Abstract
While sarcomas account for approximately 1% of malignant tumors of adults, they are particularly more common in children and adolescents affected by cancer. In contrast to malignancies that occur in later stages of life, childhood tumors, including sarcoma, are characterized by a striking paucity of somatic mutations. However, entity-defining fusion oncogenes acting as the main oncogenic driver mutations are frequently found in pediatric bone and soft-tissue sarcomas such as Ewing sarcoma (EWSR1-FLI1), alveolar rhabdomyosarcoma (PAX3/7-FOXO1), and synovial sarcoma (SS18-SSX1/2/4). Since strong oncogene-dependency has been demonstrated in these entities, direct pharmacological targeting of these fusion oncogenes has been excessively attempted, thus far, with limited success. Despite apparent challenges, our increasing understanding of the neomorphic features of these fusion oncogenes in conjunction with rapid technological advances will likely enable the development of new strategies to therapeutically exploit these neomorphic features and to ultimately turn the “undruggable” into first-line target structures. In this review, we provide a broad overview of the current literature on targeting neomorphic features of fusion oncogenes found in Ewing sarcoma, alveolar rhabdomyosarcoma, and synovial sarcoma, and give a perspective for future developments.
Graphical abstract.
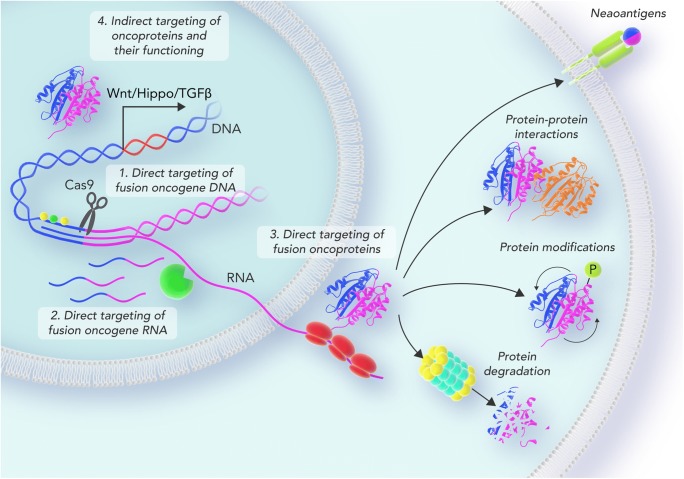
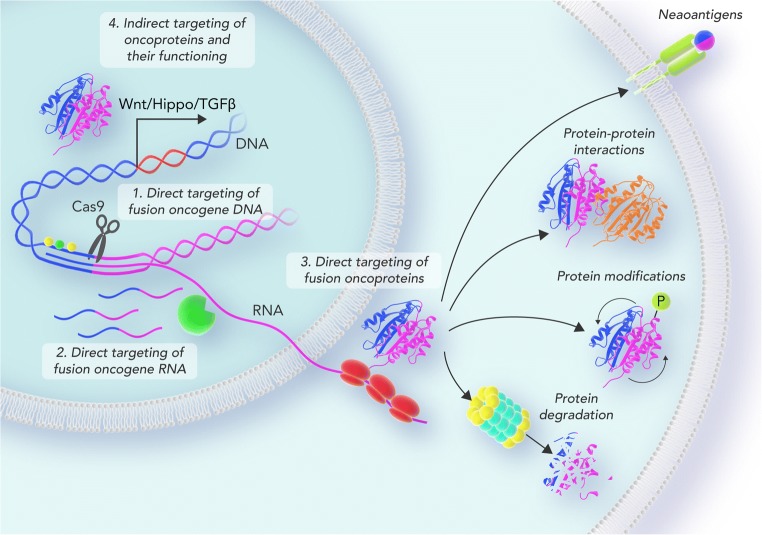
Scheme depicting the different targeting strategies of fusion oncogenes in pediatric fusion-driven sarcomas. Fusion oncogenes can be targeted on their DNA level (1), RNA level (2), protein level (3), and by targeting downstream functions and interaction partners (4).
Keywords: Ewing sarcoma, Synovial sarcoma, Rhabdomyosarcoma, Fusion oncogene, Targeted therapy
Introduction
Decades after the discovery of BCR-ABL1 t(9;22)(q34;q11) in chronic myeloid leukemia [1, 2] and EWSR1-FLI1 t(22;11)(q24;12) in Ewing sarcoma (EwS) [3], more than 20,000 fusion genes have been identified in human malignancies [4]. In cancer entities with high numbers of somatic mutations, the vast majority of gene fusions are supposed passenger mutations, i.e., byproducts of spontaneous genomic rearrangements, which accumulate progressively over time [5, 6]. However, in oligo-mutated childhood cancers, such as EwS, fusion genes are acquired early in tumorigenesis either through balanced chromosomal translocations [3] or through a complex, but well-orchestrated, genomic rearrangement called chromoplexy [7]. Once the respective fusion has occurred, oligo-mutated cancer cells show a strong oncogene addiction toward their disease-defining fusion oncogene (e.g., EWSR1-FLI1 in EwS [3], PAX3/7-FOXO1 in alveolar rhabdomyosarcoma (ARMS) [8, 9] and SS18-SSX1/2/4 in synovial sarcoma (SS) [10–12]) in terms of tumor progression and metastasis.
EwS is the second most common bone cancer in children and was first described by James Ewing in 1921 [13]. It is characterized by a small-round-blue cell phenotype and mostly arises in the metadiaphyseal bones of the lower extremities and in the pelvic region [14]. While EwS is curable in localized disease by radical surgery, radiotherapy, and chemotherapy (5-year survival rate: around 83%), patients with disseminated disease have dismal outcome (5-year survival rate: 37% or less) [15]. Molecularly, EwS is defined by the fusion of the EWSR1 gene and a transcription factor of the ETS-family, such as EWSR1-FLI1 (85%) or EWSR1-ERG (10%) [14]. This fusion event rewires the affinity of the DNA-binding domain of FLI1 and enables it to bind to GGAA-microsatellites (GGAA-mSats) in addition to its physiological binding to the ETS-specific DNA motif ACCGGAAGT. Interestingly, the affinity to those mSats correlates strongly with an increasing number of GGAA-repeats [16]. In turn, binding of EWSR1-FLI1 to such GGAA-mSats converts them into de novo enhancers and super-enhancers [16, 17]. Subsequently, EWSR1-ETS fusion oncoproteins deregulate the gene expression of hundreds of genes, such as MYBL2 [18] and PPP1R1A [19], which contribute to the malignant phenotype of EwS. Notably, germline variants of GGAA-mSats that affect the repeat length have recently been reported to contribute to EwS susceptibility and tumor progression in EwS [18, 20].
Alveolar rhabdomyosarcoma (ARMS) is a subtype of rhabdomyosarcoma that is believed to originate from precursor cells in musculoskeletal tissue or mesenchymal stem cells (MSCs) [21] and affects mostly children and adolescents [22]. Histologically, ARMS resembles the architecture of lung tissue by forming fibrovascular septa segregating the small, often discohesive growing tumor cells in an alveoli-like pattern [22]. Approximately two thirds of ARMS harbor a fusion oncogene, whereas one third is fusion oncogene negative [23]. In fusion-positive ARMS, the FOXO1 transactivation domain is fused to the DNA-binding domain of either PAX3 or PAX7 [8, 9]. In analogy to EwS, the generated fusion oncoprotein binds a unique DNA motif (ACCGTGACTAATTNN for PAX3-FOXO1) and hijacks this sequence as a de novo enhancer [24], which drives the expression of pro-tumorigenic genes. While PAX3- and PAX7-FOXO1-positive ARMS have the same prognosis in their localized states, PAX3-FOXO1-positive tumors show a more aggressive phenotype once they have metastasized [25].
Among fusion oncogene-driven sarcomas, SS is one of the most prevalent ones accounting for approximately 7–8% of all sarcomas [26]. Historically, SS has been believed to originate from synovial cells due to its histopathological appearance and its mostly joint-associated localization [27]. Although the incidence of this tumor entity peaks at the age of 30 years, 30% of the patients are children or adolescents. On a molecular level, SS is characterized by the SS18-SSX1, SSX2, or -SSX4 fusion gene (hereafter collectively referred to as SS18-SSX) [10–12]. These fusion proteins have been found to interfere with normal BRG1-associated factor (BAF) complex (alias SWI/SNF complex) formation by competing with wild-type SS18, thereby causing concomitant loss of BAF47, a tumor suppressive subunit of the BAF complex [28, 29]. Subsequently, the altered BAF complex binds broad polycomb domains, opposes PRC-mediated repression of gene sets, and activates transcription of a SS-specific gene signature [29, 30]. Furthermore, it was shown that SS18-SSX fusion proteins directly interact with the Wnt-associated transcription factor family TCF/LEF and histone deacetylases (HDACs) allowing Wnt-ligand independent Wnt-activation in SS cells [31].
While the respective driver mutations in each of the entities outlined above have long been identified, targeted therapies exploiting (fusion) oncogene addiction in these tumors remain to be integrated into standard of care treatment regimens. Given the young age of disease onset, effective yet gentle alternatives to mutilating surgeries and aggressive radio-chemotherapies are urgently needed. In this article, we will review the current literature on fusion oncogene specific therapeutic targets, evaluate their clinical potential, and propose new strategies targeting oncogene dependence in EwS, ARMS and SS.
Direct targeting of fusion oncogene DNA
CRISPR-CAS9-based strategies targeting the breakpoint of fusion oncogenes and regulatory elements bound by fusion oncoproteins
Gene engineering experiences a renaissance since the development of the clustered regularly interspaced short palindromic repeats (CRISPR) associated protein 9 (CRISPR-CAS9) technology. CRISPR-CAS9 has been successfully employed in basic and preclinical research for the last couple of years and faces its first phase I clinical trial at the moment [32, 33]. Besides monogenic disorders, malignant tumors driven by a single-driver mutation represent favorable future indications for the therapeutic use of CRISPR-CAS9 [34]. Among them, tumors harboring a fusion oncogene constitute excellent candidates for oligonucleotide-based treatment strategies due to the sequence specificity of their breakpoint region. Recently, Chen et al. showed the efficacy of a CRISPR-CAS9-based strategy inserting a suicide gene specifically into the breakpoint region of two fusion oncogenes in prostate cancer [35]. Similar strategies have been employed in fusion oncogene addicted sarcomas as well. Mitra et al. have recently shown subtotal tumor clearance of subcutaneous EwS xenografts in mice treated with CD99-targeting nanoparticles carrying EWSR1-FLI1-specific sgRNA-RNPs [36]. In a human myoblast model, the CRISPR-CAS9 mediated knockout of PAX3-FOXO1 abrogated colony formation in vitro [37]. Another elegant strategy was pursued by Johnson et al., who deleted the GGAA-mSat regulating the expression of NR0B1 using CRISPR-CAS9 and thereby impaired the proliferation and oncogenic transformation of EwS cell lines [19]. Moreover, the advent of CRISPRi/a technology has enabled researchers to silence or activate DNA regions, respectively, in a highly specific way. Boulay et al. have employed this method to directly silence various GGAA-mSats and could abrogate tumor growth in vivo by targeting a SOX2-regulating GGAA-mSat-enhancer using CRISPRi [38]. Taken together, CRISPR-CAS9-based strategies represent novel, highly specific therapeutic approaches to hijack oncogene activity by targeting either the fusion gene itself or its respective DNA-binding motif.
Pharmacologic targeting the epigenetic regulation of fusion oncogene expression
The concept of epigenetic regulation of gene expression and its implications on health and disease, especially cancer, has been widely accepted in the field of basic and translational research [39, 40]. Epigenomic alterations are common in cancer and have become therapeutically accessible by the advent of epigenome-modifying drugs such as HDAC inhibitors [39]. Although the functional principles of the fusion oncogenes in the above-mentioned sarcoma entities have been studied extensively, their transcriptional regulation has not been fully elucidated yet. However, in EwS, a strong dependence on the presence of BRD4 (a member of the bromodomain and extraterminal domain (BET) family) for efficient transcription of EWSR1-FLI1 has been observed [41]. BRD4 is known to bind acetylated lysine residues and promote transcription by interaction with P-TEFb and RNA-polymerase II [42]. Notably, BET inhibition by JQ1 (+) impaired cell viability, clonogenic, and migratory capacity and induced G1-blockage [41]. In ARMS, Entinostat, an HDAC-inhibitor, which has been found to reduce PAX3-FOXO1 expression and induce subsequent chemotherapy sensitization in preclinical models [43, 44], is currently being evaluated in combination with nivolumab in a phase I/II trial (INFORM2) [45] after it was tolerated well in an earlier phase I study (ADVL1513) [46]. As mentioned above, CRISPR-interference, a variant of the CRISPR-CAS9 system that allows the selective silencing of genes and intergenic regions, has been employed to silence GGAA-mSats and thereby disrupt their enhancer capacity in EwS cell lines [38]. This site-specific technology might represent a promising strategy to target fusion oncogenes themselves, not only their binding regions.
Direct targeting of fusion oncogene RNA
Targeting mRNA transcripts of fusion oncoproteins using RNA interference (RNAi)
With the advent of the RNAi technique, knockdown of transcripts including mRNAs of fusion oncogenes has become a standard method to investigate their biological role in cell and animal models. Due to their driver function in oncogenesis, direct targeting of fusion oncogene transcripts has been the Holy Grail of fusion-positive pediatric sarcoma treatment. Indeed, knockdown of fusion oncoproteins in EwS [47, 48], ARMS [49, 50], and SS [51, 52] consistently causes G0/G1 cell cycle arrest and cell death in vitro, which supports the potential therapeutic usefulness of this approach. Furthermore, it has been observed that suppression of the fusion oncogene or direct downstream target genes can facilitate differentiation of fusion-positive sarcoma cells in combination with adequate differentiation-stimulating media in EwS [53], SS [29, 54], and ARMS [55]. Hence, differentiation induction by RNAi and differentiation inducing drugs (such as LSD1 and HDAC inhibitors [56, 57]) could be therapeutically implemented in pediatric sarcoma treatment regimens, as exemplified by treatment of acute promyelocytic leukemia with all-trans-retinoic acid [58].
Conversely, the therapeutic strategy of direct targeting of fusion oncoprotein transcripts may also have some drawbacks. Indeed, EwS cells in which EWSR1-FLI1 expression was suppressed by RNAi showed higher metastatic potential than those without EWSR1-FLI1 knockdown in vitro and in vivo, which undermines the rationale for complete therapeutic suppression of fusion oncoprotein transcripts [59]. This phenomenon, however, has been demonstrated, thus far, only in EwS, whereas in SS and ARMS suppression of the respective fusion oncoprotein or their downstream target genes reduces invasion and migration in vitro [60–62]. Moreover, high expression of SLFN1, one of the EWSR1-FLI1 upregulated genes, has been shown to be associated with higher sensitivity to chemotherapeutics and superior patient survival [63], suggesting the possibility that suppression of EWSR1-FLI1 may lead to the emergence of chemo-resistant phenotype in EwS.
