Abstract
Oncogene-targeted and immune checkpoint therapies have revolutionized the clinical management of malignant melanoma and now offer hope to patients with advanced disease. Intimately connected to patients’ overall clinical risk is whether the initial primary melanoma lesion will metastasize and cause advanced disease, but underlying mechanisms are not entirely understood. A subset of melanomas display heightened peroxisome proliferator–activated receptor γ coactivator 1-α (PGC1α) expression that maintains cell survival cues by promoting mitochondrial function, but also suppresses metastatic spread. Here, we show that PGC1α expression in melanoma cells was silenced by chromatin modifications that involve promoter H3K27 trimethylation. Pharmacological EZH2 inhibition diminished H3K27me3 histone markers, increased PGC1α expression, and functionally suppressed invasion within PGC1α-silenced melanoma cells. Mechanistically, PGC1α silencing activated transcription factor 12 (TCF12), to increase expression of WNT5A, which in turn stabilized YAP protein levels to promote melanoma migration and metastasis. Accordingly, inhibition of components of this transcription-signaling axis, including TCF12, WNT5A, or YAP, blocked melanoma migration in vitro and metastasis in vivo. These results indicate that epigenetic control of melanoma metastasis involved altered expression of PGC1α and an association with the inherent metabolic state of the tumor.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Cell migration/adhesion, Melanoma
Introduction
Recent breakthroughs in the clinical management of malignant melanoma, which include treatment using BRAF(V600E)-targeted drugs and immune checkpoint–blocking antibodies, now improve survival for patients with advanced stage disease (1). While these remarkable achievements provide some patients with lasting benefit, others will not respond at all, and many will progress after a temporal respite. Similarly, early detection and surgical resection improves the outcome for patients with primary disease, yet some will find their disease recurring with some latency. A mechanistic understanding of who has a heightened risk of having their primary disease recur would help improve clinical management and patient outcomes by the use of increased surveillance and adjuvant stage treatment. Now replaced by the American Joint Committee on Cancer (AJCC) staging system, the pathologically assessed measures of Breslow thickness and Clark level were classical clinical prognostic factors of primary melanoma patient risk (2). These measures specifically address the vertical growth by depth (Breslow thickness) and cutaneous level engagement (Clark level) of the primary melanoma lesion and hence describe the local invasive characteristics. The underlying mechanisms that functionally contribute to vertical melanoma growth, and ultimately metastasis, are, however, not fully understood. Melanoma metastatic spread is believed to involve switching of melanoma cell behaviors from a proliferative program toward migratory and invasive characteristics (3). Intimately connected to the proliferative state is expression of differentiation-associated melanocyte markers, and conversely, their loss correlates with metastatic spread (4). Increased Rho GTPase, integrin, and TGF-β signaling have also been shown functionally to promote metastasis (5–9). There are additional regulatory mechanisms that contribute to melanoma metastasis, including the Wnt and Hippo/YAP signaling pathways (10). To this end, WNT5A is a secreted ligand that activates both canonical and alternative WNT signaling to promote malignant tumor traits (11–13). Specifically, WNT5A acts through the noncanonical pathway and activates YAP/TAZ to improve cell migration, and YAP has been associated with melanoma lymph node metastasis (14). Interestingly, through increasing survival of rare circulating tumor cells, oxidative stress resistance has recently been shown to facilitate metastatic spread in melanomas (15). However, it is still incompletely understood how these components contribute to metastatic spread and their regulatory hierarchy.
A subset of human melanomas are endowed with heightened levels of the transcriptional coactivator peroxisome proliferator–activated receptor γ coactivator 1-α (PGC1α), a protein that supports mitochondrial activity, resistance to oxidative stress, and cellular survival (16, 17). In melanoma cells specifically, PGC1α expression is regulated by the microphthalmia-associated transcription factor (MITF), the master regulator of melanocyte differentiation and survival (18, 19). Interestingly, PGC1α maintains melanoma cell survival in response to pharmacological inhibition of oncogenic BRAF(V600E) (17), but it also acts as a potent suppressor of cell invasion and metastatic spread (20). Suppression of this invasive phenotype was linked to activation of the ID2/TCF4 axis and increased downstream integrin expression and signaling (20), but was mechanistically distinct from its ERRα-dependent canonical control of bioenergetic functions (21).
In this work, we show that PGC1α expression in melanoma cells is regulated through changes in EZH2-mediated H3K27me3 levels at the regulatory PPARGC1A promoter region. PGC1α expression levels inversely correlate with YAP activity in melanoma cell lines and human specimens. Targeted suppression of PGC1α leads to an increase in transcription factor 12–dependent (TCF12-dependent) WNT5A expression that activates RhoA GTPase to block proteasome-mediated degradation of YAP. TCF12, WNT5A, or YAP inhibition suppresses melanoma cell invasion. These results show that the WNT5A/YAP melanoma invasive capacity is regulated through altered epigenetic histone markers at the PPARGC1A locus.
Results
H3K27me3 suppresses PGC1α gene expression in human melanoma cell lines.
