Abstract
Objective:
Anti-citrullinated protein antibodies (ACPAs) and rheumatoid factor (RF) are commonly present in rheumatoid arthritis without clear rationale for their coexistence. Moreover, autoantibodies develop against proteins with different post-translational modifications and native proteins without obvious unifying characteristics of the antigens. Our objective was to broadly evaluate autoantibody binding in seronegative and seropositive rheumatoid arthritis to identify novel features of reactivity.
Methods:
An array was created with 172,828 native, citrulline-containing, and homocitrulline-containing peptides derived primarily from proteins citrullinated in the rheumatoid joint. IgG and IgM binding to peptides were compared for CCP+RF+, CCP+RF−, CCP−RF+, and CCP−RF− rheumatoid arthritis versus controls (n=12). IgG-bound and endogenously citrullinated peptides were analyzed for amino acid patterns and predictors of intrinsic disorder, i.e. unstable three-dimensional structure. Binding to IgG-derived peptides was specifically evaluated. ELISA confirmed key results.
Results:
Broadly, CCP+RF+ subjects had high and CCP+RF− and CCP−RF+ subjects had modest citrulline-specific IgG binding to array peptides (median z-scores: 3.02, 1.42, 0.75, respectively, p<0.0001). All rheumatoid arthritis groups had low homocitrulline-specific binding. CCP+RF+ subjects had moderate IgG binding to native peptides (median z-score 2.38, p<0.0001). The highest IgG binding was to citrulline-containing peptides, irrespective of protein identity, especially if citrulline was adjacent to glycine or serine, motifs also seen for endogenous citrullination in the rheumatoid joint. Highly bound peptides had multiple features predictive of disorder. IgG from CCP+RF+ subjects targeted citrulline-containing IgG-derived peptides.
Conclusion:
Disordered antigens, which are frequently citrullinated, and common epitopes for ACPAs and RF are potentially unifying features for rheumatoid arthritis autoantibodies.
In rheumatoid arthritis, autoantibodies are both pathologic (1–3) and diagnostic (4). Patients with rheumatoid arthritis produce a variety of anti-citrullinated protein antibodies (ACPAs) with overlapping reactivity (5–8) that underlie the diagnostic anti-cyclic citrullinated peptide antibody (CCP) tests. They also generate rheumatoid factor (RF), antibodies of any isotype that bind to the Fc portion of IgG, which is also used for diagnosis. Additionally, patients with rheumatoid arthritis make autoantibodies that target homocitrulline, called anti-homocitrullinated protein antibodies (AHCPAs) or anti-carbamylated protein antibodies (9). There appears to be some cross-reactivity between AHCPAs and ACPAs (7, 10–12), but this issue has not been completely resolved. Additionally, rheumatoid arthritis patients make autoantibodies against malondialdehyde-acetaldehyde adducted (13) and acetylated proteins (14), suggesting that autoantibodies in rheumatoid arthritis may primarily bind post-translationally modified proteins (15). However, native proteins also can be targeted in rheumatoid arthritis (16–18) and autoantibodies against post-translationally modified proteins often coexist with RF. Why these seemingly unrelated antigens are targeted in rheumatoid arthritis is a mystery.
Although the majority of patients with rheumatoid arthritis generate ACPAs and RF, about 25% are seronegative for both CCP and RF (19). People with seronegative rheumatoid arthritis may lack autoantibodies in general or common autoantibodies for this subset simply may not have been discovered yet. Additionally, some patients are seropositive for only RF or CCP. Little is known about autoantibody reactivity in single seropositive disease. However, an understanding of autoantibodies in these groups could shed light on the spectrum of disease in rheumatoid arthritis.
Here we use a high density peptide array to evaluate autoantibodies against citrulline-containing, homocitrulline-containing and native peptides in seropositive and seronegative subjects in order to identify unifying and novel features of autoantibody reactivity in rheumatoid arthritis.
MATERIALS AND METHODS
Human Subjects:
Human subjects research was carried out in compliance with the Helsinki Declaration and was approved by the University of Wisconsin Institutional Review Board. Serum from age- and sex-matched control and rheumatoid arthritis subjects were selected from the University of Wisconsin Rheumatology Biorepository first described in (20, 21). Briefly, rheumatoid arthritis subjects were identified by having 2+ outpatient visits with rheumatoid arthritis-associated ICD codes within 24 months (22) or one visit and a positive CCP test. Rheumatoid arthritis diagnosis was confirmed by manual review of the three most recent rheumatologist progress notes. Anti-CCP was assessed by generation II anti-CCP or anti-CCP3 ELISA (Inova, San Diego, USA) and RF was assessed by latex or polystyrene agglutination in the UW clinical lab. Rheumatoid arthritis subjects were included in the following groups if CCP and/or RF titers were negative or >2x the upper limit of normal: CCP+RF+, CCP-RF+, CCP+RF-, and CCP-RF-. Controls were excluded if they had any of the following as determined by verbal screen and manual review of the medical record: rheumatoid arthritis, lupus, Sjogren’s Syndrome, scleroderma, multiple sclerosis, type I diabetes, psoriasis, spondyloarthropathy, inflammatory bowel disease, or hematologic malignancy. A total of 48 rheumatoid arthritis and 12 control subjects were included in array studies and 40 CCP+RF+ rheumatoid arthritis and 40 control subjects in confirmatory ELISAs.
High density peptide array:
Twelve amino acid peptides from 224 UniProt sequences (Supplementary Table 1) for 122 unique proteins (includes variants) were tiled at 1 amino acid to generate an array as previously by Roche Nimblegen (Madison, USA) (23). The majority of selected proteins were previously found to contain at least one citrulline in the rheumatoid joint (24–26) with some family members of those proteins included as well as a few known targets of ACPAs (3, 8, 27). Peptides containing arginine or lysine were included as native peptides and as peptides with all arginines or all lysines replaced with citrullines or homocitrullines, respectively. Redundant peptides (found in >1 protein) were included once. Peptides from five proteins were included in duplicate. Each chip contained 12 copies of the array, which each included 172,828 peptides: 35,459 citrulline-containing, 41,608 homocitrulline-containing, and 95,761 native. Sera were provided to Roche Nimblegen to apply to the array to detect IgA, IgM, and IgG that bound to each peptide as previously (23) with sera diluted 1:100 in binding buffer (0.01M Tris-HCl pH 7.4, 1% alkali-soluble casein, 0.05% Tween 20). Six serum samples were run in duplicate on different chips. Roche Nimblegen provided raw signal intensities and a key for placing redundant peptides into appropriate proteins.