A major obstacle of implementing RNAi technique in the clinic may reside in the difficulty of sufficiently and systemically introducing constructs into tumor cells. Incipient in vivo studies using intratumoral application of siRNA demonstrated certain anti-tumor effect in EwS [64, 65]. However, this approach requires frequent intratumoral injection or prior application of siRNA, which is obviously not feasible in the clinical setting. Development of nanoparticles or liposomes enabled systemic application of siRNA in murine xenograft models with mild anti-tumor effects, specifically promoting cytostaticity rather than cytotoxicity, which may be attributed to unsatisfactory suppression of fusion oncoproteins [66, 67]. A more specific approach with siRNA encapsulated into transferrin-conjugated cyclodextrin-containing polycations was tested in a murine metastatic EwS xenograft model, in which the authors exploited the observation that cell-surface transferrin receptors are highly expressed in the TC71 EwS cell line [68]. This study underlines the importance of tumor-specific delivery strategies for efficient RNAi-based therapeutic approaches. Recent advances in understanding exosome functioning and engineering of recombinant exosomes could overcome current limitations by increasing siRNA delivery efficacy [69].
Taken together, we believe that there is still ample room to investigate potential applications of directly targeting fusion oncoprotein transcripts in sarcomas via RNAi techniques, which could be facilitated by developing more specific delivery systems and utilizing combinatorial approaches with other therapeutic modalities.
Targeting the splicing machinery involved in post-transcriptional modification of fusion oncoproteins
The first association of post-transcriptional modifications of transcripts with fusion oncoproteins was identified through investigations on EWSR1-FLI1 function [70]. EWSR1-FLI1 was found to modulate the splicing machinery by interacting with RNA polymerase II [71], non-fused (wild type) EWSR1 [72], and YB1 [73], creating atypical mRNA isoforms. Selvanathan et al. gave an overview for potential isoforms alternatively spliced by EWSR1-FLI1 [74]. Since isoform variations could be a distinct feature of tumor cells, tumor-specific isoform signatures could be therapeutically implemented.
Increased VEGF165/VEGF189 ratio by alternative splicing has been reported to be caused by EWSR1-FLI1 inhibition of CAPER-alpha (RBM39) [75], which confers a more neoangiogenic phenotype to EwS by recruiting bone marrow-derived progenitor cells in a murine xenograft model [76]. Moreover, selective intratumoral suppression of VEGF165 expression by RNAi significantly reduced tumor growth in vivo [77]. Although specific protein isoforms might represent druggable targets using isoform-specific antibodies, the clinical relevance of this observation remains unclear.
Wild-type EWSR1 has been shown to alternatively splice the transmembrane domain coding exon of FAS/CD95 receptor [78]. In EwS, EWSR1-FLI1 interferes with alternative splicing of FAS/CD95 by direct interaction with non-fused (wild-type) EWSR1, resulting in exon 6 exclusion and production of a soluble isoform, which exhibits lower FAS-mediated apoptosis compared to the transmembrane form [78]. This observation may imply that suppression of EWSR1-FLI1 by RNAi could promote a shift toward a FAS-mediated apoptotic phenotype.
Moreover, EWSR1-FLI1 reduces the elongation speed of RNA polymerase II on CCND1 leading to preferential alternative splicing of the D1b isoform, which has a higher oncogenic potential than the D1a isoform [79]. Thus, suppression of EWSR1-FLI1 may lower the D1b/D1a ratio, which could sensitize EwS cells toward CDK4/6 inhibitors.
One of the subunits of the chromatin remodeling BAF complex, ARID1A, has been recently shown to be a splicing target of EWSR1-FLI1 [80]. ARID1A-L generated by alternative splicing in EwS cells plays an important role in tumor growth and promotes the stability of EWSR1-FLI1. Although this observation clearly demonstrated that complex mechanisms of (alterative) splicing are operative in EwS, it remains to be illuminated if and how they can be addressed therapeutically.
Evidence indicating a direct association of fusion oncoproteins with alternative splicing in SS and ARMS is still missing. However, the ribonucleoprotein SYT-interacting protein/co-activator activator (SIP/CoAA), which functions as a RNA splicing modulator, was found to bind to the SS18-SSX2 fusion oncoprotein, implying a potential role of SS18-SSX in alternative splicing in SS [81]. Although comprehensive data demonstrating potential tumor-specific isoform variations created by fusion oncoproteins is still missing, recent advances in transcriptome analysis may uncover tumor-specific isoform signatures, which could be further exploited to identify potential oncogenic “isoform-addiction”. In turn, researchers may be able to take a new approach in revealing potential therapeutic targets in terms of tumor vulnerability caused by isoform-addiction.
Direct targeting of fusion oncoproteins
Targeting post-translational modification, protein folding, and degradation of fusion oncoproteins
Post-translational modifications, such as phosphorylation, acetylation, or ubiquitination, are a key aspect of the regulation of protein function. These modifications can allow or inhibit the activity of a protein by changing its structure, inducing intracellular redistribution, or marking the protein for degradation [82]. In the case of the above-mentioned fusion oncoproteins, pharmacological alteration of their post-translational modifications is an interesting strategy to therapeutically influence these oncoproteins, as they are difficult to target directly.
In ARMS, phosphorylation of PAX3-FOXO1 at multiple sites has been shown to be important for stability of the fusion oncoprotein and its transcriptional activity [83]. In accordance with this, the multikinase inhibitor PKC412 has been reported to reduce the phosphorylation of certain sites at the PAX3 domain, to decrease the transcriptional activity of PAX3-FOXO1, and to exert an anti-tumorigenic effect in vitro and in xenograft models [83, 84]. As a more specific example, Polo-like kinase 1 (PLK1), a serine/threonine kinase, also phosphorylates and stabilizes the fusion protein [85]. In contrast, the authors could demonstrate that phosphorylation of wild-type FOXO1 at the equivalent serine residue triggers nuclear export of the transcription factor [85]. Inhibition of PLK1 with the small molecule BI 2536 causes increased proteasomal degradation of PAX3-FOXO1 and tumor regression in a xenograft mouse model, an effect that could be reproduced in a different study using the PLK1 inhibitor Volasertib [85, 86]. Furthermore, multiple studies have shown PAX3-FOXO1 to be a substrate of GSK3β, inhibition of which reduces transcriptional activity of the fusion protein and proliferation of tumor cells in a PAX3-FOXO1 dependent manner [87, 88]. Additionally, PAX3-FOXO1 is phosphorylated by cyclin-dependent kinase 4 (CDK4) in the ARMS cell line RH30 and inhibition of CDK4 by facsaplysin reduces the transcriptional activity and increases its cytoplasmatic levels, indicating redistribution [89]. In contrast, evidence has been published that phosphorylation of PAX3-FOXO1 by AKT reduces transcription of PAX3-FOXO1 target genes [90]. Concordantly, indirect activation of AKT via the SERCA inhibitor thapsigargin has been shown to reduce binding of PAX3-FOXO1 to regulatory DNA elements and to reduce proliferation in vitro and in vivo [91].
EWSR1-FLI1, the most common fusion oncoprotein in EwS, has also been found to be subject to various post-translational modifications. Evidence of phosphorylation of EWSR1-FLI1 was first gathered in 2001 by Olsen and Hinrichs, who identified a PKC phosphorylation site and reported this phosphorylation to be essential for transcriptional function of the oncoprotein [92]. Interestingly, PKC-β has been recognized to be necessary for EwS growth in xenograft experiments and a decrease in PKC-β activity might be a factor responsible for the reduced DNA-binding and transcriptional activity of EWSR1-FLI1 and the cytotoxic effect observed after treatment with Englerin A [93, 94]. Additionally, a different site of EWSR1-FLI1 has been shown to be a target of MAPK/ERK and JNK (c-Jun N-terminal kinase) after DNA damage but the functional and potential therapeutic relevance of these phosphorylations remains to be elucidated [95]. Similarly, evidence of acetylation, methylation by PRMT1/PRMT8, and O-GlcNAcylation of EWSR1-FLI1 has been reported with yet mostly unclear functional and therapeutic implications [96–99]. Recently, Gierisch et al. identified EWSR1-FLI1 to be subject to ubiquitination and subsequent proteasomal degradation [100]. Furthermore, they recently demonstrated that the ubiquitin-specific protease 19 (USP19) deubiquitinates the fusion oncoprotein [101]. Depletion of USP19 was shown to decrease EWSR1-FLI1 expression levels, but neither the expression of wild-type of EWSR1 nor FLI1, and to reduce tumor growth in vitro and in vivo indicating a potential therapeutic value for USP19 inhibition [101]. In addition to that, EWSR1-FLI1 has been reported to be a substrate of the chaperone HSP90 and inhibition of HSP90 by the small molecule PU-H71 reduced tumor cell growth in vitro and tumor burden in xenograft experiments [102]. On a similar note, it has been shown that reduced levels of EWSR1-FLI1 can also be achieved by treatment with the HDAC1/3 inhibitor Entinostat, as HDAC1/3 deacetylates HSP90 and thereby increases its activity [103].
Even though the fusion oncoprotein in SS was identified over 25 years ago and there are multiple sites of post-translational modification present in the domains of wild-type SS18 and SSX1/2/4, little has been published about the impact of post-translational modifications on SS18-SSX and their therapeutic value [10, 11]. Nevertheless, Patel et al. have recently shown that SS18-SSX is ubiquitinated and thereby marked for degradation by the MCL-1 ubiquitin ligase E3 (MULE) at the lysine K23 of SS18, which is not a known ubiquitination site in wild-type SS18 [104]. Furthermore, they showed that MULE is a substrate of the E3 ubiquitin-ligase MDM2, which is only active in its deacetylated state that is ensured by the histone deacetylase 2 (HDAC2) [104–106]. As a reduced activity of HDAC2 thus leads to increased ubiquitination and degradation of SS18-SSX, this might be an additional mechanism responsible for the cytotoxic effect of HDAC inhibition in SS cells [107].
Targeting fusion oncoprotein-specific neoantigens by immunotherapy
The successful clinical implementation of checkpoint inhibitors and CAR-T cells in the routine treatment of various tumor entities has heralded a new era of immunotherapies in cancer [108, 109]. Tumor-specific neoantigens derived from fusion oncogene breakpoint regions have attracted great scientific interest for many years, especially as an alternative to single-nucleotide variant-derived tumor-specific antigens [110]. Recently, gene-fusion-derived neoantigens were found to induce tumor-specific T cells and facilitated a complete response in a head and neck-cancer patient treated with immune checkpoint inhibitors despite an overall low tumor mutational burden [111]. Hence, fusion oncogene-driven pediatric sarcomas might represent promising candidates for immunotherapies targeting breakpoint-derived neoantigens.
Indeed, breakpoint-specific neoantigens have been predicted and in part validated for EWSR1-FLI1 [112], PAX3-FOXO1 [112], and SS18-SSX1 [113]. For EWSR1-FLI1, it was shown that a modified 9-mer peptide of the fusion site (YLNPSVDS) was able to induce a robust immune response in vitro and in vivo facilitating prolonged survival of xenograft-bearing mice after treatment with YLNPSVDS-specific cytotoxic T lymphocytes (CTLs) [114]. This is of special interest, since native peptides of the breakpoint region have failed to provoke significant CD8+ CTL responses before and show weak MHC-binding affinities [114, 115]. Moreover, fusion-restricted immune responses seem to highly depend on the HLA alleles. For instance, two highly similar peptides of the PAX3-FOXO1 breakpoint region showed strikingly different results depending on the HLA alleles used in the respective study: The RS10 peptide (SPQNSIRHNL) was able to induce highly effective CTLs in HLA-B7+ donor peripheral blood mononuclear cells and allowed killing of HLA-B7+ ARMS cells [116]. In contrast, the modified 9-mer peptide GLSPQNSIK, which shares 6 amino acids with RS10 and was optimized for HLA binding, failed to induce a potent CTL response in HLA-A3+ CD8+ T cells [117]. Targeting neoantigens derived from the breakpoint region of fusion oncogenes represents a sophisticated, yet challenging approach. The development of CAR-T cells specific for the peptide-MHC-complex might represent a promising strategy to tackle the low antigenicity of fusion peptides in the future.
Indirect targeting of fusion oncoproteins and their function
Targeting protein-protein interactions of fusion oncoproteins
Fusion oncogenes and their function have been extensively characterized over the past decades. By now, it has been well accepted that fusion oncogenes orchestrate a tumorigenic re-programming comprised of a plethora of interaction partners rather than acting in isolated systems. This holds especially true for SS18-SSX oncoproteins that exert their deleterious effects by interacting with the BAF complex, a multimember epigenetic regulatory machinery [29]. Such interaction partners might represent druggable targets that could indirectly compromise fusion oncoprotein functioning.
Apart from their direct DNA-binding capability, EWSR1-FLI1 and PAX3-FOXO1 also act through direct protein-protein interactions. For instance, it has been shown that EWSR1-FLI1, but not wild-type EWSR1, directly binds RNA helicase A [118] and reduces helicase activity [119]. RNA helicase A is involved in many cellular processes such as re-organizing RNA secondary structures, spliceosome assembly, and initiation of translation. In turn, blocking this interaction using the compound YK-4-279 translated in increased apoptosis and reduced growth of EwS orthotopic xenografts and might represent an elegant strategy to target this neomorphic feature of the fusion oncogene [120]. Moreover, Li et al. found a direct link between p53 inhibition and EWSR1-FLI1 [121]. Mutations in p53 have been found in as little as 5–7% of EwS [14]; however, even in wild-type sarcomas, p53 function seems to be compromised in analogy to other fusion-positive malignancies such as fusion-driven leukemia [121–123]. In EwS, the N-terminal domain of EWSR1-FLI1 was identified to form a complex with HDAC1 and p53 leading to subsequent deacetylation of p53 and inhibition of its downstream signaling [124]. However, it remains unclear whether this interaction is indeed a neomorphic feature of the fusion oncogene and not intrinsic to wild-type EWSR1, especially since the interaction seems to involve the N-terminal region of EWSR1-FLI1 [121, 124].
As outlined before, a recent study revealed a direct interaction of EWSR1-FLI1 with the BAF complex via its subunit ARID1A [80]. The authors could show that ARID1A was differentially spliced and accumulated in its long isoform ARID1A-L in the presence of EWSR1-FLI1. Notably, modulation of wild-type EWSR1 levels did not affect the isoform switch, pointing toward a neomorphic feature of the fusion gene [80]. ARID1A-L was shown to stabilize EWSR1-FLI1 and bolstered its oncogenic capacity. Interestingly, the above-mentioned compound YK-4-279 also blocked this interaction and prevented binding of EWSR1-FLI1 to the BAF complex via ARID1A. Hence, TK216, a slightly improved version of YK-4-279 that has been approved for a phase I clinical trial in relapsed or refractory EwS, represents a very promising candidate for future targeted therapies of this devastating disease [125].
Lastly, Embree et al. could demonstrate the interaction of EWSR1-FLI1 with wild-type EWSR1 in a zebrafish model and in human cell lines [72]. The researchers observed mitotic defects in EWSR1-FLI1 expressing zebrafish embryos and HeLa cells. Since the observed phenotype closely resembled one that was observed in EWSR1-deficient embryos, a direct interaction of EWSR1-FLI1 with EWSR1 was suspected. Indeed, it was found that the fusion oncoprotein bound wild-type EWSR1 and inhibited its function resulting in spindle malformation and mis-localization of Aurora kinase B [72]. Anyhow, the authors did not expand on how these findings might translate into therapeutic consequences and the effects of targeting EWSR1 in EwS still remain to be investigated.
