Abstract
Grey mould is an important necrotrophic fungal pathogen that causes huge economic losses in agriculture. Many types of bacteria are used for biological control of grey mould via competition for space or nutrients and/or the production of antifungal metabolites. Oxalate is a key component of virulent necrotic fungal pathogens. In this study, we isolated non-antifungal oxalate-degrading bacteria (ODB) from the surfaces of oxalate-rich spinach and strawberries to investigate their ability to control necrotic fungal pathogens such as grey mould. A total of 36 bacteria grown on oxalate minimal (OM) agar plates were tested for oxalate-degrading activity. Five isolates exhibiting the highest oxalate degradation activity were subjected to molecular identification using 16S rRNA gene sequencing. Two isolates exhibiting non-antifungal activity were subjected to disease suppression assays using Arabidopsis–Botrytis systems. The isolate Pseudomonas abietaniphila ODB36, which exhibited significant plant protective ability, was finally selected for further investigation. Based on whole-genome information, the pseudomonad oxalate degrading (podA) gene, which encodes formyl-CoA transferase, was analysed. The podA− mutant did not inhibit Botrytis infection and oxalate toxicity; the defects were recovered by podA complementation. Purified PodA–His converted oxalate to formate and eliminated oxalate toxicity. These results indicate that P. abietaniphila ODB36 and PodA enzyme are associated with various aspects of grey mould disease inhibitory effects.
Subject terms: Applied microbiology, Bacterial techniques and applications
Introduction
Botrytis cinerea, which causes grey mould, is an important necrotrophic pathogen that causes huge economic losses in agriculture1. Grey mould is known to attack over 200 crop hosts including vegetables, fruits, flowers, and post-harvest crops1. Grey mould can cause soft rotting of all aerial plant parts and produce prolific gray conidiophores and conidia typical of the disease1. B. cinerea causes seed contamination in flax, sunflower, and lettuce2. In Australia, seed contamination by B. cinerea has caused total crop failure in chickpea3.
Although agricultural systems typically rely on chemical pesticides to inhibit grey mould, fungicides are avoided in sustainable agriculture due to their effects on the well-being of humans and the surrounding environment4. Fungicide overuse has led to the emergence of resistant fungal strains5. During the past few decades, these issues have led many researchers to develop biological control methods using antagonistic bacteria. The modes of antagonistic bacterial activity include competition, antibiosis, lytic enzyme production, interference with pathogen activity and growth, volatility, and host resistance induction. However, most biological controls evaluated under laboratory conditions have tended to fail in the field6. Sulfur dioxide fumigation is usually used to control postharvest grey mould; however, because it decreases berry quality by absorption in detached berries, safe, effective, and economical alternatives are needed7.
Many necrotrophic pathogens including grey mould are known to infect various host plants via the production of oxalate, which is toxic to cellular organisms including mammals and plants8,9. Oxalate acts as a strong chelator of cations; it greatly oxidizes organic compounds in necrotrophic pathogens10, helps to reduce pH, thereby facilitating plant infection by pectolytic enzymes, and impedes defense signaling pathways in the host11. Therefore, degrading oxalate at the onset of host pathogen interaction should block the infection process. Oxalate-degrading microorganisms are useful alternatives to chemicals or fungicides. Oxalate-quenching Pseudomonas fluorescens PfMDU2 was shown to inhibit the mycelial growth of Rhizoctonia solani, which causes rice sheath blight12. The oxalate-degrading bacterial strain Cupriavidus campinensis was reported to protect Arabidopsis thaliana and crop species against B. cinerea13.
In human, oxalates are consumed when eating foods high in oxalates such as spinach, strawberries, coffee, tea, and chocolate, or are produced by intestinal microorganisms from metabolic precursors. In the human intestine, oxalate can combine with calcium, magnesium, potassium, or sodium to form less-soluble salts, which can lead to pathophysiological disorders, including hyperoxaluria, urolithiasis, and kidney failure14,15. To overcome these disorders, studies on the use of oxalate-degrading enzymes of intestinal microorganisms such as Oxalobacter formigenes and Lactobacillus acidophilus have been conducted15,16.
The objective of this study was to isolate, characterize, and evaluate oxalate-degrading bacteria (ODB) for the control of grey mould. We performed protein overexpression of the pseudomonad oxalate degrading enzyme (PodA) from non-antifungal Pseudomonas abietaniphila. Our aim was to provide a new approach for the control of grey mould via the application of oxalate-degrading genetic resources.
Results
ODB isolation and oxalate-degrading activity
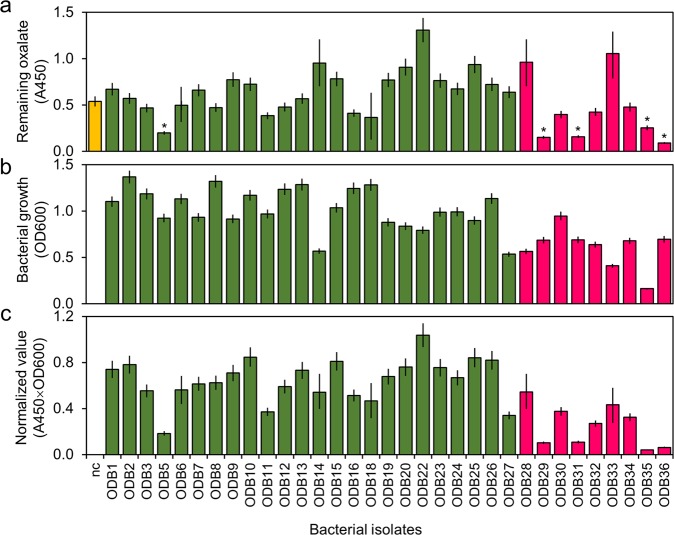
A total of 36 bacteria grown on oxalate minimal (OM) agar plates (2 g Na2C2O4, 2.7 g K2HPO4, 0.9 g NaH2PO4, 0.9 g NH4Cl, 0.27 g MgSO4·7H2O, 0.009 g CaCl2·2H2O, and 0.0024 g FeSO4·7H2O in 1 L distilled water with 1.5% agar) were isolated from the surfaces of spinach (27 isolates) and strawberries (9 isolates). The oxalate-degrading activity of these 36 bacteria was tested in OM broth supplemented with 1/10 Bacto tryptic soy broth (TSB). Oxalate in the culture supernatants was determined using an oxalate colorimetric assay kit (Bio Vision Inc., San Francisco, CA, USA). Therefore, the lower the absorbance value at 450 nm (A450), the higher the degradation activity. When the non-inoculated media control (nc) supplemented with 15 mM sodium oxalate showed a value of 0.55, five isolates (ODB5, ODB29, ODB31, ODB35, and ODB36) exhibited significantly reduced values < 0.5 (Fig. 1a). Isolates with values higher than the control (nc) were suspected to have the ability to synthesize oxalate by themselves, and thus exhibited values > 0.55 (Fig. 1a). Each isolate showed a different growth pattern; in particular, the growth of ODB35 was relatively slow compared to other isolates (Fig. 1b). Because the growth of each isolate directly affects oxalate degradation, it is necessary to normalize values. Five isolates (ODB5, ODB29, ODB31, ODB35, and ODB36) had normalized values < 0.2, indicating that they had considerable oxalate-degrading activity (Fig. 1c). Three isolates (ODB4, ODB17, and ODB21) that did not grow in liquid culture were excluded.
