Abstract
Therapy by adoptive transfer of ex vivo-expanded tumor-infiltrating or genetically modified T cells may lead to impressive clinical responses. However, there is a need to improve in vivo persistence and functionality of the transferred T cells, in particular, to face the highly immunosuppressive environment of solid tumors. Here, we investigate the potential of miR-155, a microRNA known to play an important role in CD8+ T cell fitness. We show that forced expression of miR-155 in tumor antigen-specific T cells improves the tumor control of B16 tumors expressing a low-affinity antigen ligand. Importantly, miR-155-transduced T cells exhibit increased proliferation and effector functions associated with a higher glycolytic activity independent of exogenous glucose. Altogether, these data suggest that miR-155 may optimize the antitumor activity of adoptively transferred low-affinity tumor-infiltrating lymphocytes (TILs), in particular, by rendering them more resistant to the glucose-deprived environment of solid tumors. Thus, transgenic expression of miR-155 may enable therapeutic targeting of self-antigen-specific T cells in addition to neoantigen-specific ones.
Keywords: microRNA-155, CD8+ T cells, TCR antigen affinity, therapeutic vaccine, adoptive cell transfer, oncoimmunology
Introduction
Harnessing the immune system to fight cancer has proven clinically successful in the past few years. Indeed, following the introduction in 2011 of therapeutic antibodies blocking immune checkpoint receptors and ligands, the US Food and Drug Administration (FDA) approved, in 2017, the adoptive transfer of genetically modified autologous T cells as an effective therapy against B cell malignancies. Two adoptive cell transfer approaches have shown promise in the clinical setting. On one hand, the isolation, expansion, and reinjection of tumor-infiltrating lymphocytes (TILs) proved useful in metastatic melanoma.1 On the other hand, adoptive transfer of peripheral blood T cells expressing chimeric tumor antigen-specific receptors resulted in objective clinical responses in refractory leukemia.2 However, there is still a need to improve the survival of transferred T cells in solid tumors. For instance, genetically engineered T cells with the ability to produce their own survival signal have been investigated in order to cope better with the deleterious tumor microenvironment.3, 4, 5, 6, 7, 8
microRNAs are small RNAs that regulate protein levels via specific binding to mRNAs and blocking of their translation.9 The microRNA miR-155 was first identified as an oncomir, responsible for driving B cell malignancy.10 Its importance in a variety of immune cells has subsequently been demonstrated, including dendritic cell (DC)-dependent T cell activation and response to lipopolysaccharide (LPS),11,12 as well as macrophage differentiation13,14 and CD4+ T cell immune fitness and differentiation.15, 16, 17, 18, 19 More recently, we and others have shown the importance of miR-155 for the functionality of cytotoxic CD8+ T cells.8,20, 21, 22 Upon T cell activation, the expression of miR-155 is strongly upregulated, which is crucial for CD8+ T cell expansion and survival. Indeed, CD8+ T cells lacking miR-155 can develop normally in homeostatic conditions but are impaired in their proliferation and survival ability during an antiviral response.8,21 Moreover, deletion of miR-155 was intrinsically detrimental to a potent CD8+ T cell antitumor response.8,22 Conversely, adoptive transfer of miR-155-overexpressing gp100-specific CD8+ T cells induced a superior antitumor response upon vaccinia virus–human gp100 (hgp100) infection and systemic injection of interleukin-2 (IL-2).8 Mechanistically, miR-155 expressed in T cells blocks the translation of several mRNAs, including interferon-γ receptor-α (IFN-γRα),20 suppressor of cytokine signaling 1 (SOCS-1),8 tyrosine-protein phosphatase non-receptor type 2 (PTPN2),23 and inositol polyphosphate 5-phosphatase 1 (INPP5D),24,25 which are mostly negative regulators of T cell activation, survival, expansion, and cytokine response. Interestingly, adoptive transfer of miR-155-overexpressing pmel T cells, followed by hgp100 viral infection, was proven to circumvent the need of host immunodepletion before adoptive transfer.23 Hence, miR-155 is a good candidate to improve CD8+ T cell antitumor efficiency for adoptive cell transfer.
The present study further demonstrates the potential of miR-155 overexpression to modulate T cell fitness and metabolism. Importantly, our results highlight the capacity of miR-155 overexpression to improve the antitumor activity of CD8+ T cells of low affinity to tumor antigens upon peptide-CpG vaccination. This method might prove useful for naturally occurring tumor-specific T cells in cancer patients, which are usually of low affinity to tumor antigens.
Results
Transduced OT-1 T Cells Stably Overexpress miR-155 In Vitro
We used miR-155 retroviral transduction, as described previously.8,26 Purified CD45.1 H-2Kb/ovalbumin (OVA) OT-1 T cells were activated and transduced with retrovirus to express constitutively either the green fluorescent protein (GFP) alone or fused to the microRNA miR-155 (provided by the group of Luca Gattinoni, NIH). Three days later, the percentage of transduced cells was, on average, 90%, as measured by GFP expression (Figure 1A). miR-155 expression levels, measured by qPCR, were increased 30- to 50-fold compared to control-transduced OT-1 cells (Figure 1B). As miR-155 is strongly upregulated following T cell activation,20,27,28 we compared the levels of the endogenous miR-155 over its retroviral-mediated overexpression. Indeed, as expected, activation with αCD3/αCD28-coated magnetic beads resulted in comparable 30-fold increases of miR-155 in OT-1 controls (Figure 1C). As a consequence, the level of miR-155 in the miR-155-transduced cells, 1 day after activation, was only 2-fold higher when compared to the control-transduced cells, suggesting that the transgene expression only minimally contributed to the total amount of microRNA in activated T cells. This was true for both exposure to CD3/CD28 magnetic beads (Figure 1C) and antigen-specific activation upon coculture with OVA-expressing tumor cells (Figure 1D).
Figure 1.
Transduced OT-1 Cells Stably Overexpress miR-155 In Vitro
(A) Histogram showing GFP expression in OT-1 cells transduced with the control vector (black) or the miR-155 overexpression vector (blue) 2 days after transduction. Untransduced cells are pictured in grey. (B) qPCR measurement of miR-155 levels in OT-1 control or OT-1 miR-155 resting cells (N = 3). (C and D) qPCR of miR-155 levels before and 1, 2, and 4 days following coculture with (C) anti-CD3 anti CD28-coated beads (1:1 ratio) (N = 3) and (D) B16-OVA cells (5:1 ratio) (N = 3). Pooled data extracted from two independent experiments. Plots show mean ± SD.
miR-155 Overexpression Increases In Vivo Expansion of OT-1 Cells upon Infection or Vaccination
To evaluate the impact of miR-155 overexpression on the in vivo functionality of adoptively transferred OT-1 CD8+ T cells, we first used the bacterial infection model of transgenic Listeria monocytogenes expressing the ovalbumin protein (Lm-OVA). mir-155-transduced OT-1 cells, measured as the percentage of CD45.1+ GFP+ double-positive cells out of total CD8+ T cells, were indeed increased at the peak of expansion compared to control-transduced cells (Figure 2A). Although the overexpression of miR-155 did not have an impact on the early expansion phase, the proliferation of OT-1 miR-155 cells was increased compared to OT-1 control cells, 6 days after infection, as measured by bromodeoxyuridine (BrdU) incorporation (Figure 2B). Whereas we and others20,23 showed that miR-155 overexpression results in increased accumulation of CD8+ T cells upon viral and bacterial infection (Figure 2A), such observations were not confirmed in vaccination settings. Therefore, we compared the potential of miR-155 overexpression for the functionality of CD8+ T cells in Lm-OVA-infected and OVA peptide/CpG-vaccinated mice. Seven days after infection (Figures 2C and 2D) or vaccination (Figure 2E), OT-1 miR-155 cell frequencies were increased in the blood of mice compared to OT-1 control cells. With regard to T cell differentiation, we showed in a previous publication that OT-1 cells lacking miR-155 expression expressed higher levels of CD62 ligand (CD62L) and CD127.8 Conversely, we show here that two weeks post CpG-OVA vaccination, miR-155-overexpressing OT-1 cells exhibited significantly reduced CD62L (Figure 2F) and CD127 (Figure 2G) as compared to control cells. These data suggest that miR-155 might also be a good candidate molecule to improve T cell response to cancer vaccines.
