Abstract
Triglycerides are the major form of stored fat in all animals. One important determinant of whole-body fat storage is whether an animal is male or female. Here, we use Drosophila, an established model for studies on triglyceride metabolism, to gain insight into the genes and physiological mechanisms that contribute to sex differences in fat storage. Our analysis of triglyceride storage and breakdown in both sexes identified a role for triglyceride lipase brummer (bmm) in the regulation of sex differences in triglyceride homeostasis. Normally, male flies have higher levels of bmm mRNA both under normal culture conditions and in response to starvation, a lipolytic stimulus. We find that loss of bmm largely eliminates the sex difference in triglyceride storage and abolishes the sex difference in triglyceride breakdown via strongly male-biased effects. Although we show that bmm function in the fat body affects whole-body triglyceride levels in both sexes, in males, we identify an additional role for bmm function in the somatic cells of the gonad and in neurons in the regulation of whole-body triglyceride homeostasis. Furthermore, we demonstrate that lipid droplets are normally present in both the somatic cells of the male gonad and in neurons, revealing a previously unrecognized role for bmm function, and possibly lipid droplets, in these cell types in the regulation of whole-body triglyceride homeostasis. Taken together, our data reveal a role for bmm function in the somatic cells of the gonad and in neurons in the regulation of male–female differences in fat storage and breakdown and identify bmm as a link between the regulation of triglyceride homeostasis and biological sex.
An investigation of the genetic and physiological mechanisms underlying sex differences in fat storage and breakdown in the fruit fly Drosophila identifies previously unrecognized sex- and cell type-specific roles for the conserved triglyceride lipase brummer.
Introduction
Triglycerides are the main form of stored fat in animals and are stored in lipid droplets within specialized fat storage organs, such as the adipose tissue in mammals or the fat body in insects [1–5]. One important but often overlooked factor that affects fat storage is whether the animal is male or female [6,7]. In mammals, females store approximately 10% more body fat than males [7–9], whereas in some insect species, females store up to four times more fat than males [10]. An extensive literature has revealed the important role of sex hormones and sex chromosomes in establishing this male–female difference in fat storage [7,11,12]. For example, the female sex steroid estrogen and the presence of two X chromosomes both contribute to the increased fat storage in female mice [7,11]. Although these sex-determining factors in mice, and in other animals, have been shown to promote extensive sex-biased expression of many genes, including genes involved in fat storage [13–16], the downstream metabolic genes that contribute to the sex difference in fat storage are only beginning to be uncovered.
Over the past 15 years, Drosophila has emerged as a powerful model to investigate the in vivo function of genes that are involved in the regulation of triglyceride synthesis, storage, and breakdown [4,17–19]. The main pathway of triglyceride synthesis in flies begins with the acylation of glycerol-3-phosphate to produce lysophosphatidic acid, a reaction that is catalyzed by glycerol-3-phosphate acyltransferases (GPATs) [4]. Flies have several genes that encode putative GPATs: minotaur (mino; FBgn0027579), Gpat4 (FBgn0034971), and the testis-specific CG15450 (FBgn0031132). Although previous studies confirmed a role for mino in triglyceride synthesis by demonstrating that mino overexpression leads to large lipid droplets in the larval salivary gland [20], the functional roles of Gpat4 and CG15450 in triglyceride metabolism remain largely unconfirmed. The second step in triglyceride synthesis is catalyzed by 1-acylglycerol-3-phosphate O-acyltransferases (AGPATs), which acylate lysophosphatidic acid to produce phosphatidic acid [4]. Drosophila has four genes that encode potential AGPAT proteins: Agpat1 (FBgn0030421), Agpat2 (FBgn0026718), Agpat3 (FBgn0036623), and Agpat4 (FBgn0036622). At present, these predicted AGPAT enzymes are largely uncharacterized [4]; however, Agpat2, Agpat3, and Agpat4 are all expressed in the fat body, a critical lipid-storing organ [21]. Once phosphatidic acid is produced, it is dephosphorylated in the third step of triglyceride synthesis by the phosphatase Lipin into diglyceride. The Drosophila genome contains a single Lipin gene (Lpin; FBgn0263593) that is expressed in fat-storing organs in flies [21], and Lpin loss alters lipid droplet size and impairs whole-body triglyceride storage [22].
The final step in triglyceride synthesis is the acylation of diglyceride into triglyceride by diacylglycerol O-acyltransferases (DGATs) [4]. In Drosophila, the only characterized DGAT family member is called midway (mdy; FBgn0004797) [4]. Studies have shown that loss of mdy significantly impairs triglyceride synthesis and reduces whole-body triglyceride levels [23,24], whereas mdy overexpression increases the number of small lipid droplets in the larval salivary gland [20]. In addition to mdy, the Drosophila genome also contains genes that encode proteins from a related family of enzymes that show DGAT activity, which is called the DAGAT family [4]. In flies, there are three members of this DAGAT family: CG1941 (FBgn0033214), Dgat2 (FBgn0033215), and CG1946 (FBgn0033216), all of which are functionally and biochemically uncharacterized. Once triglyceride synthesis is complete, triglycerides accumulate between the two leaflets of the phospholipid bilayer in the endoplasmic reticulum to form a lipid lens [5]. This lipid lens eventually buds off from the endoplasmic reticulum to form an organelle called a lipid droplet. The neutral lipid core of the lipid droplet is separated from the cytoplasmic contents of the cell by a phospholipid monolayer. Studies show that many proteins associated with this monolayer play key roles in regulating lipid droplet size, as well as cellular and organismal levels of triglyceride storage [5,25–28]. For example, members of the perilipin (PLIN) family of proteins in Drosophila associate with lipid droplets and influence lipid droplet size in vivo [29–31]. The Drosophila genome encodes two PLIN family members: lipid storage droplet-1 (lsd-1/PLIN1; FBgn0039114) and lipid storage droplet-2 (lsd-2/PLIN2; FBgn0030608). Importantly, altered expression of either lsd-1/PLIN1 or lsd-2/PLIN2 impacts lipid droplet size and affects whole-body triglyceride storage [29–31].
Triglyceride breakdown in Drosophila occurs in a fixed series of enzymatic reactions [4]. The first step in Drosophila triglyceride breakdown is triglyceride hydrolysis, which produces a free fatty acid and diglyceride [4]. In the Drosophila genome, the best-characterized triglyceride lipase is brummer (bmm; FBgn0036449), a member of the Patatin-like domain-containing family that catalyzes triglyceride hydrolysis in vitro and in vivo [32]. Importantly, loss of bmm function increases lipid droplet size and augments whole-body triglyceride storage, whereas bmm overexpression decreases lipid droplet size and depletes triglyceride levels [32], demonstrating a key role for bmm in regulating triglyceride homeostasis in vivo. Other than bmm, the Drosophila genome contains more than 50 predicted lipases, only a few of which have been characterized [33]. For example, larvae with loss of hormone-sensitive lipase (hsl; FBgn0034491) have larger lipid droplets and higher triglyceride levels than controls [30]. Additional genes with potential effects on triglyceride mobilization include another Patatin-like domain-containing family member called doppelganger von brummer (dob; FBgn0030607) and CG5966 (FBgn0029831). In addition to the essential role that lipases such as bmm have in promoting triglyceride breakdown, lipid droplet–associated proteins also make important contributions to lipolysis. For example, lsd-1/PLIN1 and lsd-2/PLIN2 influence triglyceride breakdown by regulating the access of key lipases such as bmm to their triglyceride substrate [4]. Together, these studies highlight the important contribution of Drosophila to our current knowledge of the molecular mechanisms underlying the regulation of whole-body triglyceride levels in vivo.
In addition to revealing the mechanisms underlying the regulation of cellular and organismal triglyceride levels, studies in Drosophila have significantly advanced our knowledge of how triglyceride homeostasis impacts life span, starvation resistance, and fertility [4]. For example, studies have shown that flies with reduced function of mdy, the enzyme that catalyzes the final step of triglyceride synthesis, have impaired egg chamber development and female sterility [23,29,34,35]. Another gene with well-studied effects on cellular and organismal phenotypes is triglyceride lipase bmm: male flies with reduced bmm function have increased starvation resistance, exaggerated sleep rebound following sleep deprivation [36], and a modest reduction in life span [32,34]. Finally, several phenotypes have been associated with lsd-1/PLIN1 and lsd-2/PLIN2, such as starvation resistance [29–31] and sleep rebound after sleep deprivation [36]. Thus, the correct regulation of triglyceride storage and breakdown impacts many aspects of Drosophila development, physiology, and life history.
In the present study, we aimed to improve our knowledge of the metabolic genes and physiological mechanisms that contribute to male–female differences in Drosophila triglyceride homeostasis. Although whole-body triglyceride storage is known to differ between mated female and male flies [37–39], most studies on triglyceride synthesis and breakdown use male flies or mixed-sex groups of larvae to determine how individual genes affect these processes. As a result, the downstream genes and mechanisms that contribute to the sex difference in triglyceride storage, and possibly other aspects of triglyceride homeostasis, remain incompletely understood. Our detailed examination of triglyceride storage and breakdown in adult male and female flies revealed significant sexual dimorphism in both aspects of triglyceride homeostasis and identified a role for one gene, triglyceride lipase bmm, in regulating sex differences in triglyceride storage and breakdown. Normally, females have more triglyceride storage than males and slower triglyceride breakdown in response to a lipolytic stimulus. Loss of bmm largely abolishes the sex difference in triglyceride storage and eliminates the sex difference in triglyceride breakdown via strongly male-biased effects. Importantly, we discovered that bmm function in the somatic cells of the gonad and in neurons, two cell types previously unknown to require bmm function, plays a role in regulating sex differences in triglyceride homeostasis. Because we show that lipid droplets, the intracellular fat storage organelle, are present in both cell types under normal physiological conditions, our findings illuminate an unexpected role for bmm function, and possibly lipid droplets, in these two cell types in the regulation of sex differences in triglyceride storage and breakdown.
Results
Sexual dimorphism in triglyceride storage and breakdown
Adult mated females have increased levels of triglyceride storage compared with males [37–39]. To determine whether this increased triglyceride storage in females reflects a mating-induced change to female physiology or a sexual dimorphism in triglyceride storage, we measured whole-body triglyceride storage in Canton-S (CS) virgin females and males. In 5-day-old adults, virgin females have increased levels of triglyceride storage compared with virgin males (Fig 1A; see S1 Table for all p-values). This difference was also present when we compared whole-body triglyceride storage in white (w)1118 virgin males and females (Fig 1B). Because we observed no significant differences in triglyceride storage between 5-day-old CS and w1118 virgin females or between 5-day-old CS and w1118 virgin males (S2 Table) our findings show that the sexual dimorphism in triglyceride storage persists in multiple genetic backgrounds. Although one obvious explanation for the sexual dimorphism in triglyceride storage is triglyceride contained within the male and female gonads, we confirm previous findings that ovary triglyceride levels represent only a fraction of the whole-body triglyceride level in females (S1A Fig) [37] and show that triglyceride levels in the testis do not significantly contribute to the whole-body triglyceride level in males (S1B Fig). Furthermore, we found that the sexual dimorphism in triglyceride storage was preserved between w1118 virgin male and female carcasses devoid of gonads (S1C Fig). Thus, the sex difference in triglyceride storage cannot be solely attributed to the triglyceride stored in the male and female gonads.
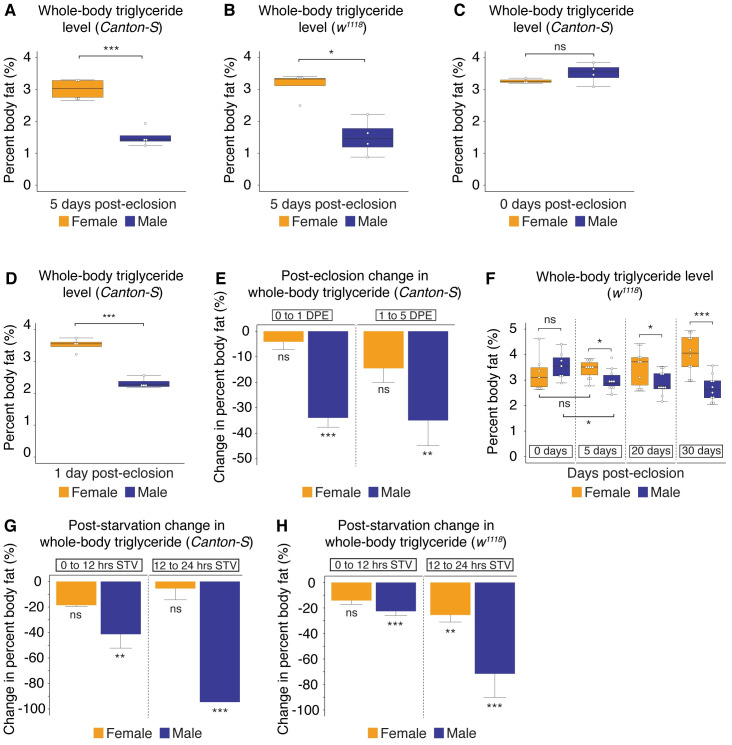
Fig 1. Sexual dimorphism in Drosophila triglyceride storage and breakdown.
(A) Whole-body triglyceride storage in 5-day-old Canton-S virgin females was significantly higher than in age-matched Canton-S virgin male flies (p = 5.4 × 10−4; Student t test). (B) Whole-body triglyceride storage in 5-day-old w1118 virgin female flies was significantly higher than in age-matched w1118 virgin male flies (p = 2.3 × 10−2; Student t test). (C) No significant difference in whole-body triglyceride storage was found between newly eclosed virgin Canton-S females and age-matched virgin males (p = 0.73; Student t test). (D) Whole-body triglyceride storage in 1-day-old Canton-S virgin females was significantly higher than in age-matched virgin males (p = 1.2 × 10−4; Student t test). (E) In females, whole-body triglyceride levels were not significantly different between newly eclosed flies and flies collected at 1 DPE or between flies collected at 1 DPE and 5 DPE (p = 0.91 and 0.38, respectively; one-way ANOVA followed by Tukey HSD test). In males, whole-body triglyceride storage was significantly lower at 1 DPE than in newly eclosed flies, with a further reduction in triglyceride storage between 1 DPE and 5 DPE (p = 4.2 × 10−4 and 5.7 × 10−3, respectively; one-way ANOVA followed by Tukey HSD test). (F) Whole-body triglyceride storage in w1118 virgin females was not significantly higher than males at eclosion, but it was significantly higher by 5 DPE, a sex difference that was maintained in 20- and 30-day-old males and females (p = 0.38, 0.024, 0.029, 1.5 × 10−4, respectively; Student t test at each time point). (G) In 5-day-old Canton-S virgin females, there was no significant difference in whole-body triglyceride levels between 0 and 12 hours STV or between 12 and 24 hours STV (p = 0.097, 0.92, respectively; one-way ANOVA followed by Tukey HSD test). In males, there was a significant decrease in whole-body triglyceride storage between 0 and 12 hours STV and a further decrease in triglyceride levels between 12 and 24 hours STV (p = 2.2 × 10−3, 1.7 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (H) In 5-day-old w1118 virgin females, there was no significant difference in whole-body triglyceride levels between 0 and 12 hours STV and a modest difference between 12 and 24 hours STV (p = 0.11, 2.2 × 10−3, respectively; one-way ANOVA followed by Tukey HSD test). In 5-day-old w1118 virgin males, we observed a significant decrease in triglyceride levels between 0 and 12 hours STV and a further decrease between 12 and 24 hours STV (p = 3.0 × 10−5, 0.0, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. COE, coefficient of error; DPE, days post-eclosion; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
To determine when this sexual dimorphism in triglyceride storage was established, we examined triglyceride levels in adult virgin male and female flies at several times post-eclosion. In newly eclosed flies, where larval fat is still present [40], there was no significant difference in triglyceride levels between CS virgin females and males (Fig 1C). By 1 day post-eclosion (DPE), however, triglyceride levels in CS virgin females were significantly higher than in age-matched males (Fig 1D), and by 5 DPE, triglyceride levels in CS virgin females were approximately 2.2 times higher than in CS virgin males (Fig 1A). When we examined triglyceride storage during early adult life within CS flies of each sex, we found that whole-body triglyceride storage at 1 DPE in CS males was significantly lower than in newly eclosed CS males and significantly lower in males at 5 DPE compared with males at 1 DPE (Fig 1E). In females, we found no significant changes to whole-body triglyceride levels during this 5-day period (Fig 1E). Thus, the sexual dimorphism in CS triglyceride storage was established over the first 5 days of adult life by a progressive reduction in whole-body triglyceride storage in males, a finding we also confirm in w1118 (Fig 1F and S2 Table). Given that previous studies showed that, in females, approximately 50% of the larval fat cells disappear within 9 hours post-eclosion [40], one possible explanation for the reduction in triglyceride levels in males post-eclosion is a male–female difference in the persistence of larval fat cells. We therefore counted the number of larval fat cells in CS and w1118 males and females at 12-hour intervals post-eclosion. We found that larval fat cells were largely eliminated in both sexes between 0 and 24 hours post-eclosion (S1D and S1E Fig); however, there was no obvious sex difference in the timing of larval fat cell loss that would explain the male–female difference in triglyceride storage that is established over the first 5 days of adult life. Once this difference is established, we show that the sexual dimorphism in triglyceride storage persists until at least 30 DPE (Fig 1F).
In addition to sexual dimorphism in triglyceride storage, male–female differences in fat breakdown have also been reported in mammals [7,41]. We therefore examined changes to whole-body triglyceride levels in response to starvation, a lipolytic stimulus, in virgin males and females. In CS 5-day-old virgin males, we observed a 41% decrease in whole-body triglyceride levels between 0 (fed flies) and 12 hours post-starvation and a further reduction in triglyceride levels between 12 and 24 hours post-starvation (Fig 1G). As a result of this substantial reduction in whole-body triglyceride levels post-starvation, whole-body triglyceride stores were largely depleted in virgin males by 24 hours post-starvation (S1F Fig), a finding that is in line with previous studies [29,30,32]. In contrast, we found no significant change in whole-body triglyceride levels in 5-day-old virgin CS females between either 0 and 12 hours or between 12 and 24 hours post-starvation (Fig 1G). Indeed, whole-body triglyceride levels in starved females remained at 77% of the levels found in fed female flies by 24 hours post-starvation, a time when triglyceride levels in starved males were at only 4% of the levels found in fed males (S1F Fig). Consistent with our data from CS flies, we observed a rapid drop in triglyceride levels post-starvation in w1118 virgin males compared with females (Fig 1H), demonstrating that the sexual dimorphism in triglyceride breakdown exists in multiple genetic backgrounds. To determine whether male and female gonads play a role in the sexual dimorphism in triglyceride breakdown, we first measured triglyceride levels post-starvation in ovaries and testes dissected from 5-day-old w1118 males and females. We found that triglyceride levels were unchanged by starvation in both organs (S1A and S1B Fig), suggesting that triglyceride levels in the gonads do not fully account for the male–female difference in triglyceride breakdown. Moreover, when we measured triglyceride levels post-starvation in w1118 male and female carcasses devoid of gonads, we found that triglyceride levels did not change in female carcasses between 0 and 12 hours post-starvation, whereas there was a significant decrease in triglyceride levels in male carcasses during this same interval (S1G Fig). Thus, in addition to the male–female difference in triglyceride storage, our findings reveal a sexual dimorphism in triglyceride breakdown.
Sexual dimorphism in metabolic rate and macronutrient utilization
One potential explanation for increased triglyceride storage and reduced triglyceride breakdown post-starvation in females is a lower demand for energy from physical activity or from basal metabolic processes. Because previous studies have shown that female flies are active over a larger portion of the day than males [42,43], we used indirect calorimetry to determine whether females have a lower energy demand due to basal metabolic processes under normal culture conditions and in response to starvation. In 5-day-old adults, mass-corrected CO2 production and O2 consumption were significantly higher in virgin females than in age-matched virgin males throughout the 24-hour monitoring window (Fig 2A and 2B; see also S2A and S2B Fig for non-mass-corrected data). This sex difference in CO2 production and O2 consumption persisted post-starvation (Fig 2C and 2D; see S2C and S2D Fig for non-mass-corrected data): although both females and males demonstrated a significant reduction in metabolic rate from 4 hours post-starvation until the end of the 24-hour observation period (S3A–S3D Fig), a change that was independent of any change in mass (S3E–S3H Fig), CO2 production and O2 consumption in virgin females remained significantly higher post-starvation than in virgin males. Taken together, these results do not support a model in which sexual dimorphism in triglyceride storage and breakdown are caused by lower energy demand in females.
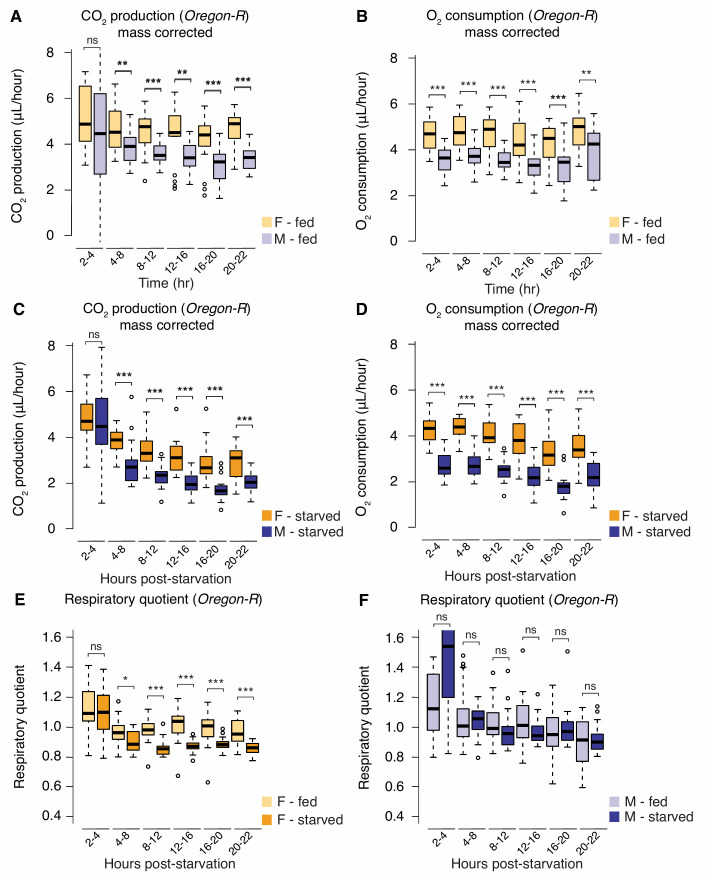
Fig 2. Sex differences in metabolic rate and macronutrient utilization.
(A) Mass-corrected CO2 production was significantly higher in fed Oregon-R virgin females than in virgin males for most intervals during the observation period (p = 0.18, 4.5 × 10−3, 5.2 × 10−5, 4.9 × 10−3, 6.6 × 10−4, 2.43 × 10−7, respectively; Student t test at each time point). (B) Mass-corrected O2 consumption was significantly higher at each interval in fed Oregon-R females than in males during the observation period (p = 9.8 × 10−6, 1.8 × 10−6, 2.2 × 10−6, 5.2 × 10−4, 7.9 × 10−4, 4.3 × 10−3, respectively; Student t test at each time point). (C) Mass-corrected CO2 production post-starvation was significantly higher in females than in males for most intervals during the observation period (p = 0.55, 3.5 × 10−4, 6.4 × 10−7, 2.7 × 10−6, 8.0 × 10−6, 5.9 × 10−5, respectively; Student t test at each time interval). (D) Mass-corrected O2 consumption post-starvation was significantly higher in females at all intervals during the observation period (p = 2.4 × 10−10, 3.4 × 10−11, 1.4 × 10−10, 1.9 × 10−8, 1.1 × 10−7, 2.5 × 10−6, respectively; Student t test at each time interval). (E) The RQ was calculated as the ratio between CO2 production to O2 consumption at defined intervals over a 24-hour observation period in 5-day-old Oregon-R virgin females and males that were placed on either standard media or starvation media. In starved females, we observed a significant reduction in RQ compared with control females on standard media from 4 to 8 hours post-starvation onward (p = 0.85, 0.014, 6.5 × 10−6, 1.3 × 10−5, 8 × 10−4, 2.2 × 10−5, respectively; Student t test at each time point). (F) In male flies, we observed no significant change in RQ compared with control males on standard medium at any time during the observation period (p = 0.066, 0.89, 0.24, 0.079, 0.39, 0.62, respectively; Student t test at each time point). For indirect calorimetry measurements, the p-values are listed in the following order: difference between fed and starved animals at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; RQ, respiratory quotient.