Figure 1.
Oxalate degradation by bacterial isolates. (a) Absorbance measurement at A450. Oxalate in the supernatants was determined using an oxalate colorimetric assay kit. Bacterial isolates were grown in oxalate minimal (OM) medium supplemented with 1/10 TSB at 28 °C for 3 days. Cell-free supernatants were subjected to the sample preparation protocols described in Methods. The values report the amount of oxalate remaining in the OM medium. A total of 36 bacteria growing on OM agar plates were isolated from the surfaces of spinach (27 isolates; green) and strawberries (9 isolates; red). (b) Optical density (OD600) of the bacterial growth. (c) Normalised values calculated by multiplying the A450 value of the oxalate amount by the OD600 of the culture. The lower the value, the higher the degradation activity. Bacterial isolates ODB5, 29, 31, 35, and 36 exhibited significant oxalate degradation. Values are averages of triplicate assays and error bars represent the range. nc denotes the non-inoculated medium control. Asterisks denote significant differences from the control (*p < 0.05; Student’s t test).
Bacterial identification and antifungal activity tests
To confirm the identities of the ODB, we amplified and sequenced the 16S rRNA genes of ODB5, ODB29, ODB31, ODB35, and ODB36, which exhibited significant oxalate-degrading activity. BLAST analysis of the 16S rRNA gene sequence of ODB5 showed 99% identity with P. fluorescens; those of ODB29, ODB31, and ODB36 showed 99% identity with P. abietaniphila; and that of ODB35 showed 99% identity with Methylobacterium zatmanii.
Isolates ODB5, ODB35, and ODB36 were selected for further antifungal activity testing. ODB5 exhibited antifungal activity against B. cinerea, Alternaria alternata, and Saccharomyces cerevisiae, whereas ODB35 and ODB36 showed no antifungal activity (see Supplementary Fig. S1).
Disease suppression assay
To investigate disease suppression of ODB35 and ODB36, which exhibited no antifungal activity, we used the standard A. thaliana–B. cinerea (plant–pathogen) system. After 16 days of inoculation, water-sprayed plants exhibited a 65% disease rate against grey mould, whereas ODB36-sprayed plants exhibited significant disease suppression depending on the treatment concentration (Fig. 2). However, ODB35 exhibited no significant disease suppression compared with ODB36 (Fig. 2).
Figure 2.
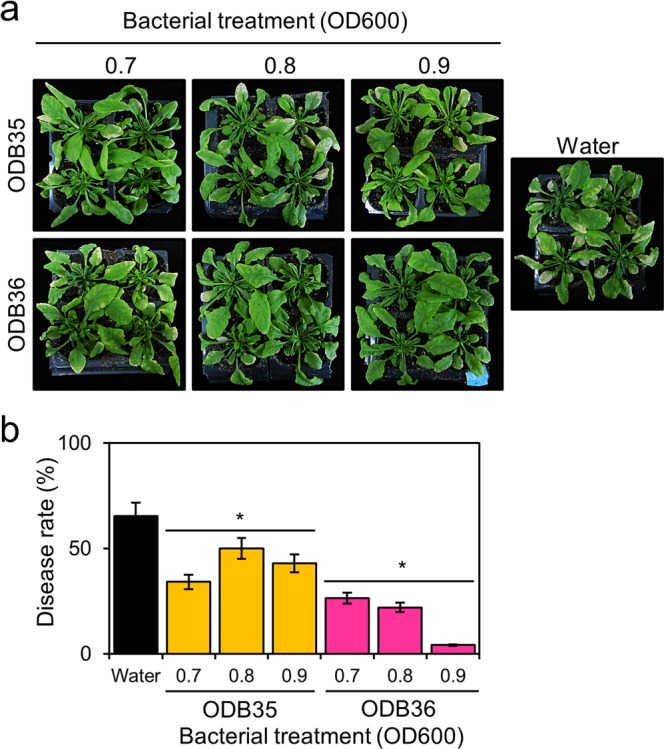
Grey mould disease suppression by ODB35 and ODB36. (a) Image captured 16 days after inoculation; (b) disease rate (%). Three-week-old Arabidopsis plants were sprayed with the ODB suspension. After incubation for 2 days, a B. cinerea spore suspension (5 × 105 spores/mL) was prepared using sterilised water. The spore suspension was sprayed until run-off occurred. Inoculated plants were placed in an opaque plastic box lined with saturated paper towels, the lids were removed after 2 days, and disease development was observed for 16 days and measured. ODB36 exhibited significant disease suppression depending on treatment concentration. Values are averages of triplicate assays; error bars represent the range. Asterisks denote significant differences from the water control (*p < 0.05; Student’s t test).
