IL-7.
(A) Evaluation of STAT5A and STAT5B expression by Western blotting in independent scrambled control (S1–S4) and STAT5 single-KO (A1–A4 or B1–B4) and double-KO (AB1–AB4) CCRF-CEM cell clones (n = 4 per genotype). (B) Viability of independent scrambled control and STAT5-KO CCRF-CEM cell clones (n = 4 per genotype) treated with 100 nM DEX with or without 25 ng/mL IL-7 for 72 hours. (C) Venn diagram depicting the overlap between the top differentially expressed genes among scrambled control CCRF-CEM cell clones (n = 4) treated with DEX versus DEX plus IL-7 and STAT5B target genes. (D and E) Fold change in the fragments per kilobase per million mapped reads (FPKM) values for (D) BCL2 transcript and (E) ARHGEF3 transcript levels as determined by RNA-Seq analysis of scrambled control cells (n = 4) and STAT5A/B double-KO (n = 4) CCRF-CEM cell clones treated in the absence or presence of 100 ng/mL IL-7 and/or 1 μM DEX for 16 hours. (F) ΔMFI of BCL-2 protein expression in scrambled control (n = 4) and STAT5-KO (n = 4) CCRF-CEM cell clones treated with 100 ng/mL IL-7 and 1 μM DEX relative to 100 ng/mL IL-7 alone for 48 hours. (G) BCL2L11 and BCL2 transcript expression in CCRF-CEM cells cultured in the absence or presence of 1 μM DEX, 100 ng/mL IL-7, and/or 10 μg/mL CHX for 16 hours as determined by qPCR performed in technical triplicate. *P < 0.05, **P < 0.01, and ****P < 0.0001, by paired t test (B), 2-sample t test (D and E), or 1-way ANOVA with Tukey’s method for multiple comparisons adjustment (F and G). With the exception of the RNA-Seq experiment, all data are representative of 3 independent experiments.
Given the antiapoptotic function of BCL-2 and the importance of downregulation of BCL-2 for DEX-induced apoptosis in T-ALL cells (24), we focused subsequent analyses on BCL-2 expression and function. Interestingly, we found that other antiapoptotic BCL-2 family members were not regulated in a similar manner in response to DEX and IL-7 (Supplemental Figure 3D), indicating a BCL-2–specific effect. To determine whether the induction of BCL-2 expression upon exposure to DEX and IL-7 is mediated specifically by STAT5B, we assessed BCL-2 protein expression in scrambled control and STAT5 single- and double-KO CCRF-CEM cell clones. This analysis revealed upregulation of BCL-2 with the combination of DEX and IL-7 in scrambled control and STAT5A single-KO clones, but not in the STAT5B single-KO or STAT5A/B double-KO clones, consistent with a central role for STAT5B in the regulation of BCL-2 expression (Figure 3F and Supplemental Figure 3E).
Given that this increased STAT5 transcriptional activity depends first on the upregulation of IL-7Rα as a primary transcriptional target of the GR, we reasoned that STAT5-mediated upregulation of BCL-2 must occur as a secondary transcriptional effect following exposure to DEX and IL-7. To test this, we performed quantitative PCR (qPCR) to measure the changes in expression over time of BCL2 and primary GR target genes in CCRF-CEM cells cultured in the presence of DEX and IL-7. This analysis revealed that although primary GR transcriptional targets were upregulated as early as 2 hours after DEX treatment, BCL2 was not significantly upregulated until the 8-hour time point (Supplemental Figure 3F). To further confirm that BCL2 was upregulated as a secondary transcriptional target, we exposed CCRF-CEM cells to DEX with or without IL-7 in the presence or absence of CHX and measured transcript and protein expression of BCL2 and the primary GR transcriptional target BCL2L11 (BIM) (24). We found that CHX was sufficient to inhibit the upregulation of BIM and BCL-2 protein expression, indicating effective inhibition of translation and de novo protein synthesis (Supplemental Figure 3G). BCL2L11 transcript expression was induced by DEX in both the absence and presence of CHX, consistent with this being a primary transcriptional target of the GR, the upregulation of which was not dependent on intermediary de novo protein synthesis. In contrast, BCL2 transcript expression was upregulated only in the absence of CHX, suggesting a dependence on de novo protein synthesis, which is consistent with this being a secondary transcriptional event (Figure 3G).
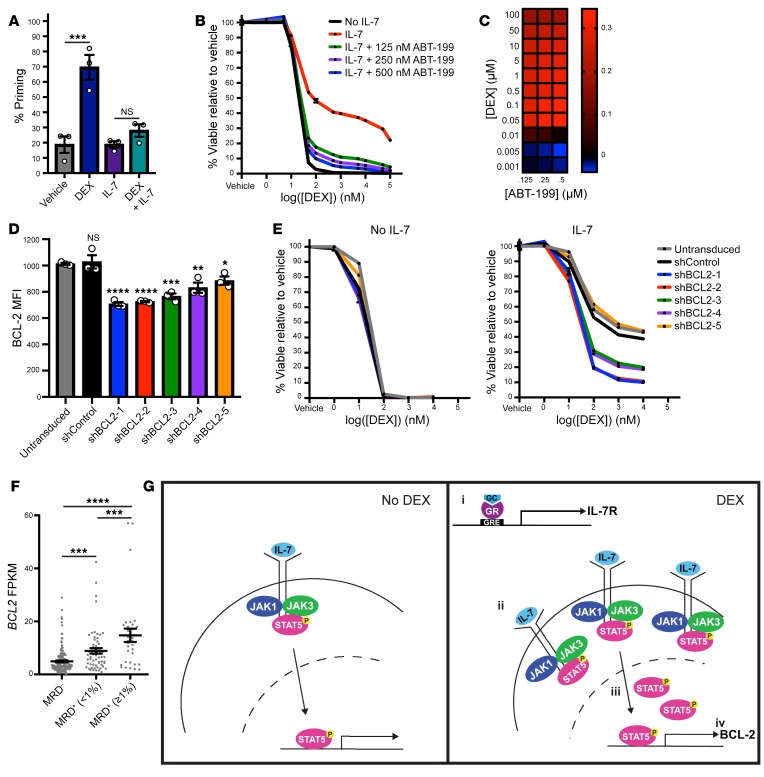
To establish the functional significance of BCL-2 upregulation, we first performed BH3 profiling with the BCL-2 inhibitor ABT-199 in CCRF-CEM cells treated with DEX with or without IL-7. Under these conditions, DEX alone produced a significant increase in apoptotic priming (P = 0.0007). This effect was attenuated in the presence of IL-7 (P = 0.64), suggesting that the increase in BCL-2 expression with the combination of DEX and IL-7 was sufficient to oppose the induction of the apoptotic program (Figure 4A). To determine whether ABT-199 may have a therapeutic role in enhancing DEX sensitivity in the presence of IL-7, we exposed CCRF-CEM cells to DEX in the presence of IL-7 and increasing concentrations of ABT-199. This analysis demonstrated that ABT-199 potently sensitized cells to DEX in the presence of IL-7 in a synergistic manner (Figure 4, B and C). In addition, we used a series of shRNAs to knock down BCL-2 expression in CCRF-CEM cells and found that loss of BCL-2 expression increased sensitivity to DEX in the presence of IL-7 in a manner that correlated with the degree of BCL-2 knockdown (Supplemental Figure 4A and Figure 4, D and E). To assess the importance of high BCL-2 expression in a patient cohort for which clinical outcome data were available, we next analyzed published gene expression data from 265 diagnostic T-ALL samples