Abstract
RNA-binding motif protein 10 (RBM10) is an RNA-binding protein frequently deleted or mutated in lung cancer cells. Recent reports showed that knockdown of RBM10 in human cancer cells enhances growth of mouse tumor xenografts, suggesting that RBM10 acts as a tumor suppressor. RBM10 also regulates alternative splicing and controls cancer cell proliferation. However, the underlying molecular mechanisms for its tumor suppression role remain largely unclear. Here, we for the first-time report that RBM10 can induce apoptosis and inhibit cancer cell proliferation by activating p53. Our analysis of cancer genomic databases showed that patients with wild type RBM10 and p53 survive longer than do those with mutated p53 or less RBM10. RBM10 overexpression markedly inhibited mitochondrial respiration, cell migration and proliferation of various cancer cells that harbor wild type p53. Also, RBM10 overexpression elongated p53’s half-life by disrupting MDM2-p53 interaction and subsequently repressing p53 ubiquitination, whereas knockdown of RBM10 decreased p53 stability. Altogether, our results demonstrate that RBM10 inhibits cancer cell proliferation and induces apoptosis in part by blocking the MDM2-p53 feedback loop.
Keywords: RBM10, p53, MDM2, apoptosis, cell proliferation, mitochondrial respiration
Introduction
The tumor suppressor p53 is frequently mutated in various human cancers as it can prevent tumorigenesis by inducing cellular senescence, regulating energy metabolism, blocking metastasis, stopping cell proliferation, and inducing apoptosis mainly via its transcriptional activity [1]. In response to various stressors, p53 is activated to induce or repress transcription of numerous target genes important for multiple biological functions [2]. For example, the p53 target gene CDKN1A (p21) is involved in p53-dependent cell cycle arrest, while the BH3-only-encoding target genes BBC3 (Puma) and PMAIP1 (Noxa) play key roles in p53-mediated apoptosis [3]. Since activated p53 is generally cytotoxic, it is subjected to tight regulation by MDM2 [4], which is encoded by a transcriptional target gene of p53. Via its N-terminal domain, MDM2 directly binds to N and C termini of p53 and mediates its ubiquitin-dependent proteolysis, as it possesses intrinsic E3 ubiquitin ligase activity, forming a negative feedback loop [5-7]. Thus, direct or indirect inhibition of the MDM2 activity is fundamental for p53 activation.
RBM10, also called S1–1, is a member of the RNA-binding motif (RBM) gene family, known to be involved in pre-mRNA (messenger RNA) splicing and post-transcriptional regulation [8]. RBM10 can be alternatively spliced to produce RBM10 RNA variant 1 and variant 2. These two RNA variants encode nuclear RNA-binding proteins containing zinc finger motifs, a G-patch, and two RNA Recognition Motif (RRMs), which regulate gene transcription, mRNA alternative splicing, and stabilization of various genes, including the apoptosis-related Fas gene [9]. Previous studies have shown that RBM10 overexpression inhibits lung adenocarcinoma malignant behaviors, such as cell viability and cell cycle progression [10]. However, the underlying molecular mechanisms remain largely unclear.
In attempt to dissect the mechanism(s) underlying the possible tumor suppression function of RBM10, we identified RBM10 as a new regulator of p53 dependent apoptosis, proliferation and migration of some cancer cells. We found that overexpression of RBM10 induced apoptosis and inhibited cell growth and migration more apparently in p53 wild-type-containing than p53-deficient cancer cells. Biochemical analysis of this action of RBM10 on p53 revealed that RBM10 can inhibit MDM2-mediated p53 ubiquitination and thus increase the half-life of p53. Interestingly, the N-terminal domain of RBM10 was required for inhibiting cancer cell growth by p53. Therefore, our results unveil RBM10 as a tumor suppressor in part by activating p53 and inducing p53-dependent apoptosis and cell growth arrest.
Results
Overexpression of RBM10 inhibits cancer cell proliferation and promotes apoptosis by inducing p53
Although RBM10 has been shown to control cancer cell proliferation by differentially regulating NUMB alternative splicing [11] and its expression is associated with advanced tumor stage [12], much more need to learn about its role in cancer development. In attempt to address this question, we first searched genomic and gene expression databases for RBM10 status. Interestingly, TCGA genome database revealed that patients with lower RBM10 expression display a much shorter survival rate than do patients with higher RBM10 expression (Supplementary Fig. 1a). Also, the expression of the RBM10 gene at its RNA level was significantly lower in breast and head-neck cancers than that in their corresponding normal tissues (Supplementary Figs. 1b and c). By performing biochemical fractionation and immunofluorescence staining assays, we found that the most of RBM10 proteins reside in the nucleus (Supplementary Figs. 2a, b and c). Intriguingly, the upregulation of RBM10 was significantly correlated with TP53 (Supplementary Fig. 1d). These results suggest that RBM10 might play a tumor suppressor role in human cancer development, and this role might be related to p53.