Phylogenetic analysis and sequence alignment
Using whole-genome information from GenBank (accession no. SDG16549), we analyzed a gene encoding an oxalate-degrading enzyme in P. abietaniphila ATCC 70068917. Translated amino acid sequences of PodA (42 kDa) showed 98% similarity with P. abietaniphila ATCC 700689 and 82% with P. fluorescens BBc6R8. Figure 3 shows the phylogenetic relationships of several putative formyl-CoA transferases from organisms whose protein sequences were available. As expected, the formyl-CoA transferases from P. abietaniphila and P. fluorescens clustered together. Sequence analysis showed that PodA, a formyl-CoA transferase, belongs to pfam02515, the CaiB-BaiF family of enzymes with diverse functions including fatty acid racemases, carnitine dehydratase (CaiB), bile acid inducible operon protein F (BaiF), benzylsuccinate CoA-transferase (BbsF), and anaerobic toluene catabolic protein in the presence of toluene. Identical residues, shown in red, are located in half of the N-termini of the proteins and partially in the C-termini (Fig. 4).
Figure 3.
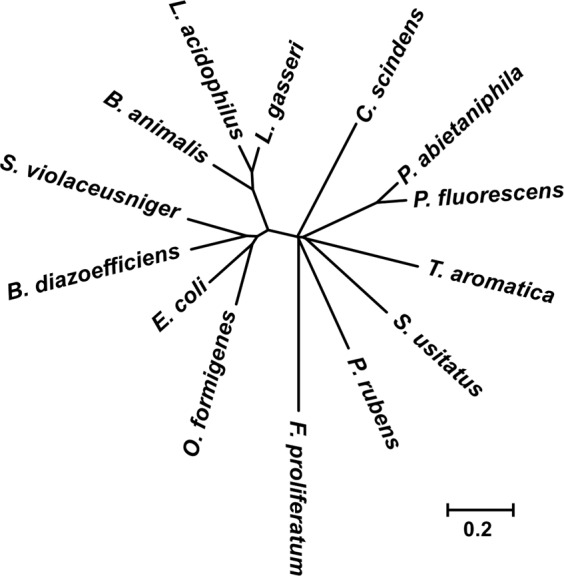
Unrooted phylogenetic tree of formyl-CoA transferase sequences. Proteins were aligned using CLUSTALX. Alignments were used for tree reconstruction. The organisms used were Bifidobacterium animalis (GenBank accession number WP004219152), Bradyrhizobium diazoefficiens USDA110 (NP769796), Clostridium scindens (AAC45415), Escherichia coli CFT073 (WP000106774), Fusarium proliferatum (AMB48876), Lactobacillus acidophilus NCK1728 (AAV42286), Lactobacillus gasseri ATCC 3323 (WP144231654), Oxalobacter formigenes Ox-B (AAC45298), Penicillium rubens Wisconsin 54–1255 (CAP92691), Pseudomonas fluorescens BBc6R8 (WP003209938), Solibacter usitatus Ellin 6076 (ABJ84069), Streptomyces violaceusniger Tu 4113 (AEM82356), and Thauera aromatica (AAF89841).
Figure 4.
Sequence alignment of formyl-CoA transferases. Pfam02515, CoA transferase (WP003209938); Baif, putative cholate CoA transferase (AAC45415); CaiB, (R)-carnitine CoA transferase (CAA52112); Frc, formyl-CoA transferase (AAC45298); and BbsF, (R)-benylsuccinate CoA transferase (AAF89841). Identical residues are shown in red; residues suggested to be involved in catalysis are shown in blue. CoA-contacting residues proposed in Frc are marked green.
PodA is required for the inhibition of Botrytis infection
To investigate the effect of the wild-type ODB36, podA− (podA–lacZY) mutant, and podA complementation strains on Botrytis infection, we performed disease suppression assays. Wild-type strain exhibited significant disease suppression. The podA− mutant did not inhibit Botrytis infection; the defect was recovered by podA complementation (Fig. 5). These results indicated that podA is critical for the inhibition of Botrytis infection in P. abietaniphila.
Figure 5.
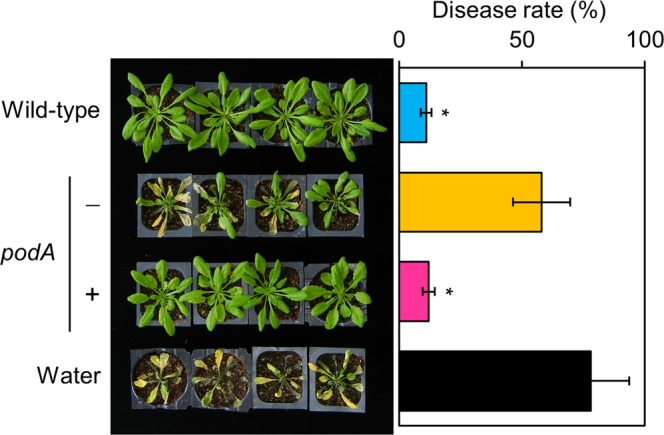
PodA is required for the inhibition of Botrytis infection. Four-week-old Arabidopsis plants were sprayed with suspensions (OD600 value 1.0) of the wild-type ODB36, podA− mutant (−), and podA complementation (+) strains. After incubation for 2 days, a B. cinerea spore suspension (5 × 105 spores/mL) was prepared using sterilised water. The spore suspension was sprayed until run-off occurred. Inoculated plants were placed in an opaque plastic box lined with saturated paper towels, the lids were removed after 2 days, and disease development was observed for 12 days and measured. Wild-type ODB36 strain exhibited significant disease suppression. The podA− mutant did not inhibit Botrytis infection; the defect was recovered by podA complementation. Values are averages of duplicate assays; error bars represent the range. Asterisks denote significant differences from the water control (*p < 0.05; Student’s t test).
