Abstract
An elevated prevalence of cryptococcal infection is a tendency in low-income countries and constitutes a global public health problem due to factors such as the limited efficacy of antifungal therapy and the AIDS/transplant immunocompromised patients. The fungus Cryptococcus neoformans, implicated in this burden, has had several genes validated as drug targets. Among them, the thioredoxin system is one of the major regulators of redox homeostasis and antioxidant defense acting on protein disulfide bonds. Thioredoxin 1 from C. neoformans (CnTrx1) was cloned and expressed in E. coli and the recombinant protein was purified and crystallized. Functional assay shows that CnTrx1 catalyzes the reduction of insulin disulfide bonds using dithiothreitol, while acting as a monomer in solution. The crystal structure of oxidized CnTrx1 at 1.80 Å resolution presents a dimer in the asymmetric unit with typical Trx-fold. Differences between the monomers in the asymmetric unit are found specially in the loop leading to the Cys-Gly-Pro-Cys active-site motif, being even larger when compared to those found between reduced and oxidized states of other thioredoxins. Although the thioredoxins have been isolated and characterized from many organisms, this new structural report provides important clues for understanding the binding and specificity of CnTrx1 to its targets.
Keywords: Thioredoxin, Cryptococcus neoformans, Heterologous expression, X-ray structure, Active site conformational plasticity
Abbreviations: AIDS, Acquired immune deficiency syndrome; CSS, Complex formation significance score
Highlights
-
•
The crystal structure of the thioredoxin 1 enzyme of the pathogenic fungus Cryptococcus neoformans is presented at 1.8 Å resolution.
-
•
The active site detailed description shows unexpected plasticity when comparing the two monomers found in the asymmetric unit.
-
•
The dimer interface is scrutinized, leading to the proposal of a crystallographic dimer with few common features found in previously stablished natural dimers.
-
•
The expression, purification and crystallization protocols are presented, as well as a enzymatic assay to show the activity of the produced thioredoxin.
1. Introduction
Thioredoxins (Trxs) are the major ubiquitous heat-stable disulfide reductases (12 kDa average) responsible for maintaining proteins in their reduced state inside cells. They are reduced by electrons from NADPH via the flavin enzyme Trx reductase (TrxR) [1]. Trxs are involved in a wide variety of fundamental biological functions such as, protection against oxidative stress [2], acts as a hydrogen donor for reductive enzymes such as ribonucleotide reductase, thioredoxin peroxidases [3,4] and methionine sulfoxide reduction [5]. They also modulate transcription factors involved in DNA binding [6,7], apoptosis [8] and important roles in the life cycles of viruses and phages [9], among other processes. The wide variety of Trx reactions is based on its broad substrate specificity and potent capacity to reduce multiple cellular targets [1].
Thioredoxins are found in all living cells from archaebacteria to mammals. All Trx proteins possess an active site made up of two cysteine residues embedded in a conserved motif, Cys-Gly-Pro-Cys [10]. Trxs use these cysteine residues to break disulfide bonds found in oxidized substrate proteins [11]. Then, reconstitution of the Trx oxidized cysteines by TrxR completes the cycle. Trxs are found in several cellular locations such as the cytosol [6], the nucleus [12] and the mitochondria [13] or are secreted to the extracellular environment [14]. In plants, thioredoxins also regulate enzymatic activities in the chloroplasts [15].
Immunocompromised patients, cancer and transplant patients show increased susceptibility to fungal pathogens [[16], [17], [18]]. Of particular concern is the high rate of mortality associated with invasive fungal infections, which often exceeds 50% despite the availability of several antifungal drugs [19]. In resource-limited regions, cryptococcosis is an important opportunistic fungal infection that represents more deaths than those caused by tuberculosis or malaria as a result of global HIV/AIDS pandemic [18]. In Cryptococcus neoformans, the Trx system is composed of two small Trx proteins and one TrxR, the latter being essential for this pathogen [20]. The redox control of the thioredoxin system is thought to regulate the expression of many stress defense enzymes and protect cells against oxidative stress. The Trx1 and Trx2 from C. neoformans are cytoplasmic isoforms that are important not only to stress resistance, but also to virulence. It was demonstrated that the Δtrx1 mutant has a severe growth defect and is sensitive to multiple stresses, while the Δtrx2 mutant is only sensitive to nitric oxide stress [21]. It appears that Trx1 is more important than Trx2 for the normal functioning of C. neoformans.
Trxs sequences vary from 105 to 110 amino acids and share between 27 and 69% sequence identity [22]. Trx1 of C. neoformans (CnTrx1) exhibits 52% identity with Trx from the skin-colonizing yeast Malassezia sympodialis, and 43% with H. sapiens Trx. In addition, since the first Trx three-dimensional structure was achieved in 1975, namely, the Escherichia coli protein with a maximum resolution of 2.8 Å [23], the structures of several Trxs have been solved such as M. sympodialis [24], Saccharomyces cerevisiae [25], Plasmodium falciparum [26], spinach chloroplasts [27], Litopenaeus vannarrei [28] and humans [29].
Despite the extensive literature regarding Trxs, new structures are subject of investigation as they contribute to understand the structural properties, the molecular mechanisms and the catalytic activity that dictate species and isoforms Trx specificities. Hence, we report the expression, purification, enzymatic assay and X-ray crystal structure of recombinant oxidized CnTrx1 solved at 1.80 Å resolution (PDB ID: 5JY5). This is the second thioredoxin structure of a human fungal pathogen, which reveals subtle variations.
2. Material and methods
2.1. Cloning and expression
A synthetic codon-optimized gene of Trx1 from Cryptococcus neoformans var. grubii H99 (GeneBank CNAG_02801) was designed for its expression in E. coli as a C-terminal six-histidine fusion protein for affinity purification. The PCR product was cloned into NdeI and XhoI restriction sites of the pET21a expression vector (Epoch Life Science, inc.). The recombinant protein with 6 × His-tag was expressed in E. coli strain BL21 (λDE3) pLysE (Novagen, San Diego, CA, USA). E. coli cells were grown in 1 L of Luria Bertani (LB) medium which was supplemented with ampicillin (100 μg/mL) and chloramphenicol (20 μg/mL) at 37 °C until reaching an OD600nm of 0.6, at which point protein expression was induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) with a shake rate of 220 rpm for 6 h at 37 °C. The strain with pLysE was chosen from different expression tests that showed higher levels of soluble expression.