Enzyme-linked immunosorbent assay (ELISA):
High binding ELISA plates (Costar 3590, Corning Inc., Corning, USA) were coated with 5 μg/ml streptavidin (Thermo Scientific, Madison, USA) in phosphate buffered saline (PBS), incubated overnight at 4°C, washed thrice with 0.2% Tween-20 in PBS (wash buffer), incubated overnight at 4°C with 20 μg/ml C-terminal biotinylated peptides (Peptide 2.0, Chantilly, USA) in PBS. Peptides are EIFDSRGNPTVEK-biotin and EIFDScitGNPTVEK-biotin derived from alpha-enolase and VFPLAPCSRSTSK-biotin, VFPLAPCScitSTSK-biotin, KPREEQYNSTYRK-biotin, KPcitEEQYNSTYcitK-biotin, FLYSRLTVDKSRK-biotin, and FLYScitLTVDKScitK-biotin derived from IgG heavy chain. Next, at room temperature, plates were washed thrice, blocked with 5% non-fat dry milk for 2 hours, incubated for 2 hours with serum diluted 1:100 in wash buffer with 5% milk, washed 4 times, incubated for 2 hours with goat anti-human IgG conjugated to horse radish peroxidase (Southern Biotech, Birmingham, USA) diluted 1:5000 in wash buffer with 5% milk, washed thrice, developed with 3,3’,5,5’-tetramethylbenzidine, and read at 450 nm with 540 nm correction. Absorbance values were normalized to blank wells and sample-specific uncoated wells. Positive results were in the linear range.
Statistical Methods:
Computations on peptide array data were performed using built-in functions and custom scripts within the R system for statistical computing (28). Raw intensity levels were transformed using a double logarithm (log[log(x)]) to stabilize variation and improve sensitivity (29). Comparisons between rheumatoid arthritis and control subjects were done via a novel distribution analysis that considers all quantitative scores and does not threshold to identify “positive” peptides. First, for each peptide or peptide-matched pair (example: citrulline-containing peptide minus corresponding arginine-containing peptide) signal intensity, we computed the Student t-statistic to measure differential abundance between a rheumatoid arthritis group and controls. We converted each t-statistic to a z-score, essentially the number of standard deviations for the rheumatoid arthritis group from controls, using the probability integral transform and then presented each set of z-scores as an empirical cumulative distribution function (ecdf). Statistical significance was determined by sample-label permutation followed by repeated regeneration of z-score ecdfs; a measure of distributional distance was computed each time between the synthetic ecdf and the null integrated bell curve (>10,000 permuted data sets). The same inference results were obtained for different distance measures: mean difference, Kullback-Leibler divergence, Hellinger distance. By this Monte Carlo testing on sets of peptides, we could infer large-scale differential antibody abundance between rheumatoid arthritis and control groups while respecting potential dependencies between abundance measures in the same subject without heavy multiple-comparison penalties associated with single-peptide testing. Comparisons between ELISA and array data or between proportion of arginine or lysine and average binding was made using distance covariance (30). ELISA data were compared between groups using a Mann-Whitney test or Wilcoxon matched-pairs signed rank test with p<0.05 considered significant.
RESULTS
Broad evaluation of autoantibody binding in rheumatoid arthritis.
A peptide array was designed with overlapping peptides primarily from proteins with at least one citrulline detected in the rheumatoid joint (24–26) including native peptides, peptides with all arginines replaced by citrullines, and peptides with all lysines replaced by homocitrullines. Sera from control and rheumatoid arthritis subjects, divided into CCP+RF+, CCP+RF−, CCP−RF+, and CCP−RF− groups, were subjected to the array to quantify IgM and IgG binding to each peptide. As shown in Supplementary Table 2, subjects were similar across groups. Additionally, the array methodology was accurate and reproducible as determined by cluster analysis, duplicate sample analysis, duplicate peptide analysis, and confirmatory ELISA (Supplementary Figures 1 and 2).
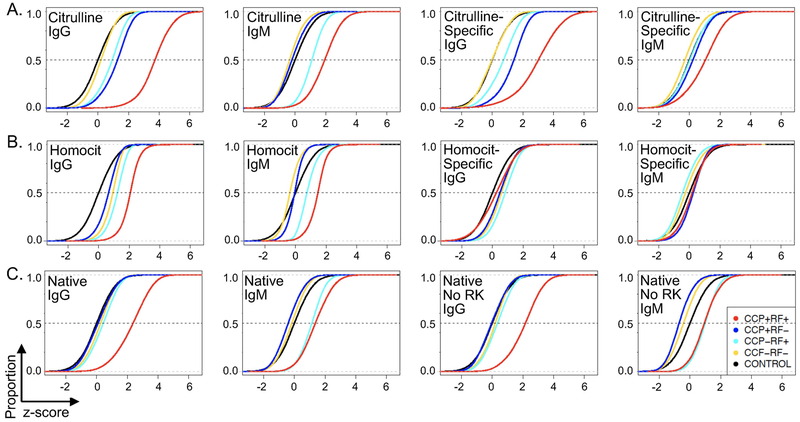
Following these confirmatory tests, we evaluated general antibody binding patterns in the four rheumatoid arthritis groups focusing first on citrulline. As expected, CCP+RF+ subjects had very high IgG binding to citrulline-containing peptides (Figure 1A). CCP+RF− and CCP−RF+ groups had modest IgG binding. For IgM, the CCP+RF+ group had high binding to citrulline-containing peptides with modest binding for the CCP-RF+ group. We next quantified citrulline-specific binding (binding to citrulline-containing peptides minus corresponding arginine-containing peptides) and found very high citrulline-specific IgG binding in CCP+RF+ subjects and modest citrulline-specific IgG binding for CCP-RF+ and CCP+RF− groups. For IgM, there was modest citrulline-specific binding in the CCP+RF+ group. We also performed similar analyses of IgA binding (Supplementary Figure 3). Although there appears to be a small increase in citrulline-specific IgA binding in CCP+RF+ subjects, we could detect only low IgA binding in general, limiting our analyses. Of note, similar to most studies evaluating antibody binding in rheumatoid arthritis, for CCP+RF+ subjects, we cannot determine how much detected binding is direct binding of Ig to citrulline-containing peptides versus indirect binding of RF to IgG bound to citrulline-containing peptides.
Figure 1. Broad patterns of antibody reactivity to peptides in rheumatoid arthritis.
IgG and IgM binding was measured for all samples for each peptide and the z score for each peptide calculated for each rheumatoid arthritis group (CCP+RF+, CCP+RF-, CCP-RF+, and CCP-RF-) compared to control. The proportion (y axis) of z scores less than the specific z value on the x axis was graphed as an empirical cumulative distribution function (ecdf) plot. The median z score for each rheumatoid arthritis group is where its ecdf curve crosses the dotted line at 0.5. Statistical significance was assessed by permutation analysis. A. Citrulline-containing peptides and citrulline-specific binding (binding values for citrulline-containing peptides minus corresponding arginine-containing peptides), n=35,459 peptides or peptide pairs. B. Homocitrulline-containing peptides and homocitrulline-specific binding (binding values for homocitrulline-containing minus corresponding lysine-containing peptides), n=41,608 peptides or peptide pairs. C. All native peptides and native peptides excluding peptides with arginine or lysine, n=95,761 native peptides, 18,694 native peptides without arginine or lysine. For all panels, n=12 samples per group, p<0.0001 for all comparisons despite some small visual differences, except citrulline-specific IgM binding for CCP-RF+ versus control, which was not significant.
We next evaluated reactivity to homocitrulline-containing peptides (Figure 1B). CCP+RF+ subjects had increased IgG binding to homocitrulline-containing peptides with small increases in binding for the other three groups. For IgM binding, the CCP+RF+ group had an increase in binding with a small increase in the CCP-RF+ group. Overall, there was low homocitrulline-specific IgM and IgG binding in all rheumatoid arthritis groups (i.e. binding for homocitrulline-containing peptides minus corresponding lysine-containing peptides).
