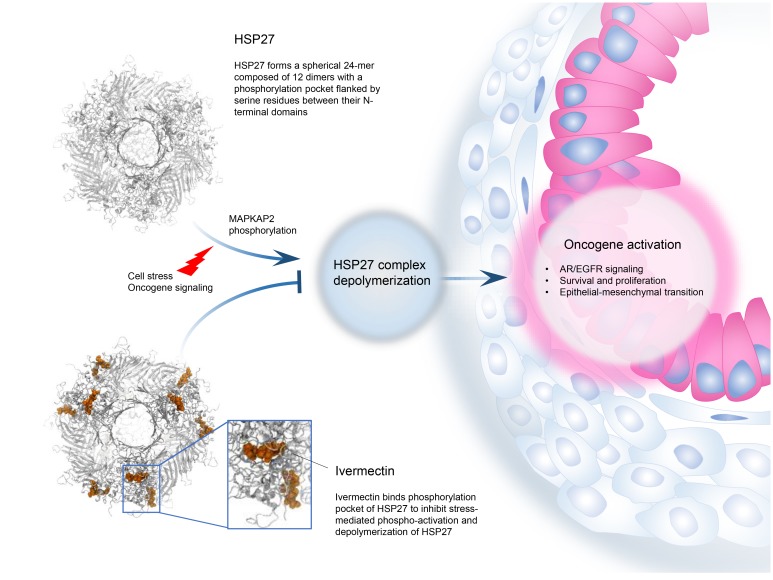
Abstract
HSP27 is highly expressed in, and supports oncogene addiction of, many cancers. HSP27 phosphorylation is a limiting step for activation of this protein and a target for inhibition, but its highly disordered structure challenges rational structure-guided drug discovery. We performed multistep biochemical, structural, and computational experiments to define a spherical 24-monomer complex composed of 12 HSP27 dimers with a phosphorylation pocket flanked by serine residues between their N-terminal domains. Ivermectin directly binds this pocket to inhibit MAPKAP2-mediated HSP27 phosphorylation and depolymerization, thereby blocking HSP27-regulated survival signaling and client-oncoprotein interactions. Ivermectin potentiated activity of anti–androgen receptor and anti-EGFR drugs in prostate and EGFR/HER2-driven tumor models, respectively, identifying a repurposing approach for cotargeting stress-adaptive responses to overcome resistance to inhibitors of oncogenic pathway signaling.
Keywords: Oncology
Keywords: Drug therapy
Introduction
Small heat shock proteins (sHSPs) are ATP-independent molecular chaperones characterized by a conserved α-crystallin domain located between a highly flexible, variable C-terminal region and a poorly conserved, disorganized N-terminal domain (NTD) (1). Like other chaperones, sHSPs bind proteins to facilitate substrate refolding, serving as ubiquitous components of protein quality control networks. HSP27 (also referred to as HSPB1) assembles into oligomers that interact with misfolded or specific client proteins to regulate proteostasis and cytoskeleton organization, as well as prosurvival signaling and transcriptional pathway activation (2). Assembly and depolymerization of HSP27 subunits are dynamic and context-dependent processes, mediated by phosphorylation of 3 serine residues (Ser15, Ser78, Ser82) in its NTD by kinases (e.g., MAPKAP2, p38 MAPK, p90RSK) activated by varied stress stimuli (e.g., hypoxia, heat shock, nutrient deprivation, anticancer treatments) (3). High levels of HSP27 are linked to neurodegenerative (Alzheimer, Parkinson) and other aggregopathies (e.g., cystic fibrosis) as well as inflammatory diseases (4, 5). HSP27 also regulates many hallmarks of cancer and is associated with poor outcome and treatment resistance. Notably, HSP27 phospho-activation facilitates proteostasis, prosurvival, and antiapoptotic pathways, as well as specific oncoprotein (e.g., BRAF, MET, AR, EGFR) interactions (6, 7). In many model systems, HSP27 suppression sensitizes cancer cells to therapy (8). Collectively, these broad effects of HSP27 on cancer cell proliferation and survival position HSP27 as a potential therapeutic target.
Since phospho-activation of HSP27 modulates its dynamic oligomer assembly and defines its affinity to client proteins (9), targeted inhibition of HSP27 phosphorylation would suppress many oncogenic pathways that facilitate treatment resistance. However, because HSP27 is intrinsically disordered and lacks an ATP-binding domain (1), rational discovery of small-molecule inhibitors of HSP27 poses significant challenges. Apatorsen (OGX-427) is a second-generation antisense oligonucleotide targeting HSP27 mRNA with anticancer activity in many preclinical models (7, 10, 11). Randomized phase II trials in castration-resistant prostate cancer reported higher response rates with apatorsen plus prednisone compared with prednisone alone, consistent with inhibition of HSP27 shuttling of androgen receptor (AR) into the nucleus (7, 12). Another phase II trial randomized 200 patients with advanced urothelial carcinoma to docetaxel plus apatorsen versus docetaxel alone and reported survival benefit for patients receiving apatorsen (13). While these studies report the first signals of activity for an HSP27 inhibitor in cancer, antisense drugs have variable intratumor distribution, and a more potent and convenient orally available small molecule would be preferable. To identify small-molecule inhibitors of HSP27, we followed a course of drug screening coupled with structural modeling approaches to resolve a targetable phosphorylation pocket in its NTD, identify ivermectin (IVM) as an inhibitor of HSP27 phospho-activation, and delineate its biochemical and functional mechanisms of action in cancer.
Results
Screening for HSP27 small-molecule inhibitors
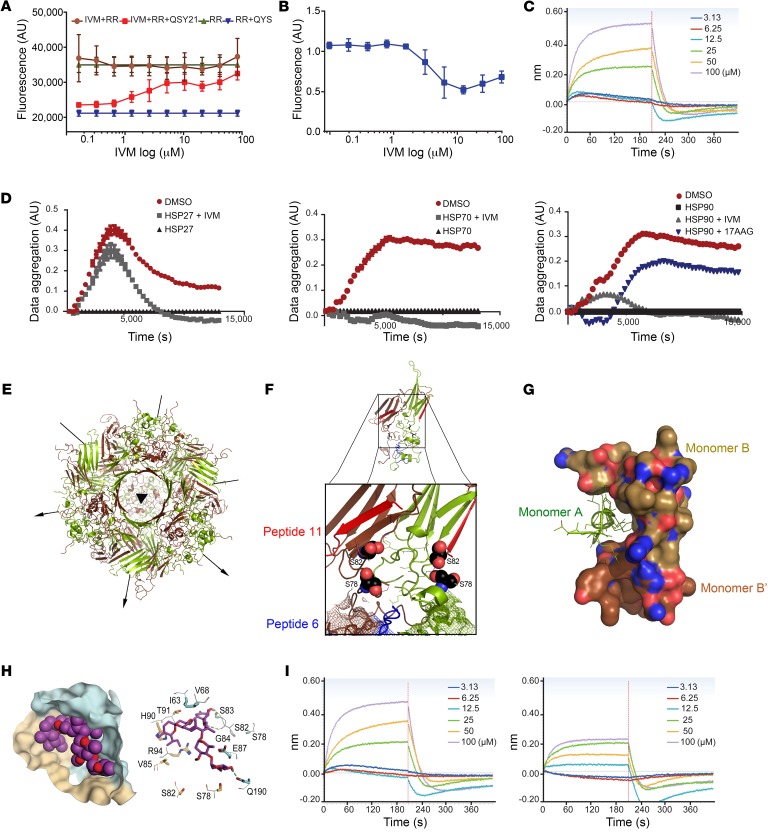
Combined screening using fluorescence resonance energy transfer (FRET) and fluorescence polarization (FP) assays was initially used to identify drugs in the Prestwick Chemical Library (Prestwick Chemical) that inhibit HSP27 functions (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI130819DS1); compounds passing both FRET and FP screening criteria were further analyzed in secondary assays (described below). Of 1280 Prestwick molecules evaluated, 6 passed initial FRET and FP screening (Supplemental Table 1), but only IVM passed confirmatory secondary assays, and was subsequently found to functionally inhibit HSP27. IVM is a member of the avermectin family of macrocyclic lactones (herein called IVM analogs) that similarly inhibit HSP27 in FRET and FP assays (Supplemental Table 2). Since rapid exchange of HSP27 subunits is essential for HSP27 chaperone activity (14), FRET, which relies on the distance-dependent transfer of energy between donor and acceptor molecule, was used to investigate drug-HSP27 interaction and effect on subunit exchange kinetics. IVM interfered with HSP27 fluorescence quenching when pools of Rhodamine Red–labeled HSP27 (HSP27-RR, electron donor) and QSY21-labeled HSP27 (HSP27-QSY21, electron acceptor) were mixed (Figure 1A). IVM also dose-dependently inhibited HSP27 interaction with unfolded insulin (Figure 1B), an established HSP27-client protein used to study HSP27 inhibition of misfolded protein precipitation, as shown by the FP assay (1, 15).
Figure 1. Ivermectin binds to HSP27 phosphorylation pocket.
(A) FRET assay using HSP27 labeled with QSY21 as negative control (blue triangles), and HSP27-RR alone (green triangles) or combined with IVM (brown circles) as positive controls. Red squares represent IVM effect on subunit exchange. (B) FITC-insulin precipitation in the presence of DMSO or increasing concentrations of IVM. (C) BLI dose-response curves reflect direct binding of IVM to purified HSP27 protein. (D) Protein aggregation in HSF1–/– MEF cells incubated with HSP27 (left) or HSP70 and HSP90 (middle and right) in the presence of IVM (gray) or DMSO (red). Protein lysate without chaperones was used as negative control (black). Results were normalized to HSP values alone. For HSP90 the effect of IVM was compared with that of 17AAG (blue). (E) Ribbon drawing of the HSP27 24-mer down its 3-fold symmetry axis. Arrows represent bisecting 2-fold axes. Monomers A are shown in green and monomers B in brown. (F) Magnified view of the interface between monomers in a dimer unit showing the phosphorylation pocket. Spheres indicate S78 and S82, and the mesh represents the WDPF motif of each monomer. Peptides 6 and 11 conferring chaperone activity are shown in blue and red, respectively. (G) Ribbon representation of the NTD of monomer A (green) fitting into a pocket created by its related monomer B and a neighboring monomer B NTD. (H) Left: Predicted binding pocket between the 2 NTDs of an HSP27 dimer: monomer A (gold), monomer B (cyan), and IVM (violet). Right: S78 and S82 and other amino acids involved in IVM interaction. Hydrogen bonds are shown in green. (I) BLI reported binding of IVM to the HSP27 phosphorylation pocket mutants S78A-S82A (left) and S78D-S82D (right).
These screening assays identify IVM analogs as possible HSP27-interacting molecules; secondary assays were subsequently used to evaluate inhibitory effects of IVM on HSP27 client–protein interactions.
Ivermectin directly binds HSP27 and prevents its interaction with misfolded proteins
Direct binding of IVM to HSP27 was confirmed by biolayer interferometry (BLI). HSP27 was expressed and purified with a biotinylated tag and immobilized on streptavidin biosensors. Sensors were then dipped in successive wells containing increasing concentrations of IVM. Binding of IVM to protein changes the spectral pattern of reflected light. BLI indicates dose-response binding of IVM to HSP27 (Figure 1C), while the AR antagonist enzalutamide, used as a negative control, did not bind HSP27 at any concentration (Supplemental Figure 1B).