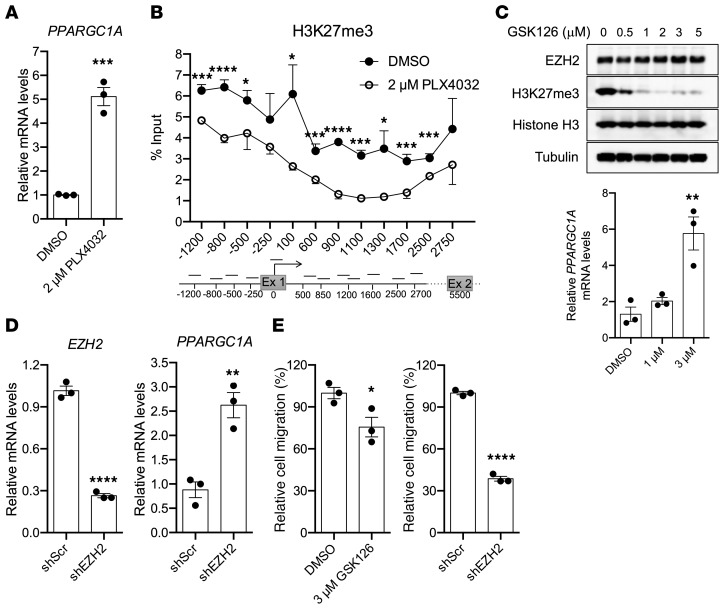
A subset of melanoma cells express high levels of PGC1α, which promote increased mitochondrial biogenesis and resistance to oxidative stress, but correspondingly also compromise their invasive characteristics (16, 17, 20, 22). Interestingly, in response to acute BRAF(V600E) inhibition, there is a marked increase in PGC1α gene expression that functionally promotes melanoma cell survival (17, 20). These observations prompted us to investigate how PGC1α expression is transcriptionally controlled in melanoma. Specifically, during conditions in which pharmacological inhibition of BRAF(V600E) using PLX4032 potently leads to upregulation of PGC1α transcripts (Figure 1A), we used ChIP to analyze alterations in the repressive histone mark H3K27me3 along the PGC1α promoter region (Figure 1B). Using this approach, PLX4032 treatment potently reduced the repressive H3K27me3 marks within the PGC1α promoter compared with treatment with vehicle. Since H3K27me3 is an epigenetic mark associated with polycomb repressive complex-2 function, wherein EZH2 is one of the main methyltransferases, and more importantly, PGC1α expression has been shown to inversely correlate with EZH2 activity in migratory cells (23), we analyzed the regulation of PGC1α expression by EZH2. Treatment with an EZH2 small molecule inhibitor GSK126 led to a dose-dependent decrease in H3K27me3 levels that was mirrored in PGC1α mRNA transcript increases (Figure 1C). Similarly, targeted silencing of EZH2 using shRNA also elevated PGC1α expression levels in human melanoma cells (Figure 1D). Because PGC1α regulates cellular invasion, we sought to determine whether EZH2 inhibition could also affect this facet of PGC1α function. Indeed, either pharmacological or genetic suppression of EZH2 decreased melanoma cell invasion (Figure 1E). Collectively, these data suggest that altered H3K27me3 histone marks deposited by EZH2 decrease PGC1α expression and increase melanoma cell invasion.
Figure 1. EZH2-mediated H3K27me3 suppresses PGC1a expression in melanoma cells.
(A) mRNA expression levels of PGC1α gene (PPARGC1A) in A375P melanoma cells upon treatment with 5 μM of BRAF(V600E) inhibitor PLX4032 overnight (n = 3). (B) H3K27me3 ChIP-qPCR analysis of PGC1α locus in A375P melanoma cells treated with or without 5 μM PLX4032 overnight (n = 3). (C) Inhibition of EZH2 by chemical inhibitor GSK126 dose dependently reduces H3K27me3 levels, leading to increased expression of PGC1α in A375 melanoma cells (n = 3). GSK126 was treated at the indicated concentrations overnight for 14 to 18 hours, followed by Western blotting and qPCR. (D) Knocking down of EZH2 by shRNA induces PGC1α expression in A375 cells (n = 3). (E) Concomitant with increased PGC1α expression, suppression of EZH2 results in impaired migration in the highly invasive A375 melanoma cells (n = 3). Quantitative results were analyzed by Student’s t test (A, D, E) or 1-way ANOVA (B and C). Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
PGC1α expression negatively correlates with Hippo/YAP gene signatures in melanomas.
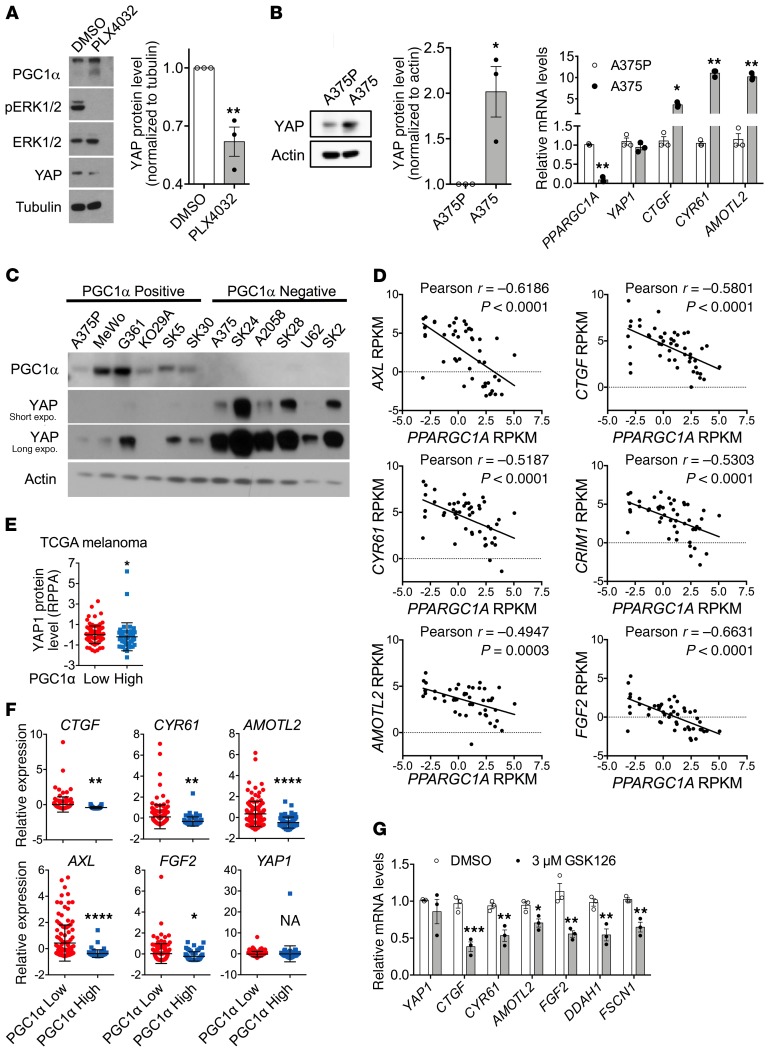
The Hippo/YAP pathway has been linked to the effects of pharmacological BRAF(V600E) inhibition (24, 25), and consistent with these observations, we verified that PLX4032 treatment indeed suppressed YAP protein expression (Figure 2A). We compared YAP levels and canonical YAP transcription target genes (CTGF, CYR61, AMOTL2) across the A375P (high PGC1α expressing) and A375 (low PGC1α expressing) melanoma cell line pair (Figure 2B). YAP protein levels and transcriptional targets inversely related to PGC1α levels. Similarly, across a panel of melanoma cell lines, high YAP protein levels correlated with low PGC1α protein levels (Figure 2C). In agreement with these results, analysis of the Cancer Cell Line Encyclopedia (CCLE) (26) showed that, among human melanoma cell lines, PGC1α gene expression inversely correlated with the YAP target genes, including AMOTL2, CTGF, CYR61, AXL, CRIM1, and FGF2 (Figure 2D). Similar results were observed in The Cancer Genome Atlas (TCGA) human skin cutaneous melanoma specimens (27), where low PGC1α gene expression was associated with higher levels of YAP protein (Figure 2E) and activity, which was reflected by expression of target genes (Figure 2F). Consistent with YAP being downstream of PGC1α, treatment with EZH2 inhibitor GSK126, which elevated PGC1α expression, led to a decrease in YAP target gene expression (Figure 2G). Taken together, these results indicate that PGC1α gene expression is related (inversely) to a presumed downstream-acting Hippo/YAP pathway.