Finally, we assessed binding to native peptides (Figure 1C). Moderate binding of IgG to native peptides in the CCP+RF+ group was observed as well as modest IgM binding to native peptides in both RF+ groups. To reduce the possibility of cross-reactivity with citrulline- and homocitrulline-containing peptides, antibody binding to native peptides that did not contain arginine or lysine was assessed. Again, we observed moderate IgG binding to native peptides only in the CCP+RF+ group and modest IgM binding in both RF+ groups.
Together, these data suggest that citrulline is a major driver of autoantibody reactivity in our doubly seropositive rheumatoid arthritis subjects, as expected, with little specific reactivity to homocitrulline and moderate binding to native peptides. Moreover, seronegative subjects had little antibody binding in general with intermediate binding for single seropositive rheumatoid arthritis, primarily in the case of citrulline.
A dominant target of IgG in rheumatoid arthritis is citrulline, irrespective of protein identity, particularly if the citrulline is adjacent to glycine or serine.
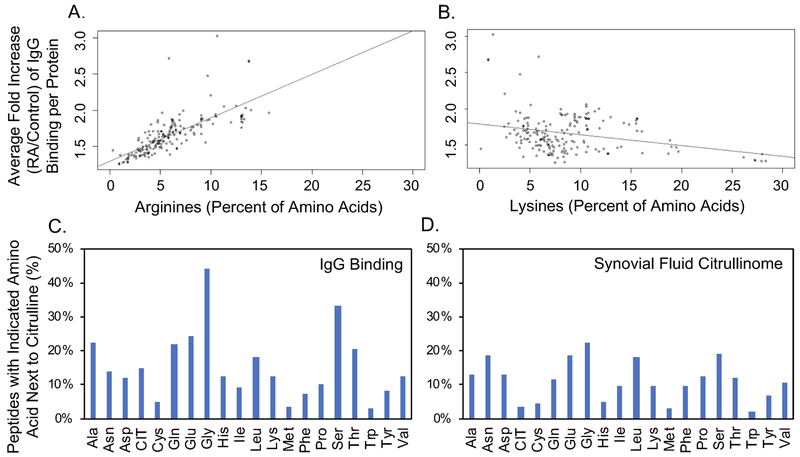
Next, we evaluated if peptides from specific proteins were preferentially targeted in rheumatoid arthritis. We identified peptides bound by IgG at a level 10X greater in rheumatoid arthritis than controls (Supplementary Table 3) and found that every protein whose peptides were included on the array had at least one peptide that met this criterion. Moreover, every peptide with >10x increased binding in rheumatoid arthritis (9098 unique peptides) contained citrulline except 2 native peptides from fibrinogen, which lacked arginine. Some proteins had more peptides highly bound by IgG than others. Given the high reactivity to citrulline overall in our subjects (Figure 1), we determined if the amount of arginine in a protein (citrullines on our array) correlated with the level of IgG binding for its peptides. We determined the fold increase in IgG binding for each peptide for rheumatoid arthritis over controls. We then averaged the fold increase in IgG binding for all peptides in each protein and compared this average to the percent of amino acids in each protein encoded as arginine. There was a strong correlation between the proportion of encoded arginines and binding of serum IgG in rheumatoid arthritis (Figure 2A). In contrast, there was no positive correlation between the proportion of encoded lysines and binding of IgG in rheumatoid arthritis (Figure 2B). Together, these data suggest that a major determinant of IgG binding in rheumatoid arthritis is citrulline, not protein identity, similar to findings for monoclonal antibodies derived from patients with rheumatoid arthritis (7).
Figure 2. Amino acids that correlate with increased IgG binding in rheumatoid arthritis.
A. The average fold increase in IgG binding for rheumatoid arthritis (RA) over control sera was determined for each peptide (including native, citrulline-containing, and homocitrulline-containing peptides) and then was averaged for each protein to generate the average fold increase of IgG binding per protein. The percent of amino acids in each protein encoded as arginine was then compared to the average fold increase of IgG binding per protein. Pearson correlation 0.74, p<0.0001. B. Average fold increase of IgG binding per protein was compared to the percent of amino acids in each protein encoded as lysine. C. Percent of peptides that contain each indicated amino acid next to citrulline for all citrulline-containing peptides for which there is >10x more IgG binding in RA compared to control. D. Percent of peptides that contain each indicated amino acid next to citrulline for citrulline-containing peptides reported in the rheumatoid joint.
We then determined if there was a motif related to high IgG binding in our rheumatoid arthritis sera. Using MEME software (31) and the above highly bound citrulline-containing peptides, no strong motifs could be identified. Rather, certain amino acids appeared frequently in multiple, minimal motifs. Thus, we determined how often each amino acid was adjacent to citrulline in the highly bound peptides. Some amino acids, most strikingly glycine and serine, were frequently next to citrulline in highly bound peptides (Figure 2C), suggesting either a preference for antibody development against citrullines next to these amino acids or preferred citrullination of arginines next to these amino acids. Of note, these amino acids are not simply more commonly encoded next to arginine, since the lowest bound citrulline-containing peptides by IgG did not have a high frequency of glycine or serine next to citrulline.
To differentiate between antibody development and citrullination preference, we evaluated the 451 citrulline-containing peptides detected in rheumatoid synovial fluid (24–26) and determined the percent of peptides that contained each amino acid next to citrulline. The citrullinome peptides differ from the array peptides in that the citrullinome includes all peptides with detected endogenous citrullines whereas the highly bound array peptides are derived from a subset of proteins identified in the citrullinome with all arginines replaced by citrullines. We found that the overall pattern of amino acids frequently next to endogenous citrulline was roughly similar to the pattern of amino acids frequently next to citrulline in highly bound peptides (Figure 2D). Again, both glycine and serine were commonly next to citrulline. Together these data suggest that arginine next to serine or glycine is frequently citrullinated and frequently targeted by IgG in rheumatoid arthritis.
Peptides bound by IgG in rheumatoid arthritis have multiple features of intrinsic structural disorder.
Given the moderate binding of native peptides in our rheumatoid arthritis subjects, we further evaluated these peptides. We identified native peptides bound by IgG >2x more in rheumatoid arthritis as compared to controls, excluding peptides with arginine or lysine to reduce potential effects from cross-reactivity with citrulline- or homocitrulline-containing peptides. As shown in Table 1, a total of 49 peptides met these criteria. Interestingly, several of the proteins from which the peptides were derived are related to complement. Additionally, many of the peptides contained repeated amino acids or repeated short motifs, hallmarks of intrinsically disordered regions of proteins, i.e. regions that lack stable three-dimensional structure. We were intrigued by this observation, since arginine-glycine and arginine-serine, which we determined were minimal motifs for both citrullination and autoantibody binding in rheumatoid arthritis (Figure 2), are found in intrinsically disordered regions of proteins (32, 33). Thus, we determined if bound native peptides were located in disordered regions by identifying regions of predicted disorder in the parent proteins using Protein DisOrder prediction System (PrDOS) (34). 35% of the native peptides bound >2x more in rheumatoid arthritis than controls were predicted to be within disordered regions. In contrast, only 18% of the 49 native peptides with the least binding in rheumatoid arthritis were in disordered regions. We also calculated the median distance from disorder for the 49 highest and lowest bound native peptides resulting in 4 and 32 amino acids, respectively (p=0.0003). Finally, PrDOS predicted that 67% of the 100 highest bound citrulline-containing peptides were in regions of disorder compared to only 38% of the lowest bound citrulline-containing peptides with median distances from disorder of 0 and 12 amino acids, respectively (p<0.0001).