To determine effects of IVM on HSP27 inhibition of protein aggregation, HSF1–/– MEF cells negative for HSP27, HSP70, and HSP90 were used as a source of cellular proteins (16). IVM reduced HSP27-mediated (but not HSP70- or HSP90-mediated) inhibition of protein aggregation in HSF1–/– MEF cell protein extracts (Figure 1D). IVM effects on HSP27-mediated inhibition of protein aggregation were similar to 17AAG suppression of HSP90-mediated inhibition of protein aggregation.
Since actin is an established interaction partner of HSP27 (17), an actin-pyrene assay was used to evaluate HSP27-regulated actin polymerization and its functional inhibition by IVM (Supplemental Figure 1C). IVM also reduced trypsin degradation of HSP27 (Supplemental Figure 1D). Collectively, these data demonstrate that IVM inhibits HSP27 interaction with specific client proteins.
Determination of oligomeric structure of HSP27
To facilitate study of HSP27 function and specifics of its inhibition by IVM, x-ray crystallography was used to resolve HSP27 structure. These studies define an oligomeric complex of HSP27 formed by 24 monomers in a spherical configuration (Figure 1E). Polypeptide chains form a layer of approximately 40 Å thickness about the surface of this spheroid, which has a central void of approximately 67 Å diameter. Notably, this complex is composed of 12 dimers wherein the interactions between 2 identical monomers (A and B) are distinct from the interactions formed with other dimer units. Overall the spheroid formed has a central 3-fold axis bisected by perpendicular 2-fold axes. Four open surface channels of approximately 28 Å diameter each are found about the surface of this complex and could potentially allow for access by small molecules.
The HSP27 monomer.
The HSP27 protein is composed of 205 amino acids constituent in a single polypeptide chain. At its structural core (residues 94–168) there is a beta-sandwich immunoglobulin-like domain that bears a close relationship to the structure and primary sequence of α-crystallin (99% similarity) (18). Emanating from this core and projected to the outer surface are the N- and C-terminal regions, consisting of residues 1–93 and 169–205, respectively. The α-crystallin domain of HSP27 is well resolved in our structural determination, whereas the remaining polypeptide chain demonstrates greater mobility. Residues found to be particularly mobile are the 55–93 and 190–205 positions. Some loop segments of the HSP27 protein were not built into our model because of positional disorder in those regions, and an average of 39 residues per chain were missing.
Dimer relationships.
Within the overall 24-meric structure of HSP27, its dimer units are involved in 2 types of interactions. Within the dimer itself there is a 2-fold relationship between identical monomers that leads to very compact structural packing. It is at this interface that much of the functionality of HSP27 would seem to reside. Notably, S78 and S82 in each monomer of HSP27 are positioned in a common surface cavity, suitable for small-molecule binding (Figure 1F). An increased degree of phosphorylation at these serine residues as well as at S15 drives dissociation of oligomeric HSP27 (9). Conversely, oligomeric complex formation is favored by a lack of phosphorylation at these residues (19). Coincident with oligomeric dissociation is the exposure of the hydrophobic surfaces of 2 nearby peptide strands composed of residues 42–56 (peptide 6) and residues 87–102 (peptide 11), which contributes to enhanced HSP27 chaperone activity with insulin and actin (17) (Figure 1B and Supplemental Figure 1C).
Modeling studies also established that the floor of the intersubunit cavity within HSP27 dimers is flanked on either side by the corresponding WDPF motif (residues 16–19) of both monomers and its associated S15 phosphorylation site. Our modeling does not reveal a direct contribution of S15 to the cavity formation (Figure 1F). The N-terminal extension of this polypeptide chain region is of particular relevance, as it forms interactions with not only the adjacent monomer, but also nearby dimer units (Figure 1G), and therefore appears to form a significant part in dimer-dimer association and oligomerization interactions. The presence of the WDPF motif along with the phosphorylation state of above-mentioned serine residues has also been reported to be a key factor in HSP27 oligomerization (20).
Placement of ivermectin binding between the NTDs of an HSP27 dimer
Our structural studies support the hypothesis that chemicals binding the phosphorylation pocket within HSP27 dimers comprising S78 and S82 of each monomer would inhibit phosphorylation and oligomer dissociation events and therefore block HSP27 function. Docking studies used to define specific interaction of IVM in this pocket indicate favorable hydrophobic interactions with H90, V85, I63, and V68 of monomer A and formation of hydrogen bonds with G54, S78, and Q190 residues of monomer B (Figure 1H). This binding model predicts that IVM would block phosphorylation activity about S78 and S82.
Further evidence of the propensity for IVM to bind at this site is apparent from 3 independent molecular dynamic (MD) simulations (Supplemental Figure 1E and Supplemental Videos 1 and 2) of the HSP27 24-mer in complex with 12 docked IVM molecules over 300 nanoseconds. These computations indicate that during each trial IVM contacted at least 1 of the 4 serine residues (Figure 1H and Supplemental Figure 1F).
Direct binding of IVM to the phosphorylation pocket of HSP27 was confirmed by BLI experiments using HSP27 double mutants S78D-S82D (chemically mimicking phosphorylated HSP27 and stabilizing HSP27 as dimers) and S78A-S82A (unphosphorylatable HSP27, stabilizing HSP27 as a 24-mer). These HSP27 double mutants were expressed and purified with a biotinylated tag and then exposed to increasing concentrations of IVM. Binding of IVM to S78A-S82A unphosphorylatable mutant was similar to its binding to wild-type (WT) protein (Figure 1I, left); in contrast, IVM binding affinity was significantly lower with the phospho-mimic double mutant S78D-S82D (Figure 1I), indicating that IVM binds to HSP27 in its non-phosphorylated form.
Ivermectin inhibits HSP27 phosphorylation and depolymerization
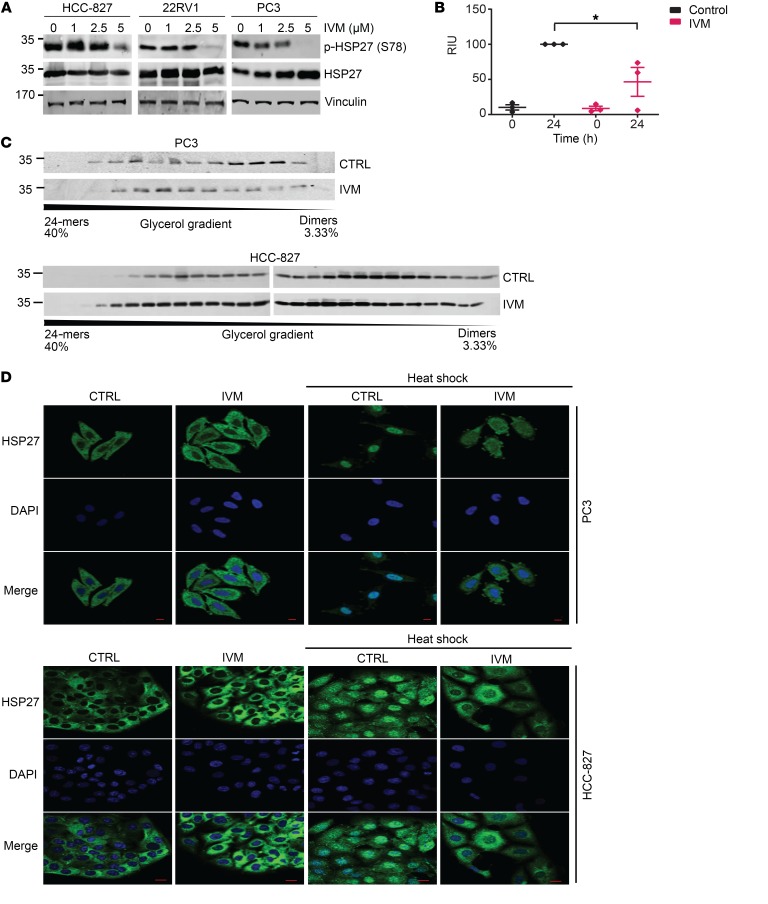
Consistent with predicted computational modeling described above, IVM potently inhibited HSP27 phosphorylation in lung and prostate cancer (Figure 2A) as well as bladder and breast cancer (Supplemental Figure 2A) cells. Notably, IVM affected mainly phosphorylated HSP27 (p-HSP27), while total HSP27 protein levels were marginally affected. Of the avermectins, IVM most potently suppressed p-HSP27 levels (Supplemental Figure 2B). Since IVM similarly inhibits phosphorylation of S78 and S82, while levels of S15 were only marginally altered, the effect of drug on S78 was selected as a readout in subsequent experiments.
Figure 2. Ivermectin inhibits phosphorylation and oligomerization of HSP27 in cancer cells.
(A) Protein expression of phosphorylated (S78) and total HSP27 was evaluated by Western blotting after heat shock (42°C for 1 hour) in lung HCC-827 and prostate 22RV1 and PC3 cancer cells after treatment with increasing doses of IVM for 24 hours. (B) Quantification of HSP27 phosphorylation expression assessed by Western blotting after in vitro incubation with MAPKAP2 kinase in the presence of DMSO or IVM, at time 0 and after 24 hours. Results were averaged and normalized as a percentage of the control at 24 hours. Data shown as mean ± SEM; *P < 0.05 by unpaired 2-tailed Student’s t test with Welch’s correction; n = 3 independent experiments. RIU, relative intensity units. (C) Expression of HSP27 protein fractions of glycerol gradient from PC3 (top panel) and HCC-827 (bottom panel) cells treated with DMSO (CTRL) or IVM (5 μM for 24 hours). (D) PC3 (top panel) and HCC-827 (bottom panel) cells treated with DMSO (CTRL) or IVM (5 μM for 24 hours) were exposed to heat shock (42°C for 1 hour), fixed using cold methanol, and stained for HSP27 (green). DAPI (blue) was used for nuclear staining. Scale bars: 20 μm.
p38 MAPK phospho-activates the serine/threonine MAPKAP2 kinase that directly phosphorylates serine and threonine residues in HSP27 (20). IVM did not alter protein levels of phospho–p38 MAPK (Supplemental Figure 2C), supporting direct suppression of HSP27 phosphorylation rather than suppression via upstream modulation of kinases that phosphorylate HSP27. Indeed, in vitro phosphorylation of purified WT-HSP27 by MAPKAP2 kinase was significantly decreased by IVM (Figure 2B), supporting direct inhibition of HSP27 by blocking of MAPKAP2-mediated phosphorylation.
Phosphorylation of HSP27 facilitates disassembly of HSP27 into smaller oligomers and enhances subunit exchange to promote HSP27 chaperone activity (9, 19). Consistent with its effects on phosphorylation, IVM shifted HSP27-oligomer equilibrium in prostate and lung cancer cells, stabilizing HSP27 in larger (less active) complexes (Figure 2C).
Ivermectin inhibits HSP27 nuclear translocation
Upon heat shock, HSP27 is phosphorylated and traffics to the nucleus to chaperone proteins required for mRNA processing during cellular recovery from stress. Consistent with its inhibitory effects on HSP27 phosphorylation and depolymerization, IVM clearly prevented heat shock–induced nuclear translocation of HSP27 in prostate and lung cancer cells (Figure 2D).
Collectively, the above screening, biochemical, structural, and computational studies identify IVM as an inhibitor of the HSP27 NTD, blocking access of MAPKAP2 kinase to S78 and S82 to prevent HSP27 phosphorylation and depolymerization. Additional biochemical studies were performed to define IVM effects on HSP27-mediated cellular and oncogenic pathways.