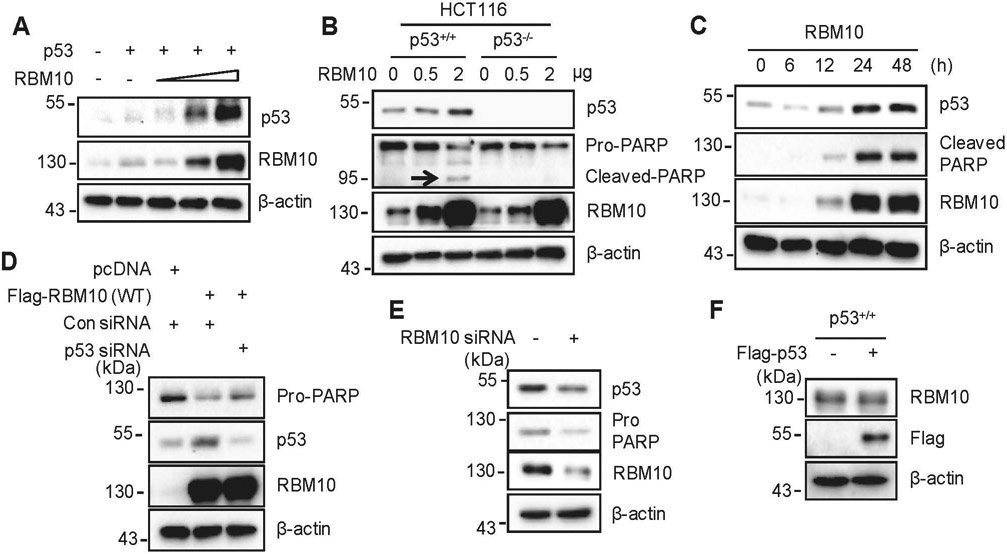
To determine if RBM10 plays a role in regulation of p53, we overexpressed both RBM10 and p53 plasmids in p53-null cancer cells and found that RBM10 increases the p53 protein level in a dose-dependent fashion (Fig. 1a). Consistently, RBM10 induced the expression of endogenous p53 in various wild-type p53-containing cancer cells as well as of its target gene p21 and apoptosis marker cleaved-PARP, but not in p53-null cancer cells dose-dependently and time-dependently (Figs. 1b, c, Supplementary Figs. 3a-d). However, overexpression of RBM10 did not change the p53 mRNA level (Supplementary Fig. 4), suggesting that RBM10 might only regulate the protein, but not mRNA, level of p53. While overexpression of RBM10 induced p53 and the cleavage of PARP, knockdown of p53 by its specific siRNA partially reversed this induction in HCT116 p53+/+ cells (Fig. 1d), suggesting that apoptosis as indicated by PARP cleavage is p53-dependent. Furthermore, knockdown of RBM10 using siRNA or shRNA in HCT116 p53+/+ cells resulted in marked decrease of p53 (Fig. 1e and Supplementary Fig. 3e). Of note, RBM10 was not induced after overexpressed p53 even in HCT116 p53+/+ cells (Fig. 1f), suggesting that RBM10 might not be the target of p53. Next, we tested if RBM10 can inhibit cell proliferation dependently of p53. As shown in Figs. 2a-c, overexpression of RBM10 led to a more dramatic reduction of colony formation in p53-positive colon cancer cells than in p53-deficient cells, though a significant reduction of cell colony formation was also observed in p53-deficient HCT116 cells. The RBM10-induced apoptosis was caspases-dependent as this effect was inhibited by a pan-caspase inhibitor, carbobenzoxy-valyl-alanyl-aspartyl (Z-VAD) (Supplementary Fig. 3f). Collectively, these results demonstrate that RBM10 induces apoptosis partly by inducing p53 and activating its activity.
Figure 1. RBM10 induces apoptosis of colon cancer cells by inducing p53.
(A) HCT116p53−/− cells were co-transfected with p53 and RBM10 for 48 h. Proteins were analyzed by IB. (B) HCT116p53+/+ and HCT116p53−/− cells were transfected with pcDNA or RBM10 plasmids in different amounts and harvested 48 h post transfection for IB analysis with indicated antibodies. (C) HCT116p53+/+ cells were transfected with RBM10 plasmid and harvested at different time points post transfection for IB analysis with indicated antibodies. (D) HCT116p53+/+ cells were transfected with Flag-RBM10 with or without p53 siRNA, and harvested 48 h post transfection for IB analysis. (E) Cells were transfected with scramble siRNA or RBM10 siRNA and harvested 72 h post transfection for IB analysis with indicated antibodies. (F) HCT116p53+/+ cells were transfected with pcDNA or Flag-p53 and harvested 48 h post transfection for IB analysis with indicated antibodies.
Figure 2. RBM10 inhibits cell proliferation and migration by inducing p53.
(A and B) HCT116p53+/+ and HCT116p53−/− cells were transfected with RBM10 and seeded in 6-well plates the next day. Colonies were fixed by methanol and stained with crystal violet solution. Quantification of colonies is shown in the right panel. (C) The relative quantification of colonies is shown in this graph. (D and E) The effect of RBM10 overexpression on cell migration of HCT116 cells. HCT116p53+/+ (D) and HCT116p53−/− (E) cells were transfected with pcDNA or RBM10 plasmid, or scramble siRNA or RBM10 siRNA, as indicated. 24 hours after transfection, the cells were re-seeded onto 24 well-plate and allowed to attach overnight followed by scratching using P200 tips and image taking at indicated time points. IB analysis was performed to confirm protein expression. The wound gaps were measured at each time point and compared with 0 h to determine the percentage of wound healed as presented at lower panels of D and E. Wherever applicable, data represent mean ± s.e.m. of triplicate experiments.
RBM10 inhibits cancer cell migration and mitochondrial respiration by inducing p53
p53 has been shown to inhibit metastatic progression, such as cell migration and invasion [13]. Next, we investigated whether overexpression of RBM10 affects cancer cell migration. As shown in Figs. 2d, e, overexpression of RBM10 inhibits colon cancer cell migration more remarkably in HCT116 p53+/+ cells than in HCT116 p53−/− cells. Because epithelial-messenchymal transition (EMT) affects cell migration, we next sought to determine if RBM10 could affect EMT markers or not. Consistently, overexpression of RBM10 inhibited the mRNA levels of vimentin and VEGFA1 in HCT116 p53+/+ cells, but not in HCT116 p53−/− cells (Supplementary Figs. 5b and c). Also, RBM10 overexpression induced the expression of a differentiation marker CREBBP [14, 15] and of Ferredoxin reductase (FDXR) [16], both of which are the targets of p53, and the latter of which can modulate p53-dependent apoptosis and is necessary for steroidogenesis and biogenesis of iron–sulfur clusters as measured by their mRNA levels (Supplementary Figs. 5d and e). And, RBM10 expression was confirmed (Supplementary Fig. 5f). To determine if RBM10 might affect the mitochondria respiration in cancer cells, we conducted Seahorse assays. As shown in Supplementary Figs. 6a-d, overexpression of RBM10 inhibited the mitochondrial respiration in both p53-containing and in p53-deficient HCT116 cells, but more pronounced in p53-positive colon cancer cells. These results demonstrate that RBM10 can inhibit cancer cell migration and mitochondrial respiration in part by activating p53.