PodA is required for the inhibition of oxalate toxicity
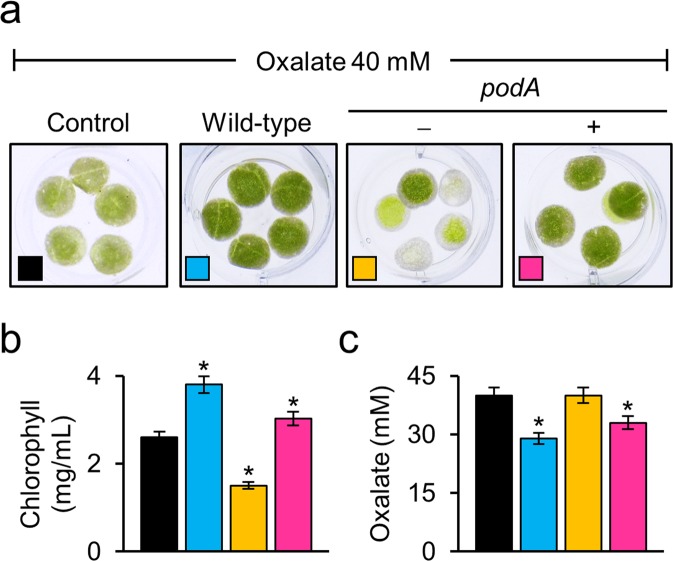
In tobacco leaf disc assays, oxalate toxicity, such as bleaching of leaf discs, was observed for the podA− (podA–lacZY) mutant but was weak for the wild-type and podA complementation strains (Fig. 6a). This result was confirmed by the quantification of chlorophyll. Tobacco leaf discs treated with the podA− mutant showed bleaching or a low chlorophyll level. These defects were restored to near wild-type levels by podA complementation (Fig. 6b). Also, oxalate degradation was induced by treatment of the wild-type and podA complementation strains (Fig. 6c).
Figure 6.
PodA is required for the inhibition of oxalate toxicity. (a) Tobacco leaf disc assays. Leaf discs were placed in suspensions of the wild-type, podA− mutant (−), and podA complementation (+) strains supplemented with 40 mM sodium oxalate. After incubation for 5 days, oxalate toxicity was observed on the leaf discs in the podA− mutant suspension but was reversed by podA complementation. (b) Chlorophyll in the leaf discs (a) was measured as described in the Methods section. (c) Remaining oxalate in suspensions (a) of the wild-type ODB36, podA− mutant, and podA complementation strains. Values are averages of duplicate assays; error bars represent the range. Asterisks denote significant differences from the control (*p < 0.05; Student’s t test).
Overexpression, purification, and oxalate-degrading and formate conversion activities of PodA protein
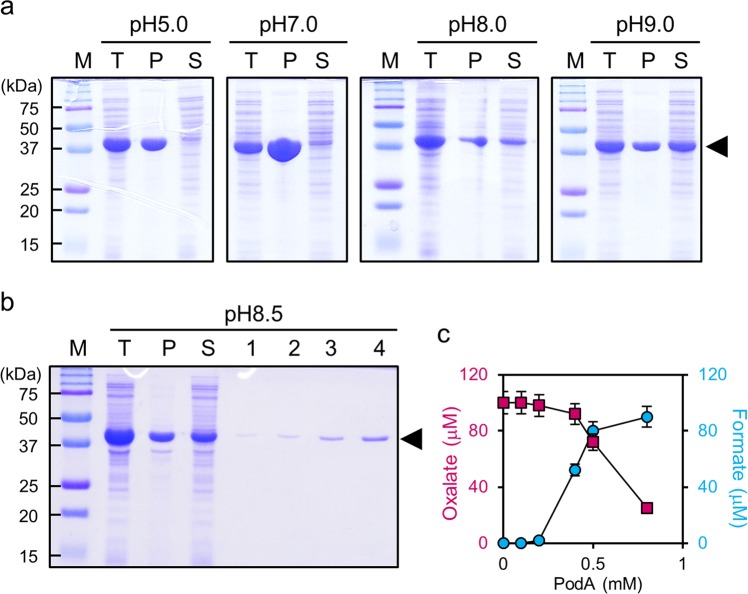
We overexpressed and purified P. abietaniphila ODB36 PodA from Escherichia coli BL21(DE3) carrying pLY205 (pET21b::podA). The podA gene encodes a protein of 396 amino acid residues with a calculated molecular weight of 42 kDa. To confirm that the podA gene encodes an oxalate-degrading enzyme, it was expressed under control of the T7 promoter in E. coli. Upon induction of E. coli BL21(DE3) harbouring pLY205 with IPTG, a His-tagged protein with a molecular mass of around 42 kDa was produced; approximately half of the PodA–His protein was expressed in soluble bodies at pH 8.0–9.0 (Fig. 7a). PodA–His was purified to near homogeneity using Ni-NTA affinity chromatography (Fig. 7b). The oxalate colorimetric assay kit revealed that PodA–His (0.8 mM) exhibited 75% oxalate-degrading activity (Fig. 7c). Using a formate colorimetric assay kit, we measured the formate conversion activity of PodA–His; the amount of degradation confirmed that oxalate was consistently converted to formate (Fig. 7c).
Figure 7.
Overexpression, purification, and oxalate-degrading and formate conversion activities of PodA–His. (a) Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) of PodA–His expression. The centrifuged cells of E. coli BL21(DE3) expressing PodA–His protein were subjected to sonic oscillation in cold 200 mM potassium phosphate buffer at pH 5.0–9.0. Each 10 µL sample was dissolved in 10 µL of 2 × loading buffer and boiled for 3 min at 100 °C. Final volumes of 10 µL were loaded in each lane. M, molecular mass standards; T, total cell extracts; P, pellet; S, supernatant of BL21(DE3) harbouring pLY205 after IPTG induction. (b) SDS-PAGE of PodA–His purification. Cell free sonic extract was worked up for purification using a Ni-NTA spin column. The purified protein eluted with 0.5 M imidazole was dialysed. Lanes 1–4: 1–, 2–, 5–, and 10–μL final elutions, respectively, containing PodA–His. Separation was conducted by 10% SDS-PAGE. Bands were visualised after staining with Coomassie Blue. The band corresponding to PodA–His is indicated. (c) Oxalate-degrading (closed square) and formate conversion (closed circle) activities of PodA–His determined using an oxalate colorimetric assay kit and a formate colorimetric assay kit, respectively. Values are averages of duplicate assays; error bars represent the range.
Inhibition of oxalate toxicity by PodA–His protein
To investigate the inhibitory effect of PodA–His protein on oxalate toxicity, we used a standard A. thaliana–oxalate (plant–toxin) system. Oxalate-sprayed plants exhibited an 81% oxalate toxicity rate, while PodA–His-sprayed plants showed significant and concentration-dependent inhibition of oxalate toxicity (Fig. 8).
Figure 8.