Figure 2. Expression of PGC1α inversely correlates with Hippo-YAP activity in melanoma.
(A) Treatment with 5 μM PLX4032 overnight increases PGC1α expression, but suppresses YAP protein abundance in A375P melanoma cells (n = 3). (B) Comparison of YAP protein, mRNA transcript, and transcriptional activity between PGC1α-positive A375P and PGC1α-negative A375 melanoma cells (n = 3). (C) YAP protein abundance is higher in a panel of human melanoma cell lines with low PGC1α expression. (D) Inverse correlation between PGC1α expression and various YAP target genes in different human melanoma cell lines according to CCLE data set. (E) In TCGA skin melanoma data set, samples with higher PGC1α mRNA transcript display lower levels of YAP protein measured by RPPA. Analysis was done by Mann-Whitney U test. (F) In TCGA skin melanoma data set, higher PGC1α mRNA expression correlates with lower YAP activity reflected by the expression level of its target genes. Of note, YAP1 mRNA transcript is comparable between the 2 groups. (G) Inhibition of EZH2 by GSK126 that increases PGC1α expression also suppresses YAP activity in A375 melanoma cells (n = 3). Quantitative results, if not otherwise indicated, were analyzed by Student’s t test. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
PGC1α posttranscriptionally controls YAP protein levels.
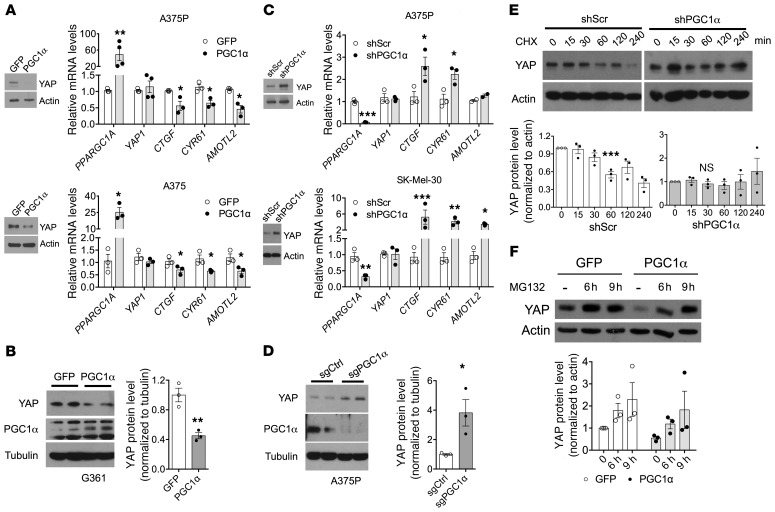
The inverse correlation between PGC1α expression and YAP protein levels in melanoma cells prompted us to further investigate whether and how PGC1α controls YAP protein amounts. Ectopic expression of PGC1α reduced YAP protein without changing mRNA expression levels (Figure 3, A and B). Suppression of PGC1α using shRNA (Figure 3C and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI130038DS1) or CRISPR/Cas9-mediated gene editing (Figure 3D) increased YAP protein amounts without altering its mRNA levels. Importantly, these effects were specific to PGC1α1 full-length isoform, as ectopic expression of PGC1α4, a splicing form that contains the N-terminal transcriptional activation region, but lacks half of the C-terminus region (28), did not affect YAP protein levels (Supplemental Figure 1B). In addition, depletion of ERRα, a PGC1α-coactivated transcription factor that increases nuclear-encoded mitochondrial gene expression (21), did not alter YAP protein amounts (Supplemental Figure 1C). To further test how PGC1α controlled YAP amounts, we used cycloheximide (CHX) to block protein synthesis and found that YAP protein turnover was accelerated when PGC1α was present (Figure 3E). Consistent with these effects on YAP protein stability, MG132-mediated proteasome inhibition abrogated the PGC1α-mediated reduction in YAP protein (Figure 3F). Collectively, these results indicate that PGC1α promotes accelerated YAP protein degradation in melanoma cells.
Figure 3. PGC1α compromises YAP stability via a proteasome-mediated pathway.
(A and B) Overexpression of PGC1α by adenovirus suppresses YAP protein expression and activity without affecting its mRNA transcript in A375P, A375 (A), and G361 (B) melanoma cells (n = 3). (C and D) Depletion of PGC1α by shRNA (C) or CRISPR (D) enhances YAP protein expression and activation without altering its mRNA level in multiple melanoma cell lines (n = 3). (E) YAP protein is more stable in A375P cells with PGC1α depletion. Cells were treated with 50 mg/mL of CHX for the indicated period and harvested for immunoblotting analysis (n = 3). (F) PGC1α-induced YAP degradation in A375P cells can be blocked by treatment with 10 μM of proteasome inhibitor MG132 overnight (n = 3). Quantitative results were analyzed by Student’s t test or 1-way ANOVA. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005.
PGC1α-dependent decrease of YAP inhibits melanoma migration in vitro and metastasis in vivo.
