Abstract
G protein-coupled receptors (GPCRs) are the largest family of cell-surface receptors in humans and regulate numerous physiological processes through the activation of heterotrimeric G proteins. GPCR kinases (GRKs) selectively phosphorylate active GPCRs, which promotes arrestin binding, receptor internalization, and initiation of alternative signaling pathways. GRK5 is a representative member of one of three GRK subfamilies that does not need post-translational lipidation or other binding partners to exhibit full activity against GPCRs, rendering it a useful tool for biophysical studies directed at characterizing GRK function. However, recombinant expression of GRK5 has thus far been limited to insect and mammalian systems. Here, we describe the expression of functional GRK5 in E. coli and its purification and biochemical characterization. Bacterially expressed GRK5 is hyperphosphorylated, primarily in regions known to be flexible from prior crystal structures, which slightly decreases its catalytic activity toward receptor substrates. Mutation of a single phosphorylation site, Thr10, restores kinetic parameters to those of GRK5 purified from insect cells. Consequently, bacterial expression will allow for production of GRK5 at a reduced cost and faster pace and would facilitate production of isotopically labeled kinase for NMR studies or for the incorporation of unnatural amino acids.
Keywords: G protein-coupled receptor kinase 5, phosphorylation, mass spectrometry, bacterial expression, autophosphorylation
Introduction
G protein-coupled receptors (GPCRs) play critical roles in the regulation of many physiological processes through the activation of heterotrimeric G proteins. They are in turn regulated by a family of seven GPCR kinases (GRKs) which initiate desensitization through the phosphorylation of the third intracellular loop and/or C-terminus of the active GPCR [1]. Phosphorylation promotes the binding of arrestins, which sterically occludes the receptors from interacting with heterotrimeric G proteins and also targets the receptors for clathrin-mediated endocytosis [2].
Like GRK2, GRK5 is upregulated following heart failure and contributes to decreased cardiac output and the progression of cardiac hypertrophy making GRK5 an attractive therapeutic target [3]. Indeed, recent inhibitor development campaigns have yielded small molecule inhibitors for GRK2 that display efficacy in preclinical animal models of heart failure [4–6]. However, small molecule inhibitors selective for GRK5 have not yet been reported. Thus, efficient expression and purification of GRK5 would be useful to supply high-quality protein not only for biophysical analysis but also for pharmacological screens. Recombinant GRK5 is almost exclusively purified from insect cells using engineered baculovirus [7,8], which is more expensive and time consuming than bacterial expression systems. Protein kinases are often not purified from bacterial systems due to cytotoxicity, however, considerable success has been achieved using truncated or catalytically inactive kinases [9]. Such variants however often have limited use in biochemical assays and structural characterization. To date, there is only one report of a GRK being expressed and purified from a bacterial system. Gan et al. reported purification of soluble human GRK2 [10], but multiple attempts in our lab could not reproduce the result.
To facilitate future biophysical and drug discovery studies of GRK5, we sought to develop a method for rapid and cost-effective purification of full-length human GRK5 from E. coli. In addition to lower cost, expression in bacterial systems affords other advantages. The production of isotopically labeled proteins for nuclear magnetic resonance (NMR) studies or incorporation of unnatural amino acids via amber stop codon methods are significantly easier in bacterial expression systems [11]. Additionally, the production of numerous protein variants for functional analysis in bacteria is much more efficient because recombinant baculovirus does not need to be produced for each variant. Herein, we describe a GRK5 construct and purification protocol to produce full-length, human GRK5 in E. coli with comparable yields and activity to insect expression systems. Characterization of the bacterially expressed kinase reveals that it undergoes extensive autophosphorylation that leads to a reduction in its catalytic efficiency when compared to GRK5 purified from insect cells. Mutation of one of the phosphorylation sites in the N-terminus of the protein rescues full activity.
Materials and methods
GRK5 cloning, expression, and purification
The human GRK5 gene was amplified with the addition of a C-terminal Val-Asp residues directly followed by a hexahistidine tag (Figure 1A). 5´ NdeI and 3´ EcoRI restriction sites were also introduced to the gene fragment via PCR. The bacterial expression vector pMalC2H10T was digested with NdeI and EcoRI to remove the MBP-polyhistidine-TEV features and the GRK5 gene ligated into the modified vector to produce the plasmid pEL17.