We next asked whether the sex differences in triglyceride homeostasis might be due to male–female differences in the preferential use of macronutrients to fuel basal metabolic processes. We therefore calculated the respiratory quotient (RQ) from the ratio of CO2 production to O2 consumption in each sex. An RQ of 1 normally indicates the use of carbohydrates as the primary fuel for metabolic processes, and an RQ below 1 indicates a shift toward fat and protein utilization [44]. Under normal culture conditions, the RQ was approximately 1 in both virgin males and females (S4A Fig), indicating that both sexes are using similar macronutrients to fuel basal metabolic processes. Thus, the sexual dimorphism in triglyceride storage was not caused by a male–female difference in overall macronutrient usage under normal conditions. When we calculated the RQ at several time points post-starvation, we saw a significant difference between males and females (S4B Fig). In starved virgin females, we observed a significant decrease in RQ compared with fed virgin females from as early as 4 to 8 hours post-starvation, a change that persisted throughout our 24-hour observation period (Fig 2E). In contrast, the RQ in starved virgin males was not significantly different from fed control virgin males at any time throughout the 24-hour starvation period (Fig 2F). Interestingly, the decreased RQ in virgin females indicates a shift from carbohydrate fuel toward either fat and/or protein catabolism; however, we found no sexual dimorphism in protein breakdown and negligible differences in other macronutrients over the 24-hour starvation period (S5A–S5C Fig). The strong shift in RQ, indicating higher lipid catabolism in females, is surprising in light of our finding that triglyceride breakdown is lower in virgin female flies post-starvation. One possible explanation for this finding is that the amount of ATP generated by one fatty acid molecule is higher than for one molecule of glucose. Because females display a shift toward lipid as the main source of energy post-starvation, this may allow for sufficient ATP production post-starvation despite less overall triglyceride breakdown compared with males. Together, these findings highlight a significant difference in energy physiology between males and females and support a model in which there is a male–female difference in lipid catabolism post-starvation.
Sex-biased gene expression of triglyceride metabolism genes
In order to identify genes that contribute to the sexual dimorphisms in triglyceride storage and breakdown, we used quantitative real-time PCR (qPCR) to measure mRNA levels in a subset of genes known or predicted to be involved in lipid synthesis, breakdown, and storage [4,17,19]. Our investigation revealed sex-specific regulation of many genes in 5-day-old w1118 virgin female and male flies cultured under normal conditions: 23 out of 31 (74%) genes we examined showed a sex difference in mRNA levels (Fig 3A). For example, GPAT enzyme mino, AGPAT enzyme Agpat4, and lipase hsl had strongly female-biased expression, whereas triglyceride lipase bmm and lsd-1/PLIN1 mRNA levels were approximately 1.8- and 4-fold higher in males than in females, respectively. Some genes, such as AGPAT enzyme Agpat1, lsd-2/PLIN2, and CG5966 showed no significant difference in mRNA level between the sexes (Fig 3A). Thus, under normal culture conditions, many genes known or predicted to affect triglyceride metabolism display strongly sex-biased expression, trends that persisted when a subset of genes was normalized to a different housekeeping gene (S6A and S6B Fig).
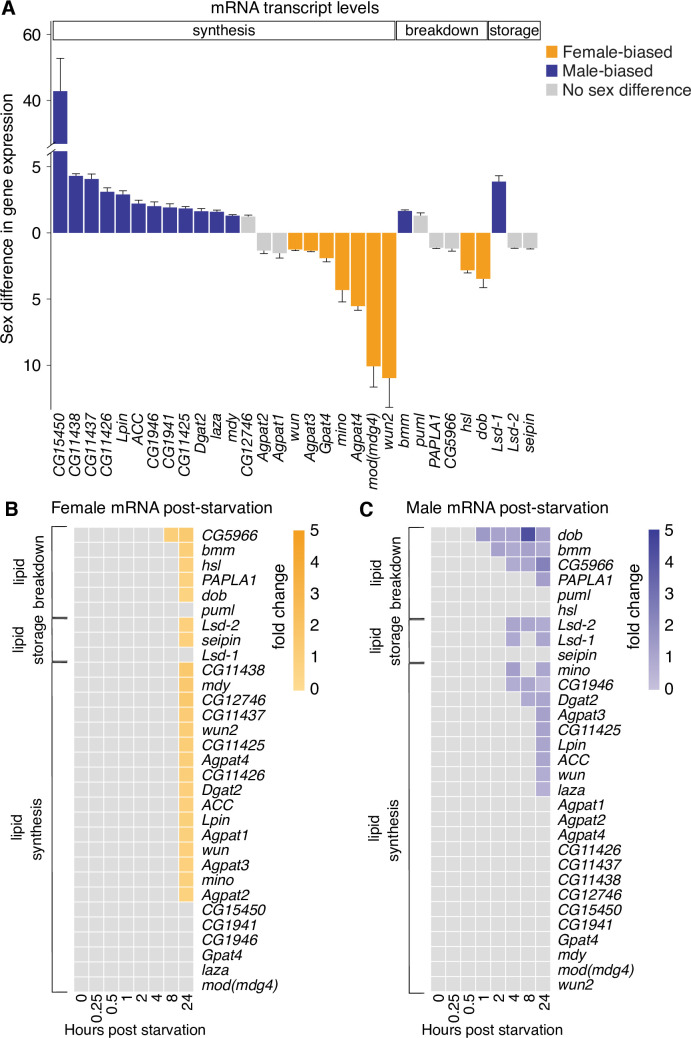
Fig 3. Extensive sex-biased expression of genes involved in maintaining triglyceride homeostasis.
(A) Sex-biased mRNA levels of a panel of 31 genes known or predicted to be involved in triglyceride metabolism in 5-day-old virgin w1118 females and males. Gray-colored bars indicate no significant difference in mRNA level between the sexes. Orange-colored bars indicate that mRNA levels are significantly higher in virgin females than in virgin males. Purple-colored bars indicate that mRNA levels are significantly higher in virgin males than in virgin females. (B, C) mRNA levels of a panel of genes involved in triglyceride metabolism in virgin 5-day-old female w1118 flies (B) and virgin 5-day-old male w1118 flies (C) measured at different times post-starvation. Gray boxes indicate that mRNA levels were not significantly different from sex-matched, fed controls; colored boxes indicate that mRNA levels were significantly different from age-matched fed flies, and the intensity of the color corresponds to the fold change in mRNA level (refer to legend). Error bars on graphs represent SEM. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying gene expression data are available in S3 Data. w, white.
To gain insight into genes that may contribute to the sexual dimorphism in triglyceride breakdown, we measured mRNA levels in virgin males and females at various time points post-starvation. Because we observed a sex difference in phenotype by 12 hours post-starvation, we predicted that the majority of gene expression changes in males and females would precede this critical time point. In females, with the exception of CG5966, a gene that may be involved in triglyceride breakdown, no genes showed significant changes in mRNA expression until 24 hours post-starvation, a time at which most genes were significantly different from fed control females (Fig 3B). In males, we found significant changes to mRNA levels starting as early as 1 hour post-starvation (Fig 3C). For example, mRNA levels of predicted triglyceride lipase dob were significantly increased from 1 hour post-starvation onward; bmm mRNA levels were significantly up-regulated from 2 hours post-starvation onward; and lsd-1/PLIN1, DAGAT family member CG1946, and GPAT enzyme mino were significantly increased from 4 hours post-starvation onward (Fig 3C). Importantly, these trends persisted when we normalized a subset of genes to an additional housekeeping gene (S7A–S7L Fig). Taken together, these results demonstrate a marked sex difference in the transcriptional response to starvation: in males, there was a rapid transcriptional response as early as 1–2 hours post-starvation; in females, the transcriptional response was delayed: mRNA levels of most genes were not significantly different until 24 hours post-starvation.
A role for triglyceride lipase bmm in the regulation of sex differences in fat storage and breakdown
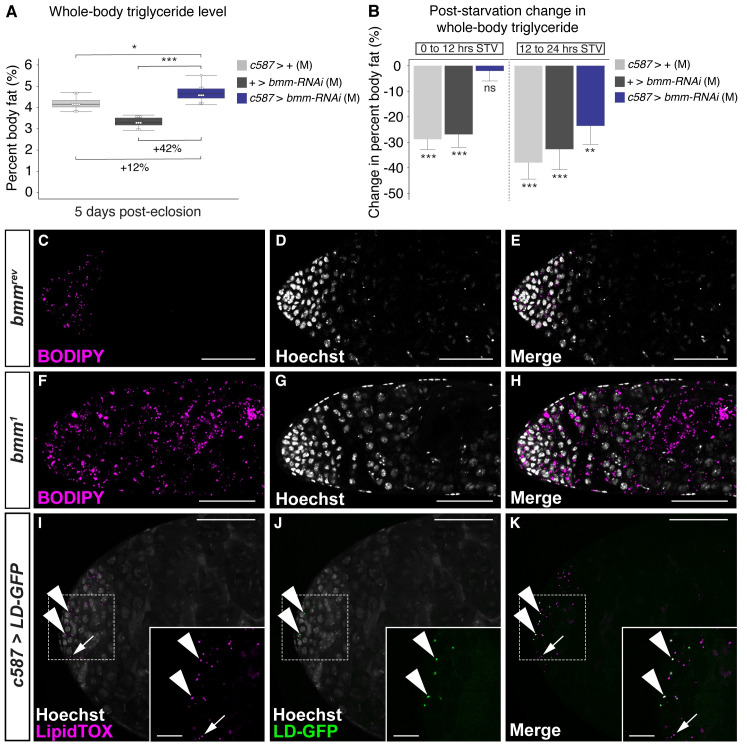
To determine whether any triglyceride metabolism genes contribute to the male–female differences in triglyceride storage and breakdown, we wanted to investigate how individual genes contribute to the sex differences in triglyceride homeostasis. For most genes with sex-biased expression under normal culture conditions (Fig 3A), the magnitude of the sex bias in gene expression remained largely consistent throughout the starvation period (see S8A–S8D Fig for graphs of representative genes). One exception to this trend was triglyceride lipase bmm, which showed sex-specific regulation during normal culture conditions (1.8-fold higher in males) and a strong male-specific increase in mRNA levels post-starvation (3.1-fold higher in males by 8 hours post-starvation) (Fig 4A). Given that bmm regulation is highly sex specific under both normal culture conditions and post-starvation and that changes to bmm expression are known to influence whole-body triglyceride homeostasis [32], we reasoned that bmm may play a role in regulating sex differences in triglyceride storage and breakdown. Therefore, we compared triglyceride homeostasis in bmm1 mutant flies to bmmrev control flies. Because the sex differences in triglyceride storage in bmmrev flies were perfectly in line with our observations in CS and w1118 flies (S2 Table) and because bmmrev flies and bmm1 mutant flies were derived from the same parental strain [32], this will allow us to investigate whether there is a role for bmm in the regulation of sex differences in triglyceride storage and breakdown. In accordance with previous reports [32], triglyceride levels in bmm1 homozygous mutant males were approximately 2.5 times higher than in bmmrev control males (Fig 4B). In bmm1 mutant females, however, triglyceride storage was increased by only 1.4 times compared with bmmrev control females (Fig 4B), demonstrating a strongly male-biased effect of bmm loss on triglyceride storage. Given that 5-day-old bmm1 mutant females fed a high-fat diet had significantly higher triglyceride levels than bmm1 females fed a normal diet (S9A Fig), the mild increase in triglyceride storage in bmm1 mutant females cannot be attributed to these females achieving a physiological limit of triglyceride storage. In fact, the remaining difference between bmm1 mutant females and males was due to the modest amount of triglyceride stored in the ovary, as no sex difference in triglyceride storage remained between bmm1 mutant females lacking ovaries and bmm1 mutant males (S9B Fig). Importantly, we reproduced these male-biased effects of reduced bmm function on triglyceride storage in flies with ubiquitous overexpression of two different upstream activation sequence (UAS)-bmm-RNAi lines driven by daughterless (da)-GAL4 (S10A–S10F Fig). These strongly male-biased effects of bmm loss on triglyceride storage reduced the sexual dimorphism in triglyceride storage in bmm1 mutants compared with bmmrev control flies (Fig 4C) and in da>UAS-bmm-RNAi flies compared with da>+ and +>UAS-bmm-RNAi controls (S10C and S10F Fig). Together, these data suggest that normal bmm function plays a role in the regulation of sexual dimorphism in Drosophila triglyceride storage.
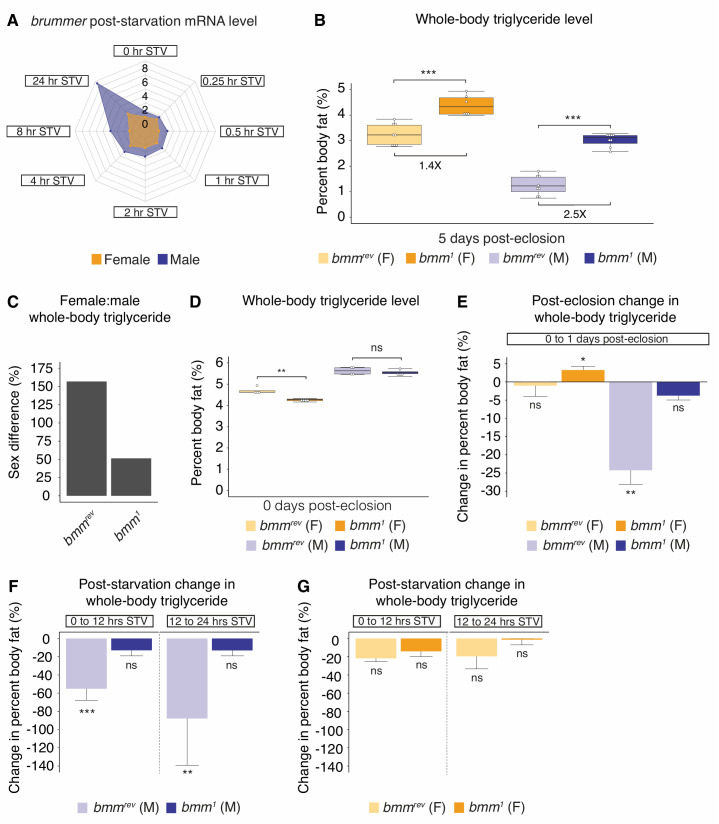
Fig 4. A role for bmm in the regulation of sex differences in triglyceride homeostasis.
(A) Radar plot showing sex-specific regulation of bmm mRNA levels STV in 5-day-old virgin w1118 females and males STV. bmm mRNA levels were 1.8-fold higher in 5-day-old fed virgin males than in age-matched virgin females (p = 0.016; Student t test). At 4 hours STV, bmm mRNA levels were 1.6-fold higher in males than females (p = 0.019; Student t test). By 8 hours STV, mRNA levels were 3.1-fold higher in males than females (p = 8.6 × 10−4; Student t test). (B) Whole-body triglyceride storage was significantly higher in 5-day-old bmm1 homozygous mutant males compared with bmmrev control males (p = 0; one-way ANOVA followed by Tukey HSD). Whole-body triglyceride storage was significantly increased in bmm1 homozygous mutant females compared with bmmrev female controls (p = 1.9 × 10−6; one-way ANOVA followed by Tukey HSD). (C) The male-biased effect of bmm loss on triglyceride storage reduced the sexual dimorphism in triglyceride storage. (D) Whole-body triglyceride storage at eclosion was not significantly different between bmm1 mutant males compared with bmmrev control males (p = 0.84; Student t test). (E) Triglyceride levels were lower in 1-day-old bmmrev males compared with newly eclosed bmmrev males (p = 0.0013; Student t test); however, there was no significant difference in whole-body triglyceride levels between 1-day-old bmm1 males and newly eclosed bmm1 males (p = 0.0793; Student t test). (F) In 5-day-old bmmrev males, whole-body triglyceride storage significantly decreased between 0 and 12 hours STV, with a further reduction between 12 and 24 hours STV (p = 6.2 × 10−5 and 2.4 × 10−3, respectively; one-way ANOVA followed by Tukey HSD test). No significant change in whole-body triglyceride levels was observed in bmm1 mutant males between 0 and 12 hours STV, or between 12 and 24 hours STV (p = 0.244 and 0.349, respectively; one-way ANOVA followed by Tukey HSD test). (G) There was no significant change in whole-body triglyceride levels in either bmmrev females or bmm1 females between 0 and 12 hours STV or between 12 and 24 hours STV (p = 0.0503 and 0.171 [0–12 hours], 0.244 and 0.998 [12–24 hours], respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements for all data presented are available in S1 and S3 Datas. bmm, brummer; COE, coefficient of error; F, female; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
bmm is an essential gene for embryogenesis [32], in which maternally provided bmm allows the survival of bmm1 mutants past this critical period in development. We identified two ways that loss of bmm may influence the sex difference in triglyceride storage: first, by increasing the amount of larval fat stored in males during development; or second, by blocking the progressive reduction in body fat over the first 5 days of adult life (Fig 1E). To distinguish between these possibilities, we measured triglyceride levels in bmm1 mutants compared with bmmrev controls at eclosion and in 1-day-old flies. We found no significant difference in whole-body triglyceride storage in newly eclosed flies between bmm1 mutants and bmmrev control males (Fig 4D), suggesting that bmm does not contribute to the sex difference in triglyceride storage by enhancing fat storage in males during larval development. Instead, the rapid decrease in triglyceride storage normally observed in bmmrev control males between 0 DPE and 1 DPE was blocked in bmm1 mutant males (Fig 4E). In further support of a role for bmm in mediating the male-specific decrease in triglyceride levels over the first few days post-eclosion, we observed an increase in bmm mRNA levels between 0- and 5-day-old flies in males but not in females (S10G Fig). We therefore propose that bmm plays a role in regulating the sex difference in triglyceride storage by promoting lipolysis in males during the first 5 days of adult life.
To determine whether bmm also plays a role in regulating the sex difference in triglyceride breakdown, we measured triglyceride breakdown post-starvation in 5-day-old bmm1 mutant virgin males and females and in bmmrev control males and females. In control bmmrev male flies, there was a significant decrease in whole-body triglyceride levels between both 0 and 12 hours post-starvation and between 12 and 24 hours post-starvation (Fig 4F), consistent with our findings in CS and w1118 virgin males. In bmm1 mutant males, however, the reduction in triglyceride levels between 0 and 12 hours post-starvation and between 12 and 24 hours post-starvation was abolished (Fig 4F), indicating that bmm promotes lipolysis post-starvation in males, as previously reported [32]. In contrast, there was no significant reduction in triglyceride levels between either 0 and 12 hours post-starvation or between 12 and 24 hours post-starvation in either bmmrev or bmm1 mutant females (Fig 4G). These male-specific effects on triglyceride breakdown were reproduced in flies with da-GAL4-mediated expression of two independent UAS-bmm-RNAi lines compared with da>+ and +>UAS-bmm-RNAi controls (S10H–S10K Fig), further supporting a role for bmm in regulating the sex difference in triglyceride breakdown post-starvation. Because of this male-specific effect of bmm loss on triglyceride breakdown, the sex difference in triglyceride breakdown was abolished. Together, these results identify novel roles for triglyceride lipase bmm in the regulation of sex differences in Drosophila triglyceride storage and breakdown.
bmm function in the somatic cells of the gonad plays a role in regulating the sexual dimorphism in whole-body triglyceride homeostasis
Given that sex differences exist in many tissues throughout the fly [45–55], we wanted to determine the anatomical focus of bmm’s effects on the male–female differences in triglyceride homeostasis. bmm is highly expressed in the fat body, and previous studies have demonstrated a central role for this tissue in the regulation of triglyceride storage and breakdown in males [29,32]. We therefore compared triglyceride storage and breakdown in virgin males and females with bmm inhibition in the fat body. We chose collagen (cg)-GAL4 and r4-GAL4 to overexpress UAS-bmm-RNAi in the fat body because these drivers have been used in many fat body studies. We confirm that both GAL4 lines drive strong green fluorescent protein (GFP) expression in the abdominal fat body and have very weak expression in the gonad (S3 Table). In line with previous studies [56], triglyceride storage in cg>UAS-bmm-RNAi males was significantly higher than in cg>+ and +>UAS-bmm-RNAi control males (S11A Fig). In females, triglyceride levels in cg>UAS-bmm-RNAi females were significantly higher than in cg>+ and +>UAS-bmm-RNAi controls (S11B Fig). Because the increase in triglyceride storage upon bmm loss in the abdominal fat body was similar in both sexes, the sex difference in triglyceride storage between cg>UAS-bmm-RNAi males and females was unchanged. Likewise, because bmm inhibition in the abdominal fat body significantly delayed triglyceride breakdown in both sexes, the sex difference in triglyceride breakdown post-starvation remained (S11C and S11D Fig). When we repeated these experiments with r4-GAL4, we observed a male-specific increase in triglyceride storage in r4>UAS-bmm-RNAi males compared with r4>+ and +>UAS-bmm-RNAi controls (S11E and S11F Fig); however, r4-GAL4-mediated loss of bmm in the fat body significantly delayed triglyceride breakdown in both sexes (S11G and S11H Fig). bmm loss in the abdominal fat body, therefore, does not fully account for the strongly male-biased effects of whole-body bmm loss on the sex differences in triglyceride storage and breakdown. Thus, despite a role for fat body bmm in maintaining triglyceride homeostasis in each sex, bmm function in additional cell types or tissues must also contribute to the sex differences in triglyceride homeostasis.
In addition to the fat body, bmm mRNA is present in the Drosophila intestine, central nervous system (CNS), muscle, neurons, glia, ovary, and testis [55]. To identify additional tissues in which bmm function is required to maintain triglyceride homeostasis, we measured triglyceride storage and breakdown in virgin females and males with RNAi-mediated inhibition of bmm in several cell types and tissues. Loss of bmm in the gut, muscles, and glia had no effect on triglyceride levels in either sex under normal culture conditions (S12A–S12F Fig). However, we identified two additional cell types in which loss of bmm function caused significant changes to whole-body triglyceride homeostasis. Using c587-GAL4, a driver with strong expression in the somatic cells of the gonad and in a limited number of neurons (S3 Table), we observed a change in whole-body triglyceride storage. Triglyceride levels in c587>UAS-bmm-RNAi males were significantly higher than in c587>+ and +>UAS-bmm-RNAi control males (Fig 5A). In addition, triglyceride breakdown was also significantly delayed in c587>UAS-bmm-RNAi males compared with c587>+ and +>UAS-bmm-RNAi control males (Fig 5B). These effects on male triglyceride storage and breakdown were specific to bmm, as we observed similar results when we used an additional RNAi line to knock down bmm function (S13A and S13B Fig), and we rescued both the increased triglyceride storage and reduced triglyceride breakdown by simultaneous overexpression of UAS-bmm-RNAi and UAS-bmm in the somatic cells of the gonad (S13C and S13D Fig). In females, there were no significant effects on either triglyceride storage or triglyceride breakdown in c587>UAS-bmm-RNAi females compared with c587>+ and +>UAS-bmm-RNAi controls for multiple RNAi lines (S14A–S14D Fig), and no significant effect of c587-GAL4-mediated coexpression of the UAS-bmm-RNAi and UAS-bmm transgenes compared with controls (S14E and S14F Fig). Because of these male-specific effects on triglyceride storage and breakdown, the sex differences in triglyceride storage and triglyceride breakdown were reduced. When we used traffic jam (tj)-GAL4, a line with expression in the somatic cells of the gonad and in a small number of neurons (S15A–S15D Fig and S3 Table), we reproduced the male-specific effects of bmm inhibition on triglyceride breakdown that we observed with c587-GAL4-mediated bmm inhibition (S15C and S15D Fig). Although we observed no change in whole-body triglyceride storage between tj>UAS-bmm-RNAi animals and controls in either sex (S15A and S15B Fig), this may reflect minor differences in the strength or timing of expression between c587-GAL4 and tj-GAL4. Because neither c587-GAL4 nor tj-GAL4 drives GFP expression in the fat body (S3 Table), these results suggest a role for bmm in the somatic cells of the gonad in regulating whole-body triglyceride storage and breakdown in males.
Fig 5. A role for bmm function in the somatic cells of the gonad in the regulation of whole-body triglyceride storage and breakdown in males.
(A) Whole-body triglyceride storage in males overexpressing the UAS-bmm-RNAi transgene in the somatic cells of the male gonad (c587>UAS-bmm-RNAi) was significantly higher than in control males (c587>+ and +>UAS-bmm-RNAi) (p = 0.027 and 2 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride levels in c587>+ and +>UAS-bmm-RNAi control males showed a significant decrease between 0 and 12 hours STV (1 × 10−7 and 1.1 × 10−6, respectively; one-way ANOVA followed by Tukey HSD test), whereas triglyceride levels were not significantly different between 0 and 12 hours STV in c587>UAS-bmm-RNAi males (p = 0.997; one-way ANOVA followed by Tukey HSD test). (C–H) We detected lipid droplets in testes dissected from 0-day-old bmm1 and bmmrev virgin male flies using BODIPY, a neutral lipid stain. Dissected testis from 0-day-old virgin bmm1 mutant males (F–H) show a dramatic increase in lipid droplets compared with control bmmrev males (C–E). (I–K) Using an LD-GFP, we found that a subset of the LipidTOX-positive lipid droplets in the testis (arrowheads) represent droplets in the somatic cells of the gonad. Non-GFP-positive droplets (arrow) likely represent lipid droplets in the germline, another cell type in the testis. Scale bars = 50 μm, except for inset images for (I–K), in which scale bars = 12.5 μm. The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying data presented in the figure are available in S1 Data. Original image files corresponding to all images acquired from genotype-matched individuals presented in panels C–K are available upon request. bmm, brummer; BODIPY, boron-dipyrromethene; COE, coefficient of error; GFP, green fluorescent protein; HSD, honest significant difference; LD-GFP, lipid droplet–targeted GFP; M, male; ns indicates no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence.