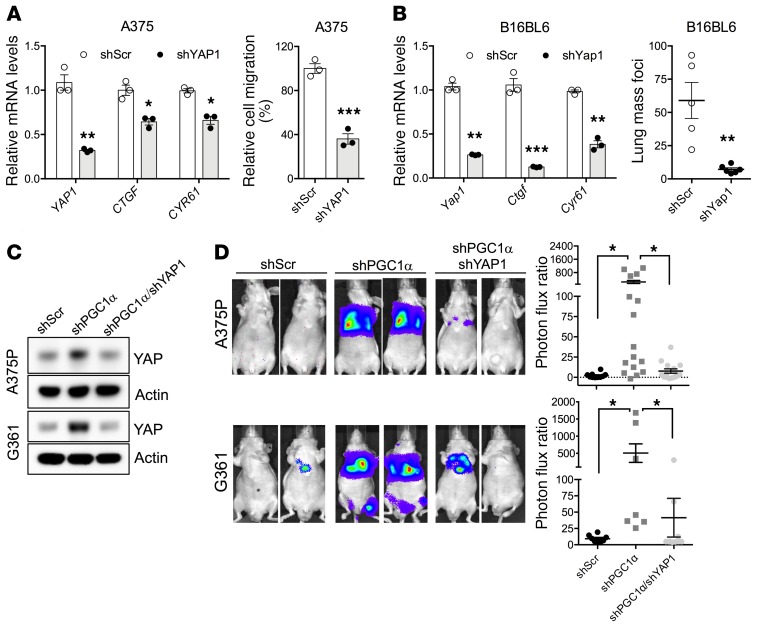
YAP protein function has been linked to cellular motility (11, 29, 30). Consistent with these findings, our data using the mouse B16BL6 melanoma cell line suggest that YAP overexpression promotes metastatic spread in congenic mice (Supplemental Figure 2A). We therefore assessed whether YAP activation contributed to the invasive melanoma phenotype promoted by reduced PGC1α levels. In low PGC1α-expressing, highly invasive melanoma cells, inactivation of YAP significantly compromised Transwell migration (Figure 4A) as well as lung metastasis after tail-vein inoculation (Figure 4B and Supplemental Figure 2B). Consistent with PGC1α acting upstream of YAP, the functional increase in migration (Figure 4C and Supplemental Figure 2, C and D) and metastasis (Figure 4D) following PGC1α suppression in melanoma could be blocked, at least partially, by YAP depletion. These results thus indicate that PGC1α-mediated changes in YAP protein levels control melanoma invasion and metastasis.
Figure 4. YAP is required for melanoma migration and metastasis instigated by loss of PGC1α.
(A) In highly invasive, PGC1α-negative A375 melanoma cells, depletion of YAP by shRNA compromises its migratory ability (n = 3). (B) In the highly migratory murine melanoma cell line B16BL6, depletion of YAP by shRNA suppresses its invasiveness in congenic mice (n = 3 for qPCR and n = 5 mice per group). (C and D) In PGC1α-positive A375P and G361 melanoma cells, the increased metastasis by loss of PGC1α can be prevented by depletion of YAP, as measured by in vivo lung colonization assay (n = 6–16 mice per group). Quantitative results were analyzed by Student’s t test (A and B) or 1-way ANOVA (D). Data are shown as mean ± SEM if not otherwise indicated. *P < 0.05; **P < 0.01; ***P < 0.005.
WNT5A mediates the regulation of YAP by PGC1α.
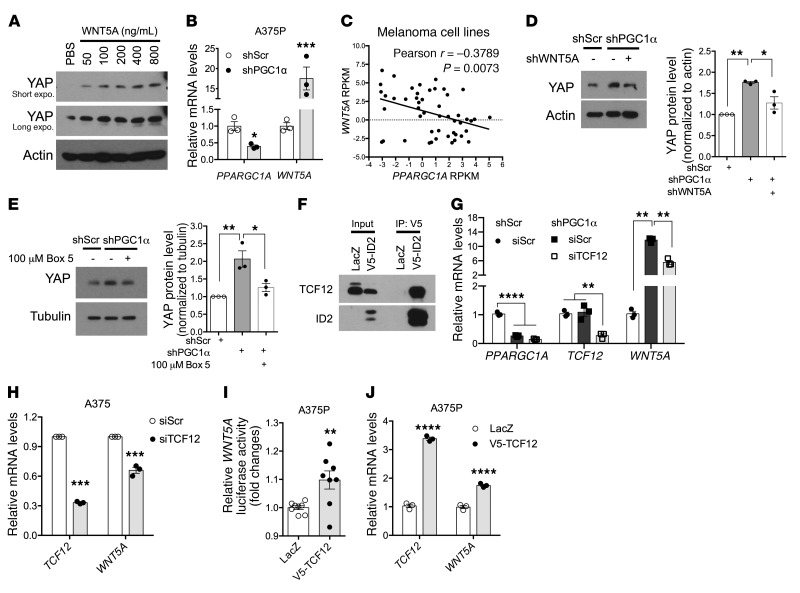
We next chose to examine the mechanism by which reducing PGC1α function stabilizes YAP protein in melanoma cells. Because secreted WNT5A was recently shown to increase YAP/TAZ signaling in breast cancer (14), we added recombinant WNT5A protein to the culture media of A375P melanoma cells and detected a dose-dependent increase in YAP protein levels (Figure 5A). Notably, WNT5A gene expression was modulated by PGC1α in melanoma cells, as shRNA-mediated suppression of PGC1α strongly induced WNT5A mRNA transcripts (Figure 5B), while ectopic expression of PGC1α suppressed WNT5A gene expression (Supplemental Figure 3A). Akin to what occurred with YAP target genes, WNT5A gene expression also demonstrated an inverse correlation with PGC1α mRNA levels across the CCLE human melanoma cell lines (Figure 5C). To determine whether WNT5A mediates the downstream effects of PGC1α on YAP levels, we knocked down WNT5A expression (Supplemental Figure 3B) and found that the observed YAP protein accumulation following PGC1α suppression was largely prevented (Figure 5D). Furthermore, interfering with WNT5A signaling by either blocking WNT5A binding to FZD5 receptor with a selective peptide antagonist Box5 (Figure 5E) (31) or inhibiting WNT5A downstream RhoA GTPase (Supplemental Figure 3C) efficiently blunted the effects of PGC1α on YAP protein stability. In aggregate, these results indicate that suppressing PGC1α leads to an increase in WNT5A expression, which in turn promotes YAP protein levels and activation in melanoma cells.
Figure 5. WNT5A mediates YAP regulation by PGC1α.