Table 1:
Native peptides (without arginine or lysine) with >2x higher IgG binding in rheumatoid arthritis than control sera
| Protein | Peptides | Disordered/ Total aa (%) | Distance from Disorder (aa) |
|---|---|---|---|
| Apolipoprotein B-100 | IPDFDVDLGTIL, VGINGEANLDFL, GINGEANLDFLN, INGEANLDFLNI, NGEANLDFLNIP | 371/4563 (8) | 23, 8, 9, 10, 11 |
| Ceruloplasmin | SSTVTPTLPGET | 183/1065 (17) | 0 |
| Collagen alpha-2(I) chain | SGGGYDFGYDGD | 471/1366 (34) | 0 |
| Complement C4A | PMPQAPALWIET, EANEDYEDYEYD, QLNDFLQEYGTQ, LNDFLQEYGTQG | 219/1744 (13) | 1, 0, 0, 0 |
| Complement C5 | DLGCGAGGGLNN, LGCGAGGGLNNA, GCGAGGGLNNAN, CGAGGGLNNANV | 217/1676 (13) | 0, 0, 0, 0 |
| Complement C7 | INNDFNYEFYNS, NNDFNYEFYNST, INNNPEFLQLAE | 88/843 (10) | 3, 2, 16 |
| Complement C8 alpha chain | DAQSVYDASYYG, GISSEFYDNAND, ISSEFYDNANDL, SSEFYDNANDLL | 115/584 (20) | 24, 3, 2, 1 |
| Complement factor B | ETIEGVDAEDGH | 24/764 (3) | 0 |
| Coronin-1A | YPPTAGPDPALT | 52/461 (11) | 22 |
| Fibrinogen gamma chain | CEIDGSGNGWTV, GDAFDGFDFGDD, DAFDGFDFGDDP, AFDGFDFGDDPS, FDGFDFGDDPSD | 63/453 (14) | 67, 98, 97, 96, 95 |
| Fibronectin | TSLSAQWTPPNV | 487/2386 (20) | 15 |
| High mobility group protein B1 | EDEEDEEDEEEE, EDEEDEEEEEDE, EDEEEEEDEEDE, EDEEDEDEEEDD | 74/215 (34) | 0, 0, 0, 0 |
| High mobility group protein B2 | EDEDEEEEDEDE | 70/209 (33) | 0 |
| Histone H2A | GGVLPNIHPELL | 87/372 (23) | 4 |
| IgM heavy chain | SILTVSEEEWNT | 54/453 (12) | 8 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | APPATSNPDPAV | 268/930 (29) | 0 |
| Myeloid cell nuclear differentiation antigen | SSVSDFNQNFEV | 143/407 (35) | 52 |
| Plasminogen | QGEPLDDYVNTQ | 35/810 (4) | 13 |
| Tenascin | VEWDPLDIAFET, LDGPSGLVTANI | 486/2201 (22) | 9, 2 |
| Vinculin | DDYEPELLLMPS | 294/1134 (26) | 0 |
| Zinc-alpha-2-glycoprotein | DIVEYYNDSNGS, VVVAVPPQDTAP, VVAVPPQDTAPY, VAVPPQDTAPYS, AVPPQDTAPYSC, VPPQDTAPYSCH | 17/298 (6) | 36, 16, 15, 14 13, 12 |
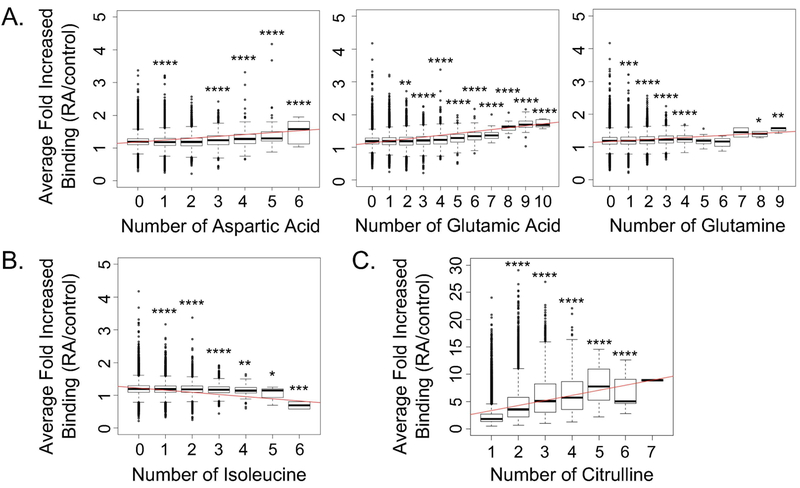
Intrinsically disordered regions also tend to contain arginine, glycine, lysine, glutamine, serine, glutamic acid, aspartic acid, and proline (32, 35). To evaluate if specific amino acids in peptides correlated with IgG binding in rheumatoid arthritis, we quantified the average binding of IgG in rheumatoid arthritis over controls for native peptides with different amounts of each amino acid. We found that with increased representation of most amino acids in peptides, there was no real change in IgG binding in rheumatoid arthritis. However, for aspartic acid, glutamic acid, and glutamine, there was higher IgG binding to peptides in rheumatoid arthritis with increasing numbers of these amino acids (Figure 3A). Smaller increases in IgG binding in rheumatoid arthritis were seen with increasing numbers of glycine or proline (data not shown). Interestingly, increasing representation of isoleucine, which is associated with ordered protein regions (35), was associated with reduced IgG binding in rheumatoid arthritis (Figure 3B). We performed a similar analysis with citrulline-containing peptides. We found a dramatic increase in IgG binding in rheumatoid arthritis as compared to controls with increasing citrulline content (Figure 3C), which vastly overshadowed the effects of any other amino acid.
Figure 3: IgG in rheumatoid arthritis targets peptides with amino acids associated with intrinsic disorder.
Peptides were grouped by the number of specific amino acids present and the average binding for all rheumatoid arthritis subjects divided by controls was plotted for each peptide group. Results are shown for amino acids in native peptides (n=95,761) that demonstrated a real increase (A) or reduction (B) in IgG binding or (C) a real increase in IgG binding for citrulline-containing peptides (n=35,459) in rheumatoid arthritis as compared to controls with increasing number of that amino acid. The difference in binding was evaluated by a t test for each peptide group compared to peptides in which that amino acid was not present (i.e. the 0 group) or, in the case of citrulline-containing peptides, compared to a single citrulline present (i.e. the 1 group). For all panels: trend line in red, n=48 rheumatoid arthritis subjects and 12 controls, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Together, these data suggest that peptides in regions of intrinsic structural disorder are targeted in rheumatoid arthritis.