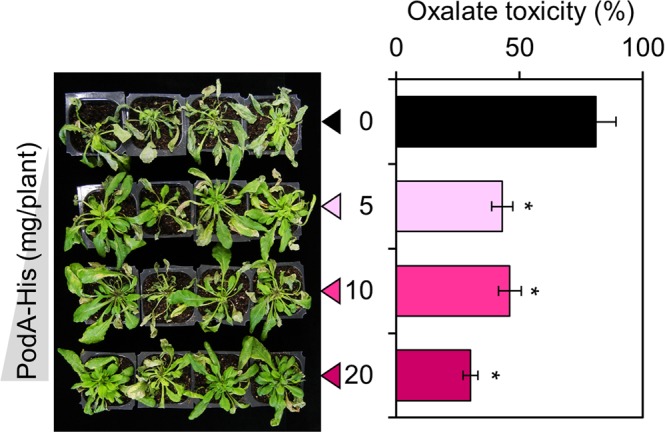
Inhibition of oxalate toxicity by PodA–His. Image captured 5 days after inoculation; oxalate toxicity rate (%). Six-week-old Arabidopsis plants were sprayed with PodA–His suspension. After incubation for 1 day, the plants were sprayed with an oxalate suspension (20 mM; 1 mL/plant). Oxalate toxicity was monitored for 5 days. PodA–His exhibited considerable and concentration-dependent inhibition of oxalate toxicity. Values are averages of duplicate assays; error bars represent the range. Asterisks denote significant differences from the control (*p < 0.05; Student’s t test).
Discussion
Grey mould causes damage during the cultivation, storage, and transportation of fresh vegetables and soft-pulling fruits1–3. Efforts have been made to control grey mould using antifungal bacteria, but this is complicated by the stability of antimicrobial substances in fresh vegetables and the emergence of resistance4,5. For this reason, we investigated the possibility of suppressing grey mould using non-antifungal ODB, similar to the use of a non-antagonistic bacterium to control Salmonella enterica, which causes food poisoning18.
In this study, we isolated ODB from the surfaces of spinach and strawberries and evaluated their disease control activity. We also explored potential correlations between oxalate-degrading ability and disease inhibitory effects of the ODB isolates. Previous studies have used antagonistic ODB13 or ODB with unknown antagonistic effects12. Therefore, unlike these previous studies, we attempted to exclude antagonistic effects by excluding P. fluorescens ODB5, which exhibited antifungal activity; we selected bacterial strains M. zatmanii ODB35 and P. abietaniphila ODB36 for further testing of dose response to disease suppression. Significant suppression of P. abietaniphila ODB36 against grey mould is shown in Fig. 2. We expect that biological control of fungal pathogens can be achieved by neutralising pathogenic factors and inhibiting the occurrence of resistant individuals by applying a method that does not kill pathogens.
The disease suppression effect of ODB35, which has the highest oxalate-degrading ability, did not reach that of ODB36. This result is likely due to the slow growth rate of ODB35. As shown in Fig. 1, oxalate-degradation activity was calculated by multiplying the A450 value by bacterial growth. ODB35 exhibited relatively slow growth, resulting in a high value. These results suggest that the growth rate of ODB on the plant surface is important to its efficacy as a disease control agent. The aim of this study was to evaluate the oxalate-degrading ability of bacteria in disease control; we finally selected ODB36 as the optimal agent among those examined. Growth on plant surfaces was not investigated in this study.
Interestingly, the wild-type and podA complementation strains eliminated the toxicity of 40 mM oxalate, such as bleaching of tobacco leaf discs, but the podA− mutant showed bleaching due to no oxalate toxicity. The remaining oxalate concentration in the wild-type and podA− complementation strains was lower than that of the podA− mutant strain. These data suggest that podA contributes to the degradation of oxalate. These results are in good agreement with the finding that grey mould control is by oxalate degradation in P. abietaniphila.
Family III CoA transferases include formyl-CoA:oxalate CoA transferase16, succinyl-CoA:(R)-benzylsuccinate CoA transferase19, (E)-cinnamoyl-CoA:(R)-phenyllactate CoA transferase20, and butyrobetainyl-CoA:(R)-carnitine CoA transferase21. These CoA transferases occur in prokaryotes and eukaryotes and catalyze CoA transfer reactions in a highly substrate- and stereo-specific manner22. Additionally, the PodA exhibited 81% identity (89% similarity) with Pfam02515, and 30% identity (47% similarity) with O. formigenes formyl-CoA transferase, which was the first member of family III of CoA transferases to be characterized22.
In this study, we did not quantify the formate concentration in ODB because P. abietaniphila strains contain several genes encoding formate dehydrogenase subunits, which catalyse the oxidation of formate to carbon dioxide. However, we performed protein overexpression and enzyme activity assays and confirmed that PodA–His converted oxalate to formate, which plays a role in the biosynthesis of many compounds in energetic metabolism and signal production related to stress in plants23.
In plants, three types of oxalate-degrading enzymes are known, namely, oxidase (OXO), decarboxylase (OXDC), and acetylases, among which four enzymes (oxalyl-CoA synthetase [AAE3], oxalyl-CoA decarboxylase [OXDE], formyl-CoA hydrolase [FXH], and formyl-CoA dehydrogenase [FXDE]) act in stages24–26. In enzymatic analysis of oxalate degradation of bacteria, formyl-CoA transferase (EC 2.8.3.16) catalyzes the chemical reaction, which is demonstrated in Fig. 915,16. The two substrates, oxalate and formyl-CoA, are converted into the two products oxalyl-CoA and formate. Formyl-CoA is required for the conversion of oxalate to oxalyl-CoA by formyl-CoA transferase16. However, the inhibitory effect of PodA on oxalate toxicity in Arabidopsis suggests the presence of formyl-CoA as a substrate. It is possibile that formyl-CoA derived from Arabidopsis-resident microorganisms acts as a substrate for PodA.
Figure 9.
