Abstract
Plant cells are encased in a semi-rigid cell wall of complex build. As a consequence, cell wall remodeling is essential for the control of growth and development as well as the regulation of abiotic and biotic stress responses. Plant cells actively sense physico-chemical changes in the cell wall and initiate corresponding cellular responses. However, the underlying cell wall monitoring mechanisms remain poorly understood. In Arabidopsis the atypical receptor kinase STRUBBELIG (SUB) mediates tissue morphogenesis. Here, we show that SUB-mediated signal transduction also regulates the cellular response to a reduction in the biosynthesis of cellulose, a central carbohydrate component of the cell wall. SUB signaling affects early increase of intracellular reactive oxygen species, stress gene induction as well as ectopic lignin and callose accumulation upon exogenous application of the cellulose biosynthesis inhibitor isoxaben. Moreover, our data reveal that SUB signaling is required for maintaining cell size and shape of root epidermal cells and the recovery of root growth after transient exposure to isoxaben. SUB is also required for root growth arrest in mutants with defective cellulose biosynthesis. Genetic data further indicate that SUB controls the isoxaben-induced cell wall stress response independently from other known receptor kinase genes mediating this response, such as THESEUS1 or MIK2. We propose that SUB functions in a least two distinct biological processes: the control of tissue morphogenesis and the response to cell wall damage. Taken together, our results reveal a novel signal transduction pathway that contributes to the molecular framework underlying cell wall integrity signaling.
Author summary
Plant cells are encapsulated by a semi-rigid and biochemically complex cell wall. This particular feature has consequences for multiple biologically important processes, such as cell and organ growth or various stress responses. For a plant cell to grow the cell wall has to be modified to allow cell expansion, which is driven by outward-directed turgor pressure generated inside the cell. In return, changes in cell wall architecture need to be monitored by individual cells, and to be coordinated across cells in a growing tissue, for an organ to attain its regular size and shape. Cell wall surveillance also comes into play in the reaction against certain stresses, including for example infection by plant pathogens, many of which break through the cell wall during infection, thereby generating wall-derived factors that can induce defense responses. There is only limited knowledge regarding the molecular system that monitors the composition and status of the cell wall. Here we provide further insight into the mechanism. We show that the cell surface receptor STRUBBELIG, previously known to control organ development in Arabidopsis, also promotes the cell’s response to reduced amounts of cellulose, a main component of the cell wall.
Introduction
Cell-cell communication is essential to regulate cellular behavior during many processes, including growth, development, and stress responses. In plants, the extra-cellular cell wall constitutes a central element of the underlying molecular mechanisms. It is mainly composed of carbohydrates, such as cellulose, hemicellulose, and pectin, and phenolic compounds, including lignin. Moreover, the cell wall also contains a plethora of different cell-wall-bound proteins [1,2]. It imposes restrictions on cell expansion and the movement of cells and serves as a barrier to pathogen attack. The cell wall counteracts turgor-driven growth and thus cell wall remodeling is required for cell expansion [3]. Cell wall fragments released by pathogen-derived lytic enzymes can act as danger signals and elicit plant immunity responses [4]. These observations imply a necessity for plant cells to monitor cell wall integrity (CWI). Such a mechanism would sense any physico-chemical alterations that occurred in the cell wall and elicit a corresponding compensatory and protective cellular response [5–8].
Little is known about the molecular mechanisms that reside at the nexus of monitoring the cell wall status and the control of development and stress responses. Only a few cell surface signaling factors are presently implicated in monitoring CWI [5,8,9]. For example, a complex between RECEPTOR-LIKE PROTEIN44 (RLP44) and BRASSINOSTEROID INSENSITIVE1 (BRI1), the brassinosteroid receptor, specifically connects BRI1-mediated signaling to the detection of pectin modifications [10,11]. In addition, members of the Catharanthus roseus Receptor-like Kinase1-like (CrRLK1L) family of receptor kinases (RKs) are implied in CWI signaling. Recent evidence indicates that a cell wall sensing mechanism depending on a complex between FERONIA (FER) [12] and extracellular leucine-rich repeat extensins (LRXs) negatively regulates vacuole size [13]. Interestingly, the extracellular domain of FER binds pectin in vitro and FER is also required to prevent cell bursting upon exposure of root cells to salt [14]. Signaling mediated by ANXUR1 (ANX1) and ANX2 appears to contribute to monitoring cell wall integrity and the prevention of the premature burst of pollen tubes [15–18]. ERULUS (ERU), a CrRLK1L family member with a role in root hair growth regulation [19,20], was shown to be involved in cell wall remodeling during root hair expansion [21].
Plant cells also respond to changes in cellulose levels in the cell wall. Cellulose is present in the form of microfibrils that constitute the main load-bearing elements resisting turgor pressure. The microfibrils are embedded in matrix polysaccharides, mainly various hemicelluloses and pectins [1,2]. Cellulose is synthesized by cellulose synthase (CESA) complexes at the plasma membrane (PM) [22]. The effects of a reduced production of cellulose on plant growth and development can be studied by analyzing mutants with defects in genes encoding CESA subunits involved in primary cell wall biosynthesis [23–27]. Alternatively, pharmacological approaches can be applied. The herbicide isoxaben is a well-characterized inhibitor of cellulose biosynthesis [28,29]. A number of findings suggest CESAs to be the direct targets of isoxaben. First, several known isoxaben-resistant mutants carry mutations near the carboxyl terminus of certain CESA subunits [30,31]. Second, isoxaben induces a rapid clearing of CESA complexes from the PM [32]. Third, isoxaben uptake or detoxification appears unaffected in resistant plants [33].
The reaction of liquid culture-grown seedlings to isoxaben-induced cellulose biosynthesis inhibition (CBI) represents a thoroughly studied stress response to cell wall damage (CWD) [34–37]. The response is sensitive to osmotic support and eventually includes the upregulation of stress response genes, the production of reactive oxygen species (ROS), an accumulation of phytohormones, such as jasmonic acid (JA), changes in cell wall composition, including the production of ectopic lignin and callose, and finally growth arrest. Similar effects were also observed when studying the phenotypes of different cesA mutants [23–27].
The mechanism controlling the CWD response to CBI is known to involve three RKs [34]. THESEUS1 (THE1), another member of the CrRLK1L family, was first implicated in this process [38]. THE1 was identified based on its genetic interaction with PROCUSTE1 (PRC1), a gene encoding a CESA6 subunit [27]. Amongst others, cellulose-deficient prc1 single mutants exhibit reduced hypocotyl length and ectopic lignin accumulation. In the1 prc1 double mutants these effects are ameliorated although cellulose levels remain reduced [38]. Moreover, THE1 is required for the altered expression levels of several stress-response genes upon exposing liquid-grown seedlings to isoxaben [39]. Recently, the leucine-rich repeat (LRR)-XIIb family RK MALE DISCOVERER1-INTERACTING RECEPTOR LIKE KINASE 2/LEUCINE-RICH REPEAT KINASE FAMILY PROTEIN INDUCED BY SALT STRESS (MIK2/LRR-KISS) [40,41] and the LRR-XIII family member FEI2 [42] have also been shown to participate in the isoxaben-induced cell wall stress response [34,39,43]. Genetic analysis revealed that THE1 and MIK2 have overlapping but also distinct functions, suggesting a complex regulation of the CBI response, with THE1 and MIK2 promoting this response via different mechanisms. FEI2 appears to be part of the THE1 genetic pathway [34].
Tissue morphogenesis in Arabidopsis requires signaling mediated by the atypical LRR-RK STRUBBELIG (SUB). SUB, also known as SCRAMBLED (SCM), is a member of the LRR-V family of RKs and controls several developmental processes, including floral morphogenesis, integument outgrowth, leaf development and root hair patterning [44–46]. SUB represents an atypical receptor kinase, as its in vivo function does not require enzymatic activity of its kinase domain [44,47]. Our previous studies indicate that SUB not only localizes to the PM but is also present at plasmodesmata (PD), channels interconnecting most plant cells [48,49], where it physically interacts with the PD-specific C2 domain protein QUIRKY (QKY) [50].
Current data also associate SUB signaling to cell wall biology. For example, whole-genome transcriptomics analysis revealed that many genes responsive to SUB-mediated signal transduction relate to cell wall remodeling [51]. Moreover, Fourier-transform infrared spectroscopy (FTIR)-analysis indicated that flowers of sub and qky mutants share overlapping defects in cell wall biochemistry [52]. Thus, apart from functionally connecting RK-mediated signal transduction and PD-dependent cell-cell communication SUB signaling also relates to cell wall biology.
Here, we report on a further exploration of the connection between the cell wall and SUB function. Our data reveal a novel role for SUB signaling in the CBI-induced CWD response. We show that SUB affects several processes, such as ROS accumulation, stress gene induction as well as ectopic lignin and callose accumulation, that are initiated upon application of exogenous isoxaben. Moreover, SUB signaling is necessary for maintaining cell shape and recovery of root growth after transient exposure to isoxaben. Our genetic data further indicate that SUB, THE1, and MIK2 act in different pathways and that not all contributions of SUB to CBI-induced CWD signaling require QKY function.
Results
In light of the connection between SUB signaling and cell wall biology, we set out to address if SUB plays a role in the seedling responses to cell wall stress. In particular, we focused on the possible role of SUB in the isoxaben-induced CWD response.