In CCP+RF+ rheumatoid arthritis, IgG binds citrulline-containing peptides derived from IgG, suggesting epitope overlap between RF and ACPAs.
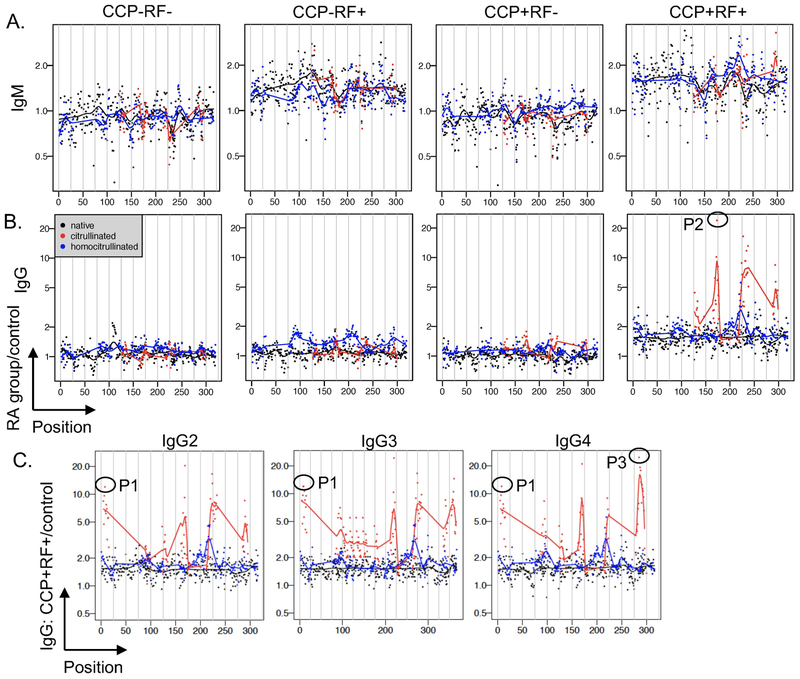
One of the most well-known antigens in rheumatoid arthritis is the Fc portion of IgG, the target of RF. Native peptides derived from IgG heavy chain were not identified as bound by IgG 2x more in rheumatoid arthritis than controls (Table 1), potentially because known RF epitopes are discontinuous (36) and not included in our array or because bound epitopes contain arginine or lysine, which were excluded from the analysis. Therefore, we examined the recognition of peptides derived from IgG more closely. RF+ subjects had modest IgM binding to all peptide types derived from the constant region of IgG1 heavy chain (Figure 4A), with similar binding to peptides derived from IgG2, IgG3, and IgG4 (data not shown). IgG binding in CCP-RF+ rheumatoid arthritis subjects compared to controls was modestly greater for homocitrulline-containing than citrulline-containing or native peptides derived from IgG1 (p<0.0001, Figure 4B), similar to IgG2, IgG3, and IgG4 (data not shown). More dramatically, the CCP+RF+ group demonstrated very high IgG binding predominantly to citrulline-containing peptides in the constant region of IgG1 (Figure 4B) with similar results for peptides derived from IgG2, IgG3, and IgG4 (Figure 4C).
Figure 4. IgM binds IgG-derived peptides at a low level and IgG binds primarily citrulline-containing IgG peptides at a high level in rheumatoid arthritis.
Binding of IgM (A) and IgG (B) for rheumatoid arthritis (RA) subjects divided by controls is graphed for each peptide according to its position in the constant region of IgG1 for CCP-RF-, CCP-RF+, CCP+RF-, and CCP+RF+ subjects (n=12 subjects per group). C. IgG binding for CCP+RF+ RA subjects divided by controls is graphed for each peptide of the constant regions of IgG2, IgG3, and IgG4 (n=12). Peptides (native and citrulline-containing versions) circled in (B) and (C) were evaluated by ELISA in Figure 5.
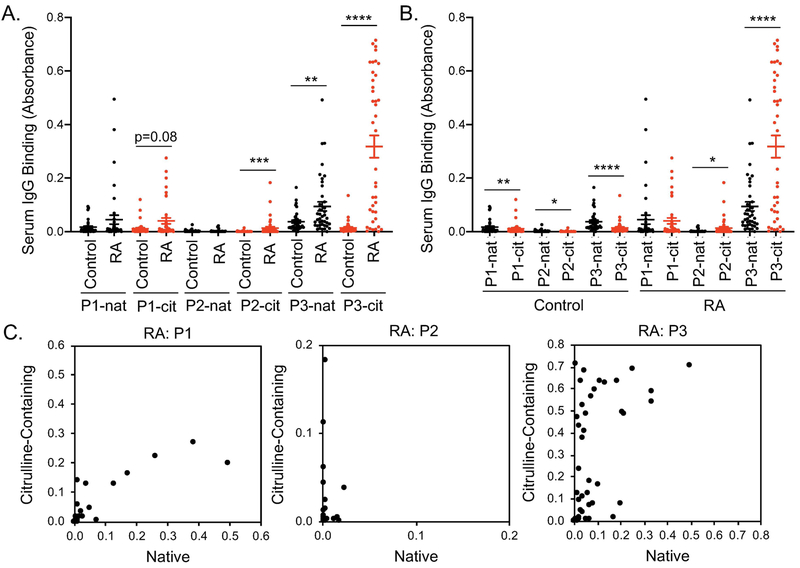
We then selected 3 highly bound IgG-derived peptides with strong sequence homology across IgG isoforms and quantified IgG binding to those peptides by ELISA using a larger number of subjects. For one native and two citrulline-containing peptides, CCP+RF+ sera had increased IgG binding compared to controls (Figure 5A). For the one native peptide with increased IgG binding in CCP+RF+ sera, binding to the citrulline-containing peptide was >3x greater than the native version in rheumatoid arthritis (Figure 5B). Additionally, for individual rheumatoid arthritis subjects, IgG binding to two of the three IgG-derived peptides appeared to favor citrulline (Figure 5C). Interestingly, although there was overall low binding to IgG-derived peptides in controls, controls had slightly higher binding to native than citrulline-containing peptides (Figure 5B). Together, these data suggest that IgG can target citrulline-containing peptides of IgG in rheumatoid arthritis, identifying epitopes that could be bound by an antibody defined as an ACPA or a RF.
Figure 5. IgG binds to citrulline-containing peptides of IgG in rheumatoid arthritis.
Peptides (native and citrulline-containing versions) circled in Figure 4 were used in ELISA to detect IgG binding in CCP+RF+ and control sera (n=40). Results are compared for control versus rheumatoid arthritis (RA) using a Mann-Whitney test (A) and native (nat) versus citrulline-containing (cit) peptides by Wilcoxon matched-pairs signed rank test (B). C. For each subject, IgG binding is graphed for the citrulline-containing peptide (y axis) versus native peptide (x axis). For all panels, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
DISCUSSION
Using high density peptide array and 4 serogroups of rheumatoid arthritis, we revealed interesting features of rheumatoid arthritis autoantibody reactivity. First, and not surprisingly, we found that citrulline was a dominant autoantibody target in the polyclonal repertoire of rheumatoid arthritis. The highest level of IgG binding in rheumatoid arthritis was to citrulline-containing peptides and the encoded arginine content (citrullines on our array) directly correlated with average peptide IgG binding for each protein. Also, at least one citrulline-containing peptide from every protein on the array was bound by IgG. These findings are consistent with a similar lack of protein specificity reported for monoclonal autoantibodies in rheumatoid arthritis using a different set of peptide antigens (7) as well as observations that ACPAs are cross-reactive (5–8) and recognize citrulline largely independently of surrounding peptides (37). Thus, our data support the theory that citrulline itself is a major driver of IgG reactivity in rheumatoid arthritis.