obtained from patients enrolled in the prior COG T-ALL trial AALL0434 (1). Consistent with our in vitro findings, we found that patients who were minimal residual disease (MRD) positive at the end of induction therapy had significantly higher BCL-2 expression levels relative to those in patients who were MRD negative (P = 0.0009 for patients with MRD <1% and P < 0.0001 for patients with MRD >1%) (Figure 4F), suggesting a relationship between high BCL-2 expression and relative GC resistance. In contrast, shRNA-mediated knockdown of the other candidate resistance gene, ARHGEF3, had no effect that would overcome IL-7–induced DEX resistance (Supplemental Figure 4, B and C), and ARHGEF3 expression did not differ according to MRD status in the AALL0434 patient cohort (Supplemental Figure 4D). Taken together, these data support a model (Figure 4G) in which DEX (a), through upregulation of IL-7Rα expression (b), paradoxically induces its own resistance by augmenting JAK/STAT5 signaling (c) and activation of STAT5B target genes (d), including BCL2. This upregulation of BCL-2 in turn is sufficient to antagonize the proapoptotic effects of DEX.
Figure 4. BCL-2 mediates IL-7–induced DEX resistance.
(A) Percentage of priming of CCRF-CEM cells treated in the absence or presence of 1 μM DEX and/or 100 ng/mL IL-7 in technical triplicate for 16 hours followed by BH3 profiling with 0.5 μM ABT-199 for 90 minutes. (B) Viability of CCRF-CEM cells treated with DEX in the absence or presence of 25 ng/mL IL-7 and increasing concentrations of ABT-199 for 72 hours in technical triplicate. The no IL-7 (black line) and the 25 ng/mL IL-7 (red line) conditions were replotted from Figure 1C. (C) Heatmap of Bliss independence scores calculated as the average of technical triplicates for the combination of DEX and ABT-199 in the presence of 25 ng/mL IL-7. (D) MFI of BCL-2 protein expression assessed in technical triplicate in untransduced CCRF-CEM cells and CCRF-CEM cells transduced with a nontargeting shRNA control (shControl) or a BCL2-targeting shRNA (shBCL2-1-5). Statistical significance is relative to the untransduced cells. (E) Viability of untransduced or shRNA-transduced CCRF-CEM cells treated with DEX in the absence or presence of 25 ng/mL IL-7 in technical triplicate for 72 hours. (F) FPKM values for BCL2 transcript levels according to published RNA-Seq data from diagnostic samples from patients enrolled in the COG AALL0434 trial, stratified on the basis of day-29 bone marrow MRD. (G) Schematic of the proposed model for the mechanism by which DEX paradoxically induces steroid resistance in T-ALL cells in the presence of IL-7. In the presence of DEX (right), the GR induces an increase in IL-7R expression (i), leading to an increase in IL-7R at the cell surface (ii). This in turn leads to an increase in STAT5 transcriptional activity (iii) that ultimately results in the upregulation of BCL-2 (iv). GRE, glucocorticoid response element. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA with Tukey’s method for multiple comparisons adjustment (A, D, and F). All CCRF-CEM cell data are representative of 3 independent experiments.
IL-7 induces DEX resistance in subsets of developing thymocytes.
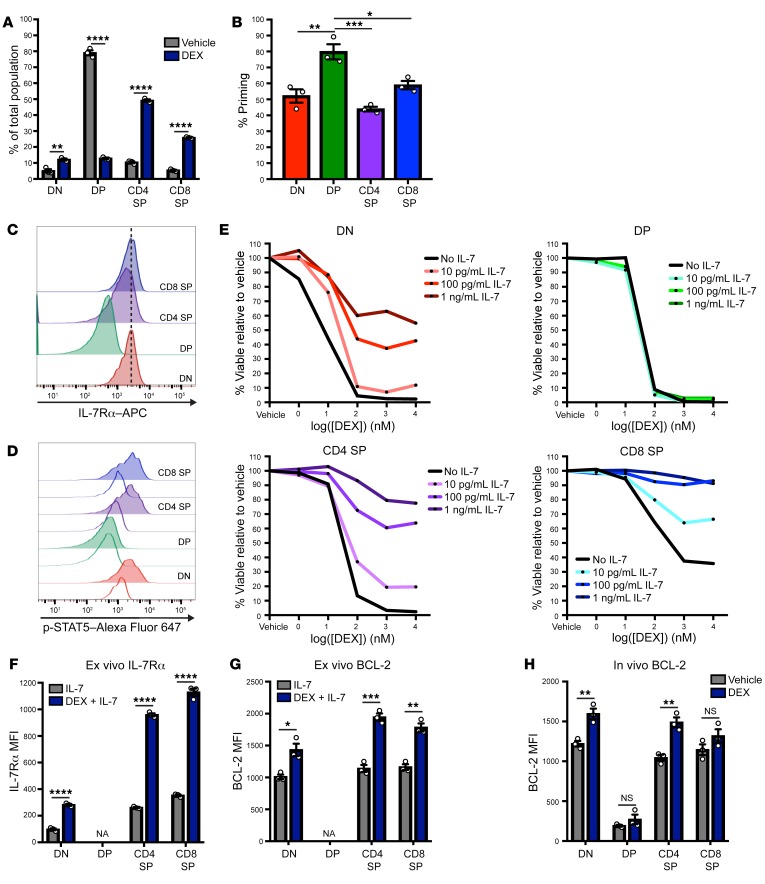
We next sought to determine why IL-7–mediated DEX resistance was found only in a subset of patients’ primary T-ALL samples. In our previous patient cohort, we demonstrated that 64% of samples with IL-7–mediated DEX resistance did not have activating mutations in the IL-7R/JAK/STAT5 pathway (14), suggesting that this phenotype is not dictated by the mutational status of a T-ALL sample. Given these findings, an alternative hypothesis is that IL-7–mediated DEX resistance might reflect a physiologic mechanism of GC resistance that occurs in normal populations of developing thymocytes, as both GC sensitivity and IL-7R expression are known to vary throughout development (10, 25). To test this hypothesis, we evaluated normal murine thymocytes to determine whether IL-7R/JAK/STAT5 signaling modulates DEX sensitivity. We first exposed mice to DEX in vivo and assessed the relative sensitivity of the major thymocyte subpopulations. In this analysis, DEX induced a significant reduction in overall thymic cellularity (P = 0.02; Supplemental Figure 5A) and resulted in a dramatic shift in distribution of the major thymocyte subpopulations. Consistent with previous reports (25), we found that DEX induced a significant reduction in the proportion of CD4/CD8 double-positive (DP) thymocytes (P < 0.0001), with a compensatory increase in the percentage of the earlier double-negative (DN) thymocytes (P = 0.005) and later single-positive (SP) thymocytes (P < 0.0001 for both CD4 and CD8 SP thymocytes) (Figure 5A and Supplemental Figure 5, B and C). Importantly, we recapitulated the findings by other investigators (26) that GR expression is paradoxically lowest at the DP stage of development, despite the fact that these cells are highly DEX sensitive, suggesting that GR expression is insufficient to explain this pattern of differential sensitivity (Supplemental Figure 5D). To determine whether this differential DEX sensitivity reflects differences in the apoptotic potential of these thymocyte subpopulations in their basal state, we performed BH3 profiling on freshly harvested thymocytes. We found that DP thymocytes had significantly higher apoptotic potential relative to DN or SP thymocytes (P = 0.002, P = 0.0004, and P = 0.01 versus DN, CD4 SP, and CD8 SP thymocytes, respectively) (Figure 5B), consistent with the pattern of DEX sensitivity observed in vivo.