Ivermectin inhibits HSP27-regulated epithelial-mesenchymal transition and proteostasis
HSP27 plays roles in both epithelial-mesenchymal transition (EMT) (2) and proteostasis (4). Similarly to HSP27 silencing (2), IVM reduced cell migration and increased E-cadherin expression in prostate and lung cancer cells, both markers of EMT (Supplemental Figure 2, D and E). Like HSP27 silencing (10), IVM also led to endoplasmic reticulum (ER) stress with increased PERK and ATF4 expression (Supplemental Figure 2F). The inhibition of HSP27-regulated EMT and ER stress provides additional evidence of the on-target inhibition of IVM.
IVM inhibition of HSP27 is synergistically lethal to cells with oncogenic activation of EGFR or AR
IVM inhibits HSP27-regulated EGFR activity.
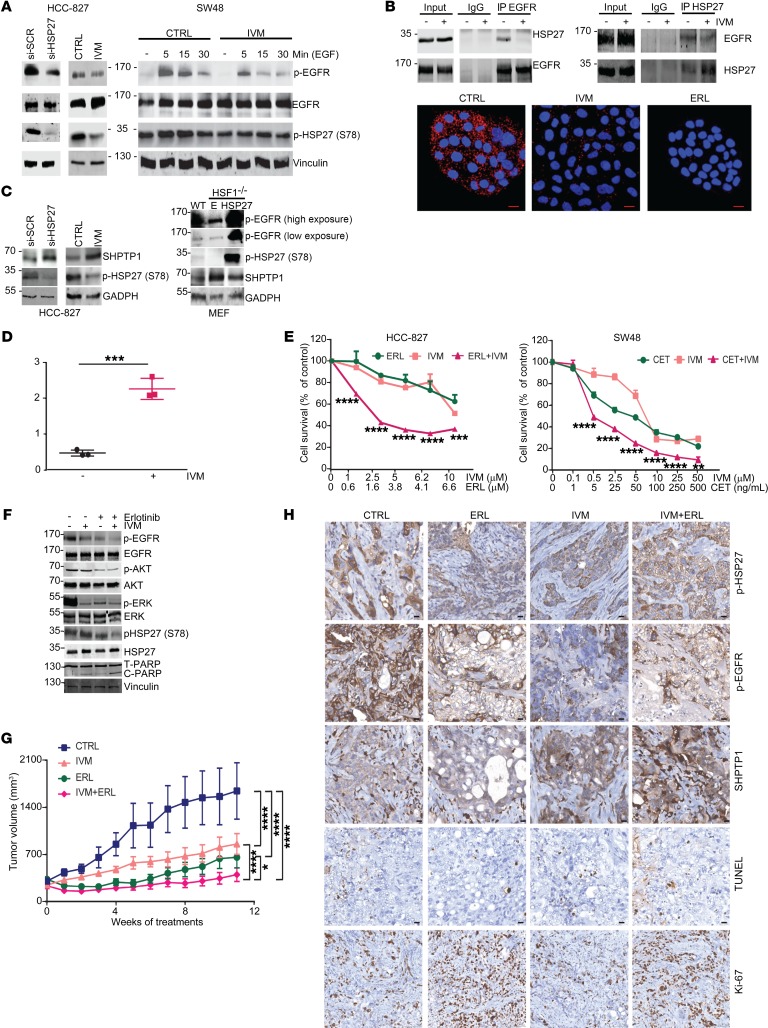
HSP27 is associated with resistance to inhibitors of EGFR and HER2, key oncogenic drivers in lung and breast cancer (11, 21). HSP27 inhibition, by IVM or si-HSP27, reduced EGFR phosphorylation in HCC-827 lung cancer cells harboring constitutionally active mutated EGFR, or after EGF stimulation in WT-EGFR SW48 colorectal cancer cells (Figure 3A). No changes were observed in EGFR mRNA or total protein levels (Supplemental Figure 3A and Figure 3A). Coimmunoprecipitation and proximity ligand assay demonstrate that HSP27 directly interacted with EGFR in HCC-827 cells and this was inhibited by IVM (Figure 3B and Supplemental Figure 3B). EGFR inhibition by erlotinib also decreased HSP27 interaction with EGFR (Figure 3B, bottom), suggesting that HSP27 interacted mostly with phosphorylated EGFR (p-EGFR); inhibition of EGFR phosphorylation, either directly with erlotinib or indirectly using IVM via HSP27, perturbed this interaction.
Figure 3. IVM reduces EGFR phosphorylation levels via HSP27 regulation of SHPTP1.
(A) Immunoblotting for p-EGFR and total and p-HSP27 after si-HSP27 (20 nM) or IVM (5 μM) in HCC-827 cells (left) or IVM (2.5 μM) before and after EGF stimulation (50 μg/mL) in SW48 cells (right). DMSO was used as control. (B) Top panels: Coimmunoprecipitation of HSP27 and EGFR with and without IVM (5 μM) for 24 hours in HCC-827 cells. IgGs were used as negative control. Bottom panels: Proximity ligation assay between HSP27 and EGFR in HCC-827 cells. Confocal microscopy was used to detect interaction (red dots). DNA was counterstained with DAPI (blue). Erlotinib was used as a negative control. Scale bars: 20 μm. (C) Left panel: Immunoblotting for SHPTP1 and p-HSP27 in HCC-827 cells as in A. Right panel: Immunoblotting for p-EGFR, SHPTP1, and p-HSP27 in wild-type and HSF1–/– MEF cells transfected with either empty (E) or HSP27 plasmid. (D) SHPTP1 phosphatase activity in HCC-827 cells after DMSO or IVM (5 μM). Data shown as mean ± SEM; ***P < 0.001 by unpaired 2-tailed Student’s t test with Welch’s correction; n = 3 independent experiments. (E) Cell survival of HCC-827 and SW48 cells measured by crystal violet assay after 4 days of treatment with DMSO, IVM, erlotinib, or cetuximab and their combination with IVM. Data shown as mean ± SEM; **P < 0.01, ***P < 0.001, ****P < 0.0001 by ANOVA with Bonferroni’s correction. (F) Immunoblotting for EGFR, p-EGFR, HSP27, p-HSP27, ERK, p-ERK, AKT, p-AKT, PARP, and cleaved PARP in HCC-827 cells treated as in E for 24 hours. (G) Tumor volume of HCC-827 xenografts treated with 15 mg/kg erlotinib, 10 mg/kg IVM, or a combination of both. Data shown as mean ± SEM; *P < 0.05, ****P < 0.0001 by ANOVA with Tukey’s correction; n = 8. (H) Immunohistochemical analysis of p-HSP27, p-EGFR, SHPTP1, and TUNEL staining of HCC-827 xenografts. Scale bars: 10 μm.
HSP27 regulates EGFR activation by modulating SHPTP1 phosphatase activity.
The activation of EGFR is tightly regulated by specific phosphatases that stop its intracellular signaling by dephosphorylating its tyrosine kinase domain (22). SHPTP1 has been demonstrated to significantly reduce EGFR phosphorylation in human colorectal cancer models (23). Therefore, changes in SHPTP1 levels were analyzed after HSP27 silencing or IVM to define how HSP27 regulates EGFR phosphorylation (Figure 3C, left). Low p-EGFR and high SHPTP1 levels were also detected in HSF1–/– MEF compared with wild-type MEF cells, further supporting an inverse relationship between HSP27 and SHPTP1 levels. HSP27 transfection in HSF1–/– MEF cells rescued these effects on p-EGFR and SHPTP1 (Figure 3C, right). SHPTP1 activity increased significantly in HCC-827 cells after IVM (Figure 3D), providing further evidence for HSP27-specific inhibition by IVM, as well as HSP27-mediated stabilization of p-EGFR levels via inhibition of SHPTP1.
Similarly to apatorsen (11), IVM augmented inhibitory effects of erlotinib and cetuximab on EGFR signaling, synergistically increasing activity in lung and colorectal cancer cells (Figure 3E); this synergistic effect was also confirmed in other EGFR/HER2-driven tumors (Supplemental Figure 3C). IVM-mediated inhibition of HSP27 phospho-activation enhanced erlotinib-mediated decreases in p-EGFR levels, suppressed AKT and ERK downstream of EGFR, and increased PARP cleavage (Figure 3F).
The pharmacokinetic profile of IVM was determined to support tumor growth inhibition studies in vivo. Following oral administration of 10 mg/kg, IVM serum concentrations peaked at 2 hours and remained at 2.13 μg/mL at 24 hours. After 6 days of oral daily treatment, serum concentration was 2.7 μg/mL, and calculated half-life was 26.30 hours. After 6 weeks of 10 mg/kg orally once daily, IVM intratumor concentration in PC3 xenografts was 1.93 μg/g with a serum concentration of 2.85 μg/mL, demonstrating excellent penetration of drug in tumor tissue (Supplemental Figure 3D). To evaluate effects of IVM on progression of constitutionally active EGFRmut lung cancer in vivo, mice bearing HCC-827 xenografts were randomized to vehicle, erlotinib, IVM, or combined erlotinib plus IVM. IVM was well tolerated with no apparent changes in activity and stable body weight of the mice treated with IVM alone or combined with erlotinib, in comparison with vehicle controls (Supplemental Figure 3D). IVM monotherapy reduced HCC-827 tumor p-HSP27 levels and inhibited tumor growth. More importantly, IVM most significantly inhibited tumor growth when combined with erlotinib (Figure 3G), increasing TUNEL staining, reducing p-EGFR staining, and increasing SHPTP1 staining. Although there was a trend toward reduction of Ki-67, this did not reach statistical significance (Figure 3H and Supplemental Figure 3E). p-EGFR staining was also reduced in A549 tumors treated with apatorsen plus erlotinib (11), further supporting the notion that HSP27 inhibition decreases p-EGFR levels in particular when combined with erlotinib (Supplemental Figure 3F).
Ivermectin inhibits AR pathway in prostate cancer models.