SUB does not affect cellulose production
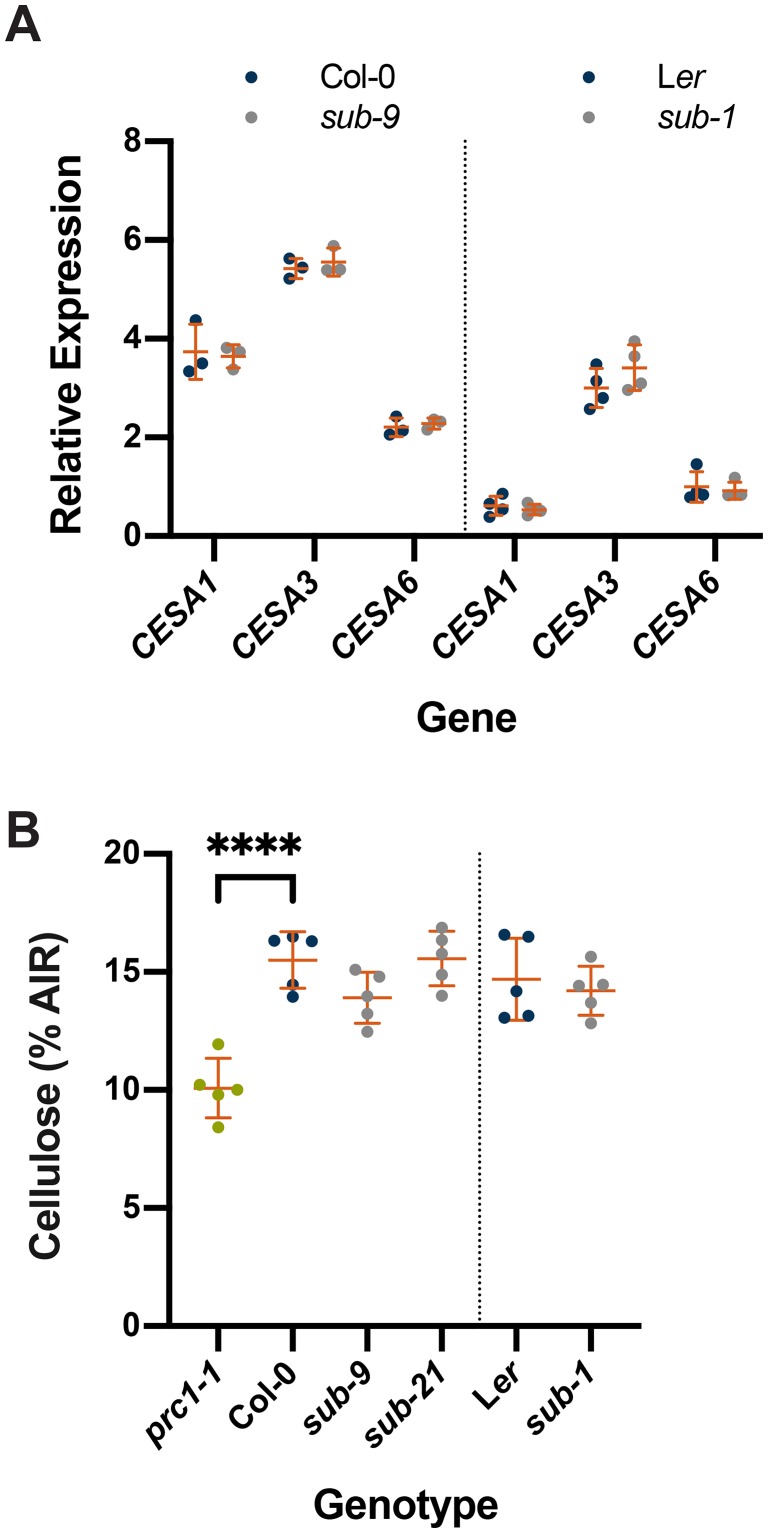
We first investigated if SUB influences cellulose biosynthesis in seven days-old seedlings. CESA1, CESA3 and CESA6 genes encode the CESA isoforms present in CSCs of the primary cell wall [53,54]. We first analyzed transcript levels of CESA1, CESA3, and CESA6 in wild-type and different sub mutants. We could not detect differences in transcript levels of CESA1, CESA3, and CESA6 between sub and wild type in quantitative real-time polymerase chain reaction (qPCR) experiments (Fig 1A). Moreover, we assessed the levels of cellulose. To this end we generated a predicted null allele of SUB, sub-21, by a CRISPR-based approach to have available two independent strong sub alleles in the Col-0 background (sub-21, sub-9; see Materials and methods). We did not detect differences between several sub mutants and wild type (Fig 1B). However, we could observe a reduction in cellulose levels in plants with a defect in CESA6 (prc1-1) that was comparable to previous findings [27] (Fig 1B). The results indicate that SUB does not play a central role in cellulose biosynthesis in seedlings.
Fig 1. Effects of SUB on cellulose content in seven-day-old seedlings.
(A) Gene expression levels for primary cell wall CESA genes in wild type and sub as assessed by qPCR. Individual CESA genes and genotypes are indicated. Mean ± SD is shown. Data points designate results of individual biological replicates. For each tested CESA gene no statistically significant deviation between wild type and mutant was observed (unpaired t tests, two-tailed P values). The experiment was performed two times with similar results. (B) Estimation of cellulose content. Genotypes are indicated. Data points indicate results of individual biological replicates. Mean ± SD is shown. Col, sub-9, sub-21, prc1-1: asterisks represent adjusted P value (**** P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). Ler, sub-1: no statistically significant difference (unpaired t test, two-tailed P values). Note absence of significant differences between wild type and different sub mutants. The prc1-1 mutant shows the expected reduction in cellulose content [27]. The experiment was repeated three times with similar results.
SUB affects the isoxaben-induced CWD response
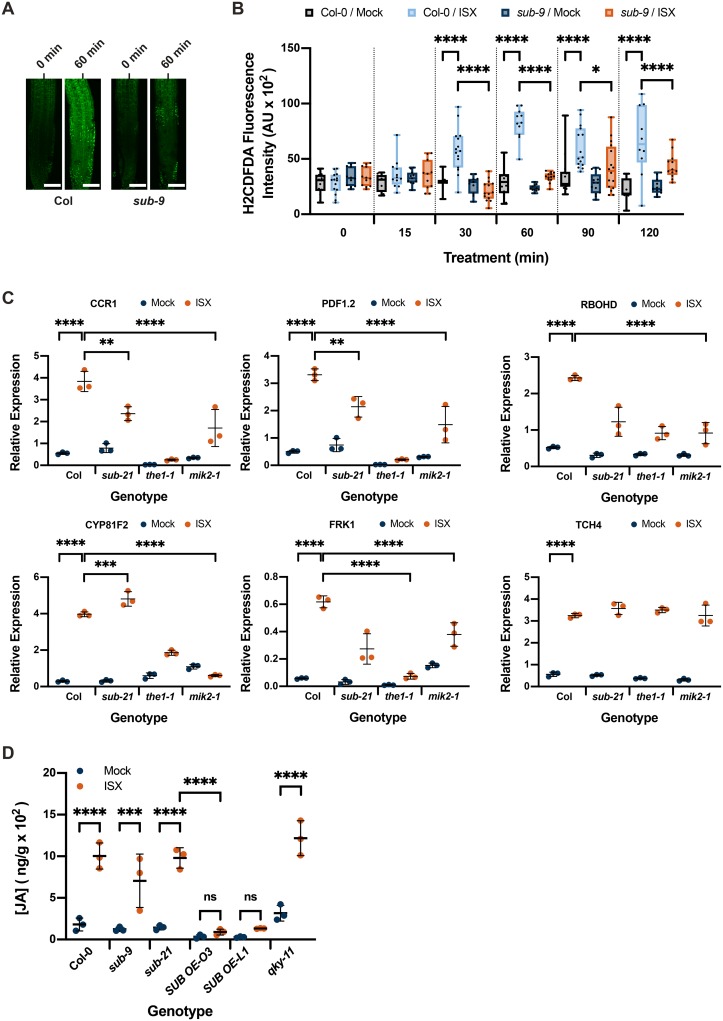
We then assessed if SUB activity is necessary for accumulation of reactive oxygen species (ROS) in response to isoxaben-induced CWD. To this end we exposed seven-day wild-type and sub-9 seedlings, grown on half-strength Murashige and Skoog (MS) plates containing one percent sucrose, to 600 nM isoxaben in a time-course experiment. Seedlings were monitored for up to 120 minutes, at 30 minutes intervals. Upon treatment we assessed fluorescence intensity of the intracellular ROS probe H2DCFDA in roots [55,56]. In wild-type Col-0 seedlings treated for 30 minutes with isoxaben, we noticed an increase in H2DCFDA signal compared with mock-treated seedlings (Fig 2A and 2B). Signal intensity of the probe increased further in seedlings exposed to isoxaben for 60 minutes. This signal intensity remained for up to 120 minutes of continuous exposure to isoxaben. In sub-9 seedlings we detected a slightly and statistically insignificant increased H2DCFDA signal after 60 to 90 minutes of exposure to isoxaben (Fig 2A and 2B). However, signal intensity was noticeably reduced in comparison to wild type. In comparison to the mock control, isoxaben-treated sub-9 seedlings continued to show slightly enhanced signal intensity for up to 120 minutes of exposure, although the relative difference to signal levels of mock-treated seedlings was never as pronounced as in wild type. Thus, in comparison to wild type, sub-9 mutants showed a delayed onset of H2CDFDA signal appearance and an overall reduced signal intensity for the time frame analyzed. The results indicate that isoxaben causes the formation of intracellular ROS in roots of treated wild-type seedlings within 30 minutes of application. Moreover, SUB affects this ROS response.
Fig 2. SUB effects on isoxaben-induced ROS production, marker gene expression and JA accumulation.
(A) Confocal micrographs showing H2CDFDA signal in root tips of six-day-old seedlings exposed to 600 nM isoxaben for the specified time. Genotypes are indicated. Note reduced signal in sub-9. (B) Quantification of results depicted in (A). Genotypes are indicated. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 10 ≤ n ≤ 15. Asterisks represent adjusted P values (**** P < 0.0001; * P < 0.05; one-way ANOVA followed by Tukey’s multiple comparison tests). No statistically significant differences were measured within or between genotypes during the first 15 minutes of mock or isoxaben treatment or between wild type and sub-9 mock treatment at any tested time point (P > 0.9995; one-way ANOVA followed by Tukey’s multiple comparison tests). Experiments were performed three times with similar results. (C) Gene expression levels of several CBI marker genes by qPCR upon exposure of seven-day-old seedlings to 600 nM isoxaben for eight hours. The results from three biological replicates are shown. Marker genes and genotypes are indicated. Mean ± SD is presented. Asterisks represent adjusted P values (*** P < 0.0033, **** P < 0.0001, one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was repeated twice with similar results. (D) JA accumulation. Mean ± SD is presented. Genotypes and treatments are indicated. The JA concentration of the isoxaben-treated Col-0, sub-9, sub-21 and qky-11 mutants did not vary significantly. Asterisks represent adjusted P values (*** P < 0.001, **** P < 0.0001, ns: not significant; one-way ANOVA followed by Tukey’s multiple comparison tests). Experiments were performed three times with similar results. Abbreviation: ISX, isoxaben. Scale bars: (A) 100 μm.
Next, we tested if SUB activity is required for the transcriptional regulation of several marker genes, known to respond to isoxaben-induced CWD within eight hours [34,37,39]. We performed quantitative real-time polymerase chain reaction (qPCR) experiments using RNA isolated from seven days-old liquid-grown seedlings that had been incubated with 600 nM isoxaben for eight hours (Fig 2C, S1 Fig). We observed that isoxaben-induced upregulation of CCR1, PDF1.2, and RBOHD was attenuated in sub-21 (Fig 2C) and sub-9 (S1 Fig) mutants compared to wild type. Moreover, we detected a somewhat enhanced upregulation of CYP81F2 in isoxaben-treated seedlings in comparison to wild type. We detected a statistically significant alteration in the upregulation of FRK1 for sub-21 but not for sub-9. We did not observe an effect of SUB on TCH4 induction by isoxaben (Fig 2C, S1 Fig). These results indicate that SUB affects the isoxaben-induced upregulation of expression of several marker genes.