Figure 5. IL-7 induces DEX resistance in subpopulations of normal developing thymocytes.
(A) Percentage of thymocyte subpopulations in thymi isolated from mice treated with vehicle control (n = 3) or DEX (n = 3) at 2 mg/kg/day for 3 days. (B) Percentage of priming of thymocytes in the basal state following BH3 profiling with 1 μM synthetic BIM peptide in technical triplicate for 90 minutes. (C) Histograms of the basal expression of IL-7Rα in the major murine thymocyte subpopulations. (D) Histograms of p-STAT5 in the major murine thymocyte subpopulations in the basal state (white histograms) and following a 15-minute stimulation with 100 ng/mL IL-7 (colored histograms). (E) Viability of murine thymocyte subpopulations following ex vivo treatment for 24 hours with DEX in the absence or presence of increasing concentrations of IL-7. (F) MFI of IL-7Rα in murine thymocytes treated ex vivo in the presence of 100 pg/mL IL-7 with or without 1 μM DEX in technical triplicate for 24 hours. DP cells could not be analyzed (NA) because of a lack of viable cells remaining after DEX exposure. (G) MFI of BCL-2 in murine thymocytes treated ex vivo in the presence of 100 pg/mL IL-7 with or without 1 μM DEX in technical triplicate for 24 hours. DP cells could not be analyzed (NA) because of a lack of viable cells remaining after DEX exposure. (H) MFI of BCL-2 in thymocytes isolated from mice treated with vehicle control (n = 3) or DEX (n = 3) at a dose of 2 mg/kg/day for 3 days. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-sample t test (A, F, G, and H) or 1-way ANOVA with Tukey’s method for multiple comparisons adjustment (B). All data are representative of 3 independent experiments.
We next evaluated basal IL-7Rα expression and signaling capacity across the major thymocyte subpopulations. Consistent with previous reports (10), we detected a reduction in IL-7Rα expression and in IL-7–induced p-STAT5 in the DP thymocytes relative to the DN and SP thymocytes (Figure 5, C and D). Given these findings and the pattern of DEX sensitivity we observed in vivo, we hypothesized that the presence of IL-7 in the in vivo microenvironment might activate JAK/STAT5 signaling in DN and SP thymocytes, which could in turn confer protection against the GC surges that occur during a physiologic stress response (27) and against pharmacologic concentrations of GCs. To test this hypothesis, we first exposed thymocytes to vehicle or DEX ex vivo in the absence or presence of increasing concentrations of IL-7. DN and SP thymocytes demonstrated profound DEX resistance specifically in the presence of IL-7, whereas DP thymocytes remained highly sensitive to DEX regardless of IL-7 (Figure 5E), consistent with their reduced IL-7Rα expression (Figure 5C). To determine whether the mechanism of JAK/STAT5-mediated DEX resistance that we elucidated in CCRF-CEM cells is applicable in these thymocyte subpopulations, we exposed thymocytes to DEX ex vivo and assessed cell-surface IL-7Rα expression and BCL-2 expression. Consistent with the observed pattern of DEX resistance in the presence of IL-7, DN and SP thymocytes significantly upregulated the expression of both IL-7R (P < 0.0001 for DN, CD4 SP and CD8 SP thymocytes) (Figure 5F) and BCL-2 (P = 0.01, P = 0.0005, and P = 0.001 for DN, CD4 SP, and CD8 SP thymocytes, respectively) (Figure 5G) following exposure to DEX in the presence of IL-7. Finally, to determine whether this mechanism is applicable in vivo under normal physiologic conditions, we treated mice with DEX and assessed BCL-2 protein expression in the major thymocyte subpopulations. We found that DN and CD4 SP thymocytes, but not DP thymocytes, significantly upregulated BCL-2 expression in response to DEX (P = 0.007 and P = 0.004 for DN and CD4 SP thymocytes, respectively) (Figure 5H). Finally, to determine whether human thymocytes demonstrate a similar pattern of IL-7Rα expression and IL-7–induced DEX resistance throughout development, we performed ex vivo analysis of healthy human thymocytes. Similar to the pattern observed in murine thymocytes, DN and SP thymocytes had the most significant increase in cell-surface IL-7Rα expression following exposure to DEX (Supplemental Figure 5E) and had the most profound IL-7–induced DEX resistance (Supplemental Figure 5F).
Developmental stage correlates with IL-7–induced DEX resistance in T-ALL.