2.2. Extraction and purification
Cells were harvested by centrifugation, suspended in 30 mM Tris-HCl, pH 8.5, 300 mM NaCl and 10 mM imidazole. The cells were lysed by 10 cycles of 50 s sonication in an ice bath with a 50% amplitude and 50 s rest. After lyses, the sample was centrifuged at 9000 g for 40 min at 4 °C. The recombinant protein obtained from the cytoplasmic fraction was filtered through a 0.45 μm disposable filter and applied to a 1 mL immobilized metal affinity chromatography (nickel IMAC) column (Hi-Trap, GE Healthcare) according to the standard protocols. Increasing the imidazole concentration from 10 to 500 mM in a stepwise manner eluted the recombinant protein. Eluted CnTrx1 protein was further purified by gel filtration using a Superdex 75TM 10/300 GL column on an ÄKTA-FPLC (GE Healthcare Biosciences, USA) pre-equilibrated against the buffer containing 30 mM Tris-HCl pH 8.5 and 300 mM NaCl. The proteins used as standards in the calibration curve were: aprotinin 6.5 kDa (1), ribonuclease A 13.7 kDa (2), carbonic anhydrase 29.0 kDa (3), ovalbumin 43.0 kDa (4) and conalbumin 75.0 kDa (5). Blue dextran 2000 was employed to determine the column void volume. All the fractions were analyzed by 15% SDS-PAGE with Coomassie blue staining [30] and also by western blot employing an anti-poly-His monoclonal antibody in a 1:1000 dilution conjugated to alkaline phosphatase (Sigma-Aldrich). The colorimetric detection of protein bands was achieved by staining with the 4-nitro-blue-tetrazolium-chloride/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP) substrate. Fractions containing pure thioredoxin were pooled and dialyzed against 10 mM Tris-HCl pH 8.5 and 300 mM NaCl, using an Amicon ultracentrifuge 2 kDa cut off concentrator (Millipore), concentrated and frozen at −80 °C. Concentration was estimated using the sample absorption at 280 nm measured with a UV-1800 spectrophotometer (Shimadzu) and an extinction coefficient of 21,095 M−1 cm-1.
2.3. Protein crystallization
Prior to crystallization, the protein was submitted to dynamic light scattering (DLS) to assess its polydispersity in a Malvern Zetasizer Nano ZS90 equipped with a 633 nm He–Ne laser and operating at an angle of 173°. Each sample, 500 μL at 0.3 mg/mL protein concentration, was measured in quartz cuvettes at 18 °C. Samples were diluted in 10 mM tris buffer (pH 8.0–8.5). A minimum of three measurements was performed for each sample. The crystallization was performed using solutions from the commercial screening kits 1 and 2 of Hampton research. Crystallization was carried out in 24-well plates using the vapor diffusion technique in sitting or hanging drop mode. The CnTrx1 crystals were obtained in drops containing 2 μL of protein sample (9 mg/mL) and 2 μL of reservoir solution number 32 of kit 1 (2.0 M ammonium sulfate). The drops were equilibrated against 500 μL reservoir solution. After optimization, crystals with hexagonal morphology grew to a maximal size of ~(380 x 330 x 210) μm3 within two weeks by hanging drop vapor-diffusion method at 294 K using 1.7 M ammonium sulfate and 0.02 M tris hydrochloride pH 8.5.
2.4. Data collection, structure determination and refinement
The X-ray diffraction data were collected using synchrotron radiation at the protein crystallography MX2 beamline by the Brazilian Synchrotron Light Laboratory (LNLS) in Campinas-Brazil, using a PILATUS2M detector from Dectris to record the oscillation data with Δϕ = 0.5°, covering a total oscillation range of 360°. Data were collected at 100 K in a nitrogen gas stream. CnTrx1 were soaked in cryoprotectant solution (reservoir solution supplemented with 25% glycerol for 15 min). The data processing for the determination of the unit-cell parameters, integration, merging and scaling of reflections was performed using the programs DENZO/SCALEPACK of the HKL2000 program package [31]. The CnTrx1 crystals diffracted to 1.8 Å and the wavelength was 1.2398 Å. The crystal structure was determined using the molecular replacement method as implemented in the program MOLREP of CCP4i [32]. The thioredoxin crystallographic model from Malassezia sympodialis (PDB ID: 2J23) [24], that shares 52% sequence identity with CnTrx1, was used as the searching template. Two Trx molecules were found in the asymmetric unit in agreement with Matthews coefficient prediction [33]. The structure was revised and modeled using COOT [34] and refined with REFMAC5 [35]. Protein atoms with double conformation or poor density had their occupancy set to 0.5. MolProbity was used to assess the quality of the final model [36]. Coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under the accession code 5JY5. The data collection/processing and refinement statistics are summarized in Table 1. Calculation of solvent-accessible surface (SAS) and other analysis were performed with chain A, unless otherwise mentioned.
Table 1.
Data collection and structure refinement statistics.
| Parameter | Oxidized CnTrx1 |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.2398 |
| Space group | P6422 |
| Cell dimensions (Å) | |
|
a, b, c α, β, γ |
110.706, 110.706, 91.761 90.00, 90.00, 120.00 |
| Resolution (Å)a | 25.00–1.80 (1.86–1.80) |
| Mosaicity (°) | 0.6 |
| No. of total reflections | 1,157,390 |
| No. of unique reflections | 31,224 |
| Redundancy | 37.1 (34.7) |
| Completeness (%) | 100.0 (100.0) |
| Rmergeb | 0.092 (0.581) |
| Rpimb | 0.015 (0.100) |
| CC1/2 | (0.976) |
| I/σ(I) | 53.9 (6.2) |
| Refinement | |
| Resolution (Å) | 25.00–1.80 |
| Model composition | |
| No. of protein residues | 105 |
| No. of water molecules | 347 |
| Ligands | 1 glycerol, 3 sulfate ions, 1 TRIS |
| Mean B-factor (Å2): all atoms | 37.27 |
| Rwork/Rfree | 0.155/0.198 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.023 |
| Bond angles (°) | 2.193 |
| Side chain double conformationc | Ile5A, Ser16A, Val20A, Val21A, Thr28A, Met35A,B, Ser37A, Lys42B, Met72A,B and Ile95B |
| PDB code | 5JY5 |
The values in parentheses refer to the highest resolution shell of 1.86–1.80 Å.