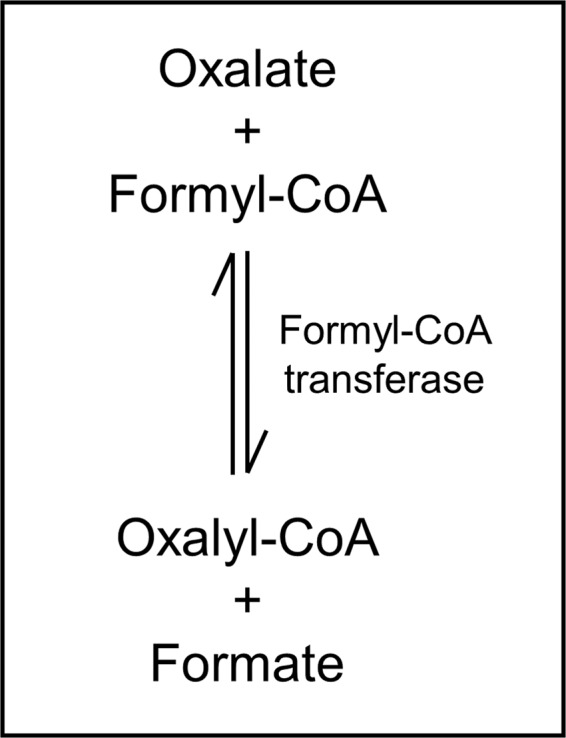
Oxalate degradation by formyl-CoA transferase. The two substrates, oxalate and formyl-CoA, are converted into the two products formate and oxalyl-CoA.
Previous studies have explored disease control in transgenic crops using plant-derived oxalate-degrading genetic resources. Barley oxalate oxidase transgenic peanuts showed enhanced resistance against Sclerotinia minor27. Soybean plants expressing wheat oxalate oxidase were resistant to Sclerotinia sclerotiorum28,29. These preliminary studies suggest that developed oxalate degradative genetic resources can be introduced into crops and inspire the development of transgenic plants with related abilities.
In conclusion, to overcome the competing processes of biological control and occurrence of resistant species, we investigated non-antimicrobial genetic resources for biological control of plant pathogens. Oxalate, a key pathogenic factor of necrotrophic fungi, was targeted to disarm, but not kill, plant pathogens. Oxalate-degrading bacteria were isolated and identified, and their activity was evaluated. Using disease suppression assays, P. abietaniphila ODB36 was finally selected for further investigation. PodA from ODB36, which encodes formyl-CoA transferase, was overexpressed and assayed using oxalate degradation tests. Bacterial cells of both ODB36 and enzyme PodA were found to be directly applicable in the field to control grey mould. The podA gene can also be applied for the development of transgenic resistant cultivars against grey mould.
Methods
Isolation of ODB
Surfaces of strawberries and spinach were swabbed with sterilised swabs, which were then streaked onto OM medium with 1.5% agar and incubated at 28 °C. After 7 days, single colonies were re-streaked onto OM agar medium for purification.
Bacterial oxalate-degrading activity
Oxalate-degrading activity was evaluated using an oxalate colorimetric assay kit according to the supplier’s protocol. Optical density (OD600) was measured in bacterial cultures grown in OM broth plus 1/10 TSB at 28 °C for 3 days. After centrifugation, the supernatants were transferred into 96-well plates and mixed with 2 μL oxalate converter, which was supplied with the kit. The mixtures were incubated at 37 °C for 1 h, and 50 μL prepared reaction mix (46 μL buffer; 2 μL oxalate enzyme mix, and 2 μL oxalate probe; all reagents were supplied with the kit) was added to each well containing standard or samples. Then the plates were incubated at 37 °C for 1 h and absorbance was measured at 450 nm (A450). The values reporting the amount of oxalate remaining in the sample were normalized to culture growth by multiplying the A450 value for the non-degraded oxalate amount by the OD600 of the culture. The lower the value, the higher the degradation activity.
16S rRNA gene sequencing
To confirm the identities of the ODB, the 16S rRNA gene was amplified and sequenced using the primers 27mF (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1429mR (5′-GGYTACCTTGTTACGACTT-3′). Total DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed using a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) with PCR polymerase (AccurPower PCR Premix; Bioneer, Daejeon, South Korea), 1 μL target DNA, and 1 mM each primer at 98 °C for 2 min, followed by 30 cycles of denaturation at 98 °C for 30 s, annealing at 55 °C for 30 s, and extension at 70 °C for 1 min, followed by a final extension at 72 °C for 4 min. The amplified products were separated by electrophoresis in 0.8% (w/v) agarose gels. PCR amplification yielded a single visible DNA product, whose band was cleaved from the ethidium bromide (EtBr)-stained gel and purified using a 200-p Expin Gel SV kit (GeneAll biotechnology, Seoul, Korea), following the manufacturer’s instructions. Purified PCR products were sequenced by Macrogen Services (Daejeon, Korea) in both directions using previously described primers30. DNA sequences were analyzed using the BLASTn program. DNA sequences of the 16S rRNA gene were compared with those in the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm.nih.gov/blast/).
Antifungal activity test
Among the ODB, the antifungal activity of the three isolates (ODB5, ODB35, and ODB36) showing the highest oxalate-degrading activity was tested against B. cinerea, A. alternata, and S. cerevisiae. Botrytis cinerea and A. alternata spores were spread on half potato dextrose agar with half protease peptone (PDP) and 1.5% agar. Saccharomyces cerevisiae cells were embedded in PDP agar. The ODB suspension (10 μL) was dropped onto the plate, which was then incubated at 28 °C for 48 h and the magnitude of inhibition of fungal growth was monitored.
Disease suppression assay
Arabidopsis thaliana Columbia (Col-0) was grown in a growth chamber at 20 °C under a 16-h photoperiod. Bacterial suspensions were prepared by resuspending the strains using sterilised water to adjust the OD600 values to the corresponding ranges (0.7–1.0). We sprayed 3-week-old Arabidopsis plants with the ODB suspensions (5 mL/plant). After 2 days of incubation, a B. cinerea spore suspension (5 × 105 spores/mL) was prepared using sterilised water. The spore suspension was sprayed until run-off. Inoculated plants were placed in an opaque plastic box lined with saturated paper towels, the lids were removed after 2 days, and disease development was observed for 16 days and measured according to the following calculation: (no. diseased leaves/no. inoculated leaves) × 100.
Sequence alignment and phylogenetic analysis
Protein sequences obtained from NCBI (https://www.ncbi.nlm.nih.gov/) were aligned and utilized to generate an unrooted phylogenetic tree using the neighbor-joining method (CLUSTALX software).