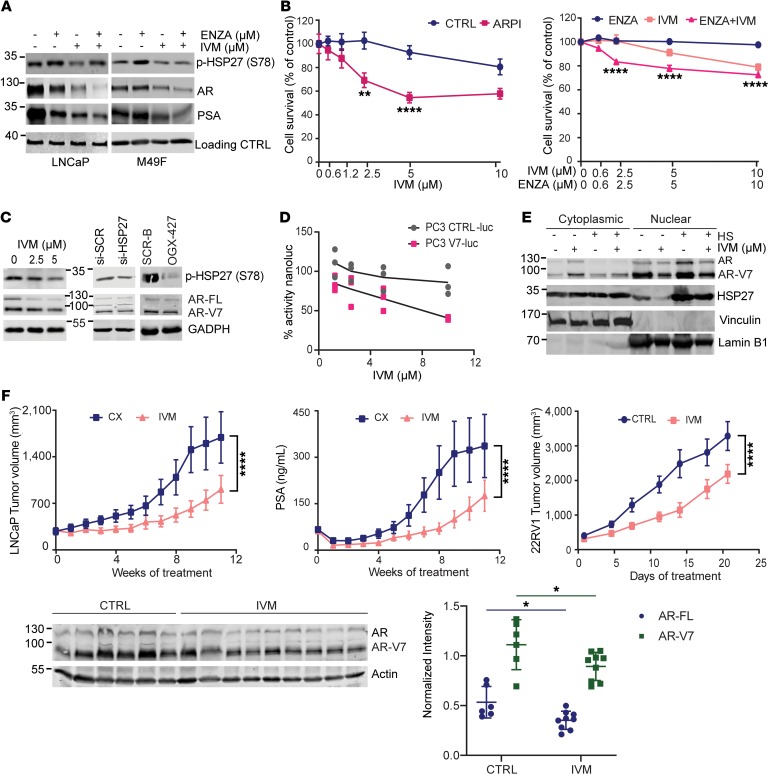
Resistance to AR pathway inhibitors in prostate cancer is associated with AR amplification, mutations, and expression of truncated AR variants, the latter characterized by loss of AR’s ligand-binding domain and constitutively active transcription (24). HSP27 has an established role in AR trafficking and stability, and its inhibition by either apatorsen or si-HSP27 reduces AR protein levels and activity, thereby delaying progression of castration-resistant prostate cancer (CRPC) (7, 8, 25). Similarly to apatorsen (7), IVM significantly reduced AR and prostate-specific antigen (PSA) protein expression in castration-sensitive LNCaP cells and ARF876L protein expression in enzalutamide-resistant M49F cells, an effect that was enhanced in combination with androgen deprivation therapy (in LNCaP cells) or enzalutamide (in M49F cells) (Figure 4A). IVM increased sensitivity of LNCaP cells to androgen deprivation therapy and of enzalutamide-resistant ARF876L M49F cells to enzalutamide (Figure 4B). Interestingly, IVM also decreased AR variant 7 (AR-V7) protein levels in 22RV1 cells, similarly to either si-HSP27 or apatorsen (Figure 4C). The reduction of AR or AR-V7 protein expression was not related to modulation of mRNA levels (Supplemental Figure 4A). The functional effect of IVM on AR-V7 transcriptional activity was also assessed using a PC3V7_3TKNLuc system that incorporates a doxycycline-inducible AR-V7 and probasin-based ARR3tk-Nanoluciferase reporter construct. As shown in Figure 4D, IVM significantly inhibited AR-V7 transcriptional activity compared with PC3_3TKNLuc control. AR-V7 nuclear translocation in 22RV1 cells was also inhibited by IVM, decreasing nuclear AR-V7 levels, with a corresponding increase in the cytoplasmic fraction (Figure 4E).
Figure 4. Ivermectin inhibits AR and AR-V7 to potentiate AR pathway inhibitors in vivo.
(A) Immunoblot for p-HSP27, AR, and PSA in LNCaP (left) and enzalutamide-resistant M49F (right) cells after treatment with DMSO, enzalutamide (ENZA) (10 μM), IVM (5 μM), or both for 24 hours. (B) Left panel: LNCaP cell survival in charcoal-stripped serum with or without IVM. Right panel: M49F cells treated with enzalutamide, IVM, or the combination of both for 4 days. Data shown as mean ± SEM; **P < 0.01,****P < 0.0001 by 1-way ANOVA with Bonferroni’s correction. ARPI, androgen receptor pathway inhibitors. (C) AR variant 7 (AR-V7) and full-length (AR-FL) protein expression was analyzed in 22RV1 cells after treatment with DMSO, IVM, si-HSP27, or apatorsen (OGX-427). Scrambled sequences were used as control (si-SCR and SCR-B). Levels of p-HSP27 were used to confirm on-target effect. (D) Dose-dependent inhibition of AR-V7 transcriptional activity measured by luminescence in PC3V7_3TKNLuc cells compared with PC3_3TKNLuc empty control, normalized to DMSO vehicle in each group. n = 3 independent experiments. (E) HSP27 and AR-V7 protein expression was evaluated in cytoplasmic and nuclear fractions of 22RV1 cells treated with DMSO or IVM (2.5 μM) for 24 hours with or without heat shock (42°C for 1 hour). (F) Top panels: Tumor volume and serum PSA quantification of LNCaP xenografts after castration treated with vehicle (CX) or IVM (10 mg/kg). PSA measurement was assessed weekly by ELISA. Lower panels: Western blotting with relative protein expression quantification for AR-FL and AR-V7 of 22RV1 xenografts treated with vehicle (CTRL) or IVM (10 mg/kg). Data shown as mean ± SEM; *P < 0.0001. Tumor volume was compared by ANOVA followed by Tukey’s correction, n = 10 and 6–9 per group for LNCaP and 22RV1, respectively.
LNCaP and 22RV1 xenograft models were used to determine effects of IVM on CRPC progression in vivo. Male mice bearing LNCaP xenografts (26) were castrated when tumors exceeded 300 mm3 and randomized to vehicle or IVM administered 10 mg/kg orally 3 times per week. IVM significantly reduced tumor volume and serum PSA levels after castration, delaying time to CRPC (Figure 4F). Moreover, IVM significantly reduced full-length AR (AR-FL) and AR-V7 protein expression and 22RV1 tumor volume growth in vivo (Figure 4F).
Collectively these data are consistent with IVM inhibition of HSP27-mediated nuclear transport of AR and its variants to delay prostate cancer progression, defining a novel approach to suppress AR genomic alterations that drive resistance in CRPC.
Ivermectin potentiates taxane chemotherapy.
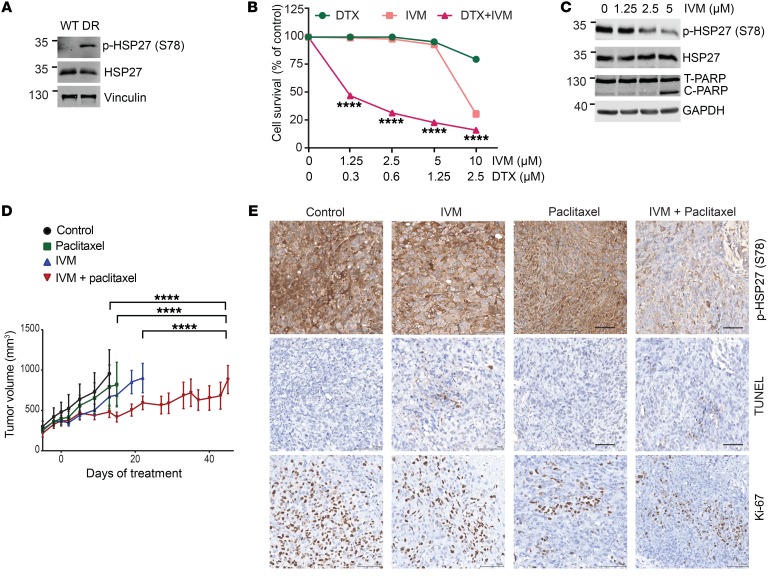
HSP27 is linked to resistance to chemotherapy in several cancers (27), and apatorsen plus docetaxel was reported to prolong survival, over docetaxel alone, in a randomized trial of 200 patients with advanced urothelial carcinoma (28). p-HSP27 levels are higher in docetaxel-resistant PC3 cells compared with parental PC3 cells (Figure 5A). IVM sensitized PC3 prostate (Figure 5, B and C) and UC14 urothelial cancer cells (data not shown) to docetaxel and reduced taxane-induced p-HSP27 protein levels. In mice, IVM significantly delayed docetaxel-resistant PC3 tumor progression compared with either drug alone (Figure 5D), lowering p-HSP27 and Ki-67 levels, and increasing TUNEL staining in tumor tissue in the combination arm (Figure 5E and Supplemental Figure 4B).
Figure 5. Ivermectin reverses taxane resistance in prostate cancer cells.
(A) Immunoblotting analysis of p-HSP27 and total HSP27 in PC3 parental (WT) and PC3 docetaxel-resistant (DR) cells. (B) Cell survival of PC3-DR treated with docetaxel (DTX), IVM, and the combination of both drugs for 4 days. Data shown as mean ± SEM; ****P < 0.0001 by ANOVA with Bonferroni’s correction. (C) Immunoblotting analysis of p-HSP27 and cleaved PARP in PC3-DR cells treated with increasing doses of IVM for 24 hours. (D) Tumor volume of PC3-DR xenografts treated with control (normal saline), IVM (10 mg/kg orally, every other day), micellar paclitaxel (0.5 mg i.v., 3 times per week, every other week), IVM (10 mg/kg by gavage, 3 times per week), and the combination of both drugs. Data shown as mean ± SEM; ****P < 0.0001; n = 8–14; compared by ANOVA followed by Tukey’s correction. (E) Representative images of immunohistochemical analysis of p-HSP27, TUNEL, and Ki-67 on PC3-DR xenografts treated as specified in D. Scale bars: 100 μm.
Discussion
Cancer cells co-opt key homeostatic stress responses supporting cell survival and therapy resistance, and molecular chaperones like sHSPs play prominent roles in many stress signaling and transcriptional regulatory networks. In particular, HSP27 is a stress-activated multifunctional chaperone highly expressed in many cancers that regulates misfolded protein and ER stress responses (2, 4), as well as cell survival and oncogenic pathways implicated in cancer progression and treatment resistance (8, 29).
HSP27 inhibition, before now only possible by nucleotide-based approaches, induces proteotoxic stress and apoptosis in many varied cancers (27, 30), downregulating oncogenic client proteins and signaling pathways that drive proliferation and EMT (10). Preclinical studies using HSP27 siRNA (25) or antisense-based therapy (10, 11, 27, 30) sensitize cancer cells to combinatorial regimens, and phase II trials of apatorsen in metastatic CRPC and bladder cancer (12, 28) report the first signals of activity for HSP27 inhibitors. While these reports are promising, more potent small-molecule inhibitors are likely to yield improved cancer outcomes.
To address challenges of drugging a highly disordered chaperone like HSP27, we integrated structural modeling approaches with biochemical screening of a library of small molecules and specific functional assays. Following initial identification of IVM in FRET and FP screening assays, interaction with and inhibition of HSP27 were confirmed using various complementary biochemical studies. BLI assays of wild-type and mutated forms of HSP27 confirmed direct binding by IVM and contributed to identification of the specific binding region within the NTD.
X-ray diffraction data and computer modeling of HSP27 structure defined a spherical 24-mer oligomeric complex with a 3-fold axis and perpendicular 2-fold axis. Each monomer is constructed about a structurally conserved α-crystallin domain exhibiting a 6-stranded beta-sandwich as previously described (31, 32). This conserved domain is flanked by more mobile and variable N- and C-termini that form flexible loops with some sections undefined owing to a lack of electron density as a result of high mobility. The overall structure is similar to an oligomeric lens α-crystallin cryoelectron microscopy structure (18).
Each dimer unit in the oligomeric assembly forms a pocket that appears to regulate HSP27 function (Figure 1F). S78 and S82 in the NTD of each monomer are positioned in a common surface pocket, the floor of which is flanked on either side by the corresponding WDPF motif (residues 16–19) of both monomers and its associated S15 phosphorylation site. This portion of polypeptide chain forms interactions with not only the adjacent monomer, but also nearby dimer units, and is likely key to promoting dimer-dimer association and oligomerization. S78 and S82 of the constituent subunits, when phosphorylated, promote dissociation of the HSP27 complex into active-state dimers and the affinity of HSP27 for the spectrum of its context-dependent interactome (9, 15). On the basis of our structural modeling studies, we conclude that IVM binds a pocket comprising S78 and S82 phosphorylation sites between the 2 NTDs of an HSP27 dimer, preventing both phosphorylation and oligomer dissociation events, and therefore HSP27 function.