Rmerge = ∑hkl ∑i |Ii − <I>|/∑hkl ∑i Ii, Rpim = ∑hkl [1/(N-1)]1/2 ∑i |Ii − <I>|/∑hkl ∑i Ii where Ii is the intensity of a reflection i of the group represented by the unique reflection hkl, and <I> is the average intensity of the group represented by reflection hkl, and N is the total number of reflections in the hkl group.
Capital letters in superscript indicate the monomer.
2.5. Enzymatic activity assay for CnTrx1
Enzymatic activity of the recombinant CnTrx1 was assessed using insulin (Sigma-Aldrich) as described previously by Holmgren [37], with modifications. The assay is based on the dithiol-disulfide oxidoreductase activity that catalyzes the reduction of insulin and is restored by dithiothreitol (DTT). Three fractions of CnTrx1 at 2.0 μM, 4.1 μM and 8.2 μM were prepared by dilution in deionized water. The final reaction mixture contains: 0.1% w/v bovine insulin in 50 mM tris HCl pH 7.5, 63 mM sodium phosphate pH 7.0, 2.0 mM EDTA, zero (control) or 80 μL of the CnTrx1 fractions and 1.0 mM DTT. The final volume was adjusted to 1 mL by addition of deionized water. The precipitation of insulin was monitored as an increase in absorbance at 650 nm (ΔA650) using a UV-1800 spectrophotometer (Shimadzu) in the absence (control) or presence of different amounts of CnTrx1. The activity (ΔA650/min) was considered as the ΔA650 during a period of 13....3 min taken in a region of linear increase: between 25.7 and 39.0 min for 2.0 μM CnTrx1, 17.7 and 31.0 min for 4.1 μM CnTrx1 and 12,1 and 25.4 min for 8.2 μM CnTrx1. The corresponding coefficient of determination (R2) for fitted lines to these linear regions are 0.994, 0.999 and 0.999, respectively. Thioredoxin concentrations were determined from their absorption at 280 nm applying molar extinction coefficients.
3. Results and discussion
3.1. Expression, purification and enzymatic assay of CnTrx1
CnTrx1 tagged with six histidines was expressed successfully in E. coli cells as shown by the ~12 kDa band in the soluble fraction SDS-PAGE and its anti-poly-His Western-blot (Supplementary Fig. 1a). Purification followed in two steps via IMAC and size exclusion chromatography (Supplementary Fig. 1b and 1c). CnTrx1 was eluted from the nickel-affinity column in standard buffer containing 300 mM imidazole. Size exclusion chromatography (elution volume of 13.16 mL) and dynamic light scattering (DLS) experiments indicate that CnTrx1 behaves as a monomer in solution. DLS mass estimation was 13.5 kDa with a polydispersity index of 14.7%, compatible with monomers as reported for thioredoxins from other species. The yield of CnTrx1 was ~9 mg/L of bacteria culture.
The enzymatic activity of CnTrx1 was assayed by its capacity to reduce the disulfide bonds of insulin (Supplementary Fig. 2), while the nonphysiological reductant dithiothreitol (DTT) was used to restore the enzyme. In the control cuvette, without CnTrx1, no precipitation was observed. The addition of 2.0, 4.1 and 8.2 μM CnTrx1 resulted in precipitation appearing after 25, 17 and 12 min, respectively. The activity of CnTrx1 (ΔA650/min) was 0.007, 0.012 and 0.017, respectively. Previous work shows that using slightly different conditions, Trxs from other organisms presented a higher relative specific activity (data not shown) [37]. This difference might be due to the experiments being conducted at different pHs and Trx concentrations.
3.2. Overall structure
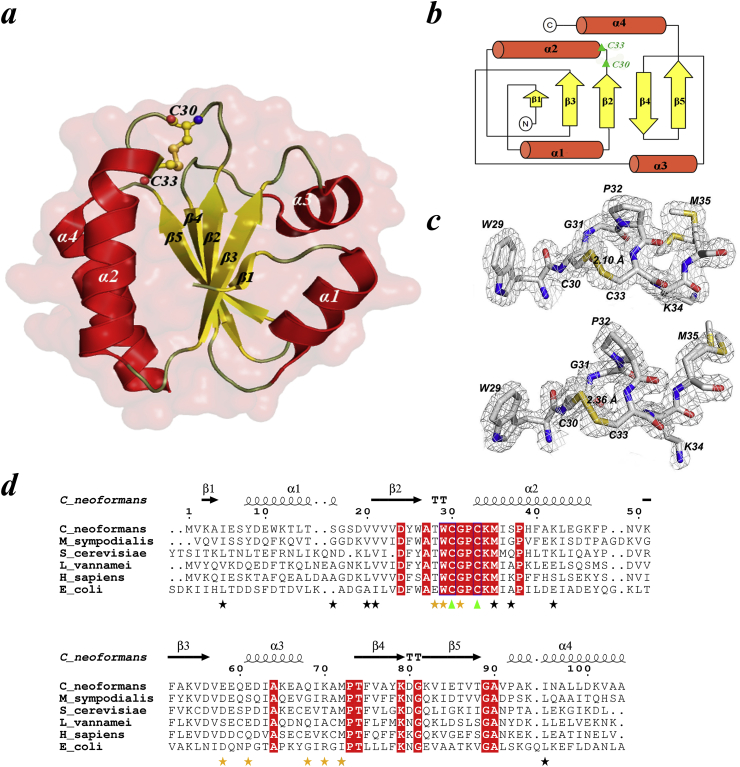
Single crystals were obtained using CnTrx1 at 9 mg/mL in 1.7 M ammonium sulfate and 0.02 M tris hydrochloride pH 8.5. The space group was P6422, with unit-cell parameters a and b = 110.706 Å, c = 91.761 Å, α and β = 90.00°, and γ = 120.00°. A Matthews coefficient of 3.54 Å3 Da-1 for two molecules in the asymmetric unit and a solvent fraction of 65.31% represented the second highest probable asymmetric unit content. The CnTrx1 structure has been determined to maximum resolution of 1.80 Å by molecular replacement. The refined model presented a final Rwork/Rfree of 0.155/0.198 with general stereochemistry parameters within acceptable limits. The data processing and refinement statistics are shown in Table 1. There are no Ramachandran plot outliers, with 98.4% of the residues in the most favored regions [38]. All residues could be located in the electron density, except for Met1, His107, His108, His109 and His110 (last four histidines from the His-tag) and the side chains of solvent-exposed Lys34, Lys94 and Lys101.