Although the somatic cells of the gonad have not previously been shown to be an important site for triglyceride storage, high-throughput data sets have detected bmm mRNA in the Drosophila male testis [55], and bmm’s mammalian homolog adipose triglyceride lipase (ATGL) is present in the murine testis [57,58]. In addition, we show that lipid droplets, a cytoplasmic organelle in which triglycerides are stored and bmm protein localizes [32], are present in the Drosophila testis (Fig 5C–5E). To determine whether loss of bmm affects triglyceride homeostasis in this tissue, we examined lipid droplets in the testis and found a clear increase in the number of lipid droplets in testes isolated from bmm1 males (Fig 5F–5H) compared with control bmmrev males (Fig 5C–5E). The Drosophila testis contains two cell types: the germline cells and the somatic support cells [59]. To determine which cell type contains the lipid droplets, we first took high-magnification images of lipid droplets in the testis of males in which the somatic cells express membrane-bound GFP (tj>UAS-mCD8::GFP). Given that some of the lipid droplets are present within the GFP boundary of the cell (arrowheads), this suggests that lipid droplets are present in somatic cells (S15E–S15G Fig). To further confirm the presence of lipid droplets in somatic cells, we overexpressed a transgene encoding a lipid droplet–targeted GFP (LD-GFP) in the somatic cells of the gonad [60]. Previous work identified the lipid droplet–targeting domain in transport regulator protein Klarsicht (Klar; FBgn0001316) [60]. When this Klar lipid droplet–targeting domain was fused to GFP (LD-GFP), the LD-GFP fusion protein localized to the surface of lipid droplets [60]. Thus, by expressing UAS-LD-GFP in a cell type of interest, lipid droplets in that cell type will be positively marked with LD-GFP [60]. In testes dissected from c587>UAS-LD-GFP males, we detected several GFP-positive punctae that were also positive for LipidTOX Red, a neutral lipid dye that marks lipid droplets (Fig 5I–5K). Because c587-GAL4 does not drive transgene expression in the germline [61], the presence of punctae that are both GFP and LipidTOX Red positive in the testis confirms that lipid droplets are present in the somatic cells of the Drosophila gonad. Together, our results identify a new role for bmm in regulating lipid droplets in the Drosophila testis and suggest a role for bmm in the somatic cells of the male gonad in the regulation of whole-body triglyceride storage and breakdown.
A role for bmm function in neurons in the regulation of sex differences in triglyceride breakdown
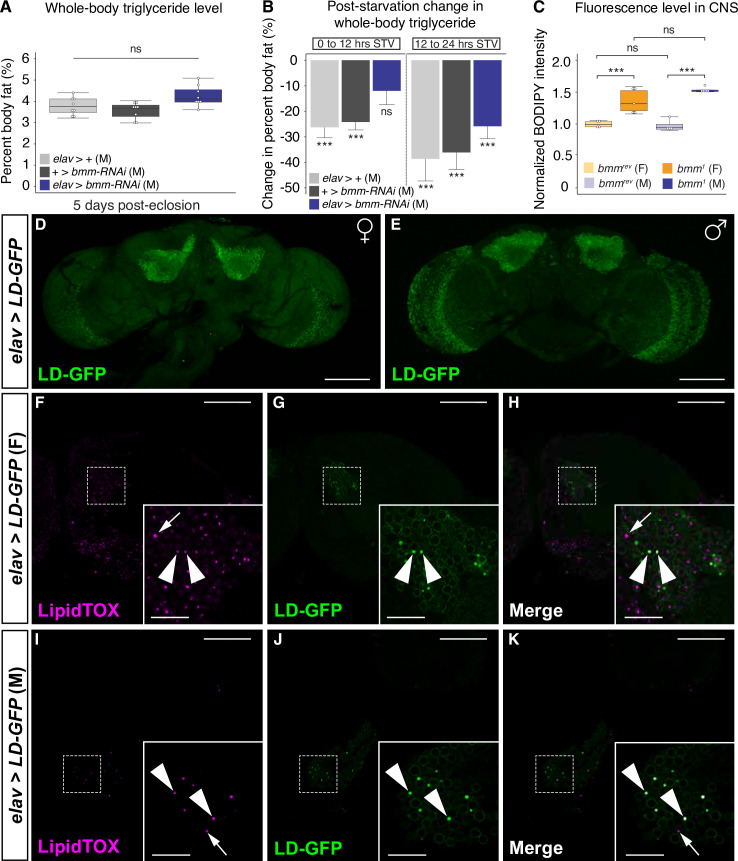
In addition to the somatic cells of the gonad, we found a role for bmm function in neurons in regulating the sex difference in triglyceride breakdown. We used embryonic lethal abnormal vision (elav)-GAL4 to overexpress the UAS-bmm-RNAi transgene in postmitotic neurons. Under normal culture conditions, we saw no significant increase in whole-body triglyceride storage in elav>UAS-bmm-RNAi males compared with elav>+ and +>UAS-bmm-RNAi control males (Fig 6A), a finding we confirmed using an independent UAS-bmm-RNAi line (S16A Fig) and an independent GAL4 line for neurons (neuronal Synaptobrevin [nSyb]-GAL4) (S16B Fig). When we measured triglyceride levels post-starvation, however, we observed a significant delay in triglyceride breakdown. In elav>+ and +>UAS-bmm-RNAi control males, triglyceride levels were significantly lower by 12 hours post-starvation compared with genotype-matched, fed control males (Fig 6B). In contrast, there was no significant reduction in whole-body triglyceride levels between 0 and 12 hours post-starvation in elav>UAS-bmm-RNAi males (Fig 6B), a finding we confirmed using an independent UAS-bmm-RNAi line (S16C Fig) and an independent neuronal GAL4 line (nSyb-GAL4) (S16D Fig). Moreover, we show that bmm transcript levels are nearly undetectable in dissected brains from elav-GAL4>UAS-bmm-RNAi males compared with control males (S16E Fig) and that simultaneous overexpression of UAS-bmm together with UAS-bmm-RNAi in neurons rescued the delay in triglyceride breakdown between 0 and 12 hours post-starvation (S16F and S16G Fig). This delay in triglyceride breakdown was largely restricted to early time points post-starvation because the decrease in triglyceride levels in elav>UAS-bmm-RNAi males between 12 and 24 hours post-starvation was in line with the reduction we observed in control males during this interval (Fig 6B). Moreover, the delay in triglyceride breakdown was specific to neurons, as triglyceride breakdown in males with loss of bmm in glia was indistinguishable from control males (S17A and S17B Fig). Together, these data provide strong evidence of a role for bmm in male neurons in regulating triglyceride breakdown. In females, neither triglyceride storage nor triglyceride breakdown was significantly different between elav>UAS-bmm-RNAi females and elav>+ and +>UAS-bmm-RNAi control females for either of our RNAi lines (S18A–S18D Fig), between nSyb>UAS-bmm-RNAi females and controls (S18E and S18F Fig), or between elav>UAS-bmm;UAS-bmm-RNAi females and controls (S18G and S18H Fig). Overall, these male-specific effects reduced the sex difference in triglyceride breakdown, identifying a novel role for bmm in male neurons in the regulation of lipolysis post-starvation.
Fig 6. A role for bmm function in neurons in the regulation of whole-body triglyceride breakdown in males.
(A) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in postmitotic neurons (elav>UAS-bmm-RNAi) was not significantly different from age-matched control males (elav>+ and +>UAS-bmm-RNAi) (p = 0.095 and 0.011; one-way ANOVA followed by Tukey HSD test). (B) There was a significant reduction in whole-body triglyceride levels in 5-day-old elav>+ and +>UAS-bmm-RNAi control males between 0 and 12 hours STV (p = 1 × 10−5 and 9 × 10−6, respectively; one-way ANOVA followed by Tukey HSD test); however, no significant decrease in triglyceride levels was observed between 0 and 12 hours STV in elav>UAS-bmm-RNAi males (p = 0.13; one-way ANOVA followed by Tukey HSD test). (C) In both sexes, lipid droplet–derived fluorescence in dissected Drosophila brains was significantly higher in 5-day-old bmm1 mutants compared with bmmrev controls (p = 2.5 × 10−5 and 0.002 in males and females, respectively; one-way ANOVA followed by Tukey HSD). (D, E) Expression of an LD-GFP transgene in neurons revealed GFP-positive punctae throughout the Drosophila CNS in females (D) and males (E). Maximum Z-projections, dorsal view, anterior up. Scale bars = 100 μm. (F–K) Expression of LD-GFP in neurons revealed that a subset of the LipidTOX-positive lipid droplets found in the CNS of 5-day-old adult females (F–H) and males (I–K) represent droplets in neurons (arrowheads). (F–K) Non-GFP-positive droplets likely represent lipid droplets in glia, another cell type in the CNS (arrow). White boxes indicate area magnified in inset. Single confocal slice through the Drosophila brain, dorsal view, anterior up. Scale bars = 50 μm; scale bars = 12.5 μm in magnified inset images. The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat or BODIPY intensity represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements for all data are available in S1 Data. Original image files corresponding to all images acquired from genotype-matched individuals presented in panels D–K are available upon request. bmm, brummer; BODIPY, boron-dipyrromethene; CNS, central nervous system; COE, coefficient of error; elav, embryonic lethal abnormal vision; F, female; GFP, green fluorescent protein; HSD, honest significant difference; LD-GFP, lipid droplet–targeted GFP; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence.
Although the Drosophila CNS is not a major site for triglyceride storage, lipids play an essential role in neuronal and glial function [62–64]. Furthermore, a recent single-cell RNA-sequencing analysis of the Drosophila brain confirms that bmm mRNA is present within both neurons and glia [65], and previous reports identified lipid droplets in the retina of adult flies [66]. To determine whether bmm contributes to the regulation of lipid droplets in this tissue, we measured the intensity of lipid droplet–derived fluorescence in the CNS of bmm1 mutants compared with bmmrev controls. We found a significant increase in fluorescence in bmm1 mutant males and females compared with bmmrev controls (Fig 6C), suggesting that bmm function normally limits neutral lipid accumulation in this tissue. Because the CNS is composed of multiple cell types that are not easily distinguished based on morphological characteristics, we used elav-GAL4 to drive LD-GFP expression exclusively in neurons to determine whether lipid droplets are present in this cell type [60]. We found punctae that were both GFP and LipidTOX Red positive throughout the CNS in both males and females (Fig 6D–6K). Because the LD-GFP protein is present only in neurons, these GFP-positive punctae represent lipid droplets in neurons, confirming that lipid droplets are normally present in adult Drosophila neurons in both sexes. Thus, lipid droplets are present in neurons under normal physiological conditions, and bmm function in neurons plays a previously unrecognized role in stimulating whole-body triglyceride breakdown post-starvation in males.
Loss of bmm affects life span and the sex difference in starvation resistance
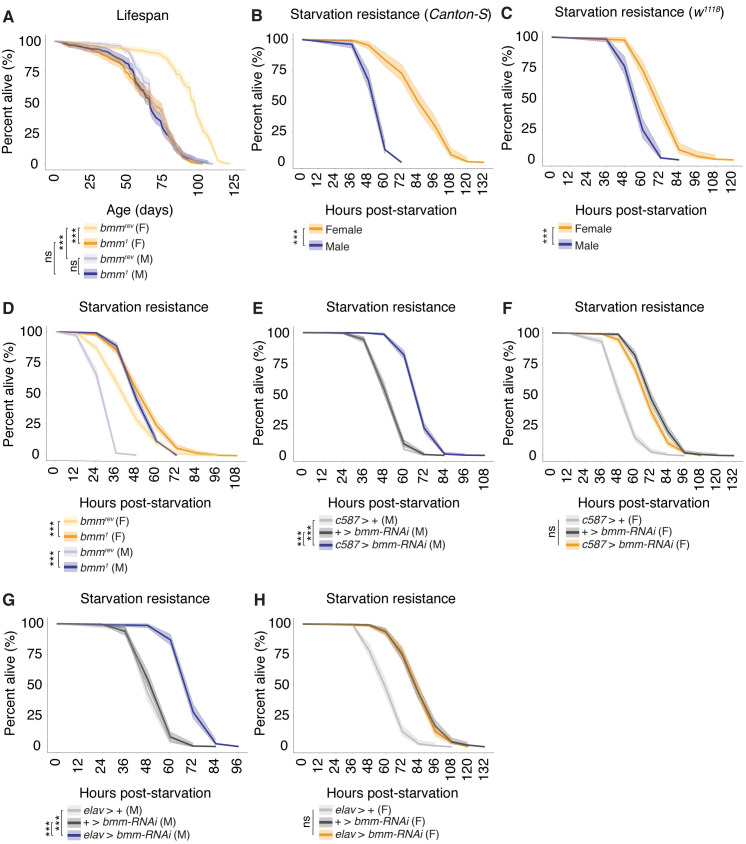
Previous studies have shown that the correct regulation of triglyceride homeostasis is important for a normal life span [32]. For example, life span was significantly reduced in bmm1 mutant males compared with bmmrev control males [32]. In light of our findings that loss of bmm has male-biased effects on triglyceride homeostasis, we wanted to examine life span in both sexes because previous studies used only male flies [32]. In bmmrev control females, median life span was 96 days, whereas in bmm1 mutant females, the median life span was only 68 days, a reduction of 29% (Fig 7A). This significant reduction in female life span was unexpected given the relatively modest increase in whole-body triglyceride level in bmm1 mutant females (Fig 4B). In bmm1 mutant males, we observed no significant reduction in life span in males (Fig 7A) despite a 2.5× increase in triglyceride storage (Fig 4B). Although this finding differs from the previously reported 10% reduction in life span in male bmm1 mutants [32], a difference likely due to minor interlaboratory variations in aging regime or diet [67], our study identifies an unexpected female-biased reduction in life span upon loss of bmm.
Fig 7. bmm-mediated regulation of triglyceride homeostasis affects life span and contributes to the sex difference in starvation resistance.
(A) Median life span was significantly higher in bmmrev virgin females than in bmmrev virgin males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons; n > 297 for all sexes and genotypes). Median life span was significantly reduced in bmm1 mutant females compared with bmmrev control females (28-day reduction in survival, p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons). No significant decrease was found in bmm1 mutant males compared with control males (p = 0.17; Log-rank test with Bonferroni correction for multiple comparisons). (B) Median survival post-starvation was significantly higher in 5-day-old virgin Canton-S females than in virgin Canton-S males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparison; n > 154). (C) Median survival post-starvation was significantly higher in 5-day-old w1118 virgin females compared with w1118 virgin males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons; n > 123). (D) Median survival post-starvation was significantly higher in 5-day-old bmmrev virgin females than in bmmrev virgin males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons; n > 454 for both sexes and genotypes). Median survival post-starvation was significantly increased in male bmm1 mutants compared with bmmrev control males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons) and in bmm1 mutant females compared with bmmrev controls (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparisons). The male-biased effects of bmm loss on starvation resistance reduced the sex difference in median survival. (E) Median survival was significantly higher in virgin males with bmm inhibition in somatic cells of the male gonad (c587>UAS-bmm-RNAi) compared with control males (c587>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 326 for all genotypes). (F) Median survival post-starvation was not significantly different in c587>UAS-bmm-RNAi virgin females compared with c587>+ and +>UAS-bmm-RNAi control females (p = 2 × 10−16 and 1.5 × 10−5, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 408 for all genotypes). (G) Median survival post-starvation was significantly higher in virgin males with bmm inhibition in postmitotic neurons (elav>UAS-bmm-RNAi) compared with elav>+ and +>UAS-bmm-RNAi control males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 178 for all genotypes). (H) Median survival post-starvation was not significantly higher in elav>UAS-bmm-RNAi virgin females compared with elav>+ and +>UAS-bmm-RNAi control females (p = 2 × 10−16 and 1, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 253 for all genotypes). The male-specific effects of bmm inhibition in neurons reduced the sex difference in median survival. The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements corresponding to all data presented in the figure are available in S4 Data. bmm, brummer; elav, embryonic lethal abnormal vision; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence; w, white.
Another phenotype that is closely associated with the regulation of triglyceride homeostasis is starvation resistance [29–32,56,68,69]. In line with previous studies showing increased starvation resistance in mated females compared with males [38,70], we demonstrate that starvation resistance is significantly higher in 5-day-old CS virgin females compared with virgin males (Fig 7B). Similar results were obtained with w1118 (Fig 7C), Oregon-R (S19A Fig), Country Mill Winery (CMW) flies (S19B Fig), and two isofemale lines: Mel c2.2 and Mel c2.3 (S19C and S19D Fig). Thus, the sexual dimorphism in starvation resistance persists in multiple genetic backgrounds. To determine whether the bmm-mediated regulation of triglyceride homeostasis contributes to the sex difference in starvation resistance, we examined starvation resistance in bmm1 mutants and bmmrev controls. In line with previous studies in males, survival post-starvation was significantly longer in bmm1 mutants compared with bmmrev controls (Fig 7D). Although starvation resistance was significantly higher in bmm1 mutant females compared with control females (Fig 7D), the strongly male-biased increase in starvation resistance largely eliminated the sex difference (Fig 7D), an effect we reproduced in males and females with da-GAL4-mediated global overexpression of two independent UAS-bmm-RNAi transgenes (S19E–S19H Fig). Given that bmm is a critical effector of the sex differences in triglyceride homeostasis, these data suggest that the sex-specific control of triglyceride homeostasis by bmm makes a key contribution to the sexual dimorphism in starvation resistance. Given that the enhanced starvation resistance in bmm1 mutant males represents a significant benefit to survival for contexts in which food is scarce, we next asked whether there were any disadvantages caused by bmm loss in males. Because the correct regulation of triglyceride homeostasis is essential for female fertility [23], we compared the number of offspring produced by bmm1 mutant males and bmmrev controls. We found that bmm1 mutant males produced significantly fewer offspring compared with bmmrev control males (S20A Fig): after being left with three females for 6 days, only five of 25 bmm1 mutant males produced progeny (S20A Fig). Thus, despite significant benefits in survival time after nutrient deprivation, loss of bmm significantly impairs normal male fertility, demonstrating a previously unrecognized role for bmm function in male physiology.
Because bmm function in several cell types and tissues plays a role in regulating sexual dimorphism in triglyceride homeostasis, we measured starvation resistance in male and female flies with cell- and tissue-specific bmm inhibition. In line with our finding that bmm inhibition in the abdominal fat body increased triglyceride storage and decreased triglyceride breakdown in both sexes, survival post-starvation in cg>UAS-bmm-RNAi males and females was significantly longer than cg>+ and +>UAS-bmm-RNAi control flies (S21A and S21B Fig). Given that survival post-starvation was increased similarly in both sexes, the male–female difference in starvation resistance remained (S21A and S21B Fig), a finding we confirm using an additional GAL4 driver, r4-GAL4 (S21C and S21D Fig). Therefore, bmm function in the abdominal fat body does not fully explain the sex difference in starvation resistance. In contrast, when we examined starvation resistance in c587>UAS-bmm-RNAi flies, we found that median survival post-starvation was significantly longer in c587>UAS-bmm-RNAi males (Fig 7E), but not females (Fig 7F), compared with c587>+ and +>UAS-bmm-RNAi control flies. Importantly, we confirm that these male-specific effects on starvation resistance are specific to bmm using an additional UAS-bmm-RNAi line (S22A and S22B Fig) and by rescuing the male-specific increase in starvation resistance by c587-GAL4-mediated coexpression of UAS-bmm and UAS-bmm-RNAi (S22C and S22D Fig). Moreover, we observed male-biased effects on starvation resistance in tj>UAS-bmm-RNAi flies (S22E and S22F Fig). Because of these male-biased or male-specific effects, the sex difference in starvation resistance was reduced. Similarly, when we compared survival post-starvation in animals with neuron-specific bmm inhibition, we found that starvation resistance was significantly increased in elav>UAS-bmm-RNAi males (Fig 7G), but not females (Fig 7H), compared with control elav>+ and +>UAS-bmm-RNAi control flies. This effect on starvation resistance was reproduced in flies with elav-GAL4-mediated expression of an additional UAS-bmm-RNAi line (S23A and S23B Fig) and was abolished when we used elav-GAL4 to coexpress UAS-bmm and UAS-bmm-RNAi in neurons (S23C and S23D Fig). Furthermore, we found that nSyb-GAL4-mediated bmm inhibition in neurons strongly extended starvation resistance (S23E and S23F Fig), providing strong support for neuronal bmm as a regulator of starvation resistance. Because of these male-specific effects, the sex difference in survival post-starvation was reduced. Inhibition of bmm in other tissues had either a very modest effect or no effect on starvation resistance in either sex (S24A–S24F Fig). Together, these findings reveal a previously unrecognized difference between males and females in the physiological mechanisms that govern starvation resistance.
Discussion
In this study, we used the fruit fly, Drosophila melanogaster, as a model to gain insight into the genetic and physiological mechanisms underlying sex differences in triglyceride homeostasis. We describe sexual dimorphisms in triglyceride storage and breakdown and demonstrate extensive sex-biased regulation of many genes involved in maintaining whole-body triglyceride levels. One important outcome from our study was the identification of a role for triglyceride lipase bmm in the regulation of sex differences in triglyceride homeostasis: loss of bmm largely eliminated the sex difference in triglyceride storage and abolished the sex difference in triglyceride breakdown. This represents a previously unrecognized role for bmm in regulating sexual dimorphism in triglyceride storage and breakdown. Another important finding was that bmm function in the somatic cells of the gonad and in neurons plays a role in regulating sex differences in triglyceride homeostasis. In females, bmm function in the abdominal fat body largely explains its regulation of whole-body triglyceride homeostasis. In contrast, in males, bmm acts in the fat body, the somatic cells of the gonad, and in neurons to regulate whole-body triglyceride storage and breakdown. Although we did not confirm whether the requirement for bmm function in the somatic cells of the gonad and in neurons affected the development of these important cell types, these findings reveal a previously unappreciated sex difference in the physiological mechanisms that govern the regulation of whole-body triglyceride levels. Moreover, we confirm that lipid droplets are present in two cell types in which knowledge of lipid droplet function is limited. Together with our data on how changes to triglyceride homeostasis affect starvation resistance and life span in each sex, our study highlights how including both sexes can accelerate the discovery of new insights into the regulation of whole-body physiology.
Our study identified many genes with sex-biased expression; however, our detailed analysis of one gene, bmm, the Drosophila homolog of mammalian ATGL [32,57,71], identified a previously unrecognized role for this gene in regulating sexual dimorphism in triglyceride homeostasis. bmm is a lipase that influences whole-body fat storage and breakdown in flies and other animals, and we found that under both normal culture conditions and post-starvation, males have higher levels of bmm mRNA than females. Yet what factors are responsible for this sex-specific bmm regulation? One possible explanation is a sex difference in food intake, as previous studies have shown that mated female flies consume more food than males [37,72]. Although our experiments use virgin males and females, a sex difference in food intake could trigger increased triglyceride storage in females by enhancing the activity of a nutrient-activated pathway, such as the insulin/insulin-like growth factor signaling (IIS) pathway [73–75]. In support of a possible role for food intake and IIS in establishing the sex difference in triglyceride storage via regulation of bmm, previous studies have shown that bmm mRNA levels are positively regulated by forkhead box O (foxo; FBgn0038197) [76,77], a transcription factor that is normally repressed by nutrient input and IIS activity [78–80]. Thus, in females, early food intake after eclosion may activate IIS pathway activity to inhibit Foxo, reducing bmm mRNA levels to promote triglyceride storage. In males, lower food intake would lead to less IIS signaling, increased Foxo activity, and higher levels of bmm mRNA to limit triglyceride storage. Indeed, a recent study in late third instar Drosophila larvae demonstrated increased IIS activity in females compared with males [81], and males and females show significant sex differences in gene expression in response to global IIS perturbation [82]. Future studies will therefore be important to confirm whether the sex-specific regulation of bmm mRNA under normal culture conditions and post-starvation occurs via IIS and Foxo. Furthermore, it will be important to test whether additional nutrient-responsive pathways contribute to the sex-specific regulation of bmm and the male–female difference in triglyceride storage, such as the adipokinetic hormone (AKH; FBgn0004552) pathway [68,83,84], the sterol response element binding protein (SREBP; FBgn0261283) pathway [85,86], and spargel/peroxisome proliferator–activated receptor γ coactivator 1 (srl/PGC-1; FBgn0037248) pathway [87,88], as much of our knowledge of these pathways is derived from studies using either a mixed-sex population of larvae or adult male flies.