RBM10 induces p53 by impeding its degradation
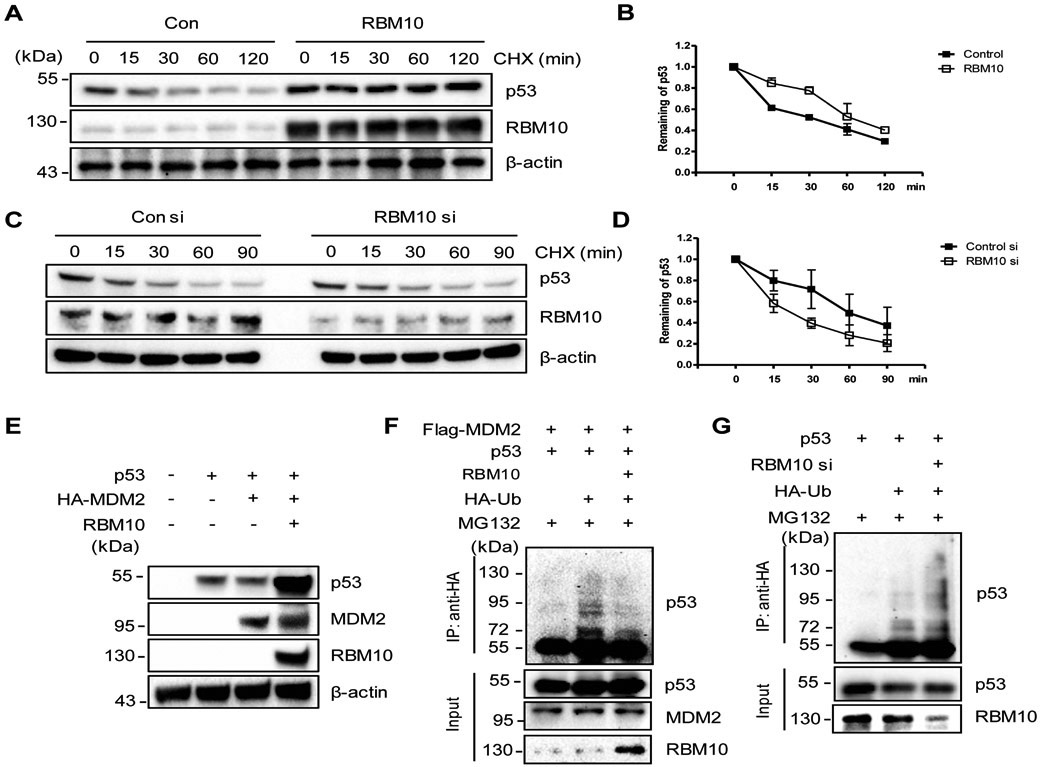
Next, we wanted to determine how RBM10 regulates p53 protein level. First, we tested the half-life of p53 in the presence or absence of ectopic RBM10. As shown in Figs. 3a, b, and Supplementary Fig. 7, overexpression of RBM10 extended the half-life of p53 from ~30 mins to more than 1.5 hours in HCT116 p53+/+ or H460 cells. By contrast, knockdown of RBM10 by siRNA led to a further reduction of p53’s half-life in HCT116 p53+/+ cells (Figs. 3c, d). Then, we determined if RBM10 might affect MDM2-mediated p53 ubiquitylation and degradation. As expected, overexpression of MDM2 dramatically reduced p53 levels. In contrast, further expression of RBM10 rescued MDM2-mediated p53 degradation (Fig. 3e). Consistent with this result, ectopic RBM10 also reduced p53 ubiquitination in HCT116 p53+/+ cells (Fig. 3f), whereas knockdown of RBM10 led to the increase of p53 ubiquitination (Fig. 3g). Taken together, these results demonstrate that RBM10 can increase p53 stability by inhibiting MDM2-mediated p53 ubiquitination and degradation.
Figure 3. RBM10 inhibits MDM2-mediated ubiquitination and degradation of p53.
(A and B) p53’s half-life is increased upon RBM10 overexpression. HCT116p53+/+ Cells were treated with pcDNA or RBM10 for 48 h, then treated with 50 μg ml−1 of CHX, and harvested at different time points as indicated for IB analysis. (C and D) p53’s half-life is decreased upon RBM10 knockdown. HCT116p53+/+ Cells were treated with scramble or RBM10 siRNA for 72 h, treated with 50 μg ml−1 of CHX, and harvested at different time points as indicated for IB analysis. (E) HCT116p53−/− cells were transfected with combinations of plasmids encoding p53, HA-MDM2 and RBM10, respectively, and harvested 48 h post transfection for IB analysis with indicated antibodies. (F) HCT116p53−/− cells were transfected with combinations of plasmids encoding p53, Flag-MDM2, HA-Ub or RBM10, and treated with MG132 (20 μM) for 6 h before being harvested for an in vivo ubiquitination assay. Bound and input proteins were detected by IB analysis using antibodies as indicated. (G) HCT116p53+/+ cells were transfected with combinations of plasmids encoding p53, HA-Ub or RBM10 siRNA, and treated with MG132 (20 μM) for 6 h before being harvested for an in vivo ubiquitination assay. Bound and input proteins were detected by IB analysis using antibodies as indicated.