IVM has known anticancer properties, including inhibition of WNT/TCF and HIPPO (33, 34) and induction of mitochondrial ROS and autophagy through degradation of PAK1 (35). Interestingly, HSP27 plays roles in WNT and proteostasis pathways, linking these reported effects to IVM-mediated inhibition of HSP27. While PAK1 can lead to phosphorylation of HSP27 via phospho–p38 MAPK, we established that IVM did not reduce phospho–p38 MAPK levels, indicating that inhibition of HSP27 phosphorylation by IVM is not mediated via PAK1. IVM also inhibits multidrug resistance and efflux of many chemotherapy drugs (36), a mechanism that in part contributes to its activity in docetaxel-resistant PC3 cells.
The current work used a broad arsenal of biochemical, structural, and computational methodologies that define a druggable pocket flanked by 2 serine residues in the NTD of HSP27 which, when bound with IVM, inhibits phosphorylation and depolymerization of HSP27 into its active oligomers. IVM inhibits HSP27-specific interaction with client proteins, nuclear translocation, and oncogenic effects similar to those seen with siRNA or antisense drugs. Several chemical compounds (e.g., hardwickiic acid, brivudine) have been reported to inhibit HSP27, but the lack of integrated structural, molecular dynamic, or functional studies fails to confirm specific and direct interaction with HSP27 (37, 38). Given its context-dependent phospho-activation and multiple interactomes, HSP27 can be indirectly affected by compounds through upstream alteration of signaling and/or kinase activity. Thus, xanthones and JCC76 downregulate HSP27 (39, 40), while KRIBB3 reduces HSP27 phosphorylation (41) through unknown mechanisms. Zerumbone and its derivatives inhibit HSP27 through induction of cysteine cross-linking of HSP27 (42), of particular relevance given its 24-mer oligomeric complex (Figure 1E). In the past, computational protocols have been used to build HSP27 monomer models for docking studies of putative HSP27 inhibitors (38, 43). These models, however, have not been published, and could not be compared with the current structural model of HSP27 (38).
HSP27 modulates cancer cell sensitivity to targeted drugs by regulating diverse survival pathways. IVM results in phenocopy effects similar to HSP27 silencing (7, 10, 27), inhibiting stress-induced HSP27 phosphorylation and functions in cancer cells, in particular in the dynamic context of oncogenic driver inhibition. IVM inhibited AR and EGFR pathways, and potentiated activity of anti-AR and -EGFR drugs in prostate and lung cancer models, respectively. We also define new molecular crosstalk between HSP27 and selective client-oncoprotein interactions. AR genomic alterations (including amplification, truncated variants, mutations) emerge under selective pressures of castration to drive CRPC progression. IVM, via inhibition of HSP27, reduces AR full-length and AR-V7 protein levels and activity, defining a novel approach to suppress AR variants that drive resistance to AR pathway inhibitors.
HSP27 is also highly expressed in, and supports oncogene addiction of, EGFR- and HER2-driven lung and breast cancer (11, 21). Despite initial responses to EGFR inhibitors, resistance-conferring EGFR mutations can emerge and drive progression, which may be overcome by cotargeting of adaptive pathways upregulated by treatment. Similarly to apatorsen (11), IVM synergistically increased activity of erlotinib in lung, cetuximab in colorectal, and trastuzumab in breast cancer cells. We define, for the first time to our knowledge, HSP27-mediated inhibition of the phosphatase SHPTP1, which increases p-EGFR levels and EGFR signaling. IVM inhibits erlotinib-induced increases in HSP27 phospho-activation, which subsequently increases SHPTP1-mediated dephosphorylation of EGFR, synergistically inhibiting tumor growth.
In summary, these studies help to define a 3D model of HSP27 with a targetable phosphorylation pocket in its NTD, and identify IVM as an inhibitor blocking access of MAPKAP2 kinase to S78 and S82 to prevent HSP27 phosphorylation, depolymerization, and activation. Inhibition of HSP27 by IVM synergistically enhances AR and EGFR pathway inhibitors, providing a novel repurposing approach to delay emergent resistance to oncogene pathway inhibition. Repurposing IVM in cancer would require higher and longer dosing than its approved use as an antiparasitic. While no significant adverse events were observed in the many in vivo experiments in this study, IVM activates GABA-A receptor activity, which can produce neurologic toxicity. To address these issues, IVM analogs have been designed with lower affinity to the GABA-A receptor while maintaining favorable oral bioavailability, pharmacokinetics, and HSP27 binding affinity, from which a lead compound will be selected for clinical development in combination with AR and EGFR/HER2 inhibitors.
Methods
Crystallization and x-ray diffraction analyses
Purified wild-type HSP27 protein in 20 mM Tris at pH 8.4 was concentrated to 20 mg/mL for crystallization trials. The sitting drop vapor diffusion method was used, with drops containing 1 μL of protein solution, 0.7 μL of 0.5 M ammonium phosphate monobasic, and 0.3 μL of 40% vol/vol tert-butanol. The well solution consisted of 130 μL of 0.5 M ammonium phosphate monobasic. Crystal growth was very slow, with only 2 small crystals (~0.002 mm/side) observed over a 6-month incubation period. These were the only crystals obtained, despite extensive attempts to crystallize this protein under these conditions or in alternative wide-ranging screening conditions. The crystals obtained were prepared for x-ray diffraction analyses by transfer to well solution, supplemented with 30% glycerol, and then flash-frozen in liquid nitrogen. Crystallographic data were collected on beamline 7-1 on an ADSC Q315R CCD detector, with an x-ray wavelength of 1.0971 Å, at the Stanford Synchrotron Radiation Lightsource. Data frames were collected using 0.5° oscillations over a 180° range at 100 Kelvin. Observed spot intensity profiles had minimal backgrounds and were well defined. Data processing was carried out with MOSFLM (44) and scaled using Scala from the CCP4 Programme Suite (45). The best crystal diffracted to 3.58 Å resolution. A molecular replacement solution was achieved using an electron microscopy model of eye lens αB-crystallin (PDB ID: 2YGD; RCSB Protein Data Bank, rcsb.org) (18), modified by CHAINSAW (45) and using the resultant polyalanine starting model in PHASER (46). Refinement was achieved by repeated alternating cycles of automated refinement in REFMAC5.8 (45) and manual building in COOT (47). REFMAC parameters used in refinement included local noncrystallographic symmetry restraints, map sharpening, and jelly-body refinement. We included an intensity-mode twin refinement in REFMAC5.8 refinement runs given that the crystal displayed 8 twin domains, including a major hkl twin domain with a fraction of 0.317. The final model was validated using the program PROCHECK (45). Related data collection and refinement statistics are given in Supplemental Table 3, and coordinates have been deposited in the Protein Data Bank (PDB ID: 6DV5; RCSB Protein Data Bank)
Molecular modeling for HSP27 oligomer
Molecular Operating Environment software (MOE2010) (48) was used to complete missing loops and determine the missing side chain orientations in the x-ray structure of HSP27 oligomer. MOE was also used to identify the serine phosphorylation sites on the HSP27 24-meric model. The binding mode of IVM within the phosphorylation site was identified by docking with GLIDE (49). All molecular dynamic (MD) simulations were carried out using the GPU implementation of pmemd (50) in the AMBER15 (51) package and were executed on Helios GPU cluster administered by Compute Canada. Detailed MD simulation protocols are provided below.
MD simulation of HSP27 24-mer and IVM-bound HSP27 24-mer complex
In order to validate the structural integrity of the predicted IVM interaction, we conducted all-atom explicit solvent MD simulation of HSP27 24-mer with 12 IVM.
Structure preparation for molecular docking and MD simulation.
The full HSP27 24-mer structure (Figure 1E) was constructed from the resolved x-ray structure with a homology modeling approach using the MOE suite of programs (52), and IVM was docked to the identified binding site between the NTDs using the GLIDE module in the Schrödinger suite of programs (53, 54).
MD simulation protocol.
All MD simulations were carried out using the GPU implementation of pmemd (50) in AMBER15 (51). ff14SB (55) was used for the HSP27 oligomer, and GAFF (56) with partial charges generated by RESP (57) was used for IVM. The solute (HSP27 oligomer or HSP27 oligomer–IVM complex) was solvated with TIP3P water molecules (58, 59) within the cubic box, ensuring a solvent shell of at least 15 Å around the solute. The system was neutralized with sodium ions.
As described in the main text, the docking-predicted binding pose of IVM interacts with the NTDs of HSP27 dimers in the phosphorylation pocket (Figure 1H), where 2 of the 3 phosphorylatable serines, S78 and S82, are located.
Three independent MD simulation runs were performed for 300 nanoseconds each at ambient conditions and showed that the IVM stayed well in the predicted phosphorylation pocket on the HSP27 24-mer.
The average root mean square deviation (RMSD) of protein backbone over 3 runs (bold lines) and the deviation of each RMSD from their average (vertical bars) were calculated as shown in Supplemental Figure 1F.
Fluorescence resonance energy transfer
Fluorescence resonance energy transfer (FRET) assays were performed in Corning 384-well black flat-bottom plates. Twenty microliters of buffer (10 mM Tris, pH 8.4), 5 μL inhibitor (200 μM), 10 μL Rhodamine Red–HSP27 (Thermo Fisher Scientific) (45 nM), 10 μL QSY21-HSP27 (0.675 μL) was successively added to wells. Samples were incubated for 20 minutes at 37°C before fluorescence measurements using TECAN INFINITE M200PRO, with an excitation wavelength of 515 nm and an emission wavelength of 590 nm.
Fluorescence polarization
After FRET measurements, 10 μL of WT-HSP27 (2.475 μM), 10 μL of insulin-FITC (1.56 μM), and 10 μM TCEP (75 mM) were added to appropriate wells with a 2-minute equilibration period between each addition. The plate was then incubated at 42°C for 20 minutes, and fluorescence polarization (FP) measurements were taken (TECAN INFINITE F500, excitation wavelength 485 nM and emission wavelength 535 nM).
Aggregation assay
HSF1–/– MEF cells were lysed using 25 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40 lysis buffer. BSA 100 g/L and 1 M DTT were prepared in advance and thawed at room temperature. In each sample well of a Corning 96-well plate, 20 μL of BSA, 100 μL of cell lysates at 1 mg/mL, and 1 μL of DMSO or IVM with or without 1 μL of 1 mg/mL WT-HSP27 (HSP70 or HSP90) were successively added, and PBS was used to complete the total volume of 320 μL. After 5 minutes of incubation at room temperature, the plate was subjected to a 3-hour kinetic cycle with reading every 5 minutes at 340 nM at 42°C.
Oligomerization assay
A 12-step glycerol gradient ranging from 3.33% to 40% was constructed with 3.33% stepwise increments with total volume of 9 mL. Cell lysates were prepared in 25 mM HEPES (pH 7.4), 1 mM EDTA, 3.33% glycerol, 0.1 mM PMSF, 1 mM DTT and sonicated for 10 seconds. An equal amount of proteins, measured by bicinchoninic acid assay analysis, were loaded on a glycerol gradient and subjected to ultracentrifugation for 18 hours at 36,000 rpm (160,000 g) using a SW41Ti swing bucket rotor. Three-hundred-microliter fractions were collected into 96-well plates using a minipump and stored at –30°C for Western blot analysis.
Trypsin degradation assay
HSP27 and DTT were dispersed into 37°C PBS at a final concentration of 1 μM HSP27 and 20 mM DTT and allowed to equilibrate in the presence or absence of 5× (wt/wt) excess IVM for 5 minutes before addition of trypsin to a final concentration of 0.126 μM. Trypsinolysis reaction was stopped at different time points by taking 100-μL aliquots and mixing it with 2 μL of 10 M hydrochloric acid. Samples were then flash-frozen and stored at –80°C until Western blotting analysis.