Figure 2.
miR-155 Overexpression Increases the Proliferation and Accumulation of OT-1 Cells In Vivo upon Infection
(A) Percentages of GFP+ OT-1 cells among total CD8 were measured in the spleen of Lm-OVA-infected mice, 4, 6, and 7 days after infection (N = 6). Plot shows mean ± SD. Sidak’s multiple comparisons test was performed. ***p ≤ 0.001; ****p ≤ 0.0001. (B) Percentage of proliferating BrdU+ cells among total OT-1 control and OT-1 miR-155 splenocytes, 2.5 h after peritoneal BrdU injection, 4 and 6 days after Lm-OVA infection. Unpaired t test was performed for each day. **p ≤ 0.01 ns p > 0.05. (C) Representative plots of (D). (D and E) Percentages of OT-1+ GFP+ cells in the CD8+ compartment, 7 days after (D) Lm-OVA infection or (E) CpG OVA vaccination. (F and G) Median fluorescence intensity (MFI) of (F) CD62L and (G) CD127 measured by flow cytometry, 2 weeks after Lm-OVA infection on OT-1 control and OT-1 miR-155 splenocytes. Results are representative of 3 independent experiments. (D–G) Unpaired t test was performed. ***p ≤ 0.001; *p ≤ 0.05
miR-155 Overexpression Is Associated with Enhanced Metabolic Activity of OT-1 Cells
miR-155 is important in controlling the signaling pathways downstream of T cell receptor (TCR) activation. Indeed, miR-155 overexpression decreases the intracellular amount of its target INPP5D and thus, augments the phosphorylation of Akt.23 We hypothesized that this regulatory mechanism could lead to an increased mechanistic target of rapamycin kinase (mTOR) activity, which would, in turn, lead to a more differentiated T cell phenotype. We thus measured the amount of phosphorylated ribosomal protein S6, which is downstream of the mTOR complex 1 (mTORC1). After 1.25 h of coculture with αCD3/αCD28, the amount of phosphorylated S6 was indeed increased in OT-1 miR-155 compared to OT-1 control cells, indicating a higher mTORC1 activity (Figure 3A).
Figure 3.
Overexpression of miR-155 Increases mTOR Signaling and the Basal and Activation-Induced Metabolic Activity of T Cells
(A) Quantity of phosphoS6 measured by flow cytometry before and 1.25 h after activation with αCD3/αCD28 beads. (B) Basal extracellular acidification rate (ECAR) of OT-1 control and miR-155 cells in the glucose-supplemented medium. Unpaired t test was performed. ***p ≤ 0.001 (N = 3). (C) Basal ECAR of OT-1 control and miR-155 cells in medium without glucose. Unpaired t test was performed. ***p ≤ 0.001 (N = 3). (D) Extracellular acidification rate (ECAR) was measured in a Seahorse machine after sequential addition of αCD3/αCD28 beads, oligomycin, FCCP, and 2DG. The starting medium contained 10 mM glucose and 2 mM glutamine. Data plots show mean ± SEM. (E) ECAR was measured using a Seahorse machine after sequential addition of αCD3/αCD28 beads, glucose, oligomycin, and 2DG. The starting medium contained 2 mM glutamine and no glucose (N = 4). Plot shows mean ± SEM. (F) Basal oxygen consumption rate (OCR) was measured in a Seahorse machine after sequential addition of αCD3/αCD28 beads, oligomycin, FCCP, and 2DG. The starting medium contained 10 mM glucose and 2 mM glutamine. Data plots show mean ± SEM. (G) OCR was measured using a Seahorse machine after sequential addition of αCD3/αCD28 beads, glucose, oligomycin, and 2DG. The starting medium contained 2 mM glutamine and no glucose (N = 4). Plot shows mean ± SEM. (E–G) Sidak’s multiple comparison test was used to compute statistical significance. *p ≤ 0.05; **p ≤ 0.01. Representative results from 1 experiment out of 2 independent experiments.
The mTOR pathway is instrumental in allowing CD8+ T cells to switch from oxidative phosphorylation (oxphos) to aerobic glycolysis.29 Moreover, this switch is crucial for the development of CD8+ T cell effector function, such as production of IFN-γ.30 We hence measured the metabolic activity of miR-155-transduced OT-1 cells, before and after activation in metabolic flow (Seahorse) assays. At the resting state, OT-1 miR-155 cells had a higher extracellular acidification rate (ECAR), both in the presence (Figure 3B) or absence of glucose (Figure 3C), as compared to control-transduced OT-1 cells. This might indicate that miR-155-overexpressing cells have a higher glucose reservoir for glycolytic needs in the absence of extracellular glucose. Next, we evaluated the capacity of OT-1 cells to upregulate their glycolytic respiration upon activation with αCD3/αCD28 beads (Figure 3D). All OT-1 cell populations could switch from the oxphos to the glycolytic pathway, regardless of the overexpression of miR-155. Since the tumor microenvironment can be deprived of glucose, due to the high consumption by cancer cells,31,32 we tested if the overexpression of miR-155 could preserve the metabolic response upon activation in the glucose-deprived conditions. OT-1 miR-155 cells displayed an increased glycolytic response compared to OT-1 control cells after being primed in the absence of glucose (Figure 3E). Additionally, OT-1 miR-155 cells showed an increased oxygen consumption rate (OCR) in the resting phase and upon activation in the presence (Figure 3F) or in the absence of extracellular glucose (Figure 3G). This demonstrates that the overexpression of miR-155 increases the metabolic activity of CD8+ T cells, independently of the presence of glucose. This faculty is a promising aspect of T cell biology, since tumor microenvironments are usually depleted from glucose and other nutrients,33 which is part of the tumor-mediated immunosuppression. In fact, the ability of T cells to switch to aerobic glycolysis has been linked to a better antitumor response.31,32
Overexpression of miR-155 in CD8+ T Cells Confers Competitive Fitness and Increased Polyfunctionality in the Tumor
In light of their higher metabolism, effector phenotype, and proliferative ability, we assessed the capacity of OT-1 miR-155 cells to mediate tumor rejection. We first tested the ability of OT-1 miR-155 T cells to infiltrate tumors and to produce cytokines following antigen recognition. To do so, OVA-expressing B16 melanoma tumor cells (B16-OVA) were inoculated in the flank of mice. To minimize the impact of intrinsic variations between animals, as well as differences in tumor sizes, we cotransferred OT-1 miR-155 and OT-1 control cells (1:1 ratio) bearing different congenic markers into the same tumor-bearing hosts. Then, mice were either vaccinated with the SIINFEKL OVA peptide/CpG or infected with Lm-OVA or PBS control (Figure 4A). Seven days after vaccination or infection, mice were sacrificed, and the percentages of OT-1 control or miR-155 was measured in the blood and in tumors. As previously shown,[8] the ability of OT-1 miR-155 CD8+ T cells to accumulate in the blood after either vaccination or infection significantly surpassed that of the OT-1 control cells (Figure 4B). Strikingly, OT-1 miR-155 CD8+ T cells were also found in higher numbers in tumors (Figures 4C and 4D), and this difference was even increased when compared to circulating cells. We then assessed the ex vivo effector functions of the OT-1 cells by restimulating them with the OVA peptide for 4 h. At the peak of the immune response, both OT-1 control and miR-155 extracted from the tumor could respond to restimulation. However, a higher percentage of OT-1 miR-155 cells produced both IFN-γ and tumor necrosis factor (TNF)-α when compared to OT-1 control cells (Figure 4E). Finally, no striking differences in levels of programmed cell death 1 (PD-1), CD62L, and CD44 were seen in the tumor, whereas circulating miR-155-transduced OT-1 cells had an increased expression of CD44 (data not shown) and decreased expression of CD62L (Figure 2F). Strikingly, we observed that the level of expression of the coreceptors CD8α and CD8β was significantly higher on OT-1 miR-155 cells as compared to OT-1 control cells, both in the blood (Figures 4F and 4G) and tumors (Figures 4H and 4I). Interestingly, OT-1 miR-155 cells had similar CD8α and CD8β expression levels as endogenous CD8 T cells (Figures 4F and 4G), suggesting that miR-155 might prevent the CD8 downregulation, which occurs upon T cell activation.