RBM10 interacts with p53
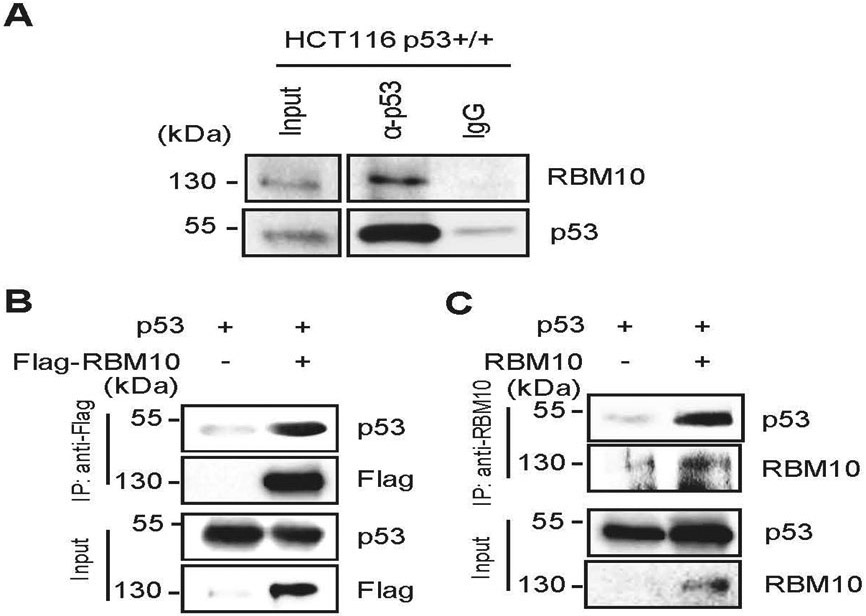
To further dissect how RBM10 regulates p53 stability, we tested if it might directly bind the latter. To do so, we performed a set of co-immunoprecipitation (co-IP) assays. Indeed, endogenous RBM10 was pulled down together with endogenous p53 by anti-p53 antibodies in HCT116 p53+/+ cells (Fig. 4a). This interaction was further verified by overexpressing both RBM10 and p53-Flag or p53-Flag alone in HCT116 p53−/− cells. By performing reciprocal co-IP assays with either anti-Flag or anti-RBM10 antibodies, we found that these two proteins are co-immunoprecipitated with each other in both of the co-IP assays (Figs. 4b, c). Interestingly, RBM10 did not appear to bind to hot spot mutant p53s, such as R249S, Y220C, R273H or R248W (Supplementary Figs. 8a and b). These results demonstrate that RBM10 mainly interacts with wild type, but not mutant, p53.
Figure 4. RBM10 interacts with p53.
(A) The association between endogenous RBM10 and p53 is detected in HCT116p53+/+ cells by co-IP-IB assays using antibodies as indicated. IgG was used as a control. (B and C) Exogenous RBM10 interacts with p53. HCT116p53−/− cells were transfected with plasmids encoding Flag-RBM10 and p53 (B) or p53 and RBM10 (C) followed by co-IP-IB assays using antibodies as indicated.
RBM10 interacts with MDM2
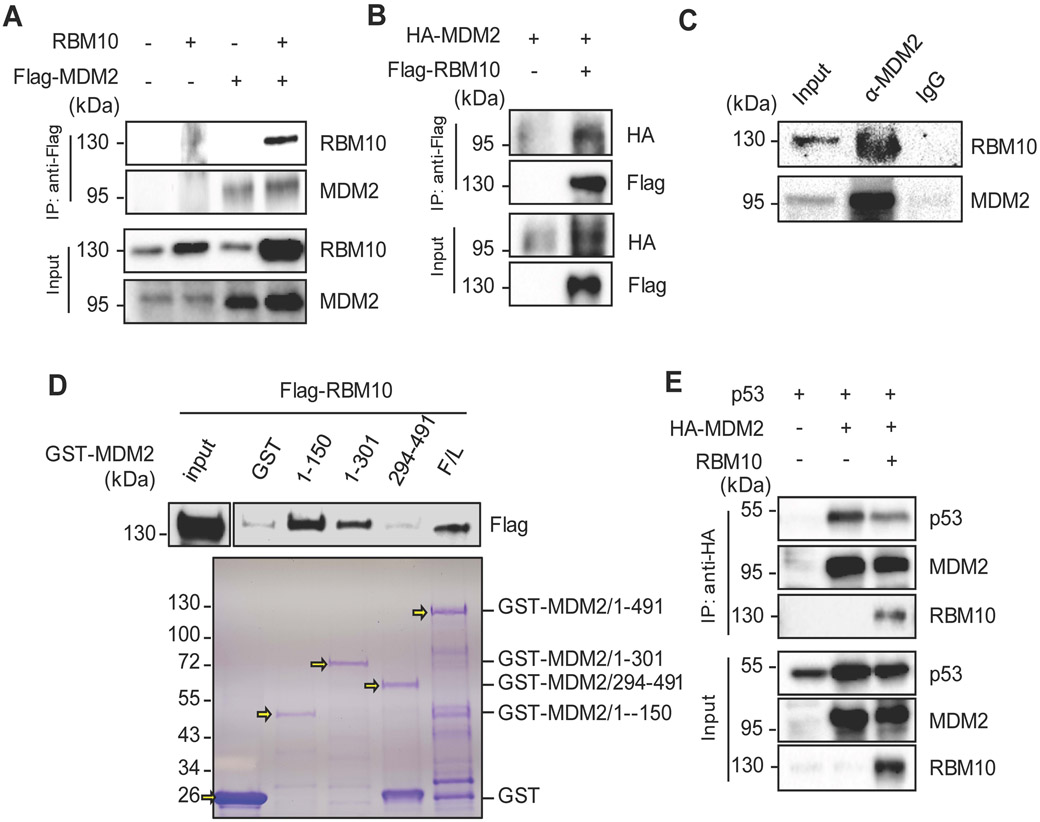
Since MDM2 is a major degrader of p53, and RBM10 can inhibit MDM2-mediated p53 degradation (Fig. 3), we then tested if RBM10 might also interact with MDM2 by conducting a set of reciprocal co-IP-IB assays. As shown in Figs. 5a, b, ectopic RBM10 bound to ectopic MDM2, and vice versa. Consistent with these results, we also detected endogenous MDM2-RBM10 complex in HCT116 p53+/+ cells by carrying out co-IP assays (Fig. 5c). This interaction was further confirmed by mapping the MDM2 binding domains of RBM10. As shown in Fig. 5d, RBM10 appeared to more preferentially bind to the N-terminal aa 1–150 fragment, but not C-terminal fragments, of MDM2 in HCT116 p53−/− cells as analyzed by GST-fusion protein-protein interaction assays. Since the N-terminal domain of MDM2 binds to p53, we then tested if RBM10 might influence the MDM2-p53 interaction by binding to this domain by co-transfecting HCT116 p53−/− cells with p53, MDM2 and RBM10 followed by a co-IP-IB assay. As shown in Fig. 5e and Supplementary Fig. 9, RBM10 indeed reduced the level of the p53-MDM2 complex in a dose dependent manner. Taken together with the results shown in Figs. 3-4, these results indicate that RBM10 can inhibit MDM2-mediated p53 ubiquitination and degradation by inhibiting MDM2-p53 binding directly.