To further address the hypothesis that IL-7R/JAK/STAT5–mediated DEX resistance may be retained from normal thymocyte development in a subset of T-ALLs, we performed RNA-Seq on 76 primary diagnostic T-ALL samples from patients enrolled in COG AALL1231. Using a gene set derived from a comparison of early versus late developing thymocytes (28), we performed unbiased hierarchical clustering of these patient samples to classify samples as developmentally “early” or “late” (Figure 6A). We next performed detailed in vitro analysis of 15 of the early T-ALL samples and 12 of the late T-ALL samples isolated from patient-derived xenografts (PDXs). Additional information about these samples is presented in Supplemental Table 4. We detected no differences in basal GR expression between these 2 groups (Supplemental Figure 6A). Upon analysis of cell-surface IL-7Rα expression, we found that the early samples tended to have higher basal IL-7Rα expression and a more robust induction of IL-7Rα upon exposure to DEX (Figure 6B). This higher basal IL-7Rα expression was also associated with an increased response to IL-7 stimulation, as measured by p-STAT5 (Supplemental Figure 6B). Consistent with this finding, only the early sample group showed a significant increase in DEX resistance in the presence of IL-7 (P = 0.0007 and P = 0.69 for early and late samples, respectively) (Figure 6C). To determine whether this resistance phenotype was associated with activating mutations in the IL-7R/JAK/STAT pathway, we performed variant calling using the RNA-Seq data and found no enrichment for IL-7R pathway mutations in the early samples (Supplemental Figure 6C and Supplemental Table 5), consistent with our previous analysis (14). Similar to the findings in CCRF-CEM cells, these early samples showed an increase in BCL-2 protein expression in the presence of IL-7, which was further augmented upon concomitant exposure to DEX and attenuated with the addition of RUX (Figure 6D). Moreover, both RUX and ABT-199 significantly sensitized early T-ALL samples to DEX in the presence of IL-7 (P < 0.0001 and P = 0.0005 for the addition of RUX or ABT-199, respectively, to DEX plus IL-7) (Figure 6E). To evaluate the utility of RUX for overcoming DEX resistance in vivo in a preclinical model, we transplanted mice with early T-ALL T24 and treated them with DEX with or without RUX, using survival as the primary endpoint. As we observed in vitro (Supplemental Figure 6D), the combination of DEX and RUX showed increased in vivo efficacy relative to either agent alone (P = 0.003 for RUX versus DEX plus RUX and P = 0.02 for DEX versus DEX plus RUX) (Figure 6F).
Figure 6. T-ALLs reflecting early stages of T cell development demonstrate DEX resistance in the presence of IL-7.
(A) Heatmap depicting the clustering of 76 primary T-ALL samples by the expression of genes that are upregulated in early developing thymocytes relative to later developing thymocytes. (B) MFI of cell-surface IL-7Rα in 15 early and 12 late T-ALL PDX samples following exposure to 1 μM DEX for 24 hours in technical triplicate. (C) Viability relative to vehicle control of 15 early and 12 late T-ALL PDX samples treated with 1 μM DEX in the absence or presence of 25 ng/mL IL-7 for 48 hours in technical triplicate. (D) MFI of BCL-2 protein expression in 10 early T-ALL PDX samples following exposure to 100 ng/mL IL-7 with or without 1 μM DEX and/or 500 nM RUX for 16 hours in technical triplicate. Some samples were not analyzed because of limited cell numbers. (E) Viability relative to vehicle control of 15 early T-ALL samples exposed to 25 ng/mL IL-7 with or without 1 μM DEX and/or 500 nM RUX or 1 μM ABT-199 for 48 hours in technical triplicate. (F) Kaplan-Meier survival analysis of mice transplanted with early T-ALL T24 and treated with vehicle control (n = 5), DEX (n = 5), RUX (n = 5), or a combination of DEX and RUX (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by paired t test (B and C), 1-way ANOVA with Tukey’s method for multiple comparisons adjustment (D and E), or a log-rank test (F).
Discussion
The poor survival rates observed in children with relapsed T-ALL (3) suggest a need for strategies to enhance the efficacy of upfront therapy as a means of improving cure rates by decreasing the likelihood of disease relapse. Although many studies have focused on understanding mechanisms of acquired drug resistance that arise during T-ALL treatment (29), the goal of our current study was instead to elucidate mechanisms of intrinsic drug resistance that dictate the initial response to therapy. In particular, the prognostic significance of the initial GC response in T-ALL (4) suggests a need for a deeper understanding of the mechanisms governing intrinsic differences in GC sensitivity. In the present study, our functional analysis of a large number of patients’ diagnostic samples revealed recurrent patterns of intrinsic GC resistance across this otherwise genetically heterogeneous patient population. We confirm in this validation cohort that over half of the diagnostic T-ALL samples analyzed exhibited intrinsic DEX resistance in vitro, which has in turn been shown to correlate with clinical outcomes (30). Furthermore, we show that within this subset, half of the samples were resistant to DEX specifically in the presence of IL-7.
Our data support a model in which GCs paradoxically induce their own resistance by upregulating IL-7Rα expression. In the presence of IL-7, this leads to increased downstream signal transduction and STAT5 transcriptional output. This ultimately results in the upregulation of BCL-2, which is sufficient to counteract the proapoptotic effect of DEX. Given the prevalence of this phenotype, our data suggest that a significant percentage of patients with T-ALL may benefit from the upfront addition of JAK or BCL-2 inhibitors as a means of improving the efficacy of GC therapy. Furthermore, we demonstrate the synergistic potential of combining DEX with these agents and propose that combination therapy may allow for a reduction in DEX dosing, thereby minimizing the numerous acute and chronic toxicities associated with steroid exposure (31) while simultaneously maximizing efficacy. In addition, our data demonstrate that STAT5B was primarily responsible for the upregulation of BCL-2 expression in this context, consistent with previous reports demonstrating that knockdown of STAT5A is insufficient to modulate IL-7–mediated regulation of BCL-2 expression (11). Interestingly, this is also consistent with the finding that activating mutations in STAT5B, but not STAT5A, commonly occur in T-ALL (1). These data support further investigation of the use of ABT-199 as a rational therapeutic strategy to enhance the efficacy of DEX in patients with STAT5B-mutated T-ALL.
In addition to mediating GC resistance in over one-third of diagnostic T-ALL samples, we demonstrate that IL-7 similarly induced GC resistance in those populations of normal thymocytes in which IL-7R signaling is important for survival and proliferation (10). Developing thymocytes are continuously exposed to endogenous GCs, but we and others (25) have demonstrated that susceptibility to GC-induced apoptosis is variable over the course of thymocyte development. These data suggest that normal thymocyte populations must possess intrinsic mechanisms of GC resistance at distinct stages of development and/or under certain environmental conditions. In particular, we found that IL-7–induced DEX resistance occurred in DN thymocytes and was enriched in T-ALL samples with an “early thymocyte” gene expression signature. Importantly, these early thymocytes go on to generate fully rearranged TCRs, which are later tested for functionality and autoreactivity in the subsequent DP stage, a key process in the generation of mature functional T cells (10). Teleologically, susceptibility to GC-induced apoptosis would be maladaptive early in development, as it would limit the availability of cells for this selection process. Our data therefore suggest that IL-7–induced GC resistance may protect these early thymocyte populations from apoptosis in the presence of endogenous GCs. Furthermore, we demonstrate that this mechanism of intrinsic resistance was retained in T-ALL cells resembling early thymocytes, where it may be exploited to enable resistance to pharmacologic concentrations of GCs.