The seedlings’ response to isoxaben also includes the accumulation of the phytohormone JA [37]. We thus tested if SUB affects the isoxaben-induced production of JA in seven days-old liquid-grown seedlings that had been incubated in 600 nM isoxaben for seven hours. We found that JA accumulation appeared largely unaffected in sub-9 or sub-21 mutants while two overexpressing lines (pUBQ::SUB:mCherry, Lines O3 and L1, (S2 Fig)) showed strongly diminished JA levels following isoxaben treatment (Fig 2D). The results indicate that SUB is not required for isoxaben-induced JA accumulation. The phenotypes of the loss-of-function and overexpressing mutants are not easy to reconcile with each other rendering an interpretation of the effects on JA accumulation seen in the SUB overexpressing lines difficult. Thus, their biological relevance needs to be assessed in further experiments.
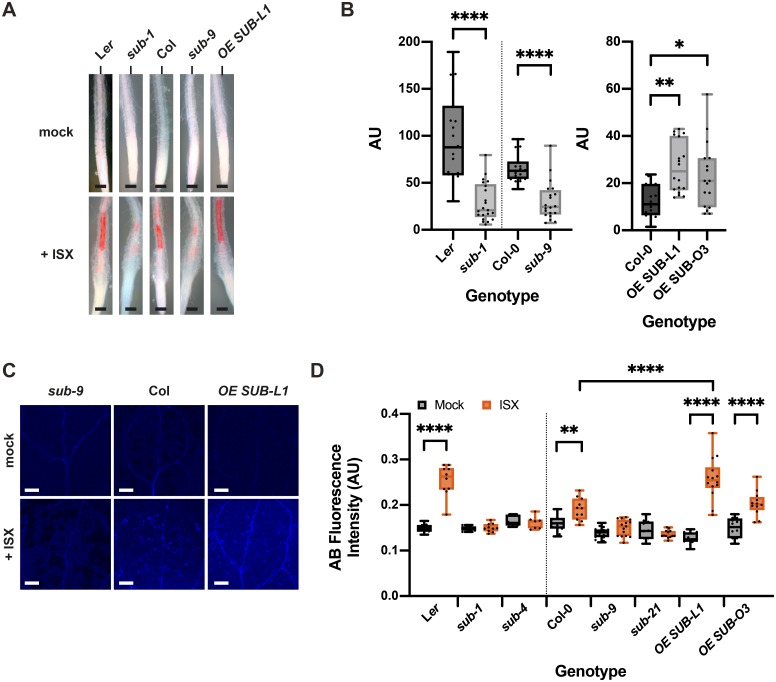
Isoxaben-induced CBI eventually results in the alteration of cell wall biochemistry as evidenced by the ectopic accumulation of lignin and callose [37]. To investigate if SUB affects lignin biosynthesis, we estimated lignin accumulation in roots using phloroglucinol staining after exposing six-day-old liquid-grown seedlings to 600 nM isoxaben for 12 hours. We observed reduced phloroglucinol staining in the root elongation zone of sub-1 seedlings in comparison to wild type Ler indicating less ectopic lignin production (Fig 3A and 3B). We also noticed reduced phloroglucinol signal in sub-9 seedlings (Col-0 background) although the effect was less prominent. However, in our hands Col-0 wild-type plants exhibited an overall weaker phloroglucinol staining indicating that isoxaben-induced lignin accumulation does not occur to the same level as in Ler (Fig 3A and 3B). We could detect increased phloroglucinol staining in two pUBQ::SUB:mCherry lines (L1, O3) (Fig 3A and 3B).
Fig 3. SUB affects isoxaben-induced lignin and callose accumulation.
(A) Phloroglucinol signal strength indicating lignin accumulation in roots of six-day-old seedlings exposed to 600 nM isoxaben for 12 hours. Genotypes: Ler, and sub-1 (Ler); Col, sub-9 (Col), pUBQ::gSUB:mCherry (line OE SUB L1). (B) Quantification of the results depicted in (A). Left panel shows results obtained from different sub mutants in the Ler or Col background. Right panel depicts results from two independent pUBQ::gSUB:mCherry transgenic lines overexpressing SUB (Col, lines OE SUB L1, O3). Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 16 ≤ n ≤ 21. Asterisks represent statistical significance. Left panel: P < 0.0001; unpaired t test, two-tailed P values. Right panel: adjusted P values; * P < 0.03, ** P < 0.002; one-way ANOVA followed by Tukey’s multiple comparison tests. The experiment was performed three times with similar results. (C) Confocal micrographs show cotyledons of seven-day-old sub-9, Col, and pUBQ::gSUB:mCherry (line L1) seedlings treated with mock or 600 nM isoxaben for 24 hours. Aniline blue fluorescence signal strength indicates callose accumulation. (D) Quantification of the results depicted in (C). Left panel shows results obtained from sub mutants in Ler background. Right panel indicates results obtained from sub mutants and two independent pUBQ::gSUB:mCherry transgenic lines overexpressing SUB (lines L1, O3) in Col background. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 7 ≤ n ≤ 18. Asterisks represent adjusted P values (** P < 0.002, **** P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed three times with similar results. Abbreviation: ISX, isoxaben. Scale bars: (A) 0.1 mm; (C) 0.2 mm.
Isoxaben-treatment for 24 hours results in the formation of callose in cotyledons of wild-type seedlings [37]. Thus, we tested if SUB is required for this process as well. To this end we transferred seven days-old plate-grown seedlings to liquid medium without isoxaben for 12 hours. Subsequently, medium was exchanged, and seedlings were kept in 600 nM isoxaben for another 24 hours followed by callose detection using aniline blue staining [57]. As expected, we observed prominent aniline blue staining in cotyledons of Ler and Col wild-type seedlings upon isoxaben treatment (Fig 3C and 3D). By contrast, we detected strongly reduced aniline blue staining in cotyledons of isoxaben-treated sub-1 and sub-4 (both in Ler) as well as sub-9 and sub-21 (both in Col). One of the two SUB overexpressors (line L1) showed statistically significant stronger aniline blue staining compared to wild type upon application of isoxaben (Fig 3C and 3D).
Taken together, the results indicate that SUB is required for the isoxaben-induced formation of lignin and callose in seedlings.
The SUB-mediated CBI response is sensitive to sorbitol
The isoxaben-induced CWD response is sensitive to turgor pressure, as indicated by the suppression of lignin or callose accumulation in the presence of osmotica, such as sorbitol [34,36,37]. To test if SUB affects a turgor-sensitive CBI response we compared isoxaben-induced accumulation of lignin and callose in six days-old Col-0, sub-9, and pUBQ::SUB:mCherry seedlings in co-treatments with 600 nM isoxaben and 300 mM sorbitol (Fig 4A–4C). We observed that simultaneous application of isoxaben and sorbitol resulted in strongly reduced phloroglucinol and aniline blue signal in all tested genotypes, including the SUB:mCherry overexpressing lines, which show hyperaccumulation of lignin or callose if solely isoxaben is applied. Lignin production was undetectable as we could not observe any phloroglucinol staining in main roots of mock/sorbitol or isoxaben/sorbitol-treated seedlings (Fig 4A). Very weak aniline blue signal could be detected in the cotyledons of mock/sorbitol or isoxaben/sorbitol-treated wild-type and mutant seedlings indicating that callose was present at low levels (Fig 4B and 4C). It even appeared that aniline blue-derived signal in the various isoxaben/sorbitol-treated sub mutants tended to be even lower when compared to the corresponding mock/sorbitol treatments. The cotyledons of the two mock/sorbitol-treated SUB:mCherry overexpressing lines exhibited low levels of aniline blue staining that that did not change upon exposure to isoxaben/sorbitol. Some of the aniline blue signal differences were statistically significant (for example sub-9, adjusted P value < 0.02, and sub-21, adjusted P value < 0.0005; one-way ANOVA followed by Tukey’s multiple comparison tests). However, given the very low signal levels it is probably unlikely that the differences reflect biological significance.
Fig 4. The effects of sorbitol on lignin and callose accumulation upon isoxaben exposure.
(A) Phloroglucinol signal strength indicating lignin accumulation in roots of six-day-old seedlings exposed to mock/300 mM sorbitol or to 600 nM isoxaben/300 mM sorbitol for 12 hours. Genotypes: Col, sub-9 (Col), pUBQ::gSUB:mCherry (line OE SUB-L1), qky-11 (Col). Note absence of detectable signal upon simultaneous application of isoxaben and sorbitol. The experiment was performed three times with similar results (n ≥ 10). (B) Confocal micrographs show cotyledons of seven-day-old sub-9, Col, and pUBQ::gSUB:mCherry (Col, line OE SUB-L1) seedlings treated with mock/300 mM sorbitol or 600 nM isoxaben/300 mM sorbitol for 24 hours. Aniline blue fluorescence signal strength indicates callose accumulation. No increase in signal intensity can be observed in isoxaben-treated seedlings. (C) Quantification of the results depicted in (B). Left panel depicts results obtained from Ler and sub-1 (Ler background). Right panel shows results obtained from Col-0 and sub mutants in Col-0 background and also depicts results from two independent pUBQ::gSUB:mCherry transgenic lines overexpressing SUB (Col, lines SUB OE-L1, SUB OE-O3). Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 5 ≤ n ≤ 10. The experiment was performed three times with similar results. Abbreviation: ISX, isoxaben. Scale bars: (A) 0.1 mm; (C) 0.2 mm.
SUB attenuates isoxaben-induced cell swelling and facilitates root growth recovery
Next, we assessed the biological relevance of SUB in the isoxaben-induced CWD response. Exposure of seedlings to isoxaben eventually results in the shortening and swelling of cells of the root epidermis, possibly a result of reduced microfibril formation in the cell wall [34]. We transferred six days-old plate-grown seedlings into a mock solution or a solution containing 600 nM isoxaben for up to seven hours. We then assessed the timing of the initial appearance of altered cellular morphology of root epidermal cells. In addition, we monitored the severity of the phenotype. We focused on cells of the elongation zone that bordered the root meristem. Notably, we did not observe any obvious morphological alterations in mock-treated wild-type or mutant seedlings (Fig 5). In Col-0 wild-type seedlings cell shortening and swelling first became noticeable during the five to six-hour interval proceeding treatment (Fig 5) (44/96 seedlings total, n = 4), as reported previously [34]. Upon isoxaben application to sub-9 or sub-21 seedlings, however, similar cellular alterations were already detected at the three to four-hour interval post treatment initiation (sub-9: 23/57, n = 2; sub-21: 20/46, n = 2). In addition, sub mutants exhibited more pronounced cellular alterations after seven hours of isoxaben treatment in comparison to wild type (Fig 5).