Apart from EWSR1-FLI1, PAX3-FOXO1 functioning has also been found to rely on direct interactions with other proteins, especially kinases, as reviewed above [85–91]. Although all of the phosphorylation sites also exist in wild-type PAX3 or FOXO1, respectively, and thereby do not represent neomorphic features of the fusion oncogene per se, kinase inhibitors showed different effects on the fusion oncogene than on wild-type PAX3 and FOXO1 [85]. Moreover, some of them directly interfered with its binding to the neomorphic binding site ACCGTGACTAATTNN that is enriched in PAX3-FOXO1 associated super-enhancers [91]. Hence, kinase inhibitors do not particularly exploit neomorphic features rather than inhibiting neomorphic functioning of PAX3-FOXO1.
In SS, the interactions of its fusion oncoprotein with the BAF complex have been widely studied and are reviewed below. Besides subunits of the BAF complex, only a few other proteins have been demonstrated to bind SS18-SSX fusion proteins directly. For instance, SS18-SSX1 was found to stabilize HDM2, a negative regulator of p53 that consecutively promoted p53 ubiquitination and degradation resulting in resistance to apoptosis-inducing drugs [126].
To conclude, targeting proteins that directly interact with fusion oncoproteins might constitute a useful addition to well-established therapies or novel treatment strategies directed against the fusion oncoprotein itself. Yet, the lower specificity of such approaches and the resulting side effects might constitute an obstacle for future clinical trials.
Targeting protein-DNA interactions of fusion oncoproteins including epigenetic rewiring and subcellular localization
While EWSR1-FLI1 and PAX3-FOXO1 are aberrant transcription factors directly binding to specific DNA sequences, SS18-SSX instead interacts with chromatin remodeling complexes [14, 22, 28, 30, 127]. Therefore, the oncogenicity of these oncoproteins is rooted in their influence on the state of chromatin and on the expression of certain genes. Preventing these oncoproteins to access the DNA might consequently be a promising strategy for more specific therapies of these sarcomas.
As an example, the small molecule lurbinectedin and trabectedin have been found to induce a nucleolar redistribution of EWSR1-FLI1 in EwS cells leading to an increase in heterochromatin-associated histone methylations at EWSR1-FLI1 target genes [128, 129]. Both compounds have preclinically shown efficacy against EwS cells, especially in combination with other compounds such as irinotecan or olaparib [128, 130]. Lurbinectedin also has shown a good safety profile and anti-cancer activity as a single agent in a phase II clinical trial [131].
As fusion-driven sarcomas are characterized by the paucity of other genetic mutations and due to the functions of their disease-defining oncogenes, epigenetic dysregulation plays an important role in the maintenance of their phenotype. Hence, epigenetic modulators, which are increasingly becoming of interest in other malignancies as well, are major potential targets in EwS, SS and ARMS.
One group of epigenetic regulators are HDACs, which remove acetyl groups from lysine residues of histone chains and thereby regulate gene expression [132]. Indeed, in preclinical models of EwS, HDAC inhibitors have been shown to induce apoptosis, promote differentiation, and reduce tumor growth in xenograft experiments [57, 133]. Antitumorigenic activity of HDAC inhibitors has also been reported in preclinical models of SS and ARMS [134–138]. Unfortunately, the few clinical trials that have been conducted to investigate the efficacy of HDAC inhibition have shown only modest response at best in these entities, when used as single agents [139–143]. HDAC inhibitors might therefore be of greater use in combination with other anti-cancer drugs. Apart from standard chemotherapeutics, possible combination partners might also include additional epigenetic drugs. Using the single agent SP2509, reversible inhibition of lysine-specific demethylase 1 (LSD1), a subunit of the Mi-2/nucleosome remodeling and deacetylase (NuRD) complex, induces apoptosis and reduces xenograft tumor growth in EwS [144]. Interestingly, this LSD1 inhibitor has a synergistic effect with the HDAC inhibitors suberoylanilide hydroxamic acid (SAHA) or romidepsin in vivo [145, 146]. Based on these preclinical studies, more investigations focusing on this and other combinations with HDAC inhibitors are justified.
Another interesting epigenetic modulator is the enhancer of zeste homolog 2 (EZH2), a Polycomb-group protein involved in DNA methylation [147]. Knockdown or pharmacological inhibition of EZH2 in ARMS cells leads to apoptosis and reduced tumor growth of ARMS xenografts in vivo [148]. In EwS, the EZH2 promoter is a direct target of EWSR1-FLI1. Here, RNAi mediated downregulation of EZH2 inhibits clonogenicity in vitro and suppresses tumor development and metastasis in xenograft experiments [149]. However, a tumor xenograft study using 4 different cell lines of ARMS and EwS showed only limited anti-tumor activity of the EZH2 inhibitor tazemetostat for both entities [150]. Therefore, further preclinical and clinical investigation into the use of EZH2 inhibitors for treatment of ARMS and EwS is of need. In SS cells, inhibition of EZH2 with the small molecule EPZ005687 has been shown to impair proliferation and migration [151]. However, in a phase II clinical trial of tazemetostat as a single agent in patients with SS, stable disease could be achieved in only 33% of treated patients indicating the possible necessity of combination partners [152].
Yet, another group of proteins involved in chromatin regulation is the bromodomain and extra terminal domain (BET) family. Its members, such as BRD4, are able to recognize and bind acetylated histones with their bromodomains and play a role in epigenetic memory and transcription regulation [153]. With the advent of specific BET bromodomain inhibitors (JQ1 and iBET) bromodomain containing proteins have entered the spotlight as attractive potential targets for a variety of different cancer entities [154, 155]. In EwS cells, BRD4 has been found to form a transcriptional complex with EWSR1-FLI1 and -ERG. RNAi mediated knockdown or JQ1 mediated inhibition of BRD4 resulted in significantly impaired oncogenic phenotype and reduced xenograft tumor growth [156]. ARMS cells have also been reported to be sensitive to JQ1 in preclinical models [157]. In addition to that, Gryder et al. could explain this sensitivity mechanistically by showing that PAX3-FOXO1 function relies on BRD4 recruitment at defined super-enhancers [24]. Furthermore, it was recently demonstrated that BRD9 is a component of the SS18-SSX containing BAF complexes in SS [158]. Inhibition or targeted degradation of BRD9 induced downregulation of oncogenic programs and reduced growth of SS xenografts [158]. In the light of these preclinical results, clinical trials to investigate the efficacy of BET inhibitors for the treatment of fusion-driven sarcomas as single agents or in combinations seem to be warranted.
Targeting specific downstream pathways of fusion oncoproteins
Malignant transformation and maintenance of tumorigenesis in EwS, SS, and ARMS depend on the expression of their specific fusion proteins to the extent that their deletion results in cell death. However, there are no available drugs that can directly target these fusion proteins, so far. Since the expression of these fusion proteins also completely rewires the expression and regulation of pathways and downstream effectors, there is great potential for alternative targeted therapeutic interventions. This section will cover a range of targeted therapies described to date against commonly altered oncogenic pathways in these fusion-driven sarcomas.
Insulin-like growth factor receptor (IGF-1R) pathway
Tumorigenesis in translocation-driven tumors is critically mediated via the IGF-1R signaling pathway [159]. Preclinical EwS models show a constant activation of the IGF-1R-mediated signaling pathway [160], which promotes drug escape [161] and seems to be essential for EwS cell viability [162]. In SS, the translocation induces the expression of IGF2, which is also required for tumor growth in vivo [163], while its immune targeting inhibits tumor growth and metastasis formation in ARMS [164]. Additionally, IGF-1R inhibition induces apoptosis of SS cells in vitro [165]. At the same time, IGF-1R is a direct target of the PAX3-FOXO1 fusion in ARMS [166].
Several antibodies and small molecules against IGF-1R pathway have been tested in clinical trials including phase II and III. However, the results have only been moderate so far due to acquired resistance through different mechanisms [167–169]. Yet, given the outstanding preclinical results obtained when blocking this pathway, significant efforts are being made to develop combined therapies that could overcome these hurdles. For instance, a recent report suggests combining heparanase inhibitors with IGF-1R antagonists for treatment of metastatic SS [170], and a combined therapy including BET inhibitors has been set forth for EwS [171]. In the case of ARMS, it has been proposed that the simultaneous inhibition of IGF-1R and additional tyrosine kinases could help to overcome resistance to treatment. For instance, concomitant inhibition of IGF-1R and YES/SRC family tyrosine kinase [172], a combination of an anti-IGF-1R antibody and the inhibition of the anaplastic lymphoma kinase (ALK) [173], or treatment with Ceritinib (ALK and IGF-1R inhibitor) with the multikinase inhibitor sorafenib have been suggested based on preclinical results [174].
Mitogen-activated protein kinase pathway
Tyrosine kinases are highly expressed in sarcomas including EGFR and PDGFRA in SS, FGFR, and EGFR in ARMS, and upstream mediators of ERK1/2 in EwS [175, 176]. Indeed, ERK1/2 is activated in the majority of EwS cell lines and is thought to hold a pivotal role in tumor development [177]. In fact, the A673 EwS cell has a constitutively activating mutation in BRAF [178]. Accordingly, the inhibition of ERK1/2 precursor MEK1/2 using U0126 impedes migration and invasion of EwS cell lines [177]. Besides, pharmacological treatment with FGFR inhibitor PD-173074 yielded a decrease in tumorigenic features in preclinical studies [179]. Therefore, mitogen-activated protein kinase (MAPK) pathway blocking agents such as RAF inhibitors have entered clinical trials [180].
In SS, the situation is partially different: although ERK1/2 MAPK signaling pathway components have been proposed as targets of SS18-SSX, few reports describing their role and druggability in SS are available so far [181, 182]. However, there is a targetable upregulation of EGFR, PDGFRA, and PDGFRB in SS that can be exploited therapeutically by using specific small molecule inhibitors [183, 184]. Additionally, and similarly to EwS, FGFR2 is also being studied as a potential therapeutic target in SS [185]. Finally, it has been recently shown that nintedanib, a triple kinase inhibitor targeting PDGFR, VEGFR, and FGFR pathways, presents promising effects in a preclinical study for SS that are beyond those shown by imatinib [186]. In ARMS, the upregulation of EGFR presents a rationale for the use of anti-EGFR monoclonal antibodies such as cetuximab that has proven to be effective in preclinical studies [187]. Notably, due to the high expression of FGFR4, the inhibitor PD-173074 has shown similar preclinical promising effects in ARMS as in EwS [176]. Moreover, sorafenib, a PDGFRA and RAF inhibitor, reduced tumor growth in a mouse model for ARMS [188].
Sonic hedgehog (SHH) pathway
In ARMS, the expression of sonic hedgehog pathway effectors like GLI1/2 is altered [189]. Inhibitory targeting of GLI1/2 using GANT-61 leads to a blockage of tumor growth in vivo, and its effect was shown to be synergistic with commonly used chemotherapeutic drugs for ARMS such as temsirolimus or vincristine [190]. Similarly, a decrease in cell viability and induced cell death was obtained in vitro by treatment with arsenic trioxide (ATO), a different GLI1 inhibitor [191], and in vivo by treatment with the sonic hedgehog pathway inhibitor foskolin [192]. Additionally, apoptosis-inducing agents like betulinic acid have been demonstrated to impair tumor growth in preclinical studies via inhibition of the hedgehog pathway in ARMS [193]. Finally, in order to avoid escape mechanisms, targeted therapies including GANT61 and the dual PI3K/mTOR inhibitor PI103 have shown a synergistic effect on ARMS cells apoptosis [194].
GLI1 was also shown to be upregulated in EwS and driven by the translocation [195], and therefore, therapeutic approaches using ATO have been attempted with promising preclinical results [196]. Thus, ATO was included in the Pediatric Preclinical Testing Program (PPTP) [197] where it unfortunately yielded negative results.
In the case of SS, since GLI and SMO were found to be overexpressed in patient’s tumors [198] and since Notch has been shown to activate GLI1, a Notch inhibitor in combination with vismodegib was tested in phase II trial that accepted SS patients to be enrolled (NCT01154452). Unfortunately, the inhibitor was discontinued and the trial had to be prematurely ended.
Wnt/β-catenin pathway
The Wnt pathway is activated in most SS [199, 200] and this activation is mediated by SS18-SSX [201]. Following this rationale, Wnt inhibitors such as pyrvinium have been preclinically tested with promising results [202]. Beyond blocking the pathways using SS18-SSX-directed small molecules, targeting some of its interacting proteins has been attempted. For instance, desatinib-a YES1 tyrosine kinase inhibitor-has been demonstrated to block the Wnt pathway leading to impairment of cell proliferation in SS [203]. Additionally, SS patients have been enrolled in a results-pending trial to test an antibody against the Wnt receptor FZD10 (NCT01469975).
In contrast, the relevance and potential for clinical intervention in the Wnt pathway in ARMS remains highly controversial [204]. So far, a single study by Annavarapu et al. found an inhibition of proliferation and self-renewal capacity after pharmacological re-activation of the Wnt pathway in vitro [205].
Interestingly, in EwS, even though Wnt activation is consistent with worse clinical outcomes and a more aggressive phenotype in vitro, cell populations present a highly heterogeneous activation of the Wnt signaling pathway, which could be at least partially mediated by the inhibition of EWSR1-ETS protein functions [206]. So far, WNT974-a modulator of the Wnt pathway at PORCN level-showed a significant delay in formation of metastasis in vivo [207].
All in all, combined targeting of oncogenic fusion proteins along with specific pharmacological intervention of one or multiple deregulated pathways may constitute a promising therapeutic approach to overcome drug resistance for these fusion-driven malignancies.
Conclusions and perspectives
Reviewing the current literature on neomorphic features of fusion oncoproteins in EwS, ARMS, and SS, we have presented four modalities of targeting fusion oncogenes in these devastating childhood sarcomas (Figure 1). First, oncogene DNA or their respective binding sites can be directly targeted by gene editing strategies, such as CRISPR-CAS9 or CRISPRi. Second, oncogene mRNA transcripts represent a favorable target for RNAi-based therapeutic approaches. Moreover, fusion oncoproteins that interfere with the splicing machinery allow isoform-specific treatment strategies. Third, oncoproteins and their subcellular localization can be directly targeted either by inhibiting or inducing their post-translational modification, respectively. Additionally, breakpoint regions constitute excellent targets for immunotherapies once they are sufficiently presented by the tumor cells. Fourth, unique interactions of fusion proteins with their respective interaction partners have already been successfully exploited in clinical trials (e.g., using YK-4-279). Furthermore, due to the lack of somatic mutations and the importance of epigenetic alterations for the malignant phenotype of fusion-driven sarcomas, targeting epigenetic regulators has been found to be promising in these entities. In a more system-based manner, tackling downstream pathways and regulatory networks of fusion oncogenes might add to the arsenal of indirect treatment options in EwS, ARMS and SS.
Fig. 1.
Scheme depicting the different targeting strategies of fusion oncogenes in pediatric fusion-driven sarcomas. Fusion oncogenes can be targeted on their DNA level (1), RNA level (2), protein level (3), and by targeting downstream functions and interaction partners (4).
Taken together, neomorphic features of EWSR1-FLI1, PAX3/7-FOXO1, and SS18-SSX fusion proteins have been therapeutically addressed in the past and represent a potent, highly specific approach worthy of further investigations in the future. New delivery strategies, advances in understanding and exploiting tumor immunity, new drugs targeting fusion oncoproteins, epigenetic regulators, and interaction partners might help to overcome the hurdles that have proved these oncogenes to be notoriously difficult to target.