The final model of CnTrx1 contains two molecules in the asymmetric unit, each comprised of residues 2–106, with a cis peptide bond between Met72 and Pro73. CnTrx1 presents a typical Trx fold, consisting of a five-stranded β-sheet surrounded by two α-helices on each side (Fig. 1a and 1b). The CnTrx1 structure was obtained in its oxidized form (CnTrx1ox, Fig. 1c). The sequence alignment (Fig. 1d) shows that CnTrx1 shares a sequence identity of 52%, 46%, 47%, 43% and 35% with Trxs from M. sympodialis (2J23) [24], S. cerevisiae (2OE0) [25,39], L. vannamei (3ZZX) [28], H. Sapiens (5DQY) [40] and E. coli (1XOA) [41], respectively. The molecular architecture of CnTrx1 is very similar to MsTrx1, which was determined at 1.4 Å maximum resolution, and the superposition of these two structures reveals great similarity with a root mean square deviation (RMSD) of 0.8 Å for all Cα atoms. The superposition revealed one conformational difference, the deletion of two residues (Gly49 and Asp50) in C. neoformans, which leads to the loss of small helical fragment after the second helix. In the case of the superposition of CnTrx1 to the human thioredoxin (HsTrx), the RMSD is higher, 1.22 Å for all Cα atoms, although the three-dimensional structure remains very similar. The most noticeable differences are at the end of helix α1 and beginning of the following loop where the HsTrx presents an insertion of 2 residues and the presence of three cysteine residues, at positions 60, 67 and 71 (62, 69 and 73 in HsTrx), in addition to the active site cysteines (positions 32 and 35 in HsTrx). Among these, Cys73A and Cys73B are involved in disulfide-linked dimerization [42].
Fig. 1.
General architecture of CnTrx1. (a) Ribbon diagram of the overall structure of CnTrx1 with secondary structure elements labelled. Catalytic cysteines Cys30 and Cys33 are shown in the oxidized conformation observed in the crystal. All structure figures were made using PyMOL [55]. (b) Topological diagram of the CnTrx1 fold, showing the five-stranded mixed beta sheet sandwiched by four alpha helices. (c) Electron density 2|Fo| –|Fc| map, contoured at 1.0 σ around the conserved active site region WCGPCKM motif of CnTrx1 chain A (top) and B (bottom). The continuous electron density presents the disulfide bond between Sγ atoms of Cys30 and Cys33. (d) Sequence alignment of CnTrx1 (5JY5) with sequences of other thioredoxin proteins of known structure from M. sympodialis (2J23), S. cerevisiae (2OE0), L. vannamei (3ZZX), H. Sapiens(5DQY) and E. coli (1XOA). Absolutely conserved active-site cysteines are indicated with green triangle. Identical residues are in red boxes. Secondary structure elements of the CnTrx1 are shown above the alignment. The yellow stars bellow the residues indicate side chains involve hydrogen bonds between monomers, while black stars indicate those that have double conformation (Thr28A and Met72A,B also have double conformation). The alignment was performed using CLUSTAL W [56] and the figure was prepared with ESPript [57]. His-tag was removed from the sequence.
3.3. Thioredoxin oligomerization state
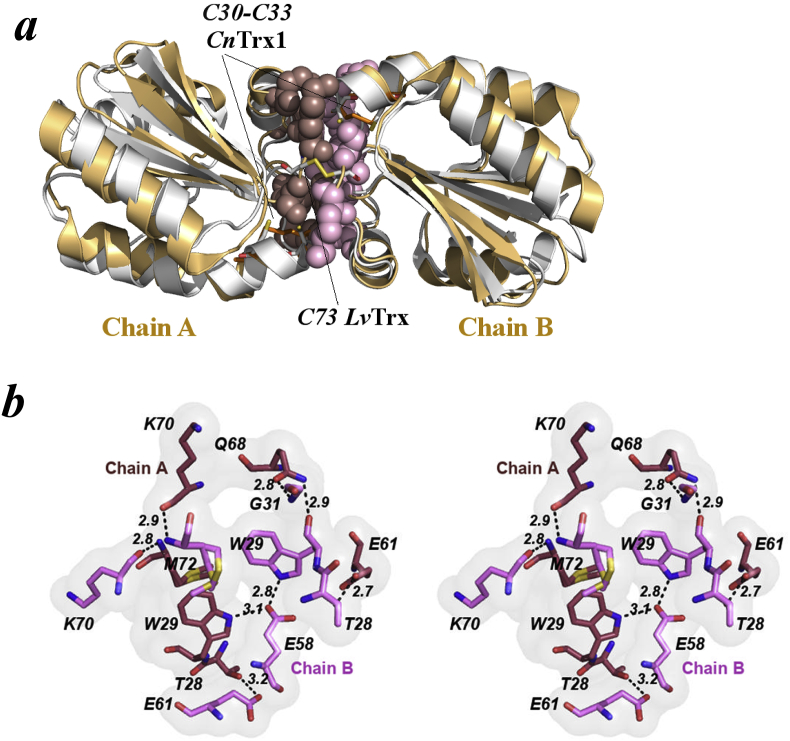
It is currently known that the crystal and solution structures of the Trxs can form monomers [24,43] and dimers [28,29,42,44]. Trx dimers are found in a small group of organisms, mostly humans and some crustaceans, that have a cysteine on its surface at the position aligned to Ala71 of CnTrx1 (Fig. 1d). This cysteine forms a disulfide bridge to its equivalent cysteine on the other monomer and this covalent bond is the main dimer-stabilizing force [28]. This dimerization seems to have some degree of functional importance as the activity of the dimer is reduced when compared to the monomer [28,42]. Other Trxs, such as ScTrx2, behaves almost exclusively as a monomer at ~40 μM with only 2% existing as dimers as observed in a gel filtration experiment [45]. The results of size exclusion chromatography (Supplementary Fig. 1c) and DLS show that CnTrx1 behaves as a monomer in solution at micromolar concentration. Thus, the two independent molecules of CnTrx1 found in the asymmetric unit can be described as a dimer formed during the crystallization procedures and not the in vivo