Our evaluation of homocitrulline reactivity revealed primarily nonspecific homocitrulline binding and none of the peptides most highly bound by IgG contained homocitrulline. These results suggest that homocitrulline may not be targeted to as great an extent as citrulline, consistent with the finding that AHCPAs have lower avidity than ACPAs (38). However, any homocitrulline-specific binding dependent upon a nearby citrulline or other modified amino acid, would not have been detected, since our peptides contained either citrulline or homocitrulline, but not both. Additionally, if homocitrulline-containing epitopes are primarily discontinuous, they would not be detected using our array of linear peptides. However, homocitrulline-containing peptides were bound by rheumatoid monoclonal antibodies using a similar array (7). Also, homocitrullination increases structural disorder (39) and thus increases the likelihood of linear epitopes. Importantly, homocitrulline reactivity may change over time in subjects (7), causing variability among studies. Future experiments with additional subjects, modifications, and time points may clarify this issue.
We also observed moderate IgG binding to native peptides in CCP+RF+ subjects. Unlike IgM binding, which was increased against native peptides in both RF+ groups, moderate IgG binding of native peptides was absent in CCP-RF+ subjects and thus unlikely due to promiscuity of RF. Further, the IgG binding was unlikely due to cross-reactivity against citrulline- and homocitrulline-containing peptides given a similar increase in IgG binding in CCP+RF+ subjects when peptides with arginine or lysine were excluded. IgG binding to native peptides could be due to an unknown endogenous post-translational modification resulting in cross-reactivity with native peptides on the array. For example, increased glutamic acid, which can mimic phosphoserine, was associated with increased IgG binding to native peptides on our array (Figure 3). If rheumatoid autoantibodies target phosphoserine in vivo, we might have simply detected cross-reactivity with glutamic acid with our array peptides. Alternatively, autoantibodies in rheumatoid arthritis may commonly target a host of native antigens as part of a polyclonal response and/or polyreactive autoantibodies.
Upon further characterization of citrulline reactivity in our experiments, we detected a preference for glycine or serine next to citrulline in highly-bound peptides as well as next to endogenous citrullines in the rheumatoid joint, suggesting that these amino acid pairs may be commonly targeted by autoantibodies due to their common presence in antigens. There were some differences between the patterns of amino acids next to citrulline in the citrullinome versus highly bound peptides, which could reflect differences in the efficacy of MHC, B cell or T cell receptor binding, other factors in generating an immune response, or simply sample size. However, our findings related to glycine are consistent with the work of others. The presence of glycine next to citrulline has been observed in the citrullinome of 30 human tissues (40), motifs targeted by the citrullinating peptidylarginine deiminases 2 and 4 (41–43), and autoantibody binding in rheumatoid arthritis (7, 44, 45). The preference for citrullination of or IgG binding to peptides with citrulline next to serine has not been previously described. Perhaps unique features of our subject groups, analyses, or array allowed this novel finding. Interestingly, arginine-serine and arginine-glycine-glycine repeats are commonly found in disordered regions of proteins (32, 33). Moreover, disordered regions of proteins are citrullinated more than ordered regions and citrullination increases disorder (39). Taken together, these observations suggest that disorder, citrullination, and the highest affinity autoantibodies in rheumatoid arthritis are profoundly linked.
We provide additional evidence that autoantibodies in rheumatoid arthritis target regions of intrinsic disorder. Native and citrulline-containing peptides bound by IgG in rheumatoid arthritis were more likely to be in regions of disorder than poorly bound peptides. Moreover, increased content of amino acids associated with intrinsic disorder (32, 35) correlated with increased IgG binding in rheumatoid arthritis. Similarly, in lupus, about a third of characteristic autoantigens contain stretches of charged amino acids (46), common in disordered regions, and nuclear autoantigens are often disordered proteins (47). Further, although many examples of structured epitopes exist, epitopes predicted to be within regions of disorder were slightly more likely to be recognized by antibodies compared to epitopes in ordered regions in a wide range of normal and pathological conditions (48, 49). Our experiments are unable to directly compare binding of disordered versus ordered regions, since ordered epitopes are often discontinuous and, thus, not present on our array. However, we provide a body of evidence that autoantibodies in rheumatoid arthritis bind intrinsically disordered regions, which are frequently citrullinated (39), a concept with potential importance for the underpinnings of rheumatoid autoantibody development. Of note, it has been hypothesized that repetition and charge lead to autoantibody reactivity (50). However, in rheumatoid arthritis, the charge may not be the driving factor since charge is reduced with citrullination. Perhaps disorder is the main driver due to the high degree of accessibility of intrinsically disordered regions of proteins (51) or due to bound DNA or RNA activating toll-like receptors.
Since the binding of citrulline-containing and native peptides was predominantly seen in CCP+RF+ rheumatoid arthritis, many of our findings related to targeting disordered regions may predominantly apply to this group. However, our subject selection also allowed an evaluation of antibody binding in seronegative and single seropositive rheumatoid arthritis. Seronegative subjects had little binding to citrulline-containing or native peptides despite evaluating 172,828 peptides, suggesting minimal autoantibody presence. Interestingly, IgG binding in CCP−RF− subjects to homocitrulline-containing peptides was greater than controls, suggesting that homocitrulline could underlie an improved diagnostic test for seronegative rheumatoid arthritis. However, despite using multiple models, we have not yet identified any peptides predictive of a diagnosis of rheumatoid arthritis in seronegative subjects. At least a subset of seronegative rheumatoid arthritis may be truly seronegative, lacking autoantibodies in general. An absence of autoantibodies in seronegative disease might suggest a different disease pathophysiology than seropositive disease given evidence that RF and ACPAs are pathologic (1, 2). Alternatively, rheumatoid arthritis may exist on a continuum. In support of this idea, levels of citrulline-specific IgG in CCP+RF− and CCP−RF+ disease fall between levels in CCP+RF+ and CCP−RF− disease. Future studies with even larger arrays that are not limited by protein selection bias or by linear epitopes, or perhaps arrays incorporating additional post-translational modifications, combinations of post-translational modifications, or full-length proteins may resolve these issues.