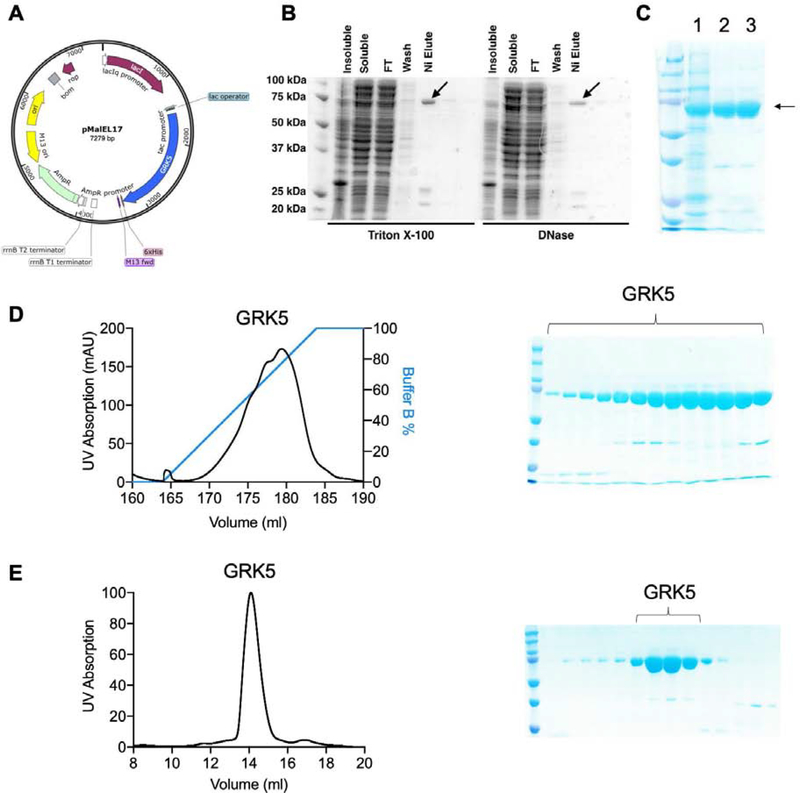
Figure 1:
Purification of full-length, human GRK5 expressed in E. coli. A) Map of human GRK5 in a modified pMalC2H10T vector (pEL17). B) Comparison of small-scale expression tests with either Triton X-100 or DNase added to the lysis buffer. GRK5 in the elution is indicated with an arrow. C) Coomassie stain showing the purity after IMAC (1), ion exchange (2) and SEC (3). GRK5 is indicated with an arrow. D) Representative ion exchange chromatogram from a 1 ml HiTrap S column. The column was eluted with a linear gradient from buffer A containing 100 mM NaCl to buffer B containing 500 mM NaCl. GRK5 is eluted with a wide salt concentration ranging from 200 mM NaCl to 500 mM NaCl. E) Representative SEC chromatogram from the indicated region of D) on an analytical S200 column.
GRK5 protein expression and purification
GRK5 was expressed in E. coli BL21 Rosetta cells containing the pRARE plasmid encoding rare tRNAs [12]. Cells were grown in TB at 37 °C until OD600 of ~0.6, cooled to 18 °C, induced with 0.2 mM IPTG at OD600 = 1.0–1.2, and grown overnight at 18 °C. Cells were then pelleted and resuspended in lysis buffer containing 20 mM HEPES pH 8.0, 200 mM NaCl, 40 mM imidazole, 5 mM MgCl2, 5 mM CaCl2, 1 mM DTT, leupeptin, lima bean trypsin protease inhibitor, and 2 μg mL−1 DNaseI. Alternatively, DNaseI can be replaced with 0.02% (v/v) Triton-X100. The cells were lysed with an EmulsiFlex-C3 cell homogenizer, and the lysate was incubated on ice for 30 min to allow for digestion of DNA prior to centrifugation at >15,000×g for 30 min. The resulting supernatant was glass filtered, passed through Ni-NTA resin, washed with lysis buffer (without DNase I or Triton-X100), and eluted with lysis buffer containing an additional 200 mM imidazole. Although either anion or cation exchange can be used to further purify GRK5, GRK5 has higher affinity for HiTrap S (GE Healthcare) than for the HiTrap Q column (GE Healthcare). When the buffer contains 100 mM NaCl, GRK5 flows through HiTrap Q column and binds to HiTrap S, whereas most of the contaminants bind to HiTrap Q (GE Healthcare). GRK5 was then eluted with a gradient of 0.1–0.5 M NaCl at pH 8.0 from a HiTrap S column (GE Healthcare). Eluted GRK5 was subsequently purified via size exclusion chromatography (SEC) in a buffer containing 20 mM HEPES pH 8.0, 200 mM NaCl, and 1 mM DTT using an analytical S200 Increase column (GE Healthcare). GRK5 concentrations were determined by absorbance at 280 nm using the calculated molecular weight and molar extinction coefficient from the primary sequence, and then the protein was flash frozen in liquid nitrogen. Typical yields of ≥95% purity GRK5 by SDS-PAGE from E. coli. range from 0.5–2 mg L−1.