Acknowledgements
Open Access funding provided by Projekt DEAL.
Funding information
This review was mainly supported through the Max-Eder program of the German Cancer Aid (70112257 to T.G.P.G). The laboratory of T.G.P.G. is further supported by grants from the “Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)”, by LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the “Mehr LEBEN für krebskranke Kinder-Bettina-Bräu-Stiftung”, the Walter Schulz Foundation, the Wilhelm Sander-Foundation (2016.167.1), the Friedrich-Baur foundation, the Barbara and Hubertus Trettner foundation, Matthias-Lackas foundation, the Dr. Leopold und Carmen Ellinger foundation, the Gert & Susanna Mayer foundation, the Dr. Rolf M. Schwiete foundation, the Deutsche Forschungsgemeinschaft (DFG 391665916), and by the German Cancer Aid (111886). T.L.B.H. was supported by a scholarship of the German Cancer Aid.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maximilian M. L. Knott and Tilman L. B. Hölting contributed equally to this work.
References
- 1.Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, de Klein A, Bartram CR, Grosveld G. Localization of the c- abl oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306(5940):239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 2.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 3.Delattre Olivier, Zucman Jessica, Plougastel Béatrice, Desmaze Chantal, Melot Thomas, Peter Martine, Kovar Heinrich, Joubert Isabelle, de Jong Pieter, Rouleau Guy, Aurias Alain, Thomas Gilles. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 4.Mitelman F, Johansson B and Mertens F (Eds.). (2019). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Retrieved from https://mitelmandatabase.isb-cgc.org. Accessed 29 October 2019
- 5.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RGW. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nature Reviews. Clinical Oncology. 2017;14(12):735–748. doi: 10.1038/nrclinonc.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson Nathaniel D., de Borja Richard, Young Matthew D., Fuligni Fabio, Rosic Andrej, Roberts Nicola D., Hajjar Simon, Layeghifard Mehdi, Novokmet Ana, Kowalski Paul E., Anaka Matthew, Davidson Scott, Zarrei Mehdi, Id Said Badr, Schreiner L. Christine, Marchand Remi, Sitter Joseph, Gokgoz Nalan, Brunga Ledia, Graham Garrett T., Fullam Anthony, Pillay Nischalan, Toretsky Jeffrey A., Yoshida Akihiko, Shibata Tatsuhiro, Metzler Markus, Somers Gino R., Scherer Stephen W., Flanagan Adrienne M., Campbell Peter J., Schiffman Joshua D., Shago Mary, Alexandrov Ludmil B., Wunder Jay S., Andrulis Irene L., Malkin David, Behjati Sam, Shlien Adam. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science. 2018;361(6405):eaam8419. doi: 10.1126/science.aam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galili Naomi, Davis Richard J., Fredericks William J., Mukhopadhyay Sunil, Rauscher Frank J., Emanuel Beverly S., Rovera Giovanni, Barr Frederic G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nature Genetics. 1993;5(3):230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 9.Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Research. 1994;54(11):2869–2872. [PubMed] [Google Scholar]
- 10.Clark Jeremy, Rocques Philippe J., Crew A. Jayne, Gill Sandra, Shipley Janet, Chan Andrew M.-L., Gusterson Barry A., Cooper Colin S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nature Genetics. 1994;7(4):502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 11.Crew A.J., Clark J., Fisher C., Gill S., Grimer R., Chand A., Shipley J., Gusterson B.A., Cooper C.S. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. The EMBO Journal. 1995;14(10):2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlén M, Larsson O. A novel fusion gene, SYT-SSX4, in synovial sarcoma. Journal of the National Cancer Institute. 1999;91(11):974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- 13.Ewing J. The classic: diffuse endothelioma of bone. Proceedings of the New York Pathological Society. 1921;12:17. Clinical Orthopaedics and Related Research. 2006;450:25–27. doi: 10.1097/01.blo.0000229311.36007.c7. [DOI] [PubMed] [Google Scholar]
- 14.Grünewald, T. G. P., Cidre-Aranaz, F., Surdez, D., Tomazou, E. M., Álava, E. de, Kovar, H., … Dirksen, U. (2018). Ewing sarcoma. Nature Reviews Disease Primers, 4(1), 1–22. 10.1038/s41572-018-0003-x [DOI] [PubMed]
- 15.Noone, A. M., Howlader, N., Krapcho, M., Miller, D., Brest, A., Yu, M., Ruhl, J., Tatalovich, Z., Mariotto, A., Lewis, D. R., Chen, H. S., Feuer, E. J., & Cronin, K. A. (Eds.). (n.d.). SEER Cancer Statistics Review, 1975–2015. Bethesda: National Cancer Institute https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018.
- 16.Johnson KM, Taslim C, Saund RS, Lessnick SL. Identification of two types of GGAA-microsatellites and their roles in EWS/FLI binding and gene regulation in Ewing sarcoma. PloS One. 2017;12(11):e0186275. doi: 10.1371/journal.pone.0186275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangwal K., Sankar S., Hollenhorst P. C., Kinsey M., Haroldsen S. C., Shah A. A., Boucher K. M., Watkins W. S., Jorde L. B., Graves B. J., Lessnick S. L. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proceedings of the National Academy of Sciences. 2008;105(29):10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musa, J., Cidre-Aranaz, F., Aynaud, M.-M., Orth, M. F., Knott, M. M. L., Mirabeau, O., … Grünewald, T. G. P. (2019). Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nature Communications, 10(1), 4128. 10.1038/s41467-019-12071-2 [DOI] [PMC free article] [PubMed]
- 19.Johnson Kirsten M., Mahler Nathan R., Saund Ranajeet S., Theisen Emily R., Taslim Cenny, Callender Nathan W., Crow Jesse C., Miller Kyle R., Lessnick Stephen L. Role for the EWS domain of EWS/FLI in binding GGAA-microsatellites required for Ewing sarcoma anchorage independent growth. Proceedings of the National Academy of Sciences. 2017;114(37):9870–9875. doi: 10.1073/pnas.1701872114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grünewald Thomas G P, Bernard Virginie, Gilardi-Hebenstreit Pascale, Raynal Virginie, Surdez Didier, Aynaud Marie-Ming, Mirabeau Olivier, Cidre-Aranaz Florencia, Tirode Franck, Zaidi Sakina, Perot Gaëlle, Jonker Anneliene H, Lucchesi Carlo, Le Deley Marie-Cécile, Oberlin Odile, Marec-Bérard Perrine, Véron Amélie S, Reynaud Stephanie, Lapouble Eve, Boeva Valentina, Frio Thomas Rio, Alonso Javier, Bhatia Smita, Pierron Gaëlle, Cancel-Tassin Geraldine, Cussenot Olivier, Cox David G, Morton Lindsay M, Machiela Mitchell J, Chanock Stephen J, Charnay Patrick, Delattre Olivier. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nature Genetics. 2015;47(9):1073–1078. doi: 10.1038/ng.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer Letters. 2009;279(2):126–136. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 22.Skapek, S. X., Ferrari, A., Gupta, A. A., Lupo, P. J., Butler, E., Shipley, J., … Hawkins, D. S. (2019). Rhabdomyosarcoma. Nature Reviews. Disease Primers, 5(1), 1. 10.1038/s41572-018-0051-2 [DOI] [PMC free article] [PubMed]
- 23.Parham DM, Barr FG. Classification of rhabdomyosarcoma and its molecular basis. Advances in Anatomic Pathology. 2013;20(6):387–397. doi: 10.1097/PAP.0b013e3182a92d0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gryder Berkley E., Yohe Marielle E., Chou Hsien-Chao, Zhang Xiaohu, Marques Joana, Wachtel Marco, Schaefer Beat, Sen Nirmalya, Song Young, Gualtieri Alberto, Pomella Silvia, Rota Rossella, Cleveland Abigail, Wen Xinyu, Sindiri Sivasish, Wei Jun S., Barr Frederic G., Das Sudipto, Andresson Thorkell, Guha Rajarshi, Lal-Nag Madhu, Ferrer Marc, Shern Jack F., Zhao Keji, Thomas Craig J., Khan Javed. PAX3–FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discovery. 2017;7(8):884–899. doi: 10.1158/2159-8290.CD-16-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorensen Poul H.B., Lynch James C., Qualman Stephen J., Tirabosco Roberto, Lim Jerian F., Maurer Harold M., Bridge Julia A., Crist William M., Triche Timothy J., Barr Frederic G. PAX3-FKHR and PAX7-FKHR Gene Fusions Are Prognostic Indicators in Alveolar Rhabdomyosarcoma: A Report From the Children’s Oncology Group. Journal of Clinical Oncology. 2002;20(11):2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 26.Stacchiotti S, Van Tine BA. Synovial sarcoma: current concepts and future perspectives. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2018;36(2):180–187. doi: 10.1200/JCO.2017.75.1941. [DOI] [PubMed] [Google Scholar]
- 27.Haagensen CD, Stout AP. Synovial sarcoma. Annals of Surgery. 1944;120(6):826–842. doi: 10.1097/00000658-194412000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride Matthew J., Pulice John L., Beird Hannah C., Ingram Davis R., D’Avino Andrew R., Shern Jack F., Charville Gregory W., Hornick Jason L., Nakayama Robert T., Garcia-Rivera Enrique M., Araujo Dejka M., Wang Wei-Lien, Tsai Jen-Wei, Yeagley Michelle, Wagner Andrew J., Futreal P. Andrew, Khan Javed, Lazar Alexander J., Kadoch Cigall. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell. 2018;33(6):1128-1141.e7. doi: 10.1016/j.ccell.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banito Ana, Li Xiang, Laporte Aimée N., Roe Jae-Seok, Sanchez-Vega Francisco, Huang Chun-Hao, Dancsok Amanda R., Hatzi Katerina, Chen Chi-Chao, Tschaharganeh Darjus F., Chandwani Rohit, Tasdemir Nilgun, Jones Kevin B., Capecchi Mario R., Vakoc Christopher R., Schultz Nikolaus, Ladanyi Marc, Nielsen Torsten O., Lowe Scott W. The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell. 2018;33(3):527-541.e8. doi: 10.1016/j.ccell.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cironi, L., Petricevic, T., Fernandes Vieira, V., Provero, P., Fusco, C., Cornaz, S., … Stamenkovic, I. (2016). The fusion protein SS18-SSX1 employs core Wnt pathway transcription factors to induce a partial Wnt signature in synovial sarcoma. Scientific Reports, 6, 22,113. 10.1038/srep22113 [DOI] [PMC free article] [PubMed]
- 32.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annual Review of Biochemistry. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 33.Cyranoski David. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535(7613):476–477. doi: 10.1038/nature.2016.20302. [DOI] [PubMed] [Google Scholar]
- 34.Khan, F. A., Pandupuspitasari, N. S., Chun-Jie, H., Ao, Z., Jamal, M., Zohaib, A., … ShuJun, Z. (2016). CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget, 7(32), 52,541–52,552. 10.18632/oncotarget.9646 [DOI] [PMC free article] [PubMed]
- 35.Chen Zhang-Hui, Yu Yan P, Zuo Ze-Hua, Nelson Joel B, Michalopoulos George K, Monga Satdatshan, Liu Silvia, Tseng George, Luo Jian-Hua. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nature Biotechnology. 2017;35(6):543–550. doi: 10.1038/nbt.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitra SA, Ravinder N, Magnon V, Nagy J, Triche TJ. Abstract 4499: Genomic editing of EWS-FLI1 and its targets, and its therapeutic potential in treatment of Ewing sarcoma. Cancer Research. 2019;79(13 Supplement):4499–4499. doi: 10.1158/1538-7445.AM2019-4499. [DOI] [Google Scholar]
- 37.Pandey PR, Chatterjee B, Olanich ME, Khan J, Miettinen MM, Hewitt SM, Barr FG. PAX3-FOXO1 is essential for tumour initiation and maintenance but not recurrence in a human myoblast model of rhabdomyosarcoma. The Journal of pathology. 2017;241(5):626–637. doi: 10.1002/path.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulay G, Volorio A, Iyer S, Broye LC, Stamenkovic I, Riggi N, Rivera MN. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes & Development. 2018;32(15–16):1008–1019. doi: 10.1101/gad.315192.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nature Reviews. Genetics. 2016;17(10):630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 40.Widschwendter Martin, Jones Allison, Evans Iona, Reisel Daniel, Dillner Joakim, Sundström Karin, Steyerberg Ewout W., Vergouwe Yvonne, Wegwarth Odette, Rebitschek Felix G., Siebert Uwe, Sroczynski Gaby, de Beaufort Inez D., Bolt Ineke, Cibula David, Zikan Michal, Bjørge Line, Colombo Nicoletta, Harbeck Nadia, Dudbridge Frank, Tasse Anne-Marie, Knoppers Bartha M., Joly Yann, Teschendorff Andrew E., Pashayan Nora. Epigenome-based cancer risk prediction: rationale, opportunities and challenges. Nature Reviews Clinical Oncology. 2018;15(5):292–309. doi: 10.1038/nrclinonc.2018.30. [DOI] [PubMed] [Google Scholar]
- 41.Jacques, C., Lamoureux, F., Baud’huin, M., Rodriguez Calleja, L., Quillard, T., Amiaud, J., … Ory, B. (2016). Targeting the epigenetic readers in Ewing sarcoma inhibits the oncogenic transcription factor EWS/Fli1. Oncotarget, 7(17), 24,125–24,140. 10.18632/oncotarget.8214 [DOI] [PMC free article] [PubMed]