Our final observation is that IgG from CCP+RF+ sera targets citrulline-containing peptides of IgG. This finding raises the possibility that IgG-RF (IgG that binds IgG) could also be an ACPA, making citrullinated IgG, which has been reported in humans (24, 26, 52), a linchpin connecting ACPAs and RF in rheumatoid arthritis. IgM did not preferentially target citrulline-containing peptides of IgG. IgM-RF, which also occurs in healthy individuals, may rise independently of citrulline due to smoking or pathogens, whereas the development of IgG-RF, which requires a break in tolerance and T cell help, may be driven by targeting citrulline like other ACPAs (53). However, it is not known if citrullinated IgG was the original antigen against which the IgG that we detected developed, or if the observed targeting of citrulline-containing IgG peptides is due to cross-reactivity of ACPAs. Also, not all IgG that bound IgG-derived peptides targeted citrulline (Figure 5), potentially pointing to different mechanisms for the break in tolerance for generating IgG-RF (53). Finally, despite having a unique name for its autoantibody, IgG may be similar to other proteins, with native epitopes targeted, but less so than citrulline-containing epitopes in rheumatoid arthritis. It will be important for future work to clearly define the reactivity of RF in rheumatoid arthritis and diseases with RF that do not have a citrulline bias in order to shed light on disease pathophysiology as well as to fully understand and optimize RF-based diagnostic tests.
In summary, using the four serogroups of rheumatoid arthritis and high density peptide array, we demonstrated very strong autoantibody targeting of citrulline, particularly if adjacent to glycine or serine, as well as a moderate binding to native peptides primarily in CCP+RF+ rheumatoid arthritis and very little antibody binding in seronegative disease. Further, we revealed disorder as a feature of IgG-targeted peptides and epitope overlap between ACPAs and RF.
Supplementary Material
Funding:
Funding was provided by the University of Wisconsin School of Medicine and Public Health from the Wisconsin Partnership Program and the Doris Duke Charitable Foundation (2015099) to MAS and the University of Wisconsin ‐ Madison Office of the Chancellor and the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation to IMO. MAS was additionally supported by NIH/NIAMS K08AR065500 and CDMRP PR170847, CLH by NIH/NHLBI T32HL07899, IMO by NIH/NCI P30CA14520 (UW Carbone Comprehensive Cancer Center’s Cancer Informatics Shared Resource), MAN by NIH/NIAID U54AI117924.
Footnotes
Conflict of Interest: After experiment completion, but before manuscript submission, MB became employed by Invenra Inc. No other authors have conflicts of interest to report.
REFERENCES
- 1.Engdahl C, Bang H, Dietel K, Lang SC, Harre U, Schett G. Periarticular Bone Loss in Arthritis Is Induced by Autoantibodies Against Citrullinated Vimentin. J Bone Miner Res. 2017;32(8):1681–91. [DOI] [PubMed] [Google Scholar]
- 2.Mathsson L, Lampa J, Mullazehi M, Ronnelid J. Immune complexes from rheumatoid arthritis synovial fluid induce FcgammaRIIa dependent and rheumatoid factor correlated production of tumour necrosis factor-alpha by peripheral blood mononuclear cells. Arthritis Res Ther. 2006;8(3):R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uysal H, Bockermann R, Nandakumar KS, Sehnert B, Bajtner E, Engstrom A, et al. Structure and pathogenicity of antibodies specific for citrullinated collagen type II in experimental arthritis. J Exp Med. 2009;206(2):449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–81. [DOI] [PubMed] [Google Scholar]
- 5.Ioan-Facsinay A, el-Bannoudi H, Scherer HU, van der Woude D, Menard HA, Lora M, et al. Anti-cyclic citrullinated peptide antibodies are a collection of anti-citrullinated protein antibodies and contain overlapping and non-overlapping reactivities. Ann Rheum Dis. 2011;70(1):188–93. [DOI] [PubMed] [Google Scholar]
- 6.Titcombe PJ, Wigerblad G, Sippl N, Zhang N, Shmagel AK, Sahlstrom P, et al. Pathogenic Citrulline-Multispecific B Cell Receptor Clades in Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70(12):1933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steen J, Forsstrom B, Sahlstrom P, Odowd V, Israelsson L, Krishnamurthy A, et al. Recognition of Amino Acid Motifs, Rather Than Specific Proteins, by Human Plasma Cell-Derived Monoclonal Antibodies to Posttranslationally Modified Proteins in Rheumatoid Arthritis. Arthritis Rheumatol. 2019;71(2):196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang B, Ge C, Lonnblom E, Lin X, Feng H, Xiao L, et al. The autoantibody response to cyclic citrullinated collagen type II peptides in rheumatoid arthritis. Rheumatology (Oxford). 2019. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Knevel R, Suwannalai P, van der Linden MP, Janssen GM, van Veelen PA, et al. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc Natl Acad Sci U S A. 2011;108(42):17372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scinocca M, Bell DA, Racape M, Joseph R, Shaw G, McCormick JK, et al. Antihomocitrullinated fibrinogen antibodies are specific to rheumatoid arthritis and frequently bind citrullinated proteins/peptides. J Rheumatol. 2014;41(2):270–9. [DOI] [PubMed] [Google Scholar]
- 11.Reed E, Jiang X, Kharlamova N, Ytterberg AJ, Catrina AI, Israelsson L, et al. Antibodies to carbamylated alpha-enolase epitopes in rheumatoid arthritis also bind citrullinated epitopes and are largely indistinct from anti-citrullinated protein antibodies. Arthritis Res Ther. 2016;18(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turunen S, Hannonen P, Koivula MK, Risteli L, Risteli J. Separate and overlapping specificities in rheumatoid arthritis antibodies binding to citrulline- and homocitrulline-containing peptides related to type I and II collagen telopeptides. Arthritis Res Ther. 2015;17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiele GM, Duryee MJ, Anderson DR, Klassen LW, Mohring SM, Young KA, et al. Malondialdehyde-acetaldehyde adducts and anti-malondialdehyde-acetaldehyde antibodies in rheumatoid arthritis. Arthritis Rheumatol. 2015;67(3):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juarez M, Bang H, Hammar F, Reimer U, Dyke B, Sahbudin I, et al. Identification of novel antiacetylated vimentin antibodies in patients with early inflammatory arthritis. Ann Rheum Dis. 2016;75(6):1099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darrah E, Andrade F. Editorial: citrullination, and carbamylation, and malondialdehyde-acetaldehyde! Oh my! Entering the forest of autoantigen modifications in rheumatoid arthritis. Arthritis Rheumatol. 2015;67(3):604–8. [DOI] [PubMed] [Google Scholar]
- 16.Hueber W, Kidd BA, Tomooka BH, Lee BJ, Bruce B, Fries JF, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52(9):2645–55. [DOI] [PubMed] [Google Scholar]
- 17.Konig MF, Giles JT, Nigrovic PA, Andrade F. Antibodies to native and citrullinated RA33 (hnRNP A2/B1) challenge citrullination as the inciting principle underlying loss of tolerance in rheumatoid arthritis. Ann Rheum Dis. 2016;75(11):2022–8. [DOI] [PubMed] [Google Scholar]