Actin pyrene assay
HSP27 was incubated at 42°C with or without IVM. Unlabeled actin was then added to HSP27 with or without IVM and incubated for 3 minutes at 42°C. HSP27 solutions with or without IVM were added separately to pyrene-labeled actin (10:1 unlabeled to labeled actin ratio). Subsequently, the exchange buffer (10 mM EGTA, pH 8, 1 mM MgCl2) was added, and the samples were balanced using buffer A (2 mM Tris-HCl, pH 8, 200 μM ATP, 0.5 mM DTT, 0.1 mM MgCl2) and transferred into a cuvette with a final actin concentration of 4 μM. Fifty microliters of 10× KMEI buffer (500 mM KCl, 100 mM imidazole, pH 7.0, 10 mM MgCl2, 10 mM EGTA) was added to each sample. The pyrene intensity was measured by fluorometry at an excitation wavelength of 365 nm and an emission wavelength of 407 nm. Actin alone was used as control.
Immunofluorescence
Prostate and lung cancer cells were plated on coverslips in 12-well plates, treated with IVM for 24 hours, and exposed to heat shock (42°C for 1 hour, no recovery time). Cells were then fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton, washed, blocked in PBS (3% BSA), and incubated with the anti-HSP27 primary antibody. Antigens were visualized using Alexa Fluor 488–conjugated antibodies. Nuclei were stained with DAPI VECTASHIELD mounting reagent (Vector Laboratories). Images were acquired on a Zeiss LSM-780 confocal microscope at ×63 magnification.
Cancer cell lines
Lung (A549 [EGFR wild-type and KRAS mutated: G12S], HCC-827 [EGFR mutated: delE746-A750]), breast (BT-474 and MCF7), prostate (22RV1, PC3), and colorectal (HCT-116, SW48) cancer cells were purchased from the American Type Culture Collection. T24 bladder cancer cells were provided by the Pathology Core of the Bladder Cancer Specialized Program of Research Excellence at MD Anderson Cancer Center (Houston, Texas, USA). Parental and HSF1–/– MEF cells were a gift from I.J. Benjamin, University of Utah (Salt Lake City, Utah, USA). Resistant HCC-827 cell lines were developed by maintenance of HCC-827 parental cells in 500 nM erlotinib for 3 months. Enzalutamide-resistant M49F cells were selected from LNCaP cells in our laboratory. Cells were maintained in RPMI 1640 medium (Invitrogen Life Technologies) containing 10% FBS (Invitrogen Life Technologies) in the presence of enzalutamide (10 μM) (11). Docetaxel-resistant PC3 cells were established in our institution, as previously reported (60). LNCaP cells were provided by Leland W.K. Chung (MD Anderson Cancer Center). PC3, MEF, MCF7, and BT-474 cell lines were maintained in DMEM (Invitrogen). LNCaP, 22RV1, A549, and HCC-827 cells were maintained in RPMI 1640 (Invitrogen Life Technologies) supplemented with 10% FBS. SW48 cells were grown in Leibovitz’s L-15 medium supplemented with 10% FBS. T24 and HCT-116 cell lines were maintained in McCoy’s 5a Medium with 5% FBS. All lines were tested for mycoplasma contamination regularly. Genomic sequencing was done to confirm cell line identity.
Reagent and antibodies
Erlotinib was purchased from LC Laboratories, docetaxel from Sanofi-Aventis, enzalutamide from Shanghai Haoyuan Chemexpress, and IVM from Aikon. Cetuximab and trastuzumab were obtained from the BC Cancer Agency, Vancouver Centre (Vancouver, British Columbia, Canada); polymeric micellar paclitaxel was supplied by Pharmaceutical Sciences; and 17AAG was purchased from AG Scientific Inc. IVM B1a and emamectin were obtained from AbovChem, milbemycin from Cayman Laboratory, and eprinomectin, abamectin, selamectin, and doramectin from AK Scientific.
The primary antibodies used in the study were anti-HSP27 (SPA-803), anti–p-HSP27 (S78) (SPA-523), and anti–p-HSP27 (S15) (SPA-524-F) from Enzo; anti–p-HSP27 (S82) (2404S), anti-EGFR (4267S), anti–p-EGFR (Tyr1068) (3056S), anti–p-ERK (Thr202/Tyr204) (9107S), anti-AKT (9102S), anti–p-AKT (S473) (4060S), anti-PARP (9541S), anti–phospho–p38 MAPK (Thr180/Tyr182) (4511S), and anti–cleaved caspase-3 (9661) from Cell Signaling Technology; anti-SHPTP1 (ab 32559) from Abcam; anti-vinculin (V9131) and anti–β-actin (A3853) from Sigma-Aldrich; and anti-AR (441), anti–lamin B1 (sc-377001), anti-GAPDH (sc-137179), anti-CREB2 (ATF4) (sc-390063), and anti-PERK (sc-377400) from Santa Cruz Biotechnology. E-cadherin (610181) was from BD Biosciences. AR-V7 (31-1109-00) was from RevMAb Biosciences. Secondary antibodies were anti-mouse IgG–HRP (sc-2314) and anti-rabbit IgG–HRP (sc-2313) from Santa Cruz Biotechnology; and anti-mouse 800 (926-32212), anti-mouse 680 (926-68072), anti-rabbit 800 (926-32213), and anti-rabbit 680 (926-68073) from LI-COR. Secondary antibodies for immunofluorescence staining were donkey anti-mouse–Alexa Fluor 594 (A-21203) and donkey anti-rabbit–Alexa Fluor 488 (A-21206) purchased from Invitrogen Life Technologies.
Transfection
Cells were transfected with siRNA and apatorsen as described previously (61). The sequence of HSP27 siRNA corresponds to the human HSP27 site (5′-GUCUCAUCGGAUUUUGCAGC-3′; Dharmacon). A scrambled siRNA (5′-CAGCGCUGACAACAGUUUCAU-3′; Dharmacon) was used as a control for RNAi experiments. HSP27 ASO and scrambled (ScrB) control oligonucleotide sequences were manufactured by Ionis Pharmaceuticals and supplied by OncoGenex Technologies. The sequence of apatorsen corresponds to the human HSP27 translation initiation site (5′-GGGACGCGGCGCTCGGTCAT-3′). pHR-CMV empty and wild-type HSP27 vectors (WT) were generated and used as previously described (61).
Immunoprecipitation and immunoblot analysis
Whole-cell extracts were obtained by lysing of the cells in an appropriate volume of ice-cold RIPA buffer composed of 50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS containing 1 mmol/L Na3VO4, 1 mmol/L NaF, 1 mmol/L PMSF, and protease inhibitor cocktail tablets (Complete, Roche Applied Science). Cellular extracts were clarified by centrifugation at 13,000 g for 10 minutes, and protein concentration was determined by a BCA protein assay kit (Thermo Fisher Scientific). Thirty micrograms of the proteins’ extracts were boiled for 5 minutes in SDS sample buffer, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Membranes were probed with dilutions of primary antibodies listed above, followed by incubation with either HRP-conjugated or fluorescent secondary antibodies. After washing, proteins were visualized by either a chemiluminescent detection system (GE Healthcare) or an Odyssey Imaging System (LI-COR). For coimmunoprecipitation the soluble proteins were precleared with protein A/G-Sepharose beads for 1 hour. Precleared lysates were incubated overnight with 5 μg antibody or IgG at 4°C with constant rotation. A/G beads were added for 1 hour, and after centrifugation, the pellet was washed twice. Proteins were eluted from the beads by incubation of the immunoprecipitates with 60 μL of 2× SDS sample buffer (24 mM Tris, pH 6.8, 10% [vol/vol] glycerol, 0.8% [vol/vol] SDS, 6 mM 2-mercaptoethanol, and 0.04% bromphenol blue) at 90°C for 3 minutes. Nuclear/cytoplasmic fractions were extracted using the CelLytic NuCLEAR Extraction Kit (Sigma-Aldrich) according to the manufacturer’s protocol. Molecular weight markers are specified in all the blots.
Migration (wound healing) assay
HCC-827 and LNCaP cells were plated in coated 96-well ImageLock plates (Essen Bioscience) at 2 × 104 cells per well. After 24 hours the wound was made in each well using Woundmaker (Essen Bioscience), and the cells were treated in the presence or absence of IVM. The wound closure was monitored for an additional 42 hours with an IncuCyte machine every 6 hours. Relative wound density was determined using IncuCyte software.
Biolayer interferometry assay
Direct reversible interactions between IVM and purified HSP27 were quantified by biolayer interferometry (BLI) using OctetRED (ForteBio). HSP27 in fusion with an N-terminal (His) 6-avidin (GLNDIFEAQKIEWHE) tag was expressed in E. coli BL21-DE3 cells cotransformed with pBirACm plasmid, which expresses a biotin ligase leading to the biotinylation of the avidin tag on HSP27. Cells were grown in LB medium at 37°C and then induced with 0.5 mM isopropyl-β-d-thiogalactoside in the presence of 0.15 mM biotin. Purified protein at 200 μg/mL was bound to the streptavidin sensors overnight at 4°C in gel filtration buffer (20 mM Tris-base pH 8.4, 100 mM NaCl, 0.2 mM TCEP, 0.1 mM PMSF). The assay was performed at 25°C in the dialysis buffer supplemented with 1% DMSO, which was also used as our reference measurement. Enzalutamide was used as negative control.
In vitro kinase assay
For in vitro kinase reactions, 35 μM wild-type HSP27 was incubated with either DMSO or 350 μM IVM for 1 hour in reaction buffer (20 mM Tris, pH 8, 150 mM NaCl, 5% glycerol, 0.2 mM TCEP, 17 mM MgCl2, 350 μM ATP, 13% DMSO). Subsequently, 35 ng/mL of MAPKAP2 kinase was added to each mixture and incubated at 30°C. Samples were taken at t = 0 and 24 hours, and the kinase reaction was stopped by addition of 5× protein loading dye and boiling for 5 minutes. Phosphorylation was determined using the HSP27 S78 antibody by Western blotting.
Sample extraction and liquid chromatography–mass spectrometry analysis
Serum samples from the in vivo studies were thawed and 20 μL transferred to individual Eppendorf tubes, followed by addition of internal standard (IS; 4 μL of 2 μg/mL abamectin) and 44 μL of PPT (methanol/acetonitrile, 25:75). Samples were vortexed, precipitated protein sedimented (5 minutes, 20,000 g), and clarified supernatant transferred to liquid chromatography vials. Standards were prepared in a similar fashion with IVM/IS–spiked blank mouse serum; parallel standards made up in solvent only were used to characterize any matrix effects. Tissue samples (200–400 mg) were weighed and 2 volumes water added, followed by homogenization using a Precellys tissue homogenizer (Bertin Technologies) with three 45-second cycles at 6000 rpm. Extraction was similar to that of serum but using 50 μL homogenate, 10 μL IS, and 110 μL PPT, and spiked serum samples were used for calibration.