Figure 4.
Overexpression of miR-155 in CD8+ T Cells Confers Competitive Fitness and Increased Polyfunctionality in the Tumor
(A) CD45.2 OT-1 cells were transduced with the miR-155 vector, and CD45.1/2 OT-1 cells were transduced with the control vector and cotransferred at a 1:1 ratio in CD45.1 tumor-bearing mice. 1 day later, mice were either infected with Lm-OVA or vaccinated with CpG and OVA. (B) 7 days after vaccination or infection, blood was collected and stained, and the percentage of GFP+ OT-1 cells was measured by flow cytometry. (C) 7 days after vaccination or infection, tumor-infiltrating lymphocytes were isolated, and the percentage of GFP+ OT-1 cells in total CD8+ T cells was measured by flow cytometry. (D) Representative fluorescence-activated cell sorting (FACS) plot of the frequency of CD45.1+ and GFP+ cells in tumors of CpG-OVA or Lm-OVA-treated mice. (E) Percentage of IFN-γ+ and TNF-α double-positive cells in total OT-1 control and OT-1 miR-155 cells isolated from tumors 7 days after vaccination or infection and ex vivo-restimulated with N4 peptide. (F and G) MFI of (F) CD8α and (G) CD8β was measured by flow cytometry on GFP+ OT-1 cells and GFP− CD8+ T cells extracted from blood. (H and I) MFI of (H) CD8α and (I) CD8β was measured by flow cytometry on GFP+ OT-1 cells extracted from tumors. (B, C, and E–I) Sidak’s multiple comparison test was performed on each pair of data. ns p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Overexpression of miR-155 in OT-1 Cells Improves Their Ability to Mediate Protection against Tumors Expressing a Low-Affinity Antigen
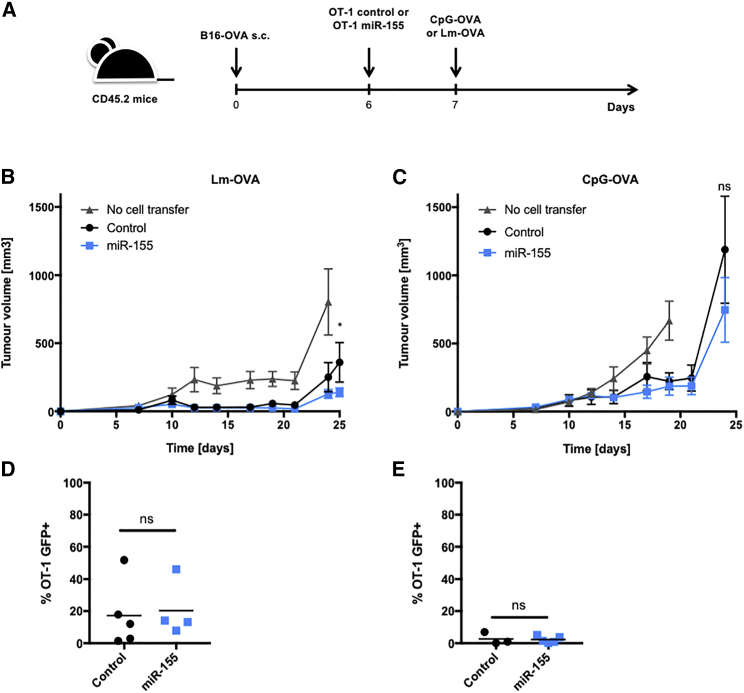
To check if the increased ability of OT-1 miR-155 cells to infiltrate tumors in a cotransfer setting translated into a better antitumor effect, we subcutaneously engrafted B16-OVA tumors in CD45.2 mice, and CD45.1 OT-1 control or miR-155 cells were transferred 6 days later. Mice were vaccinated or infected the following day (Figure 5A). As expected, the peripheral expansion of OT-1 miR-155 cells was increased as compared to OT-1 control cells in both vaccination and infection settings (data not shown). However, the antitumor protection upon Lm-OVA infection was only mildly improved when mice were transferred with miR-155 OT-1 (Figure 5B), and no difference was observed upon OVA peptide/CpG vaccination (Figure 5C). The tumors were analyzed 27 days after graft for Lm-OVA-infected mice and 25 days postvaccination. In line with the absence of improved tumor control, the infiltration of OT-1 miR-155 cells was not superior to the OT-1 control cells, whether post Lm-OVA infection (Figure 5D) or peptide vaccination (Figure 5E). These results suggested that despite an increase in peripheral expansion, as well as a more metabolically active effector phenotype, the overexpression of miR-155 did not greatly ameliorate the antitumor response in the aggressive B16-OVA melanoma model.
Figure 5.
miR-155 Overexpression in OT-1 Cells Marginally Improves Tumor Control upon Infection with Lm-OVA but Fails to Do so upon Tumor Vaccine
(A) C57BL/6 mice were engrafted subcutaneously with 1 × 105 B16-OVA (n = 5–6). 6 days later, 1 × 105 OT-1 T cells were transferred intravenously. At day 7, mice were either infected with Lm-OVA or vaccinated with CpG-OVA. (B and C) Tumor volumes were measured with a manual caliper every other day from day 7 postengraftment until the end of the experiment for (B) mice infected with Lm-OVA or (C) mice vaccinated with CpG-OVA. These figures are from one representative experiment out of three independent experiments. Sidak’s multiple comparisons test was performed on each measured day for OT-1 control versus OT-1 miR-155. ns p > 0.05; *p ≤ 0.05. (D and E) Analysis of TILs extracted from tumors of Lm-OVA-infected mice (D) 27 days post-tumor graft and (E) 25 days post-tumor graft in CpG/OVA-vaccinated mice. Unpaired t test was performed. ns p > 0.05.