Figure 5. RBM10 interacts with MDM2.
(A and B) Exogenous RBM10 interacts with p53. HCT116p53−/− cells were transfected with plasmids encoding RBM10 and Flag-MDM2 or HA-MDM2 and Flag-RBM10 as indicated followed by co-IP-IB assays using antibodies as indicated. (C) The association between endogenous RBM10 and p53 is detected in HCT116p53+/+ cells by co-IP-IB assays using antibodies as indicated. IgG was used as a control. (D) Mapping the RBM10 binding domain of MDM2 by GST-pull down assay. HCT116p53−/− cells were transfected with Flag-RBM10 encoded plasmid, and the cell lysate was incubated with GST-tagged full-length MDM2 or MDM2 fragment, aa 1-150, aa 1-301 or aa 294-491, or GST protein alone. Bound proteins were detected by IB with the anti-RBM10 antibody or using coomassie staining. (E) Exogenous RBM10 inhibits p53-MDM2 binding. HCT116p53−/− cells were transfected with combinations of plasmids encoding p53, HA-MDM2 or RBM10 as indicated followed by co-IP-IB assays using antibodies as indicated.
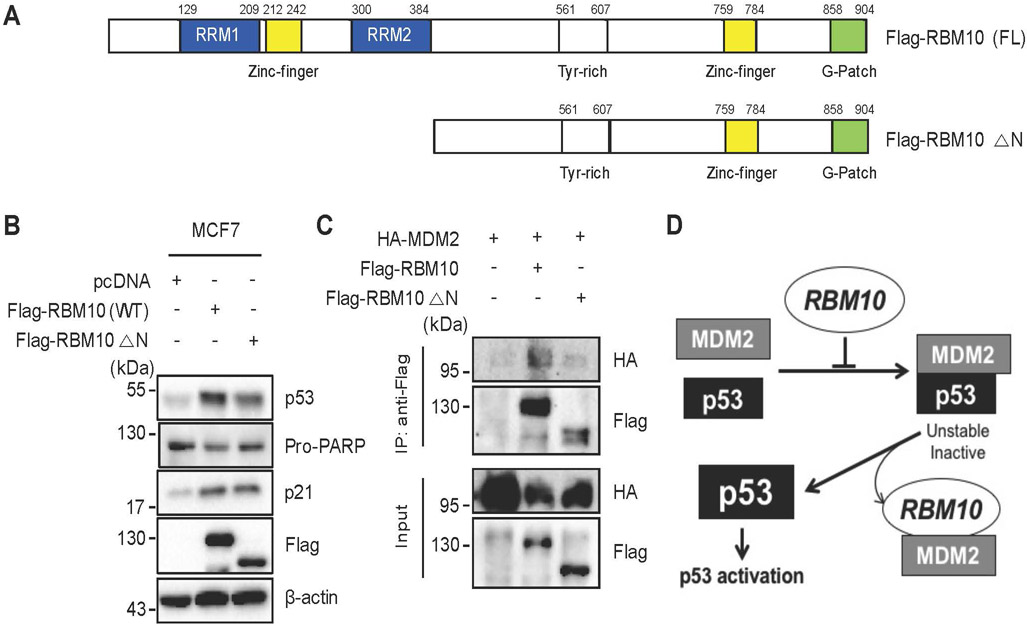
The RRM1 and RRM2 domains of RBM10 are required for induction of p53
Finally, we wanted to determine which domain(s) of RBM10 would be important for p53 activation. To do so, we generated an N-terminally deleted mutant (aa 385–904) of RBM10, which contains a zinc finger domain hanging by two RNA-binding motifs, called RRM1 and RRM2, respectively (Fig. 6a). This RRM1-Zinc Finger (ZnF) domain and RRM2 have been previously shown to play a key role in regulation of alternative splicing by recognizing RNA motifs in introns or exons of its target RNAs [17]. Hence, we wanted to test if this domain might also be important for MDM2-binding and p53 activation. As shown in Fig. 6b and Supplementary Fig. 10a, the deletion of the N-terminal RRM1-ZnF domain led to partial impairment of p53 induction by this protein when comparing the induction of p53 and p21 as well as PARP cleavage by the full length RBM10 with that by the mutant RBM10. Consistently, the ability of the mutant RBM10 to bind to MDM2 was markedly reduced as measured by co-IP-IB analysis after co-transfection of the RBM10 or its mutant plasmid with the MDM2 plasmid into HCT116p53−/− cells (Fig. 6c), though this mutant was still able to bind to p53 (Supplementary Fig. 10b). These results suggest that the N-terminal domain of RBM10 is required for MDM2-binding and consequent p53 induction.