Taken together, our data provide a strong rationale for the idea that differential sensitivity to GC therapy at the time of disease diagnosis reflects developmentally programmed mechanisms of intrinsic GC resistance that are retained during the process of leukemogenesis. This work highlights the need for further studies aimed at elucidating additional mechanisms of GC resistance at distinct stages of thymocyte development as a means of understanding the factors that contribute to intrinsic GC resistance in T-ALL.
Methods
Patient samples and PDXs.
Diagnostic blood samples were obtained from patients enrolled in the COG trial AALL1231. Immunophenotyping was performed and reviewed by immunophenotyping experts in the COG. To establish PDXs, cells were injected into NOD/SCID/Il2rgtm1wjl/Szj (NSG) mice obtained from The Jackson Laboratory. Engraftment was monitored using flow cytometric analysis of peripheral blood with antibodies against human CD45 (BD Biosciences; 560973) and CD7 (BioLegend; 343105).
CCRF-CEM cells.
CCRF-CEM cells were purchased from the UCSF Cell Culture Facility (ATCC CCL-119). Cells were authenticated via short tandem repeat DNA profiling and were routinely tested for mycoplasma contamination using the PlasmoTest Detection Kit (InvivoGen).
Preclinical trial.
Five male NSG mice (5 weeks of age) per treatment arm were randomized to receive vehicle control, DEX, RUX, or a combination of DEX and RUX once the peripheral blood blast count reached 1%. RUX was administered in chow form (Incyte) continuously over the trial duration. DEX (Fresenius Kabi and Children’s Hospital of Philadelphia Pharmacy) was administered by i.p. injection at a dose of 7.5 mg/kg/day. Mice were euthanized when they became moribund.
In vivo DEX treatment in C57BL/6x129Sv/Jae mice and isolation of human and murine thymocytes.
Six- to eight-week-old male F1 C57BL/6x129Sv/Jae mice were obtained from the UCSF Laboratory Animal Resource Center (LARC) breeding core. Mice were treated with 2 mg/kg DEX sodium phosphate (NDC 63323-516-10; UCSF pharmacy) or vehicle control (PBS) once daily for 3 days. Healthy human thymocytes were obtained from children undergoing cardiothoracic surgery at UCSF. Antibodies against murine CD4 (BioLegend; 100425) and CD8 (BioLegend; 100707) or human CD4 (BioLegend; 317420) or CD8 (BioLegend; 344706) were used to identify thymocyte subpopulations.
In vitro viability assays.
In vitro viability assays were performed by exposing cells to vehicle control or DEX (MilliporeSigma; D4902), ruxolitinib (Selleckchem; S1378), tofacitinib (Selleckchem; S5001), CHZ868 (MedKoo; 407137), or ABT-199 (ApexBio; A8194) for 72 hours (CCRF-CEM cells), 48 hours (PDX cells), or 24 hours (thymocytes) with or without recombinant human or murine IL-7 (Peprotech; 200-07 and 217-17). Cells were then stained with Hoechst 33258 (Molecular Probes; H3569) and analyzed by flow cytometry.
CRISPR/Cas9 genome editing of CCRF/CEM cells.
Cas9 protein containing a nuclear localization signal (Cas9-NLS) was purchased from the QB3 MacroLab at the University of California, Berkeley. Trans-activating CRISPR RNA (tracrRNA) and sgRNAs were purchased from Dharmacon. sgRNA sequences targeting IL-7Rα, STAT5A, and STAT5B were obtained from the Brunello sgRNA library (32) and were as follows: IL-7Rα, 5′-AAAGAGCAATATATGTGTGA-3′; STAT5A, 5′-ACATTCTGTACAATGAACAG-3′; and STAT5B, 5′-GTTCATTGTACAATATATGG-3′. The scrambled control cells were generated using a nontargeting sgRNA (5′-GGTTCTTGACTACCGTAATT-3′).
Ribonucleoproteins were prepared according to established methods (33). Electroporation was performed using the Amaxa Cell Line Nucleofector Kit C (Lonza; VACA-1004) and an Amaxa Nucleofector II Device with the electroporation code X-001. Editing was assessed by PCR amplification using the following primers: IL-7Rα, forward 5′-TGAACATGCCTCCACTCACC-3′, IL-7Rα, reverse 5′-CACACCTGGGTTTGAAGATCC-3′; STAT5A, forward 5′-TGGGGATAGTTCCTGAGGCT-3′, STAT5A, reverse 5′-TGCCACCTCTTACACTTGCC-3′; and STAT5B, forward 5′-TGTGCCCCTTAGGATGAAGC-3′, STAT5B, reverse 5′-AATCACAGGAGGCACTGTTCC-3′. The amplicons were Sanger sequenced and the sequencing traces analyzed using TIDE (Tracking of Indels by DEcomposition) analysis software (34). Clonal populations were generated using limiting dilution cell plating.
Western blot analysis.
For analysis of protein expression in whole-cell lysates, CCRF-CEM cells were resuspended in RIPA buffer. For analysis of cytoplasmic and nuclear protein, protein fractions were generated using the NE-PER Kit (Thermo Fisher Scientific; 78833). Immunoblotting was performed with the following antibodies: STAT5A (Abcam; ab32043), STAT5B (Abcam; ab178941), GR (Cell Signaling Technology; 12041), p-GR (Ser211) (Cell Signaling Technology; 4161), β-actin (Cell Signaling Technology; 3700), and p84 (Genetex; GTX70220). Donkey anti–rabbit IRDye 800 and donkey anti–mouse IRDye 680 secondary antibodies (LI-COR Biosciences) were used, and imaging was performed using the Odyssey Imaging System (LI-COR Biosciences).
qPCR.