- 42.Itzen F, Greifenberg AK, Bösken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Research. 2014;42(12):7577–7590. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham J., Nunez-Alvarez Y., Hettmer S., Carrio E., Chen H.-I. H., Nishijo K., Huang E. T., Prajapati S. I., Walker R. L., Davis S., Rebeles J., Wiebush H., McCleish A. T., Hampton S. T., Bjornson C. R. R., Brack A. S., Wagers A. J., Rando T. A., Capecchi M. R., Marini F. C., Ehler B. R., Zarzabal L. A., Goros M. W., Michalek J. E., Meltzer P. S., Langenau D. M., LeGallo R. D., Mansoor A., Chen Y., Suelves M., Rubin B. P., Keller C. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes & Development. 2014;28(14):1578–1591. doi: 10.1101/gad.238733.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharathy Narendra, Berlow Noah E., Wang Eric, Abraham Jinu, Settelmeyer Teagan P., Hooper Jody E., Svalina Matthew N., Ishikawa Yoshihiro, Zientek Keith, Bajwa Zia, Goros Martin W., Hernandez Brian S., Wolff Johannes E., Rudek Michelle A., Xu Linping, Anders Nicole M., Pal Ranadip, Harrold Alexandria P., Davies Angela M., Ashok Arya, Bushby Darnell, Mancini Maria, Noakes Christopher, Goodwin Neal C., Ordentlich Peter, Keck James, Hawkins Douglas S., Rudzinski Erin R., Chatterjee Bishwanath, Bächinger Hans Peter, Barr Frederic G., Liddle Jennifer, Garcia Benjamin A., Mansoor Atiya, Perkins Theodore J., Vakoc Christopher R., Michalek Joel E., Keller Charles. The HDAC3–SMARCA4–miR-27a axis promotes expression of thePAX3:FOXO1fusion oncogene in rhabdomyosarcoma. Science Signaling. 2018;11(557):eaau7632. doi: 10.1126/scisignal.aau7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Tilburg Cornelis Martinus, Witt Ruth, Pajtler Kristian W., Christoph Plass, Poschke Isabel, Platten Michael, Harting Inga, Sedlaczek Oliver, Freitag Angelika, Meyrath David, Taylor Lenka, Balasubramanian Gnana Prakash, Jäger Natalie, Pfaff Elke, Worst Barbara Christine, Milde Till, Pfister Stefan M., Jones David T. W., Kopp-Schneider Annette, Witt Olaf. INFORM2 exploratory multinational phase I/II combination study of nivolumab and entinostat in children and adolescents with refractory high-risk malignancies: INFORM2 NivEnt. Journal of Clinical Oncology. 2019;37(15_suppl):TPS10065–TPS10065. [Google Scholar]
- 46.Malempati Suman, Chang Bill H., Reid Joel M., Liu Xiaowei, Minard Charles G., Keller Charles, Fox Elizabeth, Weigel Brenda. ADVL1513: Results of a phase 1 trial of entinostat, an oral histone deacetylase inhibitor, in pediatric patients with recurrent or refractory solid tumors. Journal of Clinical Oncology. 2018;36(15_suppl):10556–10556. [Google Scholar]
- 47.Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. The Journal of Clinical Investigation. 1997;99(2):239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoll G, Surdez D, Tirode F, Laud K, Barillot E, Zinovyev A, Delattre O. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. Nucleic Acids Research. 2013;41(19):8853–8871. doi: 10.1093/nar/gkt678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikuchi Ken, Tsuchiya Kunihiko, Otabe Osamu, Gotoh Takahiro, Tamura Shinichi, Katsumi Yoshiki, Yagyu Shigeki, Tsubai-Shimizu Satoko, Miyachi Mitsuru, Iehara Tomoko, Hosoi Hajime. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochemical and Biophysical Research Communications. 2008;365(3):568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, 3rd, Schäfer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(23):13,164–13,169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmody Soni EE, Schlottman S, Erkizan HV, Uren A, Toretsky JA. Loss of SS18-SSX1 inhibits viability and induces apoptosis in synovial sarcoma. Clinical Orthopaedics and Related Research. 2014;472(3):874–882. doi: 10.1007/s11999-013-3065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Wenjuan, Sun Yan, Wang Wei, Han Chunrong, Ouchida Mamoru, Xia Wenbin, Zhao Xiulan, Sun Baocun. The Effect of SYT-SSX and Extracellular Signal-Regulated Kinase (ERK) on Cell Proliferation in Synovial Sarcoma. Pathology & Oncology Research. 2011;17(2):357–367. doi: 10.1007/s12253-010-9334-y. [DOI] [PubMed] [Google Scholar]
- 53.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 54.Naka, N., Takenaka, S., Araki, N., Miwa, T., Hashimoto, N., Yoshioka, K., … Itoh, K. (2010). Synovial sarcoma is a stem cell malignancy. Stem Cells (Dayton, Ohio), 28(7), 1119–1131. 10.1002/stem.452 [DOI] [PubMed]
- 55.Walters Z S, Villarejo-Balcells B, Olmos D, Buist T W S, Missiaglia E, Allen R, Al-Lazikani B, Garrett M D, Blagg J, Shipley J. JARID2 is a direct target of the PAX3-FOXO1 fusion protein and inhibits myogenic differentiation of rhabdomyosarcoma cells. Oncogene. 2013;33(9):1148–1157. doi: 10.1038/onc.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch JT, Spencer GJ, Harris WJ, Maiques-Díaz A, Ciceri F, Huang X, Somervaille TCP. Pharmacological inhibitors of LSD1 promote differentiation of myeloid leukemia cells through a mechanism independent of histone demethylation. Blood. 2014;124(21):267–267. doi: 10.1182/blood.V124.21.267.267. [DOI] [Google Scholar]
- 57.Souza Bárbara Kunzler, da Costa Lopez Patrícia Luciana, Menegotto Pâmela Rossi, Vieira Igor Araujo, Kersting Nathalia, Abujamra Ana Lúcia, Brunetto André T., Brunetto Algemir L., Gregianin Lauro, de Farias Caroline Brunetto, Thiele Carol J., Roesler Rafael. Targeting Histone Deacetylase Activity to Arrest Cell Growth and Promote Neural Differentiation in Ewing Sarcoma. Molecular Neurobiology. 2018;55(9):7242–7258. doi: 10.1007/s12035-018-0874-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warrell Raymond P., Frankel Stanley R., Miller Wilson H., Scheinberg David A., Itri Loretta M., Hittelman Walter N., Vyas Rohini, Andreeff Michael, Tafuri Agostino, Jakubowski Ann, Gabrilove Janice, Gordon Michael S., Dmitrovsky Ethan. Differentiation Therapy of Acute Promyelocytic Leukemia with Tretinoin (All-trans-Retinoic Acid) New England Journal of Medicine. 1991;324(20):1385–1393. doi: 10.1056/NEJM199105163242002. [DOI] [PubMed] [Google Scholar]
- 59.Franzetti G-A, Laud-Duval K, van der Ent W, Brisac A, Irondelle M, Aubert S, Dirksen U, Bouvier C, de Pinieux G, Snaar-Jagalska E, Chavrier P, Delattre O. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene. 2017;36(25):3505–3514. doi: 10.1038/onc.2016.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng, C., Zhao, H., Chen, W., Song, Y., Wang, X., Li, J., … Gao, C. (2016). Identification of SHCBP1 as a novel downstream target gene of SS18-SSX1 and its functional analysis in progression of synovial sarcoma. Oncotarget, 7(41), 66,822–66,834. 10.18632/oncotarget.11651 [DOI] [PMC free article] [PubMed]
- 61.Peng, C., Zhao, H., Song, Y., Chen, W., Wang, X., Liu, X., … Wang, X. (2017). SHCBP1 promotes synovial sarcoma cell metastasis via targeting TGF-β1/Smad signaling pathway and is associated with poor prognosis. Journal of experimental & clinical cancer research: CR, 36(1), 141–141. 10.1186/s13046-017-0616-z [DOI] [PMC free article] [PubMed]
- 62.Liu L, Wang Y-D, Wu J, Cui J, Chen T. Carnitine palmitoyltransferase 1A (CPT1A): a transcriptional target of PAX3-FKHR and mediates PAX3-FKHR-dependent motility in alveolar rhabdomyosarcoma cells. BMC Cancer. 2012;12:154. doi: 10.1186/1471-2407-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang S.-W., Bilke S., Cao L., Murai J., Sousa F. G., Yamade M., Rajapakse V., Varma S., Helman L. J., Khan J., Meltzer P. S., Pommier Y. SLFN11 Is a Transcriptional Target of EWS-FLI1 and a Determinant of Drug Response in Ewing Sarcoma. Clinical Cancer Research. 2015;21(18):4184–4193. doi: 10.1158/1078-0432.CCR-14-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toub Nedjma, Bertrand Jean-Rémi, Tamaddon Ali, Elhamess Hind, Hillaireau Hervé, Maksimenko Andrei, Maccario Jean, Malvy Claude, Fattal Elias, Couvreur Patrick. Efficacy of siRNA Nanocapsules Targeted Against the EWS–Fli1 Oncogene in Ewing Sarcoma. Pharmaceutical Research. 2006;23(5):892–900. doi: 10.1007/s11095-006-9901-9. [DOI] [PubMed] [Google Scholar]
- 65.Takigami Iori, Ohno Takatoshi, Kitade Yukio, Hara Akira, Nagano Akihito, Kawai Gou, Saitou Mitsuru, Matsuhashi Aya, Yamada Kazunari, Shimizu Katsuji. Synthetic siRNA targeting the breakpoint of EWS/Fli-1 inhibits growth of Ewing sarcoma xenografts in a mouse model. International Journal of Cancer. 2010;128(1):216–226. doi: 10.1002/ijc.25564. [DOI] [PubMed] [Google Scholar]
- 66.Rao Donald D., Jay Christopher, Wang Zhaohui, Luo Xiuquan, Kumar Padmasini, Eysenbach Hilary, Ghisoli Maurizio, Senzer Neil, Nemunaitis John. Preclinical Justification of pbi-shRNA EWS/FLI1 Lipoplex (LPX) Treatment for Ewing's Sarcoma. Molecular Therapy. 2016;24(8):1412–1422. doi: 10.1038/mt.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rengaswamy Venkatesh, Zimmer Doris, Süss Regine, Rössler Jochen. RGD liposome-protamine-siRNA (LPR) nanoparticles targeting PAX3-FOXO1 for alveolar rhabdomyosarcoma therapy. Journal of Controlled Release. 2016;235:319–327. doi: 10.1016/j.jconrel.2016.05.063. [DOI] [PubMed] [Google Scholar]
- 68.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering rna inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Research. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 69.Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacology & therapeutics. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 70.Knoop LL, Baker SJ. EWS/FLI alters 5′-splice site selection. Journal of Biological Chemistry. 2001;276(25):22,317–22,322. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- 71.Yang L, Chansky HA, Hickstein DD. EWS·Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. Journal of Biological Chemistry. 2000;275(48):37,612–37,618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 72.Embree LJ, Azuma M, Hickstein DD. Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines. Cancer research. 2009;69(10):4363–4371. doi: 10.1158/0008-5472.CAN-08-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chansky HA, Hu M, Hickstein DD, Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Research. 2001;61(9):3586–3590. [PubMed] [Google Scholar]
- 74.Selvanathan Saravana P., Graham Garrett T., Erkizan Hayriye V., Dirksen Uta, Natarajan Thanemozhi G., Dakic Aleksandra, Yu Songtao, Liu Xuefeng, Paulsen Michelle T., Ljungman Mats E., Wu Cathy H., Lawlor Elizabeth R., Üren Aykut, Toretsky Jeffrey A. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proceedings of the National Academy of Sciences. 2015;112(11):E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang G, Zhou Z, Wang H, Kleinerman ES. CAPER-α alternative splicing regulates the expression of vascular endothelial growth factor165 in Ewing sarcoma cells. Cancer. 2012;118(8):2106–2116. doi: 10.1002/cncr.26488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Z, Reddy K, Guan H, Kleinerman ES. VEGF(165), but not VEGF(189), stimulates vasculogenesis and bone marrow cell migration into Ewing’s sarcoma tumors in vivo. Molecular Cancer Research: MCR. 2007;5(11):1125–1132. doi: 10.1158/1541-7786.MCR-07-0174. [DOI] [PubMed] [Google Scholar]
- 77.Guan H, Zhou Z, Wang H, Jia S-F, Liu W, Kleinerman ES. A small interfering RNA targeting vascular endothelial growth factor inhibits Ewing’s sarcoma growth in a xenograft mouse model. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2005;11(7):2662–2669. doi: 10.1158/1078-0432.CCR-04-1206. [DOI] [PubMed] [Google Scholar]
- 78.Paronetto MP, Bernardis I, Volpe E, Bechara E, Sebestyén E, Eyras E, Valcárcel J. Regulation of FAS exon definition and apoptosis by the Ewing sarcoma protein. Cell Reports. 2014;7(4):1211–1226. doi: 10.1016/j.celrep.2014.03.077. [DOI] [PubMed] [Google Scholar]
- 79.Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(16):6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selvanathan, S. P., Graham, G. T., Grego, A. R., Baker, T. M., Hogg, J. R., Simpson, M., … Toretsky, J. A. (2019). EWS–FLI1 modulated alternative splicing of ARID1A reveals novel oncogenic function through the BAF complex. Nucleic Acids Research, 47(18), 9619–9636. 10.1093/nar/gkz699 [DOI] [PMC free article] [PubMed]
- 81.Perani M, Antonson P, Hamoudi R, Ingram CJE, Cooper CS, Garrett MD, Goodwin GH. The proto-oncoprotein SYT interacts with SYT-interacting protein/co-activator activator (SIP/CoAA), a human nuclear receptor co-activator with similarity to EWS and TLS/FUS family of proteins. The Journal of Biological Chemistry. 2005;280(52):42,863–42,876. doi: 10.1074/jbc.M502963200. [DOI] [PubMed] [Google Scholar]
- 82.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 83.Amstutz R., Wachtel M., Troxler H., Kleinert P., Ebauer M., Haneke T., Oehler-Janne C., Fabbro D., Niggli F. K., Schafer B. W. Phosphorylation Regulates Transcriptional Activity of PAX3/FKHR and Reveals Novel Therapeutic Possibilities. Cancer Research. 2008;68(10):3767–3776. doi: 10.1158/0008-5472.CAN-07-2447. [DOI] [PubMed] [Google Scholar]
- 84.Hecker RM, Amstutz RA, Wachtel M, Walter D, Niggli FK, Schäfer BW. p21 Downregulation is an important component of PAX3/FKHR oncogenicity and its reactivation by HDAC inhibitors enhances combination treatment. Oncogene. 2010;29(27):3942–3952. doi: 10.1038/onc.2010.145. [DOI] [PubMed] [Google Scholar]
- 85.Thalhammer V, Lopez-Garcia LA, Herrero-Martin D, Hecker R, Laubscher D, Gierisch ME, et al. PLK1 phosphorylates PAX3-FOXO1, the inhibition of which triggers regression of alveolar rhabdomyosarcoma. Cancer Research. 2015;75(1):98–110. doi: 10.1158/0008-5472.CAN-14-1246. [DOI] [PubMed] [Google Scholar]
- 86.Abbou, S., Lanvers-Kaminsky, C., Daudigeos-Dubus, E., Dret, L. L., Laplace-Builhe, C., Molenaar, J., … Committee, within the I. B. and P. E. (2016). Polo-like kinase inhibitor volasertib exhibits antitumor activity and synergy with vincristine in pediatric malignancies. Anticancer Research, 36(2), 599–609. [PubMed]