Thermal Shift Assay
Differential scanning fluorometry (DSF) was performed in an HT7900 qPCR instrument using 0.2 mg mL−1 GRK5 in assay buffer (20 mM HEPES pH 8.0, 10 mM MgCl2, and 1 mM DTT), SYPRO Orange protein gel stain (1000x stock, Sigma-Aldrich), and 200 μM ligand. Melt curves were obtained by increasing the temperature from 25–70 °C at a rate of 2 °C min−1. Data were plotted in GraphPad Prism 7 with the inflection point of the fit sigmoidal curve representing the melting point (Tm) of the protein. The experiment was performed three times in duplicate using protein from the same purification. Statistical significance was assessed via two-way ANOVA with Bonferroni correction for multiple comparisons.
Mass spectrometry (MS) and phosphosite mapping
Intact protein MS was performed on an Agilent quadrupole time-of-flight tandem mass spectrometer with a 1–3 μL injection volume of GRK5 at 1 mg mL−1 in SEC buffer containing 1% (v/v) formic acid. Protein was desalted on a C4 chromatography column prior to mass analysis with positive electrospray ionization. Phosphosite mapping was performed through the Purdue University Proteomics Facility. Briefly, purified GRK5 was digested with trypsin, the fragments analyzed via high-resolution MS without TiO2 enrichment, and phosphorylation sites identified through peptide ionization patterns compared to the nonphosphorylated primary amino acid sequence.
Kinetic analysis
Steady-state kinetic parameters were determined as previously described [13]. Briefly, 50 nM GRK5 expressed and purified from insect cells or E. coli and bovine rhodopsin in rod outer segment (ROS) membranes were combined in assay buffer containing 20 mM HEPES pH 8.0, 10 mM MgCl2. Reactions were initiated with ATP spiked with [γ−32P]-ATP and allowed to proceed for 2 min at room temperature prior to separation by SDS-PAGE and image analysis on phosphor screens. The maximum reaction velocity (Vmax) and apparent Michaelis constant (Km) were determined via GraphPad Prism using a classic Michaelis-Menten plot. Km, Vmax, and kcat for ATP were determined by varying ATP concentration from 2 to 200 μM while keeping rhodopsin constant at 5 μM. Km, Vmax, and kcat for rhodopsin were determined by varying rhodopsin concentration from 0.08 to 8 μM while keeping ATP constant at 50 μM.
Results
GRK5 associates with genomic DNA during purification
Human GRK5 was expressed with a C-terminal hexahistidine tag (Figure 1A) because N-terminal tags interfered with receptor phosphorylation. Initial attempts to purify this recombinant GRK5 from E. coli yielded insoluble protein, and co-expression with various chaperones or the use of slower promoters such as Trc also did not yield soluble protein (data not shown). Screening of lysis buffer additives revealed that addition of 2 μg mL−1 DNaseI or 0.02% (v/v) Triton-X100 yielded soluble protein (Figure 1B). Because soluble protein was obtained through the addition of DNase, it is likely that GRK5 associates with bacterial genomic DNA and pellets in the insoluble fraction during ultracentrifugation. Indeed, GRK5 has very basic surface features on its kinase domain [14] and is reported to have a DNA-binding element within the large lobe of its kinase domain (residues 388–395) [15]. Solubilized GRK5 was then purified using immobilized metal affinity chromatography (IMAC) followed by ion exchange and size exclusion chromatography (Figure 1C). For ion exchange chromatography, tandem HiTrap Q /HiTrap S columns were used resulting in >95% purity (Figure 1D). SEC indicates GRK5 elutes at a volume consistent with a monomer (Figure 1E).
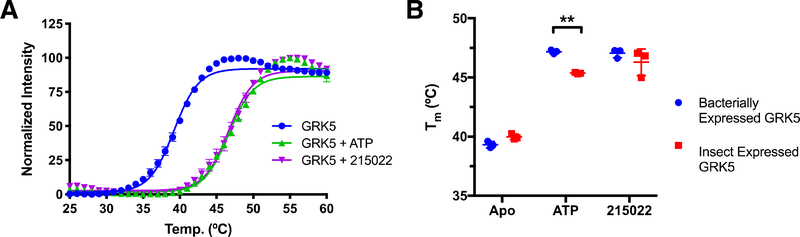
Bacterially expressed GRK5 is thermally stabilized by ATP
To test if GRK5 expressed in E. coli is properly folded, DSF was performed to measure its thermal melting point (Tm) (Figure 2, Table 1). No significant difference was observed between unliganded GRK5 purified from insect cells and that from bacteria. Bacterially expressed GRK5 was stabilized by 8 °C upon addition of either Mg2+·ATP or the pan-GRK inhibitor CCG-215022 [16]. Interestingly, GRK5 purified from insect cells was stabilized by only 5–6 ˚C with the same ligands. Because we have observed that the magnitude of ligand-induced thermal shift is correlated with binding affinity in GRKs [16], it suggests that GRK5 expressed in E. coli has higher affinity for each ligand.