An Acquity UPLC System coupled with an eLambda Photodiode Array (PDA) and a Quattro Premier mass spectrometer (Waters) was used for analysis with a 100-mm BEH C18, 1.7-μm column (Waters) and an acetonitrile gradient (0–0.5 minutes, 88%; 0.5–1.5 minutes, 88%–98%; 1.5–3.5 minutes, 98%; 3.5–3.6 minutes, 98%–88%; 0.3 mL/min) used for separation (5.5-minute run length; 0.1% formic acid present throughout). Mass spectrometry data were collected in electrospray positive slightly below unit resolution with the following instrument parameters: capillary, 3 kV; extractor and RF lens, 3 V and 0.2 V; source and desolvation temperatures, 120°C and 350°C; desolvation and cone (N2) flow, 950 L/h and 50 L/h; collision gas (Ar) flow, 0.15 mL/min (8–3-meter bar). Compounds were detected by multiple reaction monitoring with m/z 892.7>307.3 and 892.7>569.3 for IVM and m/z 890.7>305.3 and 890.7>567.3 for Adamectin (20/24 V, 20/15 V, and 20/25 V, 20/12 V, cone/collision volt combinations, respectively) with 0.1 seconds dwell each. Retention times for IVM and Adamectin were 2.6 minutes and 1.7 minutes, respectively. Extracted chromatograms of 254 nm were used for both IVM and Adamectin using the PDA data (210–500 nm).
Data were processed with Quanlynx (Waters) using Adamectin-normalized calibration for mass spectrometry and external calibration for PDA and exported to Excel for further analysis. Calibration standards ranged from 0.4 to 1200 ng/mL (6 points, extract levels). A linear fit was used, resulting in r2 > 0.98, and deviation from nominal < 15% above 1.6 ng/mL. OD data correlated well at higher concentrations. Ion suppression in both serum and tumor extracts was on the order of 30%.
Quantitative reverse transcriptase PCR
RNA extraction and reverse transcriptase PCR (RT-PCR) were carried out as described previously (42). Real-time monitoring of PCR amplification of cDNA was carried out using the following primer pairs and probes: EGFR Hs01076090_m1, CDH1 Hs1023894_m1, and GAPDH Hs03929097_g1 (Applied Biosystems), on the ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) with a TaqMan Gene Expression Master Mix (Applied Biosystems). Target gene expression was normalized to GAPDH levels in the respective samples as an internal control. The results are representative of 3 independent experiments.
Doxycycline-inducible AR-V7 PC3_3TKNLuc system
Doxycycline-inducible AR-V7 construct and probasin-based ARR3tk-Nanoluciferase reporter construct (PC3V7_3TKNLuc) were generated as previously reported (62). Nanoluciferase reporter assays were performed by culturing of cells in DMEM plus 5% charcoal-stripped serum medium in black 96-well plates. AR-V7 expression was induced by the addition of 200 ng/mL doxycycline with different doses of IVM for 24 hours, and cells were lysed with Nanoglo (Promega) lysis buffer/substrate for 5 minutes at room temperature. Luminescence was measured by a TECAN m200pro luminometer.
In vitro cell growth assay
Cells were plated in 96-well plates and treated with IVM alone or in combination with erlotinib, cetuximab, trastuzumab, or docetaxel. After 3 days, cell viability was assessed using crystal violet assay. Absorbance was determined with a microculture plate reader (Becton Dickinson Labware) at 590 nm. Changes in cell viability were calculated as percentage relative to controls. Each assay was performed in triplicate.
Tissue microarray construction and immunohistochemistry
We created the tissue microarray by taking multiple 1-mm cores from matching marked non-necrotic areas of paraffin blocks of xenograft tumors using a semiautomated tissue microarrayer (TMArrayer, Pathology Devices) attached to a Leica M50 stereo microscope. Staining was conducted using an automated staining platform (Ventana Discovery XT; Ventana Medical Systems) with an enzyme-labeled biotin streptavidin system and solvent-resistant DAB Map Detection Kit. TUNEL staining was conducted by Ventana Autostainer using protease for antigen retrieval. The mixture of terminal transferase, digoxigenin-11-dUTP, and dATP was added, followed by biotinylated anti-digoxigenin IgG, and the DAB Map Detection Kit was used. All stained slides were digitalized with the SL801 autoloader and Leica SCN400 scanning system (Leica Microsystems) and were subsequently stored in the SlidePath digital imaging hub (DIH; Leica Microsystems) of the Vancouver Prostate Centre. Using the Aperio Image Analysis IHC (Leica Biosystems), our pathologist selected the area of interest, defined the parameter, optimized the level of intensity, and selected Positive Pixel Count algorithm for each biomarker.
HSP27 antibody (SPA-803, Stressgen) was used at a concentration of 1:3000, p-HSP27 antibody (9308, Cell Signaling Technology) at 1:50, and their use for immunohistochemistry was validated as previously reported (11). The EGFR and p-EGFR antibodies (4267 and 3777, Cell Signaling Technology) were used at a concentration of 1:50 and were validated by Western blotting in NSCLC and MEF cell lines, as shown in Figure 3, A–C and F. SHPTP1 antibody (32559, Abcam) was validated in liver and lymphoid tissue, as negative and positive controls, respectively, and was used at a concentration of 1:500.
In situ proximity ligation assay
Proximity ligation assay was performed to detect the interaction between HSP27 and p-EGFR and HSP27 and EGFR. To visualize the bound antibody pairs, the Duolink Detection Kit (Duo92008) with proximity ligand assay (PLA) PLUS and MINUS probes for mouse and rabbit (Olink Bioscience) was used, according to the manufacturer’s description. Cell slides were mounted with Duolink Brightfield Mounting Medium (Olink Bioscience).
SHPTP1 phosphatase assays
In vitro SHPTP1 tyrosine phosphatase assays were performed using the Malachite Green Phosphatase Assay (Upstate Biotechnology) with phosphopeptide (RRLIEDAEpYAARG) as a substrate from Promega. Briefly, cells were lysed in NP-40 lysis buffer and incubated with anti-SHPTP1 overnight at 4°C followed by 1 hour of incubation with protein A/G-Sepharose beads. The immune complexes were washed 3 times with lysis buffer without phosphatase inhibitor and resuspended in phosphatase buffer containing the phosphopeptide. The reaction mixtures were incubated for 30 minutes at room temperature followed by the addition of Malachite Green solution. Absorbance was measured in a spectrophotometer at 620 nm. Phosphate release was determined by comparison of absorbance to the phosphate standard.
Animal manipulation and assessment of in vivo tumor growth
For the HCC-827 in vivo experiment, 6- to 8-week-old female athymic mice (Harlan Sprague Dawley Inc.) were inoculated with 2 × 106 tumor cells s.c. in the left flank via a 27-gauge needle under isoflurane anesthesia. When tumors reached 150–200 mm3, mice were randomized to receive 15 mg/kg erlotinib, 10 mg/kg IVM, or the combination of both drugs (formulated in 0.5% methylcellulose, 0.1% Tween-80) administered orally 3 days a week. For the LNCaP in vivo experiment, 2 × 106 LNCaP cells were inoculated s.c. into the left and right flanks of 6- to 8-week-old male athymic nude mice. The animals were surgically castrated when the PSA reached 50 ng/mL or the tumor volume 300 mm3 and were subsequently randomized to receive vehicle or IVM treatment, using the same schedule protocol described for the HCC-827 study.
For the 22RV1 in vivo experiment, 2 × 106 22RV1 cells were inoculated s.c. into the left flank of 6- to 8-week-old male athymic nude mice. When the tumor volume reached 300 mm3, the animals were randomized to receive vehicle or IVM, as reported above.
Six- to eight-week-old male athymic nude mice were inoculated with 6 × 106 docetaxel-resistant PC3 cells s.c. in the left flank. When tumors reached 150 mm3, mice were randomly selected for treatment with micellar paclitaxel (0.5 mg, 3 times per week) administered i.v. every other week, 10 mg/kg IVM by gavage 3 times a week, or the combination of both. For all the in vivo experiments, tumor volume measurements were performed twice weekly and calculated by the formula length × width × depth × 0.5236. Data points were expressed as average tumor volume ± SEM.
Statistical analysis
For the in vitro experiments, unpaired 2-tailed Student’s t test and ANOVA followed by Bonferroni’s multiple-comparisons test were used. For the in vivo analysis, 1-way ANOVA test with Tukey’s correction was used to compare 3 or more groups and unpaired t test with Welch’s correction to compare 2 groups. GraphPad Prism 6 software was used to calculate the statistical significance. The threshold of statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Study approval
All animal procedures were performed according to the guidelines of the Canadian Council on Animal Care and with appropriate institutional certification. Animal studies were approved by the Animal Care Committee of the University of British Columbia (protocol number A18-0118).
Author contributions
LN and AHA equally contributed as first authors. LN was chosen as first name because she had a fundamental role in all the phases of the project, from the drug discovery to the dissection of molecular mechanisms related to HSP27 inhibition. She played a key role in defining IVM antitumor activity. AHA had a primary role in the resolution of HSP27 crystal structure that informed the computational modeling for the refinement of IVM binding site. LN, AHA, GDB, MG, BLG, AZ, and EB designed the research. LN, AHA, NAN, MT, SS, EB, DB, NL, NS, SM, FZ, and LF performed the experiments. FB, NTN, ED, TY, and AC provided computational analysis and helpful discussions. LN, AHA, GDB, and MG wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by an operating grant from a Challenge Award from the Prostate Cancer Foundation; Terry Fox New Frontiers Program Project Grant 1062; a Prostate Cancer Canada Translation Acceleration Grant; and the Canadian Institutes of Health Research (FRN-111082 to GDB). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, which is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (NIGMS) (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH. LN’s salary was supported by the Michael Smith Foundation for Health Research (16089).
Version 1. 12/17/2019
Electronic publication
Version 2. 12/18/2019
Sophie Stief's name corrected in author list
Version 3. 02/03/2020
Print issue publication
Footnotes
Conflict of interest: The University of British Columbia has granted patents on the HSP27 antisense OGX-427 (PCT/CA03/01588), listing MG as an inventor. A patent application (62/756,707) has been submitted on ivermectin analogs for the treatment of cancer, listing MG as inventor.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(2):699–714.https://doi.org/10.1172/JCI130819.
Contributor Information
Lucia Nappi, Email: lnappi@prostatecentre.com.
Adeleke H. Aguda, Email: aaguda@prostatecentre.com.
Nader Al Nakouzi, Email: nalnakouzi@prostatecentre.com.
Barbara Lelj-Garolla, Email: blelj@hotmail.com.
Eliana Beraldi, Email: eberaldi@prostatecentre.com.
Nada Lallous, Email: nlallous@prostatecentre.com.
Marisa Thi, Email: mthi@prostatecentre.com.
Dulguun Battsogt, Email: dbattsogt27@gmail.com.
Fuqiang Ban, Email: fban@prostatecentre.com.
Nham T. Nguyen, Email: ntnguyen@mail.ubc.ca.
Neetu Saxena, Email: nsaxena@prostatecentre.com.
Fan Zhang, Email: Fazhang@prostatecentre.com.
Takeshi Yamazaki, Email: takeshi@ualberta.ca.
Amina Zoubeidi, Email: azoubeidi@prostatecentre.com.
Artem Cherkasov, Email: acherkasov@prostatecentre.com.
Gary D. Brayer, Email: brayer@mail.ubc.ca.
Martin Gleave, Email: m.gleave@ubc.ca.