Another possible explanation for the sex differences in triglyceride homeostasis is that sex determination genes directly establish a “male” or a “female” metabolic state via regulation of triglyceride metabolism genes such as bmm. In support of a role for sex determination genes in metabolic regulation, previous studies have shown that at least 15 triglyceride metabolism genes are putative targets of doublesex (dsx; FBgn0000504) and fruitless (fru; FBgn0004652) [49,89], two genes that direct many [90,91], but not all [50,81,92], aspects of sexual development and behavior. Indeed, one study showed that the activity of fru-expressing neurons normally represses whole-body triglyceride levels in male flies [93]. Thus, fru and/or dsx may both contribute to the sex-specific regulation of triglyceride metabolism genes, a possibility that will be easily tested in future studies given the availability of viable stocks carrying isoform-specific mutations in fru and dsx [89,91,94–100]. In addition to dsx and fru, it will be important to test whether other regulators of sexual development, such as the steroid hormone ecdysone, contribute to the sex-specific regulation of triglyceride homeostasis. Previous studies have shown that changes to ecdysone signaling affect sexual development [101], and a recent study demonstrated an important role for the ecdysone receptor (EcR; FBgn0000546) in establishing the increased triglyceride storage observed in mated females [37]. Given that ecdysone levels are higher in females than in males [102,103] and the known role of steroid hormones in mammals in creating the sex difference in fat storage [7], this represents an important area for future investigations into the sexual dimorphism in triglyceride homeostasis.
A second key finding from our study was the identification of strongly sex-biased effects of whole-body bmm deficiency on triglyceride homeostasis. Although we confirm findings from previous studies that loss of bmm dramatically increases triglyceride storage and reduces triglyceride breakdown in male flies [29,32,68], we also show that loss of bmm had only modest effects on triglyceride storage and no effect on triglyceride breakdown in female flies. As a result of these strongly male-biased effects, the sex difference in triglyceride storage was largely eliminated, and the sex difference in triglyceride breakdown was abolished in animals with whole-body bmm deficiency. This reveals a previously unrecognized role for bmm in regulating sex differences in triglyceride homeostasis. In the future, it will be important to determine how other genes with strong sex-specific regulation contribute to male–female differences in triglyceride storage and breakdown. For example, mRNA levels of lsd-1/PLIN1 were 4-fold higher in 5-day-old males compared with age-matched females. Because loss of lsd-1/PLIN1 function in males leads to a significant increase in triglyceride storage compared with control males [29], it will be interesting to determine how loss of lsd-1/PLIN1 affects triglyceride homeostasis in females. Another gene with strongly sex-biased expression was hsl, as hsl mRNA levels were approximately 3-fold higher in 5-day-old virgin females than in males. In a mixed-sex population of larvae, loss of hsl significantly increases lipid droplet size and whole-body triglyceride storage [30]; yet the adult phenotype of hsl mutants remains unclear. Future studies will be important to determine how this highly conserved lipase affects triglyceride storage and breakdown in each sex. Moreover, because our data show that loss of bmm in additional cell types, such as the somatic cells of the gonad and neurons, influences triglyceride storage and breakdown, future studies on lsd-1/PLIN1, hsl, and other genes will need to define whether there is a general requirement for triglyceride metabolism genes in these important cell types in the control of whole-body triglyceride homeostasis.
In most animals, the majority of whole-body triglycerides are contained within specialized organs dedicated to fat storage, such as the mammalian adipose tissue and insect fat body [3,104]. Interestingly, we show that some of the male-biased effects of bmm on triglyceride homeostasis were due to cell types in addition to the fat body, where bmm function has been well-described. Although we cannot rule out a role for the Drosophila head fat body in mediating some of bmm’s sex-specific effects on triglyceride storage and breakdown, as our fat body drivers were specific to the abdominal fat body (S3 Table), we found that bmm function in the somatic cells of the male gonad and in male neurons explained at least some of the male-biased effects of whole-body bmm loss on triglyceride homeostasis. Given the growing recognition that lipid droplets and triglyceride homeostasis in many cell types and tissues contribute to the normal regulation of whole-body development and physiology [25,66,105–109], our study highlights the importance of exploring how the control of triglyceride metabolism in one cell type impacts whole-body triglyceride homeostasis. For example, how does bmm function in the somatic cells of the gonad, a cell type not known to be a major site of triglyceride storage, affect whole-body triglyceride storage and breakdown? In the mammalian gonad, lipid droplets have been detected in both the Leydig and Sertoli cells [110–113]. An important function of the mammalian testis lipid droplets is the storage of cholesterol ester, which can be broken down to release free cholesterol for biosynthesis of the steroid hormone testosterone [114]. In flies, although the precise lipid composition of the testis lipid droplets is unknown, the steroid hormone ecdysone has been detected in the testis [102,115]. Thus, if lipid droplets in the Drosophila testis contribute to steroid hormone production in males, the ectopic lipid droplets in animals lacking bmm in the somatic cells of the gonad may alter neutral lipid metabolism and affect ecdysone production in male flies. In support of a model in which ectopic ecdysone production influences whole-body triglyceride storage, a previous study showed that ecdysone feeding in adult males was sufficient to enhance fat storage [37]. In the future, it will be important to directly test this model by examining changes to ecdysone titers and ecdysone signaling in males lacking bmm in the somatic cells of the gonad. Moreover, given that previous studies have shown important effects of the germline and gonad on whole-body gene expression [103,116] and phenotypes such as aging and immunity [117–120], it will be interesting to examine other aspects of development, physiology, and life span in males lacking bmm function in the gonad. Ultimately, a better mechanistic understanding of how changes to bmm function in the somatic cells of the gonad affects whole-body fat storage and breakdown will provide insight into how bmm function in diverse cell types might impact other aspects of development and physiology and suggest lines of inquiry for studies on ATGL in other models.
In addition to the somatic cells of the gonad, we also identified an important role for bmm function in Drosophila neurons in the regulation of triglyceride breakdown. In many animals, correct regulation of lipid metabolism in neurons is important for membrane synthesis and remodeling and in mediating signaling events within the neuron [62–64]. Although previous studies have detected lipid droplets in cultured neurons and brain sections [121–124] in Drosophila larval motor neuron axons [125] and have shown that neuronal dysfunction is associated with abnormal lipid droplet accumulation [126–128], more studies are needed to improve knowledge of the normal physiological roles of lipid droplets in neurons [127–129]. For example, to understand how bmm function in neurons promotes whole-body triglyceride breakdown, it will be important to identify which subsets of neurons require bmm function to control triglyceride breakdown. One obvious group of neurons are the AKH-producing cells in Drosophila [83,84]. Under normal culture conditions, ablation of AKH-producing neurons and loss of AKH peptide have no effect on development [83,84,130–132]; however, AKH neurons and AKH receptor–mediated signaling are required for starvation-mediated triglyceride breakdown [68,131]. This fits with our data that bmm function in neurons only affects triglyceride breakdown and suggests a model in which a male-specific increase in bmm within AKH neurons post-starvation triggers AKH secretion and rapid lipolysis. In the future, more studies will be needed to confirm whether AKH secretion is responsible for the sex difference in whole-body triglyceride depletion post-starvation. Moreover, because manipulation of insulin-producing cells (IPCs) in the CNS [133], fru-expressing neurons in the mushroom bodies [93,134], octopamine- and tyramine-producing neurons [135,136], Taotie neurons [137], short neuropeptide F (sNPF)-producing neurons [138], and central clock pigment dispersing factor (PDF)-positive neurons [139] affects whole-body triglyceride levels, future studies will need to examine whether bmm function in any of these neurons contributes to the sex difference in triglyceride breakdown.
Once the neuroanatomical focus of bmm’s effects on triglyceride breakdown has been identified, it will be important to investigate how loss of bmm affects neuronal development and/or function. For example, whole-body deficiency for bmm’s murine homolog ATGL causes profound changes to the composition of triglyceride-associated fatty acids in the mouse brain [140]. Loss of bmm in neurons may therefore affect fatty acid–mediated signaling in the Drosophila brain. Although the changes to fatty acid composition that accompany bmm loss have not been characterized in Drosophila, one study showed that the Drosophila homolog of nuclear hormone receptor hepatocyte nuclear factor 4 (HNF4) is positively regulated by bmm activity in the larval fat body [141]. Thus, HNF4 is a promising candidate to mediate the effects of bmm loss on neuronal development and/or function. Another possibility includes changes to the activity of SREBP because SREBP processing is normally blocked by the fatty acid palmitate in Drosophila [85]. Moreover, given that several studies have identified clear links between lipid droplets and reactive oxygen species (ROS) signaling in the Drosophila brain [66,108,109], bmm-mediated changes to ROS signaling in neurons will also need to be examined during starvation in both sexes. Overall, although the mechanism underlying the effect of bmm function in neurons on whole-body triglyceride breakdown is unknown, our identification of lipid droplets in neurons under normal culture conditions suggests Drosophila is a useful model to examine how lipid droplets contribute to the normal function of this important cell type.
In conclusion, our studies identify a role for triglyceride lipase bmm in the regulation of sex differences in Drosophila triglyceride storage and breakdown. We show that bmm function is required in the somatic cells of the gonad and in neurons to maintain whole-body triglyceride homeostasis, provide the first evidence that bmm is sex-specifically regulated, and demonstrate the role of this regulation to the male–female difference in triglyceride homeostasis and starvation resistance. Given that the correct regulation of triglyceride homeostasis has also been linked with the regulation of sleep, fertility, reproduction, and feeding [23,36,37,43,72,142–148], our studies raise the possibility that the male–female differences previously noted in at least some of these complex traits may be associated with the sex difference in Drosophila triglyceride homeostasis. Looking beyond Drosophila, it will be interesting to determine whether bmm also contributes to sex differences in triglyceride storage and breakdown in other animals because bmm homologs are found in many species [32,57,58,71]. Furthermore, given the sex-biased risk of developing diseases associated with abnormal triglyceride metabolism (e.g., cardiovascular disease, nonalcoholic fatty liver disease) [149–151], future studies on the cell- and tissue-specific regulation of triglyceride metabolism in both sexes will be essential to provide insight into the mechanisms that contribute to disease onset and progression in each sex.
Materials and methods
Fly husbandry
Fly stocks were reared at 25°C in a 12:12 light:dark cycle. All transgenic flies were backcrossed into a w1118 genetic background for a minimum of five generations. For all experiments, larvae were raised at a density of 50 larvae per 10 ml of yeast–sugar–cornmeal media [152] (see recipe in S1 Materials and Methods). Pupae were sexed either by gonad size or by the presence of sex combs and placed onto filter paper to complete pupal development. Flies eclosed into single-sex vials at a density of 20 animals per vial, and adult weight was measured by weighing groups of 10 flies in preweighed, 1.5-ml microcentrifuge tubes. Unless otherwise indicated, all assays were performed on 5- to 7-day-old adult flies. For metabolic assays, five flies were collected immediately prior to starvation and afterward at 12-hour intervals post-starvation. Flies were snap frozen on dry ice and stored at –80°C. Each biological replicate represents five flies collected into a 1.5-ml microcentrifuge tube, and each experiment includes four biological replicates for each sex and genotype. All experiments were repeated a minimum of two times for a total number of eight biological replicates per sex and per genotype.
Fly strains
The following fly strains from the Bloomington Drosophila Stock Center were used in this study: CS (#64349), w1118 (#3605), Oregon-R, y1,v1;UAS-bmm-RNAi (#25926), y1,v1;attP40 (#36304), w1118;UAS-nGFP (#4775); UAS-mCD8::GFP (#5130). The following fly strains from the Vienna Drosophila Resource Center were used in this study: UAS-bmm-RNAi (#37880), UAS-bmm-RNAi (#37877). We obtained the bmm1 mutants and bmmrev control strain as a kind gift from R. Kühnlein [32]; CMW flies, Mel c2.2, and Mel c2.3 (wild-caught isofemale lines) as a kind gift from I. Dworkin; and UAS-LD-GFP flies from M. Welte. We used the following stocks for tissue-specific overexpression of the UAS-bmm-RNAi transgene: da-GAL4 (ubiquitous), cg-GAL4 (abdominal fat body), r4-GAL4 (abdominal fat body) elav-GAL4 (postmitotic neurons), nSyb-GAL4 (neurons), repo-GAL4 (glia), Mex-GAL4 (intestinal enterocytes), c587-GAL4 (somatic cells of the gonad), tj-GAL4 (somatic cells of the gonad), and dMef2-GAL4 (muscle cells).
Starvation resistance
The 5- to 7-day-old virgin males and females raised on normal media were transferred to vials containing 2 ml of starvation media (0.7% agar in 1× PBS). Dead flies were counted at 12-hour intervals post-starvation. Each experiment used >200 flies per sex and per genotype, and was performed at least twice (total n>400 flies per sex and per genotype).
Male fertility assay
The 5-day-old virgin males were paired with three 5-day-old virgin w1118 females. On alternate days after mating, the group of four flies was transferred to fresh food vials. The old vials were kept, and male fertility was quantified by counting the number of pupae.
Triglyceride assay
Flies were collected as described above and homogenized using 100 μl of glass beads (Sigma 11079110) in 200 μl of buffer (0.1% Tween in 1× PBS) at 8.0 m/s for 5 seconds (OMNI International Bead Ruptor 24). Triglyceride concentration was measured using a coupled colorimetric assay as previously described [153]. For a detailed description of methods, refer to S1 Materials and Methods.
Protein assay
Flies were collected as described above and homogenized using 100 μl of glass beads (Sigma 11079110) in 500 μl of 1× PBS at 8.0 m/s for 5 seconds (OMNI International Bead Ruptor 24). Protein concentration was measured as previously described [154]. For a detailed description of methods, refer to S1 Materials and Methods.
Glucose and glycogen assay
Flies were collected as described above and homogenized using 100 μl of glass beads (Sigma 11079110) in 500 μl of 1× PBS at 8.0 m/s for 5 seconds (OMNI International Bead Ruptor 24). Glucose or glycogen concentration was measured as previously described [153]. For a detailed description of methods, refer to S1 Materials and Methods.
RNA extraction and cDNA synthesis
Each biological replicate represents 10 flies that were frozen in a 1.5-ml microcentrifuge tube on dry ice and stored at −80°C until processing. Each experiment contained four biological replicates per sex and per genotype, and each experiment was repeated at least twice. Total RNA was extracted as previously described [81,154]. Genomic DNA was eliminated and cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. For a detailed description of methods, refer to S1 Material and Methods.
qPCR
qPCR was performed in a 15-μL reaction volume containing 2 μL of diluted cDNA and final concentrations of 0.6 U of Platinum or 0.3 U of recombinant Taq DNA Polymerase (ThermoFisher Scientific), 0.1× SYBR Green I Nucleic Acid Gel Stain (ThermoFisher Scientific), 0.3 μM of specific primer pairs (Integrated DNA Technologies, Eurofin Genomics, ThermoFisher Scientific), 1× PCR buffer (ThermoFisher Scientific), 125 μM dNTP mix (FroggaBio), and 1.5 mM MgCl2 (ThermoFisher Scientific). qPCR was carried out in a CFX384 Touch Real-Time PCR Detection System (BioRad). Thermocycler conditions were as follows: initial denaturation for 3 minutes at 95°C and then 40 cycles of denaturation for 30 seconds at 95°C, annealing for 30 seconds at 60°C, and extension for 45 seconds at 72°C. Data were normalized to the average fold change of β-tubulin. For a full primer list, refer to S1 Material and Methods.
Sex difference in gene expression
To show the sex bias in gene expression, fold change was calculated as 2^(absolute value of ΔCT). Sex-biased gene expression graph and heat maps were generated in RStudio (version 1.0.153) using the code below.
Radar plots
The average fold change in bmm mRNA (or other mRNA) in males and females between 0 and 24 hours post-starvation was generated by 2^ΔCT. The ΔCT is the difference between the average CT value for each sex and time point compared with the average CT value of females at 0 hours post-starvation.
Metabolic rate measurements
Virgin males and females were sexed post-eclosion and aged for 5 days at 25°C on a 12:12 light:dark cycle. Two hours after lights on, single flies were lightly anaesthetized with CO2, and wet mass per individual was recorded to the nearest microgram. Individual flies were placed into 5-ml syringes that were modified to allow for gas flow along with a small cap filled with standard food (0.88% agar, 8.33% torula yeast, 10% cornmeal, 0.33% Tegosept w/v and 4.66% molasses, 1.66% ethanol (95% v/v), 0.66% propionic acid v/v dH2O) or with starvation medium (0.7% agarose in 1× PBS). At least 20 biological replicates per sex per treatment were measured for metabolic rate in a randomly assigned order. We used stop-flow respirometry to estimate metabolic rate as the volume of CO2 (VCO2) produced and the volume of O2 (VO2) consumed, which allowed for calculation of the RQ (RQ = VCO2/VO2), as previously described [155] (Sable Systems International, Las Vegas, NV). Any flies that died during the observation period were excluded from our analysis. All respirometry data were analyzed using the Expedata software package (Sable Systems) as previously described [155]. To test for differences in metabolic rates across treatments within each sex and time point, we estimated the scaling relationship between mass and VCO2 and mass and VO2 using Type II Model regression in the smatR package in R [156]. If the slopes were significantly different, then no further tests were performed; however, if the slopes were the same, we tested for effects of treatment on the elevation (i.e., on mass-specific metabolic rate) and as a shift along the x-axis (i.e., a difference in mass). In order to generate mass-corrected metabolic rates, we took residuals of these regressions and added back the grand mean for each group [155]. We also tested for differences in VCO2, VO2, and RQ between treatments within each sex and window of time using Tukey HSD tests. We also tested for differences in mass-corrected VCO2 and VO2. For a detailed description of methods, refer to S1 Material and Methods.
Lipid droplet visualization and quantification
Dissected testis from newly eclosed males and CNSs dissected from 5- to 7-day-old adult virgin males and females of the indicated genotypes were fixed for 30 minutes in 4% paraformaldehyde at room temperature. After three 10-minute washes in 1× PBS, the dissected tissues were incubated in 1× PBS with either a 1:50 dilution of HCS LipidTOX (Invitrogen, H34476) or BODIPY 493/503 (ThermoFisher, D3922) for 30 minutes. To visualize nuclei in the testis, Hoechst 33342 (ThermoFisher, 62249) was included with LipidTOX/BODIPY at a dilution of 1:1,000. Images were acquired on a Leica SP5 confocal microscope. Neutral lipid levels within each brain were measured as the sum of fluorescence using ImageJ.
Statistics
qPCR data were analyzed using one-way ANOVA paired with Tukey’s multiple comparisons test on software package Prism 6 (GraphPad). For all statistical analyses, differences were considered significant if the p-value was <0.05. All other data were analyzed using RStudio with the code described below. The lowest p-value provided by R is 2 × 10−16; therefore, many statistical tests show the same p-value. Error bars on graphs representing change in whole-body triglyceride level were calculated using the coefficient of error (COE—standard error of the mean as a percentage of the mean) as described in [157].
Log-rank test (R package “survminer”):
pairwise_survdiff(Surv(time, event) ~ genotype, data, p.adjust.method = "bonferroni")
surv_median(curve)
summary(data)
One-way ANOVA:
Results ← aov(value ~ genotype, data)
TukeyHSD(Results, conf.level = 0.95)
Student t test:
t.test(value ~ genotype, data, var.equal = TRUE)
Two-way ANOVA:
aov(percentageTG ~ genotype + sex, data)
Graphs
All graphs were prepared in R using packages “ggplot2,” “gtable,” “grid,” “survminer,” “survival,” “fmsb,” and “pheatmap” using the code described below.
Survival curve:
library (“survminer”)
library (“survival”)
curve <- survfit(Surv(time, event) ~ genotype, data)
max <- curve$time[which.max(curve$time)]
graph <- ggsurvplot(curve, size = 3, data, fun = "pct", palette = c(" "), surv.geom = geom_line, conf.int = TRUE, conf.int.style = c("step"), xlim = c(0, max), break.time.by = 12) + labs(x = NULL, y = NULL) + theme_survminer(legend = "non", font.tickslab = c(0))
Box and whisker plot:
library (“ggplot2”)
library (“gtable”)
library (“grid”)
roundUP <- ceiling(data$percentageTG[which.max(data$percentageTG)])
ggplot(data = data, aes(x = genotype, y = percentageTG, fill = genotype)) + stat_boxplot(geom = "errorbar", width = 0.5) + geom_boxplot() + scale_fill_manual (values = colours) + geom_dotplot(binaxis = 'y', stackdir = 'center', position = "dodge", pch = 21, col = "black", bg = "white", dotsize = 0.5) + facet_grid(. ~ time) + theme(legend.position = "none", panel.grid.major = element_blank(),panel.grid.minor = element_blank(), panel.background = element_blank(), axis.line = element_line(colour = "black", size = 1), axis.ticks.length = unit(0.25, "cm"), axis.ticks.y = element_line(size = 1), axis.ticks.x = element_blank(), axis.title = element_blank(), axis.text = element_blank()) + scale_y_continuous(breaks = seq(0, roundUP, by = 1), limits = c (0, roundUP)) + theme(strip.background = element_blank(), strip.text = element_blank())
Sex-biased gene expression graph:
library (“ggplot2”)
ggplot (data, aes(x = gene_name, y = average, fill = sex_bias)) + geom_errorbar(aes(ymin = average-data$sem, ymax = average+data$sem), width = 0.5) + geom_bar (position = position_dodge(), stat = "identity") + theme(axis.text.x = element_text(angle = 45, hjust = 1)) + scale_y_continuous (breaks = seq(-20, 60, by = 20), limits = c(-20, 60)) + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank(), panel.background = element_blank(), axis.line = element_line(colour = "black", size = 0.5), axis.ticks.length = unit(0.25, "cm"), axis.ticks.y = element_line(size = 0.5), axis.title = element_blank(), axis.text.y = element_blank()) + scale_fill_manual(values = c("#f39e1f", "#3a3b95", "#cdcdcd"))
Gene expression heat map:
library(“pheatmap”)
pheatmap(measurements.avg.stat[,c(2:9)], cluster_rows = F, cluster_cols = F, color = colour, border_color = “#FFFFFF”, width = 10,height = 8,filename = “heatmap_expression_F.pdf”, na_col = “#DDDDDD”, breaks = seq(0,5,0.001), annotation_row = annotation_row, cellwidth = 15, cellheight = 15)
Radar plot:
library(“fmsb”)
radarchart (data[2:7], maxmin = TRUE, axistype = 0, seg = 9, caxislabels = seq(0, 9, 1), pty = 16, pcol = "black", pfcol = fill, plty = 1, cglty = 1, cglcol = "#bebebe", vlabels = NA)
Supporting information
(A) Ovary triglyceride levels were not significantly different between fed virgin w1118 females and starved virgin w1118 females at all time points STV (p = 0.56, 0.44, 0.55, respectively; one-way ANOVA followed by Tukey HSD test). (B) The amount of triglyceride contained in the testes of 5-day-old virgin w1118 males is below the limit of detection for the coupled colorimetric assay; therefore, statistics could not be performed. (C) Triglyceride levels in 5-day-old virgin w1118 female carcasses devoid of ovaries were significantly higher than in age-matched male carcasses devoid of testes (p = 0.022; Student t test). (D) Larval fat cells in newly eclosed CS virgin females and males showed no significant change between 0 and 12 hours post-eclosion (p = 0.53 and 0.43 for females and males, respectively; one-way ANOVA followed by Tukey HSD test), but there was a significant decrease in the larval fat cell number in both sexes between 12 and 24 hours post-eclosion (p = 0.0 and 0.0 for females and males, respectively; one-way ANOVA followed by Tukey HSD test). (E) The number of larval fat cells significantly decreased in w1118 virgin females and males between 0 and 12 hours post-eclosion (p = 0.0071 and 1.0 × 10−7 for females and males, respectively; one-way ANOVA followed by Tukey HSD test) and decreased further in females but not males between 12 and 24 hours post-eclosion (p = 0.011 and 0.8 for females and males, respectively; one-way ANOVA followed by Tukey HSD test). (F) After 24 hours of starvation, triglyceride levels in virgin CS females remain at 77% of the triglyceride level in a fed virgin female (p = 0.034; one-way ANOVA followed by Tukey HSD test), whereas virgin CS males have only 4% of the triglyceride level in a fed male remaining (p = 6 × 10−7; one-way ANOVA followed by Tukey HSD test). (G) In 5-day-old virgin female w1118 carcasses devoid of ovaries, there was no significant decrease in triglyceride levels between 0 and 12 hours STV, whereas there was a significant reduction in triglyceride levels in age-matched w1118 virgin male carcasses devoid of testes (p = 0.15 and 0.013, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, there was a male-biased decrease in triglyceride levels in male and female carcasses devoid of gonads (p = 0.00065 and 0.0051, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. COE, coefficient of error; CS, Canton-S; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
(TIF)
(A) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed females compared with fed males for the majority of the intervals during the 24-hour observation period (p = 0.067, 0.0031, 2.4 × 10−4, 4.5 × 10−4, 1.4 × 10−5, 1.7 × 10−7, respectively; Student t test at each time interval). (B) Non-mass-corrected O2 consumption was significantly higher in fed females compared with fed males at all intervals during the observation period (p = 1.5 × 10−5, 1.6 × 10−5, 6.0 × 10−6, 5.8 × 10−5, 1.8 × 10−5, 3.8 × 10−5, respectively; Student t test at each time interval). (C) Non-mass-corrected CO2 production was significantly higher in starved females at every interval post-starvation from 4 hours onward (p = 0.44, 1.3 × 10−5, 5.9 × 10−13, 2.4 × 10−9, 1.9 × 10−7, 1.5 × 10−4, respectively; Student t test at each time interval). (D) Non-mass-corrected O2 consumption was significantly higher in starved females compared with starved males at all time intervals post-starvation (p = 6.0 × 10−12, 1.1 × 10−15, 1.2 × 10−14, 4.3 × 10−10, 1.7 × 10−8, 2.5 × 10−5, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the sexes at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.20, 0.0024, 4.9 × 10−5, 5.2 × 10−5, 1.4 × 10−6, 1.6 × 10−9, respectively; Student t test at each time interval). (B) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed males compared with starved males for most intervals post-starvation during the observation period (p = 0.99, 6.1 × 10−5, 4.7 × 10−10, 2.2 × 10−9, 1.3 × 10−5, 4.1 × 10−9, respectively; Student t test at each time interval). (C) Non-mass-corrected O2 consumption was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.072, 0.0080, 0.0013, 8.1 × 10−4, 7.7 × 10−6, 6.6 × 10−8, respectively; Student t test at each time interval). (D) Non-mass-corrected O2 consumption was significantly higher in fed males compared with starved males at all intervals post-starvation (p = 1.6 × 10−4, 9.6 × 10−6, 1.5 × 10−7, 3.6 × 10−6, 9.3 × 10−6, 4.8 × 10−6, respectively; Student t test at each time interval). (E) Mass-corrected CO2 production was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.55, 0.0026, 4.9 × 10−5, 0.0016, 1.3 × 10−4, 8.1 × 10−9, respectively; Student t test at each time interval). (F) Mass-corrected CO2 production was significantly higher in fed males compared with starved males for most intervals post-starvation during the observation period (p = 0.59, 4.4 × 10−4, 7.5 × 10−10, 2.0 × 10−9, 7.0 × 10−7, 3.0 × 10−10, respectively; Student t test at each time interval). (G) Mass-corrected O2 consumption was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.053, 0.014, 0.0098, 0.063, 7.6 × 10−4, 6.2 × 10−6, respectively; Student t test at each time interval). (H) Mass-corrected O2 consumption was significantly higher in fed males compared with starved males at all intervals post-starvation (p = 5.0 × 10−5, 2.6 × 10−6, 4.7 × 10−8, 1.0 × 10−6, 8.3 × 10−7, 1.1 × 10−6, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the treatments at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) In fed Oregon-R females and males, we observed no significant differences in the RQ throughout most of the observation period, with the exception of the 4- to 8-hour interval (p = 0.17, 0.031, 0.13, 0.43, 0.58, 0.15, respectively; Student t test at each time interval). (B) In starved Oregon-R females and males, starved males have a significantly higher RQ at all time intervals post-starvation (p = 0.0012, 0.0013, 7.7 × 10−4, 6.6 × 10−5, 0.0013, 0.0032, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the sexes at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points; RQ, respiratory quotient.