state. This also agrees with a more detailed analysis from the PISA server [46] that implicates the CnTrx1 dimer interface in an auxiliary role in complex formation, not able to maintain complexation by itself at physiological concentration levels (calculated CSS of 0.152). The interface area between subunits A and B is about 601 Å2 (1202 Å2 when considering the area buried in each monomer separately) corresponding to just 10.5% of the total surface of both monomers. This interface is stabilized by seven hydrogen bonds involving 8 residues: Thr28, Trp29, Gly31, Glu58, Glu61, Gln68, Lys70 and Met72. Lys70 and Met72 main chain hydrogen bonding forms a small interchain antiparallel β-sheet that includes Ala71 (Fig. 2b). This β-sheet also appears in the natural covalent dimer observed in HsTrx and L. vannamei Trx (LvTrx) in which Ala71 is substituted by Cys that forms a disulfide bridge as mentioned previously (Fig. 2a). Several other features like this small β-sheet are shared between the interface of the crystallographic CnTrx1 dimer and the LvTrx and HsTrx dimers, indicating that there shall have been other conservation reasons to maintain this interface regardless of its in vivo occurrence. The most noticeable feature is the hydrophobic pocket formed by residues Ala27, Trp29, Val57, Ala64, Ile69 and Met72 of both monomers spanning for more than 15 Å across, from Ala64A-Cβ to Ile69B-Cβ. These residues are conserved and hydrophobic amongst most Trxs. Part of this dimer interface conservation might be related to this region being also involved in TrxR heterodimeric formation while performing its oxidation/reduction function. Another important feature of this CnTrx interface is the lack of a two-fold symmetry as observed in the HsTrx. Several residues have small deviations in the interface loops. The most obvious and central break of symmetry is clearly seen in Glu58B that hydrogen bonds to Trp29A-Nε1 and Trp29B-Nε1, while Glu58A is moved away from the Trp29A/B and does not interact with them.
Fig. 2.
Molecular Interface of the crystallographic structure of CnTrx1 (a) Superposition of C. neoformans Trx1 (copper) on shrimp L. vannamei (LvTrx) (gray, PDB ID: 4AJ6) showing the residues that make contact interface between monomers as spheres, chain A (purple) and chain B (pink). CnTrx1ox auxiliary complex highlighting the positions of Cys30 and Cys33 disulfide bond (orange) and the LvTrxred dimer highlighting the positions of Cys32, Cys35, and the Cys73A–Cys73B intermolecular disulfide bond (yellow). (b) Front view of two monomers making up the intermolecular interactions, showing the 8 residues: Thr28, Trp29, Gly31B, Glu58, Glu61, Gln68A, Lys70 and Met72, involved in hydrogen bonds between inter-monomer interaction from C. neoformans Trx1. The figures were made using PyMOL.
3.4. The active site
The atoms from the active site loop, formed by amino acids WCGPC (Trp29-Cys33), were clearly defined in the electron density in both monomers. The cysteines were found in the oxidized form, with a disulfide bond length of 2.10 and 2.36 Å in chains A and B (Fig. 1c), respectively. According to the old (2.030 Å) and new (2.033 Å) expected values for S–S distance from Engh and Huber (2006) [47] and their corresponding sigma values (0.008 Å and 0.016 Å, respectively), the S–S distance in chain B is 0.330 Å and 0.327 Å longer than expected or 41.2 and 20.4 sigmas, respectively. Other structures of resolution of at least 2.0 Å have shown similar or even larger distances [48]. In particular, the crystallographic structure of Trx from Acetobacter aceti (AaTrx, 2I4A) solved to a maximum resolution of 1.00 Å presents the active site sulfurs in the oxidized state separated by 2.31 Å [49]. The geometry of the disulfide is strikingly similar in CnTrx1 chain B and AaTrx as can be deduced by comparing the χ angles, specially χ2 that has an uncommon value of −137° [48] (Supplementary Table 1). This is consistent with the observation that the atoms that define χ2 are mostly in the side-chain and that the first cysteine is just on the edge of the helix α2 (Fig. 1), while changes in χ1 and/or the second cysteine could implicate the position of N and Cα of a residue in the middle of the secondary structure.