- 87.Zeng F-Y, Dong H, Cui J, Liu L, Chen T. Glycogen synthase kinase 3 regulates PAX3–FKHR-mediated cell proliferation in human alveolar rhabdomyosarcoma cells. Biochemical and Biophysical Research Communications. 2010;391(1):1049–1055. doi: 10.1016/j.bbrc.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loupe JM, Miller PJ, Ruffin DR, Stark MW, Hollenbach AD. Inhibiting phosphorylation of the oncogenic PAX3-FOXO1 reduces alveolar rhabdomyosarcoma phenotypes identifying novel therapy options. Oncogenesis. 2015;4(3):e145–e145. doi: 10.1038/oncsis.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L, Wu J, Ong SS, Chen T. Cyclin-dependent kinase 4 phosphorylates and positively regulates PAX3-FOXO1 in human alveolar rhabdomyosarcoma cells. PLOS ONE. 2013;8(2):e58193. doi: 10.1371/journal.pone.0058193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jothi M, Nishijo K, Keller C, Mal AK. AKT and PAX3-FKHR cooperation enforces myogenic differentiation blockade in alveolar rhabdomyosarcoma cell. Cell Cycle. 2012;11(5):895–908. doi: 10.4161/cc.11.5.19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jothi M, Mal M, Keller C, Mal AK. Small molecule inhibition of PAX3-FOXO1 through AKT activation suppresses malignant phenotypes of alveolar rhabdomyosarcoma. Molecular Cancer Therapeutics. 2013;12(12):2663–2674. doi: 10.1158/1535-7163.MCT-13-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olsen RJ, Hinrichs SH. Phosphorylation of the EWS IQ domain regulates transcriptional activity of the EWS/ATF1 and EWS/FLI1 fusion proteins. Oncogene. 2001;20(14):1756–1764. doi: 10.1038/sj.onc.1204268. [DOI] [PubMed] [Google Scholar]
- 93.Surdez D., Benetkiewicz M., Perrin V., Han Z.-Y., Pierron G., Ballet S., Lamoureux F., Redini F., Decouvelaere A.-V., Daudigeos-Dubus E., Geoerger B., de Pinieux G., Delattre O., Tirode F. Targeting the EWSR1-FLI1 Oncogene-Induced Protein Kinase PKC- Abolishes Ewing Sarcoma Growth. Cancer Research. 2012;72(17):4494–4503. doi: 10.1158/0008-5472.CAN-12-0371. [DOI] [PubMed] [Google Scholar]
- 94.Caropreso V, Darvishi E, Turbyville TJ, Ratnayake R, Grohar PJ, McMahon JB, Woldemichael GM. Englerin A inhibits EWS-FLI1 DNA binding in Ewing sarcoma cells. Journal of Biological Chemistry. 2016;291(19):10,058–10,066. doi: 10.1074/jbc.M115.701375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klevernic IV, Morton S, Davis RJ, Cohen P. Phosphorylation of Ewing’s sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochemical Journal. 2009;418(3):625–634. doi: 10.1042/BJ20082097. [DOI] [PubMed] [Google Scholar]
- 96.Schlottmann, S. P., Erkizan, H. V. P., Barber-Rotenberg, J. S. P., Knights, C. P., Cheema, A. P., Üren, A. M., et al. (2012). Acetylation increases EWS-FLI1 DNA binding and transcriptional activity. Frontiers in Oncology, 2. 10.3389/fonc.2012.00107. [DOI] [PMC free article] [PubMed]
- 97.Araya N, Hiraga H, Kako K, Arao Y, Kato S, Fukamizu A. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochemical and Biophysical Research Communications. 2005;329(2):653–660. doi: 10.1016/j.bbrc.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 98.Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins: Structure, Function, and Bioinformatics. 2008;72(4):1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- 99.Bachmaier R, Aryee DNT, Jug G, Kauer M, Kreppel M, Lee KA, Kovar H. O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing’s sarcoma. Oncogene. 2009;28(9):1280–1284. doi: 10.1038/onc.2008.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gierisch ME, Pfistner F, Lopez-Garcia LA, Harder L, Schäfer BW, Niggli FK. Proteasomal degradation of the EWS-FLI1 fusion protein is regulated by a single lysine residue. Journal of Biological Chemistry. 2016;291(52):26,922–26,933. doi: 10.1074/jbc.M116.752063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gierisch ME, Pedot G, Walser F, Lopez-Garcia LA, Jaaks P, Niggli FK, Schäfer BW. USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth. Scientific Reports. 2019;9(1):1–12. doi: 10.1038/s41598-018-37264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ambati Srikanth R., Lopes Eloisi Caldas, Kosugi Kohji, Mony Ullas, Zehir Ahmet, Shah Smit K., Taldone Tony, Moreira Andre L., Meyers Paul A., Chiosis Gabriela, Moore Malcolm A.S. Pre-clinical efficacy of PU-H71, a novel HSP90 inhibitor, alone and in combination with bortezomib in Ewing sarcoma. Molecular Oncology. 2013;8(2):323–336. doi: 10.1016/j.molonc.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma Yan, Baltezor Michael, Rajewski Lian, Crow Jennifer, Samuel Glenson, Staggs Vincent S., Chastain Katherine M., Toretsky Jeffrey A., Weir Scott J., Godwin Andrew K. Targeted inhibition of histone deacetylase leads to suppression of Ewing sarcoma tumor growth through an unappreciated EWS-FLI1/HDAC3/HSP90 signaling axis. Journal of Molecular Medicine. 2019;97(7):957–972. doi: 10.1007/s00109-019-01782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel Nikita, Wang Juehong, Shiozawa Kumiko, Jones Kevin B., Zhang Yanfeng, Prokop Jeremy W., Davenport George G., Nihira Naoe T., Hao Zhenyue, Wong Derek, Brandsmeier Laurel, Meadows Sarah K., Sampaio Arthur V., Werff Ryan Vander, Endo Makoto, Capecchi Mario R., McNagny Kelly M., Mak Tak W., Nielsen Torsten O., Underhill T. Michael, Myers Richard M., Kondo Tadashi, Su Le. HDAC2 Regulates Site-Specific Acetylation of MDM2 and Its Ubiquitination Signaling in Tumor Suppression. iScience. 2019;13:43–54. doi: 10.1016/j.isci.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shmueli A, Oren M. Life, death, and ubiquitin: taming the mule. Cell. 2005;121(7):963–965. doi: 10.1016/j.cell.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 106.Kao, S.-H., Wu, H.-T., & Wu, K.-J. (2018). Ubiquitination by HUWE1 in tumorigenesis and beyond. Journal of Biomedical Science, 25(1). 10.1186/s12929-018-0470-0. [DOI] [PMC free article] [PubMed]
- 107.Laporte Aimée N., Poulin Neal M., Barrott Jared J., Wang Xiu Qing, Lorzadeh Alireza, Vander Werff Ryan, Jones Kevin B., Underhill T. Michael, Nielsen Torsten O. Death by HDAC Inhibition in Synovial Sarcoma Cells. Molecular Cancer Therapeutics. 2017;16(12):2656–2667. doi: 10.1158/1535-7163.MCT-17-0397. [DOI] [PubMed] [Google Scholar]
- 108.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory pathways in immunotherapy for cancer. Annual Review of Immunology. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 109.Jackson HJ, Rafiq S, Brentjens RJ. Driving CAR T cells forward. Nature Reviews. Clinical Oncology. 2016;13(6):370–383. doi: 10.1038/nrclinonc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS. Alternative tumour-specific antigens. Nature Reviews Cancer. 2019;19(8):465–478. doi: 10.1038/s41568-019-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Wei, Lee Ken-Wing, Srivastava Raghvendra M., Kuo Fengshen, Krishna Chirag, Chowell Diego, Makarov Vladimir, Hoen Douglas, Dalin Martin G., Wexler Leonard, Ghossein Ronald, Katabi Nora, Nadeem Zaineb, Cohen Marc A., Tian S. Ken, Robine Nicolas, Arora Kanika, Geiger Heather, Agius Phaedra, Bouvier Nancy, Huberman Kety, Vanness Katelynd, Havel Jonathan J., Sims Jennifer S., Samstein Robert M., Mandal Rajarsi, Tepe Justin, Ganly Ian, Ho Alan L., Riaz Nadeem, Wong Richard J., Shukla Neerav, Chan Timothy A., Morris Luc G. T. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nature Medicine. 2019;25(5):767–775. doi: 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chang, T.-C., Carter, R. A., Li, Y., Li, Y., Wang, H., Edmonson, M. N., … Zhang, J. (2017). The neoepitope landscape in pediatric cancers. Genome Medicine, 9(1), 78. 10.1186/s13073-017-0468-3 [DOI] [PMC free article] [PubMed]
- 113.Worley, B. S., Broeke, L. T. van den, Goletz, T. J., Pendleton, C. D., Daschbach, E. M., Thomas, E. K., … Berzofsky, J. A. (2001). Antigenicity of fusion proteins from sarcoma-associated chromosomal translocations. Cancer Research, 61(18), 6868–6875. [PubMed]
- 114.Evans C. H., Liu F., Porter R. M., O'Sullivan R. P., Merghoub T., Lunsford E. P., Robichaud K., Van Valen F., Lessnick S. L., Gebhardt M. C., Wells J. W. EWS-FLI-1-Targeted Cytotoxic T-cell Killing of Multiple Tumor Types Belonging to the Ewing Sarcoma Family of Tumors. Clinical Cancer Research. 2012;18(19):5341–5351. doi: 10.1158/1078-0432.CCR-12-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meyer-Wentrup F, Richter G, Burdach S. Identification of an immunogenic EWS-FLI1-derived HLA-DR-restricted T helper cell epitope. Pediatric Hematology and Oncology. 2005;22(4):297–308. doi: 10.1080/08880010590935194. [DOI] [PubMed] [Google Scholar]
- 116.van den Broeke LT, Pendleton CD, Mackall C, Helman LJ, Berzofsky JA. Identification and epitope enhancement of a PAX-FKHR fusion protein breakpoint epitope in alveolar rhabdomyosarcoma cells created by a tumorigenic chromosomal translocation inducing CTL capable of lysing human tumors. Cancer Research. 2006;66(3):1818–1823. doi: 10.1158/0008-5472.CAN-05-2549. [DOI] [PubMed] [Google Scholar]
- 117.Lack of effective T -lymphocyte response to the PAX3/FKHR translocation area in alveolar rhabdomyosarcoma. (n.d.). springermedizin.de. Retrieved October 23, 2019, from https://www.springermedizin.de/lack-of-effective-t-lymphocyte-response-to-the-pax3-fkhr-translo/8229164.
- 118.Toretsky Jeffrey A., Erkizan Verda, Levenson Amy, Abaan Ogan D., Parvin Jeffrey D., Cripe Timothy P., Rice Anna M., Lee Sean Bong, Üren Aykut. Oncoprotein EWS-FLI1 Activity Is Enhanced by RNA Helicase A. Cancer Research. 2006;66(11):5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 119.Erkizan Hayriye Verda, Schneider Jeffrey A., Sajwan Kamal, Graham Garrett T., Griffin Brittany, Chasovskikh Sergey, Youbi Sarah E., Kallarakal Abraham, Chruszcz Maksymilian, Padmanabhan Radhakrishnan, Casey John L., Üren Aykut, Toretsky Jeffrey A. RNA helicase A activity is inhibited by oncogenic transcription factor EWS-FLI1. Nucleic Acids Research. 2015;43(2):1069–1080. doi: 10.1093/nar/gku1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Erkizan Hayriye V, Kong Yali, Merchant Melinda, Schlottmann Silke, Barber-Rotenberg Julie S, Yuan Linshan, Abaan Ogan D, Chou Tsu-hang, Dakshanamurthy Sivanesan, Brown Milton L, Üren Aykut, Toretsky Jeffrey A. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nature Medicine. 2009;15(7):750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Yan, Tanaka Kazuhiro, Fan Xueli, Nakatani Fumihiko, Li Xu, Nakamura Tomoyuki, Takasaki Minoru, Yamamoto Shunsaku, Iwamoto Yukihide. Inhibition of the transcriptional function of p53 by EWS–Fli1 chimeric protein in Ewing Family Tumors. Cancer Letters. 2010;294(1):57–65. doi: 10.1016/j.canlet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 122.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 123.Maki K, Mitani K, Yamagata T, Kurokawa M, Kanda Y, Yazaki Y, Hirai H. Transcriptional inhibition of p53 by the MLL/MEN chimeric protein found in myeloid leukemia. Blood. 1999;93(10):3216–3224. doi: 10.11501/3163049. [DOI] [PubMed] [Google Scholar]
- 124.Li Y, Li X, Fan G, Fukushi J-I, Matsumoto Y, Iwamoto Y, Zhu Y. Impairment of p53 acetylation by EWS-Fli1 chimeric protein in Ewing family tumors. Cancer Letters. 2012;320(1):14–22. doi: 10.1016/j.canlet.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 125.Federman Noah, Meyers Paul A., Daw Najat C., Toretsky Jeffrey, Breitmeyer James Bradley, Singh Arun S., Miller Langdon L., Oltersdorf Tilman, Jezior Deborah, Jessen Katayoun A., Lannutti Brian, Ludwig Joseph A. A phase I, first-in-human, dose escalation study of intravenous TK216 in patients with relapsed or refractory Ewing sarcoma. Journal of Clinical Oncology. 2017;35(15_suppl):TPS11626–TPS11626. [Google Scholar]
- 126.D’Arcy P, Maruwge W, Ryan BA, Brodin B. The oncoprotein SS18-SSX1 promotes p53 ubiquitination and degradation by enhancing HDM2 stability. Molecular Cancer Research: MCR. 2008;6(1):127–138. doi: 10.1158/1541-7786.MCR-07-0176. [DOI] [PubMed] [Google Scholar]
- 127.Michel Brittany C., D’Avino Andrew R., Cassel Seth H., Mashtalir Nazar, McKenzie Zachary M., McBride Matthew J., Valencia Alfredo M., Zhou Qianhe, Bocker Michael, Soares Luis M. M., Pan Joshua, Remillard David I., Lareau Caleb A., Zullow Hayley J., Fortoul Nora, Gray Nathanael S., Bradner James E., Chan Ho Man, Kadoch Cigall. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nature Cell Biology. 2018;20(12):1410–1420. doi: 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Harlow Matt L., Maloney Nichole, Roland Joseph, Guillen Navarro Maria Jose, Easton Matthew K., Kitchen-Goosen Susan M., Boguslawski Elissa A., Madaj Zachary B., Johnson Ben K., Bowman Megan J., D'Incalci Maurizio, Winn Mary E., Turner Lisa, Hostetter Galen, Galmarini Carlos María, Aviles Pablo M., Grohar Patrick J. Lurbinectedin Inactivates the Ewing Sarcoma Oncoprotein EWS-FLI1 by Redistributing It within the Nucleus. Cancer Research. 2016;76(22):6657–6668. doi: 10.1158/0008-5472.CAN-16-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harlow Matt L., Chasse Maggie H., Boguslawski Elissa A., Sorensen Katie M., Gedminas Jenna M., Kitchen-Goosen Susan M., Rothbart Scott B., Taslim Cenny, Lessnick Stephen L., Peck Anderson S., Madaj Zachary B., Bowman Megan J., Grohar Patrick J. Trabectedin Inhibits EWS-FLI1 and Evicts SWI/SNF from Chromatin in a Schedule-dependent Manner. Clinical Cancer Research. 2019;25(11):3417–3429. doi: 10.1158/1078-0432.CCR-18-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ordóñez, J. L., Amaral, A. T., Carcaboso, A. M., Herrero-Martín, D., García-Macías, M. del C, Sevillano, V., … de Álava, E. (2015). The PARP inhibitor olaparib enhances the sensitivity of Ewing sarcoma to trabectedin. Oncotarget, 6(22), 18,875–18,890. 10.18632/oncotarget.4303 [DOI] [PMC free article] [PubMed]