Construction of the podA− mutant and podA complementation strains
To generate the podA− mutant, plasmid pLY201, carrying the internal fragment of the podA gene was constructed using ODB36 genomic DNA as the PCR template and PodAE (5′-AAGATACTGGGTGAGTTT-3′) and PodAK (5′-GGTACCCATCACCAGTTTCGGATT-3′) as primers. The amplified region (291 bp) was purified from an agarose gel and ligated into the pGEM-T Easy Vector System (Promega, Mannheim, Germany) to generate pLY201, which was confirmed by sequencing. pLY201 fragments generated by digestion with the restriction enzymes EcoRI and KpnI (TaKaRa Bio Inc., Kusatsu, Japan) were purified after electrophoresis from an agarose gel and inserted into the suicide vector pVIK11231, generating pLY207. The resulting construct, pLY207, was transferred into E. coli S17-1 λpir and introduced into P. abietaniphila ODB36 by conjugation, generating podA–lacZY. The lacZY reporter gene fusion insertion mutants were selected based on the kanamycin-resistance phenotype and confirmed by PCR with primers that annealed upstream of the truncated fragments of podA, PodPro (5′-ATGCAAAAGCCTTTGCAAGGG-3′) and LacFuse (5′-GGGGATGTGCTGCAAGGCG-3′).
To construct the podA complementation strain, wild-type coding sequences with Plac were cloned into the broad-host vector pLAFR3. Coding sequences were amplified by PCR from ODB36 genomic DNA, using the primers PodAH (5′-GGCAAGCTTCTGAAACAGGAAACAGCTATGCAAAAGCCTTTGCAAGGG-3′) and PodAP (5′-GGCCTGCAGTCAAATCAATCCGCGAGCGCG-3′) and Phusion polymerase. Amplicons were ligated into pGEM-T Easy, confirmed by sequencing, excised by restriction enzyme cleavage, and ligated with appropriately cleaved pLAFR3. Plasmid pLY233 harbouring Plac–podA was verified by restriction digestion and sequencing prior to conjugation with P. abietaniphilae cells.
Leaf disc assays and chlorophyll quantification
Tobacco leaf disc assays32 and chlorophyll quantification assays33 were performed as described previously.
Overexpression and purification of PodA–His protein
We overexpressed PodA in E. coli by amplifying the coding region of podA using chromosomal DNA of ODB36 as a template with the primers PodN (5′-GGCCATATGCAAAAGCCTTTGCAAGGG-3′) with initiating ATG (bold) and PodX (5′-GGCCTCGAGAATCAATCCGCGAGCGCGCCA-3′) omitting stop codon, to which NdeI and XhoI sites (underline) were added. The amplified product was cloned at NdeI/XhoI site of pET21b containing a six-His tag at C-terminal (Novagen, Darmstadt, Germany) to generate pLY205. PodA–His was overexpressed in E. coli BL21(DE3), as described by the manufacturer (Novagen, Darmstadt, Germany). Cell free extracts were prepared by subjecting a suspension of 5 g centrifuged cells (wet weight) in 40 mL cold 0.2 M potassium phosphate or 0.2 M glycine-NaOH buffer (pH 5.0–9.0) to sonic oscillation in a solicitor (VCX130; Sonics & Materials Inc., Newtown, CT, USA) for 5 min at 4 °C. The sonic extract was centrifuged at 4 °C and 10,000 × g for 10 min. The supernatant solution was worked up for purification using a Ni-NTA spin column (Qiagen, Valencia, CA, USA). The eluted protein was dialyzed with the same buffer used for sonic oscillation, and the concentration of the purified protein was measured via the Bradford method with bovine serum albumin as the standard34.
Oxalate degradation and formate conversion activities of PodA–His
The oxalate-degrading and formate conversion activities of purified PodA–His were evaluated using an oxalate colorimetric assay kit and a formate colorimetric assay kit, respectively, according to the supplier’s protocol (BioVision Inc., Milpitas, CA, USA).
Inhibition of oxalate toxicity by PodA–His protein
Arabidopsis thaliana Columbia (Col-0) was grown in a growth chamber at 20 °C under a 16-h photoperiod. Protein suspensions were prepared by resuspending PodA–His in potassium phosphate buffer (pH 8.5) to 1, 2, and 4 mg/mL. We sprayed 6-week-old Arabidopsis plants with PodA–His protein suspensions (5 mL/plant). After incubation for1 day, the plants were sprayed with a sodium oxalate suspension (20 mM; 1–mL/plant). Oxalate toxicity was observed for 5 days and calculated according to the following equation: (no. of bleached leaves/total no. of leaves) × 100.
Supplementary information
Acknowledgements
This work was supported by the Gyeongsang National University Fund for Professors on Sabbatical Leave, 2017. Additional funds were provided by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1A6A1A03031413).
Author contributions
J.K. conceived and designed research. Y.L. and O.C. conducted experiments. B.K., J.B. and S.K. analyzed data. Y.L. and J.K. wrote the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-58609-z.
References
- 1.Wiliamson B, Tudzynski B, Tudzynski P, van Kan JA. Botrytis cinerea: the cause of grey mould disease. Mol. Plant. Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 2.Maude, R. B. Disease control. In The biology of Botrytis. (eds. Coley-Smith, J. R., Verhoeff, K. & Jarvis, W. R.). 275–308 (London: Academic Press, 1980).