(A) Addition of WNT5A protein in culture media increases YAP protein in a dose-dependent manner in A375P cells (n = 3). (B) Knockdown of PGC1α induces WNT5A expression in A375P cells (n = 3). (C) The expression of PGC1α and WNT5A is inversely correlated in human melanoma cell lines according to the CCLE data set. (D and E) Silencing WNT5A expression (D) or blocking WNT5A’s binding to its receptor FZD5 (E) in A375P melanoma cells is able to prevent the induction of YAP by loss of PGC1α (n = 3). (F) ID2 is able to interact with TCF12. Cell lysate from A375P melanoma cells overexpressing V5-ID2 was subject to immunoprecipitation with V5 antibody, followed by immunoblotting with TCF12 antibody. (G) Knockdown of TCF12 is able to prevent the induction of WNT5A by loss of PGC1α in A375P cells (G, n = 3). (H) Knockdown of TCF12 suppresses WNT5A expression in PGC1α-negative A375 cells (H, n = 3). (I and J) Overexpression of TCF12 boosts the activity of WNT5A promoter–driven luciferase activity (I, n = 7) and WNT5A transcription (J, n = 3) in A375P cells. Quantitative results were analyzed by Student’s t test or 1-way ANOVA (D, E, G). Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
We recently showed that in human melanoma, PGC1α increases the expression of a basic helix-loop-helix (bHLH) transcriptional repressor ID2, which in turn suppresses the activity of a bHLH factor TCF4 on driving integrin gene expression (20). To determine whether the ID2/TCF4 complex represents the mechanism underlying how suppression of PGC1α leads to an increase in WNT5A expression, we genetically manipulated ID2 as well as TCF4 and examined changes in WNT5A expression. Although WNT5A expression was regulated by ID2, demonstrated by induced expression upon ID2 depletion, and repressed by ID2 overexpression (Supplemental Figure 3, D and E), it was refractory to TCF4 regulation (Supplemental Figure 3, F and G). This suggested that PGC1α might employ an ID2-dependent, yet TCF4-independent, mechanism to modulate WNT5A levels. Since ID2 binds and inhibits a broad number of bHLH transcription factors, we searched for other potential targets and found that the E-box TCF12 strongly interacted with ID2 in melanoma cells (Figure 5F). Deletion of TCF12 was able to reverse the induction of WNT5A by PGC1α silencing (Figure 5, G and H) and reduced YAP protein abundance (Supplemental Figure 3H), suggesting that TCF12 is a downstream component. Thus, TCF12 ectopic expression in A375P melanoma cells activated WNT5A promoter–driven luciferase activity (Figure 5I), which was followed by increased WNT5A expression (Figure 5J). Collectively, these results indicate that ID2/TCF12 mediates PGC1α-dependent WNT5A expression and signaling in melanoma.
TCF12/WNT5A controls melanoma cell invasion.
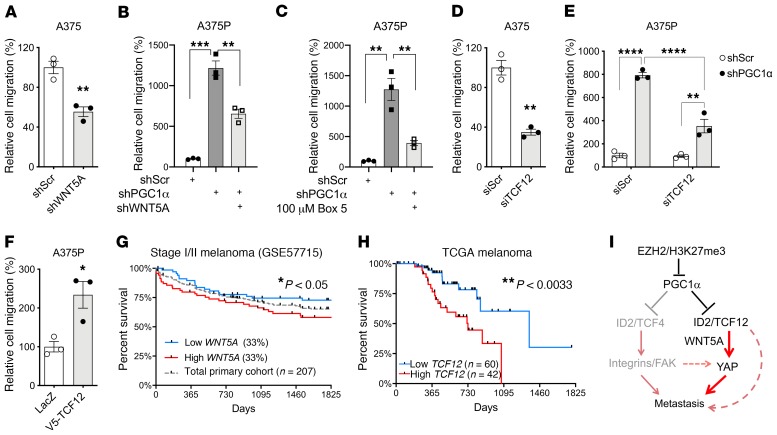
We next sought to determine whether TCF12 and WNT5A, similarly to YAP, functionally contributed to the invasive behavior of melanoma cells downstream of PGC1α. Indeed, WNT5A depletion inhibited the migratory ability of the PGC1α-negative, highly invasive A375 cells (Figure 6A and Supplemental Figure 4A) as well as the PGC1α-depleted A375P cells (Figure 6B). Consistent with these results, abrogating WNT5A signaling using the antagonist Box5 peptide (Figure 6C) or blocking the downstream RhoA activity (Supplemental Figure 4B), both of which have been shown to suppress YAP abundance (Figure 5), diminished the invasion phenotype of melanoma cells. Similarly, knockdown of TCF12, which reduced WNT5A expression, significantly impaired migration of melanoma cells with low PGC1α (Figure 6, D and E) and metastasis in vivo (Supplemental Figure 4C); on the other hand, overexpression of TCF12, which elevated WNT5A expression and YAP abundance, promoted migratory propensity of melanoma (Figure 6F). Interestingly, WNT5A and TCF12 expression both have clinical relevance, as high levels of these proteins individually predicted worse survival in patients with melanoma (Figure 6, G and H). Taken together, these results indicate that TCF12/WNT5A controls melanoma invasion downstream of PGC1α.
Figure 6. TCF12/WNT5A regulates melanoma migration.
(A) In highly invasive A375 melanoma cells, depletion of WNT5A by shRNA compromises its migratory ability (n = 3). (B and C) In nonmetastatic A375P melanoma cells, increased metastasis by loss of PGC1α can be prevented by depletion of WNT5A (B) or by blocking WNT5A interaction with its receptor (C) (n = 3). (D and E) Knocking down TCF12 by siRNA compromises migration in both A375 (D) and A375P-shPGC1α cells (E). (F) In contrast, TCF12 overexpression increases A375P cell migration (n = 3). (G) Expression levels of WNT5A predict survival of patients with early stage melanoma. (H) TCF12 expression correlates with survival of patients with metastatic melanoma. (I) Diagram depicting the regulation of melanoma metastasis by PGC1α. EZH2-dependent H3K27me3 suppresses PGC1α expression in melanoma. PGC1α, upon downregulation, can trigger ID2/TCF4-dependent activation of the integrin/FAK pathway, as previously reported, and at the same time instigate TCF12-mediated WNT5A expression to stabilize YAP. Both integrin/FAK and YAP work coordinately to drive melanoma metastasis. Quantitative results were analyzed by Student’s t test or 1-way ANOVA (B, C, E), while survival analysis was done by log-rank (Mantel-Cox) test. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Discussion
The general invasive behavior of malignant melanoma is functionally related to its rapid metastatic spread to local lymph nodes and subsequent progression to distal anatomical sites (32). Current treatment strategies include BRAF(V600E)-targeted pharmacological inhibitors and immune checkpoint–blocking antibodies (33–35). This promotes overall survival, but for many patients, their disease will return relentlessly after only some temporal respite. The molecular mechanisms and components that drive melanoma cell invasion are commonly thought to depend on a combination of tumor stage, genetic driver (and tumor suppressor) mutations, inherent metabolic and energetic status, tumor-immune microenvironment interactions, and treatment efficacy (32). Mechanistically, different canonical pathways, including integrin, TGF-β, and WNT signaling, have been shown to control melanoma cell invasion (20). Among the metabolic and energetic melanoma invasive dependencies, PGC1α defines a subset of tumors that promote survival and suppress metastasis (16, 17, 21). Thus, determining how PGC1α gene expression is dynamically controlled in melanoma cells will contribute to our understanding of the metastatic process. In these studies, we show that PGC1α gene expression is epigenetically regulated through EZH2-dependent H3K27me3 and controls a series of invasive pathways including the WNT5A/YAP axis (Figure 6I).