Fig 5. Root epidermal cell shape changes upon isoxaben treatment.
Six-day-old seedlings counter-stained with the membrane stain FM4-64 are shown. Confocal micrographs depict the region where the elongation zone flanks the root meristem. Time of exposure in hours to 600 nM isoxaben (ISX) or mock is indicated as are the genotypes. Numbers of roots showing the phenotype are indicated (n/total number of roots scored). Arrows denote aberrant cell shapes. Scale bar: 0.1 mm.
In wild-type seedlings, a transient 24-hour exposure to isoxaben results in a temporary stop of root growth followed by a rapid recovery [34]. We tested the role of SUB in root growth recovery upon a 24 hour-treatment with isoxaben. Six-day-old wild-type and mutant seedlings grown on plates were transferred onto media containing 600 nM isoxaben for 24 hours, then moved to fresh plates lacking isoxaben. Seedlings were then monitored for continued root growth at 24-hour intervals, for a total of 72 hours (Table 1). As control, we used the ixr2-1 mutant, which is resistant to isoxaben due to a mutation in the CESA6 gene [30,31,58]. We observed that 98 percent of ixr2-1 seedlings recovered root growth already within 24 hours, indicating that treatment did not generally impact the seedlings’ ability to recover root growth. We then tested wild-type seedlings. We noticed that 46 percent of Ler and 39 percent of Col seedlings had resumed root growth after 24 hours. By 72 hours, 86 percent of Ler and 90 percent of Col seedlings had recovered root growth. In contrast, a significantly reduced fraction of sub-1 and sub-4 mutants had resumed root growth when compared to wild-type Ler (Table 1). The sub-9 and sub-21 mutants also showed reduced root growth recovery in comparison to Col although sub-21 appeared less affected than sub-9. Importantly, ixr2-1 sub-9 mutants behaved identical to ixr2-1 single mutants at all time points scored. These findings indicate that ixr2-1 is epistatic to sub-9 and that the observed isoxaben-induced decrease in root growth recovery in sub-9 mutants relates to the herbicide.
Table 1. Root growth recovery after isoxaben treatment.
| Genotype | N totala | 24 hb | 48 hb | 72 hb |
|---|---|---|---|---|
| Ler | 110 | 45.5 | 69.1 | 85.5 |
| sub-1 | 82 | 24.4** | 50.0* | 64.6** |
| sub-4 | 76 | 17.1** | 39.5**** | 52.6**** |
| qky-8 | 87 | 37.9ns | 64.4ns | 88.5ns |
| Col-0 | 142 | 38.7 | 65.5 | 90.1 |
| sub-9 | 88 | 23.9* | 45.5** | 70.5** |
| sub-21 | 72 | 26.4ns | 51.4ns | 68.1** |
| qky-11 | 81 | 37.0ns | 64.2ns | 92.6ns |
| ixr2-1 | 83 | 97.6**** | 97.6**** | 97.6ns |
| sub-9 ixr2-1 | 77 | 100.0**** | 100.0**** | 100.0** |
| the1-1 | 78 | 41.0ns | 69.2ns | 88.5ns |
Percentages of root growth recovery of plate-grown, seven-day-old seedlings after transient exposure to 600 nM isoxaben for 24 hours. The top four sample rows list genotypes that are in Ler background. The bottom seven sample rows list genotypes that are in Col background.
aTotal number of samples. Cases per class and timepoint were pooled from three independent experiments. For each biological replicate 22 ≤ n ≤ 32 seedlings were analyzed per genotype and time point.
bStatistical significance (mutant vs respective wild type): na, not applicable;
ns, not significant;
* P< 0.05;
** P < 0.005;
**** P < 0.0001; (Fisher’s exact test; two-sided).
Taken together, the results suggest that SUB attenuates cell size and shape changes in root epidermal cells and facilitates root growth recovery during the isoxaben-induced CWD response.
SUB attenuates root growth in prc1-1
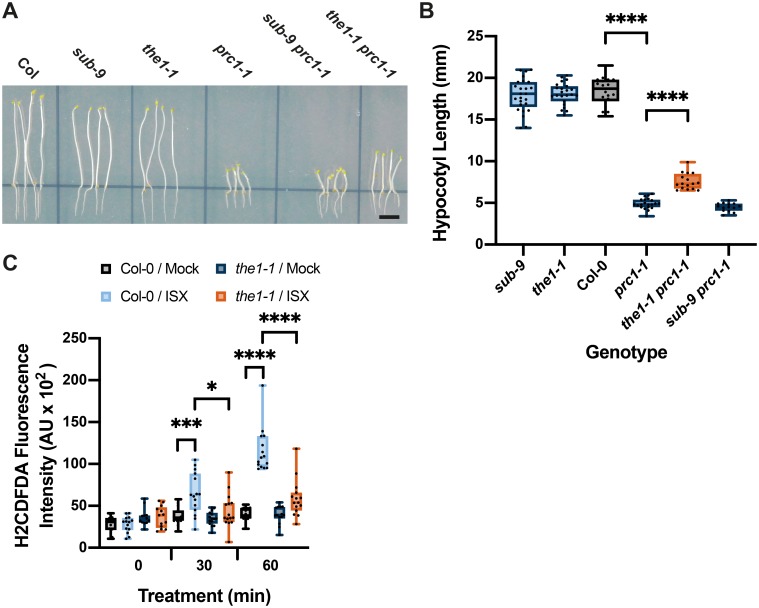
PRC1 encodes the CESA6 subunit of cellulose synthase [27] and prc1 loss-of-function mutants show reduced cellulose levels [27] (Fig 1B). In addition, prc1-1 mutants are characterized by a reduced elongation of etiolated hypocotyls and roots [27]. To test if SUB also affects a biological process in a scenario where cellulose reduction is induced genetically we compared root length in sub-9, sub-21, and prc1-1 single and sub-9 prc1-1 double mutants (Fig 6A and 6B). We found that root length of sub-9 or sub-21 did not deviate from wild type while root length of prc1-1 was markedly smaller in comparison to wild type, confirming previous results [27]. Interestingly, however, we observed that sub-9 prc1-1 exhibited a significantly longer root than prc1-1 though sub-9 prc1-1 roots were still notably smaller than wild type roots. The results indicate that SUB contributes to root growth inhibition in prc1-1.
Fig 6. SUB effect on root growth inhibition in prc1-1.
(A) Root length in seven-day-old seedlings grown on plates under long-day conditions (16 hours light). Genotypes are indicated. Note the partial rescue of root length in sub-9 prc1-1 but not in the1-1 prc1-1. (B) Quantification of the data shown in (A). Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 21 ≤ n ≤ 40. Asterisks represent adjusted P values (** P < 0.004, **** P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed three times with similar results. Scale bars: 0.5 mm.
SUB and QKY contribute differently to the CBI response
Evidence suggests that a protein complex including SUB and QKY is important for SUB-mediated signal transduction regulating tissue morphogenesis [50,59,60]. Thus, we wanted to explore if QKY is also required for the isoxaben-induced CWD response in seedlings. We first investigated if QKY affects the early isoxaben-induced changes in intracellular ROS levels by assessing H2CDFDA fluorescence in root tips of qky-11 seedlings that were treated with 600 nM isoxaben. Similar to wild type we observed an obvious increase in signal intensity between mock and isoxaben-treated qky-11 (Fig 7A). This indicates that QKY does not contribute to altered intracellular ROS levels in root tips of treated seedlings in a noticeable fashion. We then tested if QKY promotes isoxaben-induced marker gene expression in liquid-grown seedlings. Using qPCR, we observed that QKY did not noticeably affect the expression levels of the tested marker genes (Fig 7B). Next, we assessed isoxaben-induced lignin accumulation in wild-type and qky seedlings by phloroglucinol staining. We noticed reduced staining in qky-8 and qky-11 mutants compared to wild type (Fig 7C and 7D). Again, the effect was less obvious in Col-0. In addition, we noticed that qky-11 did not affect the absence of lignin accumulation in seedlings simultaneously treated with isoxaben and sorbitol (Fig 4A). We also investigated the role of QKY in isoxaben-induced callose deposition by scoring the aniline blue-derived signal in cotyledons. Similar to wild type we observed a significant difference in signal strength between mock and isoxaben-treated qky-8 and qky-11 (Fig 7E and 7F). In addition, we found that qky-11 seedlings still accumulated increased JA levels upon induction by isoxaben (Fig 2D). The results indicate that QKY is required for isoxaben-induced increase in lignin but does not play a role in the accumulation of callose or JA.
Fig 7. Role of QKY in isoxaben-induced CBI responses.
(A) Quantification of H2CDFDA signal indicating ROS accumulation in root tips of six-day-old Col-0 and qky-11 seedlings exposed to mock or 600 nM isoxaben for the indicated time. Note the high signal in qky-11 after a 30- or 60-minutes exposure to isoxaben. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 10 ≤ n ≤ 14. Asterisks represent adjusted P values (* P < 0.04, **** P < 0.0001; ns: not significant; one-way ANOVA followed by Tukey’s multiple comparison tests). No statistically significant differences were measured within or between genotypes at the zero minute time point of mock or isoxaben treatment or between wild-type and qky-11 mock treatment at the 30 and 60 minutes time points (P > 0.8557; one-way ANOVA followed by Tukey’s multiple comparison tests). Experiments were performed three times with similar results. (B) Gene expression levels of several CBI marker genes by qPCR upon exposure of seven-day-old seedlings to 600 nM isoxaben for eight hours. The results from three biological replicates are shown. Marker genes and genotypes are indicated. Mean ± SD is presented. The experiment was repeated three times with similar results. (C) Phloroglucinol signal strength indicating lignin accumulation in roots of six-day-old seedlings exposed to 600 nM isoxaben for 12 hours. Genotypes: Col, qky-11 (Col), Ler, and sub-1 (Ler). (D) Quantification of the results depicted in (C). Genotypes are indicated. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 15 ≤ n ≤ 19. Asterisks represent statistical significance (* P < 0.04, ** P < 0.01; unpaired t test, two-tailed P values). The experiment was performed three times with similar results. (E) Confocal micrographs show cotyledons of seven-day-old Ler, qky-8 (Ler), Col, and qky-11 (Col) seedlings treated with mock or 600 nM isoxaben for 24 hours. Aniline blue fluorescence signal strength indicates callose accumulation. (F) Quantification of the results depicted in (E). Left panel shows results obtained from qky-8 mutants in Ler background. Right panel indicates results obtained from qky-11 mutants in Col background. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 5 ≤ n ≤ 11. Asterisks represent adjusted P values (* P < 0.04, *** P < 0.0005, **** P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed three times with similar results. Abbreviation: ISX, isoxaben. Scale bars: (C) 0.1 mm; (E) 0.2 mm.