Figure 2:
Thermal stability of GRK5 as a function of ligand and expression host. A) Melting curves for GRK5 expressed in E. coli. Data points represent mean ± SD (n=3). B) Comparison of Tm values for human GRK5 expressed in insect vs. E. coli cells in the presence or absence of various ligands. Data reported as mean ± SD (n=3). Statistical significance determined via two-way ANOVA with Bonferroni correction for multiple comparisons between bacterially and insect expressed GRK5 (**p<0.01).
Table 1.
Thermal stability of GRK5 expressed in insect cells or E. coli
| Ligand | E. coli GRK5 (°C) | Insect GRK5 (°C) |
|---|---|---|
| none | 39.3 ± 0.3 | 40.0 ± 0.2 |
| ATP | 47.2 ± 0.2 | 45.4 ± 0.1 |
| CCG215022 | 47.1 ± 0.4 | 46.3 ± 1.1 |
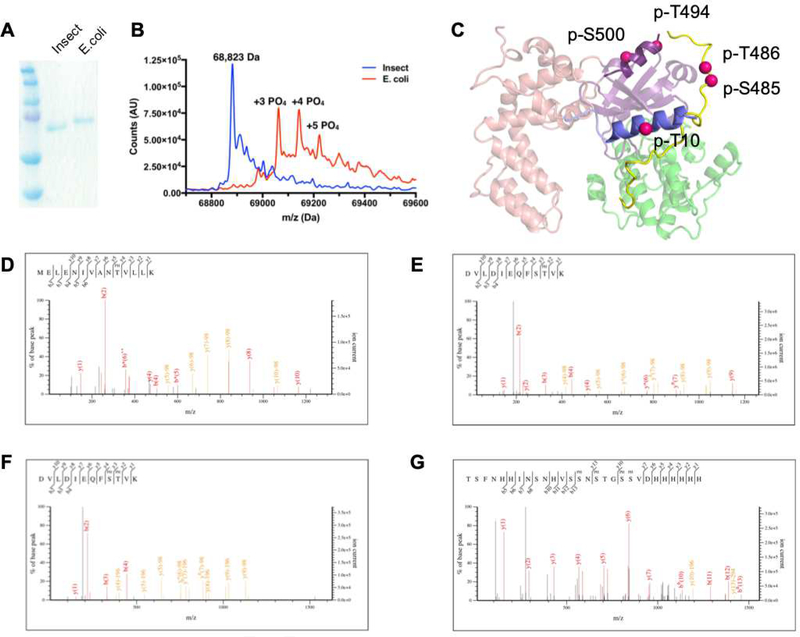
Bacterially expressed GRK5 is hyperphosphorylated
SDS-PAGE showed that bacterially expressed GRK5 exhibits slower mobility than insect cell expressed GRK5 (Figure 3A). Suspecting autophosphorylation, bacterially expressed GRK5 was analyzed via intact protein MS (Figure 3B), which revealed that the GRK5 was phosphorylated from two to eight times with the most abundant peaks corresponding to three to five phosphates. To identify the phosphorylation sites, phosphosite analysis was performed by tandem MS. The data show extensive phosphorylation, primarily in the active site tether (AST) loop and the extreme C-terminal region (residues 565–590) which has been shown via crystallography to be disordered in solution (Figure 3 C–G, Table 2). Not surprisingly, the two highest scoring phosphorylated peptides include the canonical AGC kinase autophosphorylation sites Ser484 and Thr485 in the AST (Figure 3 C,E,F). Other modified sites include Thr10 in the αN helix and Ser500 in the small lobe of the kinase domain. Several types of phosphorylation unusual for eukaryotic proteins were also observed, including phosphorylated cysteine (Table 2), perhaps as a consequence of the bacterial expression host.
Figure 3:
E. coli expressed GRK5 is extensively autophosphorylated. A) Coomassie staining shows that bacterially expressed GRK5 has slower mobility than GRK5 purified from insect cells in SDS-PAGE. B) Intact protein MS. Exact mass of unphosphorylated GRK5 is 68,824 Da. Masses consistent with 3, 4, and 5 phosphorylation sites are 69,063, 69,145, and 69,223 Da, respectively. C) GRK5 phosphorylation sites identified using tandem MS mapped on the structure of GRK5 (PDB entry 4WNK). The phosphorylation sites are represented as red spheres for the Cα positions of the corresponding residues. D-G) Representative MS peptide fragment ion spectra. Phosphorylated peptides spanning D) GRK5 residues 1–14, E-F) 476–487, and G) 571–590 + hexahistidine tag.