H3K27me3 is tightly associated with heterochromatic regions and represents an important gene-silencing marker (36). In contrast to other histone modifications, H3K27 trimethylation is mainly the product of SET domain–containing EZH2 methyltransferase and its close homolog EZH1 (37). In melanoma, EZH2 expression, gene copy number gain, and activating mutations are linked to the BRAF(V600E) mutation (38–40). EZH2 has been shown to constitute a driver for both melanoma initiation and metastatic progression (41–43). We found that BRAF(V600E) inhibition reduces H3K27me3 marks and EZH2 occupancy in the PGC1α locus, promoting PGC1α gene expression. Thus, consistent with previous reports (23), in melanoma cells, the PGC1α gene chromatin site is a direct target of EZH2 that might contribute to inhibiting invasion and metastasis. These results support the prometastatic function of EZH2 in melanoma and define a regulatory mechanism we believe is novel. An important question derived from these studies is how EZH2 is controlled in different melanomas or cell lines modifying the epigenetic state of the PGC1α locus and invasive behaviors. In addition to genetic mutations or copy numbers, EZH2 could be regulated through mechanisms involving changes in signaling pathways, such as direct phosphorylation or metabolic fluxes (44, 45). For example, 1-carbon metabolism changes cellular levels of SAM, which is an EZH2 substrate. Future studies will be needed to define how and when EZH2 activity controls PGC1α gene expression in melanoma progression or different therapies.
Spatial radial or vertical growth in skin melanomas defines the invasiveness capacity and thickness, a major clinical surrogate tightly associated with systemic disease (46). PGC1α negatively correlates with vertical growth and suppresses migration and invasion (20). We have previously shown that PGC1α inhibits invasion through transcriptional control of ID2/TCF4, which in turn increases integrin signaling and FAK activation (20). Notably, as reported in this manuscript, we found that WNT5A/YAP constitutes an additional necessary component that drives melanoma invasion in cells with low expression or depletion of PGC1α. In this case, the regulatory mechanism is through another bHLH transcription factor, TCF12, which through direct interaction with ID2 controls WNT5A gene expression. Interestingly, WNT5A has been associated with melanoma progression and, consistent with our results, inhibition of WNT5A to the FZD5 receptor using the Box5a peptide antagonist shows efficacy in blocking melanoma invasion (31). Similarly to breast cancer cells, WNT5A also signals to YAP protein stability through the RhoA GTPase in melanoma cells. The precise mechanisms of this regulation are not completely understood and might involve YAP phosphorylation by LATS1/2 at residue Ser127. This results in cytoplasmic retention and subsequent degradation of YAP, which otherwise translocates into nucleus to facilitate gene activation (47).
In parallel to suppression of the invasive program, PGC1α activates mitochondrial energy metabolism (18). Among the transcription factors involved in this energetic control are the ERRs; however, loss of these nuclear receptors is not sufficient to increase invasion (21). An intriguing question is whether PGC1α-dependent metabolic reprogramming contributes or supports invasive behavior suppression. It is conceivable that this reprogramming might confer certain advantages for blocking invasion; however, whether metabolites are sufficient to signal or control PGC1α-dependent invasion pathways is currently unknown. Interestingly, mevalonate biosynthesis and aerobic glycolysis can affect YAP activation (48, 49), contributing to its invasion function.
In summary, these studies identify regulatory mechanisms and components upstream (EZH2-dependent H3K27me3) and downstream (TCF12/WNT5A/YAP) of PGC1α that control melanoma metastasis. Detection and intervention of these mechanisms in human melanoma patients might provide novel potential clinical information and therapeutic treatments for this disease.
Methods
Tissue culture.
All melanoma cell lines were obtained from ATCC, and their authentication was confirmed by either DNA fingerprinting with small tandem repeat profiling or in-house PCR testing of melanoma marker genes and BRAF mutation status. Cells were maintained, if not otherwise indicated, in DMEM (Sigma-Aldrich) with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Reagents and antibodies.
The BRAF(V600E) inhibitor PLX4032 and EZH2 inhibitor GSK126 (used at indicated concentrations for 14 to 18 hours) were purchased from Selleck Chemicals. Recombinant mouse WNT5A protein was from R&D Systems. The RhoA inhibitor, the cell permeable C3 transferase, was obtained from Cytoskeleton Inc. The following antibodies were used: PGC1α (clone H300 and D-5, Santa Cruz Biotechnology Inc.; clone ST1202, EMD Millipore), ERRα (clone N1, GeneTex), TCF12 (clone D2C10, Cell Signaling), YAP (clone D8H1X, Cell Signaling; clone 63.7, Santa Cruz Biotechnology Inc.), EZH2 (clone D2C9, Cell Signaling), H3K27me3 (catalog 6002, Abcam), tubulin (catalog 210797, Abcam), actin (clone D6A8, Cell Signaling), histone H3 (catalog 12079, Abcam), pERK1/2 (catalog 9101, Cell Signaling), ERK1/2 (clone C-9, Santa Cruz Biotechnology Inc.), V5 (catalog R960-25, Thermo Fisher), and FLAG (clone M2, Sigma).
Lentiviral generation and transduction.