CCRF-CEM cells were cultured in the presence or absence of 1 μM DEX, 100 ng/mL IL-7, and/or 10 μg/mL CHX for 16 hours unless otherwise indicated. RNA was isolated using the RNeasy Mini Kit (QIAGEN), and cDNA was generated using the SuperScript III Kit (Thermo Fisher Scientific). TaqMan qPCR probes (Applied Biosystems) were used in conjunction with TaqMan Master Mix (Applied Biosystems) to assess transcript levels for the following genes: GAPDH (Hs02786624_g1; VIC-MGB), IL7RA (Hs00902334_m1; FAM-MGB), BCL2L11 (Hs00708019_s1; FAM-MGB), BCL2 (Hs00608023_m1; FAM-MGB), FKBP5 (Hs01561006_m1; FAM-MGB), GILZ (Hs00608272_m1; FAM-MGB), NR3C1 (H200353740_m1; FAM-MGB), MYC (Hs00153408_m1; FAM-MGB), and ARHGEF3 (Hs00989814_m1; FAM-MGB). Experiments were performed in technical triplicate and were run on a QuantStudio 5 Real-Time PCR Instrument (Applied Biosystems). The fold change in transcript expression relative to cells treated with vehicle control was calculated using the ΔΔCt method, unless otherwise indicated, and GAPDH transcript was used for normalization.
Measurement of cell-surface IL-7Rα.
For analysis of cell-surface IL-7Rα expression, cells were treated with 1 μM DEX for 24 hours. Murine thymocyte experiments were performed in the presence of 100 pg/mL recombinant murine IL-7. Antibodies against human (BioLegend; 351315) or murine (Tonbo Biosciences; 20-1271) IL-7Rα were used in conjunction with Hoechst 33258 to allow for gating on viable cells. Data are presented as the median fluorescent intensity (MFI) of the IL-7Rα signal.
Cytokine stimulation and intracellular flow cytometry.
Phosphoflow cytometry for the measurement of STAT protein phosphorylation following IL-7 stimulation was performed as previously described (14). Briefly, CCRF-CEM cells were exposed to vehicle control or 1 μM DEX for 24 hours, allowed to rest for 1 hour in serum-free media, and stimulated with IL-7 at a concentration of 100 ng/mL for 15 minutes. PDX cells were similarly allowed to rest in serum-free media for 1 hour followed by stimulation with 100 ng/mL IL-7 for 15 minutes. Cells were subsequently fixed with 2% paraformaldehyde and permeabilized with methanol. STAT protein phosphorylation was assessed using antibodies against p-STAT1 (Tyr701) (BD Biosciences; BDB612564), p-STAT3 (Tyr705) (BD Biosciences; BDB612569), p-STAT5 (Tyr694) (BD Biosciences; BDB612599), and p-STAT6 (Tyr641) (BD Biosciences; BDB612601). BIM and BCL-2 protein expression was assessed following cell fixation and permeabilization using antibodies against BIM (Cell Signaling Technology; 2933) and anti–human BCL-2 (Life Technologies, Thermo Fisher Scientific; A15796) or anti–mouse BCL-2 (BioLegend; 633509). GR expression was assessed using an anti-GR antibody. A donkey anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was used for flow cytometric detection of BIM and GR proteins.
Luciferase reporter assay.
CCRF-CEM cells were transiently transfected with the pGL4.52[luc2P/STAT5 RE/hygro] vector (Promega; E4651) using Lipofectamine 3000 Transfection Reagent (Life Technologies, Thermo Fisher Scientific). Eighteen hours after transfection, cells were treated with or without 1 μM DEX, 100 ng/mL recombinant human IL-7, and/or 500 nM ruxolitinib for 36 hours. Luciferase activity was assessed with the ONE-Glo Luciferase Assay System (Promega) and a BioTek Synergy 2 instrument. Relative luminescence was calculated by normalizing values to those obtained from cells treated with the vehicle control.
RNA-Seq analysis.
Scrambled control and STAT5-KO CCRF-CEM cell clones were cultured in vehicle control or in the presence or absence of 1 μM DEX and/or 100 ng/mL recombinant human IL-7 for 4 or 16 hours. RNA was isolated using the RNeasy Mini Kit, and cDNA was generated using the SuperScript III Kit and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA quality was assessed using an Agilent Bioanalyzer (Agilent Technologies). Libraries were prepared using 1 ng RNA and were sequenced on the HiSeq 2500 (Illumina) to generate 50-bp single-end reads.
GR-regulated genes were identified using edgeR, as previously described (35), by comparing scrambled control clones treated with vehicle with those treated with DEX for 4 hours. A gene set was created using the statistical thresholds of an absolute log fold change of greater than 1 and a FDR of less than 0.05. This analysis was then applied to perform the same comparison between scrambled control clones treated with vehicle versus DEX plus IL-7 for 4 hours. For the analysis of STAT5 target genes, GSEA was performed as previously described (36) by comparing scrambled control clones treated with IL-7 versus DEX plus IL-7 using gene sets derived from published STAT5 ChIP-Seq experiments with human CD4+ T cells (23). The default settings were used for GSEA, including permutation based on phenotype. The data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE137893).
For the fresh diagnostic T-ALL samples, total RNA was prepared using TRIzol-based (Thermo Fisher Scientific) extraction. Samples were purified and concentrated using the RNeasy Mini or the RNeasy MinElute Kit alone with the DNase Set (QIAGEN). RNA concentration was determined using a NanoDrop spectrophotometer. RNA quality was assessed using an Agilent 2200 TapeStation (Agilent Technologies). RNA (100 ng) was used to prepare libraries using the TruSeq RNA Exome RNA Kit (Illumina). For RNA samples with a DV200 (percentage of fragments >200 nucleotides) below 30%, 200 ng total RNA was used to prepare the libraries. The libraries were sequenced on an Illumina NextSeq 500 using 150-bp paired-end chemistry.
Primary T-ALL cell transcript expression was calculated via a local software pipeline built around the Bowtie2 Aligner (version 2.3.4.1) and RSEM’s (version 1.2.3.0) expectation-maximization quantification that utilized the Ensembl GRCh38 release 85 reference. After demultiplexing, which converted primary sequence data to the fastq format, and trimming adapters, the sequences were aligned against an HG38 rRNA reference using the bwa (version 0.7.12) aligner in order to screen out rRNA. Only non-rRNA aligning sequences advanced into the Bowtie2/RSEM analysis stream. These data were used to generate gene signatures associated with early versus late thymocyte development via hierarchical clustering and dendrogram analysis.
All sequencing analyses, including read alignment, quality and performance metrics, post-processing, variant calling, and variant annotation, were performed as previously described (37, 38) using the hg38 build of the human genome. Briefly, reads were aligned with the Burrows-Wheeler Aligner (39) and processed using Picard (http://broadinstitute.github.io/picard) and the Genome Analysis Toolkit (GATK) (40) to perform base quality recalibration and multiple sequence realignment. Single nucleotide variants and indels were detected with the MuTect (41) and BCFtools algorithms, respectively. Variants were negatively selected against IGSR SNP (42) and ExAC SNP (43) databases and positively selected for on the basis of recurrently mutated sites or regions within the Catalogue of Somatic Mutations in Cancer (COSMIC) (T-ALL–associated subset of mutations) (44) or as previously reported (1). Candidate somatic mutations were manually reviewed using Integrative Genomics Viewer (45). These data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE137768).