Table 2:
Phosphosite mapping of GRK5 purified from E. coli
| Peptide (# hits with PTM) | GRK5 Range | Residue Type | Phosphorylation Site Scorea |
|---|---|---|---|
| MELENIVANTVLLK (1) | 1–14 | Thr | 66 |
| DVLDIEQFSTVK (1) | 476–487 | Thr | 74 |
| DVLDIEQFSTVK (1) | 476–487 | Ser, Thr | 51 |
| DVLDIEQFSTVKGVNLDHTDDDFYSK (2) | 476–501 | Ser, Thr | 89 |
| DVLDIEQFSTVKGVNLDHTDDDFYSK (2) | 476–501 | Ser | 77 |
| DVLDIEQFSTVKGVNLDHTDDDFYSK (2) | 476–501 | Thr | 47 |
| TSFNHHINSNHVSSNSTGSSVDHHHHHH (1) | 571–590+6His | 4x Ser | 67 |
| ELFSACAQSVHEYLR (1) | 140–154 | Cys | 45 |
| LGCQEEGAAEVKR (1) | 432–444 | Cys | 79 |
Phosphorylation Site Score indicates both relative abundance and confidence in post-translational modification (PTM) assignment. The largest score for the group of peptides is shown. Phosphorylated residues of the type identified are underlined. Scores ≥40 indicate definitive assignment of the indicated PTM to the corresponding peptide. Multiple hits for the same PTM on a peptide suggest a heterogeneous population.
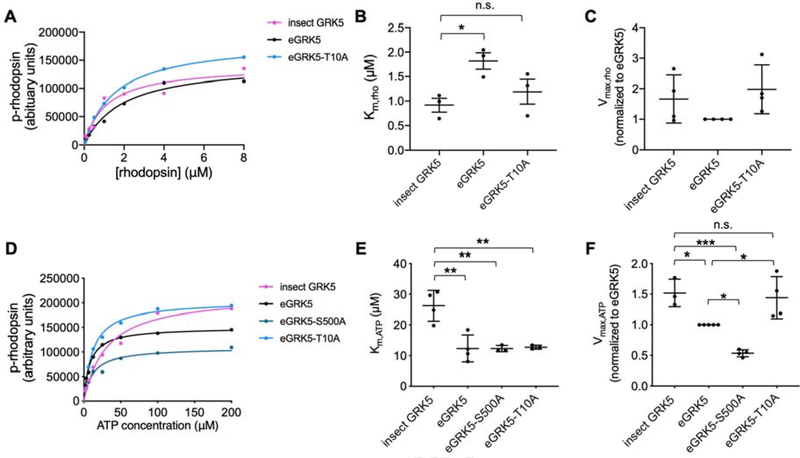
Hyperphosphorylated GRK5 has decreased kinase activity
To demonstrate that GRK5 purified from E. coli was active and to determine the effect of its hyperphosphorylation on activity, radiometric assays were used to monitor the phosphorylation of rhodopsin in native membranes. E. coli expressed GRK5 has comparable Vmax,Rho, but 2-fold higher Km,Rho compared to insect expressed GRK5 (Figure 4 A–C, Table 3). Km,ATP of the bacterially expressed GRK5 is about two times lower than that of insect expressed GRK5 (Figure 4 D–F, Table 3), whereas its Vmax,ATP is about 1.5 times lower (Figure 4 D–F, Table 3). Thr10 is located within the αN helix, and because this region has been proposed to interact with activated receptors, phosphorylation at this site may explain reductions in the ability to phosphorylate receptor substrates [17–19]. Ser500 is located in a region near the predicted membrane interface for GRKs and thus, phosphorylation at this site may repel acidic phospholipid headgroups. The GRK5-S500A however exhibited decreased Vmax,ATP and had no effect on Km,ATP (Figure 4 D–F, Table 3), whereas GRK5-T10A restored all kinetic parameters to those of insect cell expressed GRK5 with the exception of Km,ATP (Figure 4, Table 2). Elimination of both the canonical AGC kinase phosphorylation sites in the AST loop (S484A/T485A) [20] or all of the observed sites in the C-terminus 6A (S583A/S584A/S586A/T587A/S589A/S590A) dramatically reduced protein expression and prohibited kinetic analysis. The reduced expression level of 6A mutant is unlikely due to reduced solubility or misfolding since truncation of the GRK5 C-terminal tail at residue 531 expresses well in insect cells and yields soluble protein [21].