(TIF)
(A) Whole-body protein levels were not significantly different between 5-day-old virgin w1118 males and females at any time point STV (p = 0.16, 0.19, 0.37, respectively; Student t test at each time point). (B) Whole-body glucose levels were not significantly different between the sexes at 0 and 12 hours STV but were significantly higher in females compared with males by 24 hours STV (p = 0.87, 0.48, 0.034, respectively; Student t test at each time point). (C) Whole-body glycogen levels were not significantly different between the sexes at 0 or 12 hours STV but were significantly higher in females compared with males by 24 hours STV (p = 0.86, 0.063, 0.033, respectively; Student t test at each time point). The p-values are listed in the following order: difference between females and males at 0 hours, 12 hours, and 24 hours STV. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S1 Data. ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
(TIF)
(A) In normal culture conditions, Agpat4 is female-biased, mdy is male-biased, and PAPLA1 is not sex biasedly expressed when normalized to β-tubulin (p = <0.0001, 0.0079, and 0.25, respectively; Student t test for each gene). (B) In normal culture conditions, Agpat4 is female biased and mdy and PAPLA1 are male biased when normalized to both β-tubulin and β-cop (p = <0.0001, 0.0015, and 0.048, respectively; Student t test for each gene). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S3 Data. β-cop, Coat Protein (coatomer) β; Agpat, 1-acylglycerol-3-phosphate O-acyltransferase; ns, no significant difference between two sexes, two genotypes, or time points; mdy, midway; PAPLA1, phosphatidic acid phospholipase A1.
(TIF)
(A–C) In starvation conditions, Agpat4, mdy, and PAPLA1 female gene expression is significantly increased at 24 hours post-starvation when normalized to β-tubulin (p = <0.0001, <0.0001, and <0.0001, respectively at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (D–F) In starvation conditions, Agpat4 and PAPLA1 female gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin and β-cop (p = >0.05, >0.05, and <0.01 respectively at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (G–I) In starvation conditions, Agpat4 and PAPLA1 male gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin (p = >0.05, >0.05, and <0.001, respectively, at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (J–L) In starvation conditions, Agpat4 and PAPLA1 male gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin and β-cop (p = >0.05, >0.05, and <0.05, respectively, at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for list of all comparisons and p-values; quantitative measurements underlying all graphs are available in S3 Data. β-cop, Coat Protein (coatomer) β; Agpat, 1-acylglycerol-3-phosphate O-acyltransferase; HSD, honest significant difference; mdy, midway; ns, no significant difference between two sexes, two genotypes, or time points; PAPLA1, phosphatidic acid phospholipase A1.
(TIF)
Radar plots demonstrating gene expression in males and females throughout the starvation period for representative genes. (A, B) mRNA levels of two genes with male-biased expression throughout the starvation period in both males and females (Lpin, CG1941). (C, D) mRNA levels of two genes with female-biased expression throughout the starvation period in both males and females (hsl, wun2). Sex-biased expression of these genes remains consistent throughout the starvation period. See S1 Table for list of all comparisons and p-values; quantitative measurements underlying all graphs are available in S3 Data. hsl, hormone-sensitive lipase; Lpin, Lipin; STV, post-starvation; wun2, wunen-2.
(TIF)
(A) Triglyceride levels in 5-day-old bmm1 mutant females fed a high fat diet (HFD) were significantly higher than in bmm1 mutant females fed standard fly food (p = 3.9 × 10−6; Student t test). (B) Triglyceride storage was significantly higher in 5-day-old virgin bmmrev female carcasses lacking ovaries compared with age-matched bmmrev males, whereas there was no significant difference in whole-body triglyceride levels in 5-day-old bmm1 mutant virgin female carcasses devoid of ovaries compared with age-matched bmm1 mutant virgin males (p = 0.00013 in bmmrev animals, 0.12 in bmm1 mutants; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; F, female; HFD, high-fat diet; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) In 5-day-old virgin da>UAS-bmm-RNAi males, triglyceride levels were significantly higher than in da>+ or +>UAS-bmm-RNAi control males (p = 0.012 and 6.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (B) Triglyceride levels in 5-day-old virgin da>UAS-bmm-RNAi females were not significantly different from da>+ or +>UAS-bmm-RNAi control females (p = 0.98 and 0.16, respectively; one-way ANOVA followed by Tukey HSD test). (C) The male-biased effects of da>UAS-bmm-RNAi on triglyceride storage reduced the sexual dimorphism in triglyceride storage compared with da>+ or +>UAS-bmm-RNAi controls. (D) In 5-day-old virgin da>UAS-bmm-RNAi#2 (BDSC #25926) males, triglyceride levels were significantly higher than in da>+ or +>UAS-bmm-RNAi#2 control males (p = 0.0 and 0.0, respectively; one-way ANOVA followed by Tukey HSD test). (E) Triglyceride levels in 5-day-old virgin da>UAS-bmm-RNAi#2 (BDSC #25926) females were not significantly different from da>+ or +>UAS-bmm-RNAi#2 control females (p = 5.8 × 10−5 and 0.14, respectively; one-way ANOVA followed by Tukey HSD test). (F) The male-biased effects of da>UAS-bmm-RNAi#2 (BDSC #25926) on triglyceride storage reduced the sexual dimorphism in triglyceride storage compared with da>+ or +>UAS-bmm-RNAi#2 controls. (G) Between 0 DPE and 5 DPE, mRNA expression levels for bmm were not significantly increased in virgin w1118 females but were significantly increased in virgin w1118 males (p = 0.17 and 0.048, respectively; Student t test). (H) Between 0 and 12 hours STV, we observed no triglyceride breakdown in da>+, +>UAS-bmm-RNAi, or da>UAS-bmm-RNAi males (p = 0.3, 0.053, and 0.18, respectively; one-way ANOVA followed by Tukey HSD test); however, between 12 and 24 hours STV, the magnitude of triglyceride breakdown in da>UAS-bmm-RNAi males was lower than da>+ and +>UAS-bmm-RNAi control males (p = 0.038, 3.1 × 10−6, and 0.00046, respectively; one-way ANOVA followed by Tukey HSD test). (I) Between 0 and 12 hours STV, and between 12 and 24 hours STV, we observed little change in triglyceride levels in da>+, +>UAS-bmm-RNAi, or da>UAS-bmm-RNAi females (p = 0.36, 0.0024, and 0.64, respectively, for 0–12 hours STV and 0.52, 0.046, and 0.045, respectively, for 12–24 hours STV; one-way ANOVA followed by Tukey HSD test). (J) Between 0 and 12 hours STV we observed similar magnitudes of triglyceride breakdown between da>+, +>UAS-bmm-RNAi#2, and da>UAS-bmm-RNAi#2 (BDSC #25926) males (p = 8.1 × 10−6, 1.8 × 10−6, and 7.1 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test); however, between 12 and 24 hours STV, triglyceride breakdown in da>UAS-bmm-RNAi#2 (BDSC #25926) males was blocked, whereas triglyceride levels decreased in da>+ and +>UAS-bmm-RNAi#2 control males (p = 0.069, 1.3 × 10−6, and 2 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (K) Between 0 and 12 hours STV, triglyceride breakdown in da>UAS-bmm-RNAi#2 (BDSC #25926) females was blocked, whereas triglyceride levels decreased in da>+ and +>UAS-bmm-RNAi#2 control females (p = 0.18, 0.036, and 9.9 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test), whereas we observed similar magnitudes of triglyceride breakdown between 12 and 24 hours STV in da>+, +>UAS-bmm-RNAi#2, and da>UAS-bmm-RNAi#2 (BDSC #25926) females (p = 0.032, 0.28, and 0.022, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat or mRNA expression level represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 and S3 Datas. bmm, brummer; BDSC, Bloomington Drosophila Stock Center; COE, coefficient of error; da, daughterless; DPE, days post-eclosion; F, female; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; w, white.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the fat body (cg>UAS-bmm-RNAi) was significantly higher than age-matched control males (cg>+ and +>UAS-bmm-RNAi) (p = 1.0 × 10−4 and 8.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the fat body (cg>UAS-bmm-RNAi) was significantly higher than age-matched control females (cg>+ and +>UAS-bmm-RNAi) (p = 5.5 × 10−4 and 1.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (C) There was a significant reduction in control female and male triglyceride levels (cg>+) between 0 and 12 hours post-starvation (p = 2.8 × 10−6 and 1.0 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test); however, we observed no significant reduction in whole-body triglyceride levels in 5-day-old virgin cg>UAS-bmm-RNAi females and males between 0 and 12 hours post-starvation (p = 0.54 and 0.92, respectively; one-way ANOVA followed by Tukey HSD test). (D) There was a significant reduction in triglyceride storage in control females and males (cg>+ and +>UAS-bmm-RNAi) between 12 and 24 hours post-starvation (p = 1 × 10−7 and 2.7 × 10−4 (females) and p = 4.0 × 10−4 and 2 × 10−7 (males), respectively; one-way ANOVA followed by Tukey HSD test); however, there was no significant change in whole-body triglyceride levels in 5-day-old virgin cg>UAS-bmm-RNAi females between 12 and 24 hours post-starvation (p = 1.0; one-way ANOVA followed by Tukey HSD test). In males, there was a significant but blunted decrease in triglyceride levels in 5-day-old cg>UAS-bmm-RNAi virgin males between 12 and 24 hours post-starvation (p = 0.0011; one-way ANOVA followed by Tukey HSD). (E) Whole-body triglyceride storage in 5-day-old r4>UAS-bmm-RNAi males was significantly higher than r4>+ and +>UAS-bmm-RNAi control males (p = 0.0 and 0.0, respectively; one-way ANOVA followed by Tukey HSD test). (F) r4>UAS-bmm-RNAi females did not show a significant difference in whole-body triglyceride storage compared with r4>+ and +>UAS-bmm-RNAi control females (p = 0.95 and 0.00015, respectively; one-way ANOVA followed by Tukey HSD test). (G) Between 0 and 12 hours post-starvation, there was no significant decrease in triglyceride levels in either 5-day-old r4>UAS-bmm-RNAi or +>UAS-bmm-RNAi females (p = 0.42 and 0.096, respectively; one-way ANOVA followed by Tukey HSD test) and a modest but significant reduction in triglyceride level in r4>+ during the same interval (p = 0.00014; one-way ANOVA followed by Tukey HSD test). In males, we observed similar trends for r4>+, +>UAS-bmm-RNAi, and r4>UAS-bmm-RNAi males (p = 1.0 × 10−7, 0.057, and 0.0012, respectively; one-way ANOVA followed by Tukey HSD test). (H) Between 12 and 24 hours post-starvation, we observed that the magnitude of the decrease in triglyceride levels in 5-day-old r4>UAS-bmm-RNAi females and males was blunted compared with the decrease in r4>+ and +>UAS-bmm-RNAi controls for each sex (p = 0.04, 0.00048, and 2.0 × 10−7 for females; 0.0013, 0.12, and 8.2 × 10−5 for males, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting Percent Body Fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; cg, collagen; F, female; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the gut (Mex>UAS-bmm-RNAi) was not significantly different from age-matched control females (Mex>+ and +>UAS-bmm-RNAi) (p = 0.31 and 0.0073, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the gut (Mex>UAS-bmm-RNAi) was not significantly different from age-matched control males (Mex>+ and +>UAS-bmm-RNAi) (p = 0.17 and 0.079, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the muscle (dMef2>UAS-bmm-RNAi) was not significantly different from age-matched control females (dMef2>+ and +>UAS-bmm-RNAi) (p = 0.50 and 0.70, respectively; one-way ANOVA followed by Tukey HSD test). (D) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the muscle (dMef2>UAS-bmm-RNAi) was not significantly different from age-matched control males (dMef2>+ and +>UAS-bmm-RNAi) (p = 0.54 and 0.34, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride level in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the glia (repo>UAS-bmm-RNAi) was not significantly different from age-matched control females (repo>+ and +>UAS-bmm-RNAi) (p = 3.2 × 10−5 and 0.26, respectively; one-way ANOVA followed by Tukey HSD test). (F) Whole-body triglyceride levels in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the glia (repo>UAS-bmm-RNAi) were not significantly different from age-matched control males (repo>+ and +>UAS-bmm-RNAi) (p = 0.016 and 0.8, respectively; one-way ANOVA followed by Tukey HSD test). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control and difference between the GAL4/UAS genotype and the UAS control, respectively. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; F, female; HSD, honest significant difference; M, male; Mex, midgut expression 1; Mef2, myocyte enhancer factor 2; ns, no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in males with c587-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in the somatic cells of the male gonad was significantly higher than in control males (c587-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 2.3 × 10−5 and 1.1 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). (B) The decrease in whole-body triglyceride levels in c587-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) males was blunted compared with c587-GAL4>+ and +>UAS-bmm-RNAi#2 control males between 0 and 12 hours STV (p = 0.47, 5.8 × 10−6, and 0.065, respectively; one-way ANOVA followed by Tukey HSD test) but not between 12 and 24 hours STV (p = 8.2 × 10−4, 6.0 × 10−7, and 0.0033, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride levels in 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi males were not significantly different to c587-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls, demonstrating that re-expression of UAS-bmm rescued the increased fat storage caused by loss of bmm in the somatic cells of the male gonad (p = 0.87 and 1.0, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi males and control males (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) was decreased by a similar magnitude between both 0 and 12 hours or 12 and 24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in the somatic cells of the male gonad post-starvation (p = 2.2 × 10−6, 1.7 × 10−6, and 0.0 for 0–12 hours and 0.0, 0.0, and 0.0 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (c587>UAS-bmm-RNAi) was not significantly different from age-matched control females (c587>+ and +>UAS-bmm-RNAi) (p = 0.083 and 0.96, respectively; one-way ANOVA followed by Tukey HSD test). (B) There was a modest but significant reduction in whole-body triglyceride levels in 5-day-old c587>+, +>UAS-bmm-RNAi, and c587>UAS-bmm-RNAi females between 0 and 12 hours STV (p = 2.3 × 10−4, 0.0094, and 0.0051, respectively; one-way ANOVA followed by Tukey HSD test) and between 12 and 24 hours STV (p = 0.0, 0.016, and 1.6 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in females with c587-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in the somatic cells of the gonad was not significantly different from control females (c587-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.98 and 0.78, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among c587-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) females and c587-GAL4>+ and +>UAS-bmm-RNAi#2 controls showed a modest decrease of similar magnitude between both 0 and 12 hours and 12 and 24 hours STV (p = 0.096, 1.3 × 10−4, and 0.0013 for 0–12 hours and 0.004, 0.0049, and 0.022 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride levels in 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi females were not significantly different among c587-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 8.5 × 10−4 and 0.38, respectively; one-way ANOVA followed by Tukey HSD test). (F) Triglyceride breakdown post-starvation among 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi females and control females (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) showed a modest decrease of a similar magnitude at both 0–12 hours or 12–24 hours STV (p = 0.0043, 1.7 × 10−6, and 0.0027 for 0–12 hours and 0.0, 2.6 × 10−6, and 2.9 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; F, female; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (tj>UAS-bmm-RNAi) was not significantly different from age-matched control males (tj>+ and +>UAS-bmm-RNAi) (p = 0.098 and 0.97, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (tj>UAS-bmm-RNAi) was not significantly different from age-matched control females (tj>+ and +>UAS-bmm-RNAi) (p = 0.2 and 0.66, respectively; one-way ANOVA followed by Tukey HSD test). (C) Between 0 and 12 hours STV, the magnitude of triglyceride breakdown in tj>UAS-bmm-RNAi was similar to tj>+ and +>UAS-bmm-RNAi control males; however, between 12 and 24 hours STV, there was no significant decrease in triglyceride levels in tj>UAS-bmm-RNAi males, in contrast to tj>+ and +>UAS-bmm-RNAi control males, in which we observed a significant decrease in triglyceride storage STV (p = 9.7 × 10−4, 0.0018, and 0.099 for 0–12 hours and 0.37, 1.9 × 10−5, and 0.028 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among 5-day-old virgin tj>UAS-bmm-RNAi females and tj>+ and +>UAS-bmm-RNAi control females was modestly decreased by a similar magnitude at both 0–12 hours or 12–24 hours STV (p = 0.058, 0.026, and 0.022 for 0–12 hours and 0.042, 0.0093, and 3.7 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (E–G) We used tj-GAL4 to drive the expression of a membrane-bound GFP (UAS-mCD8::GFP) in the somatic cells of the gonad. The presence of lipid droplets within the GFP-marked boundary of the somatic cell indicates that lipid droplets are present in the somatic cells of the gonad. Non-GFP-positive droplets (arrow) likely represent lipid droplets in the germline cells. The image represents a single confocal slice from the Drosophila male testis. Scale bars = 50 μm, except for inset images, in which scale bars = 12.5 μm. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. Original image files corresponding to all images acquired from genotype-matched individuals presented in panels E–G are available upon request. bmm, brummer; COE, coefficient of error; F, female; GFP, green fluorescent protein; tj, traffic jam; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in males with elav-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in neurons was not significantly different than in control males (elav-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.11 and 0.91, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in males with nSyb-GAL4-mediated overexpression of the UAS-bmm-RNAi transgene in neurons was not significantly different than in control males (nSyb-GAL4>+ and +>UAS-bmm-RNAi) (p = 0.19 and 2.1 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (C) The decrease in whole-body triglyceride levels in elav-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) males was similar to elav-GAL4>+ and +>UAS-bmm-RNAi#2 control males between 0 and 12 hours STV (p = 0.01, 4.0 × 10−7, and 1.0 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). Triglyceride breakdown between 12 and 24 hours STV was blocked in elav-GAL4>UAS-bmm-RNAi#2 males, whereas triglyceride levels in control males during this interval significantly decreased (p = 0.084, 2.6 × 10−5, and 3.3 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (D) The decrease in whole-body triglyceride levels in nSyb-GAL4>UAS-bmm-RNAi males was blunted compared with nSyb-GAL4>+ and +>UAS-bmm-RNAi control males between 0 and 12 hours STV (p = 0.86, 0.051, and 4.3 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test); however, triglyceride breakdown between 12 and 24 hours STV in nSyb-GAL4>UAS-bmm-RNAi males was similar in magnitude to control males (nSyb>+ and +>UAS-bmm-RNAi) (p = 0.002, 1.2 × 10−4, and 7.0 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (E) In dissected brains from elav>UAS-bmm-RNAi males, we found that bmm transcript levels were undetectable in three out of four samples, whereas we observed amplification at a higher cycle number in three elav>+ samples. (§) Because of this dramatic decrease in bmm transcript levels in most elav>UAS-bmm-RNAi samples, we were unable to perform a statistical analysis to quantify this effect. (F) Whole-body triglyceride levels in 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi males were not significantly different to elav>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 0.84 and 0.92, respectively; one-way ANOVA followed by Tukey HSD test). (G) Whole-body triglyceride levels post-starvation among 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi males and control males (elav>+ and +>UAS-bmm;UAS-bmm-RNA) were significantly decreased by a similar magnitude at both 0–12 hours or 12–24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in neurons STV (p = 2.3 × 10−6, 1.2 × 10−4, and 0.0 for 0–12 hours and 2.0 × 10−7, 1.0 × 10−7, and 0.0 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat or mRNA expression level represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 and S3 Datas. bmm, brummer; COE, coefficient of error; elav, embryonic lethal abnormal vision; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Triglyceride breakdown post-starvation was modestly decreased by a similar magnitude in 5-day-old virgin repo>UAS-bmm-RNAi females compared with repo-GAL4>+ and +>UAS-bmm-RNAi control females at 0–12 hours post-starvation (p = 0.047, 2.3 × 10−5, and 0.096, respectively; one-way ANOVA followed by Tukey HSD test) and between repo>UAS-bmm-RNAi males and repo-GAL4>+ and +>UAS-bmm-RNAi control males during the same interval (p = 0.085, 6.9 × 10−6, and 0.057, respectively; one-way ANOVA followed by Tukey HSD test). (B) Between 12 and 24 hours post-starvation, we observed only a modest reduction in female triglyceride levels post-starvation in all genotypes (repo>UAS-bmm-RNAi, repo-GAL4>+, and +>UAS-bmm-RNAi), and the magnitude of this reduction was similar between genotypes (p = 0.0019, 4.8 × 10−6, and 2.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). Similarly, although there was a significant decrease in triglyceride levels post-starvation in repo>UAS-bmm-RNAi males, repo-GAL4>+, and +>UAS-bmm-RNAi controls (p = 8.8 × 10−6, 0.0, and 8.2 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test), the magnitude of this decrease was similar for all genotypes. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). See S1 Table for list of all multiple comparisons and p-values. Error bars on graphs represent COE. Quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; HSD, honest significant difference; ns indicates no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the postmitotic neurons (elav>UAS-bmm-RNAi) was not significantly different from age-matched control females (elav>+ and +>UAS-bmm-RNAi) (p = 0.54 and 0.95, respectively; one-way ANOVA followed by Tukey HSD test). (B) There was a significant reduction in whole-body triglyceride levels in 5-day-old elav>+, +>UAS-bmm-RNAi, and elav>UAS-bmm-RNAi females between 0 and 12 hours STV (p = 0.026, 0.0038, and 0.0013, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, 5-day-old elav>+, +>UAS-bmm-RNAi, and elav>UAS-bmm-RNAi females modestly decreased triglyceride levels by similar magnitudes (p = 1.2 × 10−4, 0.15, and 0.0012, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in females with elav-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in neurons was not significantly different from control females (elav-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.56 and 0.035, respectively; one-way ANOVA followed by Tukey HSD test). (D) There was a modest decrease in triglyceride levels between 0 and 12 hours and 12 and 24 hours STV among elav-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) females and elav-GAL4>+ and +>UAS-bmm-RNAi#2 controls (p = 0.011, 0.0027, and 1.4 × 10−6 for 0–12 hours STV and 4.6 × 10−4, 3.4 × 10−4, and 0.002 for 12–24 hours STV, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride levels in 5-day-old virgin nSyb>UAS-bmm-RNAi females were not significantly different to nSyb-GAL4>+ and +>UAS-bmm-RNAi controls (p = 0.85 and 0.52, respectively; one-way ANOVA followed by Tukey HSD test). (F) Triglyceride breakdown post-starvation was modestly decreased by a similar magnitude among genotypes in 5-day-old virgin nSyb>UAS-bmm-RNAi females and control females (nSyb>+ and +>UAS-bmm-RNAi) between 0 and 12 hours STV (p = 0.21, 0.017, and 0.43, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, the decrease in triglyceride levels in nSyb>UAS-bmm-RNAi females was blocked, whereas triglyceride levels decreased in control nSyb-GAL4>+ and +>UAS-bmm-RNAi females during this interval (p = 0.093, 3.0 × 10−5, and 0.0019, respectively; one-way ANOVA followed by Tukey HSD test). (G) Whole-body triglyceride levels in 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi females were not significantly different to elav-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 0.41 and 0.66, respectively; one-way ANOVA followed by Tukey HSD test). (H) Whole-body triglyceride levels post-starvation among 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi females and control females (elav>+ and +>UAS-bmm;UAS-bmm-RNAi) decreased by a similar magnitude between 0 and 12 hours or 12 and 24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in neurons STV (p = 0.0023, 0.0012, and 0.0027 for 0–12 hours and 1.0 × 10−6, 5.2 × 10−6, and 2.9 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; elav, embryonic lethal abnormal vision; F, female; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Median survival post-starvation was significantly higher in 5-day-old virgin Oregon-R females than in virgin Oregon-R males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparison; n > 156). (B) Median survival post-starvation was significantly higher in 5-day-old virgin CMW wild-caught females than in virgin CMW males (p = 6 × 10−10; Log-rank test with Bonferroni correction for multiple comparison; n > 118). (C, D) Median survival post-starvation was significantly higher in 5-day-old virgin females than males in two isofemale strains (Mel c2.2: p = 4.4 × 10−11; Log-rank test with Bonferroni correction for multiple comparison; n > 55 and Mel c2.3: p = 1.4 × 10−13; Log-rank test with Bonferroni correction for multiple comparison; n > 110). (E, F) Median survival post-starvation was significantly higher in virgin males (E) and females (F), with ubiquitous overexpression of UAS-bmm-RNAi compared with control males (da>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16 respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 223) and females (p = 2 × 10−16 and 1.2 × 10−13 respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 176). (G, H) Median survival post-starvation was significantly higher in virgin males (G) and significantly lower in females (H) with ubiquitous overexpression of UAS-bmm-RNAi#2 (BDSC #25926) compared to control males (da>+ and +>UAS-bmm-RNAi#2) (p = 7.7 × 10−11 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 97) and control females (p = 9.6 × 10−9 and 4.9 × 10−5, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 91). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; BDSC, Bloomington Drosophila Stock Center; CMW, Country Mill Winery; da, daughterless; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A) Males with whole-body loss of brummer have a significantly decreased number of progeny after 2 days, 4 days, and 6 days of mating (p = 2.2 × 10−16, 9.7 × 10−12, and 4.6 × 10−6, respectively; Student t test at each time point). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). See S1 Table for list of all multiple comparisons and p-values. Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S4 Data. M, male; ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin females (A) and males (B) with fat body–specific bmm inhibition (cg>UAS-bmm-RNAi) compared with control females (cg>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 279) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 365). (C, D) Median survival post-starvation was significantly higher in virgin females (C) and males (D) with fat body–specific bmm inhibition using a second GAL4 driver (r4>UAS-bmm-RNAi) compared with control females (r4>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 314) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 195). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; cg, collagen; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin males (A) but not significantly different in virgin females (B) with bmm inhibition in the somatic cells of the gonad (c587>UAS-bmm-RNAi#2 [VDRC #37877]) compared with control males (c587>+ and +>UAS-bmm-RNAi#2) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 212) and females (p = 2 × 10−16 and 1.0, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 201). (C, D) Median survival post-starvation was not significantly changed when UAS-bmm and UAS-bmm-RNAi were simultaneously overexpressed by a driver for the somatic cells of the gonad (c587>UAS-bmm;UAS-bmm-RNAi) in both males (C) and females (D) compared with control males (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) (p = 2 × 10−16 and 0.57, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 249) and females (p = 9.6 × 10−16 and 0.0035, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 207). (E, F) Median survival post-starvation was significantly increased when UAS-bmm-RNAi was overexpressed by a second driver for the somatic cells of the gonad (tj>UAS-bmm-RNAi) in both females (E) and males (F) compared with control males (tj>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 1.3 × 10−6, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 165) and females (p = 2 × 10−16 and 0.00012, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 177). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; tj, traffic jam; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin males (A) but not significantly different in virgin females (B) with bmm inhibition in the neurons (elav>UAS-bmm-RNAi#2 [VDRC #37877]) compared with control males (elav>+ and +>UAS-bmm-RNAi#2) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 90) and females (p = 3.4 × 10−6 and 0.41, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 81). (C, D) Median survival post-starvation was not significantly changed when UAS-bmm and UAS-bmm-RNAi were simultaneously overexpressed by a neuronal driver (elav>UAS-bmm;UAS-bmm-RNAi) in both males (C) and females (D) compared with control males (elav>+ and +>UAS-bmm;UAS-bmm-RNAi) (p = 4.6 × 10−5 and 1, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 101) and females (p = 8.9 × 10−6 and 1, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 23). (E, F) Median survival post-starvation was significantly increased when UAS-bmm-RNAi was overexpressed by a second neuronal driver (nSyb>UAS-bmm-RNAi) in both males (E) and females (F) compared with control males (nSyb>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 157) and females (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 157). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; elav, embryonic lethal abnormal vision; F, female; M, male; ns indicates no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A, B) Median survival post-starvation showed no significant change in virgin females (A) and males (B) with gut-specific bmm inhibition (Mex>UAS-bmm-RNAi) compared with control females (Mex>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 1.1 × 10−7, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 444) and males (p = 2 × 10−16 and 0.13 respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 456). (C, D) Median survival post-starvation was unchanged in virgin females (C) and slightly increased in males (D) with muscle-specific bmm inhibition (dMef2>UAS-bmm-RNAi) compared with control females (dMef2>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 0.4, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 340) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 382). (E, F) Median survival post-starvation showed no significant change in virgin females (E) and males (F) with glia-specific bmm inhibition (repo>UAS-bmm-RNAi) compared with control females (repo>+ and +>UAS-bmm-RNAi) (p = 0.43 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 191) and males (p = 2 × 10−16 and 0.02, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 207). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; F, female; M, male; Mex, midgut expression 1; dMef2, myocyte enhancer factor 2, ns, no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(XLSX)
(XLSX)
GFP, green fluorescent protein.