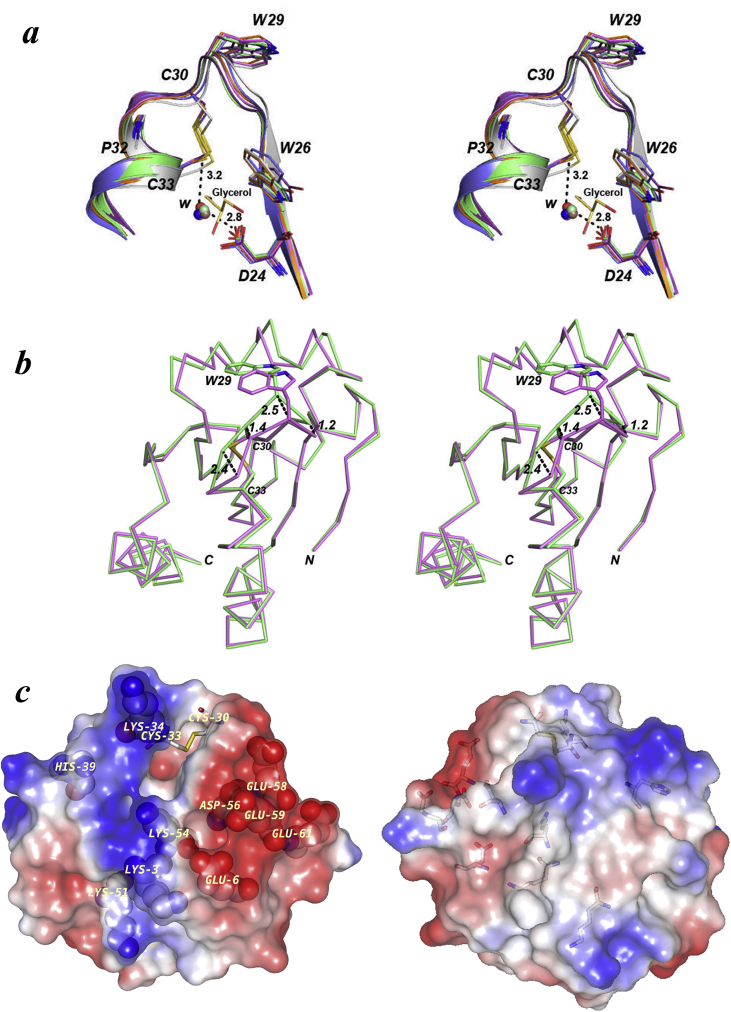
Another feature of the active site in CnTrx1 is that, despite the surface contour showing that the side chains of Cys30 and Cys33 are largely buried with less than 10% as accessible surface area (ASA) [50] (Supplementary Table 2), at least one water molecule is found close to each sulfur atom of the cysteines in both monomers at distances between 3.2 and 4.5 Å. Only the closest water at 3.2 Å is capable of forming a strong hydrogen bond to the Cys33A sulfur. This particular water occupies a similar position to waters present in other structures (Fig. 3a), bridging the cysteine to Asp24, an interaction implicated in raising the nucleophilicity of the Cys33 and facilitating the attack to the mixed disulfide [51]. In a recent NMR study of the ScTrx1, the importance of Asp24 has been reinforced by showing that it increases the rate of the cavity's hydration [52]. This layout of the active site is in agreement to the mechanism of action proposed previously [53]. Furthermore, a glycerol molecule was found in this region, only in chain A, less than 4.0 Å away from the indol of Trp26 (Fig. 3a), positioned in a similar way to a 2-mercapto-ethanol (BME) found in AaTrx.
Fig. 3.
Superposition of the active site motif from different thioredoxins and Stereo surface representation of CnTrx1 monomer. (a) Trxs C. neoformans monomer B (orange, PDB ID: 5JY5), M. sympodialys (blue, PDB ID: 2J23), S. cerevisiae (purple, PDB ID: 2OE0), L. vannamei (green, PDB ID: 4AJ6), H. Sapiens (pink, PDB ID: 5DQY) and E. coli (gray, PDB ID: 1XOA). The overlay shows the conservation around the cysteines and the position of water molecules bound to Asp. The distances indicated between the internal water molecule with Asp24 and Cys33 are for the CnTrx1. (b) Superposition of CnTrx1ox monomers A (green) and B (pink) with the side chains of Trp29, Cys30 and Cys33 side chains shown in stick. The distances between Cα atoms of the two monomers in the loop region prior to helix α2 are depicted by black dashes and given in Å. (c) Two 180° apart views of the CnTrx1 electrostatic surface potentials (red, negative; blue, positive) as calculated with the GRASP Poisson-Boltzmann [58]. The disulfide bond (Cys30-Cys33) surface was omitted to facilitate viewing. The charged amino acid residues are pointed out by their type and sequence number. The figures were made using PyMOL.
The difference in the position of Cys30A when compared to Cys30B or other equivalent cysteines in Trx structures is clearly visualized by superposing the monomers and other structures (Fig. 3a and b). A difference of 1.45 Å (Fig. 3b) is found between the Cα atoms of the two Cys30, while for the Cys33, this distance is just 0.35 Å (not shown in the figure). In fact, this difference of the backbone Cα atoms between chain A and B is greater than 1 Å for the residues spanning from Thr28 to Gly31, neighboring residues Ala27 and Pro32 are also shifted to a smaller extent. This main-chain repositioning is a consequence of the accommodation needed to form the crystallographic dimer interface, especially for Trp29A that moves its Cα atom 2.50 Å away from the position observed in chain B. Although there is no study addressing the CnTrx1red, a few studies in other organisms confirm that there are only small differences observed in the active-site loop, residues 28 to 35, and dimer interface of Trxox with respect to Trxred [42,43,54]. The differences in this loop observed in the chains A and B of C. neoformans are greater than those found in other structures and show that: (1) the plasticity of this loop is bigger than anticipated by previous Trxox and Trxred structures and (2) the alterations happen within a region that does not encompass the second cysteine in agreement with the most accepted ideas about the mechanism of action.
Fig. 3c displays the surfaces of CnTrx1ox colored according to electrostatic potential. A strongly negative charged patch was observed near the catalytic site Cys30 and Trp26, specifically involving Glu6, Asp56, Glu58, Glu59 and Glu61 residues. This distribution of the electrostatic potential is similar in thioredoxins from Drosophila melanogaster and chloroplasts, which showed a negative charge distribution near the active site [27,44] and has been recognized as a necessary feature to contact thioredoxin reductase [27]. CnTrx1 also exhibited a positively charged patch near the catalytic site Pro32 and Cys33 involving Lys34 and His39. Additional surface residues bearing a positive charge, Lys3, Lys42, Lys51 and Lys54, also contribute to the positive electrostatic potential around this area.
Lastly, since the active site changes seem insufficient to explain the differences in activity level shown by different thioredoxins, it has been proposed that the reason may be related to their interaction with target proteins [40] or differences in the layer of residues that interact directly with the active site residues. One example of the latter case in CnTrx1 is the conserved Asp58 (Fig. 1d) that is substituted by Glu58, this residue has been implicated in the active site stabilization by forming a hydrogen bond to Trp29 [26]. Although the interaction is maintained, slight variations in the activity might occur due to this change. Compared to other Trxs, a Phe is replaced by Tyr25 in CnTrx1. The hydroxyl group of Tyr25 makes hydrogen bonds to the side chains of Trp11 and Tyr78, forming a cluster that further stabilizes the protein.