Next we analyzed isoxaben-induced shortening and swelling of root epidermal cells in qky-11 mutants (Fig 4). We noticed the first defects in a three to four-hour interval (18/33, n = 3). Cell swelling after seven hours exposure to isoxaben was prominent in qky-11 but less severe in comparison to sub-9 (Fig 4). Finally, we investigated root growth recovery after transient isoxaben application. We observed that qky-8 and qky-11 mutants did not significantly deviate from wild type (Table 1).
Taken together, the results indicate that QKY and SUB contribute to the isoxaben-mediated induction of lignin accumulation. Moreover, QKY also plays a role in the suppression of isoxaben-induced alterations in cell morphology in the root epidermis. However, the results also imply that QKY is not required for isoxaben-induced early ROS accumulation in root tips, marker gene induction as well as callose accumulation in cotyledons and does not affect root growth recovery after transient exposure to isoxaben.
SUB and THE1 share partially overlapping functions
THE1 is a central regulator of the isoxaben-induced CBI response [34,39,43]. Our data indicate that SUB and THE1 have overlapping but also distinct functions in this process. For example, SUB and THE1 control isoxaben-induced lignin accumulation in roots (Fig 3) [34,39]. In contrast, we failed to detect an effect of THE1 on root growth inhibition in prc1-1, while SUB contributes to this process (Fig 6). In addition, we found that in contrast to sub root growth recovery of the1-1 seedlings upon transient exposure to isoxaben did not deviate from wild type (Table 1).
To explore further the relationship between THE1 and SUB we investigated the requirement of THE1 for isoxaben-dependent marker gene induction. We observed that like SUB THE1 is required for isoxaben-induced upregulation of CCR1, PDF1.2, and RBOHD although in the case of CCR1 and PDF1.2 the1-1 seedlings show a stronger effect than sub mutants (Fig 2C, S1 Fig). Regarding CYP81F2 we noticed a major difference between sub and the1-1. While CYP81F2 appeared slightly higher induced upon exposure to isoxaben in sub seedlings (Fig 2C, S1 Fig) we observed strongly reduced CYP81F2 transcript levels in the1-1 (Fig 2C). This observation differs from previous work where CYP81F2 induction was reported to be unaltered in the1-1 [39]. We also noticed that FRK1 induction was strongly reduced in the1-1 (Fig 2C), confirming previous data [39], while in sub seedlings isoxaben-dependent FRK1 transcript levels were either reduced or unaltered depending on the sub allele (Fig 2D, S1 Fig).
Next, we compared the role of SUB and THE1 in etiolated hypocotyl elongation. THE1 contributes to the reduction of hypocotyl length of cellulose-diminished prc1-1 seedlings grown in the dark as evidenced by the partial recovery of hypocotyl elongation in the1 prc1 double mutants [38]. We compared sub-9 prc1-1 to the1-1 prc1-1 with respect to hypocotyl elongation in six-day-old etiolated seedlings (Fig 8). We observed strongly reduced hypocotyl elongation in prc1-1 in comparison to wild type and a significant suppression of this reduction in the1-1 prc1-1 double mutants (Fig 8A and 8B), as described earlier [38]. In contrast, hypocotyl length in sub-9 mutants was not decreased, nor was there a partial reversal of reduced hypocotyl elongation in sub-9 prc1-1 double mutants (Fig 8A and 8B). The results indicate that SUB does not affect hypocotyl elongation in etiolated wild-type or prc1-1 seedlings.
Fig 8. Influence of THE1 on etiolated hypocotyl length of prc1-1 and isoxaben-induced ROS accumulation in root tips.
(A) Hypocotyl elongation in six-day-old seedlings grown on plates in the dark. Genotypes are indicated. Note the partial rescue of hypocotyl elongation in the1-1 prc1-1 but not in sub-9 prc1-1. (B) Quantification of the data shown in (A). Whisker ends mark the minimum and maximum of all the data. 14 ≤ n ≤ 25. Asterisks represent adjusted P values (**** P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed three times with similar results. (C) Quantification of H2CDFDA signal indicating ROS accumulation in root tips of six-day-old Col-0 and the1-1 seedlings exposed to mock or 600 nM isoxaben for the indicated time. Note the low signal in the1-1 after a 30- or 60-minutes exposure to isoxaben. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 12 ≤ n ≤ 15. Asterisks represent adjusted P values (* P < 0.05; *** P = 0.0005; **** P < 0.0001, one-way ANOVA followed by Tukey’s multiple comparison tests). No statistically significant differences were measured within or between genotypes at the zero minute time point of mock or isoxaben treatment or between wild-type and the1-1 mock treatment at the 30 and 60 minutes time points (P > 0.7997; one-way ANOVA followed by Tukey’s multiple comparison tests). Experiments were performed three times with similar results. Abbreviation: ISX, isoxaben. Scale bars: 0.5 mm.
Finally, we assessed the role of THE1 in isoxaben-induced early ROS accumulation in root tips. We noticed no difference in H2CDFDA signal intensity when comparing the1-1 seedlings that had been treated with either mock or isoxaben for 30 minutes (Fig 8C). By contrast, isoxaben-induced reporter signal in the root tip became slightly stronger than the mock-mediated signal at the 60 minutes time point (Fig 8C). Overall the results resemble the findings obtained with sub-9 mutants (Fig 2B).
In summary, SUB and THE1 are required for ROS and lignin accumulation in roots. The two genes exhibit partial overlap but also clear distinctions with respect to their role in stress marker gene induction. SUB and THE1 show opposite effects on growth inhibition of roots and hypocotyls in prc1 mutants and their functions differ with respect to root growth recovery.
Discussion
Cell wall signaling during plant development and stress responses relies on complex and largely unknown signaling circuitry [5,8,9,35,61]. Only a few RKs, including THE1, MIK2, and FEI2, have so far been shown to play a major role in CBI-induced CWD signaling [34,38,39,43]. Our data establish SUB signal transduction as a novel component in the molecular framework mediating the CWD response.
To date, published evidence has attributed a developmental role to SUB, particularly in the control of tissue morphogenesis and root hair pattern formation [44–46,51,62]. The evidence provided in this work identifies a novel role for SUB in CWD signaling. Application of isoxaben results in reduced levels of cellulose [28,29]. Our results indicate that SUB affects multiple aspects of the isoxaben-induced CWD response. The observation that sub-9 partially suppresses reduced root length exhibited by prc1-1 indicates that SUB also mediates a CWD response that is caused by a genetic reduction in cellulose content. Thus, the collective data support the notion that the origin of the CWD relates to the reduced production of cellulose, a major carbohydrate component of the cell wall, and that SUB contributes to the compensatory cellular response to this type of cell-wall-related stress.
The data indicate that SUB already affects the early response to isoxaben-induced CWD, in particular ROS production. Previous results had revealed that the isoxaben-induced CWD response involves THE1-dependent ROS production [37,63]. In luminol-based extracellular ROS assays involving entire seedlings ROS production could be detected after around three to four hours following the application of isoxaben [63]. We used the intracellular ROS probe H2CDFDA and a microscope-based method that enabled tissue-level resolution. H2CDFDA has for example been used to assess basal intracellular ROS levels when studying GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH) genes and root hairs [55,56]. Our time-course data indicate that isoxaben induces the formation of intracellular ROS in the root meristem within 30 minutes. To our knowledge the isoxaben-dependent change in H2CDFDA fluorescence signal represents the earliest available marker for the isoxaben-induced CWD response. It also indicates that this response occurs even earlier than previously appreciated [34].
The results suggest that SUB is required for full induction of several marker genes, such as CCR1, PDF1.2, or RBOHD. Moreover, the data indicate that SUB promotes CBI-induced accumulation of lignin and callose. The isoxaben-induced CWD response is sensitive to turgor pressure as for example induction of marker gene expression or lignin accumulation is strongly reduced upon exposing seedlings to isoxaben in combination with an osmoticum, such as sorbitol [34,37]. It has been proposed that a mechanical stimulus initiates the CWD response to the isoxaben-induced reduction in cellulose and the associated weakening of the cellulose framework counteracting turgor pressure [34,64–67]. In this model, the stimulus could derive from a displacement or distortion of the plasma membrane relative to the cell wall. Indeed, we did not observe an obvious influence of SUB on the reduced lignin and callose state that results from simultaneous application of sorbitol and isoxaben indicating that SUB mediates a response that also depends on turgor pressure. Thus, the combined results fulfil the criteria that have been established for a CBI-induced CWD response [34,37].
Interestingly, different loss-of-function (sub-1, sub-9, sub-21) or gain-of-function (pUBQ::SUB:mCherry) mutants show reciprocal effects regarding lignin and callose accumulation, with sub mutants showing less and pUBQ::SUB:mCherry lines exhibiting higher levels of lignin or callose, respectively, upon application of isoxaben. The pUBQ::SUB:mCherry O3 and L1 lines originate from independent transformation events. They differ in their combined SUB and SUB:mCherry transcript levels with line O3 showing higher levels compared to line L1 (S2 Fig). Still, upon exposure to isoxaben line L1 exhibited stronger lignin and callose accumulation than line O3. This finding may be indicative of an optimal range of SUB activity with respect to the control of lignin and callose accumulation upon isoxaben-induced CWD. Based on this evidence, we propose the model that SUB represents an important genetic regulator of isoxaben-induced lignin and callose accumulation and thus cell wall composition.
Several lines of evidence indicate that SUB plays a biologically relevant role in CWD signaling initiated by a reduction in cellulose content. Firstly, SUB attenuates isoxaben-induced cell bulging in the epidermal cells of the meristem-transition zone boundary of the root. Secondly, SUB facilitates root growth recovery upon transient exposure of seedlings to isoxaben. Thirdly, SUB is involved in root growth inhibition that is a consequence of a genetic reduction of cellulose content. In particular, root length of sub-9 prc1-1 double mutant seedlings is less diminished in comparison to the root length of prc1-1 single mutant seedlings. The results imply that SUB contributes to a compensatory response that counteracts the cellular and growth defects caused by reduced cellulose synthesis and further support the notion that SUB plays a central role in CBI-induced CWD signaling.