These results were in contrast with our previous demonstration of a significant improvement in tumor control by overexpressing miR-155 in gp100/pmel CD8+ T cells.8 However, pmel cells are self-tolerant mouse gp100 (mgp100)-specific CD8+ T cells, which do not protect against mgp100-expressing B16 tumors unless stimulated via infection with a vaccinia virus expressing the human gp100.34 The affinity of the pmel cells to their cognate antigen is relatively low, in contrast to the TCR of OT-1 cells that have a high affinity for the SIINFEKL OVA peptide, as shown by their negative thymic selection in young mice injected with OVA.35 We hypothesized that the absence of an improved inhibition of B16-N4 tumor development resulted from an already strong upregulation of endogenous miR-155 in the OT-1 control cells, which had reached a plateau and could not be further increased by constitutive miR-155 overexpression (Figure 1D). This hypothesis was indeed supported by our observation that miR-155 is less upregulated in low-affinity T cells.36 Thus, this observation would suggest that forced miR-155 overexpression may have more impact in a low-affinity T cell response.
To investigate if the affinity to the target antigen was a determining factor for the improved CD8+ T cell function upon miR-155 overexpression, we took advantage of the high-affinity SIINFEKL (N4) and low-affinity SIITFEKL (T4) OVA analog peptides. The altered peptide ligand T4 has an approximately 10-fold decreased affinity for the OT-1 TCR, which is at the limit between negative selection and self-tolerance.35,37,38 In agreement with our hypothesis, miR-155 OT-1 T cells, activated in vitro with B16-OVA T4, had a 10-fold higher miR-155 level as compared to control cells (Figure 6A), whereas the difference was only 2-fold in the presence of B16-N4 1 day after activation (Figure 1D). We subcutaneously engrafted C57BL/6J mice with 105 B16 tumor cells expressing either the native OVA epitope (B16-N4) on the right flank or the altered, low-affinity T4 peptide (B16-T4) on the left flank. OT-1 control or miR-155 cells were intravenously transferred 10 days postgraft, and vaccination was performed with CpG and the N4 peptide on the same day (Figure 6B). Similarly to the single graft of B16-OVA (N4) shown above (Figure 5C), the overexpression of miR-155 did not significantly improve the protection against B16-N4 tumors upon vaccination (Figure 6C). In contrast, 18 days after engraftment, the B16-T4 tumors treated with OT-1 miR-155 T cells were significantly smaller than the ones injected with the OT-1 control cells (Figure 6D). As expected, the OT-1 miR-155 cells were strongly enriched in the spleen at the peak of the immune response to the vaccine, illustrating their increased peripheral expansion as compared to control OT-1 cells (Figure 6E). Moreover, this increase was also reflected in the tumor-draining lymph nodes (dLNs), with more OT-1 miR-155 cells than OT-1 controls in dLNs of both N4 tumors (Figure 6F) and T4 tumors (Figure 6G). The absolute numbers of OT-1 miR-155 cells were increased in both N4 and T4 tumors but more markedly in the latter (Figure S1). However, whereas the N4 tumors contained similar frequencies of OT-1 miR-155 as compared to OT-1 control cells (Figure 6H), the T4 tumors were reproducibly enriched with miR-155 OT-1 cells, as compared to control OT-1 cells (Figure 6I), showing an increased ability of miR-155-overexpressing T cells to either survive or expand better in tumors expressing a low-affinity antigen.
Figure 6.
Overexpression of miR-155 in OT-1 Cells Improves Their Ability to Mediate Protection against Tumors Expressing a Low-Affinity Antigen
(A) qPCR of miR-155 levels before and 1, 2, and 4 days following coculture with B16-T4 cells (5:1 ratio) (N = 3). (B) B6 mice were engrafted subcutaneously on the left flank with B16-T4 and on the right flank with B16-N4. 10 days after the graft, the mice were injected intravenously with 1 × 105 OT-1 control, OT-1 miR-155 cells, or PBS. The same day, they were vaccinated with CpG, and the high-affinity OVA peptide SIINFEKL (N4). The mice were sacrificed 18 days after the graft for FACS analysis. (C) B16-N4 tumor growth was measured every 2 days with a manual caliper (N = 6). (D) B16-T4 tumor growth was also measured every 2 days (N = 6). Figures are from 1 representative experiment out of 2 independent experiments. Two-way ANOVA and Tukey’s multiple comparison test were used to compute the statistical significance at day 21 postgraft for the different treatment groups. **p ≤ 0.01; ****p ≤ 0.0001. (E–I) Percentages of GFP+ OT-1 control and OT-1 miR-155 in total CD8+ T cells in (E) spleen, (F) draining lymph nodes (dLNs) of N4 tumor, (G) dLNs of T4 tumor, (H) N4 tumor, and (I) T4 tumor. Unpaired t test were performed. ns p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001, ****p ≤ 0.0001.
Together, these results indicate that overexpression of miR-155 specifically improves the T cell response to low-affinity antigen, possibly by preventing the downregulation of the CD8 coreceptor that was shown to be particularly important in the case of lower-affinity TCR/major histocompatibility complex (MHC)/peptide interactions.39,40
Functional Responses to the Low-Affinity T4 Peptide Are Dependent on CD8α Binding in a Dose-Dependent Manner
Thus, we investigated the functional significance of the higher CD8α and CD8β levels consistently seen on the surface of OT-1 miR-155 cells after in vivo antigen stimulation. It has been previously shown that the presence or absence of CD8α could affect the ability of human T cells to respond to low-affinity antigens, whereas high-affinity antigens were usually independent of CD8α-MHC-I binding.41 Indeed, the concentration of IFN-γ after overnight stimulation with the N4 peptide was not affected by the addition of 1 μg/mL of anti-CD8α to the culture (Figures 7A and 7B), whereas the amount of IFN-γ sharply dropped when the anti-CD8α antibody was added to the cells stimulated with the low-affinity peptide T4 (Figures 7C and 7D). Moreover, miR-155 overexpression rendered the T cells more resistant to CD8α blockade.
Figure 7.
Functional Response to the Low-Affinity T4 Peptide Is Dependent on CD8 Binding
(A–D) Total IFN-γ in the supernatant after overnight coculture of control or miR-155-transduced OT-1 cells with EL4 pulsed with different concentrations of either N4 (A and B) or T4 (C and D) peptide in the absence (A and C) or presence (B and D) of 1 μg/mL blocking anti-CD8α antibody (N = 2). Statistical significance was determined by two-way ANOVA and Sidak’s multiple comparison test. (E–H) Killing assay measured via IncuCyte Live Cell Analysis. OT-1 control or OT-1 miR-155 cells were cocultured with B16-N4 (E and F) or B16-T4 cells (G and H) at a 2:1 effector:target ratio in the absence (E and G) or presence (F and H) of 1 μg/mL blocking anti-CD8α antibody. The Annexin V red signal was measured every 2 h for 2 days (N = 2). Statistical significance was determined by two-way ANOVA. Graphs are representative of 2 independent experiments.