BH3 profiling.
BH3 profiling was performed according to established methods (46). CCRF-CEM cells were treated or not with 1 μM DEX and/or 100 ng/mL recombinant human IL-7 for 16 hours prior to analysis. Thymocyte BH3 profiling was performed immediately after harvesting of thymocytes. Cytochrome C staining was performed using an anti–cytochrome C antibody (BioLegend; 612310).
shRNA-mediated knockdown of BCL2 and ARHGEF3.
The miR30-PGK-NeoR-IRES-GFP cassette from LMN-GFP (32) was subcloned into a pCDH Expression Lentivector (System Biosciences) to generate the construct pCDH-LMN-GFP. The shRNA sequences targeting human BCL2 were as follows: shBCL2-1, 5′-TTTTATTCCAATTCCTTTCGGA-3′; shBCL2-2, 5′-TAGCTGATTTGAAACTTCCCAA-3′; shBCL2-3, 5′-TACTTCATCACTATCTCCCGGT-3′; shBCL2-4, 5′-TTTAAGTACAGCATGATCCTCT-3′; and shBCL2-5, 5′-TATCAGTCTACTTCCTCTGTGA-3′. The shRNA sequences targeting human ARHGEF3 were as follows: shARHGEF3-1, 5′-TTTGATTCAACTCTTGTTCTGT-3′; shARHGEF3-2, 5′-TATATCTTGTCACACAGCTTGA-3′; and shARGHEF3-3, 5′-TATAGCTTCTTCCAAGTGCTGC-3′. 97-mer oligonucleotides were generated as previously described (47) and amplified using the following primers: forward, 5′-TACAATACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3′ and reverse 5′-ACTTAGAAGAATTCCGAGGCAGTAGGCA-3′. The following nontargeting shRNA (shControl) sequence was used as a control: 5′-TAGATAAGCATTATAATTCCTA-3′. Oligonucleotides were cloned into the EcoRI and XhoI sites of pCDH-LMN-GFP, and lentivirus was generated via calcium phosphate transfection of HEK293T cells using the packaging and envelope plasmids psPAX2 and pCMV-VSVG. Viral supernatants were collected 48 hours after transfection and concentrated using Lenti-X Concentrator (Clontech). Following lentiviral transduction, GFP-positive cells were sorted using a Sony SH800 instrument and subsequently expanded.
Flow cytometric analysis.
Flow cytometry was performed using a BD FACSVerse, and data were analyzed with FlowJo software.
Statistics.
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software). All tests were 2 sided, and the threshold for significance was a P value of 0.05 or less. Comparisons between groups were made using a 2-tailed t test, with 1-way ANOVA and Tukey’s method for multiple comparisons adjustment for comparisons of 3 or more groups. For in vivo survival analysis, the log-rank test was used to perform pairwise comparisons between survival curves. Interactions between drugs were assessed using Bliss independence analysis (48). All data indicate the mean ± SEM.
Study approval.
Written informed consent for the use of diagnostic specimens in this research was obtained from patients or their guardians at the time of sample collection as part of the COG trial AALL0434, in accordance with Declaration of Helsinki principles and with the approval of the National Cancer Institute (NCI) and the IRBs of the participating sites. All animal experiments were conducted following protocols that were approved by the IACUCs and IRBs of Children’s Hospital of Philadelphia and UCSF.
Author contributions
LKM, BJH, CDM, KMS, DTT, and MLH designed the experiments and analyzed the data. LKM, CDM, BJH, AMW, and TLV performed the experiments. BLW performed immunophenotyping of patients’ samples. PF performed the RNA-Seq of the patients’ primary samples. BJH, RPR, AH, and ABO performed bioinformatics analysis. LKM and MLH wrote the manuscript. BJH, CDM, RPR, AH, AMW, TLV, PF, ABO, BLW, TMH, KMS, and DTT reviewed the manuscript.
Supplementary Material
Acknowledgments
The authors thank Michael Adkisson, Joshua Rudolph, Andrea Barczak, and David Erle from the UCSF Functional Genomics Core Facility for their assistance with the RNA-Seq involving CCRF-CEM cells, and Anthony Letai and Jeremy Ryan for assistance with the BH3 profiling technique. This study was supported by a Genentech Foundation Research Fellowship (to LKM); a NIH Medical Scientist Training Program grant (T32GM007618, to LKM), aNCI grant (R01CA193776, to TMH, BLW, DTT, and MLH); an American Cancer Society Research Scholar grant (RSG-13-022-01-CDD, to DTT); a Buster Posey Family Pediatric Cancer Pilot Award (to MLH); the Campini Family Foundation (to MLH); and the Pepp Family Foundation (to MLH). RPR, AH, and ABO are supported by the UCSF Helen Diller Family Comprehensive Cancer Center NIH grant P30CA082103, which also supports the shared resource facilities that were used to conduct the flow cytometric work at UCSF. In addition, this work was supported by a National Clinical Trials Network (NCTN) Operations Center grant (U10CA180886), a NCTN Statistics and Data Center grant (U10CA180899) to the COG, and by Leukemia and Lymphoma Society grant (MCG 6561-18).
Version 1. 11/05/2019
In-Press Preview
Version 2. 01/13/2020
Electronic publication
Version 3. 02/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):863–876.https://doi.org/10.1172/JCI130189.
Contributor Information
Lauren K. Meyer, Email: Lauren.meyer@ucsf.edu.
Benjamin J. Huang, Email: ben.huang@ucsf.edu.
Cristina Delgado-Martin, Email: Cristina.delgado-martin@ucsf.edu.
Ritu P. Roy, Email: ritu.roy@ucsf.edu.
Aaron Hechmer, Email: aaron.hechmer@ucsf.edu.
Anica M. Wandler, Email: Anica.wandler@ucsf.edu.
Tiffaney L. Vincent, Email: vincentt@email.chop.edu.
Paolo Fortina, Email: Paolo.Fortina@jefferson.edu.
Adam B. Olshen, Email: adam.olshen@ucsf.edu.
Brent L. Wood, Email: woodbl@u.washington.edu.
Terzah M. Horton, Email: tmhorton@txch.org.
Kevin M. Shannon, Email: shannonk@peds.ucsf.edu.
David T. Teachey, Email: teacheyd@email.chop.edu.