In conclusion, although CnTrx1ox tertiary structure is very similar to other thioredoxins, this structure from an opportunistic human pathogen has revealed a non-in vivo dimer in the asymmetric unit that mimics the HsTrx in vivo dimer. The monomers that form the dimer have structural differences in the region of the catalytic cysteine (Cys30). The main-chain differences are larger than differences observed between oxidized and reduced states of different thioredoxins, indicating an impressive capacity for changes in the active site, which, in turn, might help explain, together with some point specific mutations, differences in stability, activity and most important, in the capacity for partnering with different protein targets.
CRediT authorship contribution statement
Claudia Patricia Bravo-Chaucanés: Methodology, Formal analysis, Writing - original draft, Investigation. Ana Karina Rodrigues Abadio: Methodology. Érika Seki Kioshima: Methodology. Maria Sueli Soares Felipe: Conceptualization, Formal analysis, Writing - original draft, Funding acquisition. João Alexandre Ribeiro Gonçalves Barbosa: Conceptualization, Methodology, Formal analysis, Resources, Writing - original draft, Supervision, Funding acquisition.
Declaration of competing interest
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) project 309054/2014-1 and by CNPq and FAP-DF (Fundação de Apoio à Pesquisa do Distrito Federal) Pronex projects 193.000.569/2009 and 193.001.533/2016. We thank the Brazilian Synchrotron Light Source (LNLS) for the beam time and assistance in data collection.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrep.2019.100724.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Holmgren A. Thioredoxin. Annu. Rev. Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 2.Monje-Casas F., Michán C., Pueyo C. Absolute transcript levels of thioredoxin- and glutathione-dependent redox systems in Saccharomyces cerevisiae : response to stress and modulation with growth. Biochem. J. 2004;383:139–147. doi: 10.1042/BJ20040851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurent T.C., Moore E.C., Reichard P. Enzymatic synthesis OF deoxyribonucleotides. IV. Isolation and characterization OF thioredoxin, the hydrogen donor from ESCHERICHIA coli B. J. Biol. Chem. 1964;239:3436–3444. http://www.ncbi.nlm.nih.gov/pubmed/14245400 [PubMed] [Google Scholar]
- 4.Kang S., Chae H.Z., Seo M.S., Kirn K., Baincs I.C., Rhe S.C. Mammalian peroxiredoxîn isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-o. FASEB J. 1998;12:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 5.Porqué P.G., Baldesten A., Reichard P. The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulfate. J. Biol. Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- 6.Arnér E.S.J., Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 7.Schenk H., Klein M., Erdbrügger W., Dröge W., Schulze-Osthoff K. Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-κB and AP-1. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravi D., Muniyappa H., Das K.C. Endogenous thioredoxin is required for redox cycling of anthracyclines and p53-dependent apoptosis in cancer cells. J. Biol. Chem. 2005;280:40084–40096. doi: 10.1074/jbc.M507192200. [DOI] [PubMed] [Google Scholar]
- 9.Holmgren A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 1989;254:13963–13966. [PubMed] [Google Scholar]
- 10.Holmgren A. Reduction of disulfides by thioredoxin. J. Biol. Chem. 1979;254:9113–9119. [PubMed] [Google Scholar]
- 11.Collet J.-F., Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxidants Redox Signal. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 12.Hirota K., Murata M., Nakamura H., Takeuchi J., Mori K., Yodoi J. Distinct Roles of Thioredoxin in the Cytoplasm and in the Nucleus : a two step mechanism of redox regulation of transcription factor NF- κ B. J. Biol. Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 13.Powis G., Montfort W.R. Properties and biological activities of thioredoxins. Annu. Rev. Pharmacol. Toxicol. 2001;41:261–295. doi: 10.1146/annurev.pharmtox.41.1.261. [DOI] [PubMed] [Google Scholar]
- 14.Xu S.Z., Sukumar P., Zeng F., Li J., Jairaman A., English A., Naylor J., Ciurtin C., Majeed Y., Milligan C.J., Bahnasi Y.M., Al-Shawaf E., Porter K.E., Jiang L.H., Emery P., Sivaprasadarao A., Beech D.J. TRPC channel activation by extracellular thioredoxin. Nature. 2008;451:69–72. doi: 10.1038/nature06414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai S., Schwendtmayer C., Johansson K., Ramaswamy S., Schürmann P., Eklund H. How does light regulate chloroplast enzymes? Structure-function studies of the ferredoxin/thioredoxin system. Q. Rev. Biophys. 2000;33:67–108. doi: 10.1017/s0033583500003607. [DOI] [PubMed] [Google Scholar]
- 16.Schmalzle S.A., Buchwald U.K., Gilliam B.L., Riedel D.J. Cryptococcus neoformans infection in malignancy. Mycoses. 2016;59:542–552. doi: 10.1111/myc.12496. [DOI] [PubMed] [Google Scholar]
- 17.Pappas P.G., Alexander B.D., Andes D.R., Hadley S., Kauffman C.A., Freifeld A., Anaissie E.J., Brumble L.M., Herwaldt L., Ito J., Kontoyiannis D.P., Lyon G.M., Marr K.A., Morrison V.A., Park B.J., Patterson T.F., Perl T.M., Oster R.A., Schuster M.G., Walker R., Walsh T.J., Wannemuehler K.A., Chiller T.M. Invasive fungal infections among organ transplant recipients: results of the transplant‐associated infection surveillance network (TRANSNET) Clin. Infect. Dis. 2010;50:1101–1111. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 18.Park B.J., Wannemuehler K.A., Marston B.J., Govender N., Pappas P.G., Chiller T.M. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 19.Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 20.Missall T.A., Lodge J.K. Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell. 2005;4:487–489. doi: 10.1128/EC.4.2.487-489.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missall T.A., Lodge J.K. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol. Microbiol. 2005;57:847–858. doi: 10.1111/j.1365-2958.2005.04735.x. [DOI] [PubMed] [Google Scholar]