Figure 4:
Steady-state kinetic analysis of GRK5 variants. A and D) Michaelis-Menten kinetic curves from a representative experiment with rhodopsin in rod outer segments as substrate. B) Vmax,Rho, C) Km,Rho, E) Vmax,ATP, F) Kmax,ATP values for GRK5 protein variants expressed in insect or E. coli cells (n=3–4). Statistical significance was determined via one-way ANOVA with Tukey’s correction for multiple comparisons (*p<0.05, **p<0.01, ***p<0.001, n.s.=not significant).
Table 3.
Kinetic analysis of GRK5 variants
| Vmax,Rho (fold E. coli GRK5) | Km,Rho (μM) | kcat,Rho/Km,Rho (mM−1*s−1) | Vmax,ATP (fold E. coli GRK5) | Km,ATP (μM) | kcat,ATP/Km,ATP (mM−1*s−1) | |
|---|---|---|---|---|---|---|
| E. coli GRK5 | 1 | 1.8 ± 0.3 | 2.9 ± 1.7 | 1 | 14 ± 4 | 5.9 ± 2 |
| Insect GRK5 | 1.6 ± 0.9 | 0.9 ± 0.2 | 7.3 ± 0.7 | 1.5 ± 0.2 | 26 ± 5 | 7.9 ± 3 |
| E. coli GRK5-S500A | ND | ND | ND | 0.60 ± 0.09 | 20 ± 9 | 4.1 ± 1 |
| E. coli GRK5-T10A | 1.6 ± 0.3 | 1.2 ± 0.4 | 6.5 ± 0.5 | 1.5 ± 0.3 | 14 ± 3 | 8.7 ± 4 |
Discussion
Whereas prior attempts to purify mammalian GRK5 expressed in E. coli led to insoluble protein, we show here that the simple addition of detergent or DNase is sufficient to obtain adequate amounts of high-quality and active protein for biochemical and biophysical studies. The requirement for DNase to liberate GRK5 from pellets suggests that the recombinant protein associates with genomic DNA. Indeed, GRK5 contains both a nuclear localization signal and DNA binding element, and in mammalian cells a small fraction of endogenous GRK5 is known to be nuclear and involved in the regulation of gene expression [15]. When purified from insect cells, nuclei are not typically disrupted and thus GRK5 is shielded from genomic DNA.
Interestingly, E. coli expressed GRK5 displays a pattern of phosphorylation that has a significant effect on catalytic activity toward receptor substrates. Because bacteria lack canonical Ser/Thr kinases, the phosphorylation events are likely to be the result of GRK5 autophosphorylation. GRK5 purified from insect cells lacks phosphorylation, likely through the action of endogenous phosphatases. Many of the observed autophosphorylation sites are located in the C-terminus and therefore might be expected to repel the membrane binding αC helix of GRK5 from membranes and reduce the efficiency of receptor phosphorylation. The decrease in activity of bacterially expressed GRK5 toward ROS was however not significantly different from that expressed in insect cells, possibly due to the phosphorylation sites residing in the extreme C-terminus rather than immediately adjacent to the amphipathic membrane binding αC helix. Indeed, a prior study of the C-terminus of closely-related GRK6 revealed that residues immediately following the αC helix (GRK6: 561–567) are more important for membrane association than terminal residues [22].
Mutation of selected phosphorylation sites elsewhere in GRK5 identified Thr10 in the αN helix as a contributor to the decreased activity toward ROS, because the T10A variant of E. coli expressed GRK5 exhibited the same maximal activity as GRK5 purified from insect cells, which lacks phosphorylation at this site. Rescue of catalytic activity upon mutation of this single residue is consistent with what is hypothesized by some GRK-GPCR docking models wherein the N α-helix binds directly to the GPCR [19], although this interaction is not proposed in other docking models [23].
Bacterially expressed, hyperphosphorylated GRK5 also displays a significantly decreased apparent Km for ATP, consistent with a higher affinity for ATP than unphosphorylated GRK5 from insect cells. It also shows a greater increase in thermal stability in the presence of ATP. GRK5 purified from insect cells lacks canonical AST loop phosphorylation, which generally leads to activation in AGC family kinases [7]. Autophosphorylation of these positions can however be stimulated by anionic phospholipids [24]. The lack of activating AST loop phosphorylation in GRK5 purified from insect cells may explain its higher Km for ATP (Figure 4, Table 3) and smaller change in thermal stability upon the addition of ATP (Figure 2, Table 1) relative to the bacterially expressed form. In prior studies, the S484A/T485A variant dramatically reduced phosphorylation of activated GPCRs [24]. Unfortunately, we were unable to directly test this ourselves because the S484A/T485A variant did not express at sufficient levels in bacteria to allow for purification and kinetic analysis. It is tempting to speculate that autophosphorylation of these residues may be required to facilitate production of properly folded GRK5 in bacteria.