Michelle L. Hermiston, Email: michelle.hermiston@ucsf.edu.
References
- 1.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14(6):e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 4.Gao J, Liu WJ. Prognostic value of the response to prednisone for children with acute lymphoblastic leukemia: a meta-analysis. Eur Rev Med Pharmacol Sci. 2018;22(22):7858–7866. doi: 10.26355/eurrev_201811_16411. [DOI] [PubMed] [Google Scholar]
- 5.Lauten M, et al. Prediction of outcome by early bone marrow response in childhood acute lymphoblastic leukemia treated in the ALL-BFM 95 trial: differential effects in precursor B-cell and T-cell leukemia. Haematologica. 2012;97(7):1048–1056. doi: 10.3324/haematol.2011.047613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11(Suppl 1):S45–S55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 7.Erlacher M, Knoflach M, Stec IE, Böck G, Wick G, Wiegers GJ. TCR signaling inhibits glucocorticoid-induced apoptosis in murine thymocytes depending on the stage of development. Eur J Immunol. 2005;35(11):3287–3296. doi: 10.1002/eji.200526279. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson CA, Yamamoto KR. Crosstalk pathway for inhibition of glucocorticoid-induced apoptosis by T cell receptor signaling. Proc Natl Acad Sci USA. 2000;97(13):7319–7324. doi: 10.1073/pnas.97.13.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsitoura DC, Rothman PB. Enhancement of MEK/ERK signaling promotes glucocorticoid resistance in CD4+ T cells. J Clin Invest. 2004;113(4):619–627. doi: 10.1172/JCI18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spits H. Development of alphabeta T cells in the human thymus. Nat Rev Immunol. 2002;2(10):760–772. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro D, et al. STAT5 is essential for IL-7-mediated viability, growth, and proliferation of T-cell acute lymphoblastic leukemia cells. Blood Adv. 2018;2(17):2199–2213. doi: 10.1182/bloodadvances.2018021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva A, et al. IL-7 contributes to the progression of human T-cell acute lymphoblastic leukemias. Cancer Res. 2011;71(14):4780–4789. doi: 10.1158/0008-5472.CAN-10-3606. [DOI] [PubMed] [Google Scholar]
- 13.Treanor LM, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med. 2014;211(4):701–713. doi: 10.1084/jem.20122727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado-Martin C, et al. JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias. Leukemia. 2017;31(12):2568–2576. doi: 10.1038/leu.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7(2):144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol. 2008;22(8):1754–1766. doi: 10.1210/me.2007-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe A, et al. An enhancer of the IL-7 receptor α-chain locus controls IL-7 receptor expression and maintenance of peripheral T cells. J Immunol. 2015;195(7):3129–3138. doi: 10.4049/jimmunol.1302447. [DOI] [PubMed] [Google Scholar]
- 18.Franchimont D, et al. Positive effects of glucocorticoids on T cell function by up-regulation of IL-7 receptor alpha. J Immunol. 2002;168(5):2212–2218. doi: 10.4049/jimmunol.168.5.2212. [DOI] [PubMed] [Google Scholar]
- 19.Kakal JA, Ghazawi FM, Faller EM, Sugden SM, Parmar P, MacPherson PA. Transcriptional regulation of the IL-7Rα gene by dexamethasone and IL-7 in primary human CD8 T cells. Immunogenetics. 2017;69(1):13–27. doi: 10.1007/s00251-016-0948-4. [DOI] [PubMed] [Google Scholar]
- 20.Shimba A, et al. Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity. 2018;48(2):286–298.e6. doi: 10.1016/j.immuni.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Lee HC, Shibata H, Ogawa S, Maki K, Ikuta K. Transcriptional regulation of the mouse IL-7 receptor alpha promoter by glucocorticoid receptor. J Immunol. 2005;174(12):7800–7806. doi: 10.4049/jimmunol.174.12.7800. [DOI] [PubMed] [Google Scholar]
- 22.Kruth KA, et al. Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. 2017;129(22):3000–3008. doi: 10.1182/blood-2017-02-766204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai T, et al. Identification of STAT5A and STAT5B target genes in human T cells. PLoS ONE. 2014;9(1):e86790. doi: 10.1371/journal.pone.0086790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing D, et al. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood. 2015;125(2):273–283. doi: 10.1182/blood-2014-05-576470. [DOI] [PubMed] [Google Scholar]
- 25.Berki T, Pálinkás L, Boldizsár F, Németh P. Glucocorticoid (GC) sensitivity and GC receptor expression differ in thymocyte subpopulations. Int Immunol. 2002;14(5):463–469. doi: 10.1093/intimm/14.5.463. [DOI] [PubMed] [Google Scholar]
- 26.Boldizsár F, Pálinkás L, Czömpöly T, Bartis D, Németh P, Berki T. Low glucocorticoid receptor (GR), high Dig2 and low Bcl-2 expression in double positive thymocytes of BALB/c mice indicates their endogenous glucocorticoid hormone exposure. Immunobiology. 2006;211(10):785–796. doi: 10.1016/j.imbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Gruber J, Sgonc R, Hu YH, Beug H, Wick G. Thymocyte apoptosis induced by elevated endogenous corticosterone levels. Eur J Immunol. 1994;24(5):1115–1121. doi: 10.1002/eji.1830240516. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Hanspers K, Barker CS, Korn AP, McCune JM. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol. 2004;16(8):1109–1124. doi: 10.1093/intimm/dxh112. [DOI] [PubMed] [Google Scholar]
- 29.Foo J, Michor F. Evolution of acquired resistance to anti-cancer therapy. J Theor Biol. 2014;355:10–20. doi: 10.1016/j.jtbi.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieters R, et al. Relation of cellular drug resistance to long-term clinical outcome in childhood acute lymphoblastic leukaemia. Lancet. 1991;338(8764):399–403. doi: 10.1016/0140-6736(91)91029-T. [DOI] [PubMed] [Google Scholar]
- 31.Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11(11):1096–1106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA. 2015;112(33):10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Ahmadie H, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 2014;4(9):1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke L, et al. The International Genome Sample Resource (IGSR): a worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res. 2017;45(D1):D854–D859. doi: 10.1093/nar/gkw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tate JG, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan J, Letai A. BH3 profiling in whole cells by fluorimeter or FACS. Methods. 2013;61(2):156–164. doi: 10.1016/j.ymeth.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5(6):1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Zhao W, Sachsenmeier K, Zhang L, Sult E, Hollingsworth RE, Yang H. A new Bliss independence model to analyze drug combination data. J Biomol Screen. 2014;19(5):817–821. doi: 10.1177/1087057114521867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.