- 131.Subbiah Vivek, Sankhala Kamalesh Kumar, Ratan Ravin, Sanz Garcia Enrique, Boni Valentina, Gil Thierry, Villalobos Victor Manuel, Chawla Sant P, Lardelli Pilar, Siguero Mariano, Kahatt Carmen Maria, Soto-Matos Arturo, Ferrari Stefano. Efficacy and safety of lurbinectedin (PM1183) in Ewing sarcoma: Final results from a phase 2 study. Journal of Clinical Oncology. 2018;36(15_suppl):11519–11519. [Google Scholar]
- 132.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 133.Sonnemann Jürgen, Dreyer Linn, Hartwig Maite, Palani Chithra D., Hong Le Thi Thu, Klier Ulrike, Bröker Barbara, Völker Uwe, Beck James F. Histone deacetylase inhibitors induce cell death and enhance the apoptosis-inducing activity of TRAIL in Ewing’s sarcoma cells. Journal of Cancer Research and Clinical Oncology. 2007;133(11):847–858. doi: 10.1007/s00432-007-0227-8. [DOI] [PubMed] [Google Scholar]
- 134.Ito Tatsuo, Ouchida Mamoru, Morimoto Yuki, Yoshida Aki, Jitsumori Yoshimi, Ozaki Toshifumi, Sonobe Hiroshi, Inoue Hajime, Shimizu Kenji. Significant growth suppression of synovial sarcomas by the histone deacetylase inhibitor FK228 in vitro and in vivo. Cancer Letters. 2005;224(2):311–319. doi: 10.1016/j.canlet.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 135.Lubieniecka J. M., de Bruijn D. R.H., Su L., van Dijk A. H.A., Subramanian S., van de Rijn M., Poulin N., van Kessel A. G., Nielsen T. O. Histone Deacetylase Inhibitors Reverse SS18-SSX-Mediated Polycomb Silencing of the Tumor Suppressor Early Growth Response 1 in Synovial Sarcoma. Cancer Research. 2008;68(11):4303–4310. doi: 10.1158/0008-5472.CAN-08-0092. [DOI] [PubMed] [Google Scholar]
- 136.Kutko, M. C., Glick, R. D., Butler, L. M., Coffey, D. C., Rifkind, R. A., Marks, P. A., … LaQuaglia, M. P. (2003). Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clinical Cancer Research, 9(15), 5749–5755. [PubMed]
- 137.Kurmasheva, R. T., Bandyopadhyay, A., Favours, E., Del Pozo, V., Ghilu, S., Phelps, D. A., … Houghton, P. J. (2019). Evaluation of entinostat alone and in combination with standard-of-care cytotoxic agents against rhabdomyosarcoma xenograft models. Pediatric Blood & Cancer, 66(8). 10.1002/pbc.27820 [DOI] [PMC free article] [PubMed]
- 138.Abraham J., Nunez-Alvarez Y., Hettmer S., Carrio E., Chen H.-I. H., Nishijo K., Huang E. T., Prajapati S. I., Walker R. L., Davis S., Rebeles J., Wiebush H., McCleish A. T., Hampton S. T., Bjornson C. R. R., Brack A. S., Wagers A. J., Rando T. A., Capecchi M. R., Marini F. C., Ehler B. R., Zarzabal L. A., Goros M. W., Michalek J. E., Meltzer P. S., Langenau D. M., LeGallo R. D., Mansoor A., Chen Y., Suelves M., Rubin B. P., Keller C. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes & Development. 2014;28(14):1578–1591. doi: 10.1101/gad.238733.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.van Maldegem AM, Bovée JVMG, Gelderblom H. Panobinostat-A potential treatment for metastasized Ewing sarcoma? A case report: panobinostat for metastasized Ewing sarcoma. Pediatric Blood & Cancer. 2016;63(10):1840–1843. doi: 10.1002/pbc.26077. [DOI] [PubMed] [Google Scholar]
- 140.Gore L., Holden S. N., Basche M., Raj S. K. S., Arnold I., O'Bryant C., Witta S., Rohde B., McCoy C., Eckhardt S. G. Updated results from a phase I trial of the histone deacetylase (HDAC) inhibitor MS-275 in patients with refractory solid tumors. Journal of Clinical Oncology. 2004;22(14_suppl):3026–3026. [Google Scholar]
- 141.Cassier P A, Lefranc A, Y Amela E, Chevreau C, Bui B N, Lecesne A, Ray-Coquard I, Chabaud S, Penel N, Berge Y, Dômont J, Italiano A, Duffaud F, Cadore A-C, Polivka V, Blay J-Y. A phase II trial of panobinostat in patients with advanced pretreated soft tissue sarcoma. A study from the French Sarcoma Group. British Journal of Cancer. 2013;109(4):909–914. doi: 10.1038/bjc.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chu Q. S.-C., Nielsen T.O., Alcindor T., Gupta A., Endo M., Goytain A., Xu H., Verma S., Tozer R., Knowling M., Bramwell V.B., Powers J., Seymour L.K., Eisenhauer E.A. A phase II study of SB939, a novel pan-histone deacetylase inhibitor, in patients with translocation-associated recurrent/metastatic sarcomas—NCIC-CTG IND 200. Annals of Oncology. 2015;26(5):973–981. doi: 10.1093/annonc/mdv033. [DOI] [PubMed] [Google Scholar]
- 143.Schmitt Thomas, Mayer-Steinacker Regine, Mayer Frank, Grünwald Viktor, Schütte Jochen, Hartmann Jörg T., Kasper Bernd, Hüsing Johannes, Hajda Jacek, Ottawa Gregor, Mechtersheimer Gunhild, Mikus Gerd, Burhenne Jürgen, Lehmann Lorenz, Heilig Christoph E., Ho Anthony D., Egerer Gerlinde. Vorinostat in refractory soft tissue sarcomas – Results of a multi-centre phase II trial of the German Soft Tissue Sarcoma and Bone Tumour Working Group (AIO) European Journal of Cancer. 2016;64:74–82. doi: 10.1016/j.ejca.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 144.Sankar S., Theisen E. R., Bearss J., Mulvihill T., Hoffman L. M., Sorna V., Beckerle M. C., Sharma S., Lessnick S. L. Reversible LSD1 Inhibition Interferes with Global EWS/ETS Transcriptional Activity and Impedes Ewing Sarcoma Tumor Growth. Clinical Cancer Research. 2014;20(17):4584–4597. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.García-Domínguez, D. J., Hontecillas-Prieto, L., Rodríguez-Núñez, P., Pascual-Pasto, G., Vila-Ubach, M., García-Mejías, R., … De, E. Á. (2018). The combination of epigenetic drugs SAHA and HCI-2509 synergistically inhibits EWS-FLI1 and tumor growth in Ewing sarcoma. Oncotarget, 9(59), 31,397–31,410. 10.18632/oncotarget.25829 [DOI] [PMC free article] [PubMed]
- 146.Welch D, Kahen E, Fridley B, Brohl AS, Cubitt CL, Reed DR. Small molecule inhibition of lysine-specific demethylase 1 (LSD1) and histone deacetylase (HDAC) alone and in combination in Ewing sarcoma cell lines. PLOS ONE. 2019;14(9):e0222228. doi: 10.1371/journal.pone.0222228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Viré Emmanuelle, Brenner Carmen, Deplus Rachel, Blanchon Loïc, Fraga Mario, Didelot Céline, Morey Lluis, Van Eynde Aleyde, Bernard David, Vanderwinden Jean-Marie, Bollen Mathieu, Esteller Manel, Di Croce Luciano, de Launoit Yvan, Fuks François. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2005;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 148.Ciarapica R, De Salvo M, Carcarino E, Bracaglia G, Adesso L, Leoncini P P, Dall'Agnese A, Walters Z S, Verginelli F, De Sio L, Boldrini R, Inserra A, Bisogno G, Rosolen A, Alaggio R, Ferrari A, Collini P, Locatelli M, Stifani S, Screpanti I, Rutella S, Yu Q, Marquez V E, Shipley J, Valente S, Mai A, Miele L, Puri P L, Locatelli F, Palacios D, Rota R. The Polycomb group (PcG) protein EZH2 supports the survival of PAX3-FOXO1 alveolar rhabdomyosarcoma by repressing FBXO32 (Atrogin1/MAFbx) Oncogene. 2013;33(32):4173–4184. doi: 10.1038/onc.2013.471. [DOI] [PubMed] [Google Scholar]
- 149.Richter G. H. S., Plehm S., Fasan A., Rossler S., Unland R., Bennani-Baiti I. M., Hotfilder M., Lowel D., von Luettichau I., Mossbrugger I., Quintanilla-Martinez L., Kovar H., Staege M. S., Muller-Tidow C., Burdach S. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proceedings of the National Academy of Sciences. 2009;106(13):5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kurmasheva Raushan T., Sammons Melissa, Favours Edward, Wu Jianwrong, Kurmashev Dias, Cosmopoulos Katherine, Keilhack Heike, Klaus Christine R., Houghton Peter J., Smith Malcolm A. Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the Pediatric Preclinical Testing Program. Pediatric Blood & Cancer. 2016;64(3):e26218. doi: 10.1002/pbc.26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shen JK, Cote GM, Gao Y, Choy E, Mankin HJ, Hornicek FJ, Duan Z. Targeting EZH2-mediated methylation of H3K27 inhibits proliferation and migration of Synovial Sarcoma in vitro. Scientific Reports. 2016;6:25,239. doi: 10.1038/srep25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.(n.d.). Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with synovial sarcoma (NCT02601950). Journal of Clinical Oncology. Retrieved October 27, 2019, from https://ascopubs.org/doi/abs/10.1200/JCO.2017.35.15_suppl.11057.
- 153.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Molecular biology of the cell. 2009;20(23):4899–4909. doi: 10.1091/mbc.e09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Filippakopoulos Panagis, Qi Jun, Picaud Sarah, Shen Yao, Smith William B., Fedorov Oleg, Morse Elizabeth M., Keates Tracey, Hickman Tyler T., Felletar Ildiko, Philpott Martin, Munro Shonagh, McKeown Michael R., Wang Yuchuan, Christie Amanda L., West Nathan, Cameron Michael J., Schwartz Brian, Heightman Tom D., La Thangue Nicholas, French Christopher A., Wiest Olaf, Kung Andrew L., Knapp Stefan, Bradner James E. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lovén Jakob, Hoke Heather A., Lin Charles Y., Lau Ashley, Orlando David A., Vakoc Christopher R., Bradner James E., Lee Tong Ihn, Young Richard A. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gollavilli Paradesi Naidu, Pawar Aishwarya, Wilder-Romans Kari, Natesan Ramakrishnan, Engelke Carl G., Dommeti Vijaya L., Krishnamurthy Pranathi M., Nallasivam Archana, Apel Ingrid J., Xu Tianlei, Qin Zhaohui S., Feng Felix Y., Asangani Irfan A. EWS/ETS-Driven Ewing Sarcoma Requires BET Bromodomain Proteins. Cancer Research. 2018;78(16):4760–4773. doi: 10.1158/0008-5472.CAN-18-0484. [DOI] [PubMed] [Google Scholar]
- 157.Bid H. K., Phelps D. A., Xaio L., Guttridge D. C., Lin J., London C., Baker L. H., Mo X., Houghton P. J. The Bromodomain BET Inhibitor JQ1 Suppresses Tumor Angiogenesis in Models of Childhood Sarcoma. Molecular Cancer Therapeutics. 2016;15(5):1018–1028. doi: 10.1158/1535-7163.MCT-15-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brien, G. L., Remillard, D., Shi, J., Hemming, M. L., Chabon, J., Wynne, K., … Armstrong, S. A. (2018). Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. eLife, 7, e41305. 10.7554/eLife.41305 [DOI] [PMC free article] [PubMed]
- 159.Werner, H., Meisel-Sharon, S., & Bruchim, I. (2018). Oncogenic fusion proteins adopt the insulin-like growth factor signaling pathway. Molecular Cancer, 17(1). 10.1186/s12943-018-0807-z. [DOI] [PMC free article] [PubMed]
- 160.Yee D, Favoni RE, Lebovic GS, Lombana F, Powell DR, Reynolds CP, Rosen N. Insulin-like growth factor I expression by tumors of neuroectodermal origin with the t(11;22) chromosomal translocation. A potential autocrine growth factor. The Journal of Clinical Investigation. 1990;86(6):1806–1814. doi: 10.1172/JCI114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guenther Lillian M., Dharia Neekesh V., Ross Linda, Conway Amy, Robichaud Amanda L., Catlett Jerrel L., Wechsler Caroline S., Frank Elizabeth S., Goodale Amy, Church Alanna J., Tseng Yuen-Yi, Guha Rajarshi, McKnight Crystal G., Janeway Katherine A., Boehm Jesse S., Mora Jaume, Davis Mindy I., Alexe Gabriela, Piccioni Federica, Stegmaier Kimberly. A Combination CDK4/6 and IGF1R Inhibitor Strategy for Ewing Sarcoma. Clinical Cancer Research. 2018;25(4):1343–1357. doi: 10.1158/1078-0432.CCR-18-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scotlandi, K., Benini, S., Nanni, P., Lollini, P. L., Nicoletti, G., Landuzzi, L., … Baldini, N. (1998). Blockage of insulin-like growth factor I receptor inhibits the growth of Ewing’s sarcoma in athymic mice. Cancer Research, 58(18), 4127–4131. [PubMed]
- 163.Sun Y, Gao D, Liu Y, Huang J, Lessnick S, Tanaka S. IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene. 2006;25(7):1042–1052. doi: 10.1038/sj.onc.1209143. [DOI] [PubMed] [Google Scholar]
- 164.De Giovanni C, Nanni P, Landuzzi L, Ianzano ML, Nicoletti G, Croci S, et al. Immune targeting of autocrine IGF2 hampers rhabdomyosarcoma growth and metastasis. BMC cancer. 2019;19(1):126. doi: 10.1186/s12885-019-5339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Törnkvist Maria, Natalishvili Natalia, Xie Yuntao, Girnita Ada, D’Arcy Pádraig, Brodin Bertha, Axelson Magnus, Girnita Leonard. Differential roles of SS18–SSX fusion gene and insulin-like growth factor-1 receptor in synovial sarcoma cell growth. Biochemical and Biophysical Research Communications. 2008;368(3):793–800. doi: 10.1016/j.bbrc.2008.01.162. [DOI] [PubMed] [Google Scholar]
- 166.Cao L., Yu Y., Bilke S., Walker R. L., Mayeenuddin L. H., Azorsa D. O., Yang F., Pineda M., Helman L. J., Meltzer P. S. Genome-Wide Identification of PAX3-FKHR Binding Sites in Rhabdomyosarcoma Reveals Candidate Target Genes Important for Development and Cancer. Cancer Research. 2010;70(16):6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dancsok, A. R., Asleh-Aburaya, K., & Nielsen, T. O. (2017). Advances in sarcoma diagnostics and treatment. Oncotarget, 8(4), 7068–7093. 10.18632/oncotarget.12548 [DOI] [PMC free article] [PubMed]