- 18.Demoruelle MK, Bowers E, Lahey LJ, Sokolove J, Purmalek M, Seto NL, et al. Antibody Responses to Citrullinated and Noncitrullinated Antigens in the Sputum of Subjects With Rheumatoid Arthritis and Subjects at Risk for Development of Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70(4):516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barra L, Pope JE, Orav JE, Boire G, Haraoui B, Hitchon C, et al. Prognosis of seronegative patients in a large prospective cohort of patients with early inflammatory arthritis. J Rheumatol. 2014;41(12):2361–9. [DOI] [PubMed] [Google Scholar]
- 20.Rebernick R, Fahmy L, Glover C, Bawadekar M, Shim D, Holmes CL, et al. DNA Area and NETosis Analysis (DANA): a High-Throughput Method to Quantify Neutrophil Extracellular Traps in Fluorescent Microscope Images. Biol Proced Online. 2018;20:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes CL, Peyton CG, Bier AM, Donlon TZ, Osman F, Bartels CM, et al. Reduced IgG titers against pertussis in rheumatoid arthritis: Evidence for a citrulline-biased immune response and medication effects. PLoS One. 2019;14(5):e0217221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz JN, Barrett J, Liang MH, Bacon AM, Kaplan H, Kieval RI, et al. Sensitivity and positive predictive value of Medicare Part B physician claims for rheumatologic diagnoses and procedures. Arthritis Rheum. 1997;40(9):1594–600. [DOI] [PubMed] [Google Scholar]
- 23.Bailey AL, Buechler CR, Matson DR, Peterson EJ, Brunner KG, Mohns MS, et al. Pegivirus avoids immune recognition but does not attenuate acute-phase disease in a macaque model of HIV infection. PLoS Pathog. 2017;13(10):e1006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Chen FF, Gao WB, Wang HY, Zhao NW, Xu M, et al. Identification of citrullinated peptides in the synovial fluid of patients with rheumatoid arthritis using LC-MALDI-TOF/TOF. Clin Rheumatol. 2016;35(9):2185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Bozic B, Pruijn GJ. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum. 2013;65(1):69–80. [DOI] [PubMed] [Google Scholar]
- 26.Tutturen AE, Fleckenstein B, de Souza GA. Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res. 2014;13(6):2867–73. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira L, Sebbag M, Vincent C, Arnaud M, Fournie B, Cantagrel A, et al. Performance of two ELISAs for antifilaggrin autoantibodies, using either affinity purified or deiminated recombinant human filaggrin, in the diagnosis of rheumatoid arthritis. Ann Rheum Dis. 2001;60(9):882–7. [PMC free article] [PubMed] [Google Scholar]
- 28.Team RC. R: A language and environment for statistical computing. . 2017. [cited; Available from: https://www.R-project.org/
- 29.Bartlett MS. The use of transformations. . Biometrics. 1947;3(1):39–52. [PubMed] [Google Scholar]
- 30.Székely GJR ML; Bakirov NK Measuring and testing dependence by correlation of distances. The Annals of Statistics. 2007:2769–94. [Google Scholar]
- 31.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 32.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–406. [DOI] [PubMed] [Google Scholar]
- 33.Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34(1):305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35(Web Server issue):W460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uversky VN. The alphabet of intrinsic disorder: II. Various roles of glutamic acid in ordered and intrinsically disordered proteins. Intrinsically Disord Proteins. 2013;1(1):e24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artandi SE, Calame KL, Morrison SL, Bonagura VR. Monoclonal IgM rheumatoid factors bind IgG at a discontinuous epitope comprised of amino acid loops from heavy-chain constant-region domains 2 and 3. Proc Natl Acad Sci U S A. 1992;89(1):94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge C, Xu B, Liang B, Lonnblom E, Lundstrom SL, Zubarev RA, et al. Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies. Arthritis Rheumatol. 2019;71(2):210–21. [DOI] [PubMed] [Google Scholar]
- 38.van Delft MAM, Verheul MK, Burgers LE, Rantapaa-Dahlqvist S, van der Helm-van Mil AHM, Huizinga TWJ, et al. The anti-carbamylated protein antibody response is of overall low avidity despite extensive isotype switching. Rheumatology (Oxford). 2018;57(9):1583–91. [DOI] [PubMed] [Google Scholar]
- 39.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996;271(48):30709–16. [DOI] [PubMed] [Google Scholar]
- 40.Lee CY, Wang D, Wilhelm M, Zolg DP, Schmidt T, Schnatbaum K, et al. Mining the Human Tissue Proteome for Protein Citrullination. Mol Cell Proteomics. 2018;17(7):1378–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haag S, Schneider N, Mason DE, Tuncel J, Andersson IE, Peters EC, et al. Identification of new citrulline-specific autoantibodies, which bind to human arthritic cartilage, by mass spectrometric analysis of citrullinated type II collagen. Arthritis Rheumatol. 2014;66(6):1440–9. [DOI] [PubMed] [Google Scholar]
- 42.Tanikawa C, Ueda K, Suzuki A, Iida A, Nakamura R, Atsuta N, et al. Citrullination of RGG Motifs in FET Proteins by PAD4 Regulates Protein Aggregation and ALS Susceptibility. Cell Rep. 2018;22(6):1473–83. [DOI] [PubMed] [Google Scholar]
- 43.Assohou-Luty C, Raijmakers R, Benckhuijsen WE, Stammen-Vogelzangs J, de Ru A, van Veelen PA, et al. The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochim Biophys Acta. 2014;1844(4):829–36. [DOI] [PubMed] [Google Scholar]
- 44.Burkhardt H, Koller T, Engstrom A, Nandakumar KS, Turnay J, Kraetsch HG, et al. Epitope-specific recognition of type II collagen by rheumatoid arthritis antibodies is shared with recognition by antibodies that are arthritogenic in collagen-induced arthritis in the mouse. Arthritis Rheum. 2002;46(9):2339–48. [DOI] [PubMed] [Google Scholar]
- 45.Szarka E, Aradi P, Huber K, Pozsgay J, Vegh L, Magyar A, et al. Affinity Purification and Comparative Biosensor Analysis of Citrulline-Peptide-Specific Antibodies in Rheumatoid Arthritis. Int J Mol Sci. 2018;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brendel V, Dohlman J, Blaisdell BE, Karlin S. Very long charge runs in systemic lupus erythematosus-associated autoantigens. Proc Natl Acad Sci U S A. 1991;88(4):1536–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carl PL, Temple BR, Cohen PL. Most nuclear systemic autoantigens are extremely disordered proteins: implications for the etiology of systemic autoimmunity. Arthritis Res Ther. 2005;7(6):R1360–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacRaild CA, Richards JS, Anders RF, Norton RS. Antibody Recognition of Disordered Antigens. Structure. 2016;24(1):148–57. [DOI] [PubMed] [Google Scholar]
- 49.Guy AJ, Irani V, MacRaild CA, Anders RF, Norton RS, Beeson JG, et al. Insights into the Immunological Properties of Intrinsically Disordered Malaria Proteins Using Proteome Scale Predictions. PLoS One. 2015;10(10):e0141729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plotz PH. The autoantibody repertoire: searching for order. Nat Rev Immunol. 2003;3(1):73–8. [DOI] [PubMed] [Google Scholar]
- 51.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutchinson D, Clarke A, Heesom K, Murphy D, Eggleton P. Carbamylation/citrullination of IgG Fc in bronchiectasis, established RA with bronchiectasis and RA smokers: a potential risk factor for disease. ERJ Open Res. 2017;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelef MA. New relationships for old autoantibodies in rheumatoid arthritis. Arthritis Rheumatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.