How does SUB relate to other known RK genes mediating the response to CBI-induced cell wall stress, such as THE1 and MIK2? SUB, THE1, and MIK2 all promote isoxaben-induced ectopic lignin production. The three genes are also required for full isoxaben-dependent induction of certain stress marker genes [39] (Fig 2C). However, while THE1 and MIK2 are both required for the induction of CYP81F2 SUB appears to slightly attenuate induced CYP81F2 transcript levels. THE1 and MIK2 are also required for FRK1 induction while the role of SUB in the regulation of this gene remains to be resolved. In addition, THE1 and MIK2 are necessary for isoxaben-induced JA accumulation [34,39,63], a process that apparently does not require SUB function. We also did not observe an effect of THE1 on root growth recovery upon transient exposure to isoxaben. In addition, we did not find an altered hypocotyl length of sub-9 prc1-1 in comparison to prc1-1 indicating that SUB does not affect hypocotyl growth inhibition in etiolated prc1-1 seedlings, in contrast to THE1 [38]. At the same time, our data suggest that SUB contributes to root growth inhibition in prc1-1. However, our evidence does not support a function for THE1 in this process, as root length of the1-1 prc1-1 double mutants did not deviate from the root length observed for prc1-1 single mutants. This finding also implies that the lignin accumulation in the mature root parts of prc1-1 (suppressed in prc1-1 the1-1 double mutants) [38] does not correlate with root growth inhibition. Finally, we did not observe the left-hand root skewing in sub seedlings that has been described for mik2 mutants [39], and we failed to observe floral defects in mik2 or the1 mutants (S3 Fig). Taken together, the data suggest that SUB has both overlapping and distinct functions with THE1 and MIK2. As the most parsimonious explanation of our results, we propose that SUB contributes to CBI-induced cell wall damage signaling independently from THE1 and MIK2 signaling, however the signaling pathways downstream of the different cell surface receptor kinases eventually partially converge and contribute to a subset of overlapping downstream responses.
QKY represents a central genetic component of SUB-mediated signal transduction during tissue morphogenesis and root hair patterning [51]. The expression patterns of QKY and SUB fully overlap [50] and present evidence supports the notion that SUB and QKY are part of a protein complex with QKY likely acting upstream to, or in parallel with, SUB [50,59,60]. However, genetic and whole-genome transcriptomic data suggested that SUB and QKY also play distinct roles during floral development [51]. The data presented in this study reveal that SUB and QKY both contribute to CBI-induced CWD signaling. Similar to SUB QKY is required for lignin accumulation. Moreover, QKY is also necessary for the prevention of cell bulging in the epidermal cells of the meristem-transition zone boundary of the root although the weaker phenotype of qky-11 mutants indicates a lesser requirement for QKY in comparison to SUB. However, our data also indicate that QKY is not required for early ROS accumulation, marker gene induction, ectopic callose accumulation, and root growth recovery as we did not find these processes to be noticeably affected in qky mutants. Thus, these data provide genetic evidence that the functions of SUB and QKY only partially overlap and that SUB exerts CWD signaling functions that are independent of QKY. One way to rationalize these findings is to assume that SUB can also function in isolation or in protein complexes or pathways that do not involve QKY.
The diverse functions of SUB in development and the CWD response are likely to be achieved through participation in different signaling pathways. It is not uncommon that RKs play important roles in several biological processes. In this respect, SUB resembles for example the RK BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1 (BAK1) / SOMATIC EMBROYGENESIS RECEPTOR KINASE 3 (SERK3), which functions in growth, development, and plant defense [68,69]. BAK1 interacts with a range of different LRR-RKs, including FLAGELLIN SENSING 2 (FL2) and BRASSINOSTEROID INSENSITIVE 1 (BRI1), and the discrimination between the growth and immunity functions of BAK1 was recently shown to rely on phosphorylation-dependent regulation [70,71]. In light of these considerations it is reasonable to propose that SUB is a member of different receptor complexes. As kinase activity of SUB is not required for its function [44] SUB could act as a scaffold around which the components of the various complexes assemble. A scaffold role has for example been proposed for the RK AtCERK1/OsCERK1 in chitin signaling or the RK FER in immune signaling [72,73]. It will be interesting to explore this notion in future work.
Materials and methods
Plant work, plant genetics and plant transformation
Arabidopsis thaliana (L.) Heynh. var. Columbia (Col-0) and var. Landsberg (erecta mutant) (Ler) were used as wild-type strains. Plants were grown as described earlier [51]. The sub-1, qky-8 (all in Ler), and the sub-9 and qky-11 mutants (Col) have been characterized previously [44,47,50,51]. The prc1-1 [27], the1-1 [38], the1-6 [74], mik2-1 [39], mik2-3 [75], and ixr2-1 [30] alleles were also described previously. The sub-21 (Col) allele was generated using a CRISPR/Cas9 system in which the egg cell-specific promoter pEC1.2 controls Cas9 expression [76]. The single guide RNA (sgRNA) 5’-TAATAACTTGTATATCAACTT-3’ binds to the region +478 to +499 of the SUB coding sequence. The sgRNA was designed according to the guidelines outlined in [77]. The mutant carries a frameshift mutation at position 495 relative to the SUB start AUG, which was verified by sequencing. The resulting predicted short SUB protein comprises 67 amino acids. The first 39 amino acids correspond to SUB and include its predicted signal peptide of 29 residues, while amino acids 40 to 67 represent an aberrant amino acid sequence. The pUBQ::gSUB:mCherry plasmid used to generate the SUB overexpression lines L1 and O3 was generated previously [50]. Wild-type, and sub mutant plants were transformed with different constructs using Agrobacterium strain GV3101/pMP90 [78] and the floral dip method [79]. Transgenic T1 plants were selected on Kanamycin (50 μg/ml), Hygromycin (20 μg/ml) or Glufosinate (Basta) (10 μg/ml) plates and transferred to soil for further inspection.
Cellulose quantification
Seedlings were grown on square plates with half strength MS medium and 0.3% sucrose supplemented with 0.9% agar for seven days. Cellulose content was measured following the Updegraff method essentially as described [80,81], with minor modifications as outlined here. Following the acetic nitric treatment described in [80], samples were allowed to cool at room temperature and transferred into 2 ml Eppendorf safety lock tubes. Samples were then centrifuged at 14000 rpm at 15°C for 15 min. The acetic nitric reagent was removed carefully without disturbing the pelleted material at the bottom of the tube. 1 ml of double-distilled H2O was added, and the sample was left on the bench for 10 min at room temperature followed by centrifugation at 14000 rpm at 15°C for 15 min. After aspirating off the H2O 1 ml acetone was added and the samples were incubated for another 15 min, followed by centrifugation at 14000 rpm at 15°C for 15 min. Afterwards acetone was removed, and samples were air-dried overnight. Then the protocol was continued as described in [80].
PCR-based gene expression analysis
For quantitative real-time PCR (qPCR) of CesA and stress marker genes 35 to 40 seedlings per flask were grown in liquid culture under continuous light at 18°C for seven days followed by treatment with mock or 600 nM isoxaben for eight hours or on plates (21°C, long-day conditions). With minor changes, RNA extraction and quality control were performed as described previously [82]. cDNA synthesis, qPCR, and analysis were done essentially as described [83]. Primers are listed in S1 Table.
ROS, lignin, and callose staining
Intracellular ROS accumulation in root meristems was estimated using the H2DCFDA fluorescent stain essentially as described [56]. Seeds were grown on square plates with half strength MS medium and 1% sucrose supplemented with 0.9% agar. The seeds were stratified for two days at 4°C and incubated for seven days at 22°C under long day conditions, at a 10-degree inclined position. Seven days-old seedlings were first transferred into multi-well plates containing half strength liquid MS medium supplemented with 1% sucrose for two hours. Then medium was exchanged with liquid medium containing either DMSO (mock) or 600nM isoxaben without disturbing seedlings. 10 min prior to each time point seedlings were put in the dark and the liquid medium was supplemented with 100 μM H2DCFDA staining solution. Images was acquired with a confocal microscope. For quantification a defined region of interest (ROI) located 500 μm above the root tip (excluding the root cap) was used in all samples. Staining for lignin (phloroglucinol) and callose (aniline blue) was performed as described in [39] and [57], respectively. ROS, phloroglucinol and aniline blue staining was quantified on micrographs using ImageJ software [84].
JA measurements
Chemicals
Jasmonic acid-d0 and jasmonic acid-d5 were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Formic acid was obtained from Merck (Darmstadt, Germany), ethyl acetate and acetonitrile (LC-MS grade) were obtained from Honeywell (Seelze, Germany). Water used for chromatographic separations was purified with an AQUA-Lab—B30—Integrity system (AQUA-Lab, Ransbach-Baubach, Germany).
Sample preparation
Approximately 35 to 40 seedlings per flask were grown in liquid culture (1/2 MS, 0.3% sucrose) under continuous light and 18°C for seven days followed by treatment with mock or 600 nM isoxaben for seven hours and harvesting in liquid nitrogen. The grinded plant material (100–200 mg) was placed in a 2 mL bead beater tube (CKMix-2 mL, Bertin Technologies, Montigny-le-Bretonneux, France), filled with ceramic balls (zirconium oxide; mix beads of 1.4 mm and 2.8 mm), and an aliquot (20 μL) of a solution of acetonitrile containing the internal standard (-)trans-jasmonic acid-d5 (25 μg/mL), was added. After incubation for 30 min at room temperature, the tube was filled with ice-cold ethyl acetate (1 mL). After extractive grinding (3 × 20 s with 40 s breaks; 6000 rpm) using the bead beater (Precellys Homogenizer, Bertin Technologies, Montigny-le-Bretonneux, France), the supernatant was membrane filtered (0.45 μm), evaporated to dryness (Christ RVC 2-25 CD plus, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany), resolved in acetonitrile (70 μL) and injected into the LC−MS/MS-system (2 μL).