Figure 6. The RRM1- and RRM2-containing N-terminus of RBM10 is required for p53 activation.
(A) A schematic diagram of RBM10’s functional domains. (B) MCF7 cells were transfected with combinations of plasmids encoding pcDNA, Flag-RBM10 (WT) or Flag-RBM10 (△N) and harvested 48 h post transfection for IB analysis with indicated antibodies. (C) HCT116p53−/− cells were transfected with combinations of plasmids encoding pcDNA, HA-MDM2, Flag-RBM10 (WT) or Flag-RBM10 (△N) and harvested 48 h post transfection for Co-immunoprecipitation (Co-IP) assays using antibodies as indicated. (D) A schematic for how RBM10 induces p53-dependent apoptosis and cell growth arrest by inhibiting MDM2-mediated ubiquitination and degradation of p53.
Discussion
The MDM2-p53 feedback loop plays an important role in monitoring p53 stability and activity in normal cells and tissues, while this loop is often hijacked to inactivate p53 by cancer cells for their growth advantages [18]. Hence, to combat cancerous growth, a number of protein molecules have evolved to block this feedback loop in response to various stressors in cells [19, 20]. In this report, we identified RBM10 as a novel regulator of this loop, which can activate p53 by blocking its interaction with MDM2 and thus act as a tumor suppressor in part by utilizing p53’s anti-cancer functions. To our knowledge, this is the first study to unveil the functional relationship between RBM10 and p53 as tumor suppressors. This tumor suppression role of RBM10 is also supported by available bioinformatics databases, as the lower expression profile of RBM10 was correlated with higher incidences of cancer and a lower survival rate of colorectal cancer patients (Supplementary Figs. 1a-c). Also, our cell-based studies as described here showed that RBM10 overexpression increases apoptosis (Figs. 1b, c) and decreases cell proliferation (Figs. 2a-c). Our analysis of mutual exclusivity suggested that RBM10 and p53 alterations in 240–963 cancer samples are significantly mutually exclusive. Therefore, these results strongly indicate that RBM10 could function as a tumor suppressor and suggest that this function might be p53-related.
Indeed, our further studies showed that RBM10 exerts its anti-cancer cell growth at least in part through activation of p53. First, we showed that overexpression of RBM10 induces p53-dependent apoptosis of several cancer cells (Fig. 1 and Supplementary Fig. 3). Also, we found that overexpression of RBM10 can reduce MDM2-mediated ubiquitination and degradation of p53, whereas knockdown of RBM10 leads to the reduction of p53 protein levels (Fig. 3). Mechanistically, RBM10 bound to wild type, but not mutant, p53 (Fig. 4 and Supplementary Fig. 8). RBM10 also bound to MDM2 in cancer cells (Fig. 5a-c), and this binding appeared to be through both of the N-termini of RBM10 and MDM2 (Figs. 5d, 6c). Interestingly, it was through these bindings that RBM10 prevents the interaction between MDM2 and p53 (Fig. 5e), consequently leading to the inhibition of MDM2-mediated ubiquitination and degradation of p53 as mentioned above. Further supporting this statement is that the N-terminally deleted RBM10 mutant that failed to bind to MDM2 (Fig. 6c) also lacks a strong ability to induce p53 (Fig. 6b and Supplementary 10a). We also showed that RBM10 can be coimmunoprecipitated with p53 in cancer cells (Fig. 4). Yet, it remains to determine if binding to p53 is required for RBM10 to activate p53. Regardless of this remaining question, our results strongly demonstrate that RBM10 induces apoptosis and inhibit cell growth of cancer cells in part by blocking the MDM2-p53 feedback loop and consequently activating the p53 pathway (Fig. 6d).
Previous studies showed that RBM10 possesses anti-cancer activity by regulating alternative splicing of several cancer-relevant genes [11]. Our studies as shown here reveal a new mechanism for its anti-cancer role, i.e., to activate p53 by disrupting its interaction with MDM2. However, it remains to find out under what physiological or pathological circumstance RBM10 acts to regulate the MDM2-p53 loop, as this tumor suppressor was not required for p53 activation by several DNA damaging or ribosomal stress agents, such as doxorubicin, actinomycin D, and 5-FU, as tested here (Supplementary Fig. 11). Thus, further investigation of this new finding would provide more insights into whether the p53 activation function of RBM10 might cross talk with the splicing regulation of this anti-cancer protein or respond to yet unidentified signals. Also, creating a biological model system would be necessary for our better understanding of the tumor suppression activity of this protein and its relationship with p53. These lines of information would certainly be useful for drug discovery against cancers in the future.