The described construct and purification scheme will facilitate future biochemical and biophysical studies of GRK5. Not only does bacterial expression afford AST loop phosphorylation, which is likely necessary for function in vivo, but it offers easier incorporation of unnatural amino acids via amber stop codons into GRK5 [11]. Use of photocrosslinking amino acids to capture and study novel GRK5 binding partners or substrates is now a possibility [25]. Furthermore, such selective crosslinking may aid in trapping and structure determination of a GRK5-GPCR complex by capturing the transient interaction with a greater amount of homogeneity than obtained with other crosslinking strategies [26]. NMR studies involving GRKs have also been limited due to the difficulty of producing isotopically labeled kinase in insect cells and their large size (65–80 kDa). As isotope labeling methods are well established and generally more efficient in bacterial systems, obtaining ample quantities of labeled GRK5 from E. coli for NMR studies will be easier and facilitate the study of GRK5 dynamics.
Highlights.
Expression system for G protein-coupled receptor (GPCR) kinase 5 (GRK5) in E. coli
E. coli expressed GRK5 is hyperphosphorylated, especially at C-terminus
Hyperphosphorylation seems required for efficient expression in E. coli
Some autophosphorylation sites inhibit activity towards GPCR substrates.
Represents convenient system for producing pure GRK5 for biophysical studies.
Acknowledgements
We would like to thank Meredith Skiba (University of Michigan) for assistance with collecting intact protein masses and Vicki Hedrick (Purdue University Proteomics Facility) for phosphosite mapping.
Funding
This study was supported by National Institutes of Health (NIH) Pharmacological Sciences Training Program fellowship (T32-GM007767), and U.S. Department of Education GAANN fellowship (P200A150164) to TSB, American heart association postdoctoral fellowship (19POST34450193) to QC and NIH grants HL071818, HL122416, and CA221289 (JJGT). JJGT was also supported by the Walther Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Komolov KE, Benovic JL, G protein-coupled receptor kinases: Past, present and future, Cell. Signal 41 (2018) 17–24. doi: 10.1016/j.cellsig.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mushegian A, Gurevich VV, Gurevich EV, The origin and evolution of G protein-coupled receptor kinases, PLoS One. 7 (2012) 1–12. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hullmann JE, Grisanti LA, Makarewich CA, Gao E, Gold JI, Chuprun JK, Tilley DG, Houser SR, Koch WJ, GRK5-mediated exacerbation of pathological cardiac hypertrophy involves facilitation of nuclear NFAT activity, Circ. Res 115 (2014) 976–985. doi: 10.1161/CIRCRESAHA.116.304475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Waldschmidt HV, Homan KT, Cruz-Rodríguez O, Cato MC, Waninger-Saroni J, Larimore KM, Cannavo A, Song J, Cheung JY, Kirchhoff PD, Koch WJ, Tesmer JJG, Larsen SD, Structure-Based Design, Synthesis, and Biological Evaluation of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors, J. Med. Chem 59 (2016) 3793–3807. doi: 10.1021/acs.jmedchem.5b02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schumacher SM, Gao E, Zhu W, Chen X, Kurt Chuprun J, Feldman AM, Tesmer JJG, Koch WJ, Paroxetine-mediated GRK2 inhibition reverses cardiac dysfunction and remodeling after myocardial infarction, Sci. Transl. Med 7 (2015). doi: 10.1126/scitranslmed.aaa0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Waldschmidt HV, Bouley R, Kirchhoff PD, Lee P, Tesmer JJG, Larsen SD, Utilizing a structure-based docking approach to develop potent G protein-coupled receptor kinase (GRK) 2 and 5 inhibitors, Bioorganic Med. Chem. Lett 28 (2018) 1507–1515. doi: 10.1016/j.bmcl.2018.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kunapuli P, Onorato JJ, Hosey MM, Benovic JL, Expression, purification, and characterization of the G protein-coupled receptor kinase GRK5, J. Biol. Chem 269 (1994) 1099–1105. [PubMed] [Google Scholar]