References
- 1.Lelj-Garolla B, Mauk AG. Roles of the N- and C-terminal sequences in Hsp27 self-association and chaperone activity. Protein Sci. 2012;21(1):122–133. doi: 10.1002/pro.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiota M, et al. Hsp27 regulates epithelial mesenchymal transition, metastasis, and circulating tumor cells in prostate cancer. Cancer Res. 2013;73(10):3109–3119. doi: 10.1158/0008-5472.CAN-12-3979. [DOI] [PubMed] [Google Scholar]
- 3.Giese KC, Basha E, Catague BY, Vierling E. Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc Natl Acad Sci U S A. 2005;102(52):18896–18901. doi: 10.1073/pnas.0506169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280(39):33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 5.Wettstein G, et al. Inhibition of HSP27 blocks fibrosis development and EMT features by promoting Snail degradation. FASEB J. 2013;27(4):1549–1560. doi: 10.1096/fj.12-220053. [DOI] [PubMed] [Google Scholar]
- 6.Konda JD, Olivero M, Musiani D, Lamba S, Di Renzo MF. Heat-shock protein 27 (HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol Oncol. 2017;11(6):599–611. doi: 10.1002/1878-0261.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoubeidi A, et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007;67(21):10455–10465. doi: 10.1158/0008-5472.CAN-07-2057. [DOI] [PubMed] [Google Scholar]
- 8.Rocchi P, et al. Increased Hsp27 after androgen ablation facilitates androgen-independent progression in prostate cancer via signal transducers and activators of transcription 3-mediated suppression of apoptosis. Cancer Res. 2005;65(23):11083–11093. doi: 10.1158/0008-5472.CAN-05-1840. [DOI] [PubMed] [Google Scholar]
- 9.McDonald ET, Bortolus M, Koteiche HA, Mchaourab HS. Sequence, structure, and dynamic determinants of Hsp27 (HspB1) equilibrium dissociation are encoded by the N-terminal domain. Biochemistry. 2012;51(6):1257–1268. doi: 10.1021/bi2017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumano M, et al. Cotargeting stress-activated Hsp27 and autophagy as a combinatorial strategy to amplify endoplasmic reticular stress in prostate cancer. Mol Cancer Ther. 2012;11(8):1661–1671. doi: 10.1158/1535-7163.MCT-12-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lelj-Garolla B, et al. Hsp27 inhibition with OGX-427 sensitizes non-small cell lung cancer cells to erlotinib and chemotherapy. Mol Cancer Ther. 2015;14(5):1107–1116. doi: 10.1158/1535-7163.MCT-14-0866. [DOI] [PubMed] [Google Scholar]
- 12.Yu EY, et al. A randomized phase 2 study of a HSP27 targeting antisense, apatorsen with prednisone versus prednisone alone, in patients with metastatic castration resistant prostate cancer. Invest New Drugs. 2018;36(2):278–287. doi: 10.1007/s10637-017-0553-x. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg JE, et al. Apatorsen plus docetaxel versus docetaxel alone in platinum-resistant metastatic urothelial carcinoma (Borealis-2) Br J Cancer. 2018;118(11):1434–1441. doi: 10.1038/s41416-018-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X, et al. Small heat shock protein Hsp16.3 modulates its chaperone activity by adjusting the rate of oligomeric dissociation. Biochem Biophys Res Commun. 2003;310(2):412–420. doi: 10.1016/j.bbrc.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 15.Lelj-Garolla B, Mauk AG. Self-association and chaperone activity of Hsp27 are thermally activated. J Biol Chem. 2006;281(12):8169–8174. doi: 10.1074/jbc.M512553200. [DOI] [PubMed] [Google Scholar]
- 16.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273(13):7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 17.Wieske M, et al. Defined sequence segments of the small heat shock proteins HSP25 and αB-crystallin inhibit actin polymerization. Eur J Biochem. 2001;268(7):2083–2090. doi: 10.1046/j.1432-1327.2001.02082.x. [DOI] [PubMed] [Google Scholar]
- 18.Braun N, et al. Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc Natl Acad Sci U S A. 2011;108(51):20491–20496. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jovcevski B, et al. Phosphomimics destabilize Hsp27 oligomeric assemblies and enhance chaperone activity. Chem Biol. 2015;22(2):186–195. doi: 10.1016/j.chembiol.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Kostenko S, Moens U. Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell Mol Life Sci. 2009;66(20):3289–3307. doi: 10.1007/s00018-009-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang SH, et al. Upregulated HSP27 in human breast cancer cells reduces Herceptin susceptibility by increasing Her2 protein stability. BMC Cancer. 2008;8:286. doi: 10.1186/1471-2407-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moghal N, Sternberg PW. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr Opin Cell Biol. 1999;11(2):190–196. doi: 10.1016/S0955-0674(99)80025-8. [DOI] [PubMed] [Google Scholar]
- 23.Monteleone G, et al. Silencing of SH-PTP2 defines a crucial role in the inactivation of epidermal growth factor receptor by 5-aminosalicylic acid in colon cancer cells. Cell Death Differ. 2006;13(2):202–211. doi: 10.1038/sj.cdd.4401733. [DOI] [PubMed] [Google Scholar]
- 24.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocchi P, et al. Small interference RNA targeting heat-shock protein 27 inhibits the growth of prostatic cell lines and induces apoptosis via caspase-3 activation in vitro. BJU Int. 2006;98(5):1082–1089. doi: 10.1111/j.1464-410X.2006.06425.x. [DOI] [PubMed] [Google Scholar]
- 26.Gleave ME, Hsieh JT, Wu HC, von Eschenbach AC, Chung LW. Serum prostate specific antigen levels in mice bearing human prostate LNCaP tumors are determined by tumor volume and endocrine and growth factors. Cancer Res. 1992;52(6):1598–1605. [PubMed] [Google Scholar]
- 27.Kamada M, So A, Muramaki M, Rocchi P, Beraldi E, Gleave M. Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol Cancer Ther. 2007;6(1):299–308. doi: 10.1158/1535-7163.MCT-06-0417. [DOI] [PubMed] [Google Scholar]
- 28.Choueiri TK, Hahn NM, Werner L, Regan MM, Rosenberg JE. Borealis-2: a randomized phase II study of OGX-427 (apatorsen) plus docetaxel versus docetaxel alone in platinum-resistant metastatic urothelial cancer (mUC) (Hoosier Cancer Research Network GU12-160) J Clin Oncol. 2017;35(6 suppl):289 [Google Scholar]
- 29.Parcellier A, Schmitt E, Brunet M, Hammann A, Solary E, Garrido C. Small heat shock proteins HSP27 and αB-crystallin: cytoprotective and oncogenic functions. Antioxid Redox Signal. 2005;7(3–4):404–413. doi: 10.1089/ars.2005.7.404. [DOI] [PubMed] [Google Scholar]
- 30.Rocchi P, et al. Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 2004;64(18):6595–6602. doi: 10.1158/0008-5472.CAN-03-3998. [DOI] [PubMed] [Google Scholar]
- 31.Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-dimensional structure of α-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol. 2011;411(1):110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Hochberg GK, et al. The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci U S A. 2014;111(16):E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melotti A, Mas C, Kuciak M, Lorente-Trigos A, Borges I, Ruiz i Altaba A. The river blindness drug Ivermectin and related macrocyclic lactones inhibit WNT-TCF pathway responses in human cancer. EMBO Mol Med. 2014;6(10):1263–1278. doi: 10.15252/emmm.201404084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishio M, et al. Dysregulated YAP1/TAZ and TGF-β signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice. Proc Natl Acad Sci U S A. 2016;113(1):E71–E80. doi: 10.1073/pnas.1517188113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dou Q, et al. Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer. Cancer Res. 2016;76(15):4457–4469. doi: 10.1158/0008-5472.CAN-15-2887. [DOI] [PubMed] [Google Scholar]
- 36.Korystov YN, et al. Avermectins inhibit multidrug resistance of tumor cells. Eur J Pharmacol. 2004;493(1-3):57–64. doi: 10.1016/j.ejphar.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 37.Faiella L, Piaz FD, Bisio A, Tosco A, De Tommasi N. A chemical proteomics approach reveals Hsp27 as a target for proapoptotic clerodane diterpenes. Mol Biosyst. 2012;8(10):2637–2644. doi: 10.1039/c2mb25171j. [DOI] [PubMed] [Google Scholar]
- 38.Heinrich JC, Tuukkanen A, Schroeder M, Fahrig T, Fahrig R. RP101 (brivudine) binds to heat shock protein HSP27 (HSPB1) and enhances survival in animals and pancreatic cancer patients. J Cancer Res Clin Oncol. 2011;137(9):1349–1361. doi: 10.1007/s00432-011-1005-1. [DOI] [PubMed] [Google Scholar]
- 39.Fu WM, et al. 1,3,5-Trihydroxy-13,13-dimethyl-2H-pyran [7,6-b] xanthone directly targets heat shock protein 27 in hepatocellular carcinoma. Cell Biol Int. 2014;38(2):272–276. doi: 10.1002/cbin.10193. [DOI] [PubMed] [Google Scholar]
- 40.Zhong B, et al. Synthesis and anticancer mechanism investigation of dual Hsp27 and tubulin inhibitors. J Med Chem. 2013;56(13):5306–5320. doi: 10.1021/jm4004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin KD, et al. Blocking tumor cell migration and invasion with biphenyl isoxazole derivative KRIBB3, a synthetic molecule that inhibits Hsp27 phosphorylation. J Biol Chem. 2005;280(50):41439–41448. doi: 10.1074/jbc.M507209200. [DOI] [PubMed] [Google Scholar]
- 42.Choi SH, et al. Altered cross-linking of HSP27 by zerumbone as a novel strategy for overcoming HSP27-mediated radioresistance. Int J Radiat Oncol Biol Phys. 2011;79(4):1196–1205. doi: 10.1016/j.ijrobp.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 43.Fossa P, Cichero E. In silico evaluation of human small heat shock protein HSP27: homology modeling, mutation analyses and docking studies. Bioorg Med Chem. 2015;23(13):3215–3220. doi: 10.1016/j.bmc.2015.04.070. [DOI] [PubMed] [Google Scholar]
- 44.Leslie AGW. Availability of the MOSLFM program suite for processing image plate and film data. Joint CCP4 and ESF-EACBM newsletterON protein crystallography. 1992;27:30–31. [Google Scholar]
- 45.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48. Chemical Computing Group ULC. Molecular Modeling, Simulations and Machine Learning software. Montreal, Quebec, Canada; 2018. [Google Scholar]
- 49.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 50.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 51. Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE, Darden TA, et al. AMBER 2015. University of California, San Francisco, California, USA; 2015. [Google Scholar]
- 52.Zalaudek I, et al. Flat pigmented macules on sun-damaged skin of the head/neck: junctional nevus, atypical lentiginous nevus, or melanoma in situ? Clin Dermatol. 2014;32(1):88–93. doi: 10.1016/j.clindermatol.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 53.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 54.Friesner RA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 55.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 57.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97(40):10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
- 58.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 59.Price DJ, Brooks CL. A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121(20):10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 60.Sowery RD, et al. Clusterin knockdown using the antisense oligonucleotide OGX-011 re-sensitizes docetaxel-refractory prostate cancer PC-3 cells to chemotherapy. BJU Int. 2008;102(3):389–397. doi: 10.1111/j.1464-410X.2008.07618.x. [DOI] [PubMed] [Google Scholar]
- 61.Zoubeidi A, et al. Hsp27 promotes insulin-like growth factor-I survival signaling in prostate cancer via p90Rsk-dependent phosphorylation and inactivation of BAD. Cancer Res. 2010;70(6):2307–2317. doi: 10.1158/0008-5472.CAN-09-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalal K, et al. Small molecule-induced degradation of the full length and V7 truncated variant forms of human androgen receptor. Eur J Med Chem. 2018;157:1164–1173. doi: 10.1016/j.ejmech.2018.08.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.