Materials and Methods
Plasmids and antibodies
The RBM10 expression plasmid was kindly provided by Dr. Juan Valcarcel (Centre de Regulacio Genomica, Spain). The GFP-RBM10 was kindly provided by Dr. Yongbo Wang (Fudan University, China). Flag-RBM10 (WT and ΔN) plasmid was constructed in our laboratory. The plasmids encoding HA-MDM2, Flag-MDM2, GST-MDM2 fragments, Flag-p53 fragments, p53, Flag-L5, Flag-L11, Flag-L22, Flag-p53 (R249S), Flag-p53 (Y200C), V5-p53 (R273HR), V5-p53 (R248W), and HA-Ub were described previously[10, 21, 22]. Anti-Flag (Sigma-Aldrich, catalogue no. F1804, diluted 1:3,000), anti-HA (Proteintech, catalogue no. 66006–1-Ig, diluted 1:3,000), anti-GFP (B-2, Santa Cruz Biotechnology, catalogue no. sc-9996, diluted 1:1,000), anti-RBM10 (Proteintech, catalogue no. 14423–1-AP, diluted 1:1,000), anti-Cleaved PARP (Cell Signaling Technology, catalogue no. #9541, diluted 1:1,000), anti-PARP (Cell Signaling Technology, catalogue no. #9542, diluted 1:2,000), anti-p53 (DO-1, Santa Cruz Biotechnology, catalogue no. sc-126, diluted 1:1,000), anti-p21 (CP74, Neomarkers, Fremont, catalogue no. MS-891-P0, diluted 1:1,000), anti-PUMA (H-136, Santa Cruz Biotechnology, catalogue no. sc-28226, diluted 1:1,000), anti-GAPDH (Proteintech, catalogue no. 60004–1-Ig, diluted 1:2,000), anti-Lamin A/C and anti-β-actin (C4, Santa Cruz Biotechnology, catalogue no. sc-47778, diluted 1:3,000) were commercially purchased. Antibodies against MDM2 (2A9 and 4B11) were previously described [23].
Cell culture and transient transfection
HCT116 p53+/+ and HCT116 p53−/− cells were generous gifts from Dr. Bert Vogelstein at the John Hopkins Medical Institutes. H460, U87 and MCF7 cells were purchased from American Type Culture Collection (ATCC). MEF p53−/−; Mdm2−/− cells were generous gifts from Dr. Guillermina Lozano from MD Anderson Cancer Center, the University of Texas. STR profiling was performed to ensure cell identity. No mycoplasma contamination was found. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 50Uml−1 penicillin and 0.1mgml−1 streptomycin. All cells were maintained at 37 °C in a 5% CO2 humidified atmosphere. Cells seeded on the plate overnight were transfected with plasmids as indicated in figure legends using TurboFect transfection reagent following the manufacturer’s protocol (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were harvested at 36–48 h post-transfection for future experiments.
Immunoblotting and immunoprecipitation
Cells were harvested and lysed in lysis buffer consisting of 50mM Tris/HCl (pH7.5), 0.5% Nonidet P-40 (NP-40), 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 10mM pepstatin A and 1mM leupeptin. Equal amounts of clear cell lysate (20–40 μg) were used for immunoblotting (IB) analyses. Immunoprecipitation (IP) was conducted using antibodies as indicated in the figure legends. Briefly, 300–500 μg of proteins were incubated with the indicated antibody at 4 °C in for overnight. Protein A or G beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were then added, and the mixture was incubated at 4 °C for additional 1 to 2 h. Beads were washed at least three times with lysis buffer, boiled and then resolved by 8–15% SDS-PAGE. Bound proteins were detected by IB with antibodies as indicated in the figure legends.
GST fusion protein-protein interaction assay
GST-tagged MDM2 fragment proteins were expressed in E. coli. After lysis using GST lysis buffer (PBS, glycerol, PMSF, Proteinase inhibitor Cocktail), the proteins were coupled with glutathione-Sepharose 4B beads (Sigma-Aldrich Co., St. Louis, MO, USA). For RBM10 protein preparation, HCT116 p53−/− cells were transfected with Flag-RBM10 plasmid. Cell lysates were incubated and gently rotated with the glutathione-Sepharose 4B beads containing GST-MDM2/1–491, GST-MDM2/1–301, GST-MDM2/1–150, GST-MDM2/294–491, or GST. After incubation at room temperature, the mixtures were washed 3 times in ice-cold PBS and twice in lysis buffer (50 mM Tris/HCl pH 8.0, 0.5% NP-40, 1 mM EDTA, 150 mM NaCl, 10% glycerol). Bound proteins were analyzed on a gradient gel and detected by IB analysis with the anti-RBM10 antibody.
Cell viability assay
To assess the cancer cell survival, the Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Rockville, MD, USA) was used according to the manufacturer’s instructions. Cell suspensions were seeded at 2,000 cells per well in 96-well culture plates at 24 h post-transfection. Cell viability was determined by adding WST-8 at a final concentration of 10% to each well, and the absorbance of the samples was measured at 450 nm using a Microplate Reader (Molecular Device, SpecrtraMax M5e, Sunnyvale, CA, USA) after 48 h.
Colony formation assay
Cancer cells (50% confluence) were transfected with pcDNA or RBM10 for 48 h and trypsinized. The same number of cells were seeded on each 60 mm plates. Neomycin added Media was changed every 3 days until colonies were visible. Cells were then fixed with methanol and stained with crystal violet solution at RT for 30 min.
Would healing assay
Cells transfected with either plasmids or siRNAs were seeded onto 24 well plates in triplicate and allowed to attach overnight. P200 tips were used to generate scratches in each well followed by imaging as 0 h time point. Images covering the scratches were taken every 24 hours to monitor the wound healing process.
Cell fractionation
Approximately, 1 × 106 cells were collected, washed twice with PBS, and resuspended in 0.2–0.5 mL buffer A (10 mM HEPES–KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT) for 10 min on ice. Phenylmethylsulfonyl fluoride was added to a final concentration of 0.2 mM, and the mixture was then Dounce homogenized until all cytoplasmic membranes were disrupted. For cytosolic isolation, cells were centrifuged at 1600 rpm for 5 min at 4 °C to obtain the supernatant as cytoplasm, and pellets were lysed in RIPA buffer as described previously [10].
Flow cytometry analysis
Flow cytometry analysis was done by using the methods described previously [24]. Briefly, cells transfected with RBM10 as indicated in the figure legends were fixed with ethanol overnight and stained in 1ml of propidium iodide (Sigma-Aldrich) stain buffer (50 μg ml−1 PI, 200 μg ml−1 RNase A, 0.1% Triton X-100 in phosphate-buffered saline) at 37 °C for 30 min in 4 °C. Cells were then analysed for DNA content using a BD Bioscience FACScan flow cytometer (BD Biosciences, San Jose, CA, USA). Data were analyzed using the cellQuest (BD Biosciences) and Modfit (Verity, Topsham, ME, USA) software programs.