- [8].Sterne-Marr R, Baillargeon AI, Michalski KR, Tesmer JJG, Expression, purification, and analysis of G-protein-coupled receptor kinases, in: Methods Enzymol., 1st ed., Elsevier Inc., 2013: pp. 347–366. doi: 10.1016/B978-0-12-391862-8.00019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shrestha A, Hamilton G, O’Neill E, Knapp S, Elkins JM, Analysis of conditions affecting auto-phosphorylation of human kinases during expression in bacteria, Protein Expr. Purif 81 (2012) 136–143. doi: 10.1016/j.pep.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gan XQ, Wang JY, Yang QH, Li Z, Liu F, Pei G, Li L, Interaction between the conserved region in the C-terminal domain of GRK2 and rhodopsin is necessary for GRK2 to catalyze receptor phosphorylation, J. Biol. Chem 275 (2000) 8469–8474. doi: 10.1074/jbc.275.12.8469. [DOI] [PubMed] [Google Scholar]
- [11].Chin JW, Martin AB, King DS, Wang L, Schultz PG, Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tegel H, Tourle S, Ottosson J, Persson A, Increased levels of recombinant human proteins with the Escherichia coli strain Rosetta(DE3), Protein Expr. Purif 69 (2010) 159–167. doi: 10.1016/j.pep.2009.08.017. [DOI] [PubMed] [Google Scholar]
- [13].Yao XQ, Cato MC, Labudde E, Beyett TS, Tesmer JJG, Grant BJ, Navigating the conformational landscape of G protein–coupled receptor kinases during allosteric activation, J. Biol. Chem 292 (2017) 16032–16043. doi: 10.1074/jbc.M117.807461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lodowski DT, Tesmer VM, Benovic JL, Tesmer JJG, The structure of G protein-coupled receptor kinase (GRK)-6 defines a second lineage of GRKs, J. Biol. Chem 281 (2006) 16785–16793. doi: 10.1074/jbc.M601327200. [DOI] [PubMed] [Google Scholar]
- [15].Johnson LR, Scott MGH, Pitcher JA, G Protein-Coupled Receptor Kinase 5 Contains a DNA-Binding Nuclear Localization Sequence, Mol. Cell. Biol 24 (2004) 10169–10179. doi: 10.1128/mcb.24.23.10169-10179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Homan KT, Waldschmidt HV, Glukhova A, Cannavo A, Song J, Cheung JY, Koch WJ, Larsen SD, Tesmer JJG, Crystal structure of G protein-coupled receptor kinase 5 in complex with a rationally designed inhibitor, J. Biol. Chem 290 (2015) 20649–20659. doi: 10.1074/jbc.M115.647370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boguth CA, Singh P, Huang CC, Tesmer JJG, Molecular basis for activation of G protein-coupled receptor kinases, EMBO J. 29 (2010) 3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Noble B, Kallal LA, Pausch MH, Benovic JL, Development of a yeast bioassay to characterize G protein-coupled receptor kinases: Identification of an NH 2 -terminal region essential for receptor phosphorylation, J. Biol. Chem 278 (2003) 47466–47476. doi: 10.1074/jbc.M308257200. [DOI] [PubMed] [Google Scholar]
- [19].Huang CC, Orban T, Jastrzebska B, Palczewski K, Tesmer JJG, Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain, Biochemistry. 50 (2011) 1940–1949. doi: 10.1021/bi101606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Newton AC, Regulation of the ABC kinases by phosphorylation: Protein kinase C as a paradigm, Biochem. J 370 (2003) 361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang P, Glukhova A, Tesmer JJG, Chen Z, Membrane orientation and binding determinants of G protein-coupled receptor kinase 5 as assessed by combined vibrational spectroscopic studies, PLoS One. 8 (2013) 1–11. doi: 10.1371/journal.pone.0082072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vatter P, Stoesser C, Samel I, Gierschik P, Moepps B, The variable C-terminal extension of G-protein-coupled receptor kinase 6 constitutes an accessorial autoregulatory domain, FEBS J. 272 (2005) 6039–6051. doi: 10.1111/j.1742-4658.2005.04995.x. [DOI] [PubMed] [Google Scholar]
- [23].Komolov KE, Du Y, Duc NM, Betz RM, Rodrigues JPGLM, Leib RD, Patra D, Skiniotis G, Adams CM, Dror RO, Chung KY, Kobilka BK, Benovic JL, Structural and Functional Analysis of a β 2 -Adrenergic Receptor Complex with GRK5, Cell. 169 (2017) 407–421.e16. doi: 10.1016/j.cell.2017.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kunapuli P, Gurevich VV, Benovic JL, Phospholipid-stimulated autophosphorylation activates the G protein- coupled receptor kinase GRK5, J. Biol. Chem 269 (1994) 10209–10212. [PubMed] [Google Scholar]
- [25].Dumas A, Lercher L, Spicer CD, Davis BG, Designing logical codon reassignment-Expanding the chemistry in biology, Chem. Sci 6 (2015) 50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Beyett TS, Bandekar SJ, Tesmer JJG, Molecular Basis for Targeting, Inhibition, and Receptor Phosphorylation in the G Protein-Coupled Receptor Kinase 4 Subfamily, in: 2016: pp. 59–74. doi: 10.1007/978-1-4939-3798-1_4. [DOI] [Google Scholar]