(XLSX)
(XLSX)
(CSV)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We would like to thank Dr. Ronald Kühnlein for sharing bmm1 mutants and bmmrev control strains, Dr. Ian Dworkin for providing the Drosophila melanogaster wild-caught and isofemale lines, Dr. Guy Tanentzapf for sharing the c587-GAL4 and tj-GAL4 strains, Dr. Michael Gordon for nSyb-GAL4, and Dr. Claire Thomas for sharing the Mex-GAL4 strain. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank the TriP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks and/or plasmid vectors used in this study. Transgenic fly stocks and/or plasmids were also obtained from the Vienna Drosophila Resource Center (VDRC, https://stockcenter.vdrc.at/control/main). We acknowledge critical resources and information provided by FlyBase [158].
Abbreviations
- AGPAT
1-acylglycerol-3-phosphate O-acyltransferase
- AKH
adipokinetic hormone
- ATGL
adipose triglyceride lipase
- bmm
brummer
- BODIPY
boron-dipyrromethene
- cg
collagen
- CMW
Country Mill Winery
- CNS
central nervous system
- CS
Canton-S
- da
daughterless
- DAGAT
diacylglycerol acyltransferase
- DGAT
diacylglycerol O-acyltransferase
- dob
doppelganger von brummer
- DPE
days post-eclosion
- dsx
doublesex
- EcR
ecdysone receptor
- elav
embryonic lethal abnormal vision
- foxo
forkhead box O
- fru
fruitless
- GFP
green fluorescent protein
- GPAT
glycerol-3-phosphate acyltransferase
- HNF4
hepatocyte nuclear factor 4
- hsl
hormone-sensitive lipase
- IIS
insulin/insulin-like growth factor signaling
- IPC
insulin-producing cell
- Klar
Klarsicht
- LD-GFP
lipid droplet–targeted GFP
- Lpin
Lipin
- lsd-1
lipid storage droplet-1
- lsd-2
lipid storage droplet-2
- mdy
midway
- mino
minotaur
- nSyb
neuronal Synaptobrevin
pigment dispersing factor
- PGC-1
peroxisome proliferator–activated receptor γ coactivator 1
- PLIN
perilipin
- qPCR
quantitative real-time PCR
- ROS
reactive oxygen species
- RQ
respiratory quotient
- sNPF
short neuropeptide F
- SREBP
sterol response element binding protein
- srl
spargel
- tj
traffic jam
- UAS
upstream activation sequence
- w
white
Data Availability
All relevant data are within the paper and its Supporting Information files: S1 Data, S2 Data, S3 Data, and S4 Data. Original image files corresponding to all images in the paper are available upon request.
Funding Statement
Funding for this study was provided by grants to EJR from the Canadian Institutes for Health Research (PJT-153072), Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2016-04249), Michael Smith Foundation for Health Research (16876), and the Canadian Foundation for Innovation (JELF-34879). LWW was supported by a British Columbia Graduate Scholarship Award. JWM was supported by a 4-year CELL Fellowship from UBC. HC was supported by an NSERC Undergraduate Student Research Award. MAW and MCB are supported by operating grants to MAW from the National Institutes of Health (RO1-GM102155). JLB is supported by a National Science Foundation EPSCoR Track 2 Award (#173624). FlyBase is supported by a grant from the National Human Genome Research Institute at the U.S. National Institutes of Health (U41 HG000739) and by the British Medical Research Council (MR/N030117/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Progress in lipid research. 2001;40(5):325–438. 10.1016/s0163-7827(01)00013-3 [DOI] [PubMed] [Google Scholar]
- 2.Thiele C, Spandl J. Cell biology of lipid droplets. Current opinion in cell biology. 2008;20(4):378–85. 10.1016/j.ceb.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 3.Walther TC, Farese RV Jr. Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687–714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res. 2012;53(8):1430–6. 10.1194/jlr.R024299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walther TC, Chung J, Farese RV Jr. Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol. 2017;33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Power ML, Schulkin J. Sex differences in fat storage, fat metabolism, and the health risks from obesity: possible evolutionary origins. The British journal of nutrition. 2008;99(5):931–40. 10.1017/S0007114507853347 [DOI] [PubMed] [Google Scholar]
- 7.Karastergiou K, Smith SR, Greenberg AS, Fried SK. Sex differences in human adipose tissues—the biology of pear shape. Biology of sex differences. 2012;3(1):13. 10.1186/2042-6410-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Womersley J. A comparison of the skinfold method with extent of 'overweight' and various weight-height relationships in the assessment of obesity. The British journal of nutrition. 1977;38(2):271–84. 10.1079/bjn19770088 [DOI] [PubMed] [Google Scholar]
- 9.Jackson AS, Stanforth PR, Gagnon J, Rankinen T, Leon AS, Rao DC, et al. The effect of sex, age and race on estimating percentage body fat from body mass index: The Heritage Family Study. International journal of obesity and related metabolic disorders. Journal of the International Association for the Study of Obesity. 2002;26(6):789–96. [DOI] [PubMed] [Google Scholar]
- 10.Lease HM, Wolf BO. Lipid content of terrestrial arthropods in relation to body size, phylogeny, ontogeny and sex. Physiological Entomology. 2011;36(1):29–38. [Google Scholar]
- 11.Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, et al. The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8(5):e1002709. 10.1371/journal.pgen.1002709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zore T, Palafox M, Reue K. Sex differences in obesity, lipid metabolism, and inflammation-A role for the sex chromosomes? Molecular metabolism. 2018;15:35–44. 10.1016/j.molmet.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayne BT, Bianco-Miotto T, Buckberry S, Breen J, Clifton V, Shoubridge C, et al. Large Scale Gene Expression Meta-Analysis Reveals Tissue-Specific, Sex-Biased Gene Expression in Humans. Frontiers in genetics. 2016;7:183. 10.3389/fgene.2016.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–9. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(1):218–23. 10.1073/pnas.011520898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome research. 2006;16(8):995–1004. 10.1101/gr.5217506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhnlein RP. The contribution of the Drosophila model to lipid droplet research. Progress in lipid research. 2011;50(4):348–56. 10.1016/j.plipres.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 18.Musselman LP, Kuhnlein RP. Drosophila as a model to study obesity and metabolic disease. The Journal of experimental biology. 2018;221(Pt Suppl 1). [DOI] [PubMed] [Google Scholar]
- 19.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell metabolism. 2007;6(4):257–66. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Y, Bi J, Shui G, Liu Z, Xiang Y, Liu Y, et al. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 2011;7(4):e1001364. 10.1371/journal.pgen.1001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–20. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- 22.Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol Cell Biol. 2011;31(8):1646–56. 10.1128/MCB.01335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buszczak M, Lu X, Segraves WA, Chang TY, Cooley L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics. 2002;160(4):1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschapalda K, Zhang YQ, Liu L, Golovnina K, Schlemper T, Eichmann TO, et al. A Class of Diacylglycerol Acyltransferase 1 Inhibitors Identified by a Combination of Phenotypic High-throughput Screening, Genomics, and Genetics. EBioMedicine. 2016;8:49–59. 10.1016/j.ebiom.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16(18):1783–95. 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- 26.Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, et al. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12(5):1115–26. 10.1074/mcp.M112.020230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, et al. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5(6):1082–94. 10.1074/mcp.M600011-MCP200 [DOI] [PubMed] [Google Scholar]
- 28.Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, et al. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev Cell. 2018;44(1):97-112.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell metabolism. 2010;12(5):521–32. 10.1016/j.cmet.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 30.Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kuhnlein RP, et al. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125(Pt 15):3568–77. 10.1242/jcs.101329 [DOI] [PubMed] [Google Scholar]
- 31.Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13(7):603–6. 10.1016/s0960-9822(03)00175-1 [DOI] [PubMed] [Google Scholar]
- 32.Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell metabolism. 2005;1(5):323–30. 10.1016/j.cmet.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 33.Horne I, Haritos VS, Oakeshott JG. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol. 2009;39(8):547–67. 10.1016/j.ibmb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 34.Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, et al. COPI complex is a regulator of lipid homeostasis. PLoS biology. 2008;6(11):e292. 10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–61. 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010;8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sieber MH, Spradling AC. Steroid Signaling Establishes a Female Metabolic State and Regulates SREBP to Control Oocyte Lipid Accumulation. Current biology: CB. 2015;25(8):993–1004. 10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwasinger-Schmidt TE, Kachman SD, Harshman LG. Evolution of starvation resistance in Drosophila melanogaster: measurement of direct and correlated responses to artificial selection. Journal of evolutionary biology. 2012;25(2):378–87. 10.1111/j.1420-9101.2011.02428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jehrke L, Stewart FA, Droste A, Beller M. The impact of genome variation and diet on the metabolic phenotype and microbiome composition of Drosophila melanogaster. Scientific reports. 2018;8(1):6215. 10.1038/s41598-018-24542-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguila JR, Suszko J, Gibbs AG, Hoshizaki DK. The role of larval fat cells in adult Drosophila melanogaster. The Journal of experimental biology. 2007;210(Pt 6):956–63. 10.1242/jeb.001586 [DOI] [PubMed] [Google Scholar]
- 41.Mauvais-Jarvis F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biology of sex differences. 2015;6:14. 10.1186/s13293-015-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Current biology: CB. 2010;20(13):1209–15. 10.1016/j.cub.2010.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, et al. Circadian neuron feedback controls the Drosophila sleep—activity profile. Nature. 2016;536(7616):292–7. 10.1038/nature19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bland ML. Measurement of Carbon Dioxide Production from Radiolabeled Substrates in Drosophila melanogaster. Journal of visualized experiments: JoVE. 2016(112). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millington JW, Rideout EJ. Sex differences in Drosophila development and physiology. Current Opinion in Physiology. 2018;6:46–56. [Google Scholar]
- 46.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature neuroscience. 2010;13(4):458–66. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8(5):e1000365. 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoxha V, Lama C, Chang PL, Saurabh S, Patel N, Olate N, et al. Sex-specific signaling in the blood-brain barrier is required for male courtship in Drosophila. PLoS Genet. 2013;9(1):e1003217. 10.1371/journal.pgen.1003217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clough E, Jimenez E, Kim YA, Whitworth C, Neville MC, Hempel LU, et al. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Developmental cell. 2014;31(6):761–73. 10.1016/j.devcel.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudry B, Khadayate S, Miguel-Aliaga I. The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature. 2016;530(7590):344–8. 10.1038/nature16953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regan JC, Khericha M, Dobson AJ, Bolukbasi E, Rattanavirotkul N, Partridge L. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife. 2016;5:e10956. 10.7554/eLife.10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lnenicka GA, Theriault K, Monroe R. Sexual differentiation of identified motor terminals in Drosophila larvae. Journal of neurobiology. 2006;66(5):488–98. 10.1002/neu.20234 [DOI] [PubMed] [Google Scholar]
- 53.Dworkin I, Gibson G. Epidermal growth factor receptor and transforming growth factor-beta signaling contributes to variation for wing shape in Drosophila melanogaster. Genetics. 2006;173(3):1417–31. 10.1534/genetics.105.053868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliver B. Genetic control of germline sexual dimorphism in Drosophila. International review of cytology. 2002;219:1–60. 10.1016/s0074-7696(02)19010-3 [DOI] [PubMed] [Google Scholar]
- 55.Leader DP, Krause SA, Pandit A, Davies SA, Dow JAT. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic acids research. 2018;46(D1):D809–d15. 10.1093/nar/gkx976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi S, Lim DS, Chung J. Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in Drosophila. PLoS Genet. 2015;11(5):e1005263. 10.1371/journal.pgen.1005263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–6. 10.1126/science.1100747 [DOI] [PubMed] [Google Scholar]
- 58.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312(5774):734–7. 10.1126/science.1123965 [DOI] [PubMed] [Google Scholar]
- 59.Davies EL, Fuller MT. Regulation of self-renewal and differentiation in adult stem cell lineages: lessons from the Drosophila male germ line. Cold Spring Harb Symp Quant Biol. 2008;73:137–45. 10.1101/sqb.2008.73.063 [DOI] [PubMed] [Google Scholar]
- 60.Yu YV, Li Z, Rizzo NP, Einstein J, Welte MA. Targeting the motor regulator Klar to lipid droplets. BMC cell biology. 2011;12:9. 10.1186/1471-2121-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demarco RS, Eikenes AH, Haglund K, Jones DL. Investigating spermatogenesis in Drosophila melanogaster. Methods. 2014;68(1):218–27. 10.1016/j.ymeth.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bazinet RP, Laye S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nature reviews Neuroscience. 2014;15(12):771–85. 10.1038/nrn3820 [DOI] [PubMed] [Google Scholar]
- 63.Davletov B, Montecucco C. Lipid function at synapses. Current opinion in neurobiology. 2010;20(5):543–9. 10.1016/j.conb.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 64.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. Journal of lipid research. 2004;45(8):1375–97. 10.1194/jlr.R400004-JLR200 [DOI] [PubMed] [Google Scholar]
- 65.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, et al. A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell. 2018;174(4):982-98.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160(1–2):177–90. 10.1016/j.cell.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lithgow GJ, Driscoll M, Phillips P. A long journey to reproducible results. Nature. 2017;548(7668):387–8. 10.1038/548387a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, et al. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5(6):e137. 10.1371/journal.pbio.0050137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445(7125):275–80. 10.1038/nature05382 [DOI] [PubMed] [Google Scholar]
- 70.Goenaga J, Mensch J, Fanara JJ, Hasson E. The effect of mating on starvation resistance in natural populations of Drosophila melanogaster. Evolutionary Ecology. 2011;26(4):813–23. [Google Scholar]
- 71.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457(7226):210–4. 10.1038/nature07536 [DOI] [PubMed] [Google Scholar]
- 72.Deshpande SA, Carvalho GB, Amador A, Phillips AM, Hoxha S, Lizotte KJ, et al. Quantifying Drosophila food intake: comparative analysis of current methodology. Nature methods. 2014;11(5):535–40. 10.1038/nmeth.2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Developmental cell. 2002;2(2):239–49. 10.1016/s1534-5807(02)00117-x [DOI] [PubMed] [Google Scholar]
- 74.Grewal SS. Insulin/TOR signaling in growth and homeostasis: a view from the fly world. The international journal of biochemistry & cell biology. 2009;41(5):1006–10. [DOI] [PubMed] [Google Scholar]
- 75.Teleman AA. Molecular mechanisms of metabolic regulation by insulin in Drosophila. The Biochemical journal. 2009;425(1):13–26. 10.1042/BJ20091181 [DOI] [PubMed] [Google Scholar]
- 76.Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, et al. A hormone-dependent module regulating energy balance. Cell. 2011;145(4):596–606. 10.1016/j.cell.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alic N, Andrews TD, Giannakou ME, Papatheodorou I, Slack C, Hoddinott MP, et al. Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Molecular systems biology. 2011;7:502. 10.1038/msb.2011.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC developmental biology. 2003;3:5. 10.1186/1471-213X-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & development. 2003;17(16):2006–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. Journal of biology. 2003;2(3):20. 10.1186/1475-4924-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rideout EJ, Narsaiya MS, Grewal SS. The Sex Determination Gene transformer Regulates Male–female Differences in Drosophila Body Size. PLoS Genet. 2015;11(12):e1005683. 10.1371/journal.pgen.1005683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Graze RM, Tzeng RY, Howard TS, Arbeitman MN. Perturbation of IIS/TOR signaling alters the landscape of sex-differential gene expression in Drosophila. BMC genomics. 2018;19(1):893. 10.1186/s12864-018-5308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431(7006):316–20. 10.1038/nature02897 [DOI] [PubMed] [Google Scholar]
- 84.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–23. 10.1534/genetics.167.1.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Developmental cell. 2002;2(2):229–38. 10.1016/s1534-5807(01)00119-8 [DOI] [PubMed] [Google Scholar]
- 86.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8(3):224–36. 10.1016/j.cmet.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiefenbock SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. The EMBO journal. 2010;29(1):171–83. 10.1038/emboj.2009.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mukherjee S, Duttaroy A. Spargel/dPGC-1 is a new downstream effector in the insulin-TOR signaling pathway in Drosophila. Genetics. 2013;195(2):433–41. 10.1534/genetics.113.154583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neville MC, Nojima T, Ashley E, Parker DJ, Walker J, Southall T, et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Current biology: CB. 2014;24(3):229–41. 10.1016/j.cub.2013.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: the integration of sex and pattern. Trends in genetics: TIG. 2002;18(10):510–6. 10.1016/s0168-9525(02)02769-5 [DOI] [PubMed] [Google Scholar]
- 91.Billeter JC, Villella A, Allendorfer JB, Dornan AJ, Richardson M, Gailey DA, et al. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Current biology: CB. 2006;16(11):1063–76. 10.1016/j.cub.2006.04.039 [DOI] [PubMed] [Google Scholar]
- 92.Castellanos MC, Tang JC, Allan DW. Female-biased dimorphism underlies a female-specific role for post-embryonic Ilp7 neurons in Drosophila fertility. Development. 2013;140(18):3915–26. 10.1242/dev.094714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al-Anzi B, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. Obesity-blocking neurons in Drosophila. Neuron. 2009;63(3):329–41. 10.1016/j.neuron.2009.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hildreth PE. Doublesex, recessive gene that transforms both males and females of Drosophila inot intersexes. Genetics. 1965;51:659–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nöthiger R, Leuthold M, Andersen N, Gerschwiler P, Grüter A, Keller W, et al. Genetic and developmental analysis of the sex-determining gene ‘double sex’ (dsx) of Drosophila melanogaster. Genetical Research. 1987;50(02). [Google Scholar]
- 96.Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9687–92. 10.1073/pnas.93.18.9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87(6):1079–89. 10.1016/s0092-8674(00)81802-4 [DOI] [PubMed] [Google Scholar]
- 98.Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, et al. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics. 2000;154(2):725–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121(5):785–94. 10.1016/j.cell.2005.04.027 [DOI] [PubMed] [Google Scholar]
- 100.von Philipsborn AC, Jorchel S, Tirian L, Demir E, Morita T, Stern DL, et al. Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Current biology: CB. 2014;24(3):242–51. 10.1016/j.cub.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fagegaltier D, Konig A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, et al. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics. 2014;198(2):647–68. 10.1534/genetics.114.169268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bownes M, Dübendorfer A, Smith T. Ecdysteroids in adult males and females of Drosophila melanogaster. Journal of Insect Physiology. 1984;30(10):823–30. [Google Scholar]
- 103.Parisi MJ, Gupta V, Sturgill D, Warren JT, Jallon JM, Malone JH, et al. Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC genomics. 2010;11:346. 10.1186/1471-2164-11-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annual review of entomology. 2010;55:207–25. 10.1146/annurev-ento-112408-085356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Welte MA, Cermelli S, Griner J, Viera A, Guo Y, Kim DH, et al. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Current biology: CB. 2005;15(14):1266–75. 10.1016/j.cub.2005.06.062 [DOI] [PubMed] [Google Scholar]