Next, we measured the killing efficiency of OT-1 miR-155 and OT-1 control cells against B16-N4 versus B16-T4 tumor cells, with or without the blocking the CD8α and MHC-I interaction. Similarly, CD8 blocking did not significantly affect the killing of B16-N4 tumor cells (Figures 7E and 7F), whereas B16-T4 cell killing was strictly dependent on the intact function of CD8α (Figures 7G and 7H). Of note, the killing of low-affinity B16-T4 cells in the absence of CD8 blocking was significantly higher by OT-1 cells overexpressing miR-155 compared to the controls (Figure 7G) in contrast to the B16-N4 target cells (Figure 7E). These experiments demonstrate that the ability of T cells to respond to the low-affinity antigen is dependent on the binding of CD8α to the MHC-I. Therefore, we postulate that the increased CD8α and CD8β expression observed on the surface of OT-1 miR-155 contributes to the better antitumor efficacy in a low-affinity setting.
Discussion
The main finding of this work is that miR-155 can be used to engineer T cells for improved adoptive-transfer therapy against tumors expressing a low-affinity antigen.
First, we demonstrated that the effector functions of CD8+ T cells, transduced with a retroviral vector driving miR-155 overexpression, were enhanced in vivo, as shown by higher proliferation and IFN-γ and TNF-α secretion in response to Lm-OVA infection. These results corroborate previous findings from ourselves and others, demonstrating that miR-155 is required for T cell proliferation and cytokine production.8,20,23,24 Moreover, we confirm the observation that miR-155-overexpressing cells are more terminally differentiated,24,42 which is in line with our previous observation of an increased central memory phenotype in miR-155 knockout (KO) CD8+ T cells.8
Since miR-155 targeting of INPP5D leads to Akt phosphorylation,23,43 we hypothesized an increased mTOR signaling, which was confirmed by higher phosphoS6 and glycolytic activity. mTOR activity is implicated in the ability of T cells to switch from oxidative phosphorylation to glycolysis upon activation, which is necessary for an increased production of IFN-γ.30,44 This is consistent with our observations of an increased IFN-γ production upon miR-155 overexpression. Furthermore, we observed an increased oxidative respiration and extracellular acidification rate at the resting state in the presence or absence of extracellular glucose. As it has been shown that the competition for glucose between cancer cells and T cells is an important mechanism of local tumor immunosuppression,31,32 our observations suggest that T cells overexpressing miR-155 can maintain a high metabolic activity even in the presence of low glucose levels.
Although the overexpression of miR-155 induced a strong increase of miR-155 levels at the resting state, the upregulation of the endogenous miR-155 upon activation with a high-affinity antigen failed to show an additive effect. Indeed, the relative levels of miR-155 per T cell reached an upper plateau, which may explain the lack of enhanced anti-tumor effects in vivo in the presence of strong tumor antigens. These observations suggest that there is likely a tightly regulated maximal amount of miR-155 that can be present in a cell, resulting from its expression rate, its post-transcriptional processing capacity, and its half-life. Hence, overexpression of miR-155 might be most impactful upon a weak T cell activation.
We and others had previously shown that the upregulation of the endogenous miR-155 is proportional to the affinity of the TCR to the peptide:MHC ligand, both in mouse and human CD8+ T cells.8,36,45 Together, these observations suggest that overexpressing miR-155 in low-affinity T cells would selectively support their activation and effector functions, as they are the ones with the lowest natural miR-155 increase upon antigen activation. Indeed, we demonstrated that OT-1 cells stimulated with the N4 peptide upregulated miR-155 to a higher level compared to OT-1 cells stimulation with the T4 peptide. We also have indications that OT-1 TILs in T4 tumors had a lower expression of the endogenous miR-155 compared to TILs in the N4 tumors.36 However, the number of control-transduced OT-1 TILs in T4 tumors in the present study was unfortunately too low to get reliable qPCR values of their miR-155 expression.
Although enforced miR-155 overexpression led to higher systemic expansion of OT-1 T cells, an improved antitumor response upon vaccination was only seen against tumors expressing the low-affinity OVA epitope T4. We propose two nonmutually exclusive mechanisms to explain this better effect on low-affinity tumors.
First, the higher miR-155 levels in TILs from high-affinity N4 tumors might already fall in an optimal range for the tumor context, and the addition of more miR-155 might be unnecessary. In contrast, TILs extracted from the T4 tumors had lower activation-induced miR-155 levels, which were compensated by enforced miR-155 overexpression, thus endowing the cells with all of the previously discussed functional advantages. Previous publications showing a better effect of miR-155 overexpression in CD8+ T cells were performed in the low-affinity pmel model, which supports that hypothesis. Moreover, recent work from our lab demonstrated that the levels of endogenous miR-155 in TILs are correlated with tumor control only in low-affinity tumors.36
A second mechanism that may explain the better tumor control of low-affinity tumors by miR-155 OT-1 T cells relates to their reproducibly higher CD8α and CD8β expression levels. We found that, in fact, the surface CD8 downregulation following T cell activation was significantly attenuated in miR-155-overexpressing cells, resulting in a more abundant surface expression of the coreceptor. Upon activation of CD8+ T cells, CD8α and CD8β are downregulated together with the TCR-MHC signaling synapse.46,47 A possible mechanism for attenuated CD8 downregulation might lie in the transcription factor CCAAT/enhancer-binding protein alpha (CEBPA), which is a direct target of murine48 and human miR-155.49,50 Sorting nexins are proteins that are responsible for the receptors recycling from endosomes, as well as their expression at the cell surface.51 The miR-155/CEBPA/SNX27 axis was shown to be important for protein recycling in mouse brain cells,52 and sorting nexin familymember 27 (SNX27) was shown to be important for protein recycling in T cells from endocytic vesicles.53 This absence of CD8α and CD8β downregulation might, in turn, play a key role in stabilizing a suboptimal TCR:peptide MHC-I interaction, whereas being irrelevant in the context of a high-affinity antigen. Indeed, it has been demonstrated before that low-affinity T cell function is highly dependent on the presence of the surface CD8 coreceptor, whereas high-affinity T cells are not.41,54,55 Our in vitro data corroborate these findings, as both IFN-γ expression and target cell killing were abrogated in the presence of blocking the anti-CD8α antibody upon stimulation with the T4 peptide. Further studies are necessary to tease out the targets of miR-155, which are responsible for its implication in the attenuation of activation-induced downregulation of cell-surface CD8 coreceptors.
These findings confirm and lend further support to miR-155 being an interesting candidate for immunotherapy, as circulating T cells are usually of low affinity to tumor-associated self-antigens because of central tolerance and immunoediting. The overexpression of miR-155 could potentially rescue those low-affinity T cells and make them more efficient in controlling tumor growth. We postulate that this key feature provides renewed hope to exploit effectively the repertoire of self-antigen-specific T cells for cancer immunotherapy. This is important because targeting self-tumor antigens, in contrast to neoantigens, may benefit large groups of cancer patients and avoid recurring to individualized immunotherapies.