Lentiviruses encoding shRNAs or sgRNAs (lentiCRISPRv2, Addgene, catalog 52961) were produced in HEK293T cells with packaging vectors (pMD2G and psPAX2) using Escort IV Transfection Reagent (Sigma-Aldrich). Lentivirus particles were collected 48 hours after transfection and used to infect melanoma cells, and infected cells were selected with 2 μg/ml of puromycin, or 7 μg/ml blasticidin for at least 4 days prior to experiments. The shRNA clone for individual genes are as follows: shPGC1α, TRCN0000001165; shEZH2, TRCN0000039040; shYAP1, TRCN0000107266; and shWNT5A, TRCN0000296083 (http://portals.broadinstitute.org/gpp/public/). The sgRNA sequences for individual genes are as follows: PGC1α, GACTGTCTAGTGTCTCTGTG; and WNT5A, CTATGGCTACCGCTTTGCCA.
Immunoblotting and quantitative real-time PCR.
Cells were seeded in a 6-well plate at a density of 2 × 105/well 1 day before any treatment experiment. Treatments were initiated the next day after seeding, and samples were collected at particular time points after treatment, as indicated for individual experiments. For immunoblotting, cells were lysed in RIPA buffer and quantified by the DC Protein Concentration Assay Kit (Pierce) before being subjected to SDS-PAGE gel. Total RNA was isolated with TRIzol (Invitrogen) by Direct-zol RNA MiniPrep kit (Zymo Research), and approximately 1 to 2 μg of total RNA was used for cDNA synthesis using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR (qPCR) was carried out using SYBR Green PCR Master Mix (Bio-Rad). Experimental Ct values were normalized to 36B4, and relative mRNA expression was calculated using the ΔΔCt method (50). Sequences for all primers are given in Supplemental Table 1.
In vitro migration assay.
Transwell chambers were purchased from Corning Life Science. Generally, A375P (1 × 105) or A375 (1 × 104) cells in 0.1 mL of FBS-free medium were seeded into the upper chamber and incubated overnight. Medium containing 10% FBS was used in the lower chamber as chemoattractant except for in the experiment shown in Supplemental Figure 4B, which used serum-free medium, but supplemented with 400 ng/mL WNT5A, as chemoattractant. Cells that had migrated were then fixed and stained with crystal violet. The membrane attached with migrated cells was placed on a glass slide; total cells from 3 images taken from 3 random fields under ×10 magnifications with a Nikon 80i Upright microscope were quantified. The control group was set as 100%, and the experimental groups were normalized to their respective controls.
Promoter reporter assay.
The pGL3-WNT5A promoter reporter vector was provided by Hung-Chih Kuo (Academia Sinica, Taipei, Taiwan), with the help of Chun-Ying Yu (51). The assay was performed in A375P cells stably expressing empty control or TCF12, following a protocol reported elsewhere (52) with slight modification. In general, cells were seeded in a 24-well plate, followed by transfection with 1 μg of plasmid DNA with Escort IV Transfection Reagent. After 24 hours, cells were harvested for the Luciferase assay system (Promega) with luminescence measured by a FLUOstar Omega plate reader (BMG Labtech). CMV-driven Renilla luciferase vector was cotransfected as an internal control. Firefly luciferase reporter values were normalized to Renilla values, and the fold changes related to empty vector control were presented.
Animal experiments.
Mice were housed in a controlled environment under a 12-hour dark/12-hour light cycle and provided food and water ad libitum. Melanoma cell lines were detached by 0.5 mM EDTA in PBS and washed with 1× PBS. A total of 2 × 106 A375P cells or 1 × 106 G361 cells in 0.2 mL of DMEM were injected into the tail veins of 8-week-old NCI Ath/Nu mice (Charles River). To assess the degree of tumor formation in the lung, bioluminescence imaging of living mice was performed on a Xenogen IVIS-50TM imaging system equipped with an isoflurane (approximately 1% to 3%) anesthesia system and a temperature controlled platform (performed by the Longwood Small Animal Imaging Facility at Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA). Similarly, a total of 1 or 2 × 105 B16BL6 melanoma cells were injected into the tail veins of 6-week-old male C57BL/6 mice (Jackson Laboratory), followed by lung tissue harvest 3 to 4 weeks after injection. For the subcutaneous model, a total of 1 × 105 B16BL6 melanoma cells with scramble control or Tcf12 knockdown were injected into 1 flank of 4-week-old female C57BL/6 mice (Jackson Laboratory) in 0.1 mL of PBS, followed by monitoring of tumor growth. Lung was harvested at the end point to assess the metastasis foci that were derived from the subcutaneous tumor.
ChIP.
ChIP was performed with the Millipore ChIP Kit with slight modification. Following sonication, nuclear lysates were precleared with Protein A/G-Dynabeads (Invitrogen) for 1 hour. Equal amounts of precleared lysates were incubated with IgG or specific antibodies (H3K27me3 from Abcam) overnight, followed by precipitation with protein A/G-Dynabeads for 2 hours. qPCR with SYBR green was performed to quantify the promoter occupancy. The primers for PCR are shown in Supplemental Table 1.
TCGA skin melanoma and CCLE analyses.
Publicly available RNA-Seq–based gene expression data and reverse phase protein array (RPPA) data for melanoma tumors were obtained from the TCGA portal. For expression correlation, samples were stratified based on the expression level of PPARGC1A (PGC1α), with Z score less than –0.6 (n = 96) as PGC1α low and Z score greater than 0.8 (n = 44) as PGC1α high. Their corresponding YAP1 protein and mRNA expression levels, as well as YAP target genes, including CTGF, CYR61, AMOTL2, AXL, and FGF2, were plotted. Data were presented as expression median Z score. As for TCF12 expression and survival, the mean expression level of TCF12 was calculated through all samples (FPKM 8.2); then individual samples were divided into 2 groups: those with TCF12 FPKM higher than 8.2 were considered high expression, while those with FPKM of 8.2 or less were categorized into low expression. For the analysis of the CCLE data set, the gene expression data were downloaded from the CCLE portal, and the data from 61 melanoma cell lines were used for the analysis.
Statistics.
GraphPad Prism was used for statistics. In general, for 2 experimental comparisons, 2-tailed unpaired Student’s t test was used. For multiple comparisons, 1-way ANOVA was applied. When cells were used for experiments, 3 replicates per treatment were chosen as an initial sample size. All n values defined in the legends refer to biological replicates unless otherwise indicated. A P value of less than 0.05 was considered significant.