- 106.Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell metabolism. 2009;10(6):481–90. 10.1016/j.cmet.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reiff T, Jacobson J, Cognigni P, Antonello Z, Ballesta E, Tan KJ, et al. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. eLife. 2015;4:e06930. 10.7554/eLife.06930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu L, MacKenzie KR, Putluri N, Maletic-Savatic M, Bellen HJ. The Glia-Neuron Lactate Shuttle and Elevated ROS Promote Lipid Synthesis in Neurons and Lipid Droplet Accumulation in Glia via APOE/D. Cell metabolism. 2017;26(5):719-37.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bailey AP, Koster G, Guillermier C, Hirst EM, MacRae JI, Lechene CP, et al. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell. 2015;163(2):340–53. 10.1016/j.cell.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neaves WB. Changes in testicular leydig cells and in plasma testosterone levels among seasonally breeding rock hyrax. Biology of reproduction. 1973;8(4):451–66. 10.1093/biolreprod/8.4.451 [DOI] [PubMed] [Google Scholar]
- 111.Mori H, Christensen AK. Morphometric analysis of Leydig cells in the normal rat testis. The Journal of cell biology. 1980;84(2):340–54. 10.1083/jcb.84.2.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kerr JB, De Kretser DM. Cyclic variations in Sertoli cell lipid content throughout the spermatogenic cycle in the rat. Journal of reproduction and fertility. 1975;43(1):1–8. 10.1530/jrf.0.0430001 [DOI] [PubMed] [Google Scholar]
- 113.Paniagua R, Rodriguez MC, Nistal M, Fraile B, Amat P. Changes in the lipid inclusion/Sertoli cell cytoplasm area ratio during the cycle of the human seminiferous epithelium. Journal of reproduction and fertility. 1987;80(1):335–41. 10.1530/jrf.0.0800335 [DOI] [PubMed] [Google Scholar]
- 114.Freeman DA, Ascoli M. Studies on the source of cholesterol used for steroid biosynthesis in cultured Leydig tumor cells. The Journal of biological chemistry. 1982;257(23):14231–8. [PubMed] [Google Scholar]
- 115.Handler AM. Ecdysteroid titers during pupal and adult development in Drosophila melanogaster. Developmental biology. 1982;93(1):73–82. 10.1016/0012-1606(82)90240-8 [DOI] [PubMed] [Google Scholar]
- 116.Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu J, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome biology. 2004;5(6):R40. 10.1186/gb-2004-5-6-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sgro CM, Partridge L. A delayed wave of death from reproduction in Drosophila. Science. 1999;286(5449):2521–4. 10.1126/science.286.5449.2521 [DOI] [PubMed] [Google Scholar]
- 118.Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, et al. Drosophila germ-line modulation of insulin signaling and lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6368–73. 10.1073/pnas.0709128105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Short SM, Wolfner MF, Lazzaro BP. Female Drosophila melanogaster suffer reduced defense against infection due to seminal fluid components. J Insect Physiol. 2012;58(9):1192–201. 10.1016/j.jinsphys.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Short SM, Lazzaro BP. Reproductive status alters transcriptomic response to infection in female Drosophila melanogaster. G3. 2013;3(5):827–40. 10.1534/g3.112.005306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Savage MJ, Goldberg DJ, Schacher S. Absolute specificity for retrograde fast axonal transport displayed by lipid droplets originating in the axon of an identified Aplysia neuron in vitro. Brain research. 1987;406(1–2):215–23. 10.1016/0006-8993(87)90785-2 [DOI] [PubMed] [Google Scholar]
- 122.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nature neuroscience. 2010;13(5):567–76. 10.1038/nn.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Holtta-Vuori M, Salo VT, Ohsaki Y, Suster ML, Ikonen E. Alleviation of seipinopathy-related ER stress by triglyceride storage. Human molecular genetics. 2013;22(6):1157–66. 10.1093/hmg/dds523 [DOI] [PubMed] [Google Scholar]
- 124.Renvoise B, Malone B, Falgairolle M, Munasinghe J, Stadler J, Sibilla C, et al. Reep1 null mice reveal a converging role for hereditary spastic paraplegia proteins in lipid droplet regulation. Human molecular genetics. 2016;25(23):5111–25. 10.1093/hmg/ddw315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Papadopoulos C, Orso G, Mancuso G, Herholz M, Gumeni S, Tadepalle N, et al. Spastin binds to lipid droplets and affects lipid metabolism. PLoS Genet. 2015;11(4):e1005149. 10.1371/journal.pgen.1005149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmitt F, Hussain G, Dupuis L, Loeffler JP, Henriques A. A plural role for lipids in motor neuron diseases: energy, signaling and structure. Frontiers in cellular neuroscience. 2014;8:25. 10.3389/fncel.2014.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Welte MA. Expanding roles for lipid droplets. Current biology: CB. 2015;25(11):R470–81. 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pennetta G, Welte MA. Emerging Links between Lipid Droplets and Motor Neuron Diseases. Developmental cell. 2018;45(4):427–32. 10.1016/j.devcel.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Welte MA, Gould AP. Lipid droplet functions beyond energy storage. Biochimica et biophysica acta Molecular and cell biology of lipids. 2017;1862(10 Pt B):1260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Isabel G, Martin JR, Chidami S, Veenstra JA, Rosay P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. American journal of physiology Regulatory, integrative and comparative physiology. 2005;288(2):R531–8. 10.1152/ajpregu.00158.2004 [DOI] [PubMed] [Google Scholar]
- 131.Bharucha KN, Tarr P, Zipursky SL. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. The Journal of experimental biology. 2008;211(Pt 19):3103–10. 10.1242/jeb.016451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Galikova M, Diesner M, Klepsatel P, Hehlert P, Xu Y, Bickmeyer I, et al. Energy Homeostasis Control in Drosophila Adipokinetic Hormone Mutants. Genetics. 2015;201(2):665–83. 10.1534/genetics.115.178897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(8):3105–10. 10.1073/pnas.0405775102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Al-Anzi B, Zinn K. Identification and characterization of mushroom body neurons that regulate fat storage in Drosophila. Neural development. 2018;13(1):18. 10.1186/s13064-018-0116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Erion R, DiAngelo JR, Crocker A, Sehgal A. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. The Journal of biological chemistry. 2012;287(39):32406–14. 10.1074/jbc.M112.360875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li Y, Hoffmann J, Li Y, Stephano F, Bruchhaus I, Fink C, et al. Octopamine controls starvation resistance, life span and metabolic traits in Drosophila. Scientific reports. 2016;6:35359. 10.1038/srep35359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhan YP, Liu L, Zhu Y. Taotie neurons regulate appetite in Drosophila. Nature communications. 2016;7:13633. 10.1038/ncomms13633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baumbach J, Hummel P, Bickmeyer I, Kowalczyk KM, Frank M, Knorr K, et al. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell metabolism. 2014;19(2):331–43. 10.1016/j.cmet.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 139.DiAngelo JR, Erion R, Crocker A, Sehgal A. The central clock neurons regulate lipid storage in Drosophila. PloS one. 2011;6(5):e19921. 10.1371/journal.pone.0019921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Etschmaier K, Becker T, Eichmann TO, Schweinzer C, Scholler M, Tam-Amersdorfer C, et al. Adipose triglyceride lipase affects triacylglycerol metabolism at brain barriers. Journal of neurochemistry. 2011;119(5):1016–28. 10.1111/j.1471-4159.2011.07498.x [DOI] [PubMed] [Google Scholar]
- 141.Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell metabolism. 2009;9(3):228–39. 10.1016/j.cmet.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Service PM. Physiological mechanisms of increased stress resistance in Drosophila melanogaster selected for postponed senescence. Physiological Zoology. 1987;60(3):321–6. [Google Scholar]
- 143.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods in enzymology. 2005;393:759–72. 10.1016/S0076-6879(05)93040-1 [DOI] [PubMed] [Google Scholar]
- 144.Isaac RE, Li C, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proceedings Biological sciences. 2010;277(1678):65–70. 10.1098/rspb.2009.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Catterson JH, Khericha M, Dyson MC, Vincent AJ, Callard R, Haveron SM, et al. Short-Term, Intermittent Fasting Induces Long-Lasting Gut Health and TOR-Independent Lifespan Extension. Current biology: CB. 2018;28(11):1714-24.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Harbison ST, Sehgal A. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics. 2008;178(4):2341–60. 10.1534/genetics.107.081232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Aw WC, Garvin MR, Melvin RG, Ballard JWO. Sex-specific influences of mtDNA mitotype and diet on mitochondrial functions and physiological traits in Drosophila melanogaster. PLoS ONE. 2017;12(11):e0187554. 10.1371/journal.pone.0187554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Camus MF, Wolf JB, Morrow EH, Dowling DK. Single Nucleotides in the mtDNA Sequence Modify Mitochondrial Molecular Function and Are Associated with Sex-Specific Effects on Fertility and Aging. Current biology: CB. 2015;25(20):2717–22. 10.1016/j.cub.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 149.Mendelsohn ME, Karas RH. Molecular and cellular basis of cardiovascular gender differences. Science. 2005;308(5728):1583–7. 10.1126/science.1112062 [DOI] [PubMed] [Google Scholar]
- 150.Pan JJ, Fallon MB. Gender and racial differences in nonalcoholic fatty liver disease. World journal of hepatology. 2014;6(5):274–83. 10.4254/wjh.v6.i5.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ballestri S, Nascimbeni F, Baldelli E, Marrazzo A, Romagnoli D, Lonardo A. NAFLD as a Sexual Dimorphic Disease: Role of Gender and Reproductive Status in the Development and Progression of Nonalcoholic Fatty Liver Disease and Inherent Cardiovascular Risk. Advances in therapy. 2017;34(6):1291–326. 10.1007/s12325-017-0556-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lewis EB. A new standard food medium. Drosophila Information Service. 1960;34:118–9. [Google Scholar]
- 153.Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68(1):105–15. 10.1016/j.ymeth.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1139–44. 10.1073/pnas.1113311109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hoekstra LA, Montooth KL. Inducing extra copies of the Hsp70 gene in Drosophila melanogaster increases energetic demand. BMC evolutionary biology. 2013;13:68. 10.1186/1471-2148-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Warton DI, Wright IJ, Falster DS, Westoby M. Bivariate line-fitting methods for allometry. Biological reviews of the Cambridge Philosophical Society. 2006;81(2):259–91. 10.1017/S1464793106007007 [DOI] [PubMed] [Google Scholar]
- 157.Eisenberg DT, Kuzawa CW, Hayes MG. Improving qPCR telomere length assays: Controlling for well position effects increases statistical power. American journal of human biology: the official journal of the Human Biology Council. 2015;27(4):570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, et al. FlyBase 2.0: the next generation. Nucleic Acids Research. 2019;47(D1):D759–65. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Ovary triglyceride levels were not significantly different between fed virgin w1118 females and starved virgin w1118 females at all time points STV (p = 0.56, 0.44, 0.55, respectively; one-way ANOVA followed by Tukey HSD test). (B) The amount of triglyceride contained in the testes of 5-day-old virgin w1118 males is below the limit of detection for the coupled colorimetric assay; therefore, statistics could not be performed. (C) Triglyceride levels in 5-day-old virgin w1118 female carcasses devoid of ovaries were significantly higher than in age-matched male carcasses devoid of testes (p = 0.022; Student t test). (D) Larval fat cells in newly eclosed CS virgin females and males showed no significant change between 0 and 12 hours post-eclosion (p = 0.53 and 0.43 for females and males, respectively; one-way ANOVA followed by Tukey HSD test), but there was a significant decrease in the larval fat cell number in both sexes between 12 and 24 hours post-eclosion (p = 0.0 and 0.0 for females and males, respectively; one-way ANOVA followed by Tukey HSD test). (E) The number of larval fat cells significantly decreased in w1118 virgin females and males between 0 and 12 hours post-eclosion (p = 0.0071 and 1.0 × 10−7 for females and males, respectively; one-way ANOVA followed by Tukey HSD test) and decreased further in females but not males between 12 and 24 hours post-eclosion (p = 0.011 and 0.8 for females and males, respectively; one-way ANOVA followed by Tukey HSD test). (F) After 24 hours of starvation, triglyceride levels in virgin CS females remain at 77% of the triglyceride level in a fed virgin female (p = 0.034; one-way ANOVA followed by Tukey HSD test), whereas virgin CS males have only 4% of the triglyceride level in a fed male remaining (p = 6 × 10−7; one-way ANOVA followed by Tukey HSD test). (G) In 5-day-old virgin female w1118 carcasses devoid of ovaries, there was no significant decrease in triglyceride levels between 0 and 12 hours STV, whereas there was a significant reduction in triglyceride levels in age-matched w1118 virgin male carcasses devoid of testes (p = 0.15 and 0.013, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, there was a male-biased decrease in triglyceride levels in male and female carcasses devoid of gonads (p = 0.00065 and 0.0051, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. COE, coefficient of error; CS, Canton-S; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
(TIF)
(A) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed females compared with fed males for the majority of the intervals during the 24-hour observation period (p = 0.067, 0.0031, 2.4 × 10−4, 4.5 × 10−4, 1.4 × 10−5, 1.7 × 10−7, respectively; Student t test at each time interval). (B) Non-mass-corrected O2 consumption was significantly higher in fed females compared with fed males at all intervals during the observation period (p = 1.5 × 10−5, 1.6 × 10−5, 6.0 × 10−6, 5.8 × 10−5, 1.8 × 10−5, 3.8 × 10−5, respectively; Student t test at each time interval). (C) Non-mass-corrected CO2 production was significantly higher in starved females at every interval post-starvation from 4 hours onward (p = 0.44, 1.3 × 10−5, 5.9 × 10−13, 2.4 × 10−9, 1.9 × 10−7, 1.5 × 10−4, respectively; Student t test at each time interval). (D) Non-mass-corrected O2 consumption was significantly higher in starved females compared with starved males at all time intervals post-starvation (p = 6.0 × 10−12, 1.1 × 10−15, 1.2 × 10−14, 4.3 × 10−10, 1.7 × 10−8, 2.5 × 10−5, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the sexes at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.20, 0.0024, 4.9 × 10−5, 5.2 × 10−5, 1.4 × 10−6, 1.6 × 10−9, respectively; Student t test at each time interval). (B) Non-mass-corrected CO2 production was significantly higher in Oregon-R fed males compared with starved males for most intervals post-starvation during the observation period (p = 0.99, 6.1 × 10−5, 4.7 × 10−10, 2.2 × 10−9, 1.3 × 10−5, 4.1 × 10−9, respectively; Student t test at each time interval). (C) Non-mass-corrected O2 consumption was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.072, 0.0080, 0.0013, 8.1 × 10−4, 7.7 × 10−6, 6.6 × 10−8, respectively; Student t test at each time interval). (D) Non-mass-corrected O2 consumption was significantly higher in fed males compared with starved males at all intervals post-starvation (p = 1.6 × 10−4, 9.6 × 10−6, 1.5 × 10−7, 3.6 × 10−6, 9.3 × 10−6, 4.8 × 10−6, respectively; Student t test at each time interval). (E) Mass-corrected CO2 production was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.55, 0.0026, 4.9 × 10−5, 0.0016, 1.3 × 10−4, 8.1 × 10−9, respectively; Student t test at each time interval). (F) Mass-corrected CO2 production was significantly higher in fed males compared with starved males for most intervals post-starvation during the observation period (p = 0.59, 4.4 × 10−4, 7.5 × 10−10, 2.0 × 10−9, 7.0 × 10−7, 3.0 × 10−10, respectively; Student t test at each time interval). (G) Mass-corrected O2 consumption was significantly higher in fed females compared with starved females for most intervals post-starvation during the observation period (p = 0.053, 0.014, 0.0098, 0.063, 7.6 × 10−4, 6.2 × 10−6, respectively; Student t test at each time interval). (H) Mass-corrected O2 consumption was significantly higher in fed males compared with starved males at all intervals post-starvation (p = 5.0 × 10−5, 2.6 × 10−6, 4.7 × 10−8, 1.0 × 10−6, 8.3 × 10−7, 1.1 × 10−6, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the treatments at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) In fed Oregon-R females and males, we observed no significant differences in the RQ throughout most of the observation period, with the exception of the 4- to 8-hour interval (p = 0.17, 0.031, 0.13, 0.43, 0.58, 0.15, respectively; Student t test at each time interval). (B) In starved Oregon-R females and males, starved males have a significantly higher RQ at all time intervals post-starvation (p = 0.0012, 0.0013, 7.7 × 10−4, 6.6 × 10−5, 0.0013, 0.0032, respectively; Student t test at each time interval). For indirect calorimetry measurements, the p-values are listed in the following order: difference between the sexes at 2–4 hours, 4–8 hours, 8–12 hours, 12–16 hours, 16–20 hours, and 20–22 hours. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S2 Data. ns, no significant difference between two sexes, two genotypes, or time points; RQ, respiratory quotient.
(TIF)
(A) Whole-body protein levels were not significantly different between 5-day-old virgin w1118 males and females at any time point STV (p = 0.16, 0.19, 0.37, respectively; Student t test at each time point). (B) Whole-body glucose levels were not significantly different between the sexes at 0 and 12 hours STV but were significantly higher in females compared with males by 24 hours STV (p = 0.87, 0.48, 0.034, respectively; Student t test at each time point). (C) Whole-body glycogen levels were not significantly different between the sexes at 0 or 12 hours STV but were significantly higher in females compared with males by 24 hours STV (p = 0.86, 0.063, 0.033, respectively; Student t test at each time point). The p-values are listed in the following order: difference between females and males at 0 hours, 12 hours, and 24 hours STV. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S1 Data. ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; w, white.
(TIF)
(A) In normal culture conditions, Agpat4 is female-biased, mdy is male-biased, and PAPLA1 is not sex biasedly expressed when normalized to β-tubulin (p = <0.0001, 0.0079, and 0.25, respectively; Student t test for each gene). (B) In normal culture conditions, Agpat4 is female biased and mdy and PAPLA1 are male biased when normalized to both β-tubulin and β-cop (p = <0.0001, 0.0015, and 0.048, respectively; Student t test for each gene). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S3 Data. β-cop, Coat Protein (coatomer) β; Agpat, 1-acylglycerol-3-phosphate O-acyltransferase; ns, no significant difference between two sexes, two genotypes, or time points; mdy, midway; PAPLA1, phosphatidic acid phospholipase A1.
(TIF)
(A–C) In starvation conditions, Agpat4, mdy, and PAPLA1 female gene expression is significantly increased at 24 hours post-starvation when normalized to β-tubulin (p = <0.0001, <0.0001, and <0.0001, respectively at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (D–F) In starvation conditions, Agpat4 and PAPLA1 female gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin and β-cop (p = >0.05, >0.05, and <0.01 respectively at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (G–I) In starvation conditions, Agpat4 and PAPLA1 male gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin (p = >0.05, >0.05, and <0.001, respectively, at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). (J–L) In starvation conditions, Agpat4 and PAPLA1 male gene expression is not significantly increased at 24 hours post-starvation, whereas mdy is significantly increased at 24 hours post-starvation when normalized to β-tubulin and β-cop (p = >0.05, >0.05, and <0.05, respectively, at 24 hours post-starvation; one-way ANOVA followed by Tukey HSD test for each gene). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for list of all comparisons and p-values; quantitative measurements underlying all graphs are available in S3 Data. β-cop, Coat Protein (coatomer) β; Agpat, 1-acylglycerol-3-phosphate O-acyltransferase; HSD, honest significant difference; mdy, midway; ns, no significant difference between two sexes, two genotypes, or time points; PAPLA1, phosphatidic acid phospholipase A1.
(TIF)
Radar plots demonstrating gene expression in males and females throughout the starvation period for representative genes. (A, B) mRNA levels of two genes with male-biased expression throughout the starvation period in both males and females (Lpin, CG1941). (C, D) mRNA levels of two genes with female-biased expression throughout the starvation period in both males and females (hsl, wun2). Sex-biased expression of these genes remains consistent throughout the starvation period. See S1 Table for list of all comparisons and p-values; quantitative measurements underlying all graphs are available in S3 Data. hsl, hormone-sensitive lipase; Lpin, Lipin; STV, post-starvation; wun2, wunen-2.