- 22.Eklund H., Gleason F.K., Holmgren A. Structural and functional relations among thioredoxins of different species. Proteins Struct. Funct. Bioinforma. 1991;11:13–28. doi: 10.1002/prot.340110103. [DOI] [PubMed] [Google Scholar]
- 23.Holmgren A., Soderberg B.O., Eklund H., Branden C.I. Three dimensional structure of Escherichia coli thioredoxin S2 to 2.8 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1975;72:2305–2309. doi: 10.1073/pnas.72.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limacher A., Glaser A.G., Meier C., Schmid-Grendelmeier P., Zeller S., Scapozza L., Crameri R. Cross-reactivity and 1.4-å crystal structure of Malassezia sympodialis thioredoxin (mala s 13), a member of a new pan-allergen family. J. Immunol. 2007;178:389–396. doi: 10.4049/jimmunol.178.1.389. [DOI] [PubMed] [Google Scholar]
- 25.Bao R., Zhang Y., Zhou C., Chen Y. Structural and mechanistic analyses of yeast mitochondrial thioredoxin Trx3 reveal putative function of its additional cysteine residues. BBA - Proteins Proteomics. 2009;1794:716–721. doi: 10.1016/j.bbapap.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Peng M., Cascio D., Egea P.F. Crystal structure and solution characterization of the thioredoxin-2 from Plasmodium falciparum, a constituent of an essential parasitic protein export complex. Biochem. Biophys. Res. Commun. 2015;456:403–409. doi: 10.1016/j.bbrc.2014.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capitani G., Marković-Housley Z., DelVal G., Morris M., Jansonius J.N., Schürmann P. Crystal structures of two functionally different thioredoxins in spinach chloroplasts. J. Mol. Biol. 2000;302:135–154. doi: 10.1006/jmbi.2000.4006. [DOI] [PubMed] [Google Scholar]
- 28.Campos-Acevedo A.A., Sotelo-Mundo R.R., Pérez J., Rudiño-Piñera E. Is dimerization a common feature in thioredoxins? the case of thioredoxin from Litopenaeus vannamei. Acta Crystallogr. Sect. D Struct. Biol. 2017;73:326–339. doi: 10.1107/S2059798317002066. [DOI] [PubMed] [Google Scholar]
- 29.Hwang J., Nguyen L.T., Jeon Y.H., Lee C.Y., Kim M.H. Crystal structure of fully oxidized human thioredoxin. Biochem. Biophys. Res. Commun. 2015 doi: 10.1016/j.bbrc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 33.Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall I.I.I.W.B., Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 38.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Pinheiro A.S., Amorim G.C., Soares Netto L.E., Almeida F.C.L., Valente A.P. NMR solution structure of the reduced form of thioredoxin 1 from Sacharomyces cerevisiae. Proteins Struct. Funct. Bioinforma. 2008;70:584–587. doi: 10.1002/prot.21693. [DOI] [PubMed] [Google Scholar]
- 40.Qin J., Clore G.M., Gronenborn A.M. The high-resolution three-dimensional solution structures of the oxidized and reduced states of human thioredoxin. Structure. 1994;2:503–522. doi: 10.1016/s0969-2126(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 41.Schultz L.W., Chivers P.T., Raines R.T. The CXXC motif: crystal structure of an active-site variant of Escherichia coli thioredoxin. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:1533–1538. doi: 10.1107/s0907444999008756. [DOI] [PubMed] [Google Scholar]
- 42.Weichsel A., Gasdaska J.R., Powis G., Montfort W.R. Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer. Structure. 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- 43.Amorim G.C., Pinheiro A.S., Netto L.E.S., Valente A.P., Almeida F.C.L. NMR solution structure of the reduced form of thioredoxin 2 from saccharomyces cerevisiae. J. Biomol. NMR. 2007;38:99–104. doi: 10.1007/s10858-007-9144-z. [DOI] [PubMed] [Google Scholar]
- 44.Wahl M.C., Irmler A., Hecker B., Schirmer R.H., Becker K. Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster. J. Mol. Biol. 2005;345:1119–1130. doi: 10.1016/j.jmb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Bao R., Chen Y., Tang Y.-J., Janin J., Zhou C.-Z. Crystal structure of the yeast cytoplasmic thioredoxin Trx2. Proteins Struct. Funct. Bioinforma. 2007;66:246–249. doi: 10.1002/prot.21194. [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Engh R.A., Huber R. Int. Tables Crystallogr. International Union of Crystallography; Chester, England: 2006. Structure quality and target parameters; pp. 382–392. [Google Scholar]
- 48.Bhattacharya R., Pal D., Chakrabarti P. Disulfide bonds, their stereospecific environment and conservation in protein structures. Protein Eng. Des. Sel. 2004;17:795–808. doi: 10.1093/protein/gzh093. [DOI] [PubMed] [Google Scholar]
- 49.Starks C.M., Francois J.A., MacArthur K.M., Heard B.Z., Kappock T.J. Atomic-resolution crystal structure of thioredoxin from the acidophilic bacterium Acetobacter aceti. Protein Sci. 2007;16:92–98. doi: 10.1110/ps.062519707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tien M.Z., Meyer A.G., Sydykova D.K., Spielman S.J., Wilke C.O. Maximum allowed solvent accessibilites of residues in proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chivers P.T., Prehoda K.E., Volkman B.F., Kim B.-M., Markley J.L., Raines R.T. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 52.Iqbal A., Gomes-Neto F., Myiamoto C.A., Valente A.P., Almeida F.C.L. Dissection of the water cavity of yeast thioredoxin 1: the effect of a hydrophobic residue in the cavity. Biochemistry. 2015;54:2429–2442. doi: 10.1021/acs.biochem.5b00082. [DOI] [PubMed] [Google Scholar]
- 53.Menchise V., Corbier C., Didierjean C., Saviano M., Benedetti E., Jacquot J.P., Aubry A. Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism. Biochem. J. 2001;359:65–75. doi: 10.1042/0264-6021:3590065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friemann R., Schmidt H., Ramaswamy S., Forstner M., Krauth-Siegel R.L., Eklund H. Structure of thioredoxin from Trypanosoma brucei brucei. FEBS Lett. 2003;554:301–305. doi: 10.1016/s0014-5793(03)01173-6. [DOI] [PubMed] [Google Scholar]
- 55.DeLano W. Pymol: an open-source molecular graphics tool. Newsl. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 56.Thompson J.D., Higgins D.G., Gibson T.J., CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gouet P., Courcelle E., Stuart D.I., Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 58.Nicholls A., Sharp K.A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.