- 168.Pappo Alberto S., Vassal Gilles, Crowley John J., Bolejack Vanessa, Hogendoorn Pancras C. W., Chugh Rashmi, Ladanyi Marc, Grippo Joseph F., Dall Georgina, Staddon Arthur P., Chawla Sant P., Maki Robert G., Araujo Dejka M., Geoerger Birgit, Ganjoo Kristen, Marina Neyssa, Blay Jean-Yves, Schuetze Scott M., Chow Warren A., Helman Lee J. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: Results of a Sarcoma Alliance. Cancer. 2014;120(16):2448–2456. doi: 10.1002/cncr.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kang Z, Yu Y, Zhu Y J, Davis S, Walker R, Meltzer P S, Helman L J, Cao L. Downregulation of IGFBP2 is associated with resistance to IGF1R therapy in rhabdomyosarcoma. Oncogene. 2013;33(50):5697–5705. doi: 10.1038/onc.2013.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cassinelli Giuliana, Dal Bo Laura, Favini Enrica, Cominetti Denis, Pozzi Sabina, Tortoreto Monica, De Cesare Michelandrea, Lecis Daniele, Scanziani Eugenio, Minoli Lucia, Naggi Annamaria, Vlodavsky Israel, Zaffaroni Nadia, Lanzi Cinzia. Supersulfated low-molecular weight heparin synergizes with IGF1R/IR inhibitor to suppress synovial sarcoma growth and metastases. Cancer Letters. 2018;415:187–197. doi: 10.1016/j.canlet.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 171.Loganathan SN, Tang N, Holler AE, Wang N, Wang J. Targeting the IGF1R/PI3K/AKT pathway sensitizes Ewing sarcoma to BET bromodomain inhibitors. Molecular Cancer Therapeutics. 2019;18(5):929–936. doi: 10.1158/1535-7163.MCT-18-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wan X, Yeung C, Heske C, Mendoza A, Helman LJ. IGF-1R inhibition activates a YES/SFK bypass resistance pathway: rational basis for co-targeting IGF-1R and Yes/SFK kinase in rhabdomyosarcoma. Neoplasia (New York, N.Y.) 2015;17(4):358–366. doi: 10.1016/j.neo.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Gaal J. Carlijn, Roeffen Melissa H.S., Flucke Uta E., van der Laak Jeroen A.W.M., van der Heijden Gwen, de Bont Eveline S.J.M., Suurmeijer Albert J.H., Versleijen-Jonkers Yvonne M.H., van der Graaf Winette T.A. Simultaneous targeting of insulin-like growth factor-1 receptor and anaplastic lymphoma kinase in embryonal and alveolar rhabdomyosarcoma: A rational choice. European Journal of Cancer. 2013;49(16):3462–3470. doi: 10.1016/j.ejca.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 174.Dolgikh Nadezda, Fulda Simone. Rhabdomyosarcoma cells are susceptible to cell death by LDK378 alone or in combination with sorafenib independently of anaplastic lymphoma kinase status. Anti-Cancer Drugs. 2017;28(10):1118–1125. doi: 10.1097/CAD.0000000000000552. [DOI] [PubMed] [Google Scholar]
- 175.Baird Kristin, Davis Sean, Antonescu Cristina R., Harper Ursula L., Walker Robert L., Chen Yidong, Glatfelter Arthur A., Duray Paul H., Meltzer Paul S. Gene Expression Profiling of Human Sarcomas: Insights into Sarcoma Biology. Cancer Research. 2005;65(20):9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 176.Crose LES, Etheridge KT, Chen C, Belyea B, Talbot LJ, Bentley RC, Linardic CM. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18(14):3780–3790. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.(n.d.). Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. - PubMed - NCBI. Retrieved October 28, 2019, from https://www.ncbi.nlm.nih.gov/pubmed/27487136
- 178.Davies Helen, Bignell Graham R., Cox Charles, Stephens Philip, Edkins Sarah, Clegg Sheila, Teague Jon, Woffendin Hayley, Garnett Mathew J., Bottomley William, Davis Neil, Dicks Ed, Ewing Rebecca, Floyd Yvonne, Gray Kristian, Hall Sarah, Hawes Rachel, Hughes Jaime, Kosmidou Vivian, Menzies Andrew, Mould Catherine, Parker Adrian, Stevens Claire, Watt Stephen, Hooper Steven, Wilson Rebecca, Jayatilake Hiran, Gusterson Barry A., Cooper Colin, Shipley Janet, Hargrave Darren, Pritchard-Jones Katherine, Maitland Norman, Chenevix-Trench Georgia, Riggins Gregory J., Bigner Darell D., Palmieri Giuseppe, Cossu Antonio, Flanagan Adrienne, Nicholson Andrew, Ho Judy W. C., Leung Suet Y., Yuen Siu T., Weber Barbara L., Seigler Hilliard F., Darrow Timothy L., Paterson Hugh, Marais Richard, Marshall Christopher J., Wooster Richard, Stratton Michael R., Futreal P. Andrew. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 179.Cidre-Aranaz F, Grünewald T G P, Surdez D, García-García L, Carlos Lázaro J, Kirchner T, González-González L, Sastre A, García-Miguel P, López-Pérez S E, Monzón S, Delattre O, Alonso J. EWS-FLI1-mediated suppression of the RAS-antagonist Sprouty 1 (SPRY1) confers aggressiveness to Ewing sarcoma. Oncogene. 2016;36(6):766–776. doi: 10.1038/onc.2016.244. [DOI] [PubMed] [Google Scholar]
- 180.Widemann B. C., Kim A., Fox E., Baruchel S., Adamson P. C., Ingle A. M., Bender J. G., Burke M., Weigel B., Stempak D., Balis F. M., Blaney S. M. A Phase I Trial and Pharmacokinetic Study of Sorafenib in Children with Refractory Solid Tumors or Leukemias: A Children's Oncology Group Phase I Consortium Report. Clinical Cancer Research. 2012;18(21):6011–6022. doi: 10.1158/1078-0432.CCR-11-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gouravan Sarina, Meza-Zepeda Leonardo, Myklebost Ola, Stratford Eva, Munthe Else. Preclinical Evaluation of Vemurafenib as Therapy for BRAFV600E Mutated Sarcomas. International Journal of Molecular Sciences. 2018;19(4):969. doi: 10.3390/ijms19040969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Peng Chang-Liang, Guo Wei, Ji Tao, Ren Tingting, Yang Yi, Li Da-Sen, Qu Hua yi, Li Xiao, Tang Shun, Yan Tai-Qiang, Tang Xiao-Dong. Sorafenib induces growth inhibition and apoptosis in human synovial sarcoma cells via inhibiting the RAF/MEK/ERK signaling pathway. Cancer Biology & Therapy. 2009;8(18):1729–1736. doi: 10.4161/cbt.8.18.9208. [DOI] [PubMed] [Google Scholar]
- 183.López-Guerrero JA, Navarro S, Noguera R, Carda C, Fariñas SC, Pellín A, Llombart-Bosch A. Mutational analysis of the c-KIT AND PDGFRalpha in a series of molecularly well-characterized synovial sarcomas. Diagnostic Molecular Pathology: The American Journal of Surgical Pathology, Part B. 2005;14(3):134–139. doi: 10.1097/01.pas.0000176766.33671.b6. [DOI] [PubMed] [Google Scholar]
- 184.Teng Hao-Wei, Wang Hsei-Wei, Chen Wei-Ming, Chao Ta-Chung, Hsieh Yao-Yu, Hsih Chi-Hsiu, Tzeng Cheng-Hwai, Chen Paul Chih-Hsueh, Yen Chueh-Chuan. Prevalence and prognostic influence of genomic changes of EGFR pathway markers in synovial sarcoma. Journal of Surgical Oncology. 2011;103(8):773–781. doi: 10.1002/jso.21852. [DOI] [PubMed] [Google Scholar]
- 185.Ishibe T. Disruption of Fibroblast Growth Factor Signal Pathway Inhibits the Growth of Synovial Sarcomas: Potential Application of Signal Inhibitors to Molecular Target Therapy. Clinical Cancer Research. 2005;11(7):2702–2712. doi: 10.1158/1078-0432.CCR-04-2057. [DOI] [PubMed] [Google Scholar]
- 186.Patwardhan PP, Musi E, Schwartz GK. Preclinical evaluation of nintedanib, a triple angiokinase inhibitor, in soft-tissue sarcoma: potential therapeutic implication for synovial sarcoma. Molecular Cancer Therapeutics. 2018;17(11):2329–2340. doi: 10.1158/1535-7163.MCT-18-0319. [DOI] [PubMed] [Google Scholar]
- 187.YAMAMOTO YUKI, FUKUDA KAZUMASA, FUCHIMOTO YASUSHI, MATSUZAKI YUMI, SAIKAWA YOSHIRO, KITAGAWA YUKO, MORIKAWA YASUHIDE, KURODA TATSUO. Cetuximab promotes anticancer drug toxicity in rhabdomyosarcomas with EGFR amplification in vitro. Oncology Reports. 2013;30(3):1081–1086. doi: 10.3892/or.2013.2588. [DOI] [PubMed] [Google Scholar]
- 188.Abraham Jinu, Chua Ying Xuan, Glover Jason M., Tyner Jeffrey W., Loriaux Marc M., Kilcoyne Aoife, Giles Francis J., Nelon Laura D., Carew Jennifer S., Ouyang Yongjian, Michalek Joel E., Pal Ranadip, Druker Brian J., Rubin Brian P., Keller Charles. An adaptive Src–PDGFRA–Raf axis in rhabdomyosarcoma. Biochemical and Biophysical Research Communications. 2012;426(3):363–368. doi: 10.1016/j.bbrc.2012.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ragazzini, P., Gamberi, G., Pazzaglia, L., Serra, M., Magagnoli, G., Ponticelli, F., … Benassi, M. S. (2004). Amplification of CDK4, MDM2, SAS and GLI genes in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma. Histology and Histopathology, 19(2), 401–411. 10.14670/HH-19.401 [DOI] [PubMed]
- 190.Srivastava, R. K., Kaylani, S. Z., Edrees, N., Li, C., Talwelkar, S. S., Xu, J., … Athar, M. (2014). GLI inhibitor GANT-61 diminishes embryonal and alveolar rhabdomyosarcoma growth by inhibiting Shh/AKT-mTOR axis. Oncotarget, 5(23), 12,151–12,165. 10.18632/oncotarget.2569 [DOI] [PMC free article] [PubMed]
- 191.BOEHME KAREN A., ZABORSKI JULIAN J., RIESTER ROSA, SCHWEISS SABRINA K., HOPP ULRIKE, TRAUB FRANK, KLUBA TORSTEN, HANDGRETINGER RUPERT, SCHLEICHER SABINE B. Targeting hedgehog signalling by arsenic trioxide reduces cell growth and induces apoptosis in rhabdomyosarcoma. International Journal of Oncology. 2015;48(2):801–812. doi: 10.3892/ijo.2015.3293. [DOI] [PubMed] [Google Scholar]
- 192.Yamanaka H, Oue T, Uehara S, Fukuzawa M. Hedgehog signal inhibitor forskolin suppresses cell proliferation and tumor growth of human rhabdomyosarcoma xenograft. Journal of Pediatric Surgery. 2011;46(2):320–325. doi: 10.1016/j.jpedsurg.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 193.Eichenmüller M, Hemmerlein B, von Schweinitz D, Kappler R. Betulinic acid induces apoptosis and inhibits hedgehog signalling in rhabdomyosarcoma. British Journal of Cancer. 2010;103(1):43–51. doi: 10.1038/sj.bjc.6605715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Graab U, Hahn H, Fulda S. Identification of a novel synthetic lethality of combined inhibition of hedgehog and PI3K signaling in rhabdomyosarcoma. Oncotarget. 2015;6(11):8722–8735. doi: 10.18632/oncotarget.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Beauchamp Elspeth, Bulut Gulay, Abaan Ogan, Chen Kevin, Merchant Akil, Matsui William, Endo Yoshimi, Rubin Jeffrey S., Toretsky Jeffrey, Üren Aykut. GLI1 Is a Direct Transcriptional Target of EWS-FLI1 Oncoprotein. Journal of Biological Chemistry. 2009;284(14):9074–9082. doi: 10.1074/jbc.M806233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cidre-Aranaz F, Alonso J. EWS/FLI1 target genes and therapeutic opportunities in Ewing sarcoma. Frontiers in Oncology. 2015;5:162. doi: 10.3389/fonc.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Smith MA, Kang MH, Reynolds CP, Kurmasheva RT, Alexander D, Billups CA, et al. Evaluation of arsenic trioxide by the pediatric preclinical testing program with a focus on Ewing sarcoma. Pediatric Blood & Cancer. 2012;59(4):753–755. doi: 10.1002/pbc.23391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Motoi T, Saito T, Ladanyi M. The Hedgehog signaling pathway in synovial sarcoma: mutational and functional analysis. Cancer Research. 2006;66(8 Supplement):403–403. [Google Scholar]
- 199.Ng TL, Gown AM, Barry TS, Cheang MCU, Chan AKW, Turbin DA, et al. Nuclear beta-catenin in mesenchymal tumors. Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc. 2005;18(1):68–74. doi: 10.1038/modpathol.3800272. [DOI] [PubMed] [Google Scholar]
- 200.Briski Laurence M, Thomas Dafydd G, Patel Rajiv M, Lawlor Elizabeth R, Chugh Rashmi, McHugh Jonathan B, Lucas David R. Canonical Wnt/β-catenin signaling activation in soft-tissue sarcomas: A comparative study of synovial sarcoma and leiomyosarcoma. Rare Tumors. 2018;10:203636131881343. doi: 10.1177/2036361318813431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Trautmann M, Sievers E, Aretz S, Kindler D, Michels S, Friedrichs N, Renner M, Kirfel J, Steiner S, Huss S, Koch A, Penzel R, Larsson O, Kawai A, Tanaka S, Sonobe H, Waha A, Schirmacher P, Mechtersheimer G, Wardelmann E, Büttner R, Hartmann W. SS18-SSX fusion protein-induced Wnt/β-catenin signaling is a therapeutic target in synovial sarcoma. Oncogene. 2013;33(42):5006–5016. doi: 10.1038/onc.2013.443. [DOI] [PubMed] [Google Scholar]
- 202.Barham W., Frump A. L., Sherrill T. P., Garcia C. B., Saito-Diaz K., VanSaun M. N., Fingleton B., Gleaves L., Orton D., Capecchi M. R., Blackwell T. S., Lee E., Yull F., Eid J. E. Targeting the Wnt Pathway in Synovial Sarcoma Models. Cancer Discovery. 2013;3(11):1286–1301. doi: 10.1158/2159-8290.CD-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Michels S., Trautmann M., Sievers E., Kindler D., Huss S., Renner M., Friedrichs N., Kirfel J., Steiner S., Endl E., Wurst P., Heukamp L., Penzel R., Larsson O., Kawai A., Tanaka S., Sonobe H., Schirmacher P., Mechtersheimer G., Wardelmann E., Buttner R., Hartmann W. SRC Signaling Is Crucial in the Growth of Synovial Sarcoma Cells. Cancer Research. 2013;73(8):2518–2528. doi: 10.1158/0008-5472.CAN-12-3023. [DOI] [PubMed] [Google Scholar]
- 204.Ragab, N., Viehweger, F., Bauer, J., Geyer, N., Yang, M., Seils, A., … Simon-Keller, K. (2018). Canonical WNT/β-catenin signaling plays a subordinate role in rhabdomyosarcomas. Frontiers in Pediatrics, 6, 378. 10.3389/fped.2018.00378 [DOI] [PMC free article] [PubMed]
- 205.Annavarapu Srinivas R, Cialfi Samantha, Dominici Carlo, Kokai George K, Uccini Stefania, Ceccarelli Simona, McDowell Heather P, Helliwell Timothy R. Characterization of Wnt/β-catenin signaling in rhabdomyosarcoma. Laboratory Investigation. 2013;93(10):1090–1099. doi: 10.1038/labinvest.2013.97. [DOI] [PubMed] [Google Scholar]
- 206.Pedersen E. A., Menon R., Bailey K. M., Thomas D. G., Van Noord R. A., Tran J., Wang H., Qu P. P., Hoering A., Fearon E. R., Chugh R., Lawlor E. R. Activation of Wnt/ -Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States. Cancer Research. 2016;76(17):5040–5053. doi: 10.1158/0008-5472.CAN-15-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hayashi, M., Baker, A., Goldstein, S. D., Albert, C. M., Jackson, K. W., McCarty, G., … Loeb, D. M. (2017). Inhibition of porcupine prolongs metastasis free survival in a mouse xenograft model of Ewing sarcoma. Oncotarget, 8(45), 78,265–78,276. 10.18632/oncotarget.19432 [DOI] [PMC free article] [PubMed]