(TIF)
(A) Triglyceride levels in 5-day-old bmm1 mutant females fed a high fat diet (HFD) were significantly higher than in bmm1 mutant females fed standard fly food (p = 3.9 × 10−6; Student t test). (B) Triglyceride storage was significantly higher in 5-day-old virgin bmmrev female carcasses lacking ovaries compared with age-matched bmmrev males, whereas there was no significant difference in whole-body triglyceride levels in 5-day-old bmm1 mutant virgin female carcasses devoid of ovaries compared with age-matched bmm1 mutant virgin males (p = 0.00013 in bmmrev animals, 0.12 in bmm1 mutants; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; F, female; HFD, high-fat diet; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A) In 5-day-old virgin da>UAS-bmm-RNAi males, triglyceride levels were significantly higher than in da>+ or +>UAS-bmm-RNAi control males (p = 0.012 and 6.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (B) Triglyceride levels in 5-day-old virgin da>UAS-bmm-RNAi females were not significantly different from da>+ or +>UAS-bmm-RNAi control females (p = 0.98 and 0.16, respectively; one-way ANOVA followed by Tukey HSD test). (C) The male-biased effects of da>UAS-bmm-RNAi on triglyceride storage reduced the sexual dimorphism in triglyceride storage compared with da>+ or +>UAS-bmm-RNAi controls. (D) In 5-day-old virgin da>UAS-bmm-RNAi#2 (BDSC #25926) males, triglyceride levels were significantly higher than in da>+ or +>UAS-bmm-RNAi#2 control males (p = 0.0 and 0.0, respectively; one-way ANOVA followed by Tukey HSD test). (E) Triglyceride levels in 5-day-old virgin da>UAS-bmm-RNAi#2 (BDSC #25926) females were not significantly different from da>+ or +>UAS-bmm-RNAi#2 control females (p = 5.8 × 10−5 and 0.14, respectively; one-way ANOVA followed by Tukey HSD test). (F) The male-biased effects of da>UAS-bmm-RNAi#2 (BDSC #25926) on triglyceride storage reduced the sexual dimorphism in triglyceride storage compared with da>+ or +>UAS-bmm-RNAi#2 controls. (G) Between 0 DPE and 5 DPE, mRNA expression levels for bmm were not significantly increased in virgin w1118 females but were significantly increased in virgin w1118 males (p = 0.17 and 0.048, respectively; Student t test). (H) Between 0 and 12 hours STV, we observed no triglyceride breakdown in da>+, +>UAS-bmm-RNAi, or da>UAS-bmm-RNAi males (p = 0.3, 0.053, and 0.18, respectively; one-way ANOVA followed by Tukey HSD test); however, between 12 and 24 hours STV, the magnitude of triglyceride breakdown in da>UAS-bmm-RNAi males was lower than da>+ and +>UAS-bmm-RNAi control males (p = 0.038, 3.1 × 10−6, and 0.00046, respectively; one-way ANOVA followed by Tukey HSD test). (I) Between 0 and 12 hours STV, and between 12 and 24 hours STV, we observed little change in triglyceride levels in da>+, +>UAS-bmm-RNAi, or da>UAS-bmm-RNAi females (p = 0.36, 0.0024, and 0.64, respectively, for 0–12 hours STV and 0.52, 0.046, and 0.045, respectively, for 12–24 hours STV; one-way ANOVA followed by Tukey HSD test). (J) Between 0 and 12 hours STV we observed similar magnitudes of triglyceride breakdown between da>+, +>UAS-bmm-RNAi#2, and da>UAS-bmm-RNAi#2 (BDSC #25926) males (p = 8.1 × 10−6, 1.8 × 10−6, and 7.1 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test); however, between 12 and 24 hours STV, triglyceride breakdown in da>UAS-bmm-RNAi#2 (BDSC #25926) males was blocked, whereas triglyceride levels decreased in da>+ and +>UAS-bmm-RNAi#2 control males (p = 0.069, 1.3 × 10−6, and 2 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (K) Between 0 and 12 hours STV, triglyceride breakdown in da>UAS-bmm-RNAi#2 (BDSC #25926) females was blocked, whereas triglyceride levels decreased in da>+ and +>UAS-bmm-RNAi#2 control females (p = 0.18, 0.036, and 9.9 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test), whereas we observed similar magnitudes of triglyceride breakdown between 12 and 24 hours STV in da>+, +>UAS-bmm-RNAi#2, and da>UAS-bmm-RNAi#2 (BDSC #25926) females (p = 0.032, 0.28, and 0.022, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat or mRNA expression level represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 and S3 Datas. bmm, brummer; BDSC, Bloomington Drosophila Stock Center; COE, coefficient of error; da, daughterless; DPE, days post-eclosion; F, female; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; w, white.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the fat body (cg>UAS-bmm-RNAi) was significantly higher than age-matched control males (cg>+ and +>UAS-bmm-RNAi) (p = 1.0 × 10−4 and 8.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the fat body (cg>UAS-bmm-RNAi) was significantly higher than age-matched control females (cg>+ and +>UAS-bmm-RNAi) (p = 5.5 × 10−4 and 1.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). (C) There was a significant reduction in control female and male triglyceride levels (cg>+) between 0 and 12 hours post-starvation (p = 2.8 × 10−6 and 1.0 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test); however, we observed no significant reduction in whole-body triglyceride levels in 5-day-old virgin cg>UAS-bmm-RNAi females and males between 0 and 12 hours post-starvation (p = 0.54 and 0.92, respectively; one-way ANOVA followed by Tukey HSD test). (D) There was a significant reduction in triglyceride storage in control females and males (cg>+ and +>UAS-bmm-RNAi) between 12 and 24 hours post-starvation (p = 1 × 10−7 and 2.7 × 10−4 (females) and p = 4.0 × 10−4 and 2 × 10−7 (males), respectively; one-way ANOVA followed by Tukey HSD test); however, there was no significant change in whole-body triglyceride levels in 5-day-old virgin cg>UAS-bmm-RNAi females between 12 and 24 hours post-starvation (p = 1.0; one-way ANOVA followed by Tukey HSD test). In males, there was a significant but blunted decrease in triglyceride levels in 5-day-old cg>UAS-bmm-RNAi virgin males between 12 and 24 hours post-starvation (p = 0.0011; one-way ANOVA followed by Tukey HSD). (E) Whole-body triglyceride storage in 5-day-old r4>UAS-bmm-RNAi males was significantly higher than r4>+ and +>UAS-bmm-RNAi control males (p = 0.0 and 0.0, respectively; one-way ANOVA followed by Tukey HSD test). (F) r4>UAS-bmm-RNAi females did not show a significant difference in whole-body triglyceride storage compared with r4>+ and +>UAS-bmm-RNAi control females (p = 0.95 and 0.00015, respectively; one-way ANOVA followed by Tukey HSD test). (G) Between 0 and 12 hours post-starvation, there was no significant decrease in triglyceride levels in either 5-day-old r4>UAS-bmm-RNAi or +>UAS-bmm-RNAi females (p = 0.42 and 0.096, respectively; one-way ANOVA followed by Tukey HSD test) and a modest but significant reduction in triglyceride level in r4>+ during the same interval (p = 0.00014; one-way ANOVA followed by Tukey HSD test). In males, we observed similar trends for r4>+, +>UAS-bmm-RNAi, and r4>UAS-bmm-RNAi males (p = 1.0 × 10−7, 0.057, and 0.0012, respectively; one-way ANOVA followed by Tukey HSD test). (H) Between 12 and 24 hours post-starvation, we observed that the magnitude of the decrease in triglyceride levels in 5-day-old r4>UAS-bmm-RNAi females and males was blunted compared with the decrease in r4>+ and +>UAS-bmm-RNAi controls for each sex (p = 0.04, 0.00048, and 2.0 × 10−7 for females; 0.0013, 0.12, and 8.2 × 10−5 for males, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting Percent Body Fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; cg, collagen; F, female; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the gut (Mex>UAS-bmm-RNAi) was not significantly different from age-matched control females (Mex>+ and +>UAS-bmm-RNAi) (p = 0.31 and 0.0073, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the gut (Mex>UAS-bmm-RNAi) was not significantly different from age-matched control males (Mex>+ and +>UAS-bmm-RNAi) (p = 0.17 and 0.079, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the muscle (dMef2>UAS-bmm-RNAi) was not significantly different from age-matched control females (dMef2>+ and +>UAS-bmm-RNAi) (p = 0.50 and 0.70, respectively; one-way ANOVA followed by Tukey HSD test). (D) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the muscle (dMef2>UAS-bmm-RNAi) was not significantly different from age-matched control males (dMef2>+ and +>UAS-bmm-RNAi) (p = 0.54 and 0.34, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride level in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the glia (repo>UAS-bmm-RNAi) was not significantly different from age-matched control females (repo>+ and +>UAS-bmm-RNAi) (p = 3.2 × 10−5 and 0.26, respectively; one-way ANOVA followed by Tukey HSD test). (F) Whole-body triglyceride levels in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the glia (repo>UAS-bmm-RNAi) were not significantly different from age-matched control males (repo>+ and +>UAS-bmm-RNAi) (p = 0.016 and 0.8, respectively; one-way ANOVA followed by Tukey HSD test). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control and difference between the GAL4/UAS genotype and the UAS control, respectively. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs represent SEM. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; F, female; HSD, honest significant difference; M, male; Mex, midgut expression 1; Mef2, myocyte enhancer factor 2; ns, no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in males with c587-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in the somatic cells of the male gonad was significantly higher than in control males (c587-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 2.3 × 10−5 and 1.1 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). (B) The decrease in whole-body triglyceride levels in c587-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) males was blunted compared with c587-GAL4>+ and +>UAS-bmm-RNAi#2 control males between 0 and 12 hours STV (p = 0.47, 5.8 × 10−6, and 0.065, respectively; one-way ANOVA followed by Tukey HSD test) but not between 12 and 24 hours STV (p = 8.2 × 10−4, 6.0 × 10−7, and 0.0033, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride levels in 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi males were not significantly different to c587-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls, demonstrating that re-expression of UAS-bmm rescued the increased fat storage caused by loss of bmm in the somatic cells of the male gonad (p = 0.87 and 1.0, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi males and control males (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) was decreased by a similar magnitude between both 0 and 12 hours or 12 and 24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in the somatic cells of the male gonad post-starvation (p = 2.2 × 10−6, 1.7 × 10−6, and 0.0 for 0–12 hours and 0.0, 0.0, and 0.0 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (c587>UAS-bmm-RNAi) was not significantly different from age-matched control females (c587>+ and +>UAS-bmm-RNAi) (p = 0.083 and 0.96, respectively; one-way ANOVA followed by Tukey HSD test). (B) There was a modest but significant reduction in whole-body triglyceride levels in 5-day-old c587>+, +>UAS-bmm-RNAi, and c587>UAS-bmm-RNAi females between 0 and 12 hours STV (p = 2.3 × 10−4, 0.0094, and 0.0051, respectively; one-way ANOVA followed by Tukey HSD test) and between 12 and 24 hours STV (p = 0.0, 0.016, and 1.6 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in females with c587-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in the somatic cells of the gonad was not significantly different from control females (c587-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.98 and 0.78, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among c587-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) females and c587-GAL4>+ and +>UAS-bmm-RNAi#2 controls showed a modest decrease of similar magnitude between both 0 and 12 hours and 12 and 24 hours STV (p = 0.096, 1.3 × 10−4, and 0.0013 for 0–12 hours and 0.004, 0.0049, and 0.022 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride levels in 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi females were not significantly different among c587-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 8.5 × 10−4 and 0.38, respectively; one-way ANOVA followed by Tukey HSD test). (F) Triglyceride breakdown post-starvation among 5-day-old virgin c587>UAS-bmm;UAS-bmm-RNAi females and control females (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) showed a modest decrease of a similar magnitude at both 0–12 hours or 12–24 hours STV (p = 0.0043, 1.7 × 10−6, and 0.0027 for 0–12 hours and 0.0, 2.6 × 10−6, and 2.9 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; F, female; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin males overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (tj>UAS-bmm-RNAi) was not significantly different from age-matched control males (tj>+ and +>UAS-bmm-RNAi) (p = 0.098 and 0.97, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the somatic cells of the gonad (tj>UAS-bmm-RNAi) was not significantly different from age-matched control females (tj>+ and +>UAS-bmm-RNAi) (p = 0.2 and 0.66, respectively; one-way ANOVA followed by Tukey HSD test). (C) Between 0 and 12 hours STV, the magnitude of triglyceride breakdown in tj>UAS-bmm-RNAi was similar to tj>+ and +>UAS-bmm-RNAi control males; however, between 12 and 24 hours STV, there was no significant decrease in triglyceride levels in tj>UAS-bmm-RNAi males, in contrast to tj>+ and +>UAS-bmm-RNAi control males, in which we observed a significant decrease in triglyceride storage STV (p = 9.7 × 10−4, 0.0018, and 0.099 for 0–12 hours and 0.37, 1.9 × 10−5, and 0.028 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (D) Triglyceride breakdown post-starvation among 5-day-old virgin tj>UAS-bmm-RNAi females and tj>+ and +>UAS-bmm-RNAi control females was modestly decreased by a similar magnitude at both 0–12 hours or 12–24 hours STV (p = 0.058, 0.026, and 0.022 for 0–12 hours and 0.042, 0.0093, and 3.7 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). (E–G) We used tj-GAL4 to drive the expression of a membrane-bound GFP (UAS-mCD8::GFP) in the somatic cells of the gonad. The presence of lipid droplets within the GFP-marked boundary of the somatic cell indicates that lipid droplets are present in the somatic cells of the gonad. Non-GFP-positive droplets (arrow) likely represent lipid droplets in the germline cells. The image represents a single confocal slice from the Drosophila male testis. Scale bars = 50 μm, except for inset images, in which scale bars = 12.5 μm. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05; **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. Original image files corresponding to all images acquired from genotype-matched individuals presented in panels E–G are available upon request. bmm, brummer; COE, coefficient of error; F, female; GFP, green fluorescent protein; tj, traffic jam; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; STV, post-starvation; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in males with elav-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in neurons was not significantly different than in control males (elav-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.11 and 0.91, respectively; one-way ANOVA followed by Tukey HSD test). (B) Whole-body triglyceride storage in males with nSyb-GAL4-mediated overexpression of the UAS-bmm-RNAi transgene in neurons was not significantly different than in control males (nSyb-GAL4>+ and +>UAS-bmm-RNAi) (p = 0.19 and 2.1 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (C) The decrease in whole-body triglyceride levels in elav-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) males was similar to elav-GAL4>+ and +>UAS-bmm-RNAi#2 control males between 0 and 12 hours STV (p = 0.01, 4.0 × 10−7, and 1.0 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test). Triglyceride breakdown between 12 and 24 hours STV was blocked in elav-GAL4>UAS-bmm-RNAi#2 males, whereas triglyceride levels in control males during this interval significantly decreased (p = 0.084, 2.6 × 10−5, and 3.3 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (D) The decrease in whole-body triglyceride levels in nSyb-GAL4>UAS-bmm-RNAi males was blunted compared with nSyb-GAL4>+ and +>UAS-bmm-RNAi control males between 0 and 12 hours STV (p = 0.86, 0.051, and 4.3 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test); however, triglyceride breakdown between 12 and 24 hours STV in nSyb-GAL4>UAS-bmm-RNAi males was similar in magnitude to control males (nSyb>+ and +>UAS-bmm-RNAi) (p = 0.002, 1.2 × 10−4, and 7.0 × 10−4, respectively; one-way ANOVA followed by Tukey HSD test). (E) In dissected brains from elav>UAS-bmm-RNAi males, we found that bmm transcript levels were undetectable in three out of four samples, whereas we observed amplification at a higher cycle number in three elav>+ samples. (§) Because of this dramatic decrease in bmm transcript levels in most elav>UAS-bmm-RNAi samples, we were unable to perform a statistical analysis to quantify this effect. (F) Whole-body triglyceride levels in 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi males were not significantly different to elav>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 0.84 and 0.92, respectively; one-way ANOVA followed by Tukey HSD test). (G) Whole-body triglyceride levels post-starvation among 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi males and control males (elav>+ and +>UAS-bmm;UAS-bmm-RNA) were significantly decreased by a similar magnitude at both 0–12 hours or 12–24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in neurons STV (p = 2.3 × 10−6, 1.2 × 10−4, and 0.0 for 0–12 hours and 2.0 × 10−7, 1.0 × 10−7, and 0.0 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat or mRNA expression level represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 and S3 Datas. bmm, brummer; COE, coefficient of error; elav, embryonic lethal abnormal vision; HSD, honest significant difference; M, male; ns, no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Triglyceride breakdown post-starvation was modestly decreased by a similar magnitude in 5-day-old virgin repo>UAS-bmm-RNAi females compared with repo-GAL4>+ and +>UAS-bmm-RNAi control females at 0–12 hours post-starvation (p = 0.047, 2.3 × 10−5, and 0.096, respectively; one-way ANOVA followed by Tukey HSD test) and between repo>UAS-bmm-RNAi males and repo-GAL4>+ and +>UAS-bmm-RNAi control males during the same interval (p = 0.085, 6.9 × 10−6, and 0.057, respectively; one-way ANOVA followed by Tukey HSD test). (B) Between 12 and 24 hours post-starvation, we observed only a modest reduction in female triglyceride levels post-starvation in all genotypes (repo>UAS-bmm-RNAi, repo-GAL4>+, and +>UAS-bmm-RNAi), and the magnitude of this reduction was similar between genotypes (p = 0.0019, 4.8 × 10−6, and 2.0 × 10−7, respectively; one-way ANOVA followed by Tukey HSD test). Similarly, although there was a significant decrease in triglyceride levels post-starvation in repo>UAS-bmm-RNAi males, repo-GAL4>+, and +>UAS-bmm-RNAi controls (p = 8.8 × 10−6, 0.0, and 8.2 × 10−5, respectively; one-way ANOVA followed by Tukey HSD test), the magnitude of this decrease was similar for all genotypes. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). See S1 Table for list of all multiple comparisons and p-values. Error bars on graphs represent COE. Quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; HSD, honest significant difference; ns indicates no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(A) Whole-body triglyceride storage in 5-day-old virgin females overexpressing UAS-bmm-RNAi in the postmitotic neurons (elav>UAS-bmm-RNAi) was not significantly different from age-matched control females (elav>+ and +>UAS-bmm-RNAi) (p = 0.54 and 0.95, respectively; one-way ANOVA followed by Tukey HSD test). (B) There was a significant reduction in whole-body triglyceride levels in 5-day-old elav>+, +>UAS-bmm-RNAi, and elav>UAS-bmm-RNAi females between 0 and 12 hours STV (p = 0.026, 0.0038, and 0.0013, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, 5-day-old elav>+, +>UAS-bmm-RNAi, and elav>UAS-bmm-RNAi females modestly decreased triglyceride levels by similar magnitudes (p = 1.2 × 10−4, 0.15, and 0.0012, respectively; one-way ANOVA followed by Tukey HSD test). (C) Whole-body triglyceride storage in females with elav-GAL4-mediated overexpression of an additional UAS-bmm-RNAi#2 (VDRC #37877) transgene in neurons was not significantly different from control females (elav-GAL4>+ and +>UAS-bmm-RNAi#2) (p = 0.56 and 0.035, respectively; one-way ANOVA followed by Tukey HSD test). (D) There was a modest decrease in triglyceride levels between 0 and 12 hours and 12 and 24 hours STV among elav-GAL4>UAS-bmm-RNAi#2 (VDRC #37877) females and elav-GAL4>+ and +>UAS-bmm-RNAi#2 controls (p = 0.011, 0.0027, and 1.4 × 10−6 for 0–12 hours STV and 4.6 × 10−4, 3.4 × 10−4, and 0.002 for 12–24 hours STV, respectively; one-way ANOVA followed by Tukey HSD test). (E) Whole-body triglyceride levels in 5-day-old virgin nSyb>UAS-bmm-RNAi females were not significantly different to nSyb-GAL4>+ and +>UAS-bmm-RNAi controls (p = 0.85 and 0.52, respectively; one-way ANOVA followed by Tukey HSD test). (F) Triglyceride breakdown post-starvation was modestly decreased by a similar magnitude among genotypes in 5-day-old virgin nSyb>UAS-bmm-RNAi females and control females (nSyb>+ and +>UAS-bmm-RNAi) between 0 and 12 hours STV (p = 0.21, 0.017, and 0.43, respectively; one-way ANOVA followed by Tukey HSD test). Between 12 and 24 hours STV, the decrease in triglyceride levels in nSyb>UAS-bmm-RNAi females was blocked, whereas triglyceride levels decreased in control nSyb-GAL4>+ and +>UAS-bmm-RNAi females during this interval (p = 0.093, 3.0 × 10−5, and 0.0019, respectively; one-way ANOVA followed by Tukey HSD test). (G) Whole-body triglyceride levels in 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi females were not significantly different to elav-GAL4>+ and +>UAS-bmm;UAS-bmm-RNAi controls (p = 0.41 and 0.66, respectively; one-way ANOVA followed by Tukey HSD test). (H) Whole-body triglyceride levels post-starvation among 5-day-old virgin elav>UAS-bmm;UAS-bmm-RNAi females and control females (elav>+ and +>UAS-bmm;UAS-bmm-RNAi) decreased by a similar magnitude between 0 and 12 hours or 12 and 24 hours STV, demonstrating that re-expression of UAS-bmm rescued the effects of bmm loss in neurons STV (p = 0.0023, 0.0012, and 0.0027 for 0–12 hours and 1.0 × 10−6, 5.2 × 10−6, and 2.9 × 10−4 for 12–24 hours, respectively; one-way ANOVA followed by Tukey HSD test). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars on graphs depicting percent body fat represent SEM; error bars on graphs depicting the change in percent body fat represent COE. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S1 Data. bmm, brummer; COE, coefficient of error; elav, embryonic lethal abnormal vision; F, female; HSD, honest significant difference; ns, no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; STV, post-starvation; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A) Median survival post-starvation was significantly higher in 5-day-old virgin Oregon-R females than in virgin Oregon-R males (p = 2 × 10−16; Log-rank test with Bonferroni correction for multiple comparison; n > 156). (B) Median survival post-starvation was significantly higher in 5-day-old virgin CMW wild-caught females than in virgin CMW males (p = 6 × 10−10; Log-rank test with Bonferroni correction for multiple comparison; n > 118). (C, D) Median survival post-starvation was significantly higher in 5-day-old virgin females than males in two isofemale strains (Mel c2.2: p = 4.4 × 10−11; Log-rank test with Bonferroni correction for multiple comparison; n > 55 and Mel c2.3: p = 1.4 × 10−13; Log-rank test with Bonferroni correction for multiple comparison; n > 110). (E, F) Median survival post-starvation was significantly higher in virgin males (E) and females (F), with ubiquitous overexpression of UAS-bmm-RNAi compared with control males (da>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16 respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 223) and females (p = 2 × 10−16 and 1.2 × 10−13 respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 176). (G, H) Median survival post-starvation was significantly higher in virgin males (G) and significantly lower in females (H) with ubiquitous overexpression of UAS-bmm-RNAi#2 (BDSC #25926) compared to control males (da>+ and +>UAS-bmm-RNAi#2) (p = 7.7 × 10−11 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 97) and control females (p = 9.6 × 10−9 and 4.9 × 10−5, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 91). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; BDSC, Bloomington Drosophila Stock Center; CMW, Country Mill Winery; da, daughterless; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A) Males with whole-body loss of brummer have a significantly decreased number of progeny after 2 days, 4 days, and 6 days of mating (p = 2.2 × 10−16, 9.7 × 10−12, and 4.6 × 10−6, respectively; Student t test at each time point). Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). See S1 Table for list of all multiple comparisons and p-values. Error bars on graphs represent SEM. Quantitative measurements underlying all graphs are available in S4 Data. M, male; ns, no significant difference between two sexes, two genotypes, or time points.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin females (A) and males (B) with fat body–specific bmm inhibition (cg>UAS-bmm-RNAi) compared with control females (cg>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 279) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 365). (C, D) Median survival post-starvation was significantly higher in virgin females (C) and males (D) with fat body–specific bmm inhibition using a second GAL4 driver (r4>UAS-bmm-RNAi) compared with control females (r4>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 314) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 195). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; cg, collagen; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; UAS, upstream activation sequence.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin males (A) but not significantly different in virgin females (B) with bmm inhibition in the somatic cells of the gonad (c587>UAS-bmm-RNAi#2 [VDRC #37877]) compared with control males (c587>+ and +>UAS-bmm-RNAi#2) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 212) and females (p = 2 × 10−16 and 1.0, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 201). (C, D) Median survival post-starvation was not significantly changed when UAS-bmm and UAS-bmm-RNAi were simultaneously overexpressed by a driver for the somatic cells of the gonad (c587>UAS-bmm;UAS-bmm-RNAi) in both males (C) and females (D) compared with control males (c587>+ and +>UAS-bmm;UAS-bmm-RNAi) (p = 2 × 10−16 and 0.57, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 249) and females (p = 9.6 × 10−16 and 0.0035, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 207). (E, F) Median survival post-starvation was significantly increased when UAS-bmm-RNAi was overexpressed by a second driver for the somatic cells of the gonad (tj>UAS-bmm-RNAi) in both females (E) and males (F) compared with control males (tj>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 1.3 × 10−6, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 165) and females (p = 2 × 10−16 and 0.00012, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 177). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; F, female; M, male; ns, no significant difference between two sexes, two genotypes, or time points; tj, traffic jam; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A, B) Median survival post-starvation was significantly higher in virgin males (A) but not significantly different in virgin females (B) with bmm inhibition in the neurons (elav>UAS-bmm-RNAi#2 [VDRC #37877]) compared with control males (elav>+ and +>UAS-bmm-RNAi#2) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 90) and females (p = 3.4 × 10−6 and 0.41, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 81). (C, D) Median survival post-starvation was not significantly changed when UAS-bmm and UAS-bmm-RNAi were simultaneously overexpressed by a neuronal driver (elav>UAS-bmm;UAS-bmm-RNAi) in both males (C) and females (D) compared with control males (elav>+ and +>UAS-bmm;UAS-bmm-RNAi) (p = 4.6 × 10−5 and 1, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 101) and females (p = 8.9 × 10−6 and 1, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 23). (E, F) Median survival post-starvation was significantly increased when UAS-bmm-RNAi was overexpressed by a second neuronal driver (nSyb>UAS-bmm-RNAi) in both males (E) and females (F) compared with control males (nSyb>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 157) and females (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparisons; n > 157). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; elav, embryonic lethal abnormal vision; F, female; M, male; ns indicates no significant difference between two sexes, two genotypes, or time points; nSyb, neuronal Synaptobrevin; UAS, upstream activation sequence; VDRC, Vienna Drosophila Resource Center.
(TIF)
(A, B) Median survival post-starvation showed no significant change in virgin females (A) and males (B) with gut-specific bmm inhibition (Mex>UAS-bmm-RNAi) compared with control females (Mex>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 1.1 × 10−7, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 444) and males (p = 2 × 10−16 and 0.13 respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 456). (C, D) Median survival post-starvation was unchanged in virgin females (C) and slightly increased in males (D) with muscle-specific bmm inhibition (dMef2>UAS-bmm-RNAi) compared with control females (dMef2>+ and +>UAS-bmm-RNAi) (p = 2 × 10−16 and 0.4, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 340) and males (p = 2 × 10−16 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 382). (E, F) Median survival post-starvation showed no significant change in virgin females (E) and males (F) with glia-specific bmm inhibition (repo>UAS-bmm-RNAi) compared with control females (repo>+ and +>UAS-bmm-RNAi) (p = 0.43 and 2 × 10−16, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 191) and males (p = 2 × 10−16 and 0.02, respectively; Log-rank test with Bonferroni correction for multiple comparison; n > 207). The p-values are listed in the following order: difference between the GAL4/UAS genotype and the GAL4 control/difference between the GAL4/UAS genotype and the UAS control. Asterisks indicate a significant difference between two sexes, two genotypes, or two time points (*p < 0.05, **p < 0.01, ***p < 0.001). Shaded areas represent the 95% confidence interval. See S1 Table for a list of all multiple comparisons and p-values; quantitative measurements underlying all graphs are available in S4 Data. bmm, brummer; F, female; M, male; Mex, midgut expression 1; dMef2, myocyte enhancer factor 2, ns, no significant difference between two sexes, two genotypes, or time points; repo, reversed polarity; UAS, upstream activation sequence.
(TIF)
(XLSX)
(XLSX)
GFP, green fluorescent protein.
(XLSX)
(XLSX)
(CSV)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files: S1 Data, S2 Data, S3 Data, and S4 Data. Original image files corresponding to all images in the paper are available upon request.