- 3.Burgess DR, Bretag TW, Keane PJ. Seed to seedling transmission of Botrytis cinerea in chickpea ad disinfestation of seed with moist heat. Aust. J. Exp. Agric. 1997;37:223–229. doi: 10.1071/EA96061. [DOI] [Google Scholar]
- 4.Komárek M, Čadková E, Chrastný V, Bordas F, Bollinger JC. Contamination of vineyard soils with fungicides: a review of environmental and toxicological aspects. Env. Inter. 2010;36:138–151. doi: 10.1016/j.envint.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Leroux P, et al. Mechanisms of resistance to fungicides in field of Botrytis cinerea. Pest. Manag. Sci. 2002;58:876–888. doi: 10.1002/ps.566. [DOI] [PubMed] [Google Scholar]
- 6.Haidar R, Fermaud M, Calvo-Garrido C, Roudet J, Deschamps A. Modes of action for biological control of Botrytis cinerea by antagonistic bacteria. Phytopathol. Mediterr. 2016;55:301–322. [Google Scholar]
- 7.Gabler FM, Smilanick JL. Postharvest control of table grape gray mold on detached berries with carbonate and bicarbonate salts and disinfectants. Am. J. Enol. Vitic. 2001;52:12–20. [Google Scholar]
- 8.Rollins JA, Dickman MB. pH signaling in Sclerotinia sclerotiorum: Identification of a pacC/RIMI homolog. Appl. Env. Microbiol. 2001;67:75–81. doi: 10.1128/AEM.67.1.75-81.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murru N, et al. Screening of oxalate degrading lactic acid bacteria of food origin. Ital. J. Food Saf. 2017;6:61–64. doi: 10.4081/ijfs.2017.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federici F, et al. Characerization and heterologous expression of the oxalyl coenzyme A decarboxylase gene from Bifidobacterium lactis. Appl. Env. Microbiol. 2004;70:5066–5073. doi: 10.1128/AEM.70.9.5066-5073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manteau S, Abouna S, Lambert B, Legendre L. Differential regulation by ambient pH of putative virulence factor secretion by the phytopathogenic fungus Botrytis cinerea. FEMS Microbiol. Ecol. 2003;43:359–366. doi: 10.1111/j.1574-6941.2003.tb01076.x. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajkumar M, Jayaraj J, Muthukrishnana S, Bhaskaran R, Velazhahan R. Detoxification of oxalic acid by Pseudomonas fluorescens strain PfMDU2: Implications for the biological control of rice sheath blight caused by Rhizoctonia solani. Microbiol. Res. 2005;160:291–298. doi: 10.1016/j.micres.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Schoonbeek HJ, Jacquat-Bovet AC, Mascher F, Métraux JP. Oxalate-degrading bacteria can protect Arabidopsis thaliana and crop plants against Botrytis cinerea. Mol. Plant-Microbe Interact. 2007;20:1535–1544. doi: 10.1094/MPMI-20-12-1535. [DOI] [PubMed] [Google Scholar]
- 14.Hatch M, Freel RW. Alterations in intestinal transport of oxalate in disease states. Scanning Microsc. 1995;9:1121–1126. [PubMed] [Google Scholar]
- 15.Azcarate-Peril MA, Bruno-Bárcena JM, Hassan HM, Klaenhammer TR. Transcriptional and functional analysis of oxalyl-Coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl. Env. Microbiol. 2006;72:1891–1899. doi: 10.1128/AEM.72.3.1891-1899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson S, Ricagno S, Lindqvist Y, Richards NG. Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes. J. Biol. Chem. 2004;279:36003–36012. doi: 10.1074/jbc.M404873200. [DOI] [PubMed] [Google Scholar]
- 17.Martin VJ, Mohn WW. Genetic investigation of the catabolic pathway for degradation of abietane diterpenoids by Pseudomonas abietaniphila BKME-9. J. Bacteriol. 2000;182:3784–3793. doi: 10.1128/JB.182.13.3784-3793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim W-I, et al. Inhibition of Salmonella enterica growth by competitive exclusion during early alfalfa sprout development using a seed-dwelling Erwinia persicina strain EUS78. Intl J. Food Microbiol. 2020;312:108374. doi: 10.1016/j.ijfoodmicro.2019.108374. [DOI] [PubMed] [Google Scholar]
- 19.Leutwein C, Heider J. Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J. Bacteriol. 2001;183:4288–4295. doi: 10.1128/JB.183.14.4288-4295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickert S, Pierik AJ, Linder D, Buckel W. The involvement of coenzyme A esters in the dehydration of (R)-phenyllactate to (E)-cinnamate by Clostridium sporogenes. Euro J. Biochem. 2000;267:3874–3884. doi: 10.1046/j.1432-1327.2000.01427.x. [DOI] [PubMed] [Google Scholar]
- 21.Rangarajan ES, Li Y, Iannuzzi P, Cygler M, Matte A. Crystal structure of Escherichia coli crotonobetainyl-CoA: carnitine CoA-transferase (CaiB) and its complexes with CoA and carnitinyl-CoA. Biochem. 2005;44:5728–5738. doi: 10.1021/bi047656f. [DOI] [PubMed] [Google Scholar]
- 22.Heider J. A new family of CoA-transferases. FEBS Lett. 2001;509:345–349. doi: 10.1016/S0014-5793(01)03178-7. [DOI] [PubMed] [Google Scholar]
- 23.Igamberdiex AU, Bykova NV, Kleczkowski LA. Origins and metabolism of formate in higher plants. Plant. Physiol. Biochem. 1999;37:503–513. doi: 10.1016/S0981-9428(99)00152-7. [DOI] [Google Scholar]
- 24.Anantharam V, Allison. MJ, Maloney PC. Oxalate: formate exchange. basis energy coupling Oxalobacter. J. Bio Chem. 1989;264:7244–7250. [PubMed] [Google Scholar]
- 25.Turroni S, et al. Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J. Appl. Microbiol. 2007;103:1600–1609. doi: 10.1111/j.1365-2672.2007.03388.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhao C, et al. Oxalate-degrading enzyme recombined lactic acid bacteria strains reduce hyperoxaluria. Urol. 2018;113:253.e1–253.e7. doi: 10.1016/j.urology.2017.11.038. [DOI] [PubMed] [Google Scholar]
- 27.Livingstone DM, Hampton JL, Phipps PM, Grabau EA. Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant. Physiol. 2005;137:1354–1362. doi: 10.1104/pp.104.057232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH. Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotinia sclerotiorum. Physiol. Mol. Plant. Pathol. 2001;59:297–230. doi: 10.1006/pmpp.2001.0369. [DOI] [Google Scholar]
- 29.Yang X, et al. Enhanced resistance to sclerotinia stem rot in transgenic soybean that overexpresses a wheat oxalate oxidase. Transgenic Res. 2019;28:103–114. doi: 10.1007/s11248-018-0106-x. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacterial. 1991;179:697–703. doi: 10.1128/JB.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 32.Koh S, et al. A novel light-dependent selection marker system in plants. Plant. Biotechnol. J. 2011;9:348–358. doi: 10.1111/j.1467-7652.2010.00557.x. [DOI] [PubMed] [Google Scholar]
- 33.Amon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant. physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.