Study approval.
All studies in animals were reviewed and approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
Author contributions
CL and PP conceived the project. CL, EB, and PP designed the experiments. CL, EB, EAP, JL, and CDT performed experiments. CL, EB, FV, and HRW analyzed data. All authors contributed to discussions about the data and their interpretation. The manuscript was written by CL, HRW, and PP.
Supplementary Material
Acknowledgments
We thank the Nikon Imaging Center at Harvard Medical School for the use of the microscope. We thank members of the Puigserver lab for important discussions about this project. These studies were funded in part by NIH R01CA181217 (to PP) and NCI K99CA222617 (to CL).
Version 1. 01/13/2020
Electronic publication
Version 2. 02/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):853–862.https://doi.org/10.1172/JCI130038.
Contributor Information
Chi Luo, Email: chiluo@zju.edu.cn.
Eduardo Balsa, Email: eduardo_balsamartinez@dfci.harvard.edu.
Jiaxin Liang, Email: Jiaxin_Liang@DFCI.HARVARD.EDU.
Francisca Vazquez, Email: vazquez@broadinstitute.org.
Hans R. Widlund, Email: hwidlund@bics.bwh.harvard.edu.
Pere Puigserver, Email: pere_puigserver@dfci.harvard.edu.
References
- 1.Merlino G, et al. The state of melanoma: challenges and opportunities. Pigment Cell Melanoma Res. 2016;29(4):404–416. doi: 10.1111/pcmr.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gershenwald JE, et al. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(6):472–492. doi: 10.3322/caac.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoek KS, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68(3):650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 4.Cronin JC, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22(4):435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 6.Plantefaber LC, Hynes RO. Changes in integrin receptors on oncogenically transformed cells. Cell. 1989;56(2):281–290. doi: 10.1016/0092-8674(89)90902-1. [DOI] [PubMed] [Google Scholar]
- 7.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 8.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18(9):533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malladi S, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016;165(1):45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weeraratna AT, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1(3):279–288. doi: 10.1016/S1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 12.Dissanayake SK, et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282(23):17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastas JN, et al. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J Clin Invest. 2014;124(7):2877–2890. doi: 10.1172/JCI70156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park HW, et al. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162(4):780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez F, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23(3):302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo C, Widlund HR, Puigserver P. PGC-1 coactivators: shepherding the mitochondrial biogenesis of tumors. Trends Cancer. 2016;2(10):619–631. doi: 10.1016/j.trecan.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 2015;72(7):1249–1260. doi: 10.1007/s00018-014-1791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo C, et al. A PGC1α-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537(7620):422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo C, et al. ERRα maintains mitochondrial oxidative metabolism and constitutes an actionable target in PGC1α-elevated melanomas. Mol Cancer Res. 2017;15(10):1366–1375. doi: 10.1158/1541-7786.MCR-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JH, Luo C, Vazquez F, Puigserver P. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res. 2014;74(13):3535–3545. doi: 10.1158/0008-5472.CAN-13-2893-T. [DOI] [PubMed] [Google Scholar]
- 23.Segal T, Salmon-Divon M, Gerlitz G. The heterochromatin landscape in migrating cells and the importance of H3K27me3 for associated transcriptome alterations. Cells. 2018;7(11):E205. doi: 10.3390/cells7110205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MH, et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 2016;35(5):462–478. doi: 10.15252/embj.201592081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugo W, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Redondo V, Pettersson AT, Ruas JL. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia. 2015;58(9):1969–1977. doi: 10.1007/s00125-015-3671-z. [DOI] [PubMed] [Google Scholar]
- 29.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CK, et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 2019;363(6427):644–649. doi: 10.1126/science.aav0173. [DOI] [PubMed] [Google Scholar]
- 31.Jenei V, et al. A t-butyloxycarbonyl-modified Wnt5a-derived hexapeptide functions as a potent antagonist of Wnt5a-dependent melanoma cell invasion. Proc Natl Acad Sci U S A. 2009;106(46):19473–19478. doi: 10.1073/pnas.0909409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreger P, et al. Early stem cell transplantation for chronic lymphocytic leukaemia: a chance for cure? Br J Cancer. 1998;77(12):2291–2297. doi: 10.1038/bjc.1998.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wróbel S, Przybyło M, Stępień E. The clinical trial landscape for melanoma therapies. J Clin Med. 2019;8(3):E368. doi: 10.3390/jcm8030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14(8):463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 35.Luo C, et al. Loss of ARF sensitizes transgenic BRAFV600E mice to UV-induced melanoma via suppression of XPC. Cancer Res. 2013;73(14):4337–4348. doi: 10.1158/0008-5472.CAN-12-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 37.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11(2):286–295. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiffen J, Wilson S, Gallagher SJ, Hersey P, Filipp FV. Somatic copy number amplification and hyperactivating somatic mutations of EZH2 correlate with DNA methylation and drive epigenetic silencing of genes involved in tumor suppression and immune responses in melanoma. Neoplasia. 2016;18(2):121–132. doi: 10.1016/j.neo.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan J, Gupta R, Filipp FV. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Sci Rep. 2015;5:7857. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 42.Zingg D, et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell. 2018;34(1):69–84.e14. doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Manning CS, Hooper S, Sahai EA. Intravital imaging of SRF and Notch signalling identifies a key role for EZH2 in invasive melanoma cells. Oncogene. 2015;34(33):4320–4332. doi: 10.1038/onc.2014.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha TL, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 45.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 47.Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25(9):499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16(4):357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 49.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34(10):1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo C, et al. Expression of oncogenic BRAFV600E in melanocytes induces Schwannian differentiation in vivo. Pigment Cell Melanoma Res. 2015;28(5):603–606. doi: 10.1111/pcmr.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu CY, Kuo HC. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells. 2016;34(8):2052–2062. doi: 10.1002/stem.2386. [DOI] [PubMed] [Google Scholar]
- 52.Sheng J, Luo C, Jiang Y, Hinds PW, Xu Z, Hu GF. Transcription of angiogenin and ribonuclease 4 is regulated by RNA polymerase III elements and a CCCTC binding factor (CTCF)-dependent intragenic chromatin loop. J Biol Chem. 2014;289(18):12520–12534. doi: 10.1074/jbc.M114.551762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.