MATERIALS and METHODS
Mouse Strains
OT-1 transgenic mice expressing a H-2Kb/OVA-specific TCR on the C57BL/6J background were bred in house until 6–15 weeks of age and used to obtain OT-1 CD8+ T cells. C57BL/6J mice were purchased from Envigo. C57BL/6J expressing the CD45.1 or CD45.1/2 congenic markers were bred in house. This study was approved by the Veterinary Authority of the Swiss Canton Vaud (authorization no. 1850) and performed in accordance with Swiss ethical guidelines
Cell Lines
The B16-F10 mouse melanoma cell line was used either as such or transduced to express the SIINFEKL (N4) or SIITFEKL (T4) OVA protein, as previously described.56 B16-OVA implicitly refers to the high-affinity SIINFEKL ovalbumin protein.
Production of Retroviral Particles
107 Phoenix-ECO (ATCC CRL-3214) cells were plated in a T150 flask the day before the transfection. Transfection with a mix of either pMGP-GFP or pMGP-M155 (Addgene 26527) and pCL-ECO (Addgene 12371) packaging plasmid was performed. Retrovirus was produced as previously described by B. Tschumi et. al.26
Retroviral Transduction and Maintenance of Primary Mouse T Cells
T Cell transduction was essentially done as previously described.26 Briefly, spleens of OT-1 mice were dissociated, and T cells were isolated using the mouse T cell isolation kit (STEMCELL 19851). T Cells were subsequently activated by coculturing them with αCD3/αCD28-coated magnetic beads (Life Technologies 11452D). On the following day, T cells were transduced as described.24 The percentage of transduced cells was assessed 2 days after transduction via flow cytometry detection of GFP expression. At this time, the cells were either used for in vivo tumor challenge or kept in culture, as described.24 Transduced mouse T cells could be kept in culture and used for in vitro assays until 2–3 weeks after transduction.
Tumor Challenge
C57BL/6J mice, 6–10 weeks old, were subcutaneously engrafted with 105 B16-OVA (N4) or B16-T4 tumor cells in 100 or 200 μL PBS. 6 days later, 105 miR-155 or control-transduced OT-1 cells (approximately 90% GFP+) were transferred intravenously in the tail. 7 days after the tumor graft, mice were either infected or vaccinated. Infections were performed via the intravenous injection of 2,000 plaque-forming units (PFU) L. monocytogenes, genetically modified to express the OVA protein. Vaccination was performed subcutaneously at the base of the tail. The vaccination mix incorporated 10 μg SIINFEKL (Protein and Peptide Chemistry Facility, University of Lausanne [UNIL]) peptide with 50 μg CpG (ODN 1826, U133-L01A) in PBS. Tumor growth was measured using a manual caliper from day 6 postengraftment and every 2–3 days after that until the end of the experiment. Tumor sizes were calculated in cubic millimeters as the product of length, width, and height divided by 2.
Organ Processing for In Vitro Analysis
Spleens were mashed through a 100-μm filter and treated with red blood cell lysis buffer for 5 min before staining. Lymph nodes were mashed through a 40-μm filter and stained directly. Tumors were cut in small pieces with scissors and put into a MACS C Tube with 2.5 mL of plain DMEM containing enzymes from the MACS 130-096-730 kit. Tumors were then mechanically processed using the gentleMACS dissociator (#130-093-235) and the m_impTumor_02 program. Afterward, tumors were incubated at 37°C for 20 min, rolling constantly. Finally, tumors were again mechanically processed using the program m_impTumor_03. Resuspended tumors were filtered through 40 μm, before pelleting by centrifugation. Debris were removed by Percoll purification. The tumor cell pellet was first resuspended in a 15-mL Falcon in 40% Percoll in PBS at 37°C. Then, 4 mL of 70% Percoll in PBS was carefully layered on top of the 40% Percoll. The tubes were subsequently centrifuged at 975 g for 20 min. Tumor-infiltrating lymphocytes were isolated from the middle phase of the density gradient and washed 2 times before counting and staining or restimulation for the detection of cytokine production. Both the axillary and inguinal lymph nodes on the same flank as the tumor were sampled and defined as tumor dLNs in Results.
In Vivo T Cell Proliferation
Mice were injected intraperitoneally (i.p.) with 1.8 mg of BrdU, diluted in PBS for 2.5 h before organ collection and analysis. Organs were processed as described above. After staining for surface markers, cells were fixed, and first, the cell membrane was permeabilized, and then, the nuclear membrane was permeabilized using Cytoperm Plus (Becton Dickinson [BD] 561651). DNase was added at a concentration of 0.3 μg/μL, and the samples were incubated for 1 h at 37°C. Then, anti-BrdU phycoerythrin (PE) antibody was added in diluted in Perm/Wash buffer and incubated 30 min at room temperature. Samples were washed 2 times before resuspension and acquisition via flow cytometry.
Intracellular Cytokine Staining
To assess intracellular cytokine production, 10 ng/mL final concentration phorbol 12-myristate 13-acetate (PMA) and 500 ng/mL ionomycin or 10 μM of SIINFEKL peptide were added to plated splenocytes or tumor-infiltrating lymphocytes in a 96-well plate and placed in the incubator at 37°C. After 30 min, GolgiStop (monensin; BD 554724) was added to the wells at a 1/1,500 dilution. The cells were placed back in the incubator for an additional 4 h before surface staining and intracellular staining. Fixation and permeabilization for the intracellular detection of cytokines were performed with the BD kit (554714), according to the manufacturer’s instruction.
qPCR
Cells were collected, resuspended in RNAlater (Ambion AM7020), and stored at −20°C for further RNA isolation. Subsequently, RNA was purified in 40 μL H2O using the mirVana (Ambion AM1561) kit and following the manufacturer’s instructions. miR-specific RT was performed on 5 μL of the extracted RNA and using TaqMan primer pairs (TaqMan SnoRNA202 1232, TaqMan mmu-miR155 2571) and the TaqMan RT kit (Life Technologies 4366597) following the manufacturer’s instructions. Finally, qPCR was run using 96-well optical plates (Life Technologies 4346906) in a 7500 Fast Real-Time PCR machine (Thermo Fisher Scientific 4351106). The qPCR mix volume was of 10 μL/well and comprised 2 μL of the RT reaction mixed with the primers and TaqMan Fast Universal Master Mix (Life Technologies 4352042). All qPCR reactions were performed in 2 technical replicates, and the values were excluded if the difference in cycle threshold (CT) was higher than 0.2 or if one of the CT values was higher than 33. The mean of the technical replicates of the control gene (snoRNA202) was substracted to the mean of the technical replicates of the gene of interest (miR-155) to calculate the ΔCT. Fold changes in expression were then calculated using a control sample for normalization as such:
Killing Assays
IncuCyte Live Cell Analysis was used to measure the killing of target cells. OT-1 control or OT-1 miR-155 cells were cocultured with B16-N4 or B16-T4 cells at a 2:1 effector:target ratio in the presence or absence of 1 μg/mL blocking anti-CD8α antibody. The Annexin V red signal (IncuCyte, 4641) was measured every 2 h for 2 days.