Liquid chromatography-triple quadrupole mass spectrometry (LC−MS/MS)
Phytohormone concentrations were measured by means of UHPLC-MS/MS using a QTRAP 6500+ mass spectrometer (Sciex, Darmstadt, Germany) in the multiple reaction monitoring (MRM) mode. Positive ions were detected at an ion spray voltage at 5500 V (ESI+) and the following ion source parameters: temperature (550°C), gas 1 (55 psi), gas 2 (65 psi), curtain gas (35 psi), entrance potential (10 V) and collision activated dissociation (3 V). The temperature of the column oven was adjusted to 40°C. For plant hormone analysis, the MS/MS parameters of the compounds were tuned to achieve fragmentation of the [M+H]+ molecular ions into specific product ions: (-)trans-jasmonic acid-d0 211→133 (quantifier) and 211→151 (qualifier), (-)trans-jasmonic acid-d5 216→155 (quantifier) and 216→173 (qualifier). For the tuning of the mass spectrometer, solutions of the analyte and the labelled internal standard (solved in acetonitrile:water, 1:1) were introduced into the MS system by means of flow injection using a syringe pump. Separation of all samples was carried out by means of an ExionLC UHPLC (Shimadzu Europa GmbH, Duisburg, Germany), consisting of two LC pump systems (ExionLC AD), an ExionLC degasser, an ExionLC AD autosampler, an ExionLC AC column oven– 240 V and an ExionLC controller. After sample injection (2 μL), chromatography was carried out on an analytical Kinetex F5 column (100 × 2.1 mm2, 100 Å, 1.7 μm, Phenomenex, Aschaffenburg, Germany). Chromatography was performed with a flow rate of 0.4 mL/min using 0.1% formic acid in water (v/v) as solvent A and 0.1% formic acid in acetonitrile (v/v) as solvent B, and the following gradient: 0% B held for 2 min, increased in 1 min to 30% B, in 12 min to 30% B, increased in 0.5 min to 100% B, held 2 min isocratically at 100% B, decreased in 0.5 min to 0% B, held 3 min at 0% B. Data acquisition and instrumental control was performed using Analyst 1.6.3 software (Sciex, Darmstadt, Germany).
Hypocotyl and root measurements
For measuring etiolated hypocotyl length, seedlings were grown for five days on half-strength MS agar supplemented with 0.3% sucrose. Seedlings were photographed and hypocotyl length was measured using ImageJ. For root growth assays, seedlings were grown for seven days in long-day conditions at 21°C on half-strength MS agar supplemented with 0.3% sucrose. Plates were inclined at 10 degrees. Root length was measures using ImageJ. For root growth recovery assays seedlings were grown on half-strength MS agar supplemented with 0.3% sucrose. Seeds were stratified for two days followed by incubation at 21°C in long day conditions for seven days. Plates were inclined at 10 degrees. Individual seedlings were transferred to plates containing either 0.01 percent DMSO (mock) or 600 nM isoxaben for 24 hours. After treatment, seedlings were transferred onto half-strength MS agar plates supplemented with 0.3% sucrose. The position of the root tip was marked under a dissection microscope. Root length was measured every 24 hours for up to three days. For root cell bulging assays seedlings were grown for seven days in long-day conditions at 21°C on half-strength MS agar supplemented with 0.3% sucrose. Plates were inclined at 10 degrees. Individual seedlings were first transferred into liquid medium for two hours habituation followed by treatment with 600 nM isoxaben for up to seven hours. To take images, seedlings were stained with 4 μM FM4-64 for 10 minutes and imaged using confocal microscopy. Confocal micrographs were acquired at each time point. All hypocotyl length or root measurements were performed in double-blind fashion.
Statistics
Statistical analysis was performed with PRISM8 software (GraphPad Software, San Diego, USA).
Microscopy and art work
Images of seedlings exhibiting phloroglucinol staining were taken on a Leica MZ16 stereo microscope equipped with a Leica DFC320 digital camera. Images of hypocotyl and root length were taken on a Leica SAPO stereo microscope equipped with a Nikon Coolpix B500 camera. Aniline blue-stained cotyledons and root cell bulging were imaged with an Olympus FV1000 setup using an inverted IX81 stand and FluoView software (FV10-ASW version 01.04.00.09) (Olympus Europa GmbH, Hamburg, Germany) equipped with a 10x objective (NA 0.3). For assessing cell bulging a projection of a 5 μm z-stack encompassing seven individual optical sections was used. Aniline blue fluorescence was excited at 405 nm using a diode laser and detected at 425 to 525 nm. H2DCFDA and EGFP fluorescence excitation was done at 488 nm using a multi-line argon laser and detected at 502 to 536 nm. For the direct comparisons of fluorescence intensities, laser, pinhole and gain settings of the confocal microscope were kept identical when capturing the images from the seedlings of different treatments. Scan speeds were set at 400 Hz and line averages at between 2 and 4. Measurements on digital micrographs were done using ImageJ software [84]. Images were adjusted for color and contrast using Adobe Photoshop 2020 software (Adobe, San Jose, USA).
Supporting information
Gene expression levels of several CBI marker genes by qPCR upon exposure of seven-day-old seedlings to 600 nM isoxaben for eight hours. The results from five biological replicates are shown. Marker genes and genotypes are indicated. Mean ± SD is presented. Asterisks represent adjusted P values (** P < 0.009, *** P = 0.0002, **** P < 0.0001, ns: not significant; one-way ANOVA followed by Tukey’s multiple comparison tests). For all genotypes the differences between isoxaben and mock treatments were statistically significant (adjusted P value < 0.0001). Differences between mock treated wild-type and sub-9 samples were not statistically significant. The experiment was repeated twice with similar results.
(TIF)
The results from three biological replicates are shown. Mean ± SD is presented.
(TIF)
(A) Upper panel: siliques. Bottom panel: open siliques revealing developing seeds. Genotypes are indicated. Note aberrant sub-9 silique morphology and the reduced number of developing seeds in sub-9. Arrows indicate undeveloped seeds. (B) Root angle of nine-day-old seedlings grown in an upright position (10° angle relative to direction of gravity) on half-strength MS agar medium supplemented with 1% sucrose. Pictures were taken from the front of the plate. Genotypes are indicated. Note the slight slant to the left in mik2-3. (C) Quantification of results depicted in (B) and as described in Van der Does et al (2017) PLoS Genet 13: e1006832. Genotypes are indicated. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 16 ≤ n ≤ 22. Asterisks represent adjusted P values (*** P = 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed twice. In the second experiment the root angle difference in mik2-3 was not statistically significant. Scale bars: open siliques, 1 mm; closed siliques, 0.5 cm; roots, 1 cm.
(TIF)
(DOCX)
Acknowledgments
We acknowledge Ramon Torres Ruiz and other members of the Schneitz lab for helpful discussion and suggestions. We thank Herman Höfte and Stefanie Ranf for the1 and mik2 alleles, respectively. We also thank Lynette Fulton, Silke Robatzek, Martin Stegmann and Sebastian Wolf for critical reading of the manuscript. We further acknowledge the support of the Center for Advanced Light Microscopy (CALM) at the TUM School of Life Sciences.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by the German Research Council (DFG, www.dfg.de) through grants SFB924 (TP B12) to CD and SFB924 (TP A2) to KS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lampugnani ER, Khan GA, Somssich M, Persson S (2018) Building a plant cell wall at a glance. J Cell Sci 131: [DOI] [PubMed] [Google Scholar]
- 2.Höfte H, Voxeur A (2017) Plant cell walls. Curr Biol 27: R865–R870. 10.1016/j.cub.2017.05.025 [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove DJ (2016) Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67: 463–476. 10.1093/jxb/erv511 [DOI] [PubMed] [Google Scholar]
- 4.Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- 5.Franck CM, Westermann J, Boisson-Dernier A (2018) Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69: 301–328. 10.1146/annurev-arplant-042817-040557 [DOI] [PubMed] [Google Scholar]
- 6.Herger A, Dünser K, Kleine-Vehn J, Ringli C (2019) Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr Biol 29: R851–R858. 10.1016/j.cub.2019.07.039 [DOI] [PubMed] [Google Scholar]
- 7.Vaahtera L, Schulz J, Hamann T (2019) Cell wall integrity maintenance during plant development and interaction with the environment. Nat Plants 5: 924–932. 10.1038/s41477-019-0502-0 [DOI] [PubMed] [Google Scholar]
- 8.Wolf S (2017) Plant cell wall signalling and receptor-like kinases. Biochem J 474: 471–492. 10.1042/BCJ20160238 [DOI] [PubMed] [Google Scholar]
- 9.Voxeur A, Höfte H (2016) Cell wall integrity signaling in plants: “To grow or not to grow that’s the question”. Glycobiology 26: 950–960. 10.1093/glycob/cww029 [DOI] [PubMed] [Google Scholar]
- 10.Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schurholz AK et al. (2014) A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci U S A 111: 15261–15266. 10.1073/pnas.1322979111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf S, Mravec J, Greiner S, Mouille G, Höfte H (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737. 10.1016/j.cub.2012.07.036 [DOI] [PubMed] [Google Scholar]
- 12.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC et al. (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. 10.1126/science.1143562 [DOI] [PubMed] [Google Scholar]
- 13.Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, Kleine-Vehn J (2019) Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J 38: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675.e5. 10.1016/j.cub.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boisson-Dernier A, Roy S, Kritas K, Grobei MA, Jaciubek M, Schroeder JI et al. (2009) Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288. 10.1242/dev.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franck C, Westermann J, Bürssner S, Lentz R, Lituiev DS, Boisson-Dernier A (2018) The protein phosphatases ATUNIS1 and ATUNIS2 regulate cell wall integrity in tip-growing cells. Plant Cell 30: 1906–1923. 10.1105/tpc.18.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V et al. (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603. 10.1126/science.aao5467 [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H et al. (2009) ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol 19: 1327–1331. 10.1016/j.cub.2009.06.064 [DOI] [PubMed] [Google Scholar]
- 19.Bai L, Ma X, Zhang G, Song S, Zhou Y, Gao L et al. (2014) A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 26: 1497–1511. 10.1105/tpc.114.124586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoenaers S, Balcerowicz D, Breen G, Hill K, Zdanio M, Mouille G et al. (2018) The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr Biol 28: 722–732.e6. 10.1016/j.cub.2018.01.050 [DOI] [PubMed] [Google Scholar]
- 22.Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. 10.1126/science.1102765 [DOI] [PubMed] [Google Scholar]
- 23.Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362. 10.1046/j.1365-313x.2003.01729.x [DOI] [PubMed] [Google Scholar]
- 24.Cano-Delgado AI, Metzlaff K, Bevan MW (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405. [DOI] [PubMed] [Google Scholar]
- 25.Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566. 10.1105/tpc.002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033. 10.1105/tpc.13.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G et al. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424. 10.1105/tpc.12.12.2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heim DR, Skomp JR, Tschabold EE, Larrinua IM (1990) Isoxaben inhibits the synthesis of acid insoluble cell wall materials in Arabidopsis thaliana. Plant Physiol 93: 695–700. 10.1104/pp.93.2.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefebvre A, Maizonnier D, Gaudry JC, Clair D, Scalla R (1987) Some effects of the herbicide EL-107 on cellular growth and metabolism. Weed Research 27: 125–134. [Google Scholar]