Reverse transcription and quantitative real time-PCR analysis
Reverse transcription (RT) and quantitative RT-PCR analyses. Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Total RNAs of 1.5 μg were used as templates for reverse transcription using poly-(T)20 primers and M-MLV reverse transcriptase (Promega, Madison, WI, USA). Quantitative real time-PCR (RT-qPCR) was conducted using SYBR Green Mix according to the manufacturer’s protocol (BioRad, Hercules, CA, USA). The primers for RBM10, p53, Vimentin, VEGFA1, CREBBP, FDXR and GAPDH cDNA detection are as follows: RBM10, 5’-CTCTACTATGACCCCAACTCCCA-3’ and 5’- GTCCGCCTCTCCCCATCCCA-3’; p53, 5’-CCCAAGCAATGGATGATTTGA-3’ and 5’-GGCATTCTGGGAGCTTCATCT-3’; Vimentin, 5’-GCAGGAGGCAGAAGAATGGT-3’ and 5’-CCACTTCACAGGTGAGGGAC-3’; VEGFA1, 5’-AGGAGGAGGGCAGAATCATCA-3’ and 5’-CTCGATTGGATGGCAGTAGCT-3’; CREBBP, 5’-AGGCACAACCTGTGAGACCT-3’ and 5’-ACTGAGCCCATGCTGTTCAT-3’; FDXR, 5’-AGAAACAGCCTGTGCCCTTT-3’ and 5’-GAATGGGCCGTCTGGGTAAA-3’; GAPDH, 5’-GATTCCACCCATGGCAAATTC-3’and 5’-AGCATCGCCCCACTTGATT-3’.
RNA interference
siRNAs against RBM10 (Life Technologies, Carlsbad, CA, USA) were commercially purchased. siRNA (40 nM) were introduced into cells using TurboFect transfection reagent following the manufacturer’s protocol. Cells were harvested 72 h after transfection for IB or RT-qPCR.
Statistics
All in vitro experiments were performed in biological triplicate. The student’s two-tailed t-test was used to determine mean difference among groups. P < 0.05 was considered statistically significant. Data are presented as mean ± s.e.m.
Supplementary Material
Acknowledgements
We thank Juan Valcarcel for providing us with RBM10 plasmid, Yongbo Wang for providing us with GFP-RBM10, and Xiang Zhou as well as other members of the Lu laboratory for active discussion. This work was supported in part by National Institutes of Health (NIH)-National Cancer Institute (NCI) grants (R01CA095441, R01CA172468, R01CA127724) to Hua Lu and Shelya Zeng.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- 1.Oren M. Decision making by p53: life, death and cancer. Cell death and differentiation 2003; 10: 431–442. [DOI] [PubMed] [Google Scholar]
- 2.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harbor perspectives in biology 2009; 1: a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nature reviews Cancer 2002; 2: 594–604. [DOI] [PubMed] [Google Scholar]
- 4.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res 2013; 27: 254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–299. [DOI] [PubMed] [Google Scholar]
- 6.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387: 299–303. [DOI] [PubMed] [Google Scholar]
- 7.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420: 25–27. [DOI] [PubMed] [Google Scholar]
- 8.Yang M, Sun H, Wang H, Zhang S, Yu X, Zhang L. Down-regulation of ribosomal protein L22 in non-small cell lung cancer. Med Oncol 2013; 30: 646. [DOI] [PubMed] [Google Scholar]
- 9.Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 2008; 32: 81–95. [DOI] [PubMed] [Google Scholar]
- 10.Cao B, Fang Z, Liao P, Zhou X, Xiong J, Zeng S et al. Cancer-mutated ribosome protein L22 (RPL22/eL22) suppresses cancer cell survival by blocking p53-MDM2 circuit. Oncotarget 2017; 8: 90651–90661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell 2013; 52: 720–733. [DOI] [PubMed] [Google Scholar]
- 12.Guan G, Li R, Tang W, Liu T, Su Z, Wang Y et al. Expression of RNA-binding motif 10 is associated with advanced tumor stage and malignant behaviors of lung adenocarcinoma cancer cells. Tumour Biol 2017; 39: 1010428317691740. [DOI] [PubMed] [Google Scholar]
- 13.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol 2011; 192: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma N, Jadhav SP, Bapat SA. CREBBP re-arrangements affect protein function and lead to aberrant neuronal differentiation. Differentiation 2010; 79: 218–231. [DOI] [PubMed] [Google Scholar]
- 15.Giebler HA, Lemasson I, Nyborg JK. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol Cell Biol 2000; 20: 4849–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Qian Y, Zhang J, Yan W, Jung YS, Chen M et al. Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev 2017; 31: 1243–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins KM, Kainov YA, Christodolou E, Ray D, Morris Q, Hughes T et al. An RRM-ZnF RNA recognition module targets RBM10 to exonic sequences to promote exon exclusion. Nucleic Acids Res 2017; 45: 6761–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 2004; 36: 147–150. [DOI] [PubMed] [Google Scholar]
- 19.Hunziker A, Jensen MH, Krishna S. Stress-specific response of the p53-Mdm2 feedback loop. BMC Syst Biol 2010; 4: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol 2015; 7: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao JM, Zhou X, Gatignol A, Lu H. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 2014; 33: 4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene 2013; 32: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of biological chemistry 2004; 279: 44475–44482. [DOI] [PubMed] [Google Scholar]
- 24.Jung JH, Liao JM, Zhang Q, Zeng S, Nguyen D, Hao Q et al. Inauhzin(c) inactivates c-Myc independently of p53. Cancer Biol Ther 2015; 16: 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.