Metabolic Assays
XF 96 cell-culture microplates were coated with Cell-Tak (Corning, CB40240) overnight. The next day, T cells were seeded in quadruplicate in unbuffered RPMI (Sigma-Aldrich), supplemented with 2 mM glutamine and with 10 mM glucose depending on the conditions. ECAR and OCR were measured every 5 mi in a Seahorse XF Analyzer before and after sequential addition of αCD3/αCD28 beads, oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), and 2-deoxy-glucose (2DG) or αCD3/αCD28 beads, glucose, oligomycin, and FCCP. Acquired data were normalized to protein levels.
Statistical Analysis
Flow cytometry data were analyzed with FlowJo (Tree Star), and graphs and statistical analysis were made with Prism (GraphPad Software). A two-way ANOVA, followed by a Sidak’s or Tukey’s multiple comparison, was used for simultaneous analysis of two variables among multiple groups. Single variables among multiple groups were analyzed with a two-sided t test and a 95% confidence interval. Figures show 1 representative experiment out of 2 or 3 independent experiments. The individual dots always represent biological replicates (number of mice or independent wells). When technical replicates were performed (usually in duplicates), the average of the replicates was plotted (ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Author Contributions
Conceptualization, G.C.M., P.R.; Methodology, A.D., A.M.-U., P.-C.H., E.L.; Investigation, G.C.M., A.M.-U., S.F.L., W.-C.C.; Resources, P.R., P.-C.H.; Writing – Original Draft, G.C.M., A.D., P.R.; Writing – Review & Editing, A.M.-U., P.-C.H., E.L., G.C., M.I.; Supervision, A.D., P.R.; Funding Acquisition, P.R.
Conflicts of Interest
P.R. has received speaker honoraria from Bristol-Myers Squibb and Roche and is the recipient of research grants in immune-oncology from Roche pRED, Zurich, Switzerland. G.C. has received grants, research support, and/or is a coinvestigator in clinical trials by BMS, Celgene, Boehringer Ingelheim, Roche, Iovance, and Kite; has received honoraria for consultations or presentations by Roche, Genentech, BMS, AstraZeneca, Sanofi-Aventis, Nextcure, and GeneosTx; has patents in the domain of antibodies and vaccines targeting the tumor vasculature, as well as technologies related to T cell expansion and engineering for T cell therapy; and receives royalties from the University of Pennsylvania related to T cell therapy. P.-C.H. received research support from Roche pRED and Idorsia and honorarium from Chungai and Pfizer. P.-C.H. is also a member of the Scientific Advisory Board of Elixiron Immunotherapeutics. All of the other authors declare no competing interests.
Acknowledgments
We thank Luca Gattinoni (NIH/NCI) for providing the retroviral plasmids. We thank Nina Dumauthioz, Leyder Lozano, and Candice Stoudmann for technical help, as well as Romain Bedel for technical assistance in the fluorescence-activated cell sorting (FACS). We thank Michael Hebeisen and Julien Schmidt for useful advice and discussion. We thank Francis Derouet and the staff at the animal facility of Epalinges for their work. P.R. was funded, in part, by grants from the Swiss National Foundation (Lead Agency 310030E and 310030_182735).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.12.008.
Supplemental Information
References
- 1.Rosenberg S.A., Yang J.C., Sherry R.M., Kammula U.S., Hughes M.S., Phan G.Q., Citrin D.E., Restifo N.P., Robbins P.F., Wunderlich J.R. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klebanoff C.A., Finkelstein S.E., Surman D.R., Lichtman M.K., Gattinoni L., Theoret M.R., Grewal N., Spiess P.J., Antony P.A., Palmer D.C. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2004;101:1969–1974. doi: 10.1073/pnas.0307298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno M., Ohkuri T., Kosaka A., Tanahashi K., June C.H., Natsume A., Okada H. Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J. Immunother. Cancer. 2013;1:21. doi: 10.1186/2051-1426-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegram H.J., Lee J.C., Hayman E.G., Imperato G.H., Tedder T.F., Sadelain M., Brentjens R.J. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood. 2012;119:4133–4141. doi: 10.1182/blood-2011-12-400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stromnes I.M., Blattman J.N., Tan X., Jeevanjee S., Gu H., Greenberg P.D. Abrogating Cbl-b in effector CD8(+) T cells improves the efficacy of adoptive therapy of leukemia in mice. J. Clin. Invest. 2010;120:3722–3734. doi: 10.1172/JCI41991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto T.N., Lee P.H., Vodnala S.K., Gurusamy D., Kishton R.J., Yu Z., Eidizadeh A., Eil R., Fioravanti J., Gattinoni L. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. J. Clin. Invest. 2019;129:1551–1565. doi: 10.1172/JCI121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudda J.C., Salaun B., Ji Y., Palmer D.C., Monnot G.C., Merck E., Boudousquie C., Utzschneider D.T., Escobar T.M., Perret R. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvágner G., Zamore P.D. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 10.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 11.Lu C., Huang X., Zhang X., Roensch K., Cao Q., Nakayama K.I., Blazar B.R., Zeng Y., Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceppi M., Pereira P.M., Dunand-Sauthier I., Barras E., Reith W., Santos M.A., Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann M., Barad O., Agami R., Geiger B., Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc. Natl. Acad. Sci. USA. 2010;107:15804–15809. doi: 10.1073/pnas.0915022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jablonski K.A., Gaudet A.D., Amici S.A., Popovich P.G., Guerau-de-Arellano M. Control of the Inflammatory Macrophage Transcriptional Signature by miR-155. PLoS ONE. 2016;11:e0159724. doi: 10.1371/journal.pone.0159724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L.F., Thai T.H., Calado D.P., Chaudhry A., Kubo M., Tanaka K., Loeb G.B., Lee H., Yoshimura A., Rajewsky K., Rudensky A.Y. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escobar T.M., Kanellopoulou C., Kugler D.G., Kilaru G., Nguyen C.K., Nagarajan V., Bhairavabhotla R.K., Northrup D., Zahr R., Burr P. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40:865–879. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee A., Schambach F., DeJong C.S., Hammond S.M., Reiner S.L. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur. J. Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell R.M., Kahn D., Gibson W.S., Round J.L., Scholz R.L., Chaudhuri A.A., Kahn M.E., Rao D.S., Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gracias D.T., Stelekati E., Hope J.L., Boesteanu A.C., Doering T.A., Norton J., Mueller Y.M., Fraietta J.A., Wherry E.J., Turner M., Katsikis P.D. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai C.Y., Allie S.R., Zhang W., Usherwood E.J. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J. Virol. 2013;87:2348–2351. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffaker T.B., Lee S.H., Tang W.W., Wallace J.A., Alexander M., Runtsch M.C., Larsen D.K., Thompson J., Ramstead A.G., Voth W.P. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017;292:18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y., Wrzesinski C., Yu Z., Hu J., Gautam S., Hawk N.V., Telford W.G., Palmer D.C., Franco Z., Sukumar M. miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic γc cytokines. Proc. Natl. Acad. Sci. USA. 2015;112:476–481. doi: 10.1073/pnas.1422916112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hope J.L., Stairiker C.J., Spantidea P.I., Gracias D.T., Carey A.J., Fike A.J., van Meurs M., Brouwers-Haspels I., Rijsbergen L.C., Fraietta J.A. The Transcription Factor T-Bet Is Regulated by MicroRNA-155 in Murine Anti-Viral CD8+ T Cells via SHIP-1. Front. Immunol. 2017;8:1696. doi: 10.3389/fimmu.2017.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trotta R., Chen L., Ciarlariello D., Josyula S., Mao C., Costinean S., Yu L., Butchar J.P., Tridandapani S., Croce C.M., Caligiuri M.A. miR-155 regulates IFN-γ production in natural killer cells. Blood. 2012;119:3478–3485. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tschumi B.O., Dumauthioz N., Marti B., Zhang L., Lanitis E., Irving M., Schneider P., Mach J.P., Coukos G., Romero P., Donda A. Correction to: CART cells are prone to Fas- and DR5-mediated cell death. J. Immunother. Cancer. 2018;6:92. doi: 10.1186/s40425-018-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haasch D., Chen Y.W., Reilly R.M., Chiou X.G., Koterski S., Smith M.L., Kroeger P., McWeeny K., Halbert D.N., Mollison K.W. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 28.Salaun B., Yamamoto T., Badran B., Tsunetsugu-Yokota Y., Roux A., Baitsch L., Rouas R., Fayyad-Kazan H., Baumgaertner P., Devevre E. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J. Transl. Med. 2011;9:44. doi: 10.1186/1479-5876-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollizzi K.N., Patel C.H., Sun I.H., Oh M.H., Waickman A.T., Wen J., Delgoffe G.M., Powell J.D. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gubser P.M., Bantug G.R., Razik L., Fischer M., Dimeloe S., Hoenger G., Durovic B., Jauch A., Hess C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 31.Chang C.H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho P.C., Bihuniak J.D., Macintyre A.N., Staron M., Liu X., Amezquita R., Tsui Y.C., Cui G., Micevic G., Perales J.C. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao E., Maj T., Kryczek I., Li W., Wu K., Zhao L., Wei S., Crespo J., Wan S., Vatan L. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overwijk W.W., Theoret M.R., Finkelstein S.E., Surman D.R., de Jong L.A., Vyth-Dreese F.A., Dellemijn T.A., Antony P.A., Spiess P.J., Palmer D.C. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 2003;198:569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniels M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G.A., Gascoigne N.R., Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Usatorre A., Sempere L.F., Carmona S.J., Carretero-Iglesia L., Monnot G., Speiser D.E., Rufer N., Donda A., Zehn D., Jandus C., Romero P. MicroRNA-155 expression is enhanced by T-cell receptor stimulation strength and correlates with improved tumor control in melanoma. Cancer Immunol. Res. 2019;7:1013–1024. doi: 10.1158/2326-6066.CIR-18-0504. [DOI] [PubMed] [Google Scholar]
- 37.Krummey S.M., Martinez R.J., Andargachew R., Liu D., Wagener M., Kohlmeier J.E., Evavold B.D., Larsen C.P., Ford M.L. Low-Affinity Memory CD8+ T Cells Mediate Robust Heterologous Immunity. J. Immunol. 2016;196:2838–2846. doi: 10.4049/jimmunol.1500639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehn D., Lee S.Y., Bevan M.J. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holler P.D., Kranz D.M. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 40.van den Berg H.A., Wooldridge L., Laugel B., Sewell A.K. Coreceptor CD8-driven modulation of T cell antigen receptor specificity. J. Theor. Biol. 2007;249:395–408. doi: 10.1016/j.jtbi.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Couedel C., Bodinier M., Peyrat M.A., Bonneville M., Davodeau F., Lang F. Selection and long-term persistence of reactive CTL clones during an EBV chronic response are determined by avidity, CD8 variable contribution compensating for differences in TCR affinities. J. Immunol. 1999;162:6351–6358. [PubMed] [Google Scholar]
- 42.Stelekati E., Chen Z., Manne S., Kurachi M., Ali M.A., Lewy K., Cai Z., Nzingha K., McLane L.M., Hope J.L. Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Rep. 2018;23:2142–2156. doi: 10.1016/j.celrep.2018.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lind E.F., Millar D.G., Dissanayake D., Savage J.C., Grimshaw N.K., Kerr W.G., Ohashi P.S. miR-155 Upregulation in Dendritic Cells Is Sufficient To Break Tolerance In Vivo by Negatively Regulating SHIP1. J. Immunol. 2015;195:4632–4640. doi: 10.4049/jimmunol.1302941. [DOI] [PubMed] [Google Scholar]
- 44.Chang C.H., Curtis J.D., Maggi L.B., Jr., Faubert B., Villarino A.V., O’Sullivan D., Huang S.C., van der Windt G.J., Blagih J., Qiu J. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebeisen M., Baitsch L., Presotto D., Baumgaertner P., Romero P., Michielin O., Speiser D.E., Rufer N. SHP-1 phosphatase activity counteracts increased T cell receptor affinity. J. Clin. Invest. 2013;123:1044–1056. doi: 10.1172/JCI65325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao Z., Mescher M.F., Jameson S.C. Detuning CD8 T cells: down-regulation of CD8 expression, tetramer binding, and response during CTL activation. J. Exp. Med. 2007;204:2667–2677. doi: 10.1084/jem.20062376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroger C.J., Alexander-Miller M.A. Dose-dependent modulation of CD8 and functional avidity as a result of peptide encounter. Immunology. 2007;122:167–178. doi: 10.1111/j.1365-2567.2007.02622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costinean S., Sandhu S.K., Pedersen I.M., Tili E., Trotta R., Perrotti D., Ciarlariello D., Neviani P., Harb J., Kauffman L.R. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He M., Xu Z., Ding T., Kuang D.M., Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. Cell. Mol. Immunol. 2009;6:343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worm J., Stenvang J., Petri A., Frederiksen K.S., Obad S., Elmén J., Hedtjärn M., Straarup E.M., Hansen J.B., Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullen P.J. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Zhao Y., Zhang X., Badie H., Zhou Y., Mu Y., Loo L.S., Cai L., Thompson R.C., Yang B. Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down’s syndrome. Nat. Med. 2013;19:473–480. doi: 10.1038/nm.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rincón E., Santos T., Avila-Flores A., Albar J.P., Lalioti V., Lei C., Hong W., Mérida I. Proteomics identification of sorting nexin 27 as a diacylglycerol kinase zeta-associated protein: new diacylglycerol kinase roles in endocytic recycling. Mol. Cell. Proteomics. 2007;6:1073–1087. doi: 10.1074/mcp.M700047-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Laugel B., van den Berg H.A., Gostick E., Cole D.K., Wooldridge L., Boulter J., Milicic A., Price D.A., Sewell A.K. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J. Biol. Chem. 2007;282:23799–23810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 55.Lee N.A., Loh D.Y., Lacy E. CD8 surface levels alter the fate of alpha/beta T cell receptor-expressing thymocytes in transgenic mice. J. Exp. Med. 1992;175:1013–1025. doi: 10.1084/jem.175.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Usatorre A., Donda A., Zehn D., Romero P. PD-1 Blockade Unleashes Effector Potential of Both High- and Low-Affinity Tumor-Infiltrating T Cells. J. Immunol. 2018;201:792–803. doi: 10.4049/jimmunol.1701644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.