- 30.Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E et al. (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490. 10.1104/pp.010822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci U S A 98: 10079–10084. 10.1073/pnas.191361598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. 10.1126/science.1126551 [DOI] [PubMed] [Google Scholar]
- 33.Heim DR, Skomp JR, Waldron C, Larrinua IM (1991) Differential response to isoxaben of cellulose biosynthesis by wild-type and resistant strains of Arabidopsis thaliana. Pestic Biochem Physiol 39: 93–99. [Google Scholar]
- 34.Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F et al. (2018) The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci Signal 11: eaao3070 10.1126/scisignal.aao3070 [DOI] [PubMed] [Google Scholar]
- 35.Hamann T (2015) The plant cell wall integrity maintenance mechanism—a case study of a cell wall plasma membrane signaling network. Phytochemistry 112: 100–109. 10.1016/j.phytochem.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 36.Wormit A, Butt SM, Chairam I, McKenna JF, Nunes-Nesi A, Kjaer L et al. (2012) Osmosensitive changes of carbohydrate metabolism in response to cellulose biosynthesis inhibition. Plant Physiol 159: 105–117. 10.1104/pp.112.195198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamann T, Bennett M, Mansfield J, Somerville C (2009) Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 57: 1015–1026. 10.1111/j.1365-313X.2008.03744.x [DOI] [PubMed] [Google Scholar]
- 38.Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S et al. (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17: 922–931. 10.1016/j.cub.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 39.Van der Does D, Boutrot F, Engelsdorf T, Rhodes J, McKenna JF, Vernhettes S et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet 13: e1006832 10.1371/journal.pgen.1006832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julkowska M, Koevoets IT, Mol S, Hoefsloot HC, Feron R, Tester M et al. (2017) Genetic components of root architecture remodeling in response to salt stress. Plant Cell 29: 3198–3213. 10.1105/tpc.16.00680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, Liang L, Xue Y, Jia PF, Chen W, Zhang MX et al. (2016) A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244. 10.1038/nature16975 [DOI] [PubMed] [Google Scholar]
- 42.Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079. 10.1105/tpc.108.063354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F et al. (2018) Receptor kinase THESEUS1 is a Rapid Alkalinization Factor 34 receptor in Arabidopsis. Curr Biol 28: 2452–2458.e4. 10.1016/j.cub.2018.05.075 [DOI] [PubMed] [Google Scholar]
- 44.Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M et al. (2005) STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci U S A 102: 9074–9079. 10.1073/pnas.0503526102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwak SH, Shen R, Schiefelbein J (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113. 10.1126/science.1105373 [DOI] [PubMed] [Google Scholar]
- 46.Lin L, Zhong SH, Cui XF, Li J, He ZH (2012) Characterization of temperature-sensitive mutants reveals a role for receptor-like kinase SCRAMBLED/STRUBBELIG in coordinating cell proliferation and differentiation during Arabidopsis leaf development. Plant J 72: 707–720. 10.1111/j.1365-313X.2012.05109.x [DOI] [PubMed] [Google Scholar]
- 47.Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K (2011) Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS One 6: e19730 10.1371/journal.pone.0019730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sager RE, Lee JY (2018) Plasmodesmata at a glance. J Cell Sci 131: jcs209346 10.1242/jcs.209346 [DOI] [PubMed] [Google Scholar]
- 49.Otero S, Helariutta Y, Benitez-Alfonso Y (2016) Symplastic communication in organ formation and tissue patterning. Curr Opin Plant Biol 29: 21–28. 10.1016/j.pbi.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 50.Vaddepalli P, Herrmann A, Fulton L, Oelschner M, Hillmer S, Stratil TF et al. (2014) The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141: 4139–4148. 10.1242/dev.113878 [DOI] [PubMed] [Google Scholar]
- 51.Fulton L, Batoux M, Vaddepalli P, Yadav RK, Busch W, Andersen SU et al. (2009) DETORQUEO, QUIRKY, and ZERZAUST represent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG in Arabidopsis thaliana. PLoS Genet 5: e1000355 10.1371/journal.pgen.1000355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaddepalli P, Fulton L, Wieland J, Wassmer K, Schaeffer M, Ranf S et al. (2017) The cell wall-localized atypical β-1,3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 144: 2259–2269. 10.1242/dev.152231 [DOI] [PubMed] [Google Scholar]
- 53.Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F et al. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A 104: 15572–15577. 10.1073/pnas.0706569104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P et al. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A 104: 15566–15571. 10.1073/pnas.0706592104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henry E, Fung N, Liu J, Drakakaki G, Coaker G (2015) Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ros, autophagy, and plant immune responses. PLoS Genet 11: e1005199 10.1371/journal.pgen.1005199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juárez SP, Mangano S, Estevez JM (2015) Improved ROS measurement in root hair cells. Methods Mol Biol 1242: 67–71. 10.1007/978-1-4939-1902-4_6 [DOI] [PubMed] [Google Scholar]
- 57.Schenk ST, Schikora A (2015) Staining of callose depositions in root and leaf tissues. Bio-protocol 5: e1429. [Google Scholar]
- 58.Heim DR, Roberts JL, Pike PD, Larrinua IM (1990) A second locus, Ixr B1 in Arabidopsis thaliana, that confers resistance to the herbicide isoxaben. Plant Physiol 92: 858–861. 10.1104/pp.92.3.858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trehin C, Schrempp S, Chauvet A, Berne-Dedieu A, Thierry AM, Faure JE et al. (2013) QUIRKY interacts with STRUBBELIG and PAL OF QUIRKY to regulate cell growth anisotropy during Arabidopsis gynoecium development. Development 140: 4807–4817. 10.1242/dev.091868 [DOI] [PubMed] [Google Scholar]
- 60.Song JH, Kwak SH, Nam KH, Schiefelbein J, Lee MM (2019) QUIRKY regulates root epidermal cell patterning through stabilizing SCRAMBLED to control CAPRICE movement in Arabidopsis. Nat Commun 10: 1744 10.1038/s41467-019-09715-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sampathkumar A, Yan A, Krupinski P, Meyerowitz EM (2014) Physical forces regulate plant development and morphogenesis. Curr Biol 24: R475–83. 10.1016/j.cub.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwak SH, Woo S, Lee MM, Schiefelbein J (2014) Distinct signaling mechanisms in multiple developmental pathways by the SCRAMBLED receptor of Arabidopsis. Plant Physiol 166: 976–987. 10.1104/pp.114.247288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M et al. (2011) Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol 156: 1364–1374. 10.1104/pp.111.175737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K et al. (2006) An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J 46: 307–316. 10.1111/j.1365-313X.2006.02695.x [DOI] [PubMed] [Google Scholar]
- 65.Engelsdorf T, Hamann T (2014) An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann Bot 114: 1339–1347. 10.1093/aob/mcu043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haswell ES, Verslues PE (2015) The ongoing search for the molecular basis of plant osmosensing. J Gen Physiol 145: 389–394. 10.1085/jgp.201411295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nissen KS, Willats WGT, Malinovsky FG (2016) Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci 21: 516–527. 10.1016/j.tplants.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 68.He Y, Zhou J, Shan L, Meng X (2018) Plant cell surface receptor-mediated signaling—a common theme amid diversity. J Cell Sci 131: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma X, Xu G, He P, Shan L (2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033. 10.1016/j.tplants.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 70.Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A et al. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perraki A, DeFalco TA, Derbyshire P, Avila J, Séré D, Sklenar J et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252. 10.1038/s41586-018-0471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A et al. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N et al. (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- 74.Merz D, Richter J, Gonneau M, Sanchez-Rodriguez C, Eder T, Sormani R et al. (2017) T-DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J Exp Bot 68: 4583–4593. 10.1093/jxb/erx263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coleman AD, Raasch L, Maroschek J, Ranf S, Hückelhoven R (2019) The Arabidopsis leucine-rich repeat receptor kinase MIK2 is a crucial component of pattern-triggered immunity responses to Fusarium fungi. bioRxiv 720037.
- 76.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC et al. (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7: 923–926. 10.1093/mp/ssu009 [DOI] [PubMed] [Google Scholar]
- 78.Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396. [Google Scholar]
- 79.Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 80.Kumar M, Turner S (2015) Protocol: a medium-throughput method for determination of cellulose content from single stem pieces of Arabidopsis thaliana. Plant Methods 11: 46 10.1186/s13007-015-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424. 10.1016/s0003-2697(69)80009-6 [DOI] [PubMed] [Google Scholar]
- 82.Box MS, Coustham V, Dean C, Mylne JS (2011) Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7 10.1186/1746-4811-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enugutti B, Kirchhelle C, Oelschner M, Torres Ruiz RA, Schliebner I, Leister D et al. (2012) Regulation of planar growth by the Arabidopsis AGC protein kinase UNICORN. Proc Natl Acad Sci U S A 109: 15060–15065. 10.1073/pnas.1205089109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET et al. (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression levels of several CBI marker genes by qPCR upon exposure of seven-day-old seedlings to 600 nM isoxaben for eight hours. The results from five biological replicates are shown. Marker genes and genotypes are indicated. Mean ± SD is presented. Asterisks represent adjusted P values (** P < 0.009, *** P = 0.0002, **** P < 0.0001, ns: not significant; one-way ANOVA followed by Tukey’s multiple comparison tests). For all genotypes the differences between isoxaben and mock treatments were statistically significant (adjusted P value < 0.0001). Differences between mock treated wild-type and sub-9 samples were not statistically significant. The experiment was repeated twice with similar results.
(TIF)
The results from three biological replicates are shown. Mean ± SD is presented.
(TIF)
(A) Upper panel: siliques. Bottom panel: open siliques revealing developing seeds. Genotypes are indicated. Note aberrant sub-9 silique morphology and the reduced number of developing seeds in sub-9. Arrows indicate undeveloped seeds. (B) Root angle of nine-day-old seedlings grown in an upright position (10° angle relative to direction of gravity) on half-strength MS agar medium supplemented with 1% sucrose. Pictures were taken from the front of the plate. Genotypes are indicated. Note the slight slant to the left in mik2-3. (C) Quantification of results depicted in (B) and as described in Van der Does et al (2017) PLoS Genet 13: e1006832. Genotypes are indicated. Box and whisker plots are shown. Whisker ends mark the minimum and maximum of all the data. 16 ≤ n ≤ 22. Asterisks represent adjusted P values (*** P = 0.0001; one-way ANOVA followed by Tukey’s multiple comparison tests). The experiment was performed twice. In the second experiment the root angle difference in mik2-3 was not statistically significant. Scale bars: open siliques, 1 mm; closed siliques, 0.5 cm